Abstract
Protein O-GlcNAcylation is an essential post-translational modification on hundreds of intracellular proteins in metazoa, catalyzed by O-GlcNAc transferase using unknown mechanisms of transfer and substrate recognition. Through crystallographic snapshots and mechanism-inspired chemical probes, we define how human O-GlcNAc transferase recognizes the sugar donor and acceptor peptide and employs a novel catalytic mechanism of glycosyl transfer, involving the sugar donor α-phosphate as the catalytic base, as well as an essential lysine. This mechanism appears to be a unique evolutionary solution to the spatial constraints imposed by a bulky protein acceptor substrate, and explains the unexpected specificity of a recently reported metabolic O-GlcNAc transferase inhibitor.
Modification of hundreds of intracellular proteins with O-linked β-N-acetylglucosamine (O-GlcNAc) in metazoa has been shown to affect protein stability, subcellular localization, phosphorylation and ubiquitination1,2. Dysregulation of cellular O-GlcNAc levels has been implicated in diabetes, neurodegenerative disease and cancer3. The addition of a single O-GlcNAc moiety to specific serines/threonines on nucleocytoplasmic proteins is accomplished by the enzyme uridine diphospho-N-acetylglucosamine:polypeptide β-N-acetylglucosaminyltransferase, also known as O-GlcNAc transferase or OGT. Deletion or knockdown of OGT has demonstrated its essential role in early embryogenesis4. The enzyme is part of the glycosyltransferase (GT) superfamily, a large group of enzymes that can be categorized into different families depending on their donor substrate specificity, metal dependence, structural fold and the stereochemical outcome (inverting/retaining) of the reactions they catalyze5. OGT is a metal-independent enzyme that utilizes the nucleotide sugar UDP-GlcNAc as the donor substrate to transfer β-linked O-GlcNAc on the serine or threonine of more than 1000 acceptor proteins in the human cell. The reaction proceeds with inversion of stereochemistry and formation of UDP as the second product. Inverting glycosyltransferases, like OGT, are thought to utilize an SN2-type direct displacement catalytic mechanism (reviewed in 6), which is believed to require deprotonation of the acceptor by a catalytic base at some point along the reaction coordinate. The identity of the catalytic residues is uncertain for many GTs, due to difficulties in obtaining structures of ordered complexes with donors and/or acceptor substrates. OGT is composed of a glycosyltransferase type B (GT-B) catalytic domain and thirteen N-terminal tetratricopeptide repeats (TPRs) that are thought to be required for recognition of large protein substrates, but are not essential for glycosyl transfer onto acceptor peptides7. How OGT targets specific sites on a limited subset of intracellular proteins and how it catalyzes O-GlcNAc transfer is not currently understood. Although the structures of bacterial OGT orthologs and human OGT (hOGT) have recently been reported8-10, the lack of structures of complete ternary product and substrate complexes, specifically the absence of the sugar moiety from the hOGT structures, has limited the mechanistic insights gained from these studies.
Recently, several cell penetrant OGT inhibitors have been reported that are invaluable tools for cell biological studies investigating the function of O-GlcNAc. A dicarbamate compound identified from a high throughput small molecule screen was shown to be a covalent inhibitor, targeting a conserved lysine (K842) and cysteine (C917) in the active site11. An alternative strategy has exploited a metabolic precursor, tetra acetyl-5S-GlcNAc, which in the form of the free sugar is a substrate of the UDP-GlcNAc biosynthetic pathway, resulting in intracellular synthesis of the actual inhibitor UDP-5S-GlcNAc (1, Fig. 1A)12. Strikingly, the latter appears to specifically inhibit OGT, despite many other glycosyltransferases using UDP-GlcNAc as the sugar donor, suggesting OGT may employ a mechanism different from that of other GTs.
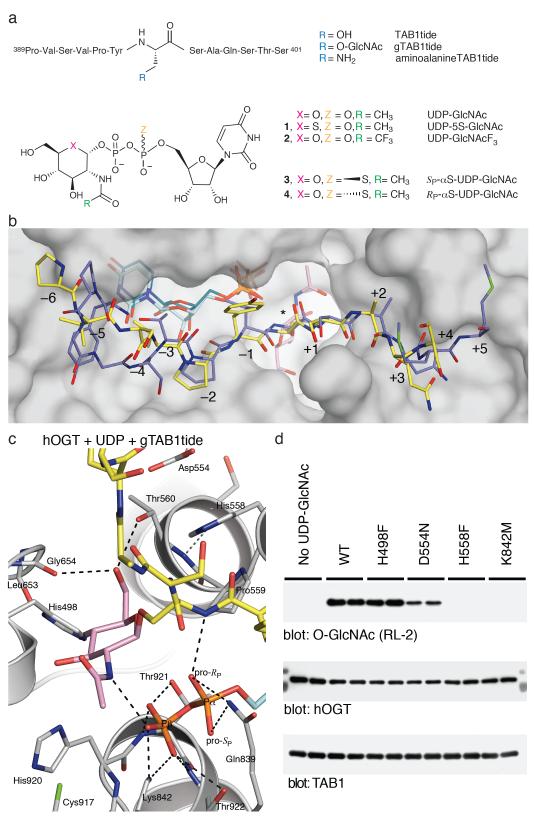
Figure 1. The structure of a ternary hOGT product complex gives insights into the peptide binding mode and participation of active site residues in O-GlcNAc transfer.
