SUMMARY
Signaling pathways are intimately involved in cellular differentiation, allowing cells to respond to their environment by regulating gene expression. While enhancers are recognized as key elements that regulate selective gene expression, the interplay between signaling pathways and actively used enhancer elements is not clear. Here, we use CD4+ T cells as a model of differentiation, mapping the acquisition of cell-type-specific enhancer elements in T-helper 1 (Th1) and Th2 cells. Our data establish that STAT proteins have a major impact on the acquisition of lineage-specific enhancers and the suppression of enhancers associated with alternative cell fates. Transcriptome analysis further supports a functional role for enhancers regulated by STATs. Importantly, expression of lineage-defining master regulators in STAT-deficient cells fails to fully recover the chromatin signature of STAT-dependent enhancers. Thus, these findings point to a critical role of STATs as environmental sensors in dynamically molding the specialized enhancer architecture of differentiating cells.
Introduction
How the extracellular environment coordinates gene transcription remains a central and largely unanswered question in biology. In bacteria, coordination of gene expression is resolved by the linear organization of the operon, a genetic entity where adjacent units are transcribed by a single regulatory region (Jacob and Monod, 1961). In metazoans, genes are regulated by the juxtaposition of promoters with enhancer regulatory regions. The latter can be located at remote distances from the transcribed units with the interactions being achieved through dynamic, long-range physical interactions. Such enhancer elements are likely to be a primary determinant of cell type specificity (Bulger and Groudine, 2011). In spite of their functional relevance, it has proven difficult to unambiguously locate enhancers.
Only recently chromatin signatures have been identified that allow genome-wide enumeration of cis-regulatory regions with enhancer properties. Specifically, the monomethylation of histone H3 lysine 4 (H3K4me1) signature is considered as the permissive enhancer signature (Heintzman et al., 2009). Other marks in combination with H4K4me1 signature have been subsequently used to differentiate active enhancer elements. These include the binding of acetyltransferase p300 (Visel et al., 2009) or deposition of H3K27ac (Creyghton et al., 2010; Rada-Iglesias et al., 2010). The predictive ability of p300-based active enhancer signature has been tested using a large series of reporter transgenic mice. In almost all cases, reproducible enhancer activity correlated with the tissue-specific p300 binding (Visel et al., 2009). In a more recent study, human cardiac enhancers have been identified using a similar approach (May et al., 2011). Mapping of p300 binding allows a refinement of the enhancer landscapes as p300 peaks offer a more discrete definition than other histone modifications leading to more precise localization of enhancers (Smale, 2010). While p300 binding constitutes a substantial portion of histone acetyltransferase activity found in the cells, other factors may also contribute to active enhancer landscapes (Krebs et al., 2011).
The new ability to profile active enhancers on a genome-wide scale raises a number of questions. Studies on differentiation of biological structures such as mammalian nervous system or immune processes explored how signaling pathways allow cells to respond to environment by regulating global gene expression patterns (Miller and Gauthier, 2007; O'Shea and Paul, 2010). However, the interplay between environment and active enhancer landscapes remains poorly understood. In particular, the contribution of exogenous inductive signals that sense the environment and endogenous tissue-specific transcription factors to the establishment of active enhancer repertoire is not clear.
Here, we chose CD4+ T cells as a model of differentiation and investigated the formation and maintenance of genome-wide enhancer signatures using H3K4me1 and p300 in two distinct T helper cell populations, Th1 and Th2 cells. T cell differentiation is a multi-step process, which through a series of progressive and alternative choices generates different cell populations dedicated to specific aspects of host defense. This process represents the integration of extrinsic cues sensed by signal transducer and activator of transcription (STATs) proteins and the induction of intrinsic master regulator transcription factors. Our genome-wide profiling of enhancers reveals that STAT proteins are pervasive effectors of active enhancer landscapes. Importantly, expression of endogenous master regulators in STAT-deficient cells fails to fully re-establish landscapes of active enhancers. In this manner, the ability of STATs to sense environmental signals is indispensable in the control of active enhancer elements and consequently alternative gene expression programs essential for specialized cells.
RESULTS
Chromatin Signature Identifies Previously Recognized and New Active Enhancers in T Cells
To begin to understand the breadth of the active enhancer repertoires in differentiating CD4+ T cells, we mapped global binding of p300, H3K4me1 and H3K4me3 histone modifications in Th1 and Th2 cells. On a genome-wide scale, we identified 25,554 statistically significant p300 peaks in Th1 and 22,534 peaks in Th2 cells. Biological repeat experiments showed the reproducibility of p300 peaks in Th1 and Th2 cells (r = 0.91 and 0.82, respectively) (Figure S1A).
To verify the utility of this approach, we first asked if this method identified known regulatory elements in CD4+ T cells. We found that the well-characterized Cd4 enhancer was marked by p300 in both Th1 and Th2 cells (Chong et al., 2010). In contrast, the CD8a and Foxp3 genes, whose products are not expressed in these cells, were devoid of p300 binding (Figure S1B). Furthermore, our genome-wide p300 profiling identified known enhancers of Ifng and Il4-Il13 genes, the signature cytokines of Th1 and Th2 cells, respectively (Figure S1B, 1A, Table S1). In addition, numerous new potential elements were identified (Figure S1B–D).
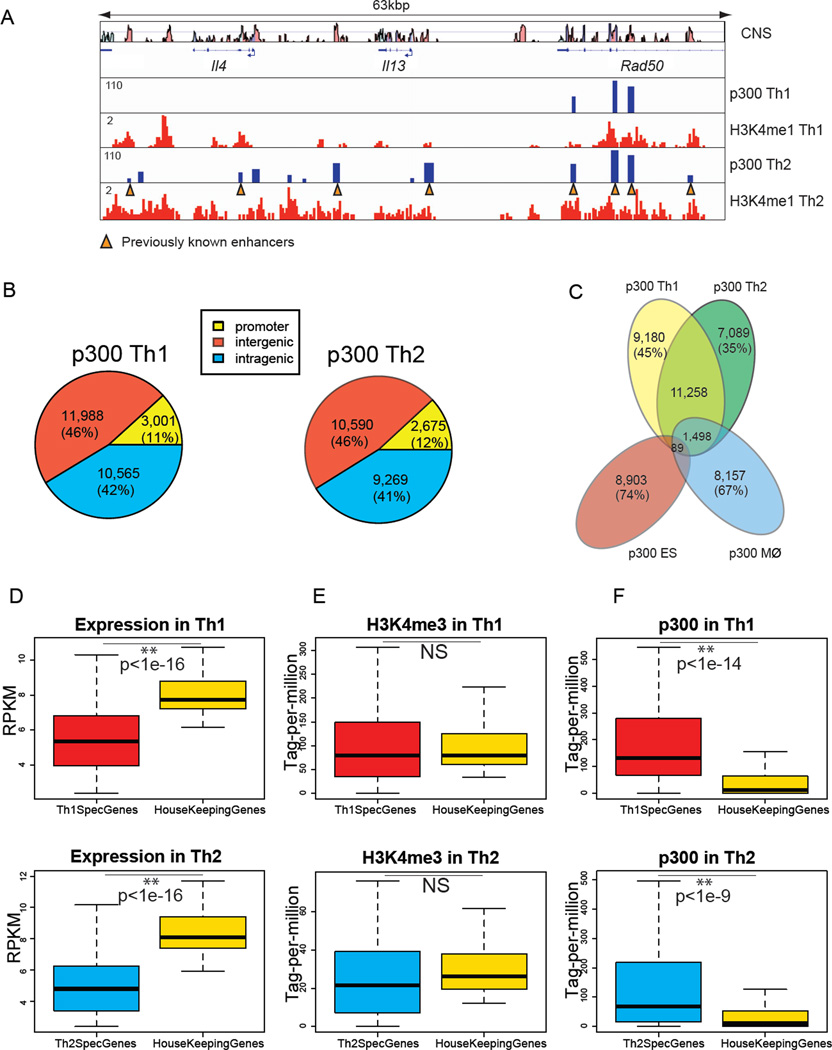
Figure 1. Active Enhancer Landscapes in Th1 and Th2 Cells Are Distinct.
