Abstract
microRNAs (miRNAs) are small non-coding RNAs that can function as endogenous silencers of target genes and play critical roles in human malignancies. To investigate the molecular pathogenesis of gastric mucosa-associated lymphoid tissue (MALT) lymphoma, the miRNA expression profile was analyzed. miRNA microarray analysis with tissue specimens from gastric MALT lymphomas and surrounding non-tumor mucosae revealed that a hematopoietic-specific miRNA miR-142 and an oncogenic miRNA miR-155 were overexpressed in MALT lymphoma lesions. The expression levels of miR-142-5p and miR-155 were significantly increased in MALT lymphomas which do not respond to Helicobacter pylori (H. pylori) eradication. The expression levels of miR-142-5p and miR-155 were associated with the clinical courses of gastric MALT lymphoma cases. Overexpression of miR-142-5p and miR-155 was also observed in Helicobacter heilmannii-infected C57BL/6 mice, an animal model of gastric MALT lymphoma. In addition, miR-142-5p and miR-155 suppress the proapoptotic gene TP53INP1 as their target. The results of this study indicate that overexpression of miR-142-5p and miR-155 plays a critical role in the pathogenesis of gastric MALT lymphoma. These miRNAs might have potential application as therapeutic targets and novel biomarkers for gastric MALT lymphoma.
Introduction
Extranodal marginal zone B-cell lymphoma of mucosa-associated lymphoid tissue (MALT) is a low-grade lymphoma characterized by histological features such as lymphoepithelial lesions (LELs). The stomach is the most common site of MALT lymphoma, accounting for almost half of all cases. Gastric MALT lymphomas are sometimes associated with chronic inflammation triggered by chronic infection with Helicobacter pylori (H. pylori), suggesting that the proliferation of MALT lymphoma cells may depend on immune responses to antigens. Indeed, H. pylori eradication therapy leads to complete remission in 60–80% of cases of gastric MALT lymphoma and has been used as a first-line treatment [1]–[3]. However, 20–40% of cases do not respond to H. pylori eradication therapy, and predictors of the response to antibiotic treatment as well as the appropriate length of the observation period before second-line treatment remain controversial.
The API2-MALT1 fusion gene, which results from a t(11;18)(q21;q21) translocation, has been identified as the most frequent chromosome translocation in MALT lymphoma cells [4], [5], and Liu et al. [6] have reported that it is a potential predictor of resistance to H. pylori eradication therapy. Since API2-MALT1 fusion transcripts lead to inhibition of apoptosis [7], [8], they may confer a survival benefit on MALT lymphoma cells. Despite these reports, the molecular mechanism underlying the initiation and progression of gastric MALT lymphoma is not fully understood.
Many previous studies have focused mainly on aberrant expression of protein-coding genes in the pathogenesis of MALT lymphoma [9]. However, it has recently become apparent that non-coding genes including microRNAs (miRNAs) play important roles as tumor suppressor genes and oncogenes during human carcinogenesis. miRNAs are small (20–25 nucleotides) non-coding RNAs that function as endogenous silencers of target genes. miRNAs are expressed in a tissue-specific manner and play critical roles in cellular proliferation, apoptosis and differentiation [10]. It has been shown that aberrant expression of miRNAs contributes to the development of human malignancies, and that miRNA expression signatures are associated with prognostic factors of human diseases [11]–[15]. Moreover, we have recently proposed that epigenetic regulation of tumor suppressor miRNAs could be a novel therapeutic approach for the treatment of human malignancies [16]–[19].
Although recent studies have shown that miR-155, a potential oncogenic miRNA, is highly expressed in diffuse large B cell lymphoma (DLBCL) [20], [21] and overexpression of miR-155 is correlated with a poor outcome in patients with DLBCL [20], the miRNA expression profiles of low-grade MALT lymphoma have not yet been described. In the present study, therefore, the miRNA expression profiles and potential miRNA target genes of gastric MALT lymphoma were analyzed to clarify the molecular pathogenesis of this malignancy.
Methods
Patients and tissue specimens
Twenty patients with primary low-grade gastric MALT lymphomas who were diagnosed and treated at Keio University Hospital (Tokyo, Japan) were enrolled. This study was approved by the ethics committee of Keio University School of Medicine (No. 18-96-3) and was registered with the Clinical Trials Registry (UMIN 000000858). Written informed consent was obtained from all the patients before examination. The clinicopathological and molecular features of the patients are shown in Table 1. The H. pylori infection status was identified using the 13C-urea breath test. Some cases were confirmed by serological or histological examination in addition to the 13C-urea breath test. Tissue specimens from gastric MALT lymphomas and the corresponding non-tumor gastric mucosae were obtained from patients during an endoscopic biopsy and were stored in RNAlater (Ambion, Austin, TX) at −80°C until RNA extraction.
