Abstract
Sirt1, an NAD-dependent protein deacetylase has emerged as important regulator of mammalian transcription in response to cellular metabolic status and stress1. Here we demonstrate that Sirt1 plays a neuroprotective role in models of Huntington’s disease (HD), an inherited neurodegenerative disorder caused by a glutamine repeat expansion in huntingtin protein2. Brain-specific knockout of Sirt1 results in exacerbation of brain pathology in HD mice, whereas overexpression of Sirt1 improves survival, neuropathology and BDNF expression in HD mice. We show that Sirt1 deacetylase activity directly targets neurons to mediate neuroprotection from mutant huntingtin. The neuroprotective effect of Sirt1 requires the presence of TORC1, a brain-specific modulator of CREB activity3. We show that under normal conditions Sirt1 deacetylates and activates TORC1 by promoting its dephoshorylation and interaction with CREB. We identified BDNF as an important target of Sirt1 and TORC1 transcriptional activity in normal and HD neurons. Mutant huntingtin interferes with the TORC1-CREB interaction to repress BDNF transcription and Sirt1 rescues this defect in vitro and in vivo. These studies suggest a key role of Sirt1 in transcriptional networks in normal and HD brain and offer an opportunity for therapeutic development.
Recent evidence suggests that Sirt1, a member of sirtuin family that has been implicated in aging and metabolism, plays a protective role in neurodegenerative disorders1. However, the exact nature of normal Sirt1 function in mammalian brain has yet to be ascertained. To clarify the function of Sirt1 in brain and determine its contribution to neurodegeneration, we focused on Huntington’s disease (HD), an autosomal dominant disease that commonly presents in adult life with personality changes, cognitive changes, psychiatric disturbances and abnormal movements2. HD belongs to a family of polyglutamine disorders that, in addition to expanded polyglutamine repeats, share other features such as adult onset, progressive neurodegeneration, inverse correlation of numbers of repeats with age of onset and presence of polyglutamine protein-containing inclusions 2. Polyglutamine diseases, including HD remain fatal as there are no effective treatments to cure the diseases or to slow their progression.
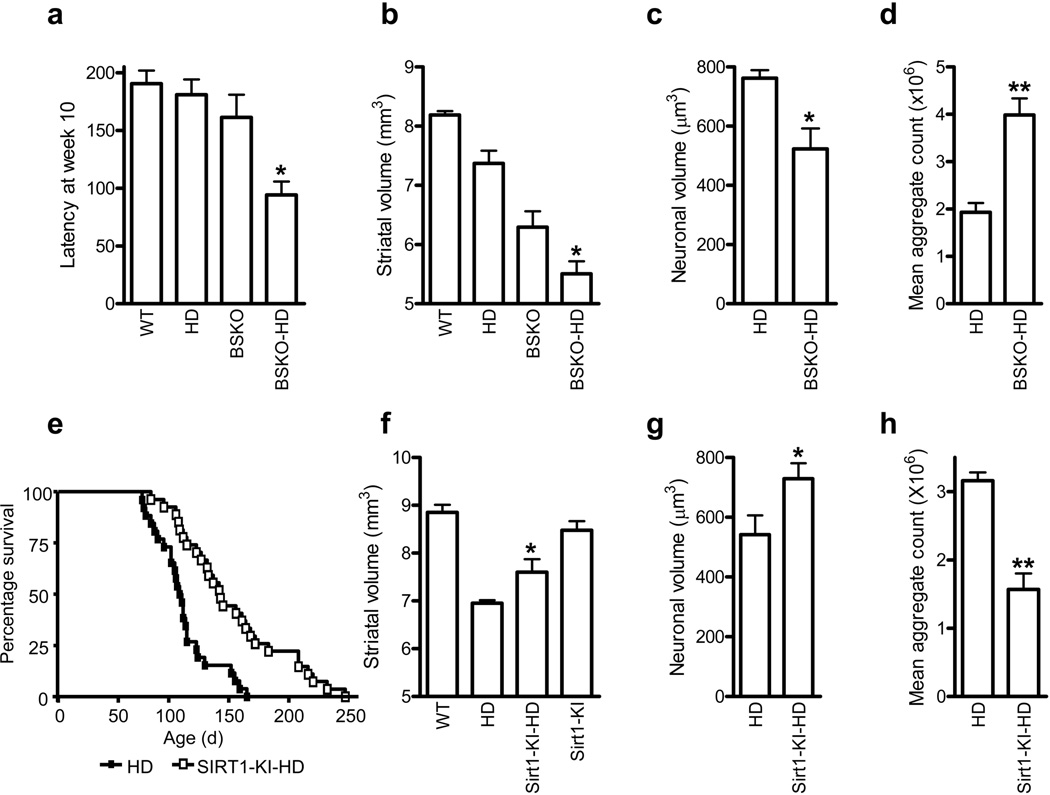
The role of Sirt1 in HD has been primarily studied in Drosophila melanogaster and in C. elegans, but the results have been contradictory. For example, in worms overexpressing Sir2 (ortholog of human Sirt1), HTT-induced neurodegeneration is suppressed, whereas overexpression in fly model of HD confers no significant protection 4,5. To begin testing the role of Sirt1 in HD mice, we first crossed a conditional allele of Sirt1 (Sirt1flox) to the Nestin-cre driver to yield Sirt1 brain-specific knockout mice (BSKO, genotype Sirt1flox/flox; nestin-cre). These mice were born at mendelian ratios and displayed no gross defects in brain development 6. To determine the impact of the ablation of neuronal Sirt1 on HD, we then crossed BSKO mice to the R6/2 model of HD 7. These crosses, which were carried out on an inbred C57BL/6 background, yielded Sirt1flox/flox controls (referred to as WT), Sirt1flox/flox; R6/2 (R6/2), Sirt1flox/flox; nestin-cre (BSKO), and Sirt1flox/flox; nestin-cre-R6/2 (BSKO-R6/2). Behavioral experiments revealed significant acceleration of motor deficits in BSKO-R6/2 compared to R6/2 mice as assessed by rotarod performance (Figs. 1a and Supplementary Fig. 1a). To determine if neuropathological alterations accompanied the behavioral phenotype, an unbiased stereological analysis of brain sections was performed. Previous studies have demonstrated that R6/2 mice exhibit progressive atrophy of striatal neurons that resembles neuropathology observed in human HD 8. As expected, R6/2 mice showed significant striatal atrophy compared to WT mice (Fig. 1b). BSKO mice, which are smaller than WT mice 6, also had smaller striata than the WT mice. BSKO-R6/2 mice showed significantly smaller striatal volume compared to BSKO or R6/2 mice (Fig. 1c). Moreover, when analyzing a series of sections spanning the striatum, a significant decrease in the mean neuronal volume was observed in striata of the BSKO-R6/2 mice compared to R6/2 mice (Fig. 1c). To examine aggregation of mutant HTT, mean aggregate count was examined in all four genotypes. While no HTT aggregates were observed in WT or BSKO mice, the number of aggregates was significantly increased in BSKO-R6/2 animals as compared to R6/2 mice (Fig. 1d). These results demonstrate that deficiency of Sirt1 accelerates neurodegeneration in HD mice.
