Abstract
BACKGROUND
Addiction susceptibility and treatment responsiveness are greatly influenced by genetic factors. Sequence variation in genes involved in the mechanisms of drug action have the potential to influence addiction risk and treatment outcome. The opioid receptor system is involved in mediating the rewarding effects of cocaine and opioids. The µ-opioid receptor (MOR) has traditionally been considered the primary target for opioid addiction. The MOR, however, interacts with and is regulated by many known MOR interacting proteins (MORIPs), including the δ-opioid receptor (DOR).
METHODS
The present study evaluated the contribution of OPRD1, the gene encoding the DOR, to the risk of addiction to opioids and cocaine. The association of OPRD1 polymorphisms with both opioid addiction (OA) and cocaine addiction (CA) was analyzed in African American (OA n=336, CA n=503) and European American (OA n=1007, CA n=336) populations.
RESULTS
The primary finding of this study is an association of rs678849 with cocaine addiction in African Americans (allelic p=0.0086). For replication purposes, this SNP was analyzed in a larger independent population of cocaine addicted African Americans and controls and the association was confirmed (allelic p=4.53×10−5; n=993). By performing a meta-analysis on the expanded populations, the statistical evidence for an association was substantially increased (allelic p=8.5 × 10 −7) (p-values non-FDR corrected).
CONCLUSION
The present study suggests that polymorphisms in OPRD1 are relevant for cocaine addiction in the African American population and provides additional support for a broad role for OPRD1 variants in drug dependence.
Keywords: addiction, association study, delta-opioid receptor, OPRD1, genetics
1. INTRODUCTION
Twin and family studies suggest that a large percentage of risk for both opioid addiction (OA; Arvidsson et al., 1995; Karkowski et al., 2000; Kendler et al., 2003; Merikangas et al., 1998; Tsuang et al., 1998) and cocaine addiction (CA; Karkowski et al., 2000; Kendler et al., 2000; Kendler and Prescott, 1998; Zhang et al., 2006) is influenced by genetic factors (reviewed in Kreek et al., 2005; Saxon et al., 2005; Yuferov et al., 2010). Understanding which genes influence addiction susceptibility could improve treatment options and patient care. However, identification of causal genes has been difficult due to the polygenic inheritance underlying addictive behavior.
Opioid receptors have been the focus of addiction research due to the involvement of these receptors in drug reward pathways. The most widely studied members of the opioid receptor family are the µ-opioid receptor (MOR), δ-opioid receptor (DOR) and κ-opioid receptor (KOR), which bind endogenous opioid peptides such as β-endorphin, endomorphines, enkephalins, and dynorphins (reviewed in Zaki et al., 1996). Stimulation of opioid receptors inhibits adenylate cyclase, increases potassium conductance, decreases calcium conductance, and activates MAP Kinase pathways (Childers, 1991, reviewed in Law et al., 2000). The rewarding effects of drug use are mediated by MOR and DOR activation, whereas KOR activation is associated with aversion (Di Chiara and Imperato, 1988 ; Herz, 1998). Furthermore, DOR is thought to be involved in analgesia, morphine tolerance and mood regulation such as anxiety and depression (Filliol et al., 2000; Perrine et al., 2006; Zhu et al., 1999).
The opioid receptor system, and particularly MOR, has been studied extensively for its role in OA (Matthes et al., 1996; Sora et al., 1997) and CA (Becker et al., 2002; Hall et al., 2004; Hummel et al., 2006). Several studies have analyzed the influence of single nucleotide polymorphisms (SNPs) in OPRM1, the gene encoding MOR, on risk for drug addiction, but the results have been inconclusive (Bart et al., 2004; Crowley et al., 2003; Hoehe et al., 2000; Smith et al., 2005; Szeto et al., 2001; Tan et al., 2003; Zhang et al., 2006). Although MOR is considered the primary target for the rewarding effects of addiction, there are many known MOR interacting proteins (MORIPs) that may modulate MOR function. One of these MORIPs is DOR (Milligan, 2005), suggesting that genetic variation in OPRD1, the gene encoding DOR, may affect susceptibility to drug addiction.
Two coding variants of OPRD1 have been studied for association with alcohol and drug addiction: rs1042114 (G80T) and rs2234918 (T921C). Mayer et al. found rs2234918 to be associated with heroin addiction in a German population (Mayer et al., 1997). rs1042114 was associated with OA and a 6 SNP haplotype which included rs1042114 and rs2234918 was associated with alcohol, cocaine, and opioid addiction in a cohort of European-Americans (Zhang et al., 2008). An additional study identified an association of three OPRD1 intronic SNPs with heroin addiction in EA (412 cases vs. 184 controls) and also reported a combined effect of OPRD1 and OPRM1 on heroin addiction (Levran et al., 2008). A recent study of 1459 case and 1495 controls found two intronic OPRD1 SNPs (rs2236857 and rs581111) to be associated with risk for heroin addiction (Nelson et al., 2012). However, no evidence was found for an association of either SNP in additional heroin- and alcohol-addicted German populations (Franke et al., 1999), heroin-addicted Chinese individuals (Xu et al., 2002), or alcohol-addicted Taiwanese Hans (Loh el et al., 2004). Xuei et al. also reported that OPRD1 variants were not associated with alcohol addiction or OA in a study of 1,923 European-American subjects from 219 multiplex alcohol-addicted families (83 individuals demonstrating OA; Xuei et al., 2007).
Currently, additional research is needed to understand the role of OPRD1 in both opioid and cocaine addiction. The present study was designed to genotype a comprehensive panel of SNPs within OPRD1 and test for association with both OA and CA populations across European-American (EA) and African-American (AA) ancestries.
2. MATERIALS AND METHODS
2.1 Population Samples
Cases
DNA samples were requested and acquired through the NIDA Center for Genetic Studies in conjunction with Washington University and Rutgers University Cell and DNA Repository (RUCDR). OA (EA: n=1007; male 65.6%; AA: n=336 male 71.4%) and CA subjects (EA: n=336; male 50.3%; AA: n=503; male 52.1%) of EA and AA descent met DSM-IV criteria for addiction (Table 1). AA CA samples from RUCDR are labeled as “Group 1” in subsequent analysis. DNA samples were transferred to 96-well stock plates and diluted to a concentration of 1 ng/µl for genotyping.
Table 1.