Table 1. Clinicopathological and molecular features of gastric MALT lymphoma cases.
| API2-MALT1 | H.pylori | infection | Remission after | Months after | |||
| No. | Sex | Age | fusion | before Tx | after Tx | eradication | eradication |
| 1 | M | 74 | + | + | − | NC | 74 |
| 2 | M | 74 | + | + | − | NC | 32 |
| 3 | M | 61 | + | − | − | NC | 10 |
| 4 | F | 75 | + | − | * | NC | * |
| 5 | F | 59 | + | − | * | NC | * |
| 6 | F | 57 | + | − | * | NC | * |
| 7 | M | 77 | − | + | − | NC | 45 |
| 8 | F | 68 | − | − | − | NC | 3 |
| 9 | F | 58 | − | + | − | CR | 24 |
| 10 | F | 46 | − | + | − | CR | 10 |
| 11 | F | 66 | − | + | − | CR | 10 |
| 12 | F | 52 | − | + | − | CR | 8 |
| 13 | M | 64 | − | + | − | CR | 6 |
| 14 | F | 71 | − | + | − | CR | 4 |
| 15 | F | 70 | − | + | − | CR | 4 |
| 16 | M | 60 | − | + | − | CR | 4 |
| 17 | F | 56 | − | + | − | CR | 4 |
| 18 | M | 54 | − | + | − | CR | 3 |
| 19 | F | 68 | − | + | − | CR | 2 |
| 20 | F | 40 | − | + | − | CR | 1 |
CR, complete remission; NC, no change.
Eradication therapy not used.
Fluorescence in situ hybridization (FISH) analysis
To detect the chromosome translocation t(11;18)(q21;q21) and the API2-MALT1 fusion gene arising from it, FISH analysis using the LSI API2-MALT1 t(11;18)(q21;q21) Dual Color, Dual Fusion Translocation Probe (Vysis/Abbot Laboratories Ltd., Maidenhead, Berkshire, United Kingdom) was performed by Mitsubishi Chemical Medience Corporation (Tokyo, Japan).
RNA extraction and microarray analysis
Total RNA, including small RNA, was extracted using a mirVana miRNA isolation kit (Ambion). The total RNAs from each of three gastric MALT lymphomas were pooled, as were the total RNAs from each of three matched samples of non-tumor gastric mucosa. The miRNA microarray analysis was performed by LC Sciences (www.lcsciences.com, Houston, TX). RNA from the MALT lymphomas was labeled using Cy5, while the RNA from the non-tumor gastric mucosa was labeled using Cy3. The microarray chip contains probe regions that detect 711 miRNA transcripts listed in Sanger miRBase Release 10.0 (http://www.sanger.ac.uk) and 5 probes for each miRNA. Detected signals greater than background plus 3 times the standard deviation were derived for each color channel; the mean and the co-variance (CV = standard deviation×100/replicate mean) of each probe having a detected signal was calculated. For two color experiments, the ratio of the two sets of detected signals and p-values of the t-test were calculated. Differentially detected signals were accepted as true when the p-values of the ratios were less than 0.01. The average values of their signal intensities are shown in Table 2. All the data were submitted to the ArrayExpress database under the accession number E-MEXP-1898.
Table 2. miRNAs differentially expressed between MALT lymphomas and non-tumor gastric mucosae.
| No. | microRNA | Non-tumor | MALT | Fold change |
| (Cy3 signal) | (Cy5 signal) | |||
| 1 | miR-142-5p | 56 | 1236.3 | 22.1 |
| 2 | miR-142-3p | 25.4 | 440.5 | 17.3 |
| 3 | miR-223 | 574.1 | 5330.9 | 9.3 |
| 4 | miR-138 | 40.9 | 200.8 | 4.9 |
| 5 | miR-155 | 3093.2 | 13503.9 | 4.4 |
| 6 | miR-572 | 1581.7 | 368.2 | 0.2 |
| 7 | miR-146b-5p | 1029.1 | 4249.4 | 4.1 |
| 8 | miR-141 | 6525.1 | 1588.8 | 0.2 |
| 9 | miR-146a | 2187.8 | 8374.4 | 3.8 |
| 10 | miR-378 | 3218.7 | 933.6 | 0.3 |
The miRNA microarray analysis was performed by LC Sciences (www.lcsciences.com, Houston, TX). All data were submitted to the ArrayExpress database; the accession number was E-MEXP-1898.
Quantitative RT-PCR of miRNAs
Levels of miRNA expression were analyzed using quantitative RT-PCR and the TaqMan microRNA assay for miR-142-5p and miR-155 (Applied Biosystems, Foster City, CA), in accordance with the manufacturer's instructions. The expression levels were normalized to that of U6 RNA and expressed as the mean ± standard deviation.
Infection of C57BL/6 mice with H. heilmannii
Ten months prior to the experiment, six-week-old C57BL/6 mice were inoculated with gastric mucosal homogenates from H. heilmannii-infected mice, as described previously [22]. Stool DNA was extracted using a QIAamp DNA Stool Mini Kit (Qiagen, Tokyo, Japan), and infection with H. heilmannii was confirmed by real-time PCR of stool DNA using specific primers (HeilF, 5′-AAGTCGAACGATGAAGCCTA-3′ and HeilR, 5′-ATTTGGTATTAATCACCATTTC-3′). RNA extraction and histological examination were performed using tissue specimens from the stomachs of H. heilmannii-infected mice and control mice. This study was approved by the Keio University Animal Research Committee (No. 08080).