Figure 1. Ablation of neuronal Sirt1 exacerbates and over-expression of Sirt1 ameliorates phenotypes in R6/2 mouse.
(a) Latency to fall from accelerating rotarod at 10 weeks of age. n = 4 - 10 per group. *P < 0.05 for HD vs. BSKO-HD by ANOVA. (b) Striatal volumes at 20 weeks of age. n = 4 per group. *P < 0.01 by ANOVA. (c) Striatal neuronal volumes at 20 weeks of age. n = 4 per group. * P < 0.05 by t-test. (d) Aggregates of HTT in striata at 20 weeks of age. ** P < 0.01 (n=6 per group) (e) Survival of male HD (closed square) and Sirt1-KI-HD (open square) mice. n = 26 - 27 per group. P < 0.001 by log-rank test. (f) Striatal volume at 100 d of age. n = 5 – 6 per group. * P < 0.05 by ANOVA. (g) Striatal neuronal volumes at 100 d of age. n = 5 - 6 per group. *P < 0.05 by t-test. (h) Aggregates of HTT in striata at 100 d of age. ** P < 0.01 (n=7 per group).
To test whether increased expression of Sirt1 might provide protection in the R6/2 model of HD, we took advantage of a transgenic mouse that over-expresses Sirt1 under the control of the endogenous β-actin promoter (Sirt1-KI) 9. These mice, which are maintained on an outbred genetic background, over-express Sirt1 in a variety of tissues, including the brain where Sirt1 is over-expressed approximately two-fold in both cortex and striatum, but the over-expression was less pronounced in female Sirt1-KI mice (Supplementary Fig. 1b,c). We crossed Sirt1 over-expressing mice (genotype Sirt1- KI/+) to R6/2 mice to generate wild-type (WT), Sirt1-KI, unmodified R6/2 and Sirt1 over-expressing R6/2 (Sirt1-KI-R6/2). These four groups were born at the expected mendelian ratios. Analysis of body weight loss and rotarod performance revealed that Sirt1 over-expression was unable to provide significant protection against these two gross phenotypes in the R6/2 mouse (Supplementary Fig. 1d,e). However, we found that Sirt1 over-expression significantly extended the survival of R6/2 animals, from an average of 100 d to an average of 130 d (Fig. 1e). Interestingly, this effect was observed only in male mice, whereas female mice showed no differences in survival (Supplementary Fig. 1f). Although the precise reasons for this gender-specific effect remain unclear, it is possible that lower levels of Sirt1 overexpression in female Sirt1-KI mice played a role (Supplementary Fig. 1c). Intriguingly, Sirt1 over-expression did not extend the lifespan of otherwise normal mice (Supplementary Fig. 2), indicating that the lifespan extension provided by this strain is specific to the pathology of HD. Neuropathological analysis revealed significant striatal and neuronal atrophy in R6/2 mice as compared to either WT or SIRT1-KI animals (Fig. 1f,g). However, the degree of both striatal and neuronal atrophy was significantly reduced in SIRT1-KI-R6/2 mice as compared to R6/2, indicating that over-expression of Sirt1 protected against striatal degeneration in this model (Fig. 1f,g). Further, an analysis of protein aggregate formation revealed that Sirt1-KI-R6/2 animals had a significantly reduced aggregate burden as compared to R6/2 animals (Figs. 1h and Supplementary Fig. 3). These results indicate that Sirt1 over-expression can provide significant protection against key neuropathological phenotypes in the HD mouse model.