Total number of CA cases, OA cases, and controls in each of the analyzed populations. The percentage of male individuals for each group is included in parentheses. Average age and standard deviation for each population is in provided in brackets.
| Population | Cocaine Addicted Cases | Opioid Addicted Cases | Controls |
|---|---|---|---|
| European Americans | 336 (50.3%)[36.1 ± 8.5] | 1007 (65.6%)[36.9 ± 11.4] | 656 (50.8%)[53.0 ± 17.6] |
| African Americans (Group 1) | 503 (52.1%)[40.9 ± 7.0] | 336 (71.4%)[48.2 ± 9.2] | 503 (38.0%)[45.9 ± 14.0] |
| African Americans (Group 2) | 993 (66.9%)[43.4 ± 6.5] | - | 875 (41.0%)[44.9 ± 12.7] |
A separate group of AA (n=993; male 66.9%) CA subjects (Group 2) were collected during clinical studies for CA treatment at the University of Pennsylvania Treatment Research Center and used for confirmation purposes (Table 1). Subjects were at least 18 years of age. All were assessed with the Structured Clinical Interview for DSM Disorders (SCID) and urine drug screens were obtained. All patients had a clinical diagnosis of CA as defined by DSM-IV. Family history was not obtained and ethnicity was determined by self-report. All psychiatric axis I disorders except anxiety disorders, major depressive disorder, alcohol addiction/abuse and nicotine addiction were used as exclusion criteria. In addition, participants were excluded if they had a history of a seizure disorder (except cocaine-induced seizures) or a severe medical illness, including a history of AIDS (but not merely of HIV+ status). Individuals currently being treated with psychotropic medications or with psychiatric symptoms, including psychosis, dementia, suicidal or homicidal ideation, mania or depression requiring antidepressant therapy were also excluded. For all samples, genomic DNA was extracted from peripheral leukocytes within obtained blood samples by standard protocols. All protocols were approved by the Institutional Review Boards at the University of Pennsylvania, and all subjects provided written informed consent before blood sample collection.
Controls
EA control individuals (n=656; male=50.8%) and AA control individuals (n=503; male= 38.0%) were acquired from the National Institute of Mental Health Genetics Initiative (NIMH-GI; www.nimhgenetics.org, Table 1). Control individuals were screened for history of substance use disorders and other psychiatric illness. For rs678849 specifically, a second AA control group (n=875; male= 41 %) consisting of NIMH-GI samples and a small number of controls samples that were collected together with the cocaine-addicted patients at the University of Pennsylvania were also genotyped (Table 1).
An additional control group was used to confirm the association of rs678849 found in AA CA. In collaboration with Dr. Hakonarson, genotype data was obtained from the pediatric control group recruited by the Children’s Hospital of Philadelphia (CHOP) clinicians, nursing and medical assistant staff within the CHOP Health Care Network. Recruitment of 12,299 AA subjects took place across the US. All control subjects were genotyped using the Illumina 550K (Illumina, San Diego, CA) SNP array which included rs678849. The Research Ethics Board of CHOP and other participating centers approved the study, and written informed consent was obtained from all subjects or their parents.
2.2 SNP selection and genotyping
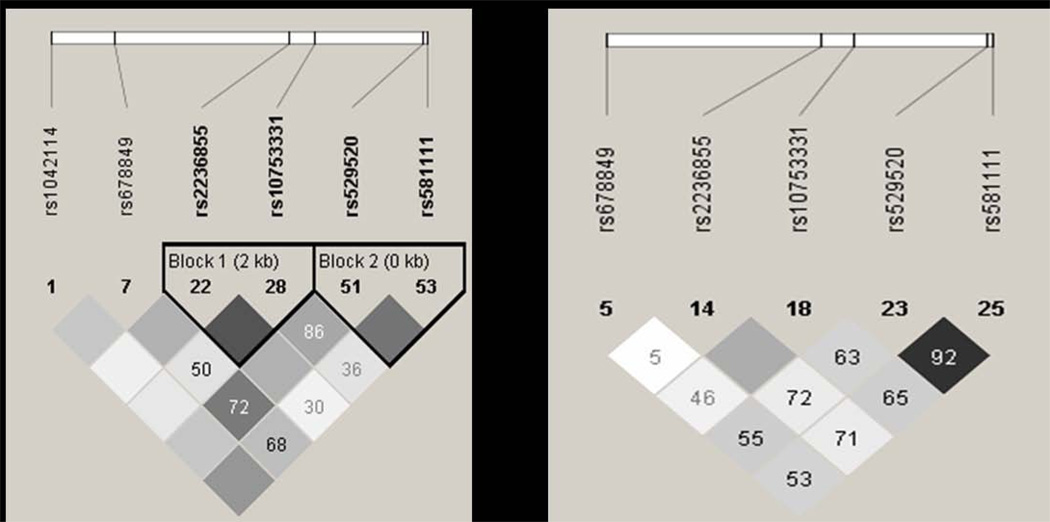
SNPs were selected using the Tagger algorithm as part of Haploview software (http://www.broadinstitute.org/haploview; Barrett et al., 2005). Using the HapMap CEU population data (HapMap data release 28 phase II and III, August 10, www.hapmap.org), 7 SNPs (rs1042114;coding non-synonymous), rs678849 (intron), rs2236855 (intron), rs10753331 (intron), rs529520 (intron), rs581111 (intron) and rs2234918; coding synonymous) capture 85% of SNPs in OPRD1 with a minor allele frequency cut-off of 10% and an r2 of 0.8 (Figure 1A). rs2234918 was not genotyped in the HapMap population and is not included in the linkage disequilibrium (LD) analysis. In the HapMap ASW population data (HapMap data phase III/rel#2 Feb09), 7 SNPs (rs533123, intron; rs678849, intron; rs2236855, intron; 10753331, intron; rs529520, intron; rs581111, intron; and rs2234918, coding synonymous) capture 33% of SNPs in OPRD1 with a minor allele frequency cut-off of 10% and an r2 of 0.8 (Figure 1B). rs2234918 and rs533123 were not included in LD analysis since they were not genotyped in the HapMap population. Two additional SNPs, rs204047 and rs797397, were also genotyped in a subset of AA CA samples.
Figure 1.