Western blotting
Protein extracts were separated using SDS/polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. The membranes were hybridized with the rabbit anti-human TP53INP1 polyclonal antibody (LifeSpan Biosciences, Seattle, WA). This antibody shows cross-reactivity with mouse TP53INP1. The signal intensities were analyzed using ImageJ software.
Immunohistochemistry
Formalin-fixed and paraffin-embedded tissues were deparaffinized and rehydrated. For antigen retrieval, the sections were treated for 20 min at 100°C in an autoclave and non-specific reactions were blocked with a blocking reagent (Protein Block Serum-Free, Dako Cytomation, Glostrup, Denmark). The sections were incubated with the rabbit anti-human TP53INP1 polyclonal antibody (diluted 1∶200; LifeSpan Biosciences) overnight at 4°C followed by horseradish peroxidase (HRP)-labeled anti-rabbit IgG (Histofine, Simple stain MAX-PO; Nichirei, Tokyo, Japan) for 30 min at room temperature. Then, the sections were treated with 3, 3′-diaminobenzidine tetrahydrochloride solution. All the sections were counterstained with HE.
Luciferase assay
Luciferase constructs were made by ligating oligonucleotides containing the wild-type or mutant target site of the TP53INP1 3′ untranslated region (UTR) into the Xba I site of the pGL3-control vector (Promega, Madison, WI). The AGS human gastric cancer cell line was used in this study. AGS was obtained from the American Type Culture Collection (Rockville, MD). Cells were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum. AGS cells were transfected with 0.4 µg of firefly luciferase reporter vector containing the wild-type or mutant target site and 0.02 µg of the control vector containing Renilla luciferase pRL-CMV (Promega) using lipofectamine 2000 (Invitrogen, Carlsbad, CA) in 24-well plates. The miR-142-5p and miR-155 precursor molecules and negative control precursor miRNAs were purchased from Ambion. The molecules were transfected into AGS cells at a final concentration of 100 nM each. The luciferase assays were performed 24 hours after transfection using the Dual Luciferase Reporter Assay System (Promega). The activity of firefly luciferase was normalized to that of Renilla luciferase.
Statistics
Differences in the miRNA levels between the groups were analyzed using paired t test and unpaired t test. Differences at p<0.05 were considered significant.
Results
Overexpression of miR-142-5p and miR-155 in gastric MALT lymphoma
To identify miRNAs that play critical roles in the development of gastric MALT lymphoma, we performed miRNA microarray analysis using RNAs obtained from three gastric MALT lymphomas and three matched samples of non-tumor gastric mucosa. The miRNAs that were differentially expressed between non-tumor gastric mucosae and MALT lymphoma lesions are listed in Table 2. The miRNA expression profile revealed that a hematopoietic-specific miRNA, miR-142 [23], [24], and an oncogenic miRNA, miR-155 [12], [25], were overexpressed in MALT lymphoma lesions, relative to the levels of expression in the corresponding non-tumor mucosae.
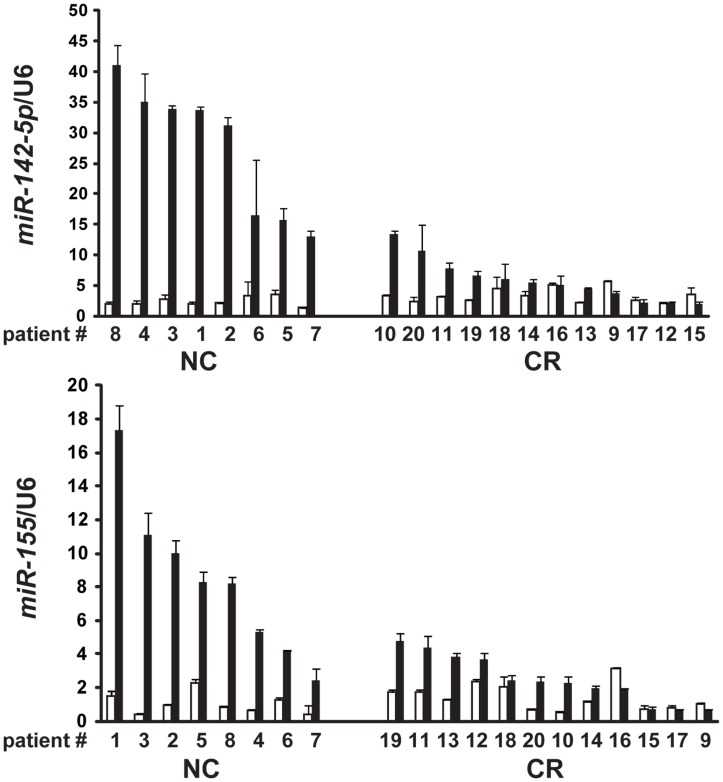
To validate the microarray data, we performed quantitative RT-PCR for miR-142-5p and miR-155 in 20 cases of gastric MALT lymphoma. The clinicopathological and molecular features of the 20 patients are shown in Table 1. All of the gastric MALT lymphoma cases in the present study were stage IE according to the Ann Arbor staging system [26]. The average age of the patients was 62.5 years (male/female: 7/13). As shown in Figure 1, the expression levels of miR-142-5p and miR-155 in the gastric MALT lymphoma lesions were significantly higher than those in the corresponding non-tumor gastric mucosae (p<0.05 and p<0.05, respectively).
