Abstract
The German Shepherd Dog (GSD) is a popular working and companion breed for which over 50 hereditary diseases have been documented. Herein, SNP profiles for 197 GSDs were generated using the Affymetrix v2 canine SNP array for a genome-wide association study to identify loci associated with four diseases: pituitary dwarfism, degenerative myelopathy (DM), congenital megaesophagus (ME), and pancreatic acinar atrophy (PAA). A locus on Chr 9 is strongly associated with pituitary dwarfism and is proximal to a plausible candidate gene, LHX3. Results for DM confirm a major locus encompassing SOD1, in which an associated point mutation was previously identified, but do not suggest modifier loci. Several SNPs on Chr 12 are associated with ME and a 4.7 Mb haplotype block is present in affected dogs. Analysis of additional ME cases for a SNP within the haplotype provides further support for this association. Results for PAA indicate more complex genetic underpinnings. Several regions on multiple chromosomes reach genome-wide significance. However, no major locus is apparent and only two associated haplotype blocks, on Chrs 7 and 12 are observed. These data suggest that PAA may be governed by multiple loci with small effects, or it may be a heterogeneous disorder.
Introduction
Shepherds and hounds were the first dog breeds developed to serve in specialized roles (Goldbecker and Hart 1967). The German Shepherd Dog (GSD), for which a breed standard was originally developed in 1899, was bred for utility and intelligence and to be a multipurpose servant of humans (Goldbecker and Hart 1967). Aptitude, temperament, structural efficiency, and other natural skills were deemed more important than aesthetic traits. Today, the GSD is among the most common breeds trained by the military, law enforcement, and service/assistance programs (Moody et al. 2006). The GSD is also an extremely popular companion dog breed, ranking second in breed registration statistics reported by the American Kennel Club in 2010. Unfortunately, despite a large population size, more than 50 hereditary diseases have been described for the GSD (Wahl et al. 2008).
Pituitary dwarfism is a rare disease of the GSD and is characterized by an abnormally small body stature because of a deficiency in pituitary growth hormone. Dogs appear to be normal at birth, but signs of stunted growth are usually evident by 2–3 months of age. There are no treatments available and life span is usually significantly shortened. Pituitary dwarfism in the GSD is inherited in an autosomal recessive fashion (Andresen and Willeberg 1976). Combined pituitary hormone deficiency (CPHD) in humans is a type of growth hormone deficiency that results from a decrease in several hormones necessary for pituitary development. Mutations that cause congenital CPHD in humans have been identified in genes encoding several transcription factors important for pituitary development. These include PIT1, PROP1, LHX3, LHX4, and HESX1 (Reynaud et al. 2004). In contrast, sequencing in affected GSDs of PIT1 (Lantinga-van Leeuwen et al. 2000b), PROP1 (Lantinga-van Leeuwen et al. 2000a), LHX4 (van Oost et al. 2002), and LIFR (Hanson et al. 2006), did not reveal any causative variants.
Degenerative myelopathy (DM) is a late-onset neurodegenerative disease characterized by ataxia and weakness in the hind limbs. Symptoms of DM worsen over time, either steadily or in phases, and eventually result in complete paraplegia. In some cases, DM may progress up the spinal cord and cause forelimb weakness or even respiratory difficulties (Barclay and Haines 1994). The age of onset of DM is generally between 5 and 14 years, with a mean age of onset of 9 years (Averill 1973). The clinical signs observed in DM are general indicators of spinal cord disorders, but a definitive diagnosis for DM can be made only by histopathological examination of spinal cord tissue postmortem (Coates et al. 2007). Most owners of affected dogs elect euthanasia within several months of symptom onset (Johnston et al. 2000). DM is most prevalent in the GSD (Coates et al. 2007). A recessive missense mutation in SOD1 is associated with DM in several breeds, including the GSD (Awano et al. 2009). DM has been proposed to be an animal model for amyotrophic lateral sclerosis (ALS). Approximately 20% of hereditary ALS cases are attributed to mutation of SOD1 (Rosen et al. 1993).
Congenital idiopathic megaesophagus (ME) is characterized by dilation and hypomotility of the esophagus. The result of these abnormalities is regurgitation several minutes to hours after eating. Regurgitation episodes may occur as often as several times a day or as infrequently as once every few days. Symptoms for congenital ME typically begin around 5 weeks of age after weaning onto solid food. Affected puppies are malnourished, show a general failure to thrive, and are at risk for aspiration pneumonia. Diagnosis of ME is achieved by standard thoracic radiographs and/or fluoroscopy (barium swallow) (Guilford 1990). ME is typically treated by feeding a high-calorie liquid diet from a raised dish (Guilford 1990). Mortality is high in affected neonates. Many survivors require lifelong management of the disorder, but other cases resolve by 4–6 months of age (Cox et al. 1980). ME is prevalent in the GSD, Great Dane, Miniature Schnauzer, Rhodesian Ridgeback, Dachshund, and Wire Fox Terrier, and is also reported to occur at lower frequencies in many other breeds (Osborne et al. 1967). Several loci are thought to be responsible for ME in the dog (Cox et al. 1980). The mode of inheritance in the GSD has not been investigated.
Pancreatic acinar atrophy (PAA) is an autoimmune disease characterized by the selective atrophy of the acinar cells of the exocrine pancreas, which synthesize and secrete pancreatic digestive enzymes under physiologic conditions (Wiberg etal. 1999). PAA is the most common cause of exocrine pancreatic insufficiency (EPI) in the dog (Westermarck et al. 1993) and is diagnosed through histopathologic examination (Pfister et al. 1980). EPI is diagnosed through the measurement of serum canine trypsin-like immunoreactivity (cTLI), with concentrations ≤2.5 μg/l considered as diagnostic (Williams and Batt 1988). By 5 years of age, 96% of affected dogs present with clinical signs of EPI, which include weight loss, increased appetite, and soft stools (Räihä and Westermarck 1989; Westermarck et al. 1993). Affected dogs can be treated with pancreatic enzyme supplements, although dogs with EPI are sometimes euthanized because of the expense associated with treatment or a poor response to treatment (Hall et al. 1991; Wiberg and Westermarck 2002). PAA is widespread in the GSD population and affects other breeds including the Collie and Eurasian (Proschowsky and Fredholm 2007; Wiberg 2004). A recent test mating in the GSD shows that EPI is likely a polygenic disorder (Westermarck et al. 2010).