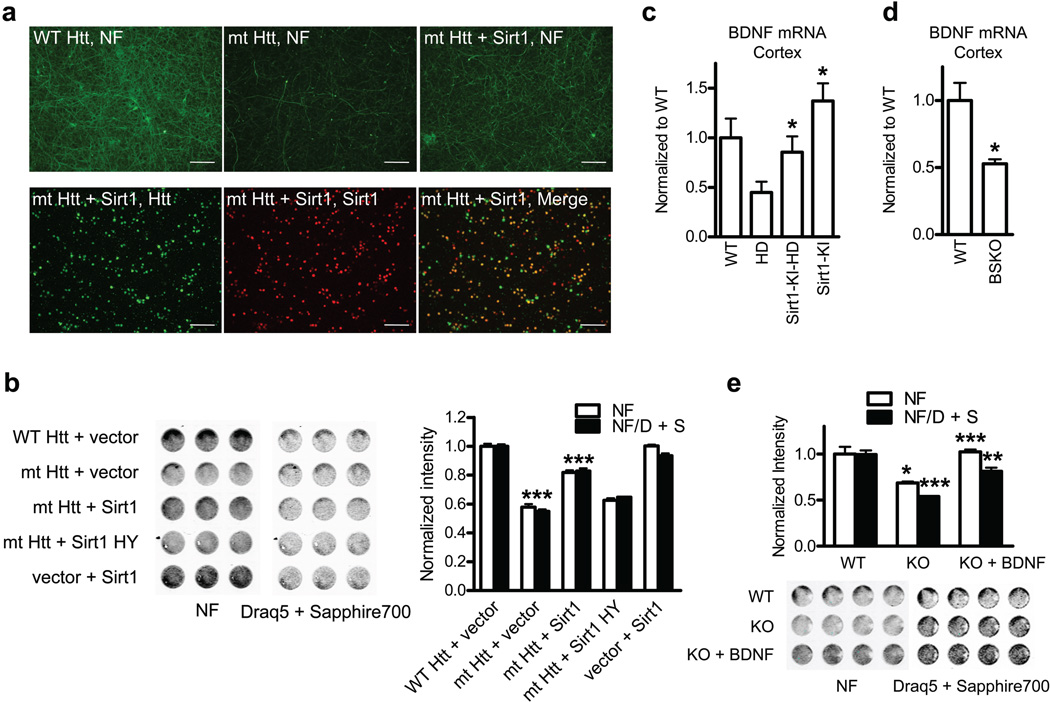
Having shown that overexpression of Sirt1 protects from mutant HTT toxicity in vivo, we asked whether this protection was due to direct effects of Sirt1 on neurons. Using lentiviral expression in primary neurons we achieved high transduction efficiency (> 90%) and predominantly neuronal expression of the transduced constructs (Supplementary Fig. 4a). Neurofilament staining (NF) was utilized to detect degeneration of neurites as an early marker of neuronal toxicity10. These experiments revealed progressive loss of NF staining that correlated with the duration of exposure of primary neurons to mutant HTT (Supplementary Fig. 4b). Lentiviral expression of mutant HTT with 72Qs resulted in significant loss of neurites compared to wild-type HTT with non-expanded polyQ tract (Fig. 2a,b). Expression of lenti-Sirt1 significantly rescued neuronal toxicity of mutant HTT despite the fact that only about 70% of neurons coexpressed both Sirt and mutant HTT (Fig. 2a,b). This protection is mediated by deacetylase activity of Sirt1 since deacetylase-deficient Sirt1 H363Y (Sirt1 HY) did not exhibit significant neuroprotection compared with wild-type Sirt1 (Fig. 2b). Using nuclear staining as readout, we found that Sirt1 also protected from mutant HTT-induced nuclear toxicity that invariably followed loss of NF staining (Supplementary Fig. 4b,c). These experiments demonstrate that Sirt1 directly protects neurons from mutant HTT toxicity and that Sirt1 deacetylase activity plays a key role in the neuroprotection.
Figure 2. Deaceylase activity of Sirt1 protects cortical neurons from mutant HTT Toxicity.
(a) Upper panels: neurofilament (NF) staining in primary cortical neurons infected with indicated lentivirus. Lower panels: double staining of mt HTT- and Sirt1-infected neurons. Scale bar, 300 µm. (b) In-cell western (ICW) for NF and Draq5 + Sapphire 700 staining of cortical neurons infected with indicated lentivirus. *** P < 0.001 for mt HTT vs. WT HTT, *** P < 0.001 for mt HTT + Sirt1 vs. mt HTT. (c) BNDF mRNA levels in mouse cortex at 100 d of age. *P < 0.05 for HD vs Sirt1-KI-HD by ANOVA. * P < 0.05 for Sirt1-KI vs. WT by ANOVA (n=4 per group). (d) BDNF mRNA levels in mouse cortex at 3 months of age. n = 5 per group. *P < 0.05 by t-test. (e) Toxicity in primary cortical neurons from Sirt1 KO mice or WT littermates. * P < 0.05 for white bars, KO vs WT; *** P < 0.001 for black bars, KO vs WT; *** P < 0.001 for white bars, KO + BDNF vs KO; ** P < 0.01 for black bars, KO + BDNF vs KO by t-test.
To examine the mechanism of Sirt1-mediated neuroprotection in HD mice, we first performed gene expression profiling in striatal samples obtained from HD mice and HD mice crossed with Sirt1 overexpressing mice. Gene Set Enrichment Analysis11 revealed that overexpresion of Sirt1 in HD mice rescued specific pathways involved in neurite development and branching (Supplementary Fig. 5). In addition, BDNF, an important regulator of these pathways12,13, was significantly decreased in HD mice as previously described 14, whereas BDNF levels were significantly higher in HD-Sirt1-KI mice compared to HD mice (Fig. 2c). By contrast, cortical brain samples from Sirt1 BSKO mice showed significantly decreased levels of BDNF mRNA compared to control littermates (Fig. 2d), further suggesting that Sirt1 might regulate BDNF expression. Importantly, experiments in primary cortical neurons isolated from the Sirt1 KO mice revealed that treatment with exogenous BDNF rescued neurotoxicity observed in the KO cultures (Fig. 2e). Taken together, these results highlight BDNF as an important target of Sirt1 function in normal and HD brain and suggest that Sirt1 regulates expression of BDNF at the level of transcription.