LD between OPRD1 SNPs genotyped in HapMap CEU population and ASW population (www.hapmap.org). A: HapMap data release 28, phase II and III, August 10. In the CEU population, 7 SNPs capture 85% of SNPs in OPRD1 with a minor allele frequency cut-off of 10% and an r2 of 0.8. (rs2234918 not included in LD analysis as HapMap data not available). B: HapMap data phase III/rel#2 Feb09. In the ASW population, 7 SNPs capture 33% of SNPs in OPRD1 with a minor allele frequency cut-off of 10% and an r2 of 0.8. (rs2234918 and rs533123 not included in LD analysis as HapMap data not available). Numbers inside the squares represent D’ and the shading is representative of r2. When no number is displayed D’ =1.
Genotyping reactions of 5 µl total volume (containing 2 µl of genomic DNA and 3 µl of Taqman Genotyping Master Mix plus Taqman Assay) were prepared in a 384-well plate format using a Biomek 3000 robotic workstation (Beckman Coulter, Inc.; Brea, CA). SNP genotyping was performed using an ABI 9700 Thermocycler and Taqman SNP Genotyping Assays (Life Technologies, Applied Biosystems Inc.; Foster City, CA, USA). Quality control was maintained by genotyping 10% duplicates for cases and controls, and the concordance rate for our genotyping was calculated to be 99.99%. Following amplification, genotypes were acquired using ABI Prism 7900 Sequence Detections System v2.2 software and Taqman Genotyper Software v1.0 (Life Technologies, Applied Biosystems Inc.; Foster City, CA, USA).
2.3 Statistical analysis
The allelic and genotypic association of SNPs with OA and CA were determined using the Chi-square test in the software package PLINK v1.07 (Purcell et al., 2007). For each SNP, deviation from Hardy-Weinberg was assessed in the total population and also in cases and controls individually. All SNPs across populations were in Hardy-Weinberg Equilibrium (p≥0.05). Allelic and genotypic association meta-analyses for rs678849 were performed on the combined samples from Group 1 and Group 2, plus an additional 35 CA samples obtained from RUCDR. All p-values were corrected for multiple comparisons using the false discovery rate (FDR) procedure (Benjamini et al., 2001). The FDR procedure allows control of the average fraction of false rejections made out of the number of false rejections performed. The cut-off for statistical significance for this study was p≤0.05 after FDR correction. Sliding-window haplotype analysis was performed in PLINK using the expectation maximization (EM) algorithm (http://pngu.mgh.harvard.edu/~purcell/plink/; Purcell et al., 2007).
3. RESULTS
3.1 European American Population Genotypic and Allelic Associations
In the EA population, no significant allelic associations were found for OA; however, rs10753331 was nominally significant for genotypic association with OA (p=0.02; Table 2). Sex-specific analyses showed nominal association in males with the non-synonymous SNPs rs1042114 (p=0.008), rs2236855 (p=0.02), and rs10753331 (p=0.01). Several haplotype blocks had nominally significant associations with OA as well (Supplemental Data Table 11).
Table 2.
Comparison of genotype and allele frequencies in an EA population of OA cases and controls. P-values represent PLINK-generated X2 tests for association between EA opioid-addicted cases and unrelated controls. C.I. =95% Confidence Interval; MA = minor allele; MAF = minor allele frequency; OR=odds ratio.
| SNP ID (MA) | Case MAF (N) | Control MAF (N) | Allelic p-value | Odds Ratio (C.I.) | Genotypic p-value |
|---|---|---|---|---|---|
| rs1042114 (G) | 0.12 (996) | 0.14 (643) | 0.09 | 0.83 (0.68–1.03) | 0.22 |
| rs678849 (C) | 0.49 (995) | 0.48 (654) | 0.32 | 1.07 (0.93–1.23) | 0.48 |
| rs2236855 (T) | 0.30 (998) | 0.28 (652) | 0.17 | 1.11 (0.95–1.30) | 0.13 |
| rs10753331 (A) | 0.37 (994) | 0.34 (647) | 0.08 | 1.14 (0.99–1.32) | 0.02 |
| rs529520 (T) | 0.48 (1001) | 0.49 (654) | 0.78 | 0.98 (0.85–1.13) | 0.29 |
| rs581111 (T) | 0.26 (998) | 0.28 (654) | 0.41 | 0.94 (0.80–1.10) | 0.70 |
| rs2234918 (C) | 0.45 (999) | 0.44 (654) | 0.88 | 1.01 (0.88–1.16) | 0.71 |
For CA, a nominally significant allelic association was observed for the synonymous SNP rs2234918 (p=0.009; Table 3). The genotypic p-values for this SNP (p=0.03), as well as rs581111 were also nominally significant (p=0.05; Table 3). Sex-specific analysis for allelic association showed nominal association in males for rs2236855 (p=0.0006), rs1075331 (p=0.002), rs529520 (p=0.02), and rs2234918 (p=0.04), and females for rs2234918 (p=0.04). None of the associations observed in the EA population were significant following FDR correction for multiple testing.
Table 3.
Comparison of genotype and allele frequencies in an EA population of CA cases and controls. P-values represent PLINK-generated X2 tests for association between EA cocaine-addicted cases and unrelated controls. C.I. =95% Confidence Interval; MA = minor allele; MAF = minor allele frequency; OR=odds ratio.
| SNP ID (MA) | Case MAF (N) | Control MAF (N) | Allelic p-value | Odds Ratio (C.I.) | Genotypic p-value |
|---|---|---|---|---|---|
| rs1042114 (G) | 0.15 (331) | 0.14 (643) | 0.69 | 1.06 (0.81–1.38) | 0.75 |
| rs678849 (C) | 0.51 (331) | 0.48 (654) | 0.19 | 1.13 (0.94–1.37) | 0.42 |
| rs2236855 (T) | 0.30 (330) | 0.28 (652) | 0.32 | 1.11 (0.90–1.37) | 0.53 |
| rs10753331 (A) | 0.37 (328) | 0.34 (647) | 0.20 | 1.14 (0.94–1.38) | 0.39 |
| rs529520 (T) | 0.52 (330) | 0.49 (654) | 0.21 | 1.13 (0.93–1.36) | 0.46 |
| rs581111 (T) | 0.31 (330) | 0.28 (654) | 0.16 | 1.16 (0.94–1.42) | 0.05 |
| rs2234918 (C) | 0.51 (329) | 0.44 (654) | 0.009 | 1.28 (1.06–1.55) | 0.03 |
3.2 African-American Population Genotypic and Allelic Associations
A nominal association was found for rs581111 in OA (allelic p=0.02; genotypic p=0.04; Table 4). In CA, nominally significant allelic associations were detected for rs2236855 in both the entire AA population (p=0.03) and AA females (p=0.003; Table 5). An association was also detected with CA for rs678849, which is located within intron one of OPRD1 (allelic p=0.009; genotypic p=0.03; Table 5). The allelic association for rs678849 was still significant after correction for multiple testing. Sex-specific analysis for allelic association also identified a nominal association in females for rs678849 (p=0.01). Furthermore, multiple haplotype blocks including the rs678849 locus were nominally associated with CA (Supplemental Data Table 22).