(A) Chromatin signatures as defined by p300 binding and H3K4me1 identify recognized and new potential enhancers in the Il4-Il13 locus. The Il4-Il13 gene track represents 13 p300 binding sites within H3K4me1 domains in Th2 cells including 8 known elements (orange triangles) (Table S1B). “CNS” lane shows conserved non-coding sequences.
(B) Genomic distribution of p300-bound elements in Th1 (total 25,554) and Th2 (total 22,534) cells at promoter (−4kbp to +500bp of TSS), intergenic (>4kbp TSS), and intragenic regions (+500bp of TSS to TES).
(C) T helper subsets have thousands of unique p300 binding sites but almost none are shared among T cells, macrophages and ES cells. Venn diagram depicts the number and percentages of shared and unique p300 binding sites in each cell type. p300 binding in ES cells and macrophages is from (Creyghton et al., 2010; Ghisletti et al., 2010).
(D–F) In contrast to differentially expressed genes in Th1 and Th2 cells, housekeeping genes have little proximal p300 binding. Boxplots show median and quartiles of (D) normalized mRNA expression levels (RPKM: reads per kilobase exon model per million reads) measured by RNA-seq, (E) normalized H3K4me3 (tag-per-million), and (F) normalized p300 binding (tag-per-million) for top 100 Th-specific genes versus 100 housekeeping genes selected from (Eisenberg and Levanon, 2003). The intensity of p300 binding was computed −20kbp to 20kbp from the TSS to capture potential enhancers. The intensity of H3K4me3 was computed −4kbp to 1kbp from the TSS to capture active promoters. (p-values wilcoxon rank-sum test).
Active Enhancer Landscapes in Th1 and Th2 Cells Are Distinct
While specialized cells are functionally distinct, they also share many key cellular processes. This raises the fundamental question of how discrete the active enhancer landscapes are in distinct cell populations. We therefore sought to evaluate the differences in the genome-wide profile of active enhancer signatures in Th1 and Th2 cells, cells that are closely related, yet functionally distinct.
The p300 binding sites in promoter regions, enriched for H3K4me3, constituted about 12% of the total binding sites and were excluded from further analysis (Figure 1B). Overall, there were 22,556 and 19,859 putative distal enhancers in Th1 and Th2 cells, respectively. Of these, 12,845 putative enhancers, 55% and 64% of Th1 and Th2 identified elements, were shared by these two T helper subsets (Figure 1C). Conversely, 9,180 (45%) and 7,089 (35%) distal p300 peaks were specific to Th1 and Th2 cells, respectively.
Given the many similarities in function in Th1 and Th2 cells, we were surprised by how different these two subsets were in their active enhancer landscapes. We therefore compared Th1 and Th2 cells to more distantly related cells, namely macrophages and embryonic stem (ES) cells (Figure 1C and S1E) (Creyghton et al., 2010; Ghisletti et al., 2010). Macrophages and T cells are both of hematopoietic origin; however, only 1,498 regions were shared among T helper cells and macrophages (~10% of the total elements). Intriguingly, the conservation of enhancer repertoire, as defined by p300 binding, did not extend to include ES cells: Only 89 DNA elements bound by p300 were shared among all four datasets (<1% of enhancer elements of each cell-type) (Figure 1C). Together, each T helper subset had many unique enhancer elements and T helper cells, macrophages and ES cells had essentially no active enhancers in common.
Housekeeping Genes Have Little or no p300 Binding
Given that many functions are shared amongst cells, we were struck by the uniqueness of the global enhancer signatures. Therefore, we wondered whether genes with a high degree of tissue-specific expression would be relatively enriched for p300 binding compared to genes that were widely expressed (e.g. “housekeeping” genes).
Therefore, we next performed genome-wide transcriptional profiling in Th1 and Th2 cells using RNA-seq (Mortazavi et al., 2008) and identified the top 100 differentially expressed genes in each subset (Figure S1F–G). In addition, we chose 100 housekeeping genes based on an earlier study (Eisenberg and Levanon, 2003) (Figure S1H). The data showed that the housekeeping genes exhibit high levels of expression and enrichment of H3K4me3 at their promoters (Figure 1 D–E). In fact, the levels of expression of housekeeping genes were significantly higher compared to T helper-specific genes. However, the pattern of p300 enrichment for T helper-specific and housekeeping genes was very different: genes selectively expressed in Th1 or Th2 cells showed significantly higher p300 binding in their extended loci (+/− 20 kbp) compared to housekeeping genes (Figure 1F). In contrast to p300 however, the distribution of H3K4me1 modifications did not distinguish between preferentially expressed and housekeeping genes to the same degree (Figure S1I). Collectively, our data demonstrate that the majority of housekeeping genes have little or no p300 binding suggesting their distinct modes of regulation compared to tissue-specific genes.
Cell-type-specific p300 Peaks Colocalize with H3K4me1 and Correlate with Cell-type-specific Gene Expression
Since the chromatin signature of active enhancers is reported as p300 positive, H3K4me1 high, H3K4me3 low, and H3K27me3 low, we characterized patterns of histone modifications around common and unique p300-bound regions in Th1 and Th2 cells. On a genome-wide scale, elements uniquely marked by p300 in Th1 or Th2 cells resided within domains of high H3K4me1, low H3K4me3, and low H3K27me3 in the corresponding cell type (Figure 2A and S2A). Of note, these T helper specific p300 elements were highly conserved among mammals and were enriched for CpG islands (Figure S2B–C). H3K4 monomethylation in regions that were differentially bound by p300 in one lineage showed relative reduction in cells of the opposite lineage (Figure 2B). Macrophages and ES cells lacked H3K4me1 across T helper p300 elements (Figure 2A). Collectively, while Th-specific p300 elements are devoid of p300 and H3K4me1 in macrophage and ES cells, these elements are marked by H3K4me1 in both T helper subsets.