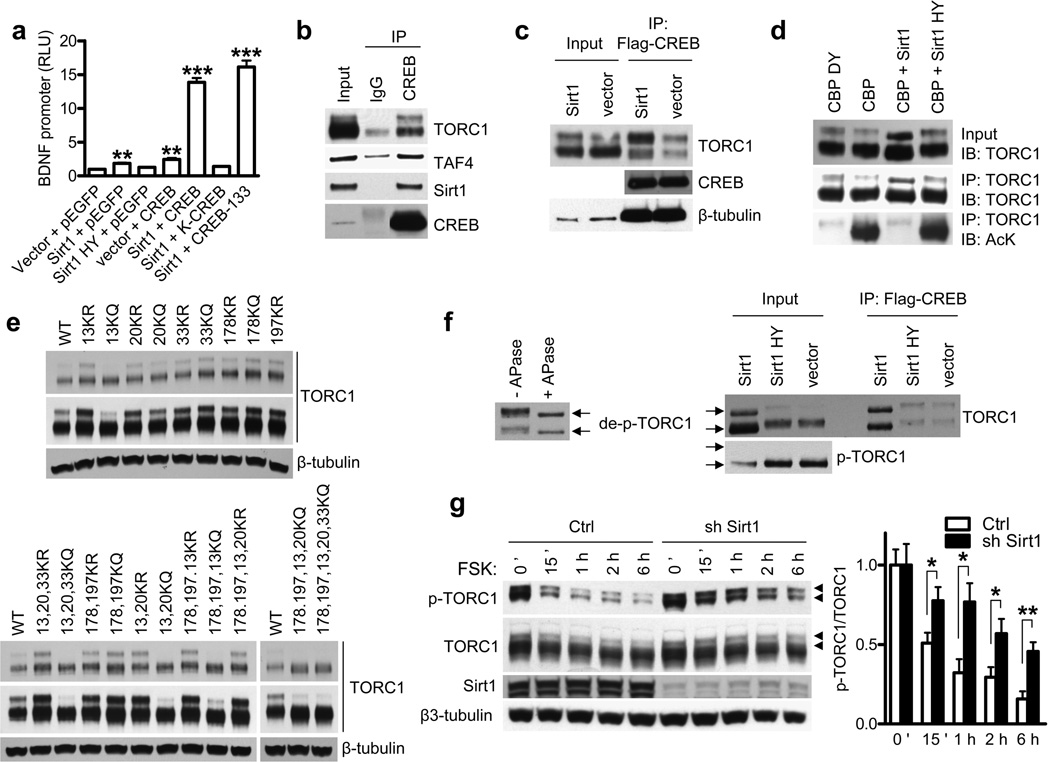
Previous studies in primary neurons demonstrated that CREB-mediated transcription from BDNF promoter IV (previous nomenclature III) accounts for most BDNF gene expression under conditions of neuronal activity 15. Co-transfection experiments revealed significant activation of the BDNF promoter IV by Sirt1 whereas deacetylase-deficient Sirt1 did not have an effect (Fig. 3a). While CREB alone also activated the promoter by about 2 fold, transfection of Sirt1 and CREB together resulted in synergistic 15-fold activation. Expression of DNA binding-incompetent form of CREB (K-CREB) did not lead to synergistic activation with Sirt1, suggesting that DNA binding of CREB is required for the synergism (Fig. 3a). Moreover, a mutant form of CREB that cannot get phosphorylated at Ser-133 still synergized with Sirt1, indicating that phosphorylation of CREB was not required for the synergism. A conserved family of coactivators designated Transducers of Regulated CREB activity (TORCs) were identified as enhancers of CREB transcription via phosphorylation-independent interaction with CREB 3. We found that activation of BDNF promoter by CREB was enhanced by co-transfection of TORC1 in a dose-dependent manner (Supplementary Fig. 6a). This effect of TORC1 was dramatically potentiated by Sirt1 (Supplementary Fig. 6a) suggesting that Sirt1 activates BDNF promoter by enhancing CREB-TORC1 transcriptional activity.
Figure 3. Sirt1 deacetylates and activates TORC1.
(a) BDNF promoter IV activity in N2a cells cotransfected with indicated plasmids at 24 h posttransfection. ** P < 0.01 vs. vector + pEGFP, ***P < 0.001 vs. vector + pEGFP by t-test. (b, c) Coimmunoprecipitations in N2a cells with indicated plasmids. (d) TORC1 immunoprecipitation and acetylation in N2a cells assessed by antibody to acetyl-lysine (AcK) (bottom panel). Western blots of TORC1 in input samples (top panel) and IP samples (middle panel). (e) Western blot of TORC1 KR or KQ mutants in N2a cells transfected with indicated TORC1 mutants. Long and short exposures are shown. (f) Left: western blot of TORC1 with or without in vitro treatment of alkaline phosphatase (APase). Right: Western blot of TORC1 and p-TORC1 and coimmunoprecipitation assay of CREB and TORC1 in N2a cells transfected with indicated plasmids. TORC1 position marked by arrows. (g) Dephosphorylation of p-TORC1 by forskolin (FSK) in the presence of Sirt1 knockdown in primary cortical neurons. Phospho-TORC1 marked with arrow heads. * P < 0.05 by t-test.
To determine how Sirt1 potentiates CREB-TORC1 activity, we next tested whether Sirt1 interacts with these factors. Previous studies suggested that TORC1 activated CREB-regulated transcription by promoting recruitment of transcriptional coactivator TAF4 3. Here we demonstrate that Sirt1 interacts with CREB, TORC1 and TAF4 (Fig. 3b and Supplementary Fig. 6b) and that expression of Sirt1 potentiated the interaction of CREB with TORC1 (Fig. 3c). Importantly, we found that Sirt1 increased the proportion of slower migrating TORC1 (upper band in Fig. 3c), which is the form of TORC1 that predominantly interacts with CREB.
Since Sirt1 is a deacetylase, we next examined whether it potentiates TORC1-CREB interaction by deacetylating TORC1. Using antibody to pan-acetyl lysine, strong acetylation of TORC1 was detected by CBP but not by catalytically inactive CBP-DY16 (Fig. 3d). Importantly, Sirt1, but not the deacetylase-deficient Sirt1 HY was able to deacetylate TORC1 (Fig. 3d, lower panel). Using mass spec analysis we identified acetylation of several lysine residues in the TORC1 protein (Supplementary Fig. 7), including lysines 13, 20, 33, 178 and 197. To study the functional consequences of lysine modifications, these residues were mutated, either individually or in various combinations, into arginine (KR) to mimic deacetylation, or glutamine (KQ) to mimic acetylation. While no obvious effect was seen with mutations of lysines 33, 178 and 197, the mutations of lysines 13 or 20 into KR and KQ stabilized and destabilized TORC1, respectively (Fig. 3e). The effect of lysines 13 and 20 was most notable on the slower-migrating form of TORC1 that interacts with CREB, suggesting that the acetylation or deacetylation status of Lys13 and to a lesser degree Lys20 plays a key role in interactions of TORC1with CREB. Since lysines 13 and 20 reside in the TORC1 tetramerization and CREB-binding domain3, we hypothesized that these lysines mediate the effect of Sirt1 on TORC1-CREB interaction and activation of BDNF promoter. While wild-type or KR mutants of TORC1 potentiated CREB-mediated BDNF reporter activity, the KQ mutations at lysines 13 or 20 exhibited significantly reduced potentiation (Supplementary Fig. 8). These results further support the notion that deacetylation of TORC1 by Sirt1 plays a role in activation of BDNF transcription.