A. Chemical structures of substrate analogs: peptides based on the O-GlcNAc site (Ser395) from TAB1 protein, and donor analogs derived from UDP-GlcNAc.
B. Substrate binding groove of hOGT. Surface representation of the hOGT active site with reaction products gTAB1tide and UDP shown as sticks (the peptide, sugar and UDP are shown with yellow, pink and turquoise carbon atoms, respectively). The previously reported10 complex with the unmodified CKII peptide and uridine diphosphate (PDB ID 3PE4) is shown as sticks with blue/dark green carbon atoms, respectively. The position of the modified serine is marked by an asterisk, and subsites −6 to +5 are numbered.
C. hOGT product complex. Close-up view of the active site of the complex between hOGT, gTAB1tide and UDP. The enzyme is shown in cartoon representation with side chains displayed as sticks with grey carbon atoms. Colors for the glycopeptide and UDP as in panel B. Hydrogen bonds are depicted by dashed lines.
D. Activity of hOGT point mutants in an in vitro O-GlcNAcylation assay of TAB1 protein. O-GlcNAc was detected by immunoblotting with a pan-O-GlcNAc antibody (RL-2). Experiments were performed in duplicate, as shown. Full-size blots are shown in Supplementary Figure 2.
Here we report the first structures of ternary product and substrate complexes of hOGT with sugar donor analogs and synthetic peptides corresponding to the O-GlcNAc site from the innate immunity signaling protein TAB1. These structures suggest that OGT substrate recognition is centered on the −3 to +3 subsites of the acceptor peptide and reveal an unusual conformation of the sugar donor analog that brings the acceptor serine hydroxyl in close proximity to the α-phosphate of the donor substrate, UDP-GlcNAc. Together with in vitro glycosylation experiments employing individual stereoisomers of the α-phosphorothioate analog of UDP-GlcNAc and site-directed mutagenesis, these data delineate a new catalytic mechanism for inverting glycosyl transfer.
Results
hOGT product complex defines peptide binding mode
Previous attempts at determining the structure of complexes between hOGT and its donor substrate UDP-GlcNAc have resulted in hydrolysis of the substrate10. In order to ascertain the precise position of GlcNAc in the active site and gain insight into OGT substrate recognition we made use of a synthetic O-GlcNAcylated peptide (gTAB1tide, Fig. 1A) derived from the regulatory O-GlcNAc site on the innate immunity signaling protein TAB113 to trap the enzyme/glycopeptide product complex. We crystallized hOGT in complex with UDP and then soaked the crystals with gTAB1tide before data collection. Electron density maps at 3.15 Å, improved by 4-fold non-crystallographic averaging, revealed unambiguous density for UDP and gTAB1tide (see Supplementary Results). UDP adopts the same conformation as observed in the recently reported hOGT-UDP-peptide complex (max. atom shift = 1.0 Å), tethered by interactions with nine residues that are all conserved in metazoan OGTs. The peptide shows ordered density for the −6 to +4 subsites (Supplementary Figure 1a), and adopts a backbone conformation in the −3 to +3 subsites similar to the previously reported complex of hOGT with a CKII-derived substrate peptide10 (VPYSSAQ for gTAB1tide, TPVSSAN for CKII, Fig. 1b). This suggests that hOGT may impose structural and/or sequence constraints on the acceptor peptide and that hOGT specificity at the peptide sequence level may be worth exploring.
The enzyme active site does not harbor the catalytic base
In the gTAB1tide-hOGT complex the β-linked sugar, adopting the 4C1 chair conformation, projects into a conserved pocket, where it is tethered by T560, H920, L653 and G654 (Fig. 1c). The methyl group of the N-acetyl moiety points between the GT-B catalytic core and the TPR repeats, into a pocket formed by C917, M501 and L502, explaining why hOGT can tolerate a diverse array of UDP-GlcNAc analogs bearing bulky amido substituents, including azido derivatives that are widely used with click-chemistry to identify and enrich for O-GlcNAc proteins14,15 (Fig. 1b,c). The identity of the OGT catalytic base that is thought to activate the Ser/Thr in acceptor proteins has been the subject of a number of studies8-10 that propose either of two histidines, H498 or H558, as candidates. Inspection of the OGT glycopeptide product complex described here reveals that both of these histidines are positioned > 4.5 Å from the acceptor hydroxyl, and lack interacting residues that would support either of them acting as a catalytic base (Fig. 1c). Indeed, when we mutated H498, most recently proposed as the catalytic base10, to phenylalanine, the enzyme retains activity (Fig. 1d). Interestingly, a phenylalanine is found in the equivalent position of the Xanthomonas campestris putative OGT8. The other candidate for the catalytic base, H558, is sandwiched between P559 and D554 with the carboxylate of D554 stacking with the H558 imidazole side chain, suggesting D554/H558 may form a catalytic dyad. However, while mutation of H558 renders the enzyme inactive, mutation of D554 does not abrogate catalysis, suggesting the effects of the H558 mutation are presumably due to structural reasons (Fig. 1c,d). Furthermore, the imidazole side chain of H558 accepts a hydrogen bond from a protein backbone amide on (deprotonated) Nδ (Fig. 1c), implying that the imidazole Nε, facing the acceptor serine, will be protonated at neutral pH, which is not compatible with a role as a general base.