The development of high-throughput genotyping technologies has facilitated the discovery of genes responsible for simple and complex phenotypes of the dog (Karlsson et al. 2007; Sutter et al. 2007). The population structure of the dog is particularly well-suited for association mapping techniques because genetic bottlenecks during breed formation created large blocks of linkage disequilibrium within breeds (Karlsson et al. 2007). Reported here is the use of a large cohort of GSDs to investigate the aforementioned four diseases that segregate in the population. Genetic profiles for 197 GSDs were generated using the Affymetrix v2 canine SNP array. Using a single data set, we carried out genome-wide association studies (GWAS) to identify risk loci for four diseases of the GSD.
Materials and methods
Sample collection
Samples were recruited from individual owners from purebred GSDs that were diagnosed with EPI, DM, or ME through standard veterinary diagnostic practices. DM cases were diagnosed by veterinary neurologists. Dogs were considered to have congenital, idiopathic ME if they did not have conditions that can cause secondary ME, such as a persistent right aortic arch, myasthenia gravis, or hypoadrenocorticism. Purebred GSDs collected as healthy controls were ≥6 years and, to the owners’ knowledge, had no immediate family members affected with EPI, DM, or ME. Owners were asked to report all known hereditary conditions as well as coat color phenotypes. Participants were recruited through breed clubs and listservs.
Whole blood or buccal swabs were submitted by dog owners. Blood samples were collected by the primary-care veterinarian of the dog. All samples were collected in accordance with protocols approved by the Clinical Research Review Committee (CRRC No. 07-04) at Texas A&M University and the Clemson University Institutional Review Board (IBC2008-17). Genomic DNA was isolated using the Gentra Puregene Blood Kit (Qiagen, Valencia, CA, USA). Serum samples were collected from all EPI cases and healthy controls and submitted to the Gastrointestinal Laboratory at Texas A&M University for measurement of serum cTLI concentration to confirm clinical status. Concentrations ≤2.5 μg/l were considered diagnostic for EPI. Control dogs had concentrations ≥5.7 μg/l. All EPI cases were assumed to result from PAA, but tissue samples were not available to differentiate EPI due to PAA or another cause of EPI.
GWAS
SNP genotypes were generated using the Affymetrix v2 canine SNP array (http://www.broadinstitute.org/mammals/dog/caninearrayfaq.html). Arrays were processed at the National Human Genome Research Institute (NHGRI) or Clemson University Genomics Institute using the GeneChip human mapping 250K Sty assay kit (Affymetrix, Santa Clara, CA, USA). All associated protocols were approved by the NHGRI Animal Care and Use Committee. The GeneChip human mapping 500K assay protocol was followed with a hybridization volume of 125 μl (Karlsson et al. 2007). Raw CEL files were analyzed with the Affymetrix Power Tools software to obtain genotypes. SNPs having >10% missing data and ≥60% heterozygosity were removed from the data set. Case/control analyses were performed on the 48,415 SNPs from the “platinum” set using PLINK (Purcell et al. 2007), with Praw <0.0001 considered significant. To determine Pgenome values (EMP2), permutation testing (100,000) was performed for PAA, DM, and ME analyses using PLINK. A less conservative correction, Benjamini–Hochberg, was also performed (Benjamini and Hochberg 1995). Analysis of population stratification in 197 GSDs was conducted using identity-by-state clustering and multidimensional scaling in PLINK. The data, in a reduced representation, were plotted in two dimensions: principal components 1 and 2.
Genotyping of SNP 12.60274687
Primers flanking the SNP 12.60274687 were designed: forward: 5′-TGAGCACACAGAGGTGAGACAT-3′ and reverse: 5′-CAGTGGGAGGGTTTAGGAAGAGAT-3′. SNP 12.60274687 was amplified for 35 additional ME dogs using Thermo ReddyMix (Thermo Fisher Scientific, Waltham, MA, USA) following the recommended protocol. Thermal cycling conditions were as follows: 95°C for 15 min; 14 cycles of 95°C for 30 s, 62°C for 1 min (decreasing 0.5°C each cycle), 72°C for 1 min; 20 cycles of 95°C for 30 s, 55°C for 1 min, 72°C for 1 min; 72°C for 10 min. PCR amplicons were analyzed by electrophoresis on agarose gels. Products were purified by adding 0.5 unit exonuclease I (New England Biolabs, Ipswich, MA, USA) and 0.25 unit of shrimp alkaline phosphatase (Promega, Madison, WI, USA) and incubating for 30 min at 37°C followed by 20 min at 80°C. Nucleotide sequencing was accomplished using the Big Dye Terminator version 3.1 Cycle Sequencing kit (Applied Biosystems, Carlsbad, CA, USA). Products were purified using Spin-50 mini columns with water (BioMax, Inc., Arnold, MD, USA) and resolved on an ABI Prism 3730 Genetic Analyzer (Applied Bio-systems). Sequences were analyzed to determine SNP genotypes. Odds ratios (OR), critical intervals, and χ2 tests were calculated to assess the differences in genotype frequencies between the two populations using 2 × 2 contingency tables.