As we evaluated TORC1 acetylation, it was noted that Sirt1 gave rise to an increase in TORC1 electrophoretic mobility, the effect that was most pronounced when TORC1 was acetylated by CBP (Fig. 3d,f, upper right panel, input). Treatment of TORC1 by alkaline phosphatase revealed that the faster electrophoretic mobility forms of TORC1 generated by Sirt1 expression represented dephosphorylated TORC1 (Fig. 3f, left panel). Previous studies showed that cytoplasmic TORC1 is phosphorylated at serine 151 and inactive, whereas its dephosphorylation leads to its nuclear translocation and activation 17,18. Using primary neurons we confirmed that the faster-migrating dephospho TORC1 represents the nuclear form of TORC1 (Supplementary Fig. 9a). Using specific antibody to phospho Ser151 (Supplementary Fig. 9b), we found that levels of phosphorylated TORC1 were greatly diminished in the presence of wild-type Sirt1, but not deacetylase-deficient Sirt1, further suggesting that Sirt1 promotes the formation of dephosphorylated, active nuclear TORC1 (Fig. 3f, lower right panel). In agreement with this conclusion, we found that knockdown of endogenous Sirt1 led to increased phosphorylation of TORC1 in primary neurons treated with forskolin19 (Fig. 3g). Interestingly, we did not observe any changes in levels of total or phospho CREB in the presence of Sirt1 knockdown (Supplementary Fig. 10), suggesting that Sirt1 does not regulate CREB directly but rather via TORC1.
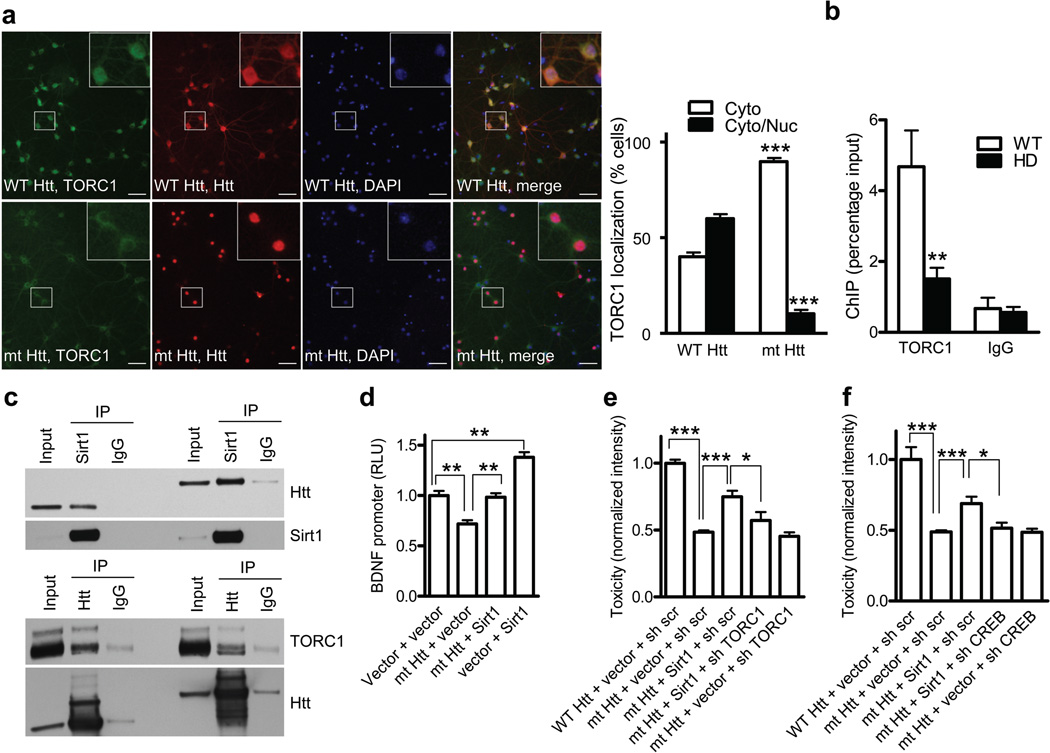
Together our data so far suggest that as part of its physiological function Sirt1 deacetylates and activates neuronal TORC1. To determine if mutant HTT interferes with this normal Sirt1 function, we first examined the localization of TORC1 in primary neurons expressing mutant or wild-type HTT. Interestingly, expression of mutant HTT resulted in predominantly cytoplasmic localization of TORC1, suggesting that TORC1 primarily exists in its inactive, phosphorylated form in the presence of mutant HTT (Fig. 4a). To substantiate this conclusion in vivo, chromatin immunoprecipitations (ChIP) revealed significantly decreased recruitment of TORC1 to BDNF promoter IV in the cortex of R6/2 mice (Fig. 4b), further suggesting that mutant HTT interferes with TORC1 nuclear localization and transcriptional activity. Moreover, we found that mutant HTT interacts with Sirt1, TORC1 (Figs. 4c and Supplementary Fig. 11b,c,e,f), TAF4 (Supplementary Fig. 11a,d)20 and it interferes with TORC1-CREB interaction (Supplementary Fig. 12a,b). Consistent with Fig. 4a, increased phosphorylation of TORC1 was noted in the presence of mutant HTT (Supplementary Fig. 12a), suggesting that mutant HTT, possibly through its interaction with Sirt1, interferes with the ability of Sirt1 to activate TORC1. Since TORC1 promotes interaction of CREB with TAF4 3, and the fact that TAF4 has been previously implicated in CREB transcription in HD20, it is also possible that interference of mutant HTT with TAF4 function contributes to transcriptional repression of BDNF gene in HD.
Figure 4. Sirt1 rescues mutant HTT-mediated interference with TORC1 activity.