Figure 1. Expression levels of miR-142-5p and miR-155 and responses to H. pylori eradication therapy in gastric MALT lymphoma cases.
The levels of miR-142-5p and miR-155 expression were evaluated using quantitative RT-PCR and normalized to the expression of U6 RNA. The filled bars and blank bars indicate MALT lymphoma lesions and non-tumor gastric mucosae, respectively. The levels of miR-142-5p and miR-155 expression in gastric MALT lymphoma lesions were significantly increased relative to the corresponding non-tumor gastric mucosae (p<0.05 and p<0.05, respectively). The levels of miR-142-5p and miR-155 expression in MALT lymphoma lesions which confers resistance to H. pylori eradication were significantly higher than those in lesions lacking the API2-MALT1 fusion gene, which exhibited CR after H. pylori eradication (p<0.0001 and p<0.005, respectively). CR, complete remission; NC, no change.
miR-142-5p and miR-155 as novel biomarkers of gastric MALT lymphoma
The patients with gastric MALT lymphoma were divided into two groups according to their response to H. pylori eradication therapy (Table 1). All the patients who were positive for H. pylori underwent eradication therapy and were subsequently confirmed to be H. pylori-negative. The use of H. pylori eradication therapy for patients with gastric MALT lymphoma who are H. pylori-negative is controversial. Nakamura et al. have recently reported the long-term clinical outcome of patients with gastric MALT lymphoma after H. pylori eradication [27]. In their study, 44 H. pylori-negative gastric MALT lymphoma patients underwent H. pylori eradication therapy, and 6 (14%) responded to H. pylori eradication. Akamatsu et al. have also reported that 1 out of 9 (11%) H. pylori-negative patients showed complete regression of gastric MALT lymphoma [28]. On the other hand, Fischbach et al. have reported that most patients with histological residuals of gastric MALT lymphoma after successful H. pylori eradication (treatment failure cases) had a favorable disease course without additional treatment (Fischbach et al. Gut 2007; 56: 1685–7). For these patients, a watch and wait strategy with regular endoscopic examinations and biopsies appears to be safe. Our policy of H. pylori eradication therapy for gastric MALT lymphoma patients who were H. pylori-negative was decided in accordance with the preference of individual patients.
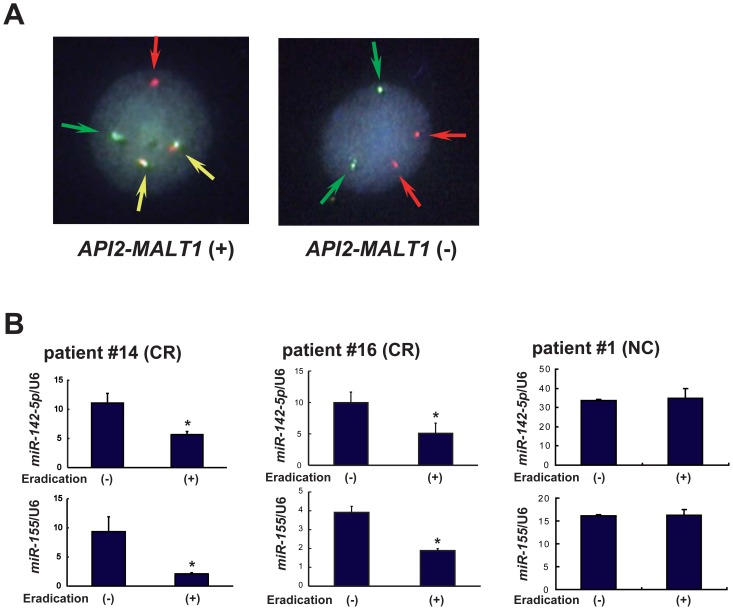
We also analyzed the chromosome translocation t(11;18)(q21;q21) with the API2-MALT1 fusion gene by FISH analysis. The probes for API2 were labeled with green signals, and the probes for MALT1 were labeled with red signals. In positive cases, the API2-MALT1 fusion genes resulting from the chromosome translocation produced yellow signals (Figure 2A).
Figure 2. Expression levels of miR-142-5p and miR-155 in gastric MALT lymphoma cases before and after H. pylori eradication therapy.
(A) Representative cases of the FISH analyses for the detection of the API2-MALT1 fusion gene are shown. The probes for API2 and MALT1 were labeled with green and red signals, respectively. The API2-MALT1 fusion gene produces a yellow signal. (B) Patients #14 and #16, positive for H. pylori and negative for the API2-MALT1 fusion gene. These patients received H. pylori eradication therapy and attained CR. The expression levels of miR-142-5p and miR-155 were significantly lower after H. pylori eradication. * p<0.05. Patient #1, positive for H. pylori and positive for the API2-MALT1 fusion gene. This patient was resistant to H. pylori eradication therapy (NC). The expression levels of miR-142-5p and miR-155 showed no significant differences.
As shown in Figure 1, the levels of expression of miR-142-5p and miR-155 in MALT lymphoma lesions that were resistant to H. pylori eradication were significantly higher than in cases showing complete remission (CR) after H. pylori eradication (p<0.0001 and p<0.005, respectively). Although the API2-MALT1 fusion gene has been identified as a potential predictor of resistance to H. pylori eradication therapy, patients #7 and #8 with gastric MALT lymphoma lacking the API2-MALT1 fusion gene were resistant to H. pylori eradication. These patients showed increased expression levels of miR-142-5p and miR-155 (Figure 1).