Results
Validation of methodology
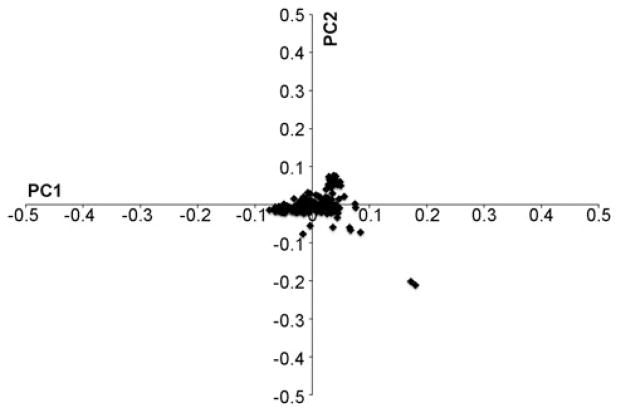
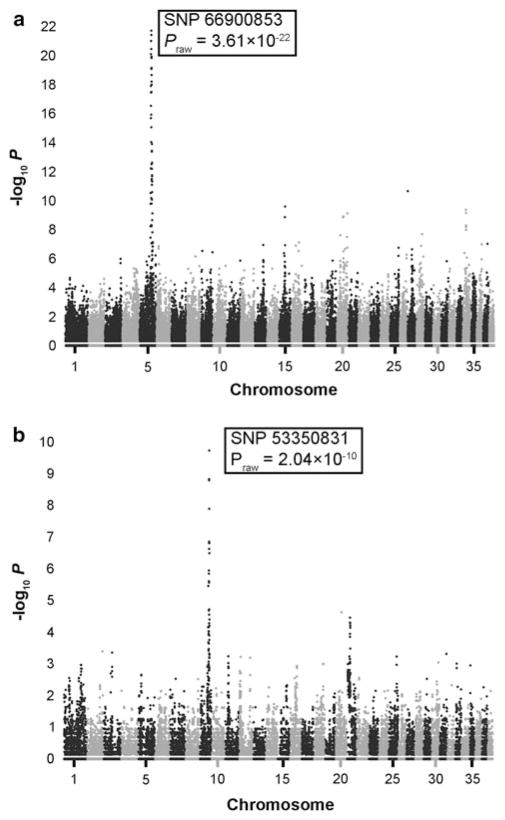
In total, 197 GSD samples were acquired from the United States, Canada, Germany, and the Netherlands. Two GSDs were affected with EPI and DM. One GSD was affected with EPI and ME. Principal component analysis with PLINK showed no evidence for population stratification within our GSD cohort (n = 197) using all “platinum” SNPs (Fig. 1). To validate both our genotype data and statistical approach, we first mapped a positive control trait. Ten dogs in the study cohort had white coats, a recessive trait caused by the e allele of MC1R in the GSD (Schmutz and Berryere 2007). GWAS using 197 GSDs (10 cases, 187 controls) reveals a strongly associated region encompassing MC1R on Chr 5 (Fig. 2a). SNP 5.66900853, the most significant result (Praw = 3.61 × 10−22), is located approximately 208 kb adjacent to the E locus. The lack of subpopulations, combined with our ability to accurately map a known locus, indicates the utility of this data set.
Fig. 1.
Principal component analysis of the GSD cohort shows all 197 dogs clustering together. The x axis is principal component 1 and the y axis is principal component 2. Two individuals that appear apart from the main grouping have the largest proportion of missing data (22.7 and 22.4% overall, compared to the average missing data rate of 12.6%)
Fig. 2.
Manhattan plots showing the results for GWAS using 48,415 SNPs. The genome-wide P values (−log10 P) for each SNP are plotted against position on each chromosome. a Ten white GSDs versus 187 nonwhite GSDs show a strong association on Chr 5. b Four pituitary dwarf GSDs versus 193 normal-sized GSDs show a strong association on Chr 9
Pituitary dwarfism
Four dogs in the control population had pituitary dwarfism. GWAS for these four cases versus 193 normal-sized controls shows a major region of association on Chr 9 (Fig. 2b). Within this region, all four pituitary dwarfs are homozygous for a haplotype extending from SNP 9.52031784 to SNP 9.54102643. The haplotype is not observed in the homozygous state among the control dogs; however, 15 control dogs were heterozygotes. The most significant association is for SNP 9.53350831 (Praw = 2.04 × 10−10), which is located approximately 764 kb from a candidate gene, LHX3.
Degenerative myelopathy
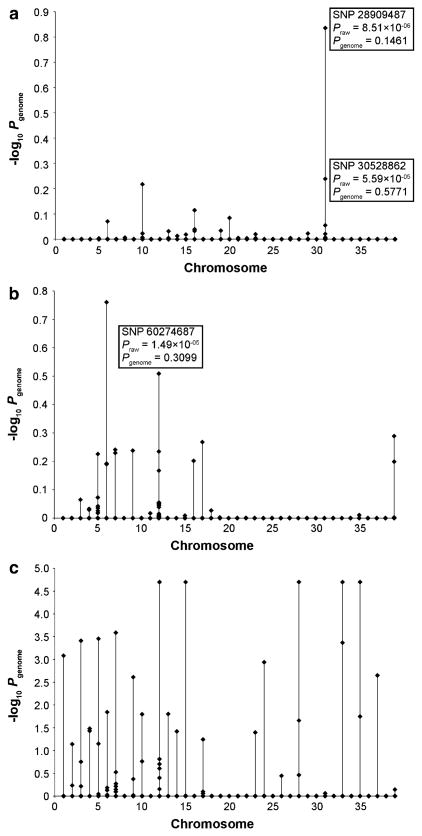
GWAS with 100,000 permutations for DM was carried out using all cases collected (n = 15) versus control dogs that were at least 8 years of age and did not exhibit any clinical signs of DM (n = 69). The two strongest associations are with SNPs 31.28909487 (Praw = 8.51 × 10−06, Pgenome = 0.1461) and 31.30528862 (Praw = 5.59 × 10−05, Pgenome = 0.5771) (Fig. 3a). These SNPs on Chr 31 are located 650 kb upstream and 965 kb downstream of SOD1, respectively. Evaluation of individual genotypes in this region reveals that only 4 of the 15 cases are homozygous for both SNPs. Five cases are homozygous for the associated allele of SNP 31.30528862, while nine cases are homozygous for the associated allele of SNP 31.28909487. In the control population, 6 of 69 GSDs are also homozygous for the same allele of SNP 31.28909487. A third noteworthy result is on Chr 10 for SNP 10.64801770 (Praw = 6.06 × 10−05, Pgenome = 0.6066).