Table 4.
Comparison of genotype and allele frequencies in an AA population of OA cases and controls. P-values represent PLINK-generated X2 tests for association between AA opioid-addicted cases and unrelated controls. C.I. =95% Confidence Interval; MA = minor allele; MAF = minor allele frequency; OR=odds ratio.
| SNP ID (MA) | Case MAF (N) | Control MAF (N) | Allelic p-value | Odds Ratio (C.I.) | Genotypic p-value |
|---|---|---|---|---|---|
| rs533123 (G) | 0.48 (333) | 0.49 (502) | 0.76 | 0.97 (0.80–1.18) | 0.24 |
| rs678849 (T) | 0.28 (332) | 0.28 (500) | 0.84 | 1.02 (0.82–1.27) | 0.25 |
| rs2236855 (T) | 0.30 (336) | 0.29 (499) | 0.88 | 1.02 (0.82–1.26) | 0.71 |
| rs10753331 (G) | 0.50 (336) | 0.45 (501) | 0.10 | 1.18 (0.97–1.44) | 0.23 |
| rs529520 (C) | 0.32 (336) | 0.28 (502) | 0.12 | 1.19 (0.96–1.47) | 0.23 |
| rs581111 (G) | 0.41 (336) | 0.35 (502) | 0.02 | 1.26 (1.03–1.54) | 0.04 |
| rs2234918 (T) | 0.37 (336) | 0.35 (493) | 0.45 | 1.08 (0.88–1.33) | 0.69 |
Table 5.
Comparison of genotype and allele frequencies in an AA population of CA cases and controls. P-values represent PLINK-generated X2 tests for association between AA cocaine-addicted cases and unrelated controls. C.I. =95% Confidence Interval; MA = minor allele; MAF = minor allele frequency; OR=odds ratio.
| SNP ID (MA) | Case MAF (N) | Control MAF (N) | Allelic p-value | Odds Ratio (C.I.) | Genotypic p-value |
|---|---|---|---|---|---|
| rs533123 (G) | 0.47 (498) | 0.49 (502) | 0.47 | 0.94 (0.79–1.12) | 0.36 |
| rs678849 (T) | 0.23 (500) | 0.28 (500) | 0.009 | 0.76 (0.62–0.93) | 0.03 |
| rs2236855 (T) | 0.34 (497) | 0.29 (499) | 0.03 | 1.24 (1.02–1.49) | 0.09 |
| rs10753331 (G) | 0.43 (499) | 0.45 (501) | 0.22 | 0.90 (0.75–1.07) | 0.48 |
| rs529520 (C) | 0.25 (492) | 0.28 (502) | 0.12 | 0.85 (0.70–1.04) | 0.28 |
| rs581111 (G) | 0.35 (497) | 0.35 (502) | 0.76 | 0.97 (0.81–1.17) | 0.36 |
| rs2234918 (T) | 0.34 (493) | 0.35 (493) | 0.60 | 0.95 (0.79–1.15) | 0.80 |
3.3 Confirmation of Allelic Association of rs678849 in Independent African-American Populations
Access to a large population of AA individuals with CA through the University of Pennsylvania Translational Research Center allowed us to confirm the initial association of rs678849 in an independent set of samples (Group 2). Group 2 (described in the methods) consists of an independent CA population and AA controls (cases n=993, controls =875). In Group 2, the significant association for rs678849 was confirmed (allelic p=4.53×10−5; genotypic p-value =7.71×10−6; Table 6). Additional CA samples (n=35) obtained from RUCDR were added to previously genotyped samples (Group 1) and a meta-analysis was performed on the combined samples from Group 1 and Group 2. The p-value for the association with CA was significantly strengthened in the meta-analysis (1527 cases vs. 1378 controls; allelic p=8.50 × 10−7; genotypic p=3.65×10−7; OR[95% C.I.]= 0.74 [.65–.83]), suggesting that the minor allele of rs678849 has a protective effect (Table 6). These p-values remained significant after FDR correction.
Table 6.
Confirmation of rs678849 association in an independent AA population of CA cases. For Group 2, P-values represent PLINK-generated X2 tests for association between AA cocaine-addicted cases and unrelated controls. C.I. =95% Confidence Interval; MAF = minor allele frequency; OR=odds ratio. Meta-analysis was conducted and the combined p-value for both genotype and association tests remained significant after correction for multiple testing.
| Case MAF (N) | Control MAF (N) | Allelic p-value | Odds Ratio (C.I.) | Genotypic p-value | |
|---|---|---|---|---|---|
| Group 2 | 0.22 (993) | 0.28 (875) | 4.53 × 10−5 | 0.73 (.63–.85) | 7.71 × 10−6 |
| Groups 1 & 2 | 0.22 (1527) | 0.28 (1378) | 8.49 × 10−7 | 0.74 (.65–.83) | 3.65 × 10−7 |
Additional control sample data was obtained from 12,299 AA patients genotyped using the Illumina 550K platform. Chi-square analysis was performed on the allele frequencies of this control population (0.254) compared to the total AA population with CA (0.223). The difference in the minor allele frequencies between the combined cases from Group1 and Group 2 and this large control population was again statistically significant (p= 0.00027), although the p-value is less significant than the previous meta-analysis. This p-value remained significant after FDR correction for multiple testing.
3.4 Analysis of Neighboring SNP
To confirm that the significant association signal was coming from the rs678849 locus, Group 1 samples were genotyped for two SNPs that are in high D’ with rs678849: rs204047 (D’=1; r2=0.092) and rs797397 (D’=.885; r2=0.638). Allelic and genotypic p-values for the association of rs204047 and rs797397 were not as low as those for rs678849, and neither SNP was significantly associated with CA after FDR correction (Table 7). Furthermore, haplotype analysis of these 3 SNPs did not increase the level of statistical significance (omnibus p=0.02).