In addition, we investigated the correlation between the levels of miR-142-5p and miR-155 expression and the clinical courses of gastric MALT lymphoma cases. The expression data for patients #14 and #16 with gastric MALT lymphoma who achieved CR after H. pylori eradication therapy showed that the levels of miR-142-5p and miR-155 expression were significantly reduced after eradication (Figure 2B, p<0.05 and p<0.05, respectively). On the other hand, in patient #1 with gastric MALT lymphoma harboring the API2-MALT1 fusion gene that was resistant to H. pylori eradication, there was no significant difference in the expression levels of miR-142-5p and miR-155 (Figure 2B). Although the H. pylori infection was eradicated by antibiotic treatment, histological examination showed no regression of the MALT lymphoma lesion. This patient was followed up by observation without treatment, and no marked changes in the endoscopic or histological findings were observed. These findings indicate that the levels of miR-142-5p and miR-155 expression have potential applicability as novel biomarkers of gastric MALT lymphoma.
Overexpression of miR-142-5p and miR-155 in an animal model of gastric MALT lymphoma
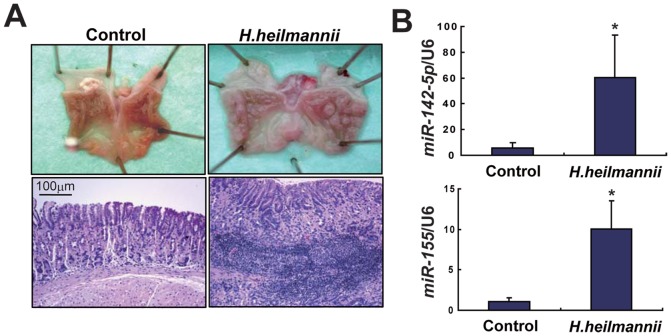
To further confirm the molecular pathogenesis of gastric MALT lymphoma, we examined the expression levels of miR-142-5p and miR-155 in an animal model of gastric MALT lymphoma. Infection with H. heilmannii induces gastric LELs that are consistent with low-grade MALT lymphomas in C57BL/6 mice [22]. Infection with H. heilmannii was confirmed using quantitative PCR analysis of stool DNA (data not shown). Ten months after infection with H. heilmannii, control (n = 3) and H. heilmannii-infected C57BL/6 mice (n = 4) were dissected, and this revealed protruding lesions in the gastric fundus in all of the latter. Light microscopic observations using hematoxylin and eosin (HE) staining revealed the presence of LELs consistent with low-grade MALT lymphomas (Figure 3A). Quantitative RT-PCR analyses showed that the levels of expression of miR-142-5p and miR-155 were significantly higher in H. heilmannii-infected C57BL/6 mice (p<0.05 and p<0.05, respectively), being similar to the results for human gastric MALT lymphomas (Figure 3B).
Figure 3. Expression levels of miR-142-5p and miR-155 in an animal model of gastric MALT lymphoma.
(A) Macroscopic and light microscopic findings with HE staining of the stomach of control and H. heilmannii-infected mice (10 months after infection). Protrusive lesions in the fundic stomach of H. heilmannii-infected mice were observed. Light microscopic observations using HE staining revealed the presence of LELs consistent with low-grade MALT lymphoma. (B) Expression levels of miR-142-5p and miR-155 in the stomach of control and H. heilmannii-infected mice. The levels of miR-142-5p and miR-155 expression normalized to the level of U6 RNA were significantly increased in H. heilmannii-infected C57BL/6 mice. * p<0.05.
miR-142-5p and miR-155 suppress the proapoptotic gene TP53INP1 as their target
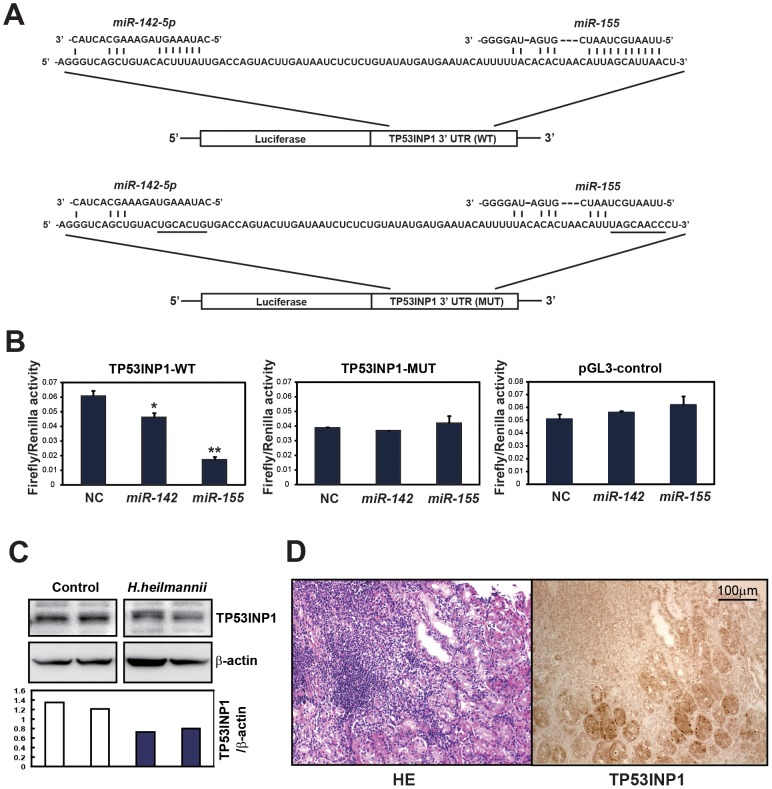
Identification of miRNA target genes is essential for determining miRNA function. Recent studies have indicated that a single miRNA may regulate more than 200 target genes. A database for predicting target genes, TargetScan (http://www.targetscan.org), revealed that both miR-142-5p and miR-155 are able to bind to the 3′ UTR of the mRNA of the proapoptotic gene TP53INP1 (Tumor Protein P53 Inducible Nuclear Protein 1). Moreover, miR-155 has been shown to repress TP53INP1, which inhibits the development of pancreatic tumors [29]. Therefore, we focused on TP53INP1 as a common target of miR-142-5p and miR-155.