Fig. 3.
GWAS using 48,415 SNPs with 100,000 permutations. The genome-wide adjusted P values (−log10 P) for each SNP are plotted by chromosome. a Fifteen GSDs with DM versus 69 healthy control GSDs. b Nineteen GSDs with ME versus 177 healthy control GSDs. c One-hundred GSDs with PAA versus 79 healthy control GSDs
Megaesophagus
GWAS with 100,000 permutations was carried out using 19 cases and 177 control dogs. Fourteen SNPs with significant Praw values are located on eight different chromosomes (Fig. 3b). Of these, three are within an 8 Mb region on Chr 12. Five other SNPs within this region are approaching significance (Praw). Further analyses of genotypes on Chr 12 reveal a haplotype block extending 4.7 Mb that is present in all affected dogs. The strongest associated of these is SNP 12.60274687 (Praw = 1.49 × 10−5, Pgenome = 0.3099). Analysis of genotypes for this SNP reveals a common allele that is present in all affected dogs (frequency = 0.83) and is less prevalent among control dogs (frequency = 0.46). To further investigate this finding, 35 additional GSDs with ME were collected and genotyped for the SNP. Genotype frequencies and odds ratios for SNP 12.60274687 are given in Table 1.
Table 1.
Frequency data for SNP 12.60274687 in GSDs with and without ME
| G/Gb (%) | G/A (%) | A/A (%) | |
|---|---|---|---|
| MEarray (n = 19) | 12 (63) | 7 (37) | 0 (0) |
| MEall (n = 54) | 33 (61) | 20 (37) | 1 (2) |
| Control (n = 172a) | 36 (21) | 84 (49) | 52 (30) |
Observed genotypes and genotypic frequencies are presented for affected and control dogs. MEarray = genotype frequencies of the 19 ME dogs run on Affymetrix SNP arrays; MEall = All ME dogs collected
Four control dogs were N/N for SNP 12.60274687 on the array and were excluded from this analysis
Genotype (G/G): P = 6.84 × 10−8, OR = 5.937 (95% CI = 3.071–11.475); Allele (G): P = 2.77 × 10−10, OR = 4.711 (95% CI = 2.817–7.878)
Pancreatic acinar atrophy
GWAS with 100,000 permutations was carried out using 100 dogs with PAA and 79 control dogs (Fig. 3c). Significant Praw values are present for 50 SNPs on 23 chromosomes (Supplementary Table 1). Chr 12 harbors seven of these SNPs, three of which are found in a cluster: 12.3781476, 12.3845215, and 12.4698937. Chr 7 harbors six SNPs and four are found in a cluster: 7.11222056, 7.12305239, 7.12310223, and 7.12424701. On ten of the chromosomes, only one SNP each is strongly linked with PAA. On the remaining 11 chromosomes, no two strongly associated SNPs are located within 5 Mb of each other and no common haplotypes are detected. Six SNPs had minor allele frequencies (MAF) <15%.
Discussion
Pituitary dwarfism
Autosomal recessive traits of the dog can be mapped using remarkably small numbers of cases (Drögemüller et al. 2009; Gill et al. 2011). In this study, a mere four cases were sufficient to detect a strong linkage to Chr 9 and define a 2 Mb pituitary dwarfism haplotype. While the critical interval harbors more than 40 genes, one is an intriguing candidate: LHX3, a LIM homeodomain transcription factor vital for pituitary and motor neuron development (Bach et al. 1995). Genetic variations within LHX3 cause CPHD in humans (Netchine et al. 2000; Pfaeffle et al. 2007). Mutations in LHX3 in humans are associated with a rigid cervical spine, and occasionally mental retardation and deafness (Bhangoo et al. 2006; Netchine et al. 2000; Rajab et al. 2008). The GSD pituitary dwarfs do not have clinical signs that indicate nervous system involvement (Kooistra et al. 2000). In the mouse, complete loss of LHX3 is lethal (Sheng et al. 1996), but mutations within the gene cause a dwarfism phenotype (Colvin et al. 2011; Lane and Dickie 1968).
Voorbij et al. (2010) announced the availability of a genetic test for pituitary dwarfism in the GSD, but information regarding the gene or mutation has not been published to date. Dwarfism in the GSD is rare, so utilization of a definitive genetic test could facilitate rapid elimination of the phenotype. Data presented here are surprising because the frequency of the 2 Mb haplotype in control dogs is 4%, suggesting a roughly 8% carrier frequency in the general population. Moreover, this estimate is likely low because only dogs having the complete 2 Mb haplotype were considered to be carriers, thereby excluding dogs with partial overlapping haplotypes that may include the causative mutation.
Degenerative myelopathy
DM is a devastating disorder of GSDs that has been difficult to eradicate from the breed, in part because of a late age of onset and poor diagnostic tools. We identified two SNPs on Chr 31 that are strongly associated with the DM phenotype. The SNPs flank SOD1, the gene in which Awano et al. (2009) identified an E40K missense mutation in DM-affected dogs from several breeds. Based on genotype frequencies, the authors propose that the mutation causes DM in an autosomal recessive fashion with incomplete penetrance (Awano et al. 2009). In the study reported here, only 9 of 15 cases were homozygous for the most tightly linked SNP, suggesting that either the relaxed diagnostic criteria used here (dogs were not diagnosed postmortem) led to the inclusion of dogs that were mis-diagnosed, or that heterozygosity for the mutation can produce an affected phenotype. Recombination between the SNP and SOD1 exon 2 mutation (located 652 kb apart) may also be a factor (5 cases are heterozygous). Six of 69 dogs in the control population were homozygous for the SNP, corresponding to approximately 9% undiagnosed controls. Similarly, Awano et al. (2009) reported that 6% of GSD breed controls were homozygous for the SOD1 mutation. The high frequency of clinically normal, aged controls that are homozygous for the mutation is consistent with reduced penetrance and suggests the involvement of modifier loci and/or environmental factors. In this analysis, a second locus was detected on Chr 10 and is supported by six proximal SNPs with Praw values approaching significance (<0.00086). Awano et al. (2009) detected weaker signals of association on Chrs 6, 18, 20, and 35. Data herein do not support regions of association on Chrs 18 and 35 but do show modest signals on Chrs 6 and 20.