(a) Immunostaining of HTT, TORC1 and DAPI in primary neurons infected with lentiviral WT HTT (upper panels) or mt HTT (lower panels). *** P < 0.001 by t-test. Scale bar, 50 µm. (b) Chromatin immunoprecipitation (ChIP) of TORC1 on BDNF in cortex of WT or HD mice. **P < 0.01 vs WT, n = 6. (c) Coimmunoprecipitations assay of indicated plasmids in N2a cells. (d) BDNF promoter IV activities in N2a cells cotransfected with indicated plasmids at 48 hooo posttransfection. ** P < 0.01. (e) NF staining intensity in primary cortical neurons infected with indicated lentivirus. * P < 0.05 by t-test. *** P < 0.001 by t-test. (f) Toxicity was assessed as in (e). * P < 0.05, *** P < 0.001 by t-test.
Since Sirt1 protected from mutant HTT toxicity and promoted the interaction of TORC1 with CREB, we examined whether expression of Sirt1 could overcome the interference of mutant HTT. As shown in Supplementary Fig. 12b, expression of Sirt1 strongly enhanced TORC1-CREB interaction in the presence of mutant HTT. Based on this finding and our in vivo data showing rescue of BDNF expression by Sirt1 (Fig. 2c), we examined whether mutant HTT and Sirt1 directly affect the BDNF promoter. Previous studies demonstrated that wild-type HTT upregulates transcription of the BDNF gene by sequestrating the neuron-restrictive silencing factor (NRSF) in the cytoplasm, suggesting that wild-type HTT function plays an important protective role in the pathogenesis of HD 14,21. While those studies examined BDNF promoter II that contains NRSF binding site, our study examined promoter IV that is regulated by the CREB-mediated mechanism15. We found that mutant HTT, but not the WT HTT interfered with Sirt1-mediated activation of BDNF promoter (Supplementary Fig. 12c). Importantly, expression of Sirt1 resulted in significant reversal of mutant HTT-mediated repression (Fig. 4d), further suggesting that Sirt1 can rescue the effect of mutant HTT on BDNF transcription.
In light of our combined results suggesting that TORC1 plays an important role in Sirt1-mediated protection from mutant HTT toxicity, we directly examined if Sirt1 protection depends on the presence of TORC1. To this end, primary cortical neurons were transduced with lentivirally-expressed mutant HTT and Sirt1 in the presence or absence of TORC1 knockdown. Using lentiviral shRNA approach, about 70% knockdown of TORC1 was achieved that was not neurotoxic by itself (Supplementary Fig. 13a,b). Importantly, Sirt1 rescue of mutant HTT toxicity was significantly compromised in the presence of TORC1 knockdown (Fig. 4e), suggesting that expression of TORC1 is at least in part required for Sirt1-mediated neuroprotection. Finally, we hypothesized that if Sirt1 primarily protects from mutant HTT toxicity by activating TORC1 and consequently CREB, the knockdown of CREB would also be expected to diminish Sirt1 rescue. Using shCREB lentivirus to achieve non-toxic knockdown of CREB at baseline, we found that Sirt1-mediated rescue of mutant HTT toxicity was completely dependent on the presence of endogenous CREB (Fig. 4f and Supplementary Fig. 13c). These results strengthen the conclusion that TORC1-CREB pathway plays a critical role in mediating Sirt1 neuroprotection in neurons expressing mutant HTT.
In sum, we demonstrate that Sirt1 plays an important neuroprotective role in the pathogenesis of HD. Using primary neurons, we confirmed that Sirt1 directly targets neurons to mediate neuroprotection from mutant HTT. Our data demonstrate that under normal conditions Sirt1 promotes deacetylation of TORC1 and its interaction with CREB which leads to activation of target genes such as BDNF. Mutant HTT interacts with Sirt1 and interferes with Sirt1-mediated activation of TORC1 that in turn results in transcriptional repression. Importantly, overexpression of Sirt1 rescued this effect of mutant HTT in vitro and in vivo.
Our finding that Sirt1 affords neuroprotection in R6/2 mice is in agreement with the accompanying manuscript where Sirt1-mediated neuroprotection was observed in another fragment model (171aa) and in full-length HD mouse model (Jiang et al.). Interestingly, Sirt1 overexpression did not affect survival or aggregation in 171aa mice despite the observed neuroprotection. Although the reasons for these discrepancies remain unknown, it is important to note that 171aa mice develop hyperinsulinemia and insulin resistance that was not seen in our model. Several published reports implicate insulin signaling in the regulation of aggregation and proteotoxicity, but its role in neurodegeneration remains controversial22. It is therefore possible that hyperinsulinemia in 171aa mice contributes to differences in survival and aggregation upon Sirt1 rescue. Although it is not precisely known why HD-like mice die, recent work23 showed that HTT fragment models (R6/2 and 171aa) develop profound hypothermia that presumably contributes to death in these animals. Since hypothermia does not seem to play a similar role in HD patients, it can be therefore argued that survival as outcome measure in HD-like mice may not directly inform about human disease. However, it is generally accepted that striatal and neuronal atrophy observed in HD-like mice strongly resemble human disease, and on this key measure, our studies are in agreement.
We identified TORC1 as a novel target of Sirt1 deacetylase, whereas Jiang et al. showed that previously known substrates of Sirt1 such as FoxO3a and p53 were also altered in HD models. Since mutant HTT partially inhibits Sirt1 deacetylase activity (Jiang et al.), it is not surprising that multiple substrates of Sirt1 exhibited increased acetylation in HD models. Importantly, overexpression of Sirt1 was able to overcome this inhibition and partially correct the acetylation status of Sirt1 substrates. In light of our data that TORC1 is required for Sirt1-mediated rescue, it will be of interest to further examine possible links between TORC1 and other Sirt1 substrates in HD. As a regulator of neuronal activity-dependent CREB transcription, TORC1 presumably regulates other CRE-regulated genes that have been implicated in the pathogenesis of HD. For example, recent studies suggest that TORC1 activates transcription of PGC-1alpha24, a master regulator of mitochondrial function that has been implicated in the pathogenesis of HD23,25 and other neurodegenerative disorders26–29. In support of this notion, we found that Sirt1 significantly rescued expression of PGC-1alpha in R6/2 mice (Supplementary Fig. 14). The regulatory regions of FoxO3 genes contain CRE sites30 suggesting that TORC1 may also regulate FoxO3a expression. It will be therefore of interest to examine whether Sirt1 deacetylation of TORC1 leads to activation of multiple target genes and as such represents an upstream target of Sirt1 in CNS. Since TORC1 is essentially brain-specific18, it represents an exciting candidate to monitor the function of Sirt1 in neurons, especially in the context of therapeutic interventions. Together, our results suggest that pharmacological targeting of Sirt1 provides an opportunity for therapeutic development in HD.