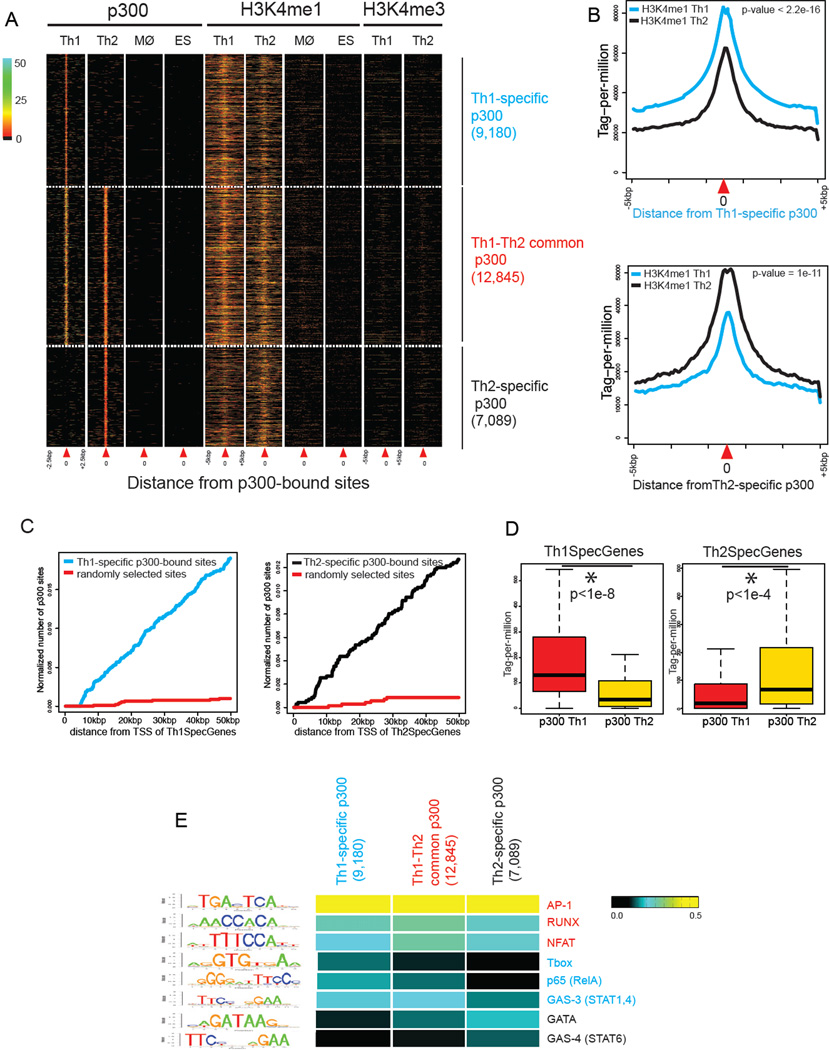
Figure 2. Properties of T Helper-specific p300-bound Elements.
(A) T helper-specific p300 elements are marked by high H3K4me1 and low H3K4me3 in both Th cells but lack p300 binding and H3K4me1 in macrophages and ES cells. Each column depicts p300 binding, H3K4me1, or H3K4me3 within a window centered on the p300-bound sites (indicated as position “0” by red triangle). Three patterns of p300 binding are shown: Th1-specific (9,180), Th1-Th2-common (12,845), and Th2-specific (7,089). Color-map corresponds to binding intensities where “black” represents no binding.
(B) H3K4me1 at Th-specific p300 sites shows enrichment in the respective lineage and relative reduction in the opposite lineage. Plots show the normalized distribution of H3K4me1 at Th1 (Th2)-specific p300 elements in Th1 and Th2 cells (+/− 5kbp) (Kolmogorov-Smirnov test).
(C) Th-specific p300 binding sites are enriched in proximity to genes selectively expressed in T helper cells. Plots depict number of Th-specific p300 binding sites within a given distance to promoters of Th-specific genes (Th1 blue, Th2 black) versus randomly generated sites (red) (wilcoxon rank sum test p-value <2.2e-16).
(D) Th-specific genes exhibit enrichment of p300 binding in the corresponding lineage and relative p300 depletion in the opposite lineage. Boxplots show median and quartiles of p300 binding in Th1 and Th2 cells around Th1- or Th2-specific genes (+/− 20kbp from the TSS) (wilcoxon rank-sum test).
(E) Th-specific p300 elements are enriched for consensus motifs of lineage-appropriate transcription factors. Consensus motifs for T cell related transcription factors were computed based on the de novo motif analysis using ChIP-seq data for each factor. A Gibbs sampling method was used to search for a motif using the genome as the background (likelihood ratio r>1000). Consensus motifs GATA and GAS-4 (STAT6) were preferentially enriched in Th2 whereas T-box, GAS-3 (STAT1,4), and p65 were enriched in Th1-specific p300 elements.
We next evaluated whether differential gene expression correlated with the presence of Th-specific p300 elements in the appropriate subset. In fact, we found that genes that were differentially expressed exhibited significant enrichment of p300 binding in the corresponding cell-type, with significantly less p300 recruitment in the opposite lineage (Figure 2C–D). Overall, Th1 and Th2-specific p300-bound regions had the chromatin characteristics of active enhancers and strongly correlated with cell-type-specific expression of proximate genes.
Enrichment of Lineage-specific Transcription Factor Binding Sites at Lineage-Specific Enhancers
The finding that the lineage-specific p300 elements correlate with differential gene expression led us to next assess whether these elements exhibited enrichment of binding sites for transcription factors that promote a lineage-specific gene expression program. We first characterized the enrichment of consensus motifs identified from publically available ChIP-seq datasets. AP-1 and NFAT, factors activated by T cell receptor engagement, and Runx are prevalent in Th cell enhancer elements but do not discriminate between Th1- and Th2-selective elements (Figure 2E). In contrast, motifs for factors that promote Th1 differentiation, such as RelA (p65) (Balasubramani et al., 2010) and T-bet (Szabo et al., 2000), were enriched in Th1- but not Th2-specific enhancers. Th1-specific enhancers were enriched for gamma-interferon activation-site with three base pair spacer (GAS-3), which is the consensus motifs for STAT1 and STAT4, but were relatively devoid of the STAT6 binding motif (GAS-4) (Wei et al., 2010). In contrast, Th2 elements exhibited enrichment of STAT6 and GATA3 motifs.
To more rigorously assess binding of relevant transcription factors to helper cell enhancers, we utilized available ChIP-seq data in CD4+ T cells (Table S2) (Nakayamada et al., 2011; Wei et al., 2011; Wei et al., 2010). Our analysis revealed that 48% of Th1-specific p300 elements were bound by STAT4 or STAT1 and 36% were bound by T-bet. Similarly, 31% and 11% of Th2-specific enhancers were bound by STAT6 and GATA3, respectively. Taken together, our findings reveal that T helper-specific p300 elements are enriched for lineage-specific transcription factors and correlate with lineage-specific gene expression programs these transcription factors promote.
STAT6 Has a Major Role in Generating Active Enhancers of Th2 Cells
Although the functional importance of enhancer elements in gene regulation is well-recognized, the factors that shape nascent enhancer landscapes of highly specialized cells are mostly unknown. Since cell-type-specific p300 elements were enriched for STAT binding sites, we asked whether chromatin signatures of active enhancers were also STAT-dependent. The ability of T cells to remain viable and retain their developmental potential in the absence of STAT proteins allowed us to assess the consequence of genetic deletion of these proteins on the enhancer repertoire of T cells.
Activated by IL-4, STAT6 is a key player in Th2 cell specification (Goenka and Kaplan, 2011; Zhu and Paul, 2008). To evaluate the contribution of STAT6 in shaping the active enhancer structure, we generated p300 and H3K4me1 profiles in wild-type and STAT6-deficient cells (Figure 3A, S3A). Focusing first on the Il4 extended locus (Ansel et al., 2006), we observed that STAT6 bound to more than half of the regulatory regions marked by p300 and p300 binding was abrogated in STAT6-deficient T cells (Figure S3B).