Megaesophagus
The mode of inheritance of ME has not been previously investigated in the GSD, but it has been suggested that the frequent occurrence of affected puppies born to clinically normal parents serves as evidence for a recessive mutation. In this work we successfully mapped two simple autosomal recessive traits with ten and four cases and a complex recessive trait with 15 cases. GWAS using 19 dogs with ME, however, did not yield a single, strong region of association. Rather, minor associations were detected on eight different chromosomes. It is, therefore, unlikely that ME is inherited in an autosomal recessive fashion in the GSD. Similarly, evaluation of inheritance patterns of ME in the Miniature Schnauzer did not suggest a simple recessive mode of inheritance (Cox et al. 1980).
A locus on Chr 12 is the only one to reach genome-wide significance and have supporting proximal SNPs (8 SNPs having Praw values <0.00014). To verify an association, 35 additional ME GSDs were collected and genotyped for the most strongly associated SNP in this region. In total, 53 of 54 ME cases were heterozygous or homozygous for the associated allele, which occurs at a significantly lower frequency in the control population. These data are consistent with an autosomal dominant mode of inheritance, with incomplete penetrance and/or modifying loci. On the other hand, it is also conceivable that the allele is fully penetrant but that the control population includes dogs with undetected ME. ME varies in severity and can resolve completely after a few months, making detection of mild cases difficult.
The dilation and lack of muscle contraction in the esophagus is thought to be a neurological defect rather than a muscular defect (Guilford 1990). The associated haplotype on Chr 12 harbors approximately 12 genes, several of which are expressed in the nervous system. Additional genome-wide SNP analyses in ME GSDs are necessary to confirm this association.
Pancreatic acinar atrophy
Fifty SNPs, representing 45 loci, exceed genome-wide significance thresholds for association with PAA. Many of these map to Chr 12, although only three SNPs on the chromosome support the same locus. These SNPs map to the dog leukocyte antigen (DLA) complex in the centromeric region of the chromosome. The association of this region with PAA is consistent with the autoimmune component of the disease. Previous work showed that pancreases from GSDs with PAA show an increased expression of DLA-88 (Clark et al. 2005). DLA-88 is the most highly polymorphic class I locus of the DLA complex. DLA class II loci are associated with several immune-related disorders, including diabetes mellitus, anal furunculosis, hypoadrenocorticism, and necrotizing meningoencephalitis (Barnes et al. 2009; Greer et al. 2010; Hughes et al. 2010; Kennedy et al. 2006). Canine systemic lupus erythematosus, a classic autoimmune disease, is also reported to be associated with alleles of the DLA class II loci, and GWAS revealed five additional associated loci (Wilbe et al. 2009, 2010). The current SNP data, together with previous expression data, suggest that the DLA region may harbor a genetic risk factor for the development of PAA. Further investigation of the DLA loci is planned. A second region of interest is located on Chr 7, which has four strongly associated SNPs located within a 1.2 Mb region having 11 genes. None of these genes is known to be involved in autoimmune disorders, although several are expressed in acinar cells.
The data reported here suggest that PAA may be governed by multiple loci with small effects, or it may be a heterogeneous disorder. Although PAA was thought to be a simple autosomal recessive disorder (Moeller et al. 2002; Westermarck 1980), the results of a recent test mating between two GSDs with PAA indicate more complex genetic underpinnings (Westermarck et al. 2010). The study monitored the cTLI scores for the resulting litter of six over the course of the dogs’ lives (8–13 years) and the pancreas for each dog was examined at necropsy. Only two of the six dogs in the litter were diagnosed with PAA (Westermarck et al. 2010). Similarly, the clinical presentation of PAA is inconsistent. The age of onset is variable within families, with diagnoses made in puppies and adult dogs alike. The severity of clinical signs also varies, with some dogs responding better to traditional treatment regimens than others. To further complicate the matter, some GSDs never exhibit clinical signs, despite serum cTLI concentrations that are diagnostic for EPI. Variability in age of onset and clinical signs may be an indicator of genetic heterogeneity. Although PAA is the predominant cause of EPI in dogs, chronic pancreatitis, pancreatic cancer, and other underlying medical conditions may also cause EPI.
Variation between processed arrays, a so-called “batch effect,” can cause spurious associations (Hong et al. 2008). The abundance of isolated SNPs detected in our GWAS for PAA is suggestive of such artifacts. However, principal component analysis does not reveal subpopulations that indicate a batch effect. Furthermore, no population stratification is detected between PAA cases and controls (Supplementary Fig. 1). In addition, we included only the robust “platinum” SNPs in our analysis to minimize artifacts from incorrect genotype calls.
In this study, we used data from a single cohort of GSDs to identify major loci associated with pituitary dwarfism, DM, and ME. Our data also reveal that the genetics underlying PAA are highly complex. Candidate loci identified herein provide immediate targets for future work.
Supplementary Material
Acknowledgments
We thank the American Kennel Club Canine Health Foundation for supporting this work and the Intramural Program of the National Human Genome Research Institute. We are grateful to the numerous dog owners and veterinarians who provided samples.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00335-011-9376-9) contains supplementary material, which is available to authorized users.
