Abstract
Appetitive behaviors require complex decision-making, involving the integration of environmental stimuli and physiological needs. C. elegans mate searching is a male-specific exploratory behavior regulated by two competing needs: food versus reproductive appetite. Here we show that the Pigment Dispersing Factor Receptor (PDFR-1) modulates the circuit that encodes the male reproductive drive promoting male exploration upon mate-deprivation. PDFR-1 and its ligand PDF-1 stimulate mate searching in the male but not in the hermaphrodite. pdf-1 is required in the gender-shared interneuron AIM and the receptor acts in internal and external environment-sensing neurons of the shared nervous system (URY, PQR and PHA) to produce mate-searching behavior. Thus, the pdf-1/pdfr-1 pathway functions in non sex-specific neurons to produce a male-specific, goal-oriented exploratory behavior. Our results indicate that secretin neuropeptidergic signaling plays an ancient role in regulating motivational internal states.
Introduction
Biological drives are internal physiological states that produce goal oriented behaviors which are critical for survival and reproduction 1. Drives shape behavioral decisions according to the animal’s physiological needs to maintain homeostasis. In turn, the physiological needs of the animal are determined by ecological niche 2,3 gender 4 and genetic make up 5. Often, animals need to choose between competing needs and prioritize one drive over another. Therefore, when faced with identical stimuli, animals may respond differently based on previous experience and physiological states. The cellular and molecular basis of behavioral choice under competing needs is only beginning to be elucidated.
C. elegans mate searching behavior is an example of such a goal-orientated behavior that is shaped by at least two competing internal physiological needs, feeding and reproduction 6,7. Well-fed isolated males leave a plentiful source of food to explore their environment. In contrast, when mates are present on the food, male exploratory behavior is suppressed and males remain at the food source. A period of starvation also suppresses mate-searching behavior in the absence of mates. Internal signals that indicate the nutritional and reproductive status of the male are conveyed through an insulin-like signaling pathway and a steroid nuclear hormone receptor respectively 6,8. External environmental cues, i.e. food and mates, are sensed through two independent circuits that include the core chemosensory amphid neurons in the head and the male specific ray neurons in the tail respectively. In the absence of mates, ray neurons promote exploration away from food, and food-sensing amphid neurons inhibit exploration away from food 7. At the mechanistic level, the switch from exploration to retention with a mate is achieved by regulating the characteristics of locomotion, such as the frequency of reversals and high angle turns, upon exiting the edge of the food lawn 7. The molecules and circuitry controlling mate-searching behavior are only partially known.
Neuropeptides are molecular indicators of internal states and important modulators of mood and appetitive behaviors in all animals. Here we show that pigment dispersing factor receptor (PDFR-1), a member of the secretin family of G protein-coupled receptors, and its neuropeptide ligand PDF-1 9,10 are major regulators of mate-searching behavior in C. elegans. PDF-1/PDFR-1 signaling modulates the circuit that conveys the male reproductive drive to explore. The lack of exploratory behavior by pdfr-1 mutant males reflects an imbalance in the relative contribution of the circuits regulating a distributed neural network for exploration. Moreover, the PDF-1/PDFR-1 pathway functions in a discrete non sex-specific neuronal circuit within a sexually dimorphic neural network for navigation.