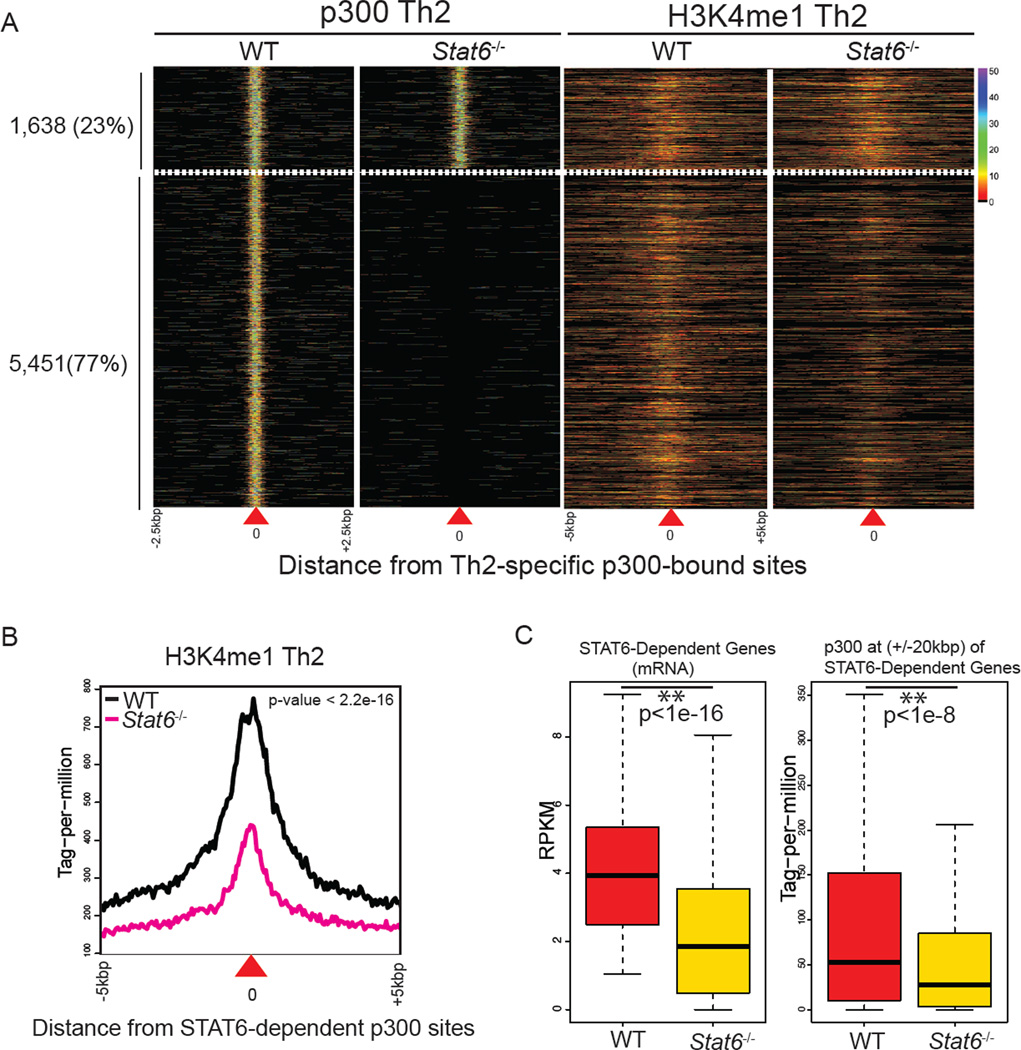
Figure 3. STAT6 Has a Major Role in Generating Active Enhancers of Th2 Cells.
(A) STAT6 is critical for the global chromatin signature of Th2-specific enhancers. Globally, p300 binding and H3K4me1 at 77% of Th2-specific p300 sites (5,451) were STAT6-dependent. The plot in each column represents the pattern of p300 binding and H3K4me1 in wild-type or Stat6−/− cells centered on the Th2-specific p300-bound sites (as indicated by position “0”). Color-map corresponds to binding intensities where “black” represents no binding.
(B) H3K4me1 at Th2-specific p300 sites is STAT6-dependent. Plot shows the normalized distribution of H3K4me1 at 5,451 STAT6-dependent p300 elements (Kolmogorov-Smirnov test).
(C) STAT6-positively regulated genes are enriched with STAT6-dependent p300 binding sites. Using RNA-seq data from wildtype Th2 and STAT6-deficient cells, we identified positively regulated genes by STAT6 (>2 fold-change). Accumulation of p300 binding at these genes in wildtype and STAT6-deficient cells was computed (+/− 20kbp from the TSS). Boxplots show median and quartiles of gene expression levels in RPKM (left) and p300 binding in tag-permillion (right) at STAT6-dependent genes in wildtype and STAT6-deficient cells (wilcoxon rank-sum test).
Globally, the impact of STAT6 deficiency on the chromatin signatures of Th2-specific active enhancers was striking: 77% of the Th2-specific p300 sites (5,451) were STAT6-dependent. Further analysis revealed that the magnitude of H3K4me1 marks was also significantly dependent on STAT6 at STAT6-dependent p300 elements (Figure 3B).
To link the effects of STAT6 on transcriptome and the active enhancer landscape, we measured global gene expression in wildtype and STAT6-deficient cells using RNA-seq. Indeed, p300 binding at the extended loci of STAT6-regulated genes showed STAT6-dependency (Figures 3C and S3C). Collectively, our data revealed a major role for STAT6 in p300 binding and H3K4me1 marks at active enhancers of Th2 cells.
STAT4 and STAT1, but not T-bet, Are Global Regulators of Active Enhancers in Th1 Cells
STAT4 and STAT1 work in concert to drive Th1 differentiation, being activated by IL-12 and IFNγ, respectively (Lighvani et al., 2001; Thieu et al., 2008). To assess the contribution of these transcription factors in shaping global enhancer structures, we generated p300 and H3K4me1 profiles using wild-type cells and cells lacking STAT4 or STAT1 (Figure 4A). Duplicate experiments showed the reproducibility of p300 peaks in these genotypes (Figure S4A).
Figure 4. STAT4 and STAT1, but not T-bet, Are Critical for Active Enhancers of Th1 Cells.
(A) STAT1 and STAT4, but not T-bet, play major roles in generating the active enhancer landscape of Th1 cells. Globally, 60% of Th1-specific enhancers were STAT-dependent whereas 17% were T-bet-dependent. Each column represents the pattern of p300 binding in wild-type, Stat4−/−, Stat1−/−, or T-bet−/− cells centered on the Th1-specific p300-bound sites. Color-map corresponds to binding intensities where “black” represents no binding.
(B) STAT4-positively regulated genes are enriched with STAT4-dependent p300 binding sites. Using RNA-seq data in wildtype Th1 and Stat4-deficient cells, positively regulated genes by STAT4 were identified (>2-fold change). Accumulation of p300 binding at these genes in wildtype and STAT4-deficient cells was computed (+/− 20kbp). Boxplots show normalized gene expression levels in RPKM (left) and p300 binding in tag-per-million (right) at STAT4-dependent genes in wildtype and STAT4-deficient cells (wilcoxon rank-sum test).
(C) p300 binding at the extended loci of positively regulated genes by T-bet is not T-bet dependent. Using RNA-seq data in wildtype Th1 and T-bet-deficient cells, we selected positively regulated genes by T-bet (>2 fold-change). Boxplots show normalized gene expression levels in RPKM (left) and p300 binding in tag-per-million (right) at T-bet-dependent genes in wildtype and T-bet-deficient cells.