A Michaelis complex suggests substrate-assisted catalysis
Surprisingly, it appears that none of the enzyme side chains closest to the acceptor serine can act as a catalytic base. To uncover its identity, we endeavored to trap a complex with intact substrates by reducing the rate of enzymatic turnover in crystallo using two artificial substrate analogues: UDP-5S-GlcNAc (1), a recently reported hOGT donor substrate analog inhibitor12, and the aminoalanine derivative of the TAB1 acceptor peptide (aaTAB1tide), where the serine hydroxyl is replaced with a primary amine (Fig. 1a, for chemical syntheses see Supplementary Methods). Notably, although turnover is reduced, OGT can utilize both UDP-5S-GlcNAc (1) (reported reduction of kcat by factor 14 compared to UDP-GlcNAc12) and aaTAB1tide as substrates for glycosyl transfer onto the aminoalanine (Supplementary Figure 3). The observed activity with these pseudo-substrates suggests that they possess catalytically competent binding modes, similar to the natural substrates. hOGT was crystallized in complex with UDP-5S-GlcNAc (1) and soaked with aaTAB1tide. Synchrotron data were collected to 3.3 Å and electron density maps (improved by 4-fold non-crystallographic averaging) revealed unambiguous density of a pseudo-Michaelis complex (Fig. 2a and Supplementary Figure 1b). The overall conformation of the enzyme is almost unchanged (RMSD on 698 Cα atoms = 0.3 Å, max. atomic shift of any active site residue after overall superposition = 0.3 Å). The UDP moiety of UDP-5S-GlcNAc adopts a conformation similar to that of UDP in the product complex (RMSD = 0.2 Å, max. atomic shift = 1.0 Å), whereas the sugar is tilted away from the acceptor compared to the product complex (angle of rotation = ~30°), and is tethered by H920, L653, G654 and T560 on the O3, O4 and O6 hydroxyls (see Supplementary Movie 1 for an interpolation illustrating the atomic shifts between the ternary substrate and product complexes).
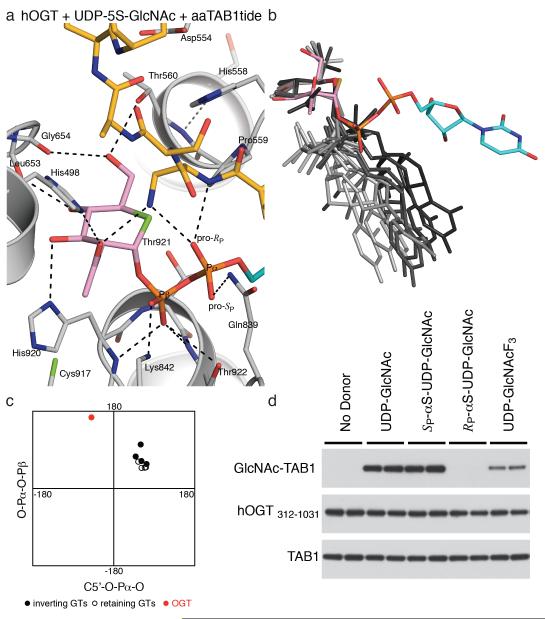
Figure 2. The unusual conformation of the sugar nucleotide in the hOGT pseudo-Michaelis complex suggests substrate-assisted catalysis.
A. hOGT pseudo-Michaelis complex: Close-up view of the active site of hOGT (grey carbons) in complex with the donor analog/inhibitor UDP-5S-GlcNAc (pink carbons for the sugar, turquoise carbons for the uridine moiety) and the synthetic peptide aaTAB1tide (acceptor serine is replaced with an aminoalanine, yellow carbons). Hydrogen bonds are depicted by dashed lines.