Table 7.
Analysis of SNPs Neighboring rs678849 in an AA population of CA cases. Two SNPs that were in high D’ with rs678849 were genotyped in the AA CA Group 1: rs204047 (D’=1; r2=.092) and rs797397 (D’=.885; r2=.638). Table shows the allele and genotypic frequencies for rs204047 and rs797397 compared to rs678849. P-values represent PLINK-generated X2 tests for association between AA cocaine-addicted cases and unrelated controls. C.I. =95% Confidence Interval; MA = minor allele; MAF = minor allele frequency; OR=odds ratio
| SNP ID (MA) | Case MAF (N) | Control MAF (N) | Allelic p-value | Odds Ratio (C.I.) | Genotypic p-value |
|---|---|---|---|---|---|
| rs204047 (T) | 0.23 (496) | 0.20 (501) | 0.11 | 1.19 (0.96–1.47) | 0.26 |
| rs678849 (T) | 0.23 (500) | 0.28 (500) | 0.009 | 0.76 (0.622–0.93) | 0.03 |
| rs797397 (T) | 0.19 (486) | 0.23 (457) | 0.02 | 0.77 (0.62–0.96) | 0.09 |
4. DISCUSSION
The present study focused on the association between OPRD1 genetic variants and cocaine and opioid addiction in EA and AA populations. In the EA population, a nominally significant association of the synonymous SNP rs2234918 with CA was observed, as well as sex-specific nominal associations for several other OPRD1 SNPs. In the EA population with OA, a nominal sex-specific association of the non-synonymous SNP rs1042114 was identified. Associations of rs581111 with OA and rs2236855 with CA in the AA population were also nominally significant. Further, the intronic SNP, rs678849, was found to be strongly associated with CA and confirmed in an independent set of samples. This is the first study to report an association of OPRD1 with CA in an AA population.
Other studies have analyzed OPRD1 polymorphisms and substance dependence. A large candidate gene study of Australian heroin addicted cases and controls analyzed SNPs in OPRD1. They found rs2236857 to be associated with heroin addiction (p=2.9×10−4, OR=1.25; Nelson et al., 2012). The present study genotyped rs2236855, which is in complete linkage disequilibrium with rs2236857. We observed no association between rs2236855 and OA in the EA population, although rs2236855 was nominally associated with CA in the AA population (allelic p=0.03, OR=1.24). The Australian study also identified an association between a haplotype block of rs2236857 and rs581111 and heroin addiction (Nelson et al., 2012). Interestingly, rs581111 was found to be nominally associated with CA in EA (genotypic p=0.05, OR=1.16) and OA in AA (allelic p=0.02, OR=1.26) in our study. Zhang et al. genotyped 1063 EA substance addicted cases and 443 controls and found a 6 SNP haplotype to be associated with alcohol, cocaine and opioid addiction (Zhang et al., 2008). This haplotype contained rs1042114 and rs2234918, both of which displayed nominal significance with opioid and cocaine addiction in our study. A large scale candidate gene study analyzing 1350 variants in 412 cases and 184 controls found 3 intronic SNPs in OPRD1 to be associated with heroin addiction (Levran et al., 2008). Although the same SNPs were not genotyped in our study, tag SNPs rs1075331 and rs2236855 are in LD with these variants and they were found to be nominally associated with cocaine and opioid addiction, respectively.
The strongest association reported in the present study was between rs678849 and CA in the AA population. The minor allele (T) was overrepresented in the controls compared to cases, indicative of a protective effect. Following the initial association of rs678849 with CA in the Group 1 samples, the association was confirmed in an independent set of samples (Group 2). Two additional SNPs in high LD with rs678849 (D’>0.88) were genotyped to determine the source of the association signal. The results revealed that rs678849 had the strongest association with CA, supporting the surrounding intronic segment as a genomic region of interest in addiction. At this time, we cannot conclude whether rs678849 is driving the association as un-typed variants genetically linked to the SNP may also be responsible for the signal. However, previous studies have implicated rs678849 in addiction susceptibility and treatment efficacy. In a pharmacogenetic study of naltrexone treatment in alcohol addiction, individuals carrying the “T” allele of rs678849 had a significantly lower relapse rate after treatment compared to matched placebo controls (Gelernter et al., 2007). Another recent study by Luo et al. reported an association between a haplotype including rs678849 and drug addiction in a mixed population of EA and AA cases (Luo et al., 2008). In conjunction with our findings, these results suggest that future studies designed to test the pharmacogenetic effects of rs678849 in AA CA populations are warranted.
We cannot exclude the possibility that associations in the AA population are due to population stratification. According to data from the International Hapmap Project (www.hapmap.org), the minor allele of rs678849 (T) has a frequency of 26% in a population with African ancestry from the Southwest USA (ASW). In a population of Utah residents with Northern and Western European ancestry (CEU), however, the ‘T’ allele has a frequency of 53%. Due to this large difference in minor allele frequency between the two populations, different degrees of population admixture between AA cases and controls may have contributed to the association found in the present study. In support of our findings, African genetic heritage was not found to be associated with OA or CA in a previous study (Ducci et al., 2009) and the significance level of the rs678849 allele remains significant when comparing our control data to a larger independent control group. Further work using ancestry informative markers is needed to determine whether rs678849 is relevant for CA in AA.
The present study, in combination with the current literature, suggests that DOR may be a potential therapeutic target for addiction. OPRD1 knock-out mice demonstrate anxiogenic-like and depressive-like behaviors, suggesting a role for DOR in modulating emotional state (Filliol et al., 2000; Konig et al., 1996; Ragnauth et al., 2001). Decreased DOR signaling resulting from treatment with the DOR antagonist naltrindole also causes anxiogenic-like effects (Marin et al., 2003). Mediation of anxiety makes DOR an appealing target for alleviation of cocaine withdrawal-induced anxiety. SNC-80, a DOR agonist, produced anxiolytic-like effects in males similar to the effects of classical therapeutics for anxiety (Perrine et al., 2006) and was able to decrease anxiety-like behavior following withdrawal from chronic cocaine in male (Perrine et al., 2008) and female rats (Ambrose-Lanci, 2009). Non-peptide DOR agonists, such as SNC-80, have greater bioavailability than peptide agonists and more readily cross the blood-brain-barrier. These non-peptide agonists may have clinical potential for the treatment of not only anxiety but also depression, pain and other neurological disorders. Despite some potential limitations related to convulsant effects, the use of DOR agonists is encouraging since they are reported to have fewer negative side effects and lower abuse potential compared to MOR agonists (reviewed in Jutkiewicz, 2006; Pradhan et al., 2011).