ONLINE METHODS
Mouse Studies
To generate BSKO-R6/2 mice, Sirt1flox/flox-nestin-cre and R6/2 were intercrossed for two generations. Nestin-cre was always passed through the male germline. The crosses were all carried out on a C57BL/6 background. R6/2 mice on the C57BL6/J background were obtained from Psychogenics, Inc. Sirt1-KI-R6/2 mice were generated by intercrossing mice heterozygous for the Sirt1-KI transgene, maintained on an outbred background 9 with R6/2 mice on the B6CBAF1/J background (Jackson Laboratories). Rotarod analysis was performed using a Rotamex-5 Rota Rod apparatus (Columbus Instruments). For neuropathological analyses, brain sections were examined stereologically as described 25. The dissector counting method was used to assess striatal neurons and HTT-positive aggregates using an unbiased random selection of serial sections in a defined volume of the neostriatum, as described 8.
Quantitative PCR
qPCR was performed using iQ SYBR Green Supermix (Bio-Rad Laboratories) on a Light Cycler 480 II (Roche). Primers used were: BDNF-F: 5'-TCATACTTCGGTTGCATGAAGG-3' BDNF-R: 5'-AGACCTCTCGAACCTGCCC-3' 18S-F: 18S-R: as obtained from the Harvard Primer Bank 31. Relative mRNA abundance was calculated using the ΔΔCT method.
Plasmids and Antibodies
Lentiviral transfer vectors HTTEx1-25Q, HTTEx1-72Q and Sirt1, BDNF-luciferase construct (pIII(170)Luc)15, Sirt1 and Sirt1 HY plasmids, Flag- CREB, TORC1, Flag-TORC1 and Flag-TORC1 S151A3 were described previously. CREB, K-CREB, CREB-133 and pEGFP were purchased from Clontech. Human HTTEx1 25Q-GFP, HTTEx1-103Q-GFP, HTT480-17stop, HTT480-68stop, HA-TAF and CBP-HAT constructs were described previously 16. Short hairpin RNA lentiviral plasmids (pLKO.1) targeting Sirt1 (5'-AAGTTGACCTCCTCATTGTTA-3') and TORC1 (5'-TTGATTCAGACCATCAGTTTC-3') were purchased from Sigma. Lentiviral Sirt1 HY was generated by mutating 363 Histidine to Tyrosine. Primary antibodies to listed next were from the following sources: neurofilament (2H3, Developmental Studies Hybridoma Bank), Flag (F7425, Sigma), Flag M2 (F3165, Sigma), HA (HA.11, Covance, #3724, Cell Signaling), TORC1 (#2587, Cell Signaling Technology, #6937 from M. Montminy), phospho S151 TORC1 (#3359, Cell Signaling Technology), Myc (#2276, Cell Signaling Technology, sc-789, Santa Cruz, MCA1929, serotec), acetyl lysine (ICP0380, Immunechem), Sirt1 (07-131, Upstate, sc-15404, Santa Cruz, #2028, Cell Signaling and custom-made Sirt1 antibody from L. Guarente), β-tubulin (T4026, Sigma), β3-tubulin (#4466, Cell Signaling Technology) and HTT (MAB5492 and MAB2166, Millipore and Ab1 from M. Difiglia).
Primary neurons
Rat and mouse embryonic primary cortical neurons prepared from E19 and E17 embryos, respectively, were infected with lentiviral vectors at moi of 3 on DIV4. Lentivirus was produced as previously described 32. Neurons fixed and double stained with anti-neurofilament antibody and Draq5 + Sapphire700 were analyzed using Odyssey Infrared Imaging System (LI-COR). Sirt1 KO and WT cultures were prepared by dissecting and plating each embryo separately from E16 littermates. BDNF was added to Sirt1 KO culture at 50 ng ml−1 every other day starting DIV5. For knockdown of Sirt1, neurons were infected with sh scrambled or sh Sirt1 lentivirus at moi of 1 on DIV8. At 5 d post infection (DIV13), neurons were treated with 25 µM forskolin (FSK).
Reporter assays and coimmunoprecipitations
Neuro2a (N2a) cells cultured in DMEM with 10% FBS were transfected using Lipofectamine 2,000. Tranfected cell were harvested at 24 h post transfection unless otherwise specified. For coimmunoprecipitation, transfected Neuro2a cells were harvested at 24 h posttransfection and coimmunoprecipitation performed as previously described 33. For luciferase reporter assays, 50 ng of reporter was used per well of 24-well plates. Cell extracts were prepared at 24 to 48 h posttransfection and luciferase assay performed by standard protocol. For alkaline phosphatase treatment, Neuro2a cells co-transfected with Flag-CREB and TORC1 was immunoprecipitated with anti-Flag antibody. Dephosphorylation reactions were carried out on coimmunoprecipitated TORC1-containing protein G beads.
Multidimensional Protein Identification Technology (MudPIT) and LTQ Orbitrap Mass Spectromtery is described in the supplement
Poor quality spectra were removed from the dataset using an automated spectral quality assessment algorithm 34. MS/MS spectra remaining after filtering were searched with the ProLuCID algorithm35. All searches were parallelized and performed on a Beowulf computer cluster consisting of 100 1.2GHz Athlon CPUs 36. ProLuCID 37 results were assembled and filtered using the DTASelect (version 2.0) program 38. Confidence for modifications was estimated from overlapping modified peptides as described previously39. The validity of peptide/spectrum matches was assessed using the SEQUEST-defined parameters, cross-correlation score (XCorr), and normalized difference in cross-correlation scores (DeltaCn).
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assay from mouse brains was performed using Magna ChIP kit (Millipore) according to manufacturer’s protocol by using TORC1 antibody (#6937, PBL-Salk Institute) and normal rabbit IgG (Santa Cruz). The primer sequences for BDNF promoter IV were published previously40.