B. Comparison of sugar nucleotide conformations from complexes with GT-B family enzymes. The structures of donor substrates in complex with active GT-B family enzymes deposited in the PDB database are shown superimposed on the sugar ring. The donor substrate from the hOGT complex is shown with pink carbons for the sugar and turquoise carbons in the uridine moiety. Sugar nucleotide coordinates were obtained from crystal structures of the following enzymes, and depicted in increasingly lighter shades of grey: Inverting enzymes; MurG (PDB ID 1NLM), UGT71G1 (2ACW), UGT72B1 (2VCE), VvGT1 (2C1Z) and retaining enzymes; AGT (1Y6F), OtsA (1UQU, 1UQT), WaaG (2IW1).
C. Selected torsion angles of the donor substrates of active glycosyltransferases belonging to the GT-B family (see E) plotted in a 2D-graph.
D. In vitro O-GlcNAcylation assay using mechanism-inspired UDP-GlcNAc analogs. O-GlcNAcylation of TAB1 by hOGT (312—1031) was detected by immunoblotting with a site-specific TAB1 anti-O-GlcNAc S395 antibody. Experiments were performed in duplicate, as shown. Full-size blots are shown in Supplementary Figure 4.
Strikingly, the observed conformation of the donor substrate is remarkably different from previously reported structures of GT-B donor complexes (Fig. 2b,c). It appears that binding in the OGT active site induces a ‘back-bent’ UDP-GlcNAc conformation, characterized by unusual torsion angles of the pyrophosphate, which positions the sugar directly opposite the α-phosphate (Fig. 2a,b,c). This unusual donor conformation brings the α-phosphate pro-RP oxygen to within 2.8 Å of the aminoalanine amino group mimicking the serine hydroxyl, suggesting that they would form a hydrogen bond. Concomitantly, the carbonyl group of the GlcNAc N-acetyl group approaches the serine analog to within 2.9 Å (Fig. 2a). In fact, these two substrate moieties approach the serine analog side chain more closely than any atom on the OGT enzyme itself. The acceptor appears to be positioned for nucleophilic attack with in-line displacement on the sugar anomeric carbon (angle nucleophile-anomeric carbon-leaving group = 151°), yet the side chains of H498 and H558 remain > 4.5 Å away from the acceptor serine as in the product complex (Fig. 1c,2a). Thus, inspection of this pseudo-Michaelis complex leads to the tentative identification of two non-enzymic functional groups, residing on the substrate, as candidates for the elusive catalytic base. We therefore hypothesized that GlcNAc transfer catalyzed by OGT proceeds without involvement of traditional enzymic general bases like aspartate or histidine. This hypothesis was tested by devising derivatives of the substrate, UDP-GlcNAc, analogous to the common practice of investigating enzymic bases by site-directed mutagenesis.
Mechanism-inspired donor analogs identify catalytic base
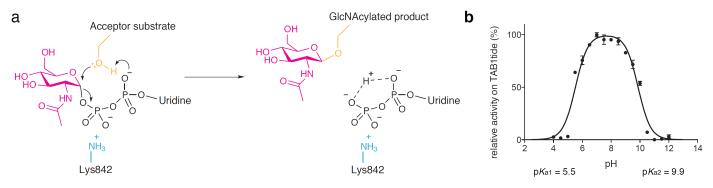
To investigate the possibility of a mechanism involving substrate-assisted catalysis, a number of UDP-GlcNAc derivatives were prepared. The two moieties considered as possible candidates for the catalytic base were the carbonyl oxygen of the N-acetyl group, as well as the non-bonding (pro-RP) oxygen of the α-phosphate, due to their proximity to the acceptor serine analog in the pseudo-Michaelis complex (Fig. 2a). While the carbonyl oxygen of the N-acetyl group has been shown to act as the catalytic nucleophile in O-GlcNAc hydrolysis16, it is not likely to act as a general base catalyst in O-GlcNAc transfer, as the extremely low pKa (approx. −0.5) of the conjugated acid makes it an unlikely proton acceptor. However, pKa values of titratable groups can be perturbed to a significant extent in the active sites of enzymes, so the previously described N-trifluoroacetyl UDP-GlcNAc derivative17 (2, UDP-GlcNAcF3, Fig. 1a) was prepared, with the aim of affecting the electronegativity of the N-acetyl carbonyl moiety in order to formally exclude it as the catalytic base. We found that the UDP-GlcNAcF3 compound bound the enzyme with a Kd similar to UDP-GlcNAc and proved to be a functional donor substrate for hOGT (Table 1, Fig. 2d), which excludes a significant catalytic role of the N-acetyl group. To investigate the potential role of the UDP-GlcNAc