Contributor Information
Kate L. Tsai, Department of Genetics and Biochemistry, College of Agriculture, Forestry and Life Sciences, Clemson University, Clemson, SC 29634, USA
Rooksana E. Noorai, Department of Genetics and Biochemistry, College of Agriculture, Forestry and Life Sciences, Clemson University, Clemson, SC 29634, USA
Alison N. Starr-Moss, Department of Genetics and Biochemistry, College of Agriculture, Forestry and Life Sciences, Clemson University, Clemson, SC 29634, USA
Pascale Quignon, Cancer Genetics Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD 20892, USA. Institut de Génétique et Développement de Rennes, CNRS-UMR6061, Université de Rennes 1, 2 avenue Prof. Léon Bernard, CS34317, Rennes Cedex 35043, France.
Caitlin J. Rinz, Department of Genetics and Biochemistry, College of Agriculture, Forestry and Life Sciences, Clemson University, Clemson, SC 29634, USA
Elaine A. Ostrander, Cancer Genetics Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD 20892, USA
Jörg M. Steiner, Department of Small Animal Clinical Sciences, College of Veterinary Medicine and Biomedical Sciences, Texas A&M University, College Station, TX 77843, USA
Keith E. Murphy, Department of Genetics and Biochemistry, College of Agriculture, Forestry and Life Sciences, Clemson University, Clemson, SC 29634, USA
Leigh Anne Clark, Email: lclark4@clemson.edu, Department of Genetics and Biochemistry, College of Agriculture, Forestry and Life Sciences, Clemson University, Clemson, SC 29634, USA.
References
- Andresen E, Willeberg P. Pituitary dwarfism in German shepherd dogs: additional evidence of simple, autosomal recessive inheritance. Nord Vet Med. 1976;28:481–486. [PubMed] [Google Scholar]
- Averill DR., Jr Degenerative myelopathy in the aging German shepherd dog: clinical and pathologic findings. J Am Vet Med Assoc. 1973;162:1045–1051. [PubMed] [Google Scholar]
- Awano T, Johnson GS, Wade CM, Katz ML, Johnson GC, Taylor JF, Perloski M, Biagi T, Baranowska I, Long S, March PA, Olby NJ, Shelton GD, Khan S, O’Brien DP, Lindblad-Toh K, Coates JR. Genome-wide association analysis reveals a SOD1 mutation in canine degenerative myelopathy that resembles amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2009;106:2794–2799. doi: 10.1073/pnas.0812297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach I, Rhodes SJ, Pearse RV, II, Heinzel T, Gloss B, Scully KM, Sawchenko PE, Rosenfeld MG. P-Lim, a LIM homeodo-main factor, is expressed during pituitary organ and cell commitment and synergizes with Pit-1. Proc Natl Acad Sci USA. 1995;92:2720–2724. doi: 10.1073/pnas.92.7.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay KB, Haines DM. Immunohistochemical evidence for immunoglobulin and complement deposition in spinal cord lesions in degenerative myelopathy in German shepherd dogs. Can J Vet Res. 1994;58:20–24. [PMC free article] [PubMed] [Google Scholar]
- Barnes A, O’Neill T, Kennedy LJ, Short AD, Catchpole B, House A, Binns M, Fretwell N, Day MJ, Ollier WE. Association of canine anal furunculosis with TNFA is secondary to linkage disequilibrium with DLA-DRB1*. Tissue Antigens. 2009;73:218–224. doi: 10.1111/j.1399-0039.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Bhangoo APS, Hunter CS, Savage JJ, Anhalt H, Pavlakis S, Walvoord EC, Ten S, Rhodes SJ. Clinical case seminar: a novel LHX3 mutation presenting as combined pituitary hormonal deficiency. J Clin Endocr Metab. 2006;91:747–753. doi: 10.1210/jc.2005-2360. [DOI] [PubMed] [Google Scholar]
- Clark LA, Wahl JM, Steiner JM, Zhou W, Ji W, Famula TR, Williams DA, Murphy KE. Linkage analysis and gene expression profile of pancreatic acinar atrophy in the German shepherd dog. Mamm Genome. 2005;16:955–962. doi: 10.1007/s00335-005-0076-1. [DOI] [PubMed] [Google Scholar]
- Coates JR, March PA, Oglesbee M, Ruaux CG, Olby NJ, Berghaus RD, O’Brien DP, Keating JH, Johnson GS, Williams DA. Clinical characterization of a familial degenerative myelopathy in Pembroke Welsh Corgi dogs. J Vet Intern Med. 2007;21:1323–1331. doi: 10.1892/07-059.1. [DOI] [PubMed] [Google Scholar]
- Colvin SC, Malik RE, Showalter AD, Sloop KW, Rhodes SJ. Model of pediatric pituitary hormone deficiency separates the endocrine and neural functions of the LHX3 transcription factor in vivo. Proc Natl Acad Sci USA. 2011;108:173–178. doi: 10.1073/pnas.1009501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox VS, Wallace LJ, Anderson VE, Rushmer RA. Hereditary esophageal dysfunction in the miniature Schnauzer dog. Am J Vet Res. 1980;41:326–330. [PubMed] [Google Scholar]
- Drögemüller C, Becker D, Brunner A, Haase B, Kircher P, Seeliger F, Fehr M, Baumann U, Lindblad-Toh K, Leeb T. A missense mutation in the SERPINH1 gene in Dachshunds with osteogenesis imperfecta. PLoS Genet. 2009;5:e1000579. doi: 10.1371/journal.pgen.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill JL, Tsai KL, Krey C, Noorai RE, Vanbellinghen JF, Garosi LS, Shelton GD, Clark LA, Harvey RJ. A canine BCAN microdeletion associated with episodic falling syndrome. Neurobiol Dis. 2011 doi: 10.1016/j.nbd.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbecker WM, Hart EH. This is the German shepherd. T.F.H Publications Inc; Jersey City: 1967. [Google Scholar]