The present study provides an important step in understanding the genetic influence of OPRD1 on both CA and OA. In the OPRD1 gene, nominally significant associations were reported as well as a highly statistically significant association an intronic SNP (rs678849) and CA in AA. In light of previous findings implicating the DOR system in both emotional state and addiction withdrawal, our data support further research to fully determine the role of DOR in cocaine addiction.
Supplementary Material
Acknowledgements
We would like to acknowledge NIDA’s Center for Genetic Studies in conjunction with Washington University and Rutgers University Cell & DNA Repository for providing DNA samples collected from the following studies and investigators: Opioid Samples: Addictions: Genotypes, Polymorphisms and Function, Mary Jeanne Kreek, M.D.; Genetics of Opioid Dependence, Joel Gelernter, M.D., Kathleen Brady, M.D., Ph.D., Henry Kranzler, M.D., Roger Weiss, M.D.; Opioid Dependence, Wade Berrettini, M.D., Ph.D. Cocaine Samples: An Introduction to the Family Study of Cocaine Dependence, Laura Bierut, M.D.; Genetics of Cocaine Induced Psychosis, Joseph F Cubells, M.D., Ph.D. We would also like to acknowledge the NIDA Clinical Trials Network (CTN) and the University of Pennsylvania CTN node (George Woody, M.D., grant: 2U10DA013043) for support and provision of additional DNA samples from opioid addicted individuals. The NIMH control subjects were collected by the NIMH Schizophrenia Genetics Initiative 'Molecular Genetics of Schizophrenia II' (MGS-2) collaboration. The investigators and co-investigators are: ENH/Northwestern University, Evanston, IL, MH059571, Pablo V. Gejman, M.D. (Collaboration Coordinator; PI), Alan R. Sanders, M.D.; Emory University School of Medicine, Atlanta, GA, MH59587, Farooq Amin, M.D. (PI); Louisiana State University Health Sciences Center; New Orleans, Louisiana, MH067257, Nancy Buccola APRN, BC, MSN (PI); University of California-Irvine, Irvine, CA, MH60870, William Byerley, M.D. (PI); Washington University, St. Louis, MO, U01,MH060879, C. Robert Cloninger, M.D. (PI); University of Iowa, Iowa, IA, MH59566, Raymond Crowe, M.D. (PI), Donald Black, M.D.; University of Colorado, Denver, CO, MH059565, Robert Freedman, M.D. (PI); University of Pennsylvania, Philadelphia, PA, MH061675, Douglas Levinson M.D. (PI); University of Queensland, Queensland, Australia, MH059588, Bryan Mowry, M.D. (PI); Mt. Sinai School of Medicine, New York, NY, MH59586, Jeremy Silverman, Ph.D. (PI).
Role of Funding Sources
This work was supported by the Center for Neurobiology and Behavior, Department of Psychiatry, University of Pennsylvania, Training Program in Neuropsychopharmacology (T32MH014654, P.I.: I.Lucki) and a NIDA Distinguished International Scientist Award (MV). Financial support is gratefully acknowledged from NIDA grant P20DA025995 (P.I.: W. Berrettini), the Veterans Administration Mental Illness Research Education and Clinical Center MIRECC) at the Philadelphia VAMC (David Oslin, MD, PI) and NIDA grants P60 DA 05186 (P.I.: Charles O’Brien) and P50 DA012756 (P.I.: H. Pettinati). NIDA had no further role in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication
Footnotes
Supplementary material can be found by accessing the online version of this paper at http://dx.doi.org and by entering doi:…
Supplementary material can be found by accessing the online version of this paper at http://dx.doi.org and by entering doi:…
Supplementary material can be found by accessing the online version of this paper at http://dx.doi.org and by entering doi:…
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
The authors LMAL, MV, TKC, GAD, and WB were responsible for the study concept and design. LMAL, MV, AZ, CY were responsible for the acquisition of genotype data. LMAL, TKC, RC, GAD conducted the bioinformatic and statistical analysis. LMAL, WHB, TNF, FWL, GAD, KMK, CPO, HMP, DWO and HH were responsible for sample acquisition and characterization. RC, LMAL and WB drafted the manuscript. All authors critically reviewed content and approved final version for publication.
Conflicts of Interest
The authors report no biomedical financial interests or potential conflicts of interest.