Results
PDF-1 neuropeptide signaling stimulates mate searching
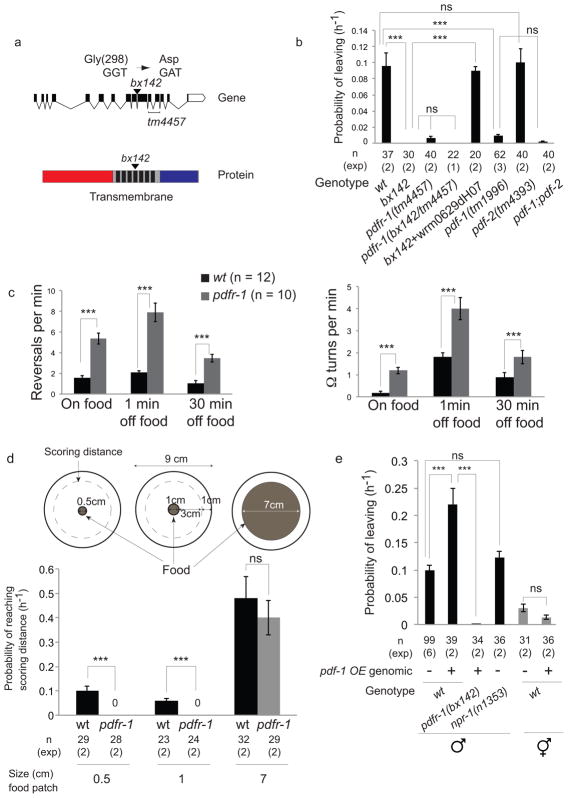
In order to identify molecular pathways that regulate male mate-searching behavior, we screened for leaving assay defective (Las) mutant males that remain on food in the absence of mates. The Las mutation bx142 was mapped to the pdfr-1 locus on chromosome III using single nucleotide polymorphisms 11, two point genetic mapping and genetic complementation tests with a series of chromosomal deficiencies 12,13 (Supplementary Fig. 1a). bx142 introduces a missense mutation, which results in a G298D substitution within the fourth trans-membrane domain of the PDFR-1 G protein-coupled receptor (Fig. 1a). A pdfr-1 deletion allele, tm4457, displayed the same Las phenotype and failed to complement bx142 (Fig. 1b). The bx142 phenotype was rescued with a genomic fragment containing the pdfr-1 locus (Fig. 1b). Thus, bx142 is a loss of function allele of pdfr-1.
Figure 1. PDF-1 neuropeptide signaling stimulates mate searching.
(a) Genomic and protein domain structures of pdfr-1. (Top) Exons (black boxes) and introns (lines) of the pdfr-1 locus; (bottom) extracellular (red), 7 trans-membrane domains (black) and intracellular (blue) regions are shown. The changes in codon and amino acid produced by the bx142 mutation are indicated.
(b) Graph shows PL (probability of leaving food) values of wild type (wt), pdfr-1 (bx142) and (tm4457) mutants and mutants in the PDFR-1 ligands pdf-1(tm1996) and pdf-2(tm4393). The pdfr-1(tm4457) allele fails to complement bx142 indicating that the two mutations affect the same locus. The fosmid wrm0629dH07 contains the pdfr-1 locus. Error bars indicate SEM. n indicates total number of animals tested; (exp) indicates the number of independent population-based experiments. Maximum likelihood statistical analysis was used to compare PL values, ***p<0.001, ns: no statistically significant difference (p≥0.05).
(c) Graphs show the frequency of reversals and high angle turns (Ω turns) performed by wt and pdfr-1(bx142) mutant males during 5 minutes. Locomotion was observed on food and at two different time points off food: 1 minute after removal from food and 30 minutes after removal from food. n indicates total number of animals tested. Error bars indicate SEM. Statistical analysis was performed using Mann-Whitney test, ***p<0.001.
(d) Graphs show the rate at which wild-type (wt) and pdfr-1(bx142) males travel 3.5 cm, from the starting point at the center of the plate within a food patch to the scoring distance 1 cm away from the edge of the assay plate. This was calculated as in the leaving assay, as PL values (probability of reaching the scoring distance). Error bars indicate SEM. n indicates total number of animals tested; (exp) indicates the number of independent population-based experiments. Maximum likelihood statistical analysis was used to compare PL values (probability of reaching the scoring distance), ***p<0.001, ns: no statistically significant difference (p≥0.05).
(e) Graph shows PL values of wild-type and pdfr-1(bx142) mutant males and wild-type hermaphrodites with or without the pdf-1 genomic overexpression (pdf-1 OE) transgene. npr-1(n1353) mutant males display rates of mate-searching behavior similar to wild-type males. Error bars indicate SEM; n indicates total number of animals tested; (exp) indicates the number of independent population-based experiments. Maximum likelihood statistical analysis was used to compare PL values, ***p<0.001, ns: no statistically significant difference (p≥0.05).