α-phosphate as the catalytic base, a pair of diastereomeric phosphorothioate analogs of UDP-GlcNAc were designed in which either of the non-bonding oxygens (pro-S or pro-R) of the α-phosphate was replaced with a sulfur atom (3 and 4, Fig. 1a). Both diastereomers were able to form a complex with the enzyme with Kd values similar to that of UDP-GlcNAc (Table 1) and underwent non-enzymatic hydrolysis at rates similar to UDP-GlcNAc (Supplementary Figure 6). The SP diastereomer (3) was found to be a functional donor in an O-GlcNAc transfer reaction (Fig. 2d). Strikingly, the RP diastereomer (4), where the sulfur replaces the oxygen pointing towards the acceptor serine (Fig. 2a), was not a substrate (Fig. 2d), and indeed inhibited the reaction with an IC50 of 25 μm (Supplementary Figure 7), while not affecting binding of the acceptor peptide (Supplementary Figure 5). These data suggest that the unusual conformation of UDP-GlcNAc positions the α-phosphate to act as the (initial) proton acceptor in hOGT-catalyzed O-GlcNAc transfer (Fig. 3a). While the pKa of UDP-GlcNAc in solution is thought to be approximately 2, our measurements suggest the pKa may be much higher (Supplementary Figure 8). Additionally, pKa perturbations by 3—4 pH units are common in the active site of enzymes18, and the acidic leg of a pH-activity profile of hOGT shows a pKa of 5.5 (Fig. 3b). Inspection of the substrate/product complexes also reveals that the α-phosphate lacks any interactions with positively charged side chains, which is likely to increase its pKa. Conversely, the β-phosphate of UDP-GlcNAc is located in a perfectly-shaped oxyanion hole, constructed by three elements: hydrogen bonds from the backbone amides of H920, T921 and T922, alignment with an α-helical electrostatic dipole and interaction with the evolutionarily conserved K842 (Fig. 1c,2a), an arrangement suited to stabilize the unusual conformation of the substrate that is required for correct positioning of the α-phosphate opposite the acceptor serine (Fig 2a), as well as stabilizing the negative charge developing on the leaving group, UDP. It is possible that this developing negative charge on the β-phosphate further contributes to the α-phosphate acting as initial proton acceptor, with the β-phosphate being the terminal proton acceptor (Fig. 3a). As expected, the K842M mutation, removing the positive charge near the β-phosphate, abrogates enzymatic activity (Fig. 1d) as well as binding of the reaction product, UDP (Table 1). With a theoretical pKa of 10.5, deprotonation of this lysine may represent the basic leg of the pH-activity profile of hOGT (Fig. 3b).
Table 1. Binding of sugar nucleotides to hOGT.
Binding affinity (Kd ± s.d.) of UDP, UDP-sugars and the α-phosphorothioate analogs of UDP-GlcNAc as determined by surface plasmon resonance (sensograms are shown in Supplementary Figure 5).
| WT | D554N | H558F | K842M | |
|---|---|---|---|---|
| UDP | 0.54 ± 0.01 | 0.60 ± 0.03 | 0.7 ± 0.1 | nb |
| UDP-GlcNAc | 16.1 ± 0.1 | 32.1 ± 0.3 | 42.3 ± 0.4 | 4.7 ± 0.1 |
| UDP-5S-GlcNac | 7.5 ± 0.1 | 6.2 ± 0.1 | 14.6 ± 0.1 | 5.7 ± 0.1 |
| SP-αS-UDP-GlcNAc | 16.2 ± 0.1 | 10.8 ± 0.1 | 19.6 ± 0.1 | 50.0 ± 0.1 |
| RP-αS-UDP-GlcNAc | 11.3 ± 0.1 | 11.5 ± 0.1 | 14.0 ± 0.1 | 2.8 ± 0.1 |
| UDP-GlcNAcF3 | 12.1 ± 0.1 | 9.5 ± 0.1 | 13.8 ± 0.1 | 29.3 ± 0.1 |
(Unit = μM, nb = no binding)
Figure 3. Proposed catalytic mechanism and pH activity profile of hOGT.
A. Schematic representation of the proposed catalytic mechanism of hOGT, showing substrate assisted catalysis involving the sugar donor phosphates.
B. Activity of wild type hOGT on N-terminally biotinylated TAB1tide in phosphate buffer of pH 4—12 was measured by scintillation proximity assay. Data points show the mean and s.e.m. of 3 observations.
Finally, the proposed mechanism explains the surprising observation that UDP-5S-GlcNAc is a potent and specific inhibitor of hOGT, but not of any other human GlcNAc transferases12. The smaller C5-S-C1 bond angle, the longer C5-S/S-C1 bonds and the larger van der Waals radius of the sulfur (Fig. 2) would all perturb the trajectory of proton transfer from the acceptor serine hydroxyl onto the UDP α-phosphate, a process that so far appears to be unique to hOGT.