REFERENCES
- Ambrose-Lanci LM. Cocaine withdrawal-induced anxiety in females: impact of circulating estrogen and potential use of delta-opioid receptor agonists for treatment. J. Neurosci. Res. 2009;88:816–824. doi: 10.1002/jnr.22259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson U, Riedl M, Chakrabarti S, Lee JH, Nakano AH, Dado RJ, Loh HH, Law PY, Wessendorf MW, Elde R. Distribution and targeting of a mu-opioid receptor (MOR1) in brain and spinal cord. J. Neurosci. 1995;15:3328–3341. doi: 10.1523/JNEUROSCI.15-05-03328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bart G, Heilig M, LaForge KS, Pollak L, Leal SM, Ott J, Kreek MJ. Substantial attributable risk related to a functional mu-opioid receptor gene polymorphism in association with heroin addiction in central Sweden. Mol. Psychiatry. 2004;9:547–549. doi: 10.1038/sj.mp.4001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, Grecksch G, Kraus J, Loh HH, Schroeder H, Hollt V. Rewarding effects of ethanol and cocaine in mu opioid receptor-deficient mice. Naunyn Schmiedebergs Arch. Pharmacol. 2002;365:296–302. doi: 10.1007/s00210-002-0533-2. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- Childers SR. Opioid receptor-coupled second messenger systems. Life Sci. 1991;48:1991–2003. doi: 10.1016/0024-3205(91)90154-4. [DOI] [PubMed] [Google Scholar]
- Crowley JJ, Oslin DW, Patkar AA, Gottheil E, DeMaria PA, Jr, O'Brien CP, Berrettini WH, Grice DE. A genetic association study of the mu opioid receptor and severe opioid dependence. Psychiatr. Genet. 2003;13:169–173. doi: 10.1097/00041444-200309000-00006. [DOI] [PubMed] [Google Scholar]
- Di Chiara G, Imperato A. Opposite effects of mu and kappa opiate agonists on dopamine release in the nucleus accumbens and in the dorsal caudate of freely moving rats. J. Pharmacol. Exp. Ther. 1988;244:1067–1080. [PubMed] [Google Scholar]
- Ducci F, Roy A, Shen PH, Yuan Q, Yuan NP, Hodgkinson CA, Goldman LR, Goldman D. Association of substance use disorders with childhood trauma but not African genetic heritage in an African American cohort. Am. J. Psychiatry. 2009;166:1031–1040. doi: 10.1176/appi.ajp.2009.08071068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filliol D, Ghozland S, Chluba J, Martin M, Matthes HW, Simonin F, Befort K, Gaveriaux-Ruff C, Dierich A, LeMeur M, Valverde O, Maldonado R, Kieffer BL. Mice deficient for delta- and muopioid receptors exhibit opposing alterations of emotional responses. Nat. Genet. 2000;25:195–200. doi: 10.1038/76061. [DOI] [PubMed] [Google Scholar]
- Franke P, Nothen MM, Wang T, Neidt H, Knapp M, Lichtermann D, Weiffenbach O, Mayer P, Hollt V, Propping P, Maier W. Human delta-opioid receptor gene and susceptibility to heroin and alcohol dependence. Am. J. Med. Genet. 1999;88:462–464. doi: 10.1002/(sici)1096-8628(19991015)88:5<462::aid-ajmg4>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Gueorguieva R, Kranzler HR, Zhang H, Cramer J, Rosenheck R, Krystal JH. Opioid receptor gene (OPRM1, OPRK1, and OPRD1) variants and response to naltrexone treatment for alcohol dependence: results from the VA Cooperative Study. Alcohol. Clin. Exp. Res. 2007;31:555–563. doi: 10.1111/j.1530-0277.2007.00339.x. [DOI] [PubMed] [Google Scholar]
- Hall FS, Goeb M, Li XF, Sora I, Uhl GR. mu-Opioid receptor knockout mice display reduced cocaine conditioned place preference but enhanced sensitization of cocaine-induced locomotion. Brain Res. 2004;121:123–130. doi: 10.1016/j.molbrainres.2003.10.024. [DOI] [PubMed] [Google Scholar]
- Herz A. Opioid reward mechanisms: a key role in drug abuse? Can. J. Physiol. Pharmacol. 1998;76:252–258. doi: 10.1139/cjpp-76-3-252. [DOI] [PubMed] [Google Scholar]
- Hoehe MR, Kopke K, Wendel B, Rohde K, Flachmeier C, Kidd KK, Berrettini WH, Church GM. Sequence variability and candidate gene analysis in complex disease: association of mu opioid receptor gene variation with substance dependence. Hum. Mol. Genet. 2000;9:2895–2908. doi: 10.1093/hmg/9.19.2895. [DOI] [PubMed] [Google Scholar]
- Hummel M, Schroeder J, Liu-Chen LY, Cowan A, Unterwald EM. An antisense oligodeoxynucleotide to the mu opioid receptor attenuates cocaine-induced behavioral sensitization and reward in mice. Neuroscience. 2006;142:481–491. doi: 10.1016/j.neuroscience.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Jutkiewicz EM. The antidepressant -like effects of delta-opioid receptor agonists. Mol. Interv. 2006;6:162–169. doi: 10.1124/mi.6.3.7. [DOI] [PubMed] [Google Scholar]
- Karkowski LM, Prescott CA, Kendler KS. Multivariate assessment of factors influencing illicit substance use in twins from female-female pairs. Am. J. Med. Genet. 2000;96:665–670. [PubMed] [Google Scholar]
- Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am. J. Psychiatry. 2003;160:687–695. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Karkowski LM, Neale MC, Prescott CA. Illicit psychoactive substance use, heavy use, abuse, and dependence in a US population-based sample of male twins. Arch. Gen. Psychiatry. 2000;57:261–269. doi: 10.1001/archpsyc.57.3.261. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA. Cocaine use, abuse and dependence in a population-based sample of female twins. Br. J. Psychiatry. 1998;173:345–350. doi: 10.1192/bjp.173.4.345. [DOI] [PubMed] [Google Scholar]
- Konig M, Zimmer AM, Steiner H, Holmes PV, Crawley JN, Brownstein MJ, Zimmer A. Pain responses, anxiety and aggression in mice deficient in pre-proenkephalin. Nature. 1996;383:535–538. doi: 10.1038/383535a0. [DOI] [PubMed] [Google Scholar]
- Kreek MJ, Bart G, Lilly C, LaForge KS, Nielsen DA. Pharmacogenetics and human molecular genetics of opiate and cocaine addictions and their treatments. Pharmacol. Rev. 2005;57:1–26. doi: 10.1124/pr.57.1.1. [DOI] [PubMed] [Google Scholar]
- Law PY, Wong YH, Loh HH. Molecular mechanisms and regulation of opioid receptor signaling. Annu. Rev. Pharmacol. Toxicol. 2000;40:389–430. doi: 10.1146/annurev.pharmtox.40.1.389. [DOI] [PubMed] [Google Scholar]
- Levran O, Londono D, O'Hara K, Nielsen DA, Peles E, Rotrosen J, Casadonte P, Linzy S, Randesi M, Ott J, Adelson M, Kreek MJ. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav. 2008;7:720–729. doi: 10.1111/j.1601-183X.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh el W, Fann CS, Chang YT, Chang CJ, Cheng AT. Endogenous opioid receptor genes and alcohol dependence among Taiwanese Han. Alcohol. Clin. Exp. Res. 2004;28:15–19. doi: 10.1097/01.ALC.0000106303.41755.B8. [DOI] [PubMed] [Google Scholar]