To confirm the target specificity of miR-142-5p and miR-155 for TP53INP1, we performed a luciferase reporter assay using a vector containing the putative TP53INP1 3′ UTR target sites downstream of the luciferase reporter gene, which was transfected into AGS cells. The base pairing between miR-142-5p and miR-155 and the wild-type (WT) or mutant (MUT) target sites in the 3′ UTR of TP53INP1 mRNA is shown in Figure 4A. The luciferase activities of the AGS cells transfected with the TP53INP1-WT construct were significantly lower after transfection with miR-142-5p and miR-155 (p<0.05 and p<0.005, respectively), whereas those of cells transfected with the TP53INP1-MUT construct and the pGL3 control vector (empty vector) showed no significant differences (Figure 4B). It has been shown that conserved perfect 6- to 8-bp matches between the 5′ end of the mature miRNA and the 3′ UTR of the predicted target mRNA (called ‘seed’ matches) are the most important factor determining miRNA targets [30]. As shown in Fig. 4A, ‘seed’ matches between the 5′ end of the miRNAs and the 3′ UTR of TP53INP1 were stronger in miR-155 than in miR-142, suggesting that miR-155 may have a more profound effect on suppression of TP53INP1.
Figure 4. miR-142-5p and miR-155 suppress TP53INP1 as their target.
(A) Luciferase reporter constructs of wild-type (WT) and mutant (MUT) target sites in the 3′ UTR of TP53INP1 mRNA. The base pairings between miR-142-5p and miR-155 and their putative target sites in the 3′ UTR of TP53INP1 mRNA are shown. Underlining indicates mutant sequences. (B) Firefly luciferase activity was normalized to the Renilla luciferase activity of TP53INP1 wild-type (WT), mutant (MUT), and pGL3 control (empty vector) in AGS cells transfected with negative control (NC), miR-142-5p, and miR-155. The luciferase activities of the AGS cells transfected with the TP53INP1-WT construct were significantly lower after transfection of miR-142-5p and miR-155, whereas those transfected with the TP53INP1-MUT construct or the pGL3 control vector (empty vector) showed no significant differences. * p<0.05 and ** p<0.005, compared with negative control. (C) Western blot analysis of TP53INP1 in control and H. heilmannii-infected mice. β-actin was used as a loading control. The expression of TP53INP1 was suppressed in H. heilmannii-infected mice, compared with the expression level in control mice. (D) Immunohistochemistry for TP53INP1 in human gastric MALT lymphoma. Sections were counterstained with HE. TP53INP1 staining was markedly reduced in the MALT lymphoma lesions.
We next examined the expression levels of TP53INP1 by Western blot analysis in H. heilmannii-infected mice and immunohistochemistry in human gastric MALT lymphoma-like lesions. The expression of TP53INP1 was suppressed in H. heilmannii-infected mice, relative to control mice (Figure 4C). We examined the expression levels of TP53INP1 in human gastric tissues by immunohistochemistry. The gastric tissue shown in Fig. 4D contains both MALT lymphoma and non-MALT lymphoma tissue. The infiltrating lymphocytic lesion shown by HE staining is consistent with MALT lymphoma. TP53INP1 staining is negative or faint in the MALT lymphoma lesion, whereas normal gastric glands around the MALT lymphoma show sufficient staining for TP53INP1. Thus TP53INP1 staining was markedly reduced in MALT lymphoma lesions (Figure 4D). These findings suggest that TP53INP1 is a common target of miR-142-5p and miR-155 and is suppressed by overexpression of both miR-142-5p and miR-155 in gastric MALT lymphoma lesions.