Discussion
OGT is an essential metazoan glycosyltransferase that targets specific sites on a large number of protein substrates, utilizing UDP-GlcNAc as the donor substrate. We report crystal structures of ternary substrate and product complexes, providing snapshots of hOGT catalysis. These data show that hOGT binds peptide substrates in a defined orientation and with similar conformations near the site of O-GlcNAc transfer. Together with synthetic probes, diastereomeric phosphorothioate derivatives of UDP-GlcNAc, these structures define a new mechanism of inverting glycosyl transfer, where the catalytic base is not provided by the enzyme, but by the α-phosphate on the donor substrate itself. This differs from other Leloir-type GTs, where this function is generally performed by a side chain carboxylate or imidazole (reviewed in 6). Participation of substrate phosphates in enzymatic catalysis is an established concept for farnesyl pyrophosphate synthases19, and the cumulative evidence of multiple crystal structures of Clostridium spp. glucosylating toxins has led to the suggestion of the β-phosphate of UDP-glucose as the base in these inverting GT-A family enzymes20. The main impediment to a mechanism involving pyrophosphodiesters as proton acceptors is the weak basicity of such moieties (pKa ~2 in solution). The general assumption about inverting glycosyl transfer is that it proceeds sequentially: a catalytic base de-protonates the acceptor, which launches a nucleophilic attack onto the anomeric carbon, with in-line displacement of the leaving group, resulting in inversion of stereochemistry. Yet there is little direct proof for this sequence of events, and enzymic bases are unlikely to achieve deprotonation of a serine side chain into an alcoholate (pKa ~16). SN2 reactions can be asynchronous or concerted events, and subject to transition-state poise the strength of the base may not be limiting. Similarly, retaining glycosyltransfer is thought to proceed via a highly dissociative transition state6,21. Lately, a catalytic role for the β-phosphate has been proposed for the GDP-fucose Protein O-fucosyltransferase (POFUT)-122, an inverting GT-B family enzyme, although a recent study reporting the structure of a GT from the same family, POFUT2, proposes an enzyme side chain as the catalytic base23. Intriguingly, the substrates for POFUT1 are specific serine residues on EGF repeats of the Notch receptor, and the clostridial toxins glucosylate Rho GTPases, so like OGT, these enzymes act on protein substrates. The involvement of the UDP-GlcNAc α-phosphate in catalysis may be what enables OGT to act on both Ser and Thr acceptors, as it provides an evolutionary solution to the simultaneous spatial constraints imposed by a large peptide acceptor, the requirement for recognition of the N-acetyl group on the UDP-GlcNAc donor and the need to accommodate the additional γ-methyl group on threonines. The mechanistic insights into hOGT catalysis are invaluable for the design of drug-like inhibitors to facilitate further research into the cell biological role of O-GlcNAc, and the targeting of human diseases such as diabetes and cancer. It is noteworthy that as a part of our mechanistic investigations, we have now identified a hOGT point mutant, K842M, that appears to be unaffected in substrate binding but is catalytically inactive and would be useful for cell biological dissection of the role of OGT as an enzyme versus a scaffolding function through its TPR repeats.
Methods
Protein Crystallography
See Supplementary Methods for expression and purification details of human OGT (312-1031). Protein was crystallized in complex with donor substrate/product, and crystals were soaked with peptide/glycopeptide prior to freezing. Vapor diffusion crystallization experiments with hanging drops containing 1 μl protein (100 μm in a buffer of 10 mm Tris-Cl pH 8.5, 50 mm NaCl, 0.5 mm THP, 1 mm UDP or UDP-5S-GlcNAc) and 0.6 μl reservoir solution (1.45 m K2HPO4, 10 mm EDTA, 1 % xylitol) gave bar-shaped crystals with maximum dimensions of 0.1 × 0.1 × 0.4 mm after 3—4 days at 20 °C. These were transferred to a drop of reservoir solution containing 2 mm of the (glyco)peptide (Ac-PVSVPYS(-β-O-GlcNAc)SAQSTS-NH2) for 30 min, then cryoprotected (1.45 m K2HPO4, 10 mm EDTA, 27 % xylitol) and flash-frozen. Data were collected at the European Synchrotron Radiation Facility (ESRF) at 100 K and wavelengths of 0.939 Å and 0.873 Å on beamlines ID14-4 and ID23-2, respectively. Crystals belonged to space group P321 and contained 4 molecules per asymmetric unit. The structure was solved by molecular replacement using the A chain of PDB ID 3PE3 as the search model. Model building was performed in Coot, and various programs of the CCP4 suite24,25 were used for structure refinement. Ligand topologies were calculated using PRODRG26. Data collection and refinement statistics are given in Supplementary Table 1.