Since these factors are important for the regulation of Ifng expression, we first assessed their roles in the generation of enhancers of this gene (Figure S4B). More than half of regulatory elements in Ifng gene extended locus were dependent on STAT4 or STAT1 (Figure S4C–G). Globally, deficiency of STAT4 and STAT1 resulted in a significant reduction in p300 marks: 60% of Th1-specific elements were dependent on either STAT1 or STAT4 (Figure 4A). More specifically, 15% and 13% were uniquely dependent on STAT1 and STAT4, respectively, whereas 30% of p300 binding disappeared in the absence of either STAT1 or STAT4. Interestingly, STAT-dependent p300 binding sites were enriched in proximity to genes whose expression levels were positively regulated by STATs (Figure 4B). In contrast, H3K4me1 at Th1-specific p300 elements were largely independent of STAT1 or STAT4 (Figure S4H).
IL-12 and IFNs acting via STAT4 and STAT1 induce the expression of Tbx21, which encodes the master regulator transcription factor T-bet (Szabo et al., 2000). Given its role in Th1 differentiation, we next asked if T-bet was also an important driver of the genomic enhancer signature. To our surprise, T-bet had a modest effect on the genomic enhancer repertoire (p-value = 0.06). While elements in proximity to some genes like the Ifng locus, were regulated by both STATs and T-bet (Figure S4E–G), 83% of Th1-specific p300-bound elements were independent of T-bet (Figure 4A). Transcriptional profiling in T-bet deficient cells revealed that p300 binding sites in proximity to genes positively regulated by T-bet were not dependent on this transcription factor (Figure 4C). Taken together, our findings indicate that STAT1 and STAT4 play major roles in generating the active enhancer landscape of Th1 cells, whereas T-bet has a modest impact.
STATs Exert Positive and Negative Effects on p300 Recruitment
Thus far, our findings indicate that STAT proteins bind to many T helper-specific p300 elements and are responsible for p300 deposition in T helper cells. We next assessed the extent to which STAT binding and recruitment of p300 were related. The integration of STAT and p300 ChIP-seq data revealed that around one-third of STAT-dependent p300 elements were also bound by the cognate STAT (Figure S5A), arguing that STAT proteins likely shape the enhancer landscape of T helper cells both directly and through deployment of other factors.
To further characterize the direct effect of STATs, we asked whether we could quantitate the role of STAT binding on cognate p300 recruitment. In particular, we explored the extent to which the accumulation of STAT binding associated with the acquisition of lineage-appropriate enhancer elements and the suppression of lineage-inappropriate marks. Our analysis revealed that the enrichment of STAT6 binding positively correlated with p300 recruitment at 22% of binding sites of this protein (3,523 of 16,079) (Figure 5A). Examples included multiple genes that contribute to Th2 differentiation such as Gata3, Nfil3, and Il24 (Figure 5A) (Kashiwada et al., 2011; Wei et al., 2010). Alternatively, the intensity of p300 binding increased more than 4-fold in STAT6-deficient cells at 10% of STAT6-bound sites (1,606 of 16,079), arguing for a substantial role of STAT6 in limiting p300 recruitment. One intriguing example was the Ifng locus where intergenic STAT6 binding sites in Th2 cells correlated with the loss of p300 binding and H3K4me1 modification in this cell type (Figure 5 A,B). Indeed, the expression of Ifng gene increased in the absence of STAT6 (Figure 5B). In general, transcriptome analysis in STAT6-deficient cells revealed that the effect of STAT6 on p300 recruitment correlated well with its role on gene expression patterns (Figure S5B). Overall, given the multiplicity of factors that influence T cell activation and differentiation, the extent of STAT6 binding sites with an effect on p300 deposition was notable.
Figure 5. Quantification of Direct Contribution of STATs to p300 Binding.
(A) Global binding of STAT6 leads to both gain and loss of cognate p300 binding. Two-dimensional histogram depicts STAT6 binding resulting in a change in p300 recruitment in wild-type versus Stat6−/− cells. Percentages of STAT6-bound sites with positive or negative effect on p300 are represented in the marked area (> 4 fold-change). The X-axis corresponds to intensity of STAT6 binding (log2). The Y-axis measures the fold-change of p300 binding in wild-type versus Stat6−/− cells (log2). Color-map corresponds to the number of binding events. Examples of genes with proximal STAT6 binding include Nfil3, Il24 and Gata3 (for positive effect) and Ifng, Xcl1, and Il18r1 (for negative effect).
(B) STAT6 has direct negative effects on Ifng enhancers in Th2 cells. Gene track shows that STAT6 binding (dotted box) in Th2 cells leads to loss of p300 binding and H3K4me1 at Ifng enhancers. RNA-seq lanes depict the expression of Ifng gene increased in the absence of STAT6 (14 to 135 RPKM).
(C) STAT4 binding correlates with gain and loss of p300 binding. Examples of genes with proximal STAT4 binding include Ifng, Nfatc2 and Il18r1 (for positive effect) and Il2 and Il4ra (for negative effect).
(D) Contrasting effect of T-bet on p300 binding. T-bet has a dominant role as a repressor rather than an activator based on p300 binding data. Examples of genes with proximal T-bet binding include Ifng, and Xcl1 (for positive effect) and Eomes and Il4-Il13 (for negative effect).
Similar to STAT6, the binding of STAT4 was associated with gain (13%, 2,646) or loss (9%, 1,723) of p300 binding (Figure 5C). STAT1 binding also demonstrated a direct effect on p300 deposition (Figure S5C). In contrast, T-bet binding positively correlated with very few p300 binding sites (6%, 1,138 of 19,152) (Figure 5D). Intriguingly, at 2,352 sites (13%), T-bet binding was associated with the inhibition of p300 recruitment and these elements were proximal to genes negatively regulated by T-bet (Figure S5B). Examples of genes with proximal T-bet binding for which p300 was negatively regulated by T-bet included Eomes and Il4-Il13 (Intlekofer et al., 2005; Thieu et al., 2008). Consistent with our earlier findings, T-bet exerts a modest role on the acquisition of Th1-specific enhancers and a more dominant role in limiting lineage inappropriate p300 deposition.
In contrast to the effect on p300 binding, the direct effects of STAT4, STAT1, or T-bet on H3K4me1 modifications were not significant (Figure S5D). However, at 7% of its binding sites (1,152 of 16,079) STAT6 binding was associated with an increase in H3K4me1 marks (Figure S5D). The major impact of STAT6 on p300 and H3K4me1 deposition suggests its crucial role in recruiting ‘writers’ complex of 'histone code' to enhancer cohorts of specialized cells.
GATA3 Expression Fails to Re-establish the STAT6-dependent Active Enhancer Landscape
While deletion of STATs significantly altered the active enhancer landscape of T helper cells, the argument could be made that without these factors, differentiated T helper cells were not efficiently generated due to the suboptimal expression of endogenous master regulators. To overcome this problem, we next asked whether overexpression of master regulators could reconstitute the chromatin signature of T helper-specific enhancers in STAT-deficient cells. Starting with Th2 cells, we found that the overexpression of GATA3 in STAT6-deficient cells was sufficient to restore IL-4 production (Figure S6A), as previously reported (Lee et al., 2001).
We next assessed the genome-wide binding of p300 in cells that lacked STAT6 but expressed GATA3 and IL-4, comparing binding patterns in wildtype and STAT6-deficient cells.
At the Il4-Il13 locus, expression of GATA3 restored many p300 binding sites; however, the most conserved non-coding region in the locus lacked p300 binding in the absence of STAT6 (Figure S6B) (Table S3). On the genome-wide scale, 51% of Th2-specific-STAT6-dependent sites (2,639 of 5,041) were recovered after GATA3 overexpression (Figure 6A, S6C). However, 2,402 sites (49% of STAT6-dependent enhancers) could not be reconstituted. Of note, around 26% (690) of the recovered elements were bound by GATA3. In contrast, only 7% (157) of non-recovered elements exhibited GATA3 binding (Wei et al., 2011).
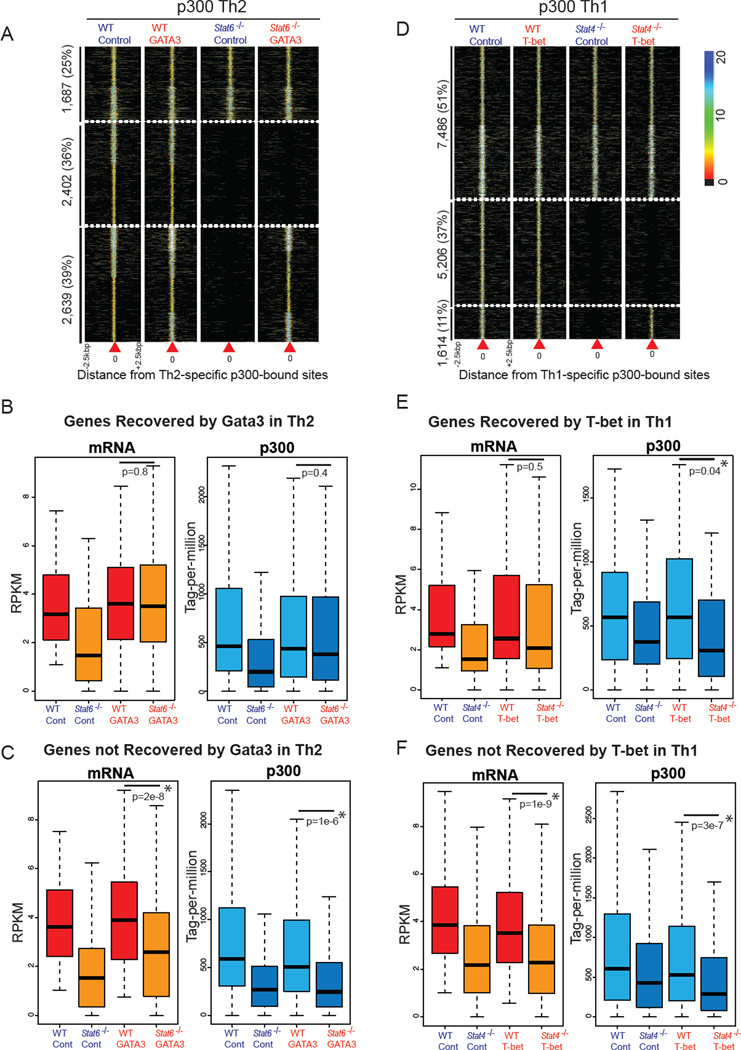
Figure 6. Overexpression of T-bet or GATA3 in STAT-deficient Cells Fails to Reconstitute STAT-dependent Active Repertoires.
(A) GATA3 expression recovers half of STAT6-dependent elements in Th2 cells. Of 5,041 Th2-specific-STAT6-dependent p300 sites, 2,639 (50%) regulatory elements are recovered in STAT6-deficient cells in which GATA3 was reconstituted. Overall, 36% of Th2-specific enhancers are STAT6-dependent, GATA3-independent. Each column represents p300 binding in wild-type (Stat6−/−) cells transduced with control or GATA3-expressing retrovirus centered on the Th2-specific p300 sites. Color-map corresponds to binding intensities where “black” represents no binding.
(B) GATA3 recovers p300 binding at genes whose expression levels are recovered by GATA3. Boxplots show median and quartiles of expression levels in RPKM (left) and normalized p300 binding in tag-per-million (right) in wild-type (Stat6−/−) cells transduced with control or GATA3-expressing retrovirus at genes recovered by GATA3.
(C) GATA3 has no effect on p300 binding at genes whose expression levels are not affected by GATA3.
(D) T-bet overexpression fails to recover the chromatin signature of STAT4-dependent enhancers. Of 6,820 Th1-specific-STAT4-dependent sites, 1,614 (23%) regulatory elements are recovered in STAT4-deficient T-bet-expressing cells. Each column represents p300 binding in wild-type (Stat4−/−) cells infected with control or T-bet-expressing retrovirus centered on the Th1-specific p300 sites.
(E) T-bet fails to recover p300 binding at genes whose expression levels are recovered by T-bet. Boxplots show median and quartiles of gene expression levels in RPKM (left) and normalized p300 binding in tag-per-million (right) in wild-type (Stat4−/−) cells transduced with control or T-bet-expressing retrovirus.
(F) T-bet has no effect on p300 binding at genes whose expression levels are not recovered by T-bet.
GATA3-mediated Transcriptome Changes Correlate With Changes in p300 Binding
To evaluate the degree to which gene expression correlates with establishment of the p300 binding repertoire, we measured global transcription in wildtype and STAT6-deficient cells in the absence or presence of GATA3 overexpression. Of 249 genes whose expression levels were reduced in STAT6-deficient cells, 42% (99) were induced by GATA3 overexpression (Figures S6D, 6B–C). To assess how transcription might correlate with the appearance of enhancer landscape, we quantitated the p300 binding in the vicinity of genes (+/− 20 kbp) whose expression were recovered by GATA3 (Figure 6B). For these genes, p300 binding was significantly increased upon GATA3 expression. In contrast, genes that were not induced by GATA3 exhibited no change in proximal p300 binding (Figure 6C). Overall, the integration of transcriptome changes and p300 binding revealed that changes in gene expression patterns mediated by GATA3 correlate with changes in p300 binding.
T-bet Expression in STAT4-deficient Cells Fails to Recover Active Enhancers in Th1 Cells
Next we asked whether T-bet would function similarly to GATA3. Consistent with previous work, we found that the overexpression of T-bet in STAT4-deficient cells was sufficient to restore IFNγ production (Figure S6E) (Afkarian et al., 2002; Mullen et al., 2001). In addition, we found that p300 binding to many sites within the Ifng locus was restored after T-bet overexpression. However, the majority of STAT4-dependent p300 binding sites were not recovered (Figure S6F, Table S3). On the genome-wide scale, only 23% of Th1-specific-STAT4-dependent p300 sites (1,614 of 6,820) were recovered after T-bet overexpression (Figure 6D).
To assess whether changes in transcription related to recovery in p300 binding, we measured global gene expression levels in wildtype and STAT4-deficient cells in the absence or presence of T-bet. Consistent with our earlier observations, T-bet expression had limited effects on the p300 binding of 37 genes where their expression levels were reconstituted by this transcription factor (Figure S6G, 6E–F). Overall, our results highlight the importance of STAT signaling in molding the active enhancer landscape and generating alternative gene expression programs of T helper cells, independent of endogenous factors such as T-bet and GATA3.
Discussion
The ability of a cell to sense and interpret environmental stimuli and appropriately modify gene expression is a fundamental tenet of evolutionary adaptation. Irrespective of the nature of the instigating stimulus and elicited signaling pathways, the final decoding of the message occurs at the genome level with a binary decision resulting in either activation or repression of transcription. Taking part in these binary decisions are chromatin structures including regulatory elements such as enhancers, which are now recognized as having major contributions to cell-type-specific gene expression programs. Enhancer elements either adopt a poised structure or upon receiving appropriate signals transition to transcriptional competency. Here, we set out to interrogate the roles of environment-sensing transcription factors and phenotype-defining master transcription factors in shaping the active enhancer landscape during cell fate specification. Our study argues for the pervasive involvement of cytokine-regulated STATs in conferring enhancer specificity and an indispensable role of environmental signals in creating the active enhancer repertoire. These are functions that are not replaced by the enforced action of phenotype-defining master regulators.
Distinct Chromatin Signatures Identify Active Enhancers in T Cell Populations
We profiled the repertoires of H3K4me1 high, p300 high regions (operationally defined as active enhancers) in Th1 and Th2 cells. Our data establish that closely related T helper cells have distinct active enhancer landscapes. The ability of p300 mapping to discriminate cell type specificity becomes more evident when our analysis included macrophages and ES cells. In this respect, it is worth pointing out that a precise understanding of what constitutes an active enhancer has not been firmly established (Natoli, 2010). While p300 binding successfully identified known enhancers of key genes in T cell populations, it is likely that p300 binding reports only a cross-section of active enhancer repertoire, and the entire view of active enhancer landscape also include mapping of other HAT complexes such as CBP (May et al., 2011) or SAGA (Krebs et al., 2011). Nevertheless, we have established that unbiased mapping of p300 binding is a powerful way to broadly interrogate enhancer activity with fine resolution and sufficient coverage in closely related cell populations.
The resulting annotations of active enhancers have implications for the interpretation of genome-wide association studies. Top-scoring disease single nucleotide polymorphisms are frequently positioned within enhancer elements specifically active in relevant cell types (Ernst et al., 2011). Global profiling of enhancers in various cell types thus provides knowledge base for the systematic investigation of such elements in health and disease.
Environment-Directed STATs Are Major Drivers of Active Enhancer Landscape of T Cells
Soluble secreted factors in the environment play key roles in cellular specification. For T cells, cytokines are the major factors that determine fate commitment, mainly through the activation and recruitment of STATs to chromatin. By comparing wild type and STAT-deficient T cells, we observed an unexpectedly large contribution of these factors to the active enhancer landscape. Clearly, transcriptomic changes mediated by STATs correlated well with STAT-dependent changes in p300 binding. While we demonstrated the direct role of STATs on p300 recruitment and STATs may directly associate with p300 (Paulson et al., 1999), the extent to which these proteins interact on a large scale will require further validation.
In contrast to the major effect of STATs on p300-bound enhancers, the impact of STATs on H3K4me1-positive enhancers was variable. The absence of STAT6 reduced but did not abrogate H3K4me1 marks, whereas the lack of STAT1 or STAT4 minimally influenced H3K4me1 distribution. The modest effect of STAT1 or STAT4 on H3K4me1 is not unexpected as both can contribute to Th1 specification and mice lacking both these factors have not been generated (Lighvani et al., 2001; Thieu et al., 2008). Considering that H3K4me1 broadly maps regions that may include both inactive and poised elements as well as active regulatory sites, “pioneering factors” other than STATs are likely responsible for deposition of this mark. Possibly the appearance of such marks occur at an earlier stage of T helper differentiation. In contrast, STATs are the major drivers of p300-bound, active enhancer landscape. This suggests a stepwise process of the enhancer firing in which the establishment of H3K4me1 modifications may precede STAT binding and HAT recruitment. In a sense the process of enhancer formation can be seen as a volleyball game: some factors “set” the play by forming a permissive enhancer landscape on which environment-sensing factors “spike” to create the productive enhancer elements.
Our study also identified potential candidates for pioneering or “setter” factors that might contribute to the poised enhancer landscape. These include transcription factors that globally determine T cell commitment or factors like AP-1 and NFAT, which sense T cell receptor engagement. Indeed, our analysis revealed that both Th1- and Th2-specific enhancers are enriched for binding sites of these transcription factors. It has been shown that transcription factor AP-1 can condition chromatin to be a basal permissive state, which then facilitates the ligand-dependent recruitment of glucocorticoid receptors (Biddie et al., 2011).
Redundant and Unique Enhancer-Shaping Properties of Master Regulators and STATs
T-bet and GATA-3 are referred to as helper T cell master regulators because they are sufficient to induce characteristic cytokine expression in Th1 and Th2 cells in the absence of STAT4 or STAT6 (Lee et al., 2001; Mullen et al., 2001). Our global enhancer profiling revealed that T-bet and GATA3 differed in their capacity to affect p300 binding in STAT-deficient cells; however, neither was sufficient to re-establish the normal active enhancer landscape.
Our data revealed that GATA3 overexpression could restore roughly half of STAT6-dependent enhancer elements. For genes induced by GATA3, proximate p300 binding correlated with gene expression. In an interesting contrast, T-bet had limited effects on global p300 maps. This was true both in T-bet-deficient cells and upon enforced T-bet expression in STAT4-deficient cells. The differential effects of T-bet and GATA3 argue that “master regulators” may have very distinct modes of action in cell specification. In the future, it will be of interest to compare and contrast the effects of these lineage defining transcription factors with other classic master regulators. In this regards, an additional unexpected observation on T-bet was its preferential repressive, rather than enabling, role in establishing enhancer competence; whether this is a general property of T-box transcription factors remains to be determined. While it is believed that silencing of genes expressed in other cell fates are the most relevant regulatory decisions in lineage commitment (Zhang et al., 2012), the direct negative role of key regulatory factors on global enhancers of opposite lineages has not been shown before. We speculate that context dependent protein-protein interaction involving transcription factors may impose alternative accretion of coactivator or corepressor complexes and, in doing so, dictate enhancer competence and transcriptional outcome.
The overlapping yet specialized contribution of STATs and master regulators to shape enhancer signature points to an interesting possibility of the formation of coherent feed-forward loops (FFL) (Shen-Orr et al., 2002). Coherent FFL have several dynamic and functional properties, one of the most relevant being the ability to protect the system from undesired responses to fluctuating inputs. For T cells, which migrate to diverse sites within the body and continually survey infectious challenges, such a scenario is likely to be advantageous. Such a perspective might also be relevant in other cells for which plastic versus static patterns of gene expression are functionally important.
An emerging view from our study is that while broad potentials exist in the poised enhancer landscape, STATs sense environmental stimuli and act upon the available repertoire of enhancers to trigger specific transcriptional responses. Beyond T cells and host defense, STATs have broad functions in programming gene expression in embryonic development, cell growth and cancer in various organisms (Horvath, 2000). The fact that STATs are involved in synaptic plasticity in the brain (Miller and Gauthier, 2007; Nicolas et al., 2012) is reminiscent of STAT action as environment sensors in T cells. Similarly in mammary tissue, STATs control different phases of cell development and involution (Watson and Neoh, 2008). It is tempting to speculate that the creation of active enhancer landscapes may be dependent upon STATs in these and other cells. In a broader perspective, there are a wide variety of transcription factors like STATs that sense environmental cues and regulate cellular differentiation. The extent to which nuclear hormones, retinoid receptors, SMAD family transcription factors and Wnt pathway act analogously to STATs will be interesting to ascertain. Clearly though, the present work establishes that environmental sensors can have profound effects on active enhancer landscapes in the process of cellular differentiation and in this manner directly link signal transduction with epigenetic regulation.
EXPERIMENTAL PROCEDURES
Mice, isolation of cells and cell culture
C57BL/6J, T-bet-, and STAT6-deficient mice were purchased from Jackson Laboratory. STAT4- and STAT1-deficient mice were provided by Dr. Mark Kaplan (Indiana University) and Dr. Joan Durbin (NYU), respectively. Animals were handled and housed in accordance with the guidelines of the NIH Animal Care and User Committee. Splenic and lymph node T cells were obtained by disrupting organs of 8- to 10-week-old mice. All cell cultures were performed in RPMI supplemented with 10% fetal calf serum, 2 mM glutamine, 100 IU/mL penicillin, 0.1 mg/mL streptomycin, and 2.5 μM b-mercaptoethanol. T cells were enriched using a CD4+ T Cell Kit and AutoMacs isolator (Miltenyi Biotec, Auburn CA). Naive CD4+ T cells were isolated by flow cytometry, staining with anti-CD4, anti-CD62L, anti-CD44, and anti-CD25 antibodies. Naïve CD4+ T cells were first cultured in the presence of plate-bound anti-CD3 and anti-CD28 (10 μg/mL each), IL-12 (10 ng/mL) and anti-IL-4 (10 μg/mL) for 3 days, followed by IL-2 (50 U/mL) and IL-12 (10 ng/mL) for 4 days (Th1) or anti-CD3 and anti-CD28, IL-4 (10 ng/mL) and anti-IFNγ (10 μg/mL) for 3 days, followed by IL-2 (50 U/mL) and IL-4 (10 ng/mL) for 4 days (Th2). Before harvesting, cells were restimulated with plate-bound anti-CD3 and anti-CD28 and cytokines for 2 hours. Cytokines were from R&D Systems (Minneapolis, MN) and antibodies were from BD Phamingen (San Jose, CA) and eBiosciences.
Computational and Statistical Analyses
Detailed description is available in the Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Closely related T helper cell subsets have distinct p300 binding profiles.
STATs generate lineage-specific enhancers and suppress those of alternative fates.
Overexpression of master regulators fails to re-establish STAT-dependent enhancers.
Changes in transcriptome correspond to changes in p300 binding.
ACKNOWLEDGEMENTS
The authors thank Drs. K. Zhao, A. Poholek, K. Ghoreschi for critically reading this manuscript. We also thank G. Gutierrez-Cruz (Biodata Mining Core Facility, NIAMS), J. Simone, and J. Lay (Flow Cytometry Section, NIAMS) for their excellent technical support. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the NIH. This work was supported by the Intramural Research Programs of NIAMS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STATI-induced regulator for IL-12R expression in naive CD4+ T cells. Nature Immunology. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annual Review of Immunology. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Balasubramani A, Shibata Y, Crawford GE, Baldwin AS, Hatton RD, Weaver CT. Modular Utilization of Distal cis-Regulatory Elements Controls Ifng Gene Expression in T Cells Activated by Distinct Stimuli. Immunity. 2010;33:35–47. doi: 10.1016/j.immuni.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddie S, John S, Sabo P, Thurman R, Johnson T, Schiltz RL, Miranda T, Sung MH, Trump S, Lightman S, et al. Transcription Factor AP1 Potentiates Chromatin Accessibility and Glucocorticoid Receptor Binding. Molecular Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MMW, Simpson N, Ciofani M, Chen G, Collins A, Littman DR. Epigenetic propagation of CD4 expression is established by the Cd4 proximal enhancer in helper T cells. Genes and Development. 2010;24:659–669. doi: 10.1101/gad.1901610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. PNAS. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends in Genetics. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011 doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and Characterization of Enhancers Controlling the Inflammatory Gene Expression Program in Macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Goenka S, Kaplan MH. Transcriptional regulation by STAT6. Immunologic Research. 2011;50:87–96. doi: 10.1007/s12026-011-8205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath CM. STAT proteins and transcriptional responses to extracellular signals. Trends in Biochemical Sciences. 2000;25:496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nature Immunology. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. Journal of molecular biology. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Kashiwada M, Cassel SL, Colgan JD, Rothman PB. NFIL3/E4BP4 controls type 2 T helper cell cytokine expression. EMBO Journal. 2011;30:2071–2082. doi: 10.1038/emboj.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs A, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Molecular Cell. 2011;44:410–423. doi: 10.1016/j.molcel.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GR, Fields PE, Flavell RA. Regulation of IL-4 gene expression by distal regulatory elements and GATA-3 at the chromatin level. Immunity. 2001;14:447–459. doi: 10.1016/s1074-7613(01)00125-x. [DOI] [PubMed] [Google Scholar]
- Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, O'Shea JJ. T-bet is rapidly induced by interferon-g in lymphoid and myeloid cells. PNAS. 2001;98:15137–15142. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, et al. Large-scale discovery of enhancers from human heart tissue. Nature Genetics. 2011 doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller FD, Gauthier AS. Timing Is Everything: Making Neurons versus Glia in the Developing Cortex. Neuron. 2007;54:357–369. doi: 10.1016/j.neuron.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, Livingston DM, Kung AL, Cereb N, Yao TP, Yang SY, Reiner SL. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292:1907–1910. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- Murphy KM, Ouyang W, Szabo SJ, Jacobson NG, Guler ML, Gorham JD, Gubler U, Murphy TL. T helper differentiation proceeds through Stat1-dependent, Stat4-dependent and Stat4-independent phases. Current Topics in Microbiology and Immunology. 1999:23–26. doi: 10.1007/978-3-662-09709-0_2. [DOI] [PubMed] [Google Scholar]
- Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu K, Johnson T, Sun HW, Vahedi G, Hakim O, Handon R, et al. Early Th1 Cell Differentiation Is Marked by a Tfh Cell-like Transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G. Maintaining Cell Identity through Global Control of Genomic Organization. Immunity. 2010;33:12–24. doi: 10.1016/j.immuni.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Nicolas C, Peineau S, Amici M, Csaba Z, Fafouri A, Javalet C, Collett V, Hildebrandt L, Seaton G, Choi SL, et al. The JAK/STAT Pathway Is Involved in Synaptic Plasticity. Neuron. 2012;73:374–390. doi: 10.1016/j.neuron.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea J, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4 + T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson M, Pisharody S, Pan L, Guadagno S, Mui AL, Levy DE. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. Journal of Biological Chemistry. 1999;274:25343–25349. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010 doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nature Genetics. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- Smale ST. Seq-ing LPS-Induced Enhancers. Immunity. 2010;32:296–298. doi: 10.1016/j.immuni.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, Sehra S, Kaplan MH. Signal Transducer and Activator of Transcription 4 Is Required for the Transcription Factor T-bet to Promote T Helper 1 Cell-Fate Determination. Immunity. 2008;29:679–690. doi: 10.1016/j.immuni.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CJ, Neoh K. The Stat family of transcription factors have diverse roles in mammary gland development. Seminars in Cell and Developmental Biology. 2008;19:401–406. doi: 10.1016/j.semcdb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Wei G, Abraham B, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup D, Tang Q, Paul W, et al. Genome-wide Analyses of Transcription Factor GATA3-Mediated Gene Regulation in Distinct T Cell Types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang CZ, et al. Discrete Roles of STAT4 and STAT6 Transcription Factors in Tuning Epigenetic Modifications and Transcription during T Helper Cell Differentiation. Immunity. 2010;32:840–851. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Paul WE. CD4 T cells: Fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.