Discussion
A recent study has demonstrated that miRNA expression profiles can be used to classify the developmental lineages and differentiation stages of tumors, and are more accurate for tumor classification than conventional mRNA profiles [31]. Furthermore, miRNA expression signatures are associated with prognostic factors and disease progression in chronic lymphocytic leukemia [11] and lung cancer [32]. Thus miRNA expression is clinically promising as both a diagnostic tool and a prognostic marker for human malignancies. The results of microarray analysis revealed a unique miRNA expression profile in gastric MALT lymphoma. Recent studies have demonstrated that the expression level of miR-155 is significantly elevated in DLBCL, which is considered to represent high-grade transformation from MALT lymphoma [20], [33]. miR-142, miR-155 and miR-223 have been reported to be hematopoiesis-specific miRNAs [24]. These findings are consistent with our results, and in the present study we focused on miR-142 and miR-155, because miR-142 is the most up-regulated miRNA and miR-155 plays a critical role in the pathogenesis of B-cell lymphoma. The levels of miR-142-5p and miR-155 expression were associated with the clinical course of gastric MALT lymphoma, including the response to H. pylori eradication. The API2-MALT1 fusion gene has been identified as a potential predictor of resistance to H. pylori eradication therapy. In the present study, two cases of gastric MALT lymphoma resistant to H. pylori eradication lacked the API2-MALT1 fusion gene (patients #7, #8). These cases showed increased expression of miR-142-5p and miR-155. Therefore, these miRNAs may be more useful markers than the API2-MALT1 fusion gene in patients with gastric MALT lymphoma.
TP53INP1 is a proapoptotic stress-induced p53 target gene. p53 activates TP53INP1 transcription, and overexpression of TP53INP1 induces cell cycle arrest and apoptosis [34]. Gironella et al. [29] have shown that oncogenic miR-155 is overexpressed in pancreatic ductal adenocarcinoma and suppresses its target, TP53INP1, resulting in cancer progression. In agreement with these findings, our results showed that TP53INP1 was suppressed by both miR-142-5p and miR-155, possibly leading to inhibition of apoptosis and acceleration of MALT lymphoma cell proliferation. These findings suggest that overexpression of miR-142-5p and miR-155 concomitant with suppression of TP53INP1 reflect the increased proliferation of MALT lymphoma cells. The distinct connection between aberrant expression of miR-142-5p and miR-155 and the progression of MALT lymphoma suggests that miRNAs could be potential therapeutic targets. A recent study has shown that chemically engineered oligonucleotides, termed ‘antagomirs,’ can work as specific inhibitors of endogenous miRNAs in mice [35], and might be potentially applicable to silence miR-142-5p and miR-155 for the treatment of gastric MALT lymphomas that are resistant to H. pylori eradication therapy.
Conclusions
Overexpression of miR-142-5p and miR-155 is presumed to play a critical role in the initiation and progression of gastric MALT lymphoma, suggesting that these miRNAs may be potentially useful as therapeutic targets and novel biomarkers for gastric MALT lymphomas. Inhibition of miR-142-5p and miR-155 might be a novel approach for the prevention and treatment of gastric MALT lymphoma. Further studies involving more patients are warranted to explore the clinical use of miR-142-5p and miR-155 for diagnosis and treatment of gastric MALT lymphoma.
Funding Statement
This work was supported by Grant-in-Aid for Young Scientists A (23680090 to Y.S.) and Grant-in-Aid for Scientific Research B (22300169 to H.S.) from Japan Society for Promotion of Science (www.jsps.go.jp), Takeda Science Foundation (www.takeda-sci.or.jp; to Y.S.), Sagawa Foundation for Promotion of Cancer Research (sagawa-gan.or.jp; to Y.S.), Smoking Research Foundation (www.srf.or.jp; to H.S.), and Keio Gijuku Academic Development Funds (www.keio.ac.jp; to Y.S. and H.S.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sackmann M, Morgner A, Rudolph B, Neubauer A, Thiede C, et al. (1997) Regression of gastric MALT lymphoma after eradication of Helicobacter pylori is predicted by endosonographic staging. MALT Lymphoma Study Group. Gastroenterology 113: 1087–1090. [DOI] [PubMed] [Google Scholar]
- 2. Ruskone-Fourmestraux A, Lavergne A, Aegerter PH, Megraud F, Palazzo L, et al. (2001) Predictive factors for regression of gastric MALT lymphoma after anti-Helicobacter pylori treatment. Gut 48: 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suzuki H, Hibi T, Marshall BJ (2007) Helicobacter pylori: present status and future prospects in Japan. J Gastroenterol 42: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Akagi T, Motegi M, Tamura A, Suzuki R, Hosokawa Y, et al. (1999) A novel gene, MALT1 at 18q21, is involved in t(11;18) (q21;q21) found in low-grade B-cell lymphoma of mucosa-associated lymphoid tissue. Oncogene 18: 5785–5794. [DOI] [PubMed] [Google Scholar]
- 5. Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez JM, et al. (1999) The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood 93: 3601–3609. [PubMed] [Google Scholar]