- Greer KA, Wong AK, Liu H, Famula TR, Pedersen NC, Ruhe A, Wallace M, Neff MW. Necrotizing meningoencephalitis of Pug dogs associates with dog leukocyte antigen class II and resembles acute variant forms of multiple sclerosis. Tissue Antigens. 2010;76:110–118. doi: 10.1111/j.1399-0039.2010.01484.x. [DOI] [PubMed] [Google Scholar]
- Guilford WG. Megaesophagus in the dog and cat. Semin Vet Med Surg (Small Anim) 1990;5:37–45. [PubMed] [Google Scholar]
- Hall EJ, Bond PM, McLean C, Batt RM, McLean L. A survey of the diagnosis and treatment of canine exocrine pancreatic insufficiency. J Small Anim Pract. 1991;32:613–619. [Google Scholar]
- Hanson JM, Mol JA, Leegwater PA, Kooistra HS, Meji BP. The leukemia inhibitory factor receptor gene is not involved in the etiology of pituitary dwarfism in German shepherd dogs. Res Vet Sci. 2006;81:316–320. doi: 10.1016/j.rvsc.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Hong H, Su Z, Ge W, Shi L, Perkins R, Fang H, Xu J, Chen JJ, Han T, Kaput J, Fuscoe JC, Tong W. Assessing batch effects of genotype calling algorithm BRLMM for the Affymetrix Gene-Chip human mapping 500 K array set using 270 HapMap samples. BMC Bioinformatics. 2008;9:S17. doi: 10.1186/1471-2105-9-S9-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AM, Jokinen P, Bannasch DL, Lohi H, Oberbauer AM. Association of a dog leukocyte antigen class II haplotype with hypoadrenocorticism in Nova Scotia duck tolling retrievers. Tissue Antigens. 2010;75:684–690. doi: 10.1111/j.1399-0039.2010.01440.x. [DOI] [PubMed] [Google Scholar]
- Johnston PE, Barrie JA, McCulloch MC, Anderson TJ, Griffiths IR. Central nervous system pathology in 25 dogs with chronic degenerative radiculomyelopathy. Vet Rec. 2000;146:629–633. doi: 10.1136/vr.146.22.629. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NH, Zody MC, Anderson N, Biagi TM, Patterson N, Pielberg GR, Kulbokas EJ, 3rd, Comstock KE, Keller ET, Mesirov JP, von Euler H, Kämpe O, Hedhammar A, Lander ES, Andersson G, Andersson L, Lindblad-Toh K. Efficient mapping of Mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Kennedy LJ, Davison LJ, Barnes A, Short AD, Fretwell N, Jones CA, Lee AC, Ollier WE, Catchpole B. Identification of susceptibility and protective major histocompatibility complex haplotypes in canine diabetes mellitus. Tissue Antigens. 2006;68:467–476. doi: 10.1111/j.1399-0039.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- Kooistra HS, Voorhout G, Mol JA, Rijnberk A. Combined pituitary hormone deficiency in German shepherd dogs with dwarfism. Domest Anim Endocrin. 2000;19:177–190. doi: 10.1016/s0739-7240(00)00074-6. [DOI] [PubMed] [Google Scholar]
- Lane PW, Dickie MM. Three recessive mutations producing disproportionate dwarfing in mice: achondroplasia, brachymorphic, and stubby. J Hered. 1968;59:300–308. doi: 10.1093/oxfordjournals.jhered.a107725. [DOI] [PubMed] [Google Scholar]
- Lantinga-van Leeuwen IS, Kooistra HS, Mol JA, Renier C, Breen M, van Oost BA. Cloning, characterization, and physical mapping of the canine Prop-1 gene (PROP1): exclusion as a candidate gene for combined pituitary hormone deficiency in German shepherd dogs. Cytogenet Cell Genet. 2000a;88:140–144. doi: 10.1159/000015507. [DOI] [PubMed] [Google Scholar]
- Lantinga-van Leeuwen IS, Mol JA, Kooistra HS, Rijnberk A, Breen M, Renier C, van Oost BA. Cloning of the canine gene encoding transcription factor Pit-1 and its exclusion as a candidate gene in a canine model of pituitary dwarfism. Mamm Genome. 2000b;11:31–36. doi: 10.1007/s003350010006. [DOI] [PubMed] [Google Scholar]
- Moeller EM, Steiner JM, Clark LA, Murphy KE, Famula TR, Williams DA, Stankovics ME, Vose AS. Inheritance of pancreatic acinar atrophy in German shepherd dogs. Am J Vet Res. 2002;63(10):1429–1434. doi: 10.2460/ajvr.2002.63.1429. [DOI] [PubMed] [Google Scholar]
- Moody JA, Clark LA, Murphy KE. Working dogs: history and applications. In: Ostrander EA, Giger U, Lindblad-Toh K, editors. The dog and its genome. Cold Spring Harbor Laboratory Press; Woodbury: 2006. pp. 1–18. [Google Scholar]
- Netchine I, Sobrier ML, Krude H, Schnabel D, Maghnie M, Marcos E, Duriez B, Cacheux V, Moers A, Goossens M, Grüters A, Amselem S. Mutations in LHX3 result in a new syndrome revealed by combined pituitary hormone deficiency. Nat Genet. 2000;25:182–186. doi: 10.1038/76041. [DOI] [PubMed] [Google Scholar]
- Osborne CA, Clifford DH, Jessen C. Hereditary esophageal achalasia in dogs. J Am Vet Med Assoc. 1967;151:572–581. [PubMed] [Google Scholar]
- Pfaeffle RW, Savage JJ, Hunter CS, Palme C, Ahlmann M, Kumar P, Bellone J, Schoenau E, Korsch E, Brämswig JH, Stobbe HM, Blum WF, Rhodes SJ. Four novel mutations of the LHX3 gene cause combined pituitary hormone deficiencies with or without limited neck rotation. J Clin Endocrinol Metab. 2007;92:1909–1919. doi: 10.1210/jc.2006-2177. [DOI] [PubMed] [Google Scholar]
- Pfister K, Rossi GL, Freudiger U, Bigler B. Morphological studies in dogs with chronic pancreatic insufficiency. Virchows Arch A. 1980;386:91–105. doi: 10.1007/BF00432647. [DOI] [PubMed] [Google Scholar]