Site-directed mutagenesis of TORC1
TORC1 mutants were generated by using QuikChange II XL site-directed mutagenesis kit (Agilent). Incorporation of mutations was verified by DNA sequencing (MGH DNA sequencing core).
Additional methods
Detailed methodology is described in the Supplementary Methods.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Montminy, Peptide Biology Laboratories, the Salk Institute, for Flag- CREB, Flag-TORC1, Flag-TORC1 S151A plasmids and antibody to TORC1 (#6937, PBL-Salk Institute) and helpful discussions; M. E. Greenberg for pIII(170)Luc plasmid15;T. Kouzarides for Sirt1 and Sirt1 HY plasmids; Z. Wu for TORC1 plasmid;M. Difiglia, Department of Nuerology, Massachusetts General Hospital, for antibody to HTT (Ab1); E. Regulier for lentiviral vectors expressing HTTEx1-25Q, HTTEx1-72Q and Sirt1; JM Park for lentiviral vector for CREB knockdown. We also acknowledge C. Whitaker for analysis of microarray data. This work was supported by R01NS051303 (D.K); CHDI, HDF (D.E.C); NIH, Glenn Foundation for Medical Research (L.G), R01MH067880-08 and P41RR011823-15 (J.R.Y).
Footnotes
AUTHOR CONTRIBUTIONS
H.J performed experiments in figures 2, 3, 4, S4, S6, S9, S9, S10, S11, S12, S13; D.C and A.S. performed experiments in figures 1a, 1e, 2c, 2d, S1, S5; L.C performed experiments in figures 1b-h, S13, S14; J.R.M helped with experiments in figure 4f; J.N.S and J.R.Y performed experiments in S7; L.B. performed experiments in S2; L.P.G and D.K provided conceptual framework and discussed the results; D.K. and H.J. wrote the paper.
COMPETING FINANCIAL INTERESTS
L. Guarente is a member of the Scientific Advisory Board of Sirtris/Glaxo Smith Kline.
REFERENCES
- 1.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes & development. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 2.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nature reviews. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 3.Conkright MD, et al. TORCs: transducers of regulated CREB activity. Molecular cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 4.Pallos J, et al. Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington's disease. Human molecular genetics. 2008;17:3767–3775. doi: 10.1093/hmg/ddn273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parker JA, et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nature genetics. 2005;37:349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 6.Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes & development. 2009;23:2812–2817. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 8.Stack EC, et al. Chronology of behavioral symptoms and neuropathological sequela in R6/2 Huntington's disease transgenic mice. The Journal of comparative neurology. 2005;490:354–370. doi: 10.1002/cne.20680. [DOI] [PubMed] [Google Scholar]
- 9.Bordone L, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 10.Zala D, et al. Progressive and selective striatal degeneration in primary neuronal cultures using lentiviral vector coding for a mutant huntingtin fragment. Neurobiology of disease. 2005;20:785–798. doi: 10.1016/j.nbd.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 11.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg ME, Xu B, Lu B, Hempstead BL. New insights in the biology of BDNF synthesis and release: implications in CNS function. J Neurosci. 2009;29:12764–12767. doi: 10.1523/JNEUROSCI.3566-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao MV, Rajagopal R, Lee FS. Neurotrophin signalling in health and disease. Clin Sci (Lond) 2006;110:167–173. doi: 10.1042/CS20050163. [DOI] [PubMed] [Google Scholar]
- 14.Zuccato C, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science (New York, N.Y. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 15.Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 16.Jeong H, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bittinger MA, et al. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr Biol. 2004;14:2156–2161. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Altarejos JY, et al. The Creb1 coactivator Crtc1 is required for energy balance and fertility. Nature medicine. 2008;14:1112–1117. doi: 10.1038/nm.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Zhang C, Takemori H, Zhou Y, Xiong ZQ. TORC1 regulates activity-dependent CREB-target gene transcription and dendritic growth of developing cortical neurons. J Neurosci. 2009;29:2334–2343. doi: 10.1523/JNEUROSCI.2296-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunah AW, et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science (New York, N.Y. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 21.Zuccato C, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nature genetics. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 22.Cohen E, Dillin A. The insulin paradox: aging, proteotoxicity and neurodegeneration. Nat Rev Neurosci. 2008;9:759–767. doi: 10.1038/nrn2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weydt P, et al. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell metabolism. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Wu Z, et al. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:14379–14384. doi: 10.1073/pnas.0606714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Beal MF. Mitochondria and neurodegeneration. Novartis Foundation symposium. 2007;287:183–192. doi: 10.1002/9780470725207.ch13. discussion 192-186. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, et al. The role of Sirt1: at the crossroad between promotion of longevity and protection against Alzheimer's disease neuropathology. Biochimica et biophysica acta. 2010;1804:1690–1694. doi: 10.1016/j.bbapap.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 28.Zhao W, et al. Peroxisome Proliferator Activator Receptor Gamma Coactivator-1alpha (PGC-1alpha) Improves Motor Performance and Survival in a Mouse Model of Amyotrophic Lateral Sclerosis. Molecular neurodegeneration. 2011;6:51. doi: 10.1186/1750-1326-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng B, et al. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson's disease. Science translational medicine. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic acids research. 2010;38:D792–D799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nature protocols. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 33.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 34.Bern M, Goldberg D, McDonald WH, Yates JR., 3rd 3rd. Automatic quality assessment of peptide tandem mass spectra. Bioinformatics (Oxford, England) 2004;20(Suppl 1):i49–i54. doi: 10.1093/bioinformatics/bth947. [DOI] [PubMed] [Google Scholar]
- 35.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. Journal of proteome research. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 36.Sadygov RG, et al. Code developments to improve the efficiency of automated MS/MS spectra interpretation. Journal of proteome research. 2002;1:211–215. doi: 10.1021/pr015514r. [DOI] [PubMed] [Google Scholar]
- 37.Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Analytical chemistry. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 38.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. Journal of proteome research. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacCoss MJ, Wu CC, Yates JR., 3rd Probability-based validation of protein identifications using a modified SEQUEST algorithm. Analytical chemistry. 2002;74:5593–5599. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]
- 40.Gao J, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.