- Luo X, Zuo L, Kranzler H, Zhang H, Wang S, Gelernter J. Multiple OPR genes influence personality traits in substance dependent and healthy subjects in two American populations. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2008;147B:1028–1039. doi: 10.1002/ajmg.b.30701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin S, Marco E, Biscaia M, Fernandez B, Rubio M, Guaza C, Schmidhammer H, Viveros MP. Involvement of the kappa-opioid receptor in the anxiogenic-like effect of CP 55,940 in male rats. Pharmacol. Biochem. Behav. 2003;74:649–656. doi: 10.1016/s0091-3057(02)01041-9. [DOI] [PubMed] [Google Scholar]
- Matthes HW, Maldonado R, Simonin F, Valverde O, Slowe S, Kitchen I, Befort K, Dierich A, Le Meur M, Dolle P, Tzavara E, Hanoune J, Roques BP, Kieffer BL. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature. 1996;383:819–823. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- Mayer P, Rochlitz H, Rauch E, Rommelspacher H, Hasse HE, Schmidt S, Hollt V. Association between a delta opioid receptor gene polymorphism and heroin dependence in man. Neuro report. 1997;8:2547–2550. doi: 10.1097/00001756-199707280-00025. [DOI] [PubMed] [Google Scholar]
- Merikangas KR, Stolar M, Stevens DE, Goulet J, Preisig MA, Fenton B, Zhang H, O'Malley SS, Rounsaville BJ. Familial transmission of substance use disorders. Arch. Gen. Psychiatry. 1998;55:973–979. doi: 10.1001/archpsyc.55.11.973. [DOI] [PubMed] [Google Scholar]
- Milligan G. Opioid receptors and their interacting proteins. Neuromolecular Med. 2005;7:51–59. doi: 10.1385/NMM:7:1-2:051. [DOI] [PubMed] [Google Scholar]
- Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand FL, Henders AK, Wallace L, Todorov AA, Schrage AJ, Madden PA, Degenhardt L, Martin NG, Montgomery GW. Association of OPRD1 polymorphisms with heroin dependence in a large case-control series. Addict Biol. 2012 doi: 10.1111/j.1369-1600.2012.00445.x. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrine SA, Hoshaw BA, Unterwald EM. Delta opioid receptor ligands modulate anxiety-like behaviors in the rat. Br. J. Pharmacol. 2006;147:864–872. doi: 10.1038/sj.bjp.0706686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrine SA, Sheikh IS, Nwaneshiudu CA, Schroeder JA, Unterwald EM. Withdrawal from chronic administration of cocaine decreases delta opioid receptor signaling and increases anxiety- and depression-like behaviors in the rat. Neuropharmacology. 2008;54:355–364. doi: 10.1016/j.neuropharm.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan AA, Befort K, Nozaki C, Gaveriaux-Ruff C, Kieffer BL. The delta opioid receptor: an evolving target for the treatment of brain disorders. Trends Pharmacol. Sci. 2011;32:581–590. doi: 10.1016/j.tips.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragnauth A, Schuller A, Morgan M, Chan J, Ogawa S, Pintar J, Bodnar RJ, Pfaff DW. Female preproenkephalin-knockout mice display altered emotional responses. Proc. Natl. Acad. Sci. U S A. 2001;98:1958–1963. doi: 10.1073/pnas.041598498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxon AJ, Oreskovich MR, Brkanac Z. Genetic determinants of addiction to opioids and cocaine. Harvard Rev. Psychiatry. 2005;13:218–232. doi: 10.1080/10673220500243364. [DOI] [PubMed] [Google Scholar]
- Smith RJ, Doyle GA, Han AM, Crowley JJ, Oslin DW, Patkar AA, Mannelli P, Demaria PA, Jr, O'Brien CP, Berrettini WH. Novel exonic mu-opioid receptor gene (OPRM1) polymorphisms not associated with opioid dependence. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2005;133B:105–109. doi: 10.1002/ajmg.b.30105. [DOI] [PubMed] [Google Scholar]
- Sora I, Takahashi N, Funada M, Ujike H, Revay RS, Donovan DM, Miner LL, Uhl GR. Opiate receptor knockout mice define mu receptor roles in endogenous nociceptive responses and morphineinduced analgesia. Proc. Natl. Acad. Sci. U S A. 1997;94:1544–1549. doi: 10.1073/pnas.94.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto CY, Tang NL, Lee DT, Stadlin A. Association between mu opioid receptor gene polymorphisms and Chinese heroin addicts. Neuroreport. 2001;12:1103–1106. doi: 10.1097/00001756-200105080-00011. [DOI] [PubMed] [Google Scholar]
- Tan EC, Tan CH, Karupathivan U, Yap EP. Mu opioid receptor gene polymorphisms and heroin dependence in Asian populations. Neuroreport. 2003;14:569–572. doi: 10.1097/00001756-200303240-00008. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch. Gen. Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- Xu K, Liu XH, Nagarajan S, Gu XY, Goldman D. Relationship of the delta-opioid receptor gene to heroin abuse in a large Chinese case/control sample. Am. J. Med. Genet. 2002;110:45–50. doi: 10.1002/ajmg.10374. [DOI] [PubMed] [Google Scholar]
- Xuei X, Flury-Wetherill L, Bierut L, Dick D, Nurnberger J, Jr, Foroud T, Edenberg HJ. The opioid system in alcohol and drug dependence: family-based association study. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2007;144B:877–884. doi: 10.1002/ajmg.b.30531. [DOI] [PubMed] [Google Scholar]
- Yuferov V, Levran O, Proudnikov D, Nielsen DA, Kreek MJ. Search for genetic markers and functional variants involved in the development of opiate and cocaine addiction and treatment. Ann. NY Acad. Sci. 2010;1187:184–207. doi: 10.1111/j.1749-6632.2009.05275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki PA, Bilsky EJ, Vanderah TW, Lai J, Evans CJ, Porreca F. Opioid receptor types and subtypes: the delta receptor as a model. Annu. Rev. harmacol. Toxicol. 1996;36:379–401. doi: 10.1146/annurev.pa.36.040196.002115. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol. Psychiatry. 2008;13:531–543. doi: 10.1038/sj.mp.4002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Luo X, Kranzler HR, Lappalainen J, Yang BZ, Krupitsky E, Zvartau E, Gelernter J. Association between two mu-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum. Mol. Genet. 2006;15:807–819. doi: 10.1093/hmg/ddl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, King MA, Schuller AG, Nitsche JF, Reidl M, Elde RP, Unterwald E, Pasternak GW, Pintar JE. Retention of supraspinal delta-like analgesia and loss of morphine tolerance in delta opioid receptor knockout mice. Neuron. 1999;24:243–252. doi: 10.1016/s0896-6273(00)80836-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.