In vitro glycosylation of hTAB1
1 mg/ml of purified hTAB1 protein (7—402)27 was incubated with 0.05 mg/ml of purified hOGT in 100 mm potassium phosphate pH 7.5 containing 1 mm UDP-GlcNAc and 1 mm DTT. The reaction was allowed to proceed for 1.5 h at 37 °C and stopped by addition of SDS loading buffer and heating to 95 °C. Samples were subjected to SDS-PAGE and transferred to PVDF membrane followed by immunological detection of OGT, TAB1 and O-GlcNAc with primary and secondary antibodies diluted 1 : 5,000 in TBS-Tween containing 3 % bovine serum albumin. Site-specific antibody against O-GlcNAc-TAB1 S395 was raised in rabbit against a keyhole limpet hemocyanin (KLH)-conjugated glycopeptide.13 The anti-TAB1 antibody28 was obtained from the Division of Signal Transduction and Therapy, University of Dundee.
hOGT activity measurements by scintillation proximity assay
Radiometric detection of OGT activity on peptide substrates was achieved through scintillation proximity technology (PerkinElmer). Assays were conducted in 20 μl format in 384-well polypropylene plates. Reactions contained 200 nm hOGT (312—1031), 2 μm biotinylated substrate peptides and 500 nm UDP-GlcNAc with 0.3 Ci/mmol UDP-[3H]-GlcNAc as a radioactive tracer, 100 mm potassium phosphate, 1 mm DTT and 0.2 mg/ml BSA. Reactions were stopped by addition of 40 μl of 0.75 m phosphoric acid, and transferred to a streptavidin-coated FlashPlate®-384 (PerkinElmer) for detection on a TopCount NXT microplate luminescence counter (PerkinElmer).
Surface Plasmon Resonance
SPR measurements were collected using a Biacore T100 instrument (GE Healthcare). Streptavidin was immobilized on a CM5 sensor chip (GE Healthcare) using standard amine coupling method. 10 mm HEPES, 150 mm NaCl, pH 7.4 was used as a running buffer for immobilization. hOGT was biotinylated by mixing of hOGT with amine-binding biotin (Pierce) in 1:1 molar ratio. The chip surface was primed with running buffer (25 mm Tris pH 7.5, 150 mm NaCl, 1 mm DTT and 0.05 % Tween 20) and biotinylated hOGT protein (312—1031) was captured on the streptavidin surface. All compounds were injected in duplicates with highest concentrations from 10—500 μm depending on affinity, followed by a 1 : 3 dilution series. Association was measured for 1 min and dissociation for 2 min. All experiments were performed at flow rate 30 μl/min and temperature 25 °C. All data were referenced for blocked streptavidin surface and blank injections of buffer. Srubber 2 (BioLogic Software) was used to process and analyze data. Affinities were calculated using 1:1 equilibrium binding fit. Sensograms and curve-fit are shown in Supplementary Figure 5.
Synthesis of UDP-GlcNAc analogues
UDP-5S-GlcNAc, αS-UDP-GlcNAc and UDP-GlcNAcF3 were synthesized using the recently published pyrophosphorylation bond forming strategy29 (see Supplementary Scheme 1 for a reaction flow diagram). Briefly, the reaction of 3,4,6-tri-O-Ac-5-S-GlcNAc-1- or 3,4,6-tri-O-Ac-GlcNAcF3-1-phosphates with 2′,3′-di-O-acetyl-5′-O-(N,N-di-isopropylamino-O-cyanoethyl)phosphoramidite in the presence of 4,5-dicyano imidazole as nucleophilic catalyst resulted in the formation of intermediate phosphite-phosphate anhydrides which were oxidized with anhydrous t-BuOOH to yield the required sugar nucleotide analogs after global deacetylation with Et3N-MeOH-water or guanidine, respectively. To synthesize αS-UDP-GlcNAc the method was elaborated to allow for the synthesis of phosphorothioates by substituting the oxidization step by sulfurization. The intermediate phosphite-phosphate anhydride obtained from 3,4,6-tri-O-Ac-GlcNAc-1-phosphate and 2′,3′-di-O-acetyl-5′-O-(N,N-di-isopropylamino-O-cyanoethyl)phosphoramidite was treated with Beaucage reagent and deprotected to give a 1:1 diastereomeric mixture of αS-UDP-GlcNAc derivatives. 31P NMR spectra of the product after size-exclusion purification showed two pairs of characteristic doublets (δ 43.26 (d, JPα,Pβ = 29.3 Hz), 43.17 (d, JPα’,Pβ’ = 27.6 Hz), −14.04 (d, JPα’,Pβ’ = 27.6 Hz), −14.06 (d, JPα,Pβ = 29.3 Hz) proving the formation of the phosphorothioate derivative. For a detailed description of the synthetic procedure see Supplementary Methods.
The mixture of RP and SP isomers of αS-UDP-GlcNAc was separated by ion-pair reverse-phase HPLC30 on a Waters XBridge C18 Peptide separation technology 19×100 column (flow rate 24 ml/min) using a 15-min linear gradient (1 to 15 % MeCN in 50 mm phosphate/2.5 mm TBAHS; pH 6.2). Retention time for the earlier eluting (SP) isomer was 6.19 min; and 7 min for the later eluting (RP) isomer. Products were desalted by anion exchange chromatography on a 25×150 mm Q FF sepharose column (flow rate 10 ml/min) using a linear gradient (0 to 0.4 m in 15 min) of NH4HCO3. Final polishing was achieved by size exclusion chromatography (Bio-Gel P2 fine; column 2.6×100 cm; flow rate 0.4 ml/min) in 0.25 m NH4HCO3.