Two genes have been identified in C. elegans that encode neuropeptide ligands for PDFR-1, pdf-1 and pdf-2 10. A pdf-1 null allele, tm1996, was Las, while pdf-2(tm4393) males displayed wild-type leaving behavior (Fig. 1b).
pdfr-1 and pdf-1 mutant males have lost their drive to explore away from a food source (Supplementary Fig. 1b). Consistent with a suppression of mate-searching behavior (i.e. male exploration away from food), pdfr-1 mutants produce an increase in the frequency of both reversals and high angle turns (omega turns) compared to wild-type males (Fig. 1c). In addition, pdfr-1 mutant males display slow movement on food due to a decrease in the frequency of body bends (propagation of the sinusoidal wave) (Supplementary Fig. 1c). The slow movement phenotype can be dissociated genetically from the defects in mate-searching behavior indicating that the slow movement phenotype is not responsible for the mate-searching defect (Supplementary Fig. 1c and d).
The lack of mate-searching behavior by pdfr-1 males is not the result of a general locomotion defect. pdfr-1 males travel as far as the food edge regardless of the size of the food lawn. If the food area is restricted to 1 or 0.5 cm diameter, pdfr-1 males, unlike wild-type males, only travel 1 or 0.5 cm, even after 24 hours of assay (Fig. 1d). In striking contrast, pdfr-1 males can travel the scoring distance of the leaving assay plate (3.5 cm radius) at a rate similar to wild-type males if the area is covered with food (Fig. 1d). Thus, pdfr-1 mutants display a specific defect in their ability to leave food and hence, in mate-searching behavior.
pdf-1 and pdfr-1 mutants can successfully mate, although they do not respond as avidly as wild-type males to mate contact (Supplementary Fig. 1e). Thus, pdfr-1 regulates appetitive, but not consummatory, reproductive behaviors in C. elegans males.
pdf-1 acts in a sexually dimorphic manner. Mate-searching behavior is a male-specific exploratory behavior 6. However, pdf-1 hermaphrodites also display similar locomotion defects, such as increased frequency of reversals and slow locomotion 10. We asked whether over-expression of pdf-1 from a transgene under the control of the endogenous promoter (pdf-1 OE) in wild-type hermaphrodites could cause them to behave like wild-type males in the leaving assay. In wild-type males, pdf-1 OE caused a two-fold increase in the rate of leaving behavior that was dependent on the receptor, pdfr-1 (Fig. 1e). In contrast, pdf-1 OE did not cause wild-type hermaphrodites to produce leaving behavior (Fig. 1e), despite rescuing the locomotion defects of pdf-1(tm1996) mutant hermaphrodites (data not shown). Male-specific pdf-1-expressing neurons are not required for the production of male mate-searching behavior (see below Fig. 5e). This indicates that pdf-1 acts in gender-shared neurons through a sexually dimorphic circuit for navigation to produce mate-searching behavior in males.
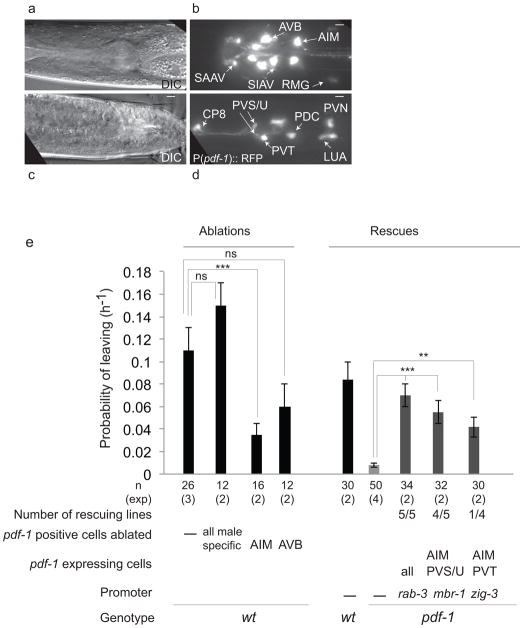
Figure 5. The interneuron AIM is a source of PDF-1 for mate-searching behavior.