- 6. Liu H, Ye H, Ruskone-Fourmestraux A, De Jong D, Pileri S, et al. (2002) T(11;18) is a marker for all stage gastric MALT lymphomas that will not respond to H. pylori eradication. Gastroenterology 122: 1286–1294. [DOI] [PubMed] [Google Scholar]
- 7. Motegi M, Yonezumi M, Suzuki H, Suzuki R, Hosokawa Y, et al. (2000) API2-MALT1 chimeric transcripts involved in mucosa-associated lymphoid tissue type lymphoma predict heterogeneous products. Am J Pathol 156: 807–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hosokawa Y, Suzuki H, Suzuki Y, Takahashi R, Seto M (2004) Antiapoptotic function of apoptosis inhibitor 2-MALT1 fusion protein involved in t(11;18)(q21;q21) mucosa-associated lymphoid tissue lymphoma. Cancer Res 64: 3452–3457. [DOI] [PubMed] [Google Scholar]
- 9. Farinha P, Gascoyne RD (2005) Molecular pathogenesis of mucosa-associated lymphoid tissue lymphoma. J Clin Oncol 23: 6370–6378. [DOI] [PubMed] [Google Scholar]
- 10. He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531. [DOI] [PubMed] [Google Scholar]
- 11. Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, et al. (2005) A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 353: 1793–1801. [DOI] [PubMed] [Google Scholar]
- 12. Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866. [DOI] [PubMed] [Google Scholar]
- 13. Saito Y, Suzuki H, Hibi T (2009) The role of microRNAs in gastrointestinal cancers. J Gastroenterol 44 Suppl 19: 18–22. [DOI] [PubMed] [Google Scholar]
- 14. Cho WC (2010) MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol 42: 1273–1281. [DOI] [PubMed] [Google Scholar]
- 15. Cho WC (2010) MicroRNAs in cancer - from research to therapy. Biochim Biophys Acta 1805: 209–217. [DOI] [PubMed] [Google Scholar]
- 16. Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, et al. (2006) Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9: 435–443. [DOI] [PubMed] [Google Scholar]
- 17. Saito Y, Jones PA (2006) Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle 5: 2220–2222. [DOI] [PubMed] [Google Scholar]
- 18. Saito Y, Friedman JM, Chihara Y, Egger G, Chuang JC, et al. (2009) Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem Biophys Res Commun 379: 726–731. [DOI] [PubMed] [Google Scholar]
- 19. Saito Y, Suzuki H, Tsugawa H, Nakagawa I, Matsuzaki J, et al. (2009) Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene 28: 2738–2744. [DOI] [PubMed] [Google Scholar]
- 20. Eis PS, Tam W, Sun L, Chadburn A, Li Z, et al. (2005) Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A 102: 3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A (2004) High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer 39: 167–169. [DOI] [PubMed] [Google Scholar]
- 22. Nakamura M, Murayama SY, Serizawa H, Sekiya Y, Eguchi M, et al. (2007) “Candidatus Helicobacter heilmannii” from a cynomolgus monkey induces gastric mucosa-associated lymphoid tissue lymphomas in C57BL/6 mice. Infect Immun 75: 1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen CZ, Li L, Lodish HF, Bartel DP (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86. [DOI] [PubMed] [Google Scholar]
- 24. Ramkissoon SH, Mainwaring LA, Ogasawara Y, Keyvanfar K, McCoy JP Jr, et al. (2006) Hematopoietic-specific microRNA expression in human cells. Leuk Res 30: 643–647. [DOI] [PubMed] [Google Scholar]
- 25. Calin GA, Croce CM (2006) MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene 25: 6202–6210. [DOI] [PubMed] [Google Scholar]
- 26. Lister TA, Crowther D, Sutcliffe SB, Glatstein E, Canellos GP, et al. (1989) Report of a committee convened to discuss the evaluation and staging of patients with Hodgkin's disease: Cotswolds meeting. J Clin Oncol 7: 1630–1636. [DOI] [PubMed] [Google Scholar]
- 27. Nakamura S, Sugiyama T, Matsumoto T, Iijima K, Ono S, et al. (2012) Long-term clinical outcome of gastric MALT lymphoma after eradication of Helicobacter pylori: a multicentre cohort follow-up study of 420 patients in Japan. Gut 61: 507–513. [DOI] [PubMed] [Google Scholar]
- 28. Akamatsu T, Mochizuki T, Okiyama Y, Matsumoto A, Miyabayashi H, et al. (2006) Comparison of localized gastric mucosa-associated lymphoid tissue (MALT) lymphoma with and without Helicobacter pylori infection. Helicobacter 11: 86–95. [DOI] [PubMed] [Google Scholar]
- 29. Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, et al. (2007) Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci U S A 104: 16170–16175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 31. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. (2005) MicroRNA expression profiles classify human cancers. Nature 435: 834–838. [DOI] [PubMed] [Google Scholar]
- 32. Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, et al. (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9: 189–198. [DOI] [PubMed] [Google Scholar]
- 33. Fang C, Zhu DX, Dong HJ, Zhou ZJ, Wang YH, et al. (2012) Serum microRNAs are promising novel biomarkers for diffuse large B cell lymphoma. Ann Hematol 91: 553–559. [DOI] [PubMed] [Google Scholar]
- 34. Tomasini R, Seux M, Nowak J, Bontemps C, Carrier A, et al. (2005) TP53INP1 is a novel p73 target gene that induces cell cycle arrest and cell death by modulating p73 transcriptional activity. Oncogene 24: 8093–8104. [DOI] [PubMed] [Google Scholar]
- 35. Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, et al. (2005) Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438: 685–689. [DOI] [PubMed] [Google Scholar]