- Proschowsky HF, Fredholm M. Exocrine pancreatic insufficiency in the Eurasian dog breed–inheritance and exclusion of two candidate genes. Anim Genet. 2007;38:171–173. doi: 10.1111/j.1365-2052.2007.01570.x. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Räihä M, Westermarck E. The signs of pancreatic degenerative atrophy in dogs and the role of external factors in the ethiology of the disease. Acta Vet Scand. 1989;30:447–452. doi: 10.1186/BF03548022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajab A, Kelberman D, de Castro SC, Biebermann H, Shaikh H, Pearce K, Hall CM, Shaikh G, Gerrelli D, Grueters A, Krude H, Dattani MT. Novel mutations in LHX3 are associated with hypopituitarism and sensorineural hearing loss. Hum Mol Genet. 2008;17:2150–2159. doi: 10.1093/hmg/ddn114. [DOI] [PubMed] [Google Scholar]
- Reynaud R, Saveanu A, Barlier A, Enjalbert A, Brue T. Pituitary hormone deficiencies due to transcription factor gene alterations. Growth Horm IGF Res. 2004;14:442–448. doi: 10.1016/j.ghir.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng H-X, Rahmani Z, Krizus A, McKenna-Yasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, Van den Bergh R, Hung W-Y, Bird T, Deng G, Mulder DW, Smyth C, Laing NG, Soriano E, Pericak–Vance MA, Haines J, Rouleau GA, Gusella JS, Horvitz HR, Brown RH., Jr Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Schmutz SM, Berryere TG. The genetics of cream coat color in dogs. J Hered. 2007;98:544–548. doi: 10.1093/jhered/esm018. [DOI] [PubMed] [Google Scholar]
- Sheng HZ, Zhadanov AB, Mosinger B, Jr, Fujii T, Bertuzzi S, Grinberg A, Lee EJ, Huang SP, Mahon KA, Westphal H. Specification of pituitary cell lineages by the LIM homeobox gene LHX3. Science. 1996;272:1004–1007. doi: 10.1126/science.272.5264.1004. [DOI] [PubMed] [Google Scholar]
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, Quignon P, Johnson GS, Parker HG, Fretwell N, Mosher DS, Lawler DF, Satyaraj E, Nordborg M, Lark KG, Wayne RK, Ostrander EA. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316:112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oost BA, Versteeg SA, Imholz S, Kooistra HS. Exclusion of the lim homeodomain gene LHX4 as a candidate gene for pituitary dwarfism in German shepherd dogs. Mol Cell Endocrinol. 2002;197:57–62. doi: 10.1016/s0303-7207(02)00292-7. [DOI] [PubMed] [Google Scholar]
- Voorbij AM, Leegwater PA, Kooistra HS. Hypopituitarism associated dwarfism in German shepherds, saarloos wolf dogs and Czechoslovakian wolf dogs. Access to genetic testing. Tijdschr Diergeneeskd. 2010;135:950–954. [PubMed] [Google Scholar]
- Wahl JM, Clark LA, Skalli O, Ambrus A, Rees CA, Mansell JL, Murphy KE. Analysis of gene transcript profiling and immunobiology in Shetland sheepdogs with dermatomyositis. Vet Dermatol. 2008;19:52–58. doi: 10.1111/j.1365-3164.2008.00655.x. [DOI] [PubMed] [Google Scholar]
- Westermarck E. The hereditary nature of canine pancreatic degenerative atrophy in the German shepherd dog. Acta Vet Scand. 1980;21:389–394. doi: 10.1186/BF03546871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermarck E, Batt RM, Valliant C, Wiberg M. Sequential study of pancreatic structure and function during development of pancreatic acinar atrophy in a German shepherd dog. Am J Vet Res. 1993;54:1088–1094. [PubMed] [Google Scholar]
- Westermarck E, Saari SA, Wiberg ME. Heritability of exocrine pancreatic insufficiency in German shepherd dogs. J Vet Intern Med. 2010;24:450–452. doi: 10.1111/j.1939-1676.2009.0461.x. [DOI] [PubMed] [Google Scholar]
- Wiberg ME. Pancreatic acinar atrophy in German shepherd dogs and rough-coated collies. Etiopathogenesis, diagnosis and treatment. A review. Vet Q. 2004;26:61–75. doi: 10.1080/01652176.2004.9695169. [DOI] [PubMed] [Google Scholar]
- Wiberg ME, Westermarck E. Subclinical exocrine pancreatic insufficiency in dogs. J Am Vet Med Assoc. 2002;220:1183–1187. doi: 10.2460/javma.2002.220.1183. [DOI] [PubMed] [Google Scholar]
- Wiberg ME, Saari SA, Westermarck E. Exocrine pancreatic atrophy in German shepherd dogs and rough coated collies: an end result of lymphocytic pancreatitis. Vet Pathol. 1999;36:530–541. doi: 10.1354/vp.36-6-530. [DOI] [PubMed] [Google Scholar]
- Wilbe M, Jokinen P, Hermanrud C, Kennedy LJ, Strandberg E, Hansson-Hamlin H, Lohi H, Andersson G. MHC class II polymorphism is associated with a canine SLE-related disease complex. Immunogenetics. 2009;61:557–564. doi: 10.1007/s00251-009-0387-6. [DOI] [PubMed] [Google Scholar]
- Wilbe M, Jokinen P, Truvé K, Seppala EH, Karlsson EK, Biagi T, Hughes A, Bannasch D, Andersson G, Hansson-Hamlin H, Lohi H, Lindblad-Toh K. Genome-wide association mapping identifies multiple loci for a canine SLE-related disease complex. Nat Genet. 2010;42:250–254. doi: 10.1038/ng.525. [DOI] [PubMed] [Google Scholar]
- Williams DA, Batt RM. Sensitivity and specificity of radioimmunoassay of serum trypsin-like immunoreactivity for the diagnosis of canine exocrine pancreatic insufficiency. J Am Vet Med Assoc. 1988;192:195–201. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.