The configuration at the α-phosphorus atom was unambiguously established by direct comparison of the retention times of the synthetic compounds with stereodefined product of enzymatic synthesis (Supplementary Methods).
Supplementary Material
Acknowledgements
This work was supported by a Wellcome Trust Senior Research Fellowship (WT087590MA) to D.M.F.v.A. Coordinates and structure factors were deposited with the PDB (entries 4ay5 and 4ay6).
Footnotes
Competing Financial Interests Statement The authors declare no competing financial interests.
References
- 1.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 2.Fujiki R, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480:557–560. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Love DC, Krause MW, Hanover JA. O-GlcNAc cycling: emerging roles in development and epigenetics. Semin. Cell Dev. Biol. 2010;21:646–654. doi: 10.1016/j.semcdb.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 7.Iyer SP, Hart GW. Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J. Biol. Chem. 2003;278:24608–24616. doi: 10.1074/jbc.M300036200. [DOI] [PubMed] [Google Scholar]
- 8.Clarke AJ, et al. Structural insights into mechanism and specificity of O-GlcNAc transferase. EMBO J. 2008;27:2780–2788. doi: 10.1038/emboj.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Fleites C, et al. Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat. Struct. Mol. Biol. 2008;15:764–765. doi: 10.1038/nsmb.1443. [DOI] [PubMed] [Google Scholar]
- 10.Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 2011;469:564–567. doi: 10.1038/nature09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang J, Lazarus MB, Pasquina L, Sliz P, Walker S. A neutral diphosphate mimic crosslinks the active site of human O-GlcNAc transferase. Nat. Chem. Biol. 2012;8:72–77. doi: 10.1038/nchembio.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gloster TM, et al. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat. Chem. Biol. 2011;7:174–181. doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pathak S, et al. O-GlcNAcylation of TAB1 modulates TAK1-mediated cytokine release. EMBO J. 2012;31:1394–1404. doi: 10.1038/emboj.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macauley MS, Whitworth GE, Debowski AW, Chin D, Vocadlo DJ. O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J. Biol. Chem. 2005;280:25313–25322. doi: 10.1074/jbc.M413819200. [DOI] [PubMed] [Google Scholar]
- 17.Sala RF, MacKinnon SL, Palcic MM, Tanner ME. UDP-N-trifluoroacetylglucosamine as an alternative substrate in N-acetylglucosaminyltransferase reactions. Carbohydr. Res. 1998;306:127–136. doi: 10.1016/s0008-6215(97)10033-7. [DOI] [PubMed] [Google Scholar]
- 18.Harris TK, Turner GJ. Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. IUBMB Life. 2002;53:85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- 19.Hosfield DJ, et al. Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis. J. Biol. Chem. 2004;279:8526–8529. doi: 10.1074/jbc.C300511200. [DOI] [PubMed] [Google Scholar]
- 20.Ziegler MO, Jank T, Aktories K, Schulz GE. Conformational changes and reaction of clostridial glycosylating toxins. J. Mol. Biol. 2008;377:1346–1356. doi: 10.1016/j.jmb.2007.12.065. [DOI] [PubMed] [Google Scholar]
- 21.Lee SS, et al. Mechanistic evidence for a front-side, SNi-type reaction in a retaining glycosyltransferase. Nat. Chem. Biol. 2011;7:631–638. doi: 10.1038/nchembio.628. [DOI] [PubMed] [Google Scholar]
- 22.Lira-Navarrete E, et al. Structural insights into the mechanism of protein O-fucosylation. PLoS ONE. 2011;6:e25365. doi: 10.1371/journal.pone.0025365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CI, et al. Structure of human POFUT2: Insights into thrombospondin type 1 repeat fold and O-fucosylation. EMBO J. 2012;31:3183–3197. doi: 10.1038/emboj.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Schüttelkopf AW, van Aalten DMF. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 27.Conner SH, et al. TAK1-binding protein 1 is a pseudophosphatase. Biochem. J. 2006;399:427–434. doi: 10.1042/BJ20061077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheung PC, Campbell DG, Nebreda AR, Cohen P. Feedback control of the protein kinase TAK1 by SAPK2a/p38alpha. EMBO J. 2003;22:5793–5805. doi: 10.1093/emboj/cdg552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gold H, et al. Synthesis of sugar nucleotides by application of phosphoramidites. J. Org. Chem. 2008;73:9458–9460. doi: 10.1021/jo802021t. [DOI] [PubMed] [Google Scholar]
- 30.Meynial I, Paquet V, Combes D. Simultaneous separation of nucleotides and nucleotide sugars using an ion-pair reversed-phase HPLC: Application for assaying glycosyltransferase activity. Anal. Chem. 1995;67:1627–1631. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.