(a–d) Expression of P(pdf-1)::RFP in adult males. (a and c) DIC images, (b and d) fluorescence images of neurons in the head (b) and the tail and posterior ventral cord (d). All images show a ventral view, anterior to the left. Scale bars, 5 μm.
(e) Effects on mate-searching behavior (PL values) of ablation of all pdf-1-expressing male-specific neurons, AVB or AIM in wild-type males. Rescue of mate-searching behavior in pdf-1(tm1996) mutants by expression of a pdf-1 cDNA in AIM. Error bars indicate SEM; n indicates total number of animals tested; (exp) indicates the number of independent population-based experiments. Maximum likelihood statistical analysis was used to compare PL values, ***p<0.001, **p<0.01, ns: no statistically significant difference (p≥0.05).
Hermaphrodites of some wild isolates of C. elegans, explore away from food albeit at much shorter distances than males 14. Hermaphrodite exploration away from food is a food-searching strategy when animal density is high and food resources are depleting 15. Hermaphrodite food-leaving behavior is associated with polymorphisms in the npr-1 and tyra-3 genes, which encode a neuropeptide Y receptor and a tyramine receptor respectively 14,16. Hermaphrodites with reduction-of-function mutations in the npr-1 gene display high rates of food-leaving behavior. We asked whether pdf-1 OE could increase the levels of exploration in wild-type hermaphrodites in the hermaphrodite food-leaving assay 14. In contrast to npr-1 mutant hermaphrodites, hermaphrodites over-expressing pdf-1 did not significantly increase exploration levels (Supplementary Fig. 1f). This indicates that the pdf-1/pdfr-1 pathway functions instructively specifically in males to produce exploration away from food. Furthermore, although reduction-in-function mutations in npr-1 have dramatic effects on hermaphrodite food-leaving behavior they have no significant effects on male mate-searching behavior (Fig. 1e). Combined, these data are consistent with the idea that food-leaving behavior in males and hermaphrodites are two distinct exploratory behaviors with different genetic contributions.
The PDFR-1 pathway does not signal nutritional status
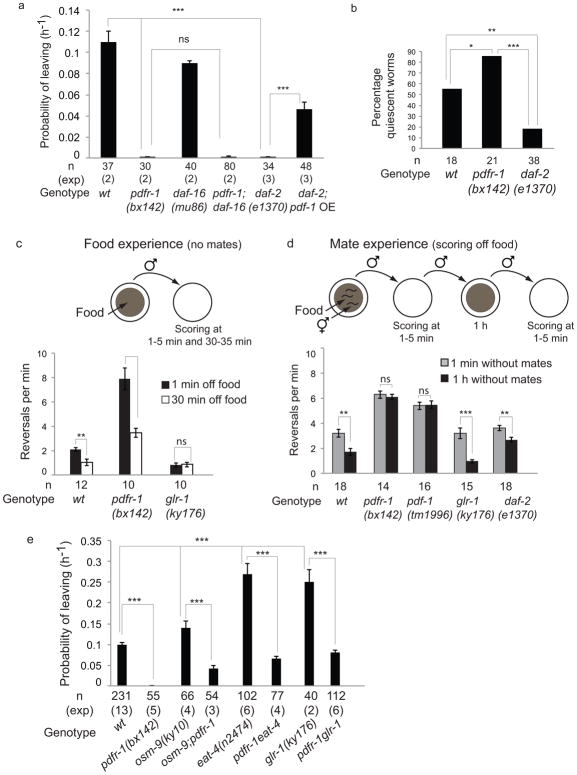
The decision to explore away from food depends on two competing needs, feeding and reproduction. A period of starvation suppresses mate-searching behavior 6. Thus, the lack of mate-searching behavior in pdfr-1 males may reflect defects in a pathway that signals the nutritional status of the animal or a pathway that signals reproductive drive. In vertebrates, orthologs of PDFR-1 have an important role regulating energy homeostasis and insulin secretion 17. Furthermore, mate-searching behavior is regulated by internal signals indicating the nutritional status of the animal and mediated in part through insulin signaling 6. DAF-2 is the main insulin receptor in C. elegans 18 and like pdfr-1 mutants, daf-2(e1370) mutant males are severely defective in mate searching behavior 6 (Fig. 2a). DAF-2 acts via inactivation of the forkhead transcription factor DAF-1619. Consistent with this, the mate-searching defects of daf-2 mutant males are suppressed by daf-16 loss of function mutations 6. If pdfr-1 exerts its effect on mate-searching behavior through the regulation of the DAF-2/DAF-16 insulin pathway, then we would expect the daf-16(mu86) null mutation to suppress the phenotype of pdfr-1(bx142) mutants. However, pdfr-1(bx142);daf-16(mu86) double mutants displayed the same mate-searching defects as pdfr-1(bx142) single mutants (Fig. 2A). Moreover, overexpression of the ligand PDF-1 in daf-2(e1370) mutants can rescue the mate searching defects of daf-2 mutants (Fig. 2a). These results indicate that the PDF-1/PDFR-1 pathway acts downstream or independently of the DAF-2/DAF-16 insulin pathway to regulate mate-searching behavior.
Figure 2. The PDF-1/PDFR-1 pathway stimulates dispersal upon mate deprivation.
(a) PL values are shown for mutants in the insulin pathway: daf-16 encodes a forkhead transcription factor inhibited by insulin signaling and daf-2 encodes an insulin receptor. Error bars indicate SEM. n indicates total number of animals tested; (exp) indicates the number of independent population-based experiments. Maximum likelihood statistical analysis was used to compare PL values, ***p<0.001, ns: no statistically significant difference (p≥0.05).
(b) Percentage of quiescent worms in nutritionally high food (E. coli HB101). n indicates number of animals tested. χ2 test was used for statistical analysis, ***p<0.001, **p<0.01, *p<0.05.
(c) Frequency of reversals (during 5 minutes) was measured 1 minute and 30 minutes after removal from food. (d) Frequency of reversals (during 5 minutes) was measured off food 1 minute and 1 hour after removal from mates. Note that values in the first column of (c) and the second column of (d) are equivalent. Variability exists due to experiments being performed in different days for (c) and (d). In (c) and (d), n indicates number of animals tested, error bars indicate SEM and Mann-Whitney test was used for statistical analysis, ***p<0.001, **p<0.01, ns: no statistically significant difference (p≥0.05).
(e) PL values are shown for mutants in the food-sensing pathway. osm-9 encodes a TRPV channel expressed in amphid chemosensory neurons; eat-4 encodes a glutamate vesicular transporter expressed in amphid neurons; glr-1 encodes an AMPA glutamate receptor expressed in amphid interneurons. Error bars indicate SEM; n indicates total number of animals tested; (exp) indicates the number of independent population-based experiments. Maximum likelihood statistical analysis was used to compare PL values, ***p<0.001.
Two pieces of evidence support a model in which PDF-1/PDFR-1 and DAF-2 act in different pathways (reproductive drive and nutritional status respectively) to regulate mate-searching behavior. First, unlike daf-2 mutants, pdfr-1 mutants underwent behavioral quiescence in nutritionally high food (Fig. 2b), a behavior associated with nutritional satiety and characterized by the cessation of both movement and pharyngeal pumping 20. Thus, unlike daf-2 mutants, pdfr-1 mutants reached satiety and did not appear nutritionally deprived. Second, daf-2 males, like wild-type males, modulated their patterns of locomotion based on previous experience with a mate (see below) (Fig. 2d). