Abstract
In the postgenomic era, proteomics has become a dominant field for identifying and quantifying the complex protein machinery of the cell. The expression levels, post-translational modifications, and specific interactions of proteins control the biology of such processes as development, differentiation, and signal transduction. Studies of the proteins involved in these processes often leads to a better understanding of biology and of human disease. Powerful separation techniques and sensitive detection methods enable researchers to untangle these complicated networks of processes. Capillary electrophoresis coupled with either mass spectrometry or laser-induced fluorescence are two of the techniques that make this possible. This review will cover proven capillary electrophoresis-based methods for proteomics on the cell and tissue level and their application in biological and clinical studies, relevant new developments in enabling technology such as microfluidic CE-MS demonstrated on model systems, and comment on the future of CE in proteomics.
Keywords: Capillary electrophoresis, Mass Spectrometry, Proteome, Proteomics
1. Introduction
Proteomics is a continually growing field that has vast potential in biology-related disciplines. MS-based proteomics has become the workhorse that facilitates investigation into cellular processes such as development, differentiation, and signal transduction through understanding of the regulation of the expression levels, interactions, and modifications of proteins [1]. Although linked to genomics, as proteins are a translation of the genetic code, proteomics entails direct analysis of the dynamic and complex functional machinery of the cell. In addition to the diversity of expressed genes from a genome, the proteome is more complex due to PTMs, protein isoforms, and differential expression, making analysis significantly more difficult. These complexities of protein translation are biologically interesting, yet at the same time are the most challenging aspect of investigation. Protein expression levels are believed to be present with a dynamic range of 7–8 orders of magnitude in cells [2] and up to 12 in plasma [3]. Thus for any proteomic analysis, a high demand is placed on the dynamic range of either optical or mass detectors and is further accentuated by the complexity of samples. Adequate separations of either proteins or peptides prior to detection is of utmost importance to probe low abundance proteins. Peak overlap often hides low intensity peaks using optical detection and, in the case of ESI-MS or MALDI-MS, can significantly suppress the signal of low abundance proteins or peptides. Adequate front-end separation techniques are necessary to alleviate these issues.
Traditionally, proteomic analysis has been performed by resolving complex protein mixtures using 2-DE followed by MS identification [4]. Excised proteins are digested with trypsin in a bottom-up approach while thoroughly purified proteins can be analyzed directly with high resolution MS. Although cost-effective and of relatively high resolution, the interest in proteomics began to outgrow the time-intensive and tedious procedure of 2-DE. Shotgun proteomics using automated, liquid-based separation methods emerged to increase the overall throughput of proteomics experiments. Gel-free shotgun proteomics methods have since become routine for protein identification, protein quantitation, protein-protein interaction studies, and post-translational modification analysis [5–9]. Gel free shotgun proteomics is enabled by the ability to search mass spectrometry data through sequence databases to identify proteins.[10–12] At the forefront are 2-D HPLC methods such as MudPIT where protein digests are loaded onto a strong-cation exchange resin which are serially bumped onto a LC column using salt pulses for subsequent RP separation and ESI-IT-MS/MS analysis [13]. Although primarily dominated by LC methods, CE has become quite promising and beneficial in many proteomics applications due to the inherent fast analysis times, low sample and reagent consumption, high efficiencies, and number of versatile separation modes [14, 15].
Since the introduction of CE by Jorgenson [16] there have been numerous advances in capillary coatings [17, 18], detection methods [19–21], separation modes [22, 23], integration to microfluidic chips [24, 25], and application to bioanalysis [26–28]. Additionally, improvements in protein [29] and peptide [30] separations using CE have significantly advanced, making application to proteomics possible. The development of CE with primarily two detection methods, MS and LIF, has come to fruition in the field of proteomics yielding results that are biologically relevant. CE-MS routinely yields comprehensive information on protein identification and quantitation of complex samples, while CE-LIF has proven most useful for monitoring changes in single cell expression profiles from cell development or external stimuli. The current impact of CE as a powerful separation method in proteomics will be reviewed, particularly in proven methods, relevant recent developments on model protein and peptide systems, and biological and clinical application, followed by comments on potential advances and applications.
2. CE-MS
2.1. ESI Interfacing
CE-MS has had the largest impact of CE-based techniques and has become a relatively routine method for proteomic analysis. Since their introduction to large biomolecules, ESI [31] and MALDI [32] have been the primary interfaces investigated for CE-MS. ESI has proven most useful in shotgun proteomic experiments where peptide digests are serially fractionated prior to an IT or TOF mass spectrometer using LC or CE. The soft ionization process is also useful for electrospraying intact proteins prior to higher resolution mass spectrometers such as the FTICR-MS [33, 34] and Orbitrap (unpublished observations). Commercial mass spectrometers are equipped with API sources which provide a sheath of gas or liquid for ESI. CE has been successfully interfaced with MS using this methodology or similar home-built versions [35]. Although the use of a sheathflow liquid is robust and versatile, the use of a sheathless configuration can improve S/N more than 10-fold through reduced dilution and background noise [36]. Thus, multiple sheathless ESI designs have been developed.
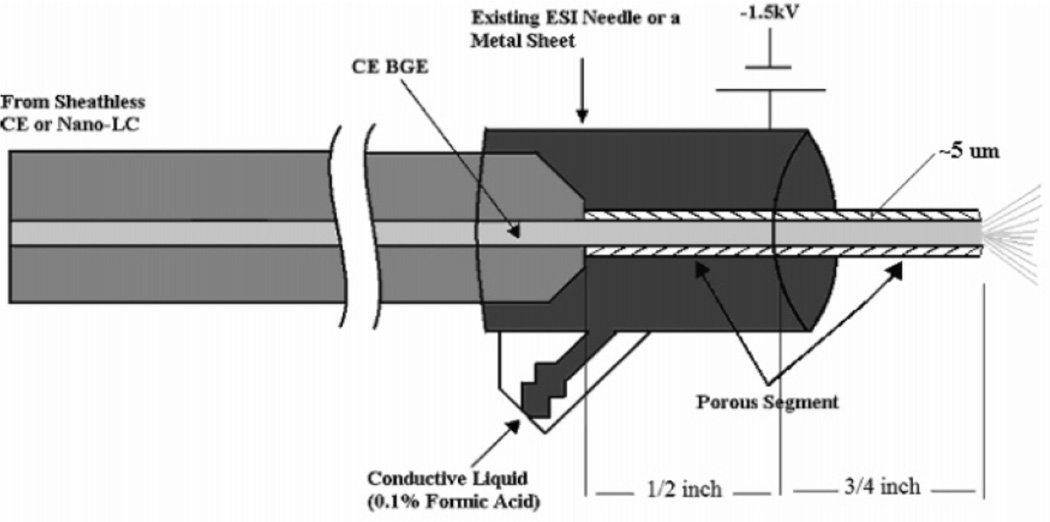
The main challenge to sheathless ESI is creating an electrical connection at the ESI tip without interrupting the CE flow. Sheathless ESI can generally be categorized by two designs. The first design entails coating an ESI tip with a conductive material such as gold [37–39] or graphite [40–43] for an electrical connection. Unfortunately, coated ESI tips have short lifetimes and frequently clog. Alternatively, the second general design requires creating a porous region near the ESI tip for electrical connection via an electrolyte [44, 45]. Porous junctions can be difficult to make reproducibly and are fragile, so other alternatives have been proposed. A pressurized liquid junction employing a ~100 µm gap between closely positioned capillaries facilitated simple application of the ESI voltage and a stable ESI spray independent of the CE effluent flow rate or composition [46–48]. A fully etched porous tip has also proven robust [49]. A 3 cm polyimide-stripped end of a capillary was etched in HF until the wall was 5–10 µm thick. The tip was inserted into an ESI needle with BGE allowing an electrical connection to generate a stable ESI spray at 1.5 kV. Figure 1 gives a schematic of the design.
Figure 1.
Schematic of the porous ESI tip for sheathless CE. The tip of a capillary is HF-etched to ~5 µm wall thickness making it porous and thus electrically conductive. The tip is inserted into a metal ESI needle or sheet and surrounded by a conductive liquid. A voltage of 1.5 kV is applied to generate a stable ESI spray. (Modified from ref. [49] with permission. © 2007 American Chemical Society.)
2.2. ESI-MS Sampling
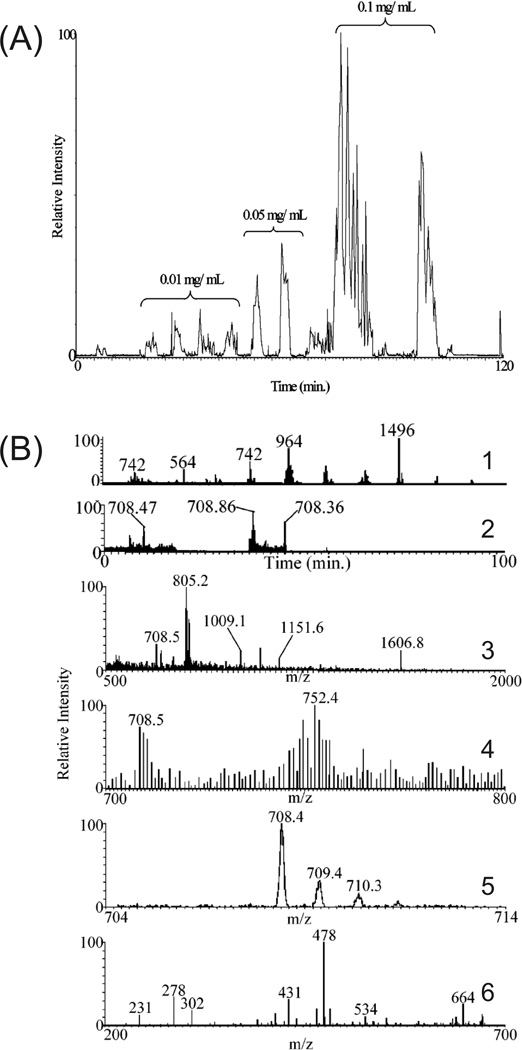
A common concern with CE-ESI-MS/MS in shotgun proteomics is the potential undersampling of peptides at the MS due to the poorly matched, slow duty cycle of the MS with the highly efficient, sharp CE peaks. A few methods have been proposed to increase protein identifications and sequence coverage while maintaining short analysis times. Tong et al performed CE-MS separations at a constant reduced voltage after elution of a neutral marker at the ESI tip, increasing peptide and protein identifications by a factor of three [50]. A similar approach used data-dependent control of the electrophoretic conditions [51]. When precursor ion intensities reached a threshold value the separation voltage was dropped from 20 kV to 5 kV to reduce the elution rate as MS/MS were acquired. The dynamic method was most effective at increasing identifications and sequence coverage for samples of intermediate complexity. No improvements were noticed for simple samples while high complexity samples ran nearly entirely at the reduced 5 kV. Alternatively, two (CE-MS/MS)n methods have been used to increase the sequence coverage of tryptic digests while using 30 kV for separations [52]. Six successive CE-MS/MS runs were performed on an identical sample. Each additional run appended precursor ions selected for MS/MS in the previous run to an exclusion list. Thus, previously detected peptides were excluded, increasing the probability with each run of detecting unsampled precursor peptide ions of low abundance. To further enhance identifications, increasing concentrations of sample were injected which, in this case, detected six more low abundance peptides and increased sequence coverage 5%; an electropherogram is shown in figure 2A. In the same report, gas-phase fractionation prior to MS/MS was also demonstrated to increase peptide identifications. Repeated injections of the same sample followed by defined MS scans (100 µm/z) effectively added another dimension of separation and allowed for identification of lower abundance peptides. Spectra are shown in figure 2B to illustrate the technique. The gas-phase fractionation method circumvents the use of dynamic exclusion where precursor ions of similar m/z can be excluded entirely. Similar results for both methods were observed, yielding a 100% increase in sequence coverage of a simple protein standard mixture over LC-MS/MS in the same analysis time.
Figure 2.
(A) (CE-MS/MS)n of increasing sample concentrations on a digested six protein mixture. Low abundance peptides are IDed in the most concentrated sample using an m/z exclusion list of high abundance peptides previously selected for MS/MS in the lower concentration samples. (B) Narrow mass range (CE-MS/MS)n analysis of the same protein mixture. (1) CE-MS/MS run with a full scan range (500–2000 m/z) followed by successive narrow mass range scans (100 µm/z). (2) CE-MS/MS electropherogram from (1) displaying only the 700–800 m/z range. Mass spectra of the 708.47 (3) and 708.86 peaks (4) from (2). The 708.86 peptide is present in (3), but its relative intensity was too low to be selected for MS/MS. The 708.86 peptide is easily detected in (4), with an expanded view in (5), from the 700–800 m/z scan and selected for MS/MS, resulting in a database-searchable peptide fragmentation pattern shown in (6). (Reprinted from ref. [52] with permission. © 2006 American Chemical Society.)
2.3. MALDI Interfacing
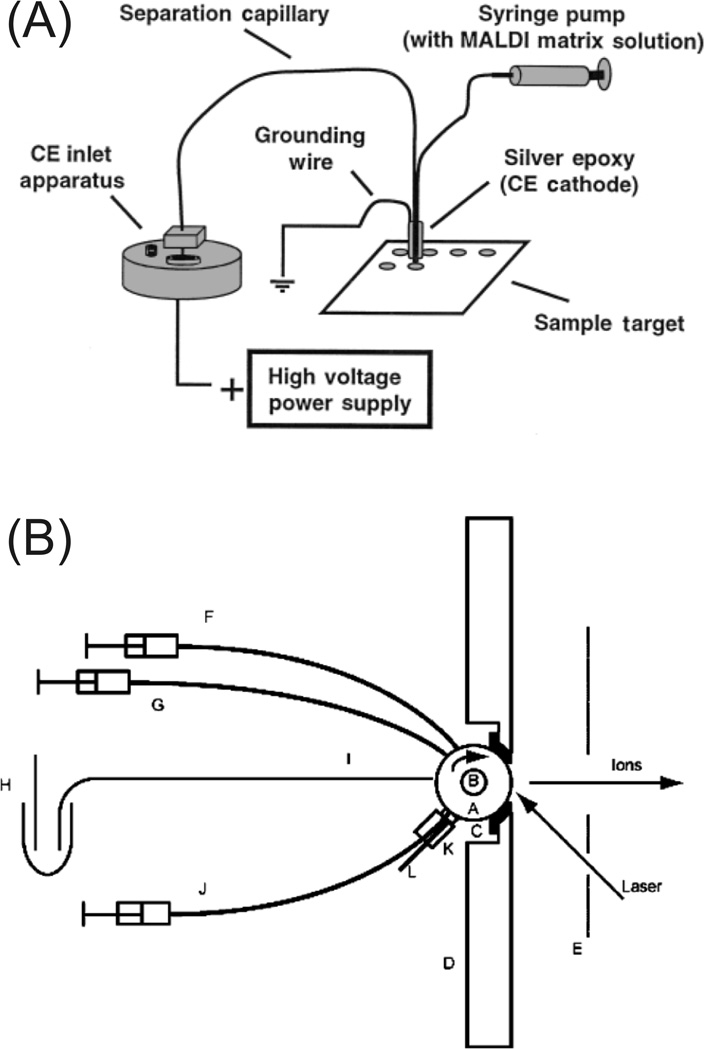
As with LC, the other prevalent alternative to ESI for CE is MALDI, although not nearly as readily used. MALDI is particularly well matched with TOF instruments, which typically have slower duty cycles than IT instruments. MALDI is commonly used to investigate intact proteins, but has recently proven useful for CE analysis of peptides. In general, MALDI is more tolerant to salts, surfactants, and other chemical interference than ESI. Unlike LC, where all fluids are pressure-driven and easily deposited onto MALDI targets, the electrical necessity of CE complicates collections onto the target. Multiple groups have demonstrated procedures for collections. A home-built CE instrument was first used to deposit CE effluent in a continuous line on a cellulose membrane-mounted MALDI target [53]. More recent developments facilitated direct spotting of CE effluent onto MALDI targets using either a sheath-flow setup containing matrix at the outlet of the capillary [54, 55] or a sheathless interface with a membrane-coated porous joint [56] . An example of a sheath-flow setup is shown in figure 3A. Collections have also been performed post-electrophoresis with a commercial CE instrument using pressure with the capillary outlet manually moved outside of the instrument above the MALDI target [57]. Beneficial to separation efficiency, a commercial CE has also been used to collect fractions directly onto the target during the separation using a 96-well Becton Dickinson MALDI sample concentrator device placed on a grounded MALDI target [58]. Collected samples were subsequently dried and dissolved in MALDI matrix. This approach was optimized by another group for the analysis of hydrophobic peptides and proteins [59].
Figure 3.
(A) Schematic of the CE-MALDI-MS fractionation apparatus. CE effluent is mixed with matrix before spotting on the MALDI target. (B) Schematic representation of the rotating ball inlet interface for continuous CE-MALDI-MS indicating (A) ball, (B) driveshaft, (C) Teflon gasket, (D) ISO-100 flange, (E) extraction lens, (F) matrix capillary, (G) buffer capillary, (H) anode and running buffer, (I) electrophoresis capillary, (J) cleaning solvent capillary, (K) felt pad and holder, and (L) solvent drain. CE effluent is continuously deposited with matrix on the rotating ball and desorbed using a laser. The ball is cleaned using solvent and a felt pad. Modified from refs. (A) [54] and (B) [61] with permission. ©2000 The Royal Society of Chemistry and © 2004 American Chemical Society, respectively.)
Interfacing CE with MALDI is still under development including methods that may rival ESI’s continuous nature. CE effluent was deposited continuously onto a moving tape under vacuum conditions and subsequently desorbed into a TOF-MS [60]. CE separation efficiency was maintained and allowed for continuous 24 hr operation. Similarly, a rotating ball MALDI target was demonstrated as a continuous alternative to ESI ionization with three model peptides [61]. The sample at the outlet of the capillary is mixed online with matrix, deposited onto the rotating ball, and desorbed by a laser to the MS after rotating 180O. The ball is subsequently cleaned by solvent-saturated felt. The rotating-ball instrument configuration is shown in figure 3B.
2.4. Sample Enrichment
Due to the nanoliter injections inherent to CE that yield poor concentration LODs, it is often necessary to concentrate peptide samples and sharpen peaks prior to separation and analysis using methods such as field amplified sample stacking. Additionally, CE-MS has been used following a fractionation step such as HPLC where samples are diluted, requiring further sample concentration. More than 10 years ago, SPE-CE-MS/MS of a tryptic digest from 2-DE separated yeast proteins using shotgun proteomics was successfully demonstrated [62, 63]. LODs of 660 amol were achieved for peptide samples with a total concentration less than 300 amol/µL. A 1000-fold increase in sensitivity from sample clean-up and concentration was possible. More recent reports have introduced SPE configurations with higher separation efficiencies and similar LODs using a SPE disk between two capillary ends [44] and an electrically insulated SPE loop valving system [64]. Field amplified sample stacking using discontinuous electric fields at the head of the capillary have also been used to enrich proteins [57] and peptides [65]. Concentration of the sample at the outlet of the capillary has been demonstrated using MALDI [66]. A simple, cheap paraffin wax film was stretched across a MALDI target. As drops were collected on the target they were concentrated during evaporation due to a solvent repellent effect to the surface, resulting in ~3-fold increase in S/N.
2.5. PTM Analysis
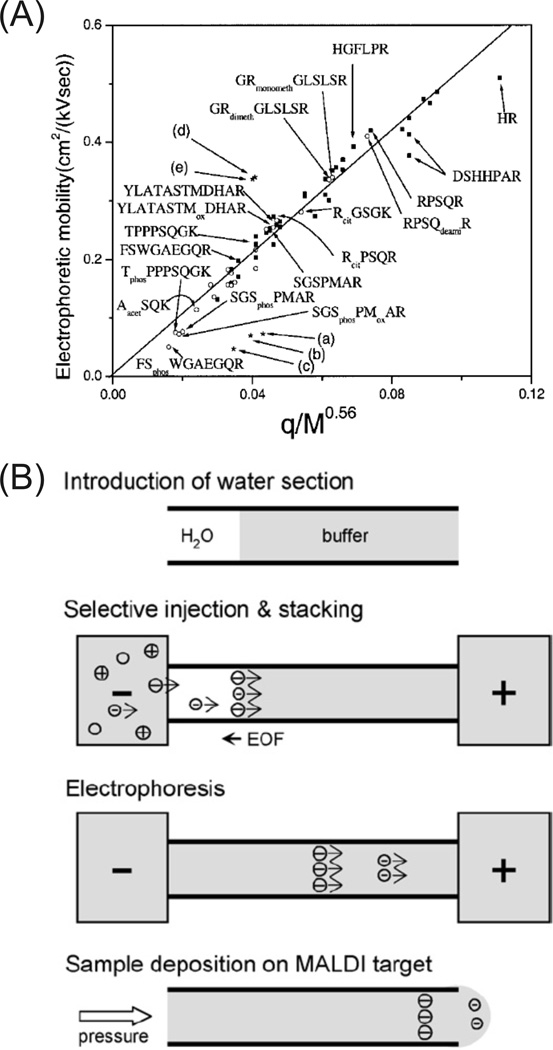
PTMs of proteins are often involved in regulation of their localization, turnover, activity, and interactions with other proteins. Protein heterogeneity created from PTMs makes proteomic analysis difficult since modified proteins, and thus peptides, are often in relative low abundance in a mixture. Thus, selective enrichment strategies are often necessary [67]. The inherent change in chemical charge to a protein or peptide by a PTM makes CE well-suited for specific PTM analysis. For instance, a peptide mobility study investigated the effect of PTMs such as phosphorylation, acetylation, and methylation. The results showed that phosphorylated peptides were grouped into a slower electrophoretic mobility window than non-phosphorylated peptides [68], illustrated in figure 4A. This methodology was exploited for selective sampling of phosphopeptides for CE-MALDI-MS [69] and CE-ESI-MS [70] and has since been improved for selective injection, stacking, and fractionation [71], shown in figure 4B. Enrichment factors of up to 600 were achieved on a synthetic phosphopeptide. Phosphopeptide analysis of a highly phosphorylated protein, osteopontin, indicated the ability to analyze multiple variable phosphorylation sites: 41 different phosphopeptides were detected with 29 previously reported phosphorylation sites. Similarly, phosphorylation of intact proteins results in a pI shift that facilitates separation using CIEF. This was initially demonstrated on mono- and diphosphoovalbumins separated by CIEF and detected by ESI-MS [72]. A common offline strategy for phosphoprotein and phosphopeptide enrichment is immobilized metal-ion affinity chromatography (IMAC). Fe(III)-IMAC resin was loaded at the head of a CE capillary for selective online phosphopeptide enrichment prior to CE-ESI-MS/MS analysis [73].
Figure 4.
(A) Correlation of electrophoretic mobility with q(charge)/Mr0.56 of peptides from a tryptic digest of myelin basic protein. Peptide mobilities with (■) and without (○) PTMs correlated well to the theoretical relationship. The phosphorylated peptides (phos) were grouped into a defined mobility window (< 0.1 cm2/kVsec). (B) An integrated procedure for phosphopeptide analysis. An injection of water is used to create a discontinuous electric field for selective injection and field amplified sample stacking of phosphopeptides. High mobility phosphopeptides (> EOF) stack into the water zone, are separated by electrophoresis, and deposited onto a MALDI target. (Modified from refs. (A) [68] and (B) [71] with permission. ©2003 Wiley-VCH and © 2006 Elsevier, respectively.)
PTMs other than phosphorylation have also been investigated using CE. CE-MALDI-MS/MS was used to indentify sialylated glycopeptides and characterize four N-glycosylation sites from bovine α1-glycoprotein [58]. Successive CE-MALDI-MS/MS runs of untreated and deglycosylated bovine α1-glycoprotein effectively separated the sialylated and asialo content of glycoprotein digests allowing for determination of the nature and location of the PTMs. Similarly, CE-ESI-MS/MS facilitated the study of non-enzymatically glycosylated PTMs on collagen [74]. A validation of CE-MS using a HPLC-MS characterized glycoprotein indicated that CE-MS was a comparable method [55].
2.6. Proteomic Analyses using CE-MS
There are multiple examples in the literature of the capabilities and potential of CE-MS for biological studies. The gentle, non-denaturing separation and ionization possible with CE-ESI-MS has proven useful for the analysis of stable protein complexes. Using a porous ESI tip capillary, zinc-carbonic anhydrase I & II and hemoglobin holoprotein complexes were separated and detected from a blood lysate [75]. Notably, no sample preparation was necessary and detection of a ~3 order of magnitude difference in concentration was achieved. Sheathflow ESI conditions for analysis of intact or dissociated myoglobin were found based on different methanol concentrations [76]. A mixture of commercially available glyceraldehyde-3-phosphate dehydrogenase (GAPDH), creatine phosphokinase (CPK), hemoglobin, β-lactoglobulin A, and lentil lectin were analyzed by CIEF-ESI-FTICR-MS [77]. Different sheathflow conditions were found which enabled analysis of either intact or dissociated protein complexes. Intact GAPDH and CPK complexes were first identified under gentle conditions and then subsequently analyzed under dissociating conditions allowing for subunit differentiation using MS. A similar, high sensitivity frontal analysis CE-ESI-MS method was used to characterize the antithrombin-heparin pentasaccharide complex [78]. The binding stoichiometry of the complex along with the Mr and sulfation features of the pentasaccharide were determined using the combination of denaturing and non-denaturing sheathflow conditions.
Identification of proteins in mixtures of low and intermediate complexity has been demonstrated in top-down and bottom-up approaches using CE-MS. The cleavage patterns of recombinant and natural bovine pepsin A were compared using CE-MS [79]. An assay to measure the acetylation pattern of human histone H4 was developed using a combined CE and MALDI-TOF-MS approach [80]. This assay was used to screen histone deacetylase inhibitors, functioning as potential anticancer drugs, in HT29 cell lines. Using ~3.5 ng of 56 E. coli ribosomal proteins, 55 of the proteins were identified using CE-MS based on Mr [81]. Purified conventional and transgenic maize proteins were analyzed using CE-MS [82] using a high organic percentage BGE (40% ACN) to minimize aggregation of zein proteins and improve separation efficiency [83]. The CE-MS method facilitated comparison of the zein protein fingerprint between the conventional and transgenic maize. More complex analyses have been performed on pathogens from microbial mixtures [84] and clinical samples [85]. Proteotypic peptides with abundances as low as 1% in the samples allowed for confident identification of the pathogens present. Peptides from degraded proteins in rat urine have been characterized as potential non-invasive diagnostics for monitoring toxicological or other physiological effects from pharmaceuticals in animal models [86]. An automated CE-MS platform has been validated to detect two-fold changes in peptide concentrations in human serum for biomarker discovery [87]. Mapping software has been created to perform easy differential comparison of CE-MS data [88].
A subset of proteomics, peptidomics, which involves direct analysis of endogenous peptides such as neuropeptides, has also benefited from improvements in CE-MS. A method for analysis of two neuropeptides, methionine-enkephalin and substance P 1–7, from Callitrix jacchus brain was demonstrated using SPE-CE-ESI-MS/MS [89]. Similarly, a SPE-CE-MALDI-TOF-MS protocol was established for proteomic analysis of single neurons of Aplysia californica [54]. With this method, numerous proteins and peptides up to 11 kDa were detected including egg-laying hormone δ/γ-bag cell peptide [90]. More recently, from a few pooled neuronal tissues from Cancer borealis the identification of 43 neuropeptides in a single run was achieved [56]. A total of 203 peaks were detected contributing information to the PTMs and neuropeptide families of the 70 total identified neuropeptides from pooled runs.
3. 2-D CE Shotgun Proteomics
Multiple instrument configurations have been proposed for performing 2-D CE shotgun proteomics experiments. Initially, CE separations were coupled to proven LC separations, adding a dimension of orthogonality. The first coupling of LC and CE-MS for peptide analysis was achieved using a plastic flow-gate interface [91], but have since been demonstrated using offline fractionation [92], a droplet transfer and gravity injection interface [93], a microinjector valve [94], a microselection valve to multiple trapping columns [95], and a PDMS flow-gate [96]. Integrating two online, orthogonal CE techniques has also been demonstrated. The necessity of multiple electrical connections has limited the CE-CE interface to flow gating [97] and a microdialysis junction [98, 99].
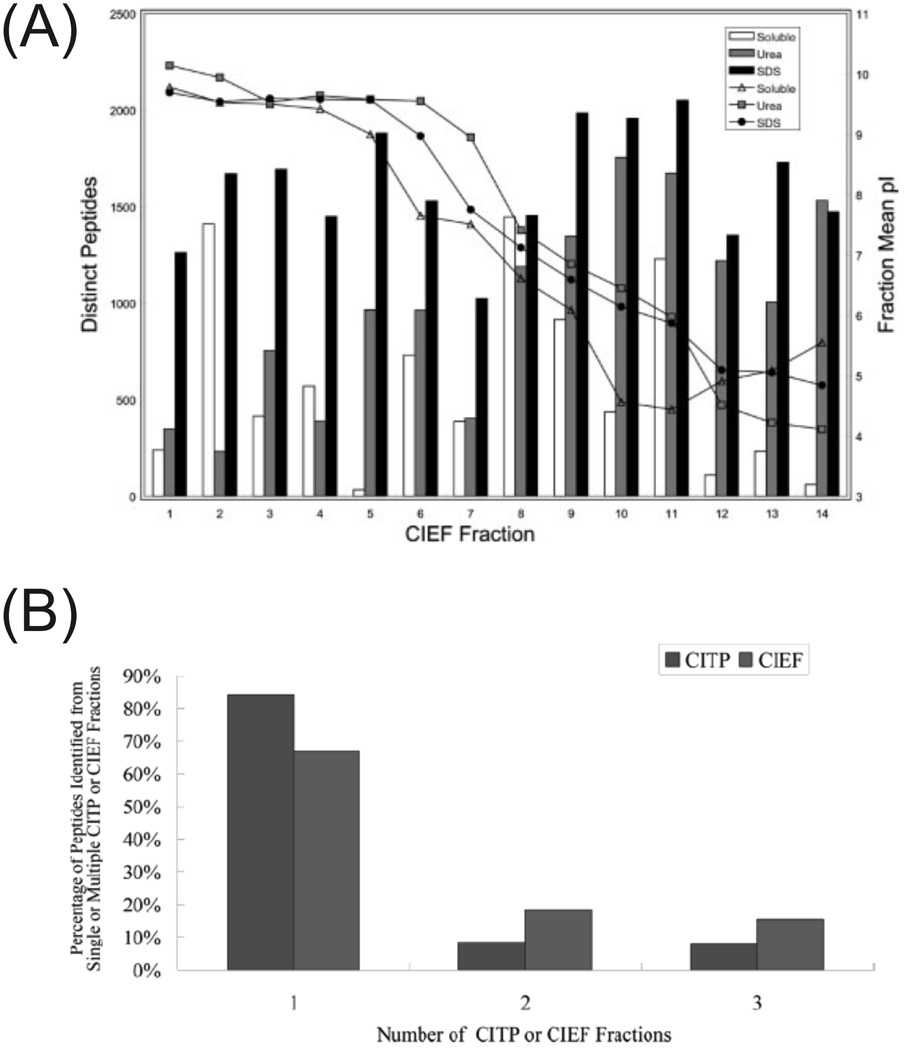
As a result of the above-mentioned developments, CE has become a viable alternative to 2-D LC for performing separations prior to MS in shotgun proteome analysis. Coupling CIEF to LC-MS/MS with a fast IT instrument yielded identification of 2513 distinct yeast proteins in a single run and 3632 proteins from three solubilization conditions, the highest yeast proteome coverage at that time [100]. The source of the high number of identifications is illustrated in figure 5A, as there are a relatively constant number of unique peptide identifications per CIEF fractionation step throughout the entire run, particularly from the SDS solubilized protein sample. Applied to the human salivary proteome, CIEF-LC-MS/MS identified 1381 distinct proteins in a single run, nearly three times more than other LC methods [101]. For a more complex sample such as the mouse brain mitochondrial proteome, 76% coverage of the MitoP2-database reference set was achieved through identification of 2191 proteins using combined CITP-CZE and CIEF-LC analyses with technical replicates [102]. A similar protocol was established for membrane proteome analysis of ovarian tumor tissue samples allowing for identification of 773 human proteins predicted to have transmembrane domains (24%), the highest reported coverage to date [103]. A recent comparison illustrated the advantages of CITP- or CIEF-CE-MS/MS over 2-D LC-MS/MS using the characterization of the human salivary proteome as a model system [104]. Of the identified peptides from the CITP method, only 8% were in two fractions and 7% in three or more; in comparison, 2-D LC methods using strong cation exchange chromatography unnecessarily burden the second separation dimension with 40–80% overlap between fractions, reducing peak capacity and the ability to identify low abundance peptides. Between CIEF and CITP methods, the CITP method identified 8% more proteins with half as many first dimension fractions and ~10% less overlap of peptide identifications between fractions as illustrated in figure 5B; additionally, 7% more proteins were identified by two or more distinct peptides, a typical criterion for protein identification.
Figure 5.
(A) Plot of distinct peptide IDs and mean pI as a function of the CIEF fraction number for three different solubilization conditions. The SDS solubilized sample had relatively consistent IDs per fraction. (B) Comparison of CITP and CIEF carryover of peptides between fractions. CITP minimizes dilution of peptides between multiple fractions more than CIEF. Both have significantly less carryover than LC-based strong cation exchange fractionations. (Modified from refs. (A) [100] and (B) [104] with permission. ©2007 Wiley-VCH and © 2007 American Chemical Society, respectively.)
It is apparent that the multiple modes of CE each have distinct advantages for 2-D peptide separations: CZE for separation efficiency and speed relative to LC; CIEF for 50- to 100-fold sample concentration [94], peptide pI information [100] for increased identification confidence [105], and highly efficient peak shapes which facilitates increased fractionation [106] and decreased overlap between fractions [99]; and CITP for enrichment and identification of trace peptides 500,000 times less abundant than the most concentrated [107]. In general, 2-D CE methods only require high nanogram amounts of protein for analysis [95, 99], at least an order of magnitude lower than 2-D LC methods.
4. Intact Protein Separations
The analysis of intact proteins may have the potential to gain otherwise difficult or unattainable information about PTMs and protein interactions using MS. Top-down proteomics is the field of intact protein analysis where high resolution instruments such as FTICR-MS [108] and the LTQ-Orbitrap [109] are used to identify proteins and PTMs based on their Mr determined from their isotopic envelope and by fragments generated with gas-phase fragmentation. Currently there are challenges to easily identifying complex mixtures of proteins with this method [110], but as MS and bioinformatics aspects advance, the ability to separate proteins with high resolution and capacity will be of utmost importance. Numerous high efficiency separations of proteins have been achieved using CE, which could have the potential to be applied in top-down proteomics. At the same time, many of these separations have already proven quite useful in their own right.
CIEF separations coupled to a FTICR-MS using ESI have already been demonstrated [111] with the ability to acquire 400–1000 putative protein masses using only ~300 ng of E. coli proteins [33]. Further improvements in separation resolution and FTICR-MS speed were cited as necessary, particularly for MS/MS to attain unambiguous protein identifications. Additionally, RPLC fractionation was performed prior to CIEF of a Shewanella oneidensis lysate [34]. Three times as many protein masses (166) were detected using RPLC fractionation compared to a dialyzed sample. A similar 2-D CIEF-LC-ESI-TOF-MS approach increased the system capacity to between 4320–7200 peaks and identified 534 distinct protein masses from 10 µg of a soluble S. cerevisiae lysate [112].
Since MS technology isn’t available to readily identify intact proteins, other groups have integrated bottom-up proteomics strategies with intact protein separations. In a fully online system, the end of a first dimension CZE protein separation capillary was packed with immobilized pepsin beads to create a microreactor prior to a second dimension CZE peptide separation [113]. A flow-gate was used to transfer digested peptides to the second dimension separation coupled to a triple quadrupole MS. A simple mixture of cytochrome C and myoglobin were used to test the system: a sequence coverage of 48% and 22% were achieved, respectively, with a peak capacity of 590. Another method was capable of incorporating two successive intact protein separations by performing offline HPLC fractionation to CIEF with on-target digestion of proteins prior to MALDI-TOF-MS [114]. CIEF eluted samples were concentrated on immobilized RP beads spotted onto a MALDI target, washed of ampholytes, digested with drops of trypsin solution, suspended in a matrix droplet, and dried. Analysis of a rat liver extract was performed and 376 unique proteins were identified. CIEF has also been directly coupled to hollow fiber flow field-flow fractionation creating a pI vs. size separation similar to 2-DE [115]. The 2-D separation was followed by LC-MS/MS on the trypsin-digested fractions. Analysis of a human urine sample yielded 114 protein identifications.
Another method has combined the high sensitivity of LIF with the repeatability and efficiency of CE to generate protein maps based on two orthogonal CE separation conditions similar to 2-DE. This concept was first introduced by Michels et al [97]. Very little sample is required and complete transfer of sample from one capillary to another is achieved using a flow-gate. 2-D CE-LIF is particularly advantageous even over MS due to the high sensitivity for monitoring zeptomoles of protein. Protein lysine groups are reacted with the fluorogenic reagent 3-(2-furoyl)quinoline-2-carboxaldehyde, creating highly fluorescent products with minimal background. Submicellar concentrations of SDS are added to the separation buffer to mask unreacted lysine heterogeneity [116]. Notably, the amount of protein injected into the instrument was equivalent to that of a single cell and was later demonstrated on a single cell [117]. Comparison of 2-D CE-LIF runs from single MCF-7 breast cancer cells presorted by ploidy demonstrated the ability to differentiate cell cycle dependence of the proteome with a degree of variability greater than 2.5 [118]. This was possible through high run-to-run and day-to-day reproducibility [119]. The initial demonstration of 2-D CE-LIF combined two CZE separations performed at different pHs, but has since employed the greater orthogonality of CSE-MEKC [120, 121] generating spot capacities of ~500 and component resolutions in the hundreds in 1 hr [122, 123]. Further instrumental design incorporated five 2-D CE-LIF instruments for analysis of five samples in parallel [124]. Although MS has not been integrated, the method has proven quite useful as described in the Applications section and yields great potential for characterization of protein expression variability such as in genetic disorders.
Other instrumental designs for intact protein separations have been evaluated using optical detection. CIEF-CEC coupled with UV detection proved adequate for the separations of human serum proteins with a theoretical peak capacity of ~54,000 [125]. Two LIF methods have also been demonstrated. A yeast lysate was derivatized using BODIPY maleimide, fractionated using RPLC, then focused and detected using CIEF with a theoretical peak capacity of 10,000 [126]. From the same group, inversion of the separation order allowed for a fully automated system using an array of CIEF capillaries [127]. The system was demonstrated on a BODIPY-derivatized rat liver lysate with an estimated peak capacity of 18,000 for a 3 hr analysis.
5. Microchip CE and ESI
Microfluidic devices hold immense potential for dispersing powerful separation techniques and integrated procedures to the non-expert. Lab-on-a-Chip devices have integrated multiple laboratory techniques such as sample preparation and enrichment, separation, and detection, all while minimizing adsorptive sample losses from transfers, sample requirements, and analysis time [128]. Microchip CE has already proven useful in genetic analysis [129]; proteomics should also benefit from Lab-on-a-Chip devices that are currently being developed. Considering the complexity and need for identification of proteomics samples, two major drives have been to integrate 2-D CE protein and peptide separations to a microfluidic chip and couple them to MS using ESI. These topics will be the focus of this section, but other reviews also provide a comprehensive overview of microfluidic technology developments for proteomics [130, 131].
Preliminary demonstrations of 2-D CE microfluidic proteome-related separations were initially performed using serial injections from the first to second dimension. The first such demonstration of this technique was accomplished by Ramsey and colleagues on a fluorescently-labeled tryptic digest of model proteins [132]. As with capillary-based 2-D CE separations, the interface between dimensions is of utmost importance and was accomplished using a standard cross-geometry gate injection etched in glass substrates. Peak capacities were estimated at 500–1000 in less than 10 minutes using orthogonal CZE and MEKC separation modes. A similar design from the same lab evaluated open-channel electrochromatography coupled to CE for tryptic digest separations [133]. Other groups have since coupled other CE techniques in cheaper substrates such as poly(dimethylsiloxane) [134–136], poly(methyl methacrylate) [137–140], and polycarbonate [141]. The Whiteside’s lab later demonstrated the potential for 2-D intact protein separations on a microchip using fluorescently-labeled proteins [136] using a previously introduced parallel analysis concept [142]. Similar to 2-DE, the separated proteins from an IEF separation were simultaneously injected onto an array of orthogonal gel-filled channels for SDS gel electrophoresis. In this case the IEF focused first dimension was physically transferred to the parallel channels, but has since been automated by multiple labs [139–141, 143]. The parallel analysis scheme minimizes sample loss and analysis time relative to analyses requiring serial injections.
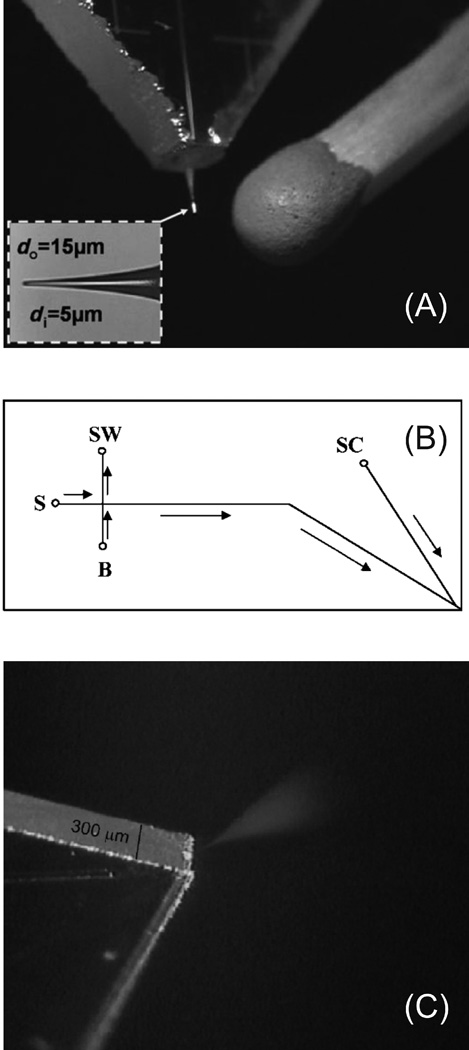
As with CE, it will be important to couple proteomic microfluidic separations to MS. This concept and the demonstration of directly electrospraying from the edge of microchip was first introduced over ten years ago [144, 145]. There have been multiple methods proposed for coupling microchip CE and ESI with better sensitivity, but most are based on manually assembled external emitters [146]. Top-down analysis of a DNA-binding domain of human TTAGGG repeat binding factor 2 was demonstrated using this configuration [147]. Recently there have been multiple demonstrations of fully-integrated ESI tips to microchip CE with sensitivities comparable to commercially available nanospray tips. As shown in figure 6A, a computer numerical control machined cone at the end of a CE separation channel at the edge of a glass microchip was drawn to an ESI tip using a home-built Pt heating coil puller [148]. SU-8, a biocompatible and chemical-resistant negative photoresist commonly used for fabricating microfluidic devices, has been used to generate a three-layer CE-ESI microfluidic chip with a sheath liquid channel for ESI electrical connection [149]. A similar all-glass design was demonstrated where a separation channel is coupled to a sheath liquid channel at the corner of the microchip which is only 300 µm thick from the two 150 µm glass substrates used [150]. Images of the design and spray from the glass microchip are shown in figure 6B and C, respectively.
Figure 6.
(A) Image of an integrated ESI tip at the edge of a glass CE microchip that was fabricated by pulling a CNC-milled cone using a Pt heating coil. (B) Schematic of a corner ESI microchip for CE-MS. Arrows indicate the direction of flow. Separations are performed by applying a separation voltage to the buffer reservoir (B) and an ESI voltage to the side channel (SC). (C) Image of a corner ESI microchip electrospray. (Modified from refs. (A) [148] and (B) and (C) [150] with permission. ©2007 Wiley-VCH and © 2008 American Chemical Society, respectively.)
6. Clinical Applications
As highlighted in this review, CE technology has reached a point where its speed and minimal sample requirements can be exploited in proteomic applications. The following are some examples which have an impact on understanding and diagnosis of diseases. Using the 2-D CE-MS instruments developed in the Lee lab described in the 2-D CE Shotgun Proteomics section, techniques have been developed to perform proteomic experiments on specific tissue samples. Glioblastoma multiforme (GBM) tumor tissue proteins were prepared and analyzed by flash freezing tissue samples, microdissection, protein isolation, trypsin digestion, and 2-D CIEF-LC-MS [151]. This technique was used to compare the proteomes of GBM with normal brain cortex tissue samples [152]. Wolf-Hirschhorn syndrome candidate 1 (WHSC1), among 103 other proteins, was found only in the GBM sample, verified by western blotting and immunohistochemsitry, and found to increase with ascending tumor proliferation activity. RNA interference on the WHSC1 gene transcripts suppressed tumor growth. A similar, wide-ranging method has been established to analyze the vast library of archived tissues, typically formalin-fixed or paraffin-embedded [153].
The 2-D CE-LIF instrumentation from the Dovichi lab described in the Intact Protein section facilitated the rapid characterization of proteins and biogenic amines in endoscopic and surgical biopsies from Barrett’s esophagus patients [154]. Barrett’s esophagus is caused by chronic gastroesophageal reflux, which damages the normal squamous epithelium, causing the development of specialized intestinal epithelium in the esophagus. These patients have a 30 to 40-fold increased risk of developing esophageal adenocarcinoma. While surgery essentially eliminates the risk of cancer, esophagectomy has a 5–20% mortality rate. Thus, accurate diagnosis of the cancer progression would be useful. Tissue biopsy samples were fixed, homogenized, fluorescently labeled, and subjected to 2-D CE-LIF. Protein expression level differences were evident, but the most convincing markers were 13 amino acids, identified by co-migration, as high as 40-fold more concentrated in the case of glycine. Although only amino acids and biogenic amines could be identified and correlated to the disease state, the potential for proteomic comparison on minimal clinical samples was demonstrated. Improved protein resolution and advances in coupling to MS were cited by the authors as means for better probing protein biomarkers using this technique.
CE-MS has been applied for the clinical follow-up of allogeneic hematopoietic peripheral blood stem cell transplantation (allo-HSCT) [155] and prediction of acute graft-versus-host disease (GVHD) [156]. Allo-HSCT has been successful for treatment of multiple hematopoietic malignancies. However, there are potential life-threatening complications that arise such as GVHD, which would benefit from early diagnosis and better control than immunosuppressive treatment. In their first CE-MS paper, more than 1000 polypeptides were detected in urine from patients and polypeptide patterns were significantly different from healthy volunteers. Specifically, 16 polypeptides were differentially expressed, forming a pattern for GVHD afflicted patients, while discriminating from sepsis using 13 other polypeptide markers. Specificities of 82 and 97% for GVHD and sepsis were found, respectively. Further studies in their second CE-MS paper validated the method to diagnose GVHD afflicted patients.
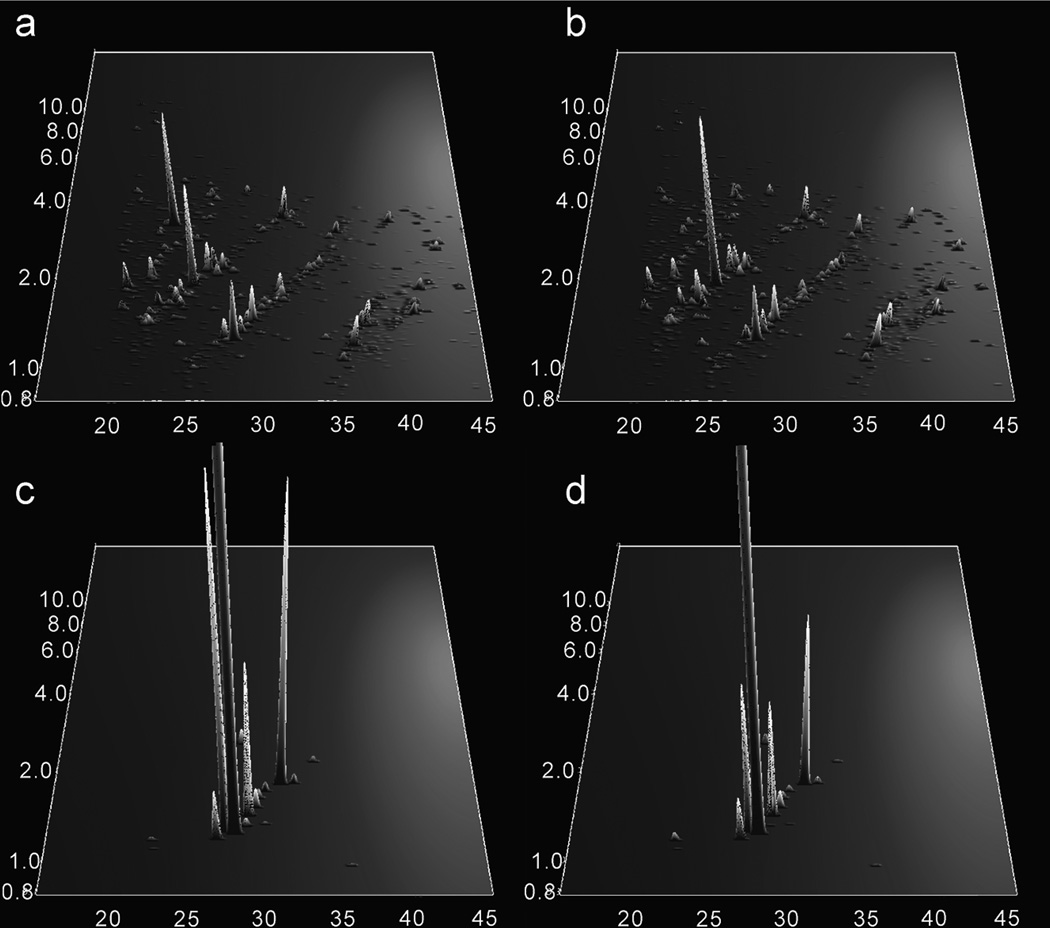
Other examples of urinary proteomics using CE-MS have also proven useful. A comparison of surface-enhanced laser desorption ionization (SELDI)-MS and CE-MS for differentiation of healthy subjects from membranous glomerulonephritis patients indicated the potential of CE-MS as three potential biomarkers were identified by SELDI and 200 by CE [157]. A more targeted study found polypeptide patterns from healthy subjects and renal disease groups from focal segmental glomerulosclerosis, minimal change disease, and membranous glomerulonephritis all had specific peptide signatures [158]. Eleven polypeptides with a given mass and migration time were used to discriminate between healthy and diseased patients, while four to nine peptides were used to discriminate among the three diseases. Two studies using a similar approach were used to diagnose coronary artery disease from urine. Polypeptide patterns from CE-MS runs of CAD and non-CAD patients that facilitated confident diagnosis in a blinded study are shown in figure 7. Most up-regulated polypeptides could be matched to collagen which positively stained in human atherosclerotic plagues [159]. In addition to confident diagnosis, the polypeptide patterns of patients were tracked after therapeutic intervention and physical activity, yielding a shift to healthy polypeptide signatures [160].
Figure 7.
Polypeptide patterns from urine samples distinguishing coronary artery disease (CAD) patients from healthy patients plotted as normalized molecular weight in kDa (y-axis) vs. CE migration time in minutes (x-axis). The averaged data sets from 24 CAD patients were processed to determine the MS signal (z-axis) of indicative peptides of (a) CAD and (b) non-CAD patients. A differentiation pattern of 17 polypeptides were found between (c) CAD and (d) non-CAD patients. Most polypeptides were found to be up-regulated and from human collagen. (Reprinted from ref. [159] with permission. © 2008 American Chemical Society.)
7. Conclusions and Outlooks
The advent of gel-free proteomics has undoubtedly had a significant impact on the speed and throughput of proteomic experiments, the analysis of low abundance proteins and difficult to analyze proteins such as transmembrane domains, the identification and localization of PTMs, and protein expression comparisons through quantitative proteomics. Most of these techniques are based on liquid chromatography of digested proteins. As highlighted in this review, CE based methods have begun to compete with LC methods and in some cases actually eclipse them in speed, protein identifications, and sequence coverage. A significant absence in current CE-MS technology is quantitative proteomics using isotopic labeling common to LC methods. There should be no reason for concern in its potential demonstration since the MS analysis is the most crucial aspect; simply, it has not been reported prior to this review. Furthermore, the reduced mass requirements of CE should yield impressive benefits as MS sensitivity improves. With lower flow rates than LC and thus less background, CE should be advantageous for sensitive MS measurements of proteins. Ultimately, the goal should be to perform MS-coupled proteomics experiments on single cells, as the Dovichi lab has already demonstrated with LIF, potentially allowing for quantitative proteomic comparison between single cells.
As the need for separations of intact proteins increases in applications such as protein-protein interactions studies or top-down proteomics, CE may bear further advantages. For protein interaction studies using gradient LC elutions [161], where organic or salt composition is varied, physiologically protein interactions may no longer be stable. On the other hand, CE has become a routine method for measuring binding interactions among purified biomolecules [162] and thus has the potential to be extended to a complex protein mixture if enough peak capacity can be generated among orthogonal separations under appropriate conditions. Considering the importance of surface area to an LC separation, the ample adsorptive surfaces in an LC column can present recovery issues for hydrophobic proteins [163, 164]. With reduced surface area to volume ratios and electrophoretic buffers often similar to physiological buffers, CE may yield the greatest benefits for these applications. However, CE is not completely free from adsorption problems. Although not covered due to the scope of this review, capillary wall coatings are an absolutely essential aspect of peptide and especially protein separations. Great efforts have been made to minimize wall adsorption using dynamic and permanent wall coatings which maintain separation efficiency and speed, recovery, and migration time repeatability, covered in numerous reviews [17, 18, 165, 166].
Slow diffusion coefficients of proteins make efficient LC separations difficult. Ultrahigh-pressure LC methods hint at circumventing some of these problems [167], but have only recently been demonstrated on a cell lysate [168]. CE is perfectly suited for protein separations since analytes with slow diffusion coefficients have the highest separation efficiency. Additionally, specific CE modes such as CIEF and CITP actively concentrate proteins during the course of the separation. As a final persuasion, intact protein separations and protein complex analysis have already been demonstrated using 2-DE. The relationship between gel-based 2-DE and solution-based 2-D CE begs to question why CE hasn’t had a larger impact in complex protein separations already. At this point the main challenges are MS related; the sensitivity necessary for coupling the low mass 2-D CE with MS hasn’t been reached and most 2-D CE separations of proteins aren’t currently MS compatible. Furthermore, the ability to identify intact proteins from a complex mixture based on Mr and fragmentation is complicated by multiple PTMs and is still currently under development. Until these problems are solved, CE can’t be used to its full potential in proteomics.
Acknowledgements
Funding from the National Institutes of Health (DK074798-02). The authors thank James Moresco and Dwight Stoll for helpful discussions.
Abbreviations
- CEC
capillary electrochromatography
- CITP
capillary isotachophoresis
- CSE
capillary sieving electrophoresis
- FTICR
Fourier transform ion cyclotron resonance
- m/z
mass-to-charge ratio
- MEKC
micellar electrokinetic capillary electrophoresis
- PTM
post-translational modification
References
- 1.Cravatt BF, Simon GM, Yates JR., 3rd Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 2.Anderson NL, Anderson NG. Electrophoresis. 1998;19:1853–1861. doi: 10.1002/elps.1150191103. [DOI] [PubMed] [Google Scholar]
- 3.Anderson NL, Anderson NG. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 4.Issaq H, Veenstra T. BioTechniques. 2008;44:697–698. 700. doi: 10.2144/000112823. [DOI] [PubMed] [Google Scholar]
- 5.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 6.Mann M, Jensen ON. Nature biotechnology. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 7.Ong SE, Mann M. Nature chemical biology. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 8.Westermann S, Cheeseman IM, Anderson S, Yates JR, 3rd, et al. The Journal of cell biology. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheeseman IM, Niessen S, Anderson S, Hyndman F, et al. Genes & development. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dongre AR, Eng JK, Yates JR., 3rd Trends in biotechnology. 1997;15:418–425. doi: 10.1016/S0167-7799(97)01110-4. [DOI] [PubMed] [Google Scholar]
- 11.Sadygov RG, Cociorva D, Yates JR., 3rd Nature methods. 2004;1:195–202. doi: 10.1038/nmeth725. [DOI] [PubMed] [Google Scholar]
- 12.Eng JK, McCormack AL, Yates JR, III, J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 13.Motoyama A, Yates JR., 3rd Analytical chemistry. 2008;80:7187–7193. doi: 10.1021/ac8013669. [DOI] [PubMed] [Google Scholar]
- 14.Cooper JW, Wang Y, Lee CS. Electrophoresis. 2004;25:3913–3926. doi: 10.1002/elps.200406154. [DOI] [PubMed] [Google Scholar]
- 15.Simpson DC, Smith RD. Electrophoresis. 2005;26:1291–1305. doi: 10.1002/elps.200410132. [DOI] [PubMed] [Google Scholar]
- 16.Jorgenson JW, Lukacs KD. Science. 1983;222:266–272. doi: 10.1126/science.6623076. [DOI] [PubMed] [Google Scholar]
- 17.Dolnik V. Electrophoresis. 2004;25:3589–3601. doi: 10.1002/elps.200406113. [DOI] [PubMed] [Google Scholar]
- 18.Lucy CA, MacDonald AM, Gulcev MD. Journal of chromatography. 2008;1184:81–105. doi: 10.1016/j.chroma.2007.10.114. [DOI] [PubMed] [Google Scholar]
- 19.Zemann AJ. Electrophoresis. 2003;24:2125–2137. doi: 10.1002/elps.200305476. [DOI] [PubMed] [Google Scholar]
- 20.Johnson ME, Landers JP. Electrophoresis. 2004;25:3513–3527. doi: 10.1002/elps.200406086. [DOI] [PubMed] [Google Scholar]
- 21.Maxwell EJ, Chen DD. Analytica chimica acta. 2008;627:25–33. doi: 10.1016/j.aca.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 22.Issaq HJ. Electrophoresis. 2000;21:1921–1939. doi: 10.1002/1522-2683(20000601)21:10<1921::AID-ELPS1921>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 23.Juvancz Z, Kendrovics RB, Ivanyi R, Szente L. Electrophoresis. 2008;29:1701–1712. doi: 10.1002/elps.200700657. [DOI] [PubMed] [Google Scholar]
- 24.Becker H, Manz A. Sens. Update. 1998;3:209–238. [Google Scholar]
- 25.McDonald JC, Duffy DC, Anderson JR, Chiu DT, et al. Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 26.Haselberg R, de Jong GJ, Somsen GW. Journal of chromatography. 2007;1159:81–109. doi: 10.1016/j.chroma.2007.05.048. [DOI] [PubMed] [Google Scholar]
- 27.Kostal V, Arriaga EA. Electrophoresis. 2008;29:2578–2586. doi: 10.1002/elps.200700917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lapainis T, Sweedler JV. Journal of chromatography. 2008;1184:144–158. doi: 10.1016/j.chroma.2007.10.098. [DOI] [PubMed] [Google Scholar]
- 29.Dolnik V. Electrophoresis. 2008;29:143–156. doi: 10.1002/elps.200700584. [DOI] [PubMed] [Google Scholar]
- 30.Kasicka V. Electrophoresis. 2008;29:179–206. doi: 10.1002/elps.200700550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka K, Waki H, Ido Y, Akita S, et al. Rapid Commun. Mass Spectrom. 1988;2:151–153. [Google Scholar]
- 33.Jensen PK, Pasa-Tolic L, Anderson GA, Horner JA, et al. Analytical chemistry. 1999;71:2076–2084. doi: 10.1021/ac990196p. [DOI] [PubMed] [Google Scholar]
- 34.Simpson DC, Ahn S, Pasa-Tolic L, Bogdanov B, et al. Electrophoresis. 2006;27:2722–2733. doi: 10.1002/elps.200600037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitt-Kopplin P, Englmann M. Electrophoresis. 2005;26:1209–1220. doi: 10.1002/elps.200410355. [DOI] [PubMed] [Google Scholar]
- 36.Kelly JF, Ramaley L, Thibault P. Anal. Chem. 1997;69:51–60. [Google Scholar]
- 37.Kele Z, Ferenc G, Klement E, Toth GK, Janaky T. Rapid Commun Mass Spectrom. 2005;19:881–885. doi: 10.1002/rcm.1866. [DOI] [PubMed] [Google Scholar]
- 38.Nilsson S, Klett O, Svedberg M, Amirkhani A, Nyholm L. Rapid Commun Mass Spectrom. 2003;17:1535–1540. doi: 10.1002/rcm.1082. [DOI] [PubMed] [Google Scholar]
- 39.Ramsey RS, McLuckey SA. J. Microcolumn Sep. 1995;7:461–469. [Google Scholar]
- 40.Chang YZ, Chen YR, Her GR. Analytical chemistry. 2001;73:5083–5087. doi: 10.1021/ac010429o. [DOI] [PubMed] [Google Scholar]
- 41.Chang YZ, Her GR. Analytical chemistry. 2000;72:626–630. doi: 10.1021/ac990535e. [DOI] [PubMed] [Google Scholar]
- 42.Nilsson S, Wetterhall M, Bergquist J, Nyholm L, Markides KE. Rapid Commun Mass Spectrom. 2001;15:1997–2000. doi: 10.1002/rcm.466. [DOI] [PubMed] [Google Scholar]
- 43.Zhu X, Thiam S, Valle BC, Warner IM. Analytical chemistry. 2002;74:5405–5409. doi: 10.1021/ac025877q. [DOI] [PubMed] [Google Scholar]
- 44.Janini GM, Conrads TP, Wilkens KL, Issaq HJ, Veenstra TD. Analytical chemistry. 2003;75:1615–1619. doi: 10.1021/ac020661+. [DOI] [PubMed] [Google Scholar]
- 45.Whitt JT, Moini M. Analytical chemistry. 2003;75:2188–2191. doi: 10.1021/ac026380j. [DOI] [PubMed] [Google Scholar]
- 46.Foret F, Zhou H, Gangl E, Karger BL. Electrophoresis. 2000;21:1363–1371. doi: 10.1002/(SICI)1522-2683(20000401)21:7<1363::AID-ELPS1363>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 47.Fanali S, D'Orazio G, Foret F, Kleparnik K, Aturki Z. Electrophoresis. 2006;27:4666–4673. doi: 10.1002/elps.200600322. [DOI] [PubMed] [Google Scholar]
- 48.Kusy P, Kleparnik K, Aturki Z, Fanali S, Foret F. Electrophoresis. 2007;28:1964–1969. doi: 10.1002/elps.200600640. [DOI] [PubMed] [Google Scholar]
- 49.Moini M. Analytical chemistry. 2007;79:4241–4246. doi: 10.1021/ac0704560. [DOI] [PubMed] [Google Scholar]
- 50.Tong W, Link A, Eng JK, Yates JR., 3rd Analytical chemistry. 1999;71:2270–2278. doi: 10.1021/ac9901182. [DOI] [PubMed] [Google Scholar]
- 51.Figeys D, Corthals GL, Gallis B, Goodlett DR, et al. Analytical chemistry. 1999;71:2279–2287. doi: 10.1021/ac9813991. [DOI] [PubMed] [Google Scholar]
- 52.Garza S, Moini M. Analytical chemistry. 2006;78:7309–7316. doi: 10.1021/ac0612269. [DOI] [PubMed] [Google Scholar]
- 53.Zhang H, Caprioli RM. J Mass Spectrom. 1996;31:1039–1046. doi: 10.1002/(SICI)1096-9888(199609)31:9<1039::AID-JMS398>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 54.Page JS, Rubakhin SS, Sweedler JV. Analyst. 2000;125:555–562. [Google Scholar]
- 55.Amon S, Plematl A, Rizzi A. Electrophoresis. 2006;27:1209–1219. doi: 10.1002/elps.200500725. [DOI] [PubMed] [Google Scholar]
- 56.Wang J, Ma M, Chen R, Li L. Analytical chemistry. 2008;80:6168–6177. doi: 10.1021/ac800382t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nesbitt CA, Jurcic K, Yeung KK. Electrophoresis. 2008;29:466–474. doi: 10.1002/elps.200700339. [DOI] [PubMed] [Google Scholar]
- 58.Snovida SI, Chen VC, Krokhin O, Perreault H. Analytical chemistry. 2006;78:6556–6563. doi: 10.1021/ac060738k. [DOI] [PubMed] [Google Scholar]
- 59.Jacksen J, Redeby T, Emmer A. Journal of separation science. 2006;29:288–295. doi: 10.1002/jssc.200500338. [DOI] [PubMed] [Google Scholar]
- 60.Preisler J, Hu P, Rejtar T, Karger BL. Analytical chemistry. 2000;72:4785–4795. doi: 10.1021/ac0005870. [DOI] [PubMed] [Google Scholar]
- 61.Musyimi HK, Narcisse DA, Zhang X, Stryjewski W, et al. Analytical chemistry. 2004;76:5968–5973. doi: 10.1021/ac0489723. [DOI] [PubMed] [Google Scholar]
- 62.Figeys D, Ducret A, Yates JR, 3rd, Aebersold R. Nature biotechnology. 1996;14:1579–1583. doi: 10.1038/nbt1196-1579. [DOI] [PubMed] [Google Scholar]
- 63.Figeys D, Ducret A, Aebersold R. Journal of chromatography. 1997;763:295–306. doi: 10.1016/s0021-9673(96)00847-3. [DOI] [PubMed] [Google Scholar]
- 64.Tempels FW, Underberg WJ, Somsen GW, de Jong GJ. Electrophoresis. 2007;28:1319–1326. doi: 10.1002/elps.200600403. [DOI] [PubMed] [Google Scholar]
- 65.Locke S, Figeys D. Analytical chemistry. 2000;72:2684–2689. doi: 10.1021/ac0003293. [DOI] [PubMed] [Google Scholar]
- 66.Wang J, Chen R, Ma M, Li L. Analytical chemistry. 2008;80:491–500. doi: 10.1021/ac701614f. [DOI] [PubMed] [Google Scholar]
- 67.Cantin GT, Yates JR., 3rd Journal of chromatography. 2004;1053:7–14. doi: 10.1016/j.chroma.2004.06.046. [DOI] [PubMed] [Google Scholar]
- 68.Kim J, Zand R, Lubman DM. Electrophoresis. 2003;24:782–793. doi: 10.1002/elps.200390098. [DOI] [PubMed] [Google Scholar]
- 69.Zhang H, Zhang C, Lajoie GA, Yeung KK. Analytical chemistry. 2005;77:6078–6084. doi: 10.1021/ac050565j. [DOI] [PubMed] [Google Scholar]
- 70.Ballard JN, Lajoie GA, Yeung KK. Journal of chromatography. 2007;1156:101–110. doi: 10.1016/j.chroma.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 71.Zhang H, Hunter GK, Goldberg HA, Lajoie GA, Yeung KK. Analytica chimica acta. 2007;581:268–280. doi: 10.1016/j.aca.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 72.Wei J, Yang L, Harrata AK, Lee CS. Electrophoresis. 1998;19:2356–2360. doi: 10.1002/elps.1150191316. [DOI] [PubMed] [Google Scholar]
- 73.Cao P, Stults JT. Journal of chromatography. 1999;853:225–235. doi: 10.1016/s0021-9673(99)00481-1. [DOI] [PubMed] [Google Scholar]
- 74.Mikulikova K, Eckhardt A, Pataridis S, Miksik I. Journal of chromatography. 2007;1155:125–133. doi: 10.1016/j.chroma.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 75.Nguyen A, Moini M. Analytical chemistry. 2008;80:7169–7173. doi: 10.1021/ac801158q. [DOI] [PubMed] [Google Scholar]
- 76.Brenner-Weiss G, Kirschhofer F, Kuhl B, Nusser M, Obst U. Journal of chromatography. 2003;1009:147–153. doi: 10.1016/s0021-9673(03)00553-3. [DOI] [PubMed] [Google Scholar]
- 77.Martinovic S, Berger SJ, Pasa-Tolic L, Smith RD. Analytical chemistry. 2000;72:5356–5360. doi: 10.1021/ac0004557. [DOI] [PubMed] [Google Scholar]
- 78.Fermas S, Gonnet F, Varenne A, Gareil P, Daniel R. Analytical chemistry. 2007;79:4987–4993. doi: 10.1021/ac070146h. [DOI] [PubMed] [Google Scholar]
- 79.Simo C, Gonzalez R, Barbas C, Cifuentes A. Analytical chemistry. 2005;77:7709–7716. doi: 10.1021/ac051067d. [DOI] [PubMed] [Google Scholar]
- 80.Olmo S, Gotti R, Naldi M, Andrisano V, et al. Analytical and bioanalytical chemistry. 2008;390:1881–1888. doi: 10.1007/s00216-008-1903-5. [DOI] [PubMed] [Google Scholar]
- 81.Moini M, Huang H. Electrophoresis. 2004;25:1981–1987. doi: 10.1002/elps.200305906. [DOI] [PubMed] [Google Scholar]
- 82.Erny GL, Marina ML, Cifuentes A. Electrophoresis. 2007;28:4192–4201. doi: 10.1002/elps.200700323. [DOI] [PubMed] [Google Scholar]
- 83.Erny GL, Marina ML, Cifuentes A. Electrophoresis. 2007;28:2988–2997. doi: 10.1002/elps.200700132. [DOI] [PubMed] [Google Scholar]
- 84.Hu A, Tsai PJ, Ho YP. Analytical chemistry. 2005;77:1488–1495. doi: 10.1021/ac0484427. [DOI] [PubMed] [Google Scholar]
- 85.Hu A, Chen CT, Tsai PJ, Ho YP. Analytical chemistry. 2006;78:5124–5133. doi: 10.1021/ac060513+. [DOI] [PubMed] [Google Scholar]
- 86.Frommberger M, Zuerbig P, Jantos J, Krahn T, et al. Proteomics: Clin. Appl. 2007;1:650–660. doi: 10.1002/prca.200700195. [DOI] [PubMed] [Google Scholar]
- 87.Sassi AP, Andel F, 3rd, Bitter HM, Brown MP, et al. Electrophoresis. 2005;26:1500–1512. doi: 10.1002/elps.200410127. [DOI] [PubMed] [Google Scholar]
- 88.Erny GL, Cifuentes A. Electrophoresis. 2007;28:1335–1344. doi: 10.1002/elps.200600357. [DOI] [PubMed] [Google Scholar]
- 89.Javerfalk-Hoyes EM, Bondesson U, Westerlund D, Andren PE. Electrophoresis. 1999;20:1527–1532. doi: 10.1002/(SICI)1522-2683(19990601)20:7<1527::AID-ELPS1527>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 90.Rubakhin SS, Page JS, Monroe BR, Sweedler JV. Electrophoresis. 2001;22:3752–3758. doi: 10.1002/1522-2683(200109)22:17<3752::AID-ELPS3752>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 91.Lewis KC, Opiteck GJ, Jorgenson JW, Sheeley DM. J. Am. Soc. Mass Spectrom. 1997;8:495–500. [Google Scholar]
- 92.Janini GM, Chan KC, Conrads TP, Issaq HJ, Veenstra TD. Electrophoresis. 2004;25:1973–1980. doi: 10.1002/elps.200405948. [DOI] [PubMed] [Google Scholar]
- 93.Zhang J, Hu H, Gao M, Yang P, Zhang X. Electrophoresis. 2004;25:2374–2383. doi: 10.1002/elps.200405956. [DOI] [PubMed] [Google Scholar]
- 94.Chen J, Lee CS, Shen Y, Smith RD, Baehrecke EH. Electrophoresis. 2002;23:3143–3148. doi: 10.1002/1522-2683(200209)23:18<3143::AID-ELPS3143>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 95.Chen J, Balgley BM, DeVoe DL, Lee CS. Analytical chemistry. 2003;75:3145–3152. doi: 10.1021/ac034014+. [DOI] [PubMed] [Google Scholar]
- 96.Bergstrom SK, Samskog J, Markides KE. Analytical chemistry. 2003;75:5461–5467. doi: 10.1021/ac030117g. [DOI] [PubMed] [Google Scholar]
- 97.Michels DA, Hu S, Schoenherr RM, Eggertson MJ, Dovichi NJ. Mol Cell Proteomics. 2002;1:69–74. doi: 10.1074/mcp.t100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 98.Mohan D, Lee CS. Electrophoresis. 2002;23:3160–3167. doi: 10.1002/1522-2683(200209)23:18<3160::AID-ELPS3160>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 99.Mohan D, Pasa-Tolic L, Masselon CD, Tolic N, et al. Analytical chemistry. 2003;75:4432–4440. doi: 10.1021/ac0342572. [DOI] [PubMed] [Google Scholar]
- 100.Wang W, Guo T, Song T, Lee CS, Balgley BM. Proteomics. 2007;7:1178–1187. doi: 10.1002/pmic.200600722. [DOI] [PubMed] [Google Scholar]
- 101.Guo T, Rudnick PA, Wang W, Lee CS, et al. Journal of proteome research. 2006;5:1469–1478. doi: 10.1021/pr060065m. [DOI] [PubMed] [Google Scholar]
- 102.Fang X, Wang W, Yang L, Chandrasekaran K, et al. Electrophoresis. 2008;29:2215–2223. doi: 10.1002/elps.200700609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang W, Guo T, Rudnick PA, Song T, et al. Analytical chemistry. 2007;79:1002–1009. doi: 10.1021/ac061613i. [DOI] [PubMed] [Google Scholar]
- 104.Fang X, Yang L, Wang W, Song T, et al. Analytical chemistry. 2007;79:5785–5792. doi: 10.1021/ac070611a. [DOI] [PubMed] [Google Scholar]
- 105.Storms HF, van der Heijden R, Tjaden UR, van der Greef J. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;824:189–200. doi: 10.1016/j.jchromb.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 106.Balgley BM, Wang W, Song T, Fang X, et al. Electrophoresis. 2008;29:3047–3054. doi: 10.1002/elps.200800050. [DOI] [PubMed] [Google Scholar]
- 107.An Y, Cooper JW, Balgley BM, Lee CS. Electrophoresis. 2006;27:3599–3608. doi: 10.1002/elps.200600093. [DOI] [PubMed] [Google Scholar]
- 108.Siuti N, Kelleher NL. Nature methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Macek B, Waanders LF, Olsen JV, Mann M. Mol Cell Proteomics. 2006;5:949–958. doi: 10.1074/mcp.T500042-MCP200. [DOI] [PubMed] [Google Scholar]
- 110.Kelleher NL. Analytical chemistry. 2004;76:197A–203A. [PubMed] [Google Scholar]
- 111.Yang L, Lee CS, Hofstadler SA, Pasa-Tolic L, Smith RD. Analytical chemistry. 1998;70:3235–3241. doi: 10.1021/ac980224o. [DOI] [PubMed] [Google Scholar]
- 112.Wang Y, Balgley BM, Rudnick PA, Evans EL, et al. Journal of proteome research. 2005;4:36–42. doi: 10.1021/pr049876l. [DOI] [PubMed] [Google Scholar]
- 113.Schoenherr RM, Ye M, Vannatta M, Dovichi NJ. Analytical chemistry. 2007;79:2230–2238. doi: 10.1021/ac061638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yu W, Li Y, Deng C, Zhang X. Electrophoresis. 2006;27:2100–2110. doi: 10.1002/elps.200500820. [DOI] [PubMed] [Google Scholar]
- 115.Kang D, Moon MH. Analytical chemistry. 2006;78:5789–5798. doi: 10.1021/ac0606958. [DOI] [PubMed] [Google Scholar]
- 116.Pinto DM, Arriaga EA, Craig D, Angelova J, et al. Anal. Chem. 1997;69:3015–3021. doi: 10.1021/ac9611677. [DOI] [PubMed] [Google Scholar]
- 117.Hu S, Michels DA, Fazal MA, Ratisoontorn C, et al. Analytical chemistry. 2004;76:4044–4049. doi: 10.1021/ac0498314. [DOI] [PubMed] [Google Scholar]
- 118.Harwood MM, Bleecker JV, Rabinovitch PS, Dovichi NJ. Electrophoresis. 2007;28:932–937. doi: 10.1002/elps.200600500. [DOI] [PubMed] [Google Scholar]
- 119.Sobhani K, Fink SL, Cookson BT, Dovichi NJ. Electrophoresis. 2007;28:2308–2313. doi: 10.1002/elps.200700017. [DOI] [PubMed] [Google Scholar]
- 120.Fazal MA, Palmer VR, Dovichi NJ. Journal of chromatography. 2006;1130:182–189. doi: 10.1016/j.chroma.2006.05.053. [DOI] [PubMed] [Google Scholar]
- 121.Chen X, Fazal MA, Dovichi NJ. Talanta. 2007;71:1981–1985. doi: 10.1016/j.talanta.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Harwood MM, Christians ES, Fazal MA, Dovichi NJ. Journal of chromatography. 2006;1130:190–194. doi: 10.1016/j.chroma.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 123.Michels DA, Hu S, Dambrowitz KA, Eggertson MJ, et al. Electrophoresis. 2004;25:3098–3105. doi: 10.1002/elps.200405939. [DOI] [PubMed] [Google Scholar]
- 124.Zhu C, He X, Kraly JR, Jones MR, et al. Analytical chemistry. 2007;79:765–768. doi: 10.1021/ac061652u. [DOI] [PubMed] [Google Scholar]
- 125.Zhang M, El Rassi Z. Journal of proteome research. 2006;5:2001–2008. doi: 10.1021/pr060185u. [DOI] [PubMed] [Google Scholar]
- 126.Mao Y, Zhang X. Electrophoresis. 2003;24:3289–3295. doi: 10.1002/elps.200305578. [DOI] [PubMed] [Google Scholar]
- 127.Mao Y, Li Y, Zhang X. Proteomics. 2006;6:420–426. doi: 10.1002/pmic.200500220. [DOI] [PubMed] [Google Scholar]
- 128.Freire SL, Wheeler AR. Lab on a chip. 2006;6:1415–1423. doi: 10.1039/b609871a. [DOI] [PubMed] [Google Scholar]
- 129.Kan CW, Fredlake CP, Doherty EA, Barron AE. Electrophoresis. 2004;25:3564–3588. doi: 10.1002/elps.200406161. [DOI] [PubMed] [Google Scholar]
- 130.Lion N, Rohner TC, Dayon L, Arnaud IL, et al. Electrophoresis. 2003;24:3533–3562. doi: 10.1002/elps.200305629. [DOI] [PubMed] [Google Scholar]
- 131.Peng Y, Pallandre A, Tran NT, Taverna M. Electrophoresis. 2008;29:157–178. doi: 10.1002/elps.200700347. [DOI] [PubMed] [Google Scholar]
- 132.Rocklin RD, Ramsey RS, Ramsey JM. Analytical chemistry. 2000;72:5244–5249. doi: 10.1021/ac000578r. [DOI] [PubMed] [Google Scholar]
- 133.Gottschlich N, Jacobson SC, Culbertson CT, Ramsey JM. Analytical chemistry. 2001;73:2669–2674. doi: 10.1021/ac001019n. [DOI] [PubMed] [Google Scholar]
- 134.Wang YC, Choi MH, Han J. Analytical chemistry. 2004;76:4426–4431. doi: 10.1021/ac0497499. [DOI] [PubMed] [Google Scholar]
- 135.Dodge A, Brunet E, Chen S, Goulpeau J, et al. Analyst. 2006;131:1122–1128. doi: 10.1039/b606394b. [DOI] [PubMed] [Google Scholar]
- 136.Chen X, Wu H, Mao C, Whitesides GM. Analytical chemistry. 2002;74:1772–1778. doi: 10.1021/ac0109422. [DOI] [PubMed] [Google Scholar]
- 137.Herr AE, Molho JI, Drouvalakis KA, Mikkelsen JC, et al. Analytical chemistry. 2003;75:1180–1187. doi: 10.1021/ac026239a. [DOI] [PubMed] [Google Scholar]
- 138.Shadpour H, Soper SA. Analytical chemistry. 2006;78:3519–3527. doi: 10.1021/ac0600398. [DOI] [PubMed] [Google Scholar]
- 139.Griebel A, Rund S, Schonfeld F, Dorner W, et al. Lab on a chip. 2004;4:18–23. doi: 10.1039/b311032j. [DOI] [PubMed] [Google Scholar]
- 140.Liu J, Yang S, Lee CS, DeVoe DL. Electrophoresis. 2008;29:2241–2250. doi: 10.1002/elps.200700608. [DOI] [PubMed] [Google Scholar]
- 141.Li Y, Buch JS, Rosenberger F, DeVoe DL, Lee CS. Analytical chemistry. 2004;76:742–748. doi: 10.1021/ac034765b. [DOI] [PubMed] [Google Scholar]
- 142.Becker H, Lowack K, Manz A. J. Micromech. Microeng. 1998;8:24–28. [Google Scholar]
- 143.Das C, Zhang J, Denslow ND, Fan ZH. Lab on a chip. 2007;7:1806–1812. doi: 10.1039/b712794d. [DOI] [PubMed] [Google Scholar]
- 144.Ramsey RS, Ramsey JM. Anal. Chem. 1997;69:1174–1178. doi: 10.1021/ac971475k. [DOI] [PubMed] [Google Scholar]
- 145.Xue Q, Foret F, Dunayevskiy YM, Zavracky PM, et al. Analytical chemistry. 1997;69:426–430. doi: 10.1021/ac9607119. [DOI] [PubMed] [Google Scholar]
- 146.Koster S, Verpoorte E. Lab on a chip. 2007;7:1394–1412. doi: 10.1039/b709706a. [DOI] [PubMed] [Google Scholar]
- 147.Akashi S, Suzuki K, Arai A, Yamada N, et al. Rapid Commun Mass Spectrom. 2006;20:1932–1938. doi: 10.1002/rcm.2541. [DOI] [PubMed] [Google Scholar]
- 148.Hoffmann P, Hausig U, Schulze P, Belder D. Angewandte Chemie (International edition) 2007;46:4913–4916. doi: 10.1002/anie.200605152. [DOI] [PubMed] [Google Scholar]
- 149.Sikanen T, Tuomikoski S, Ketola RA, Kostiainen R, et al. Analytical chemistry. 2007;79:9135–9144. doi: 10.1021/ac071531+. [DOI] [PubMed] [Google Scholar]
- 150.Mellors JS, Gorbounov V, Ramsey RS, Ramsey JM. Analytical chemistry. 2008;80:6881–6887. doi: 10.1021/ac800428w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang Y, Rudnick PA, Evans EL, Li J, et al. Analytical chemistry. 2005;77:6549–6556. doi: 10.1021/ac050491b. [DOI] [PubMed] [Google Scholar]
- 152.Li J, Yin C, Okamoto H, Mushlin H, et al. Neuro-oncology. 2008;10:45–51. doi: 10.1215/15228517-2007-036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Xu H, Yang L, Wang W, Shi SR, et al. Journal of proteome research. 2008;7:1098–1108. doi: 10.1021/pr7006768. [DOI] [PubMed] [Google Scholar]
- 154.Kraly JR, Jones MR, Gomez DG, Dickerson JA, et al. Analytical chemistry. 2006;78:5977–5986. doi: 10.1021/ac061029+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kaiser T, Kamal H, Rank A, Kolb HJ, et al. Blood. 2004;104:340–349. doi: 10.1182/blood-2004-02-0518. [DOI] [PubMed] [Google Scholar]
- 156.Weissinger EM, Schiffer E, Hertenstein B, Ferrara JL, et al. Blood. 2007;109:5511–5519. doi: 10.1182/blood-2007-01-069757. [DOI] [PubMed] [Google Scholar]
- 157.Neuhoff N, Kaiser T, Wittke S, Krebs R, et al. Rapid Commun Mass Spectrom. 2004;18:149–156. doi: 10.1002/rcm.1294. [DOI] [PubMed] [Google Scholar]
- 158.Weissinger EM, Wittke S, Kaiser T, Haller H, et al. Kidney international. 2004;65:2426–2434. doi: 10.1111/j.1523-1755.2004.00659.x. [DOI] [PubMed] [Google Scholar]
- 159.von Zur Muhlen C, Schiffer E, Zuerbig P, Kellmann M, et al. Journal of proteome research. 2008 doi: 10.1021/pr800615t. [DOI] [PubMed] [Google Scholar]
- 160.Zimmerli LU, Schiffer E, Zurbig P, Good DM, et al. Mol Cell Proteomics. 2008;7:290–298. doi: 10.1074/mcp.M700394-MCP200. [DOI] [PubMed] [Google Scholar]
- 161.Dong M, Yang LL, Williams K, Fisher SJ, et al. Journal of proteome research. 2008;7:1836–1849. doi: 10.1021/pr700624e. [DOI] [PubMed] [Google Scholar]
- 162.Schou C, Heegaard NH. Electrophoresis. 2006;27:44–59. doi: 10.1002/elps.200500516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Goheen SC, Gibbins BM. Journal of chromatography. 2000;890:73–80. doi: 10.1016/s0021-9673(00)00572-0. [DOI] [PubMed] [Google Scholar]
- 164.Greibrokk T, Pepaj M, Lundanes E, Andersen T, Novotna K. LC-GC Eur. 2005;18:355–356. 358–360. [Google Scholar]
- 165.Doherty EA, Meagher RJ, Albarghouthi MN, Barron AE. Electrophoresis. 2003;24:34–54. doi: 10.1002/elps.200390029. [DOI] [PubMed] [Google Scholar]
- 166.Horvath J, Dolnik V. Electrophoresis. 2001;22:644–655. doi: 10.1002/1522-2683(200102)22:4<644::AID-ELPS644>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 167.Eschelbach JW, Jorgenson JW. Analytical chemistry. 2006;78:1697–1706. doi: 10.1021/ac0518304. [DOI] [PubMed] [Google Scholar]
- 168.Everley RA, Croley TR. Journal of chromatography. 2008;1192:239–247. doi: 10.1016/j.chroma.2008.03.058. [DOI] [PubMed] [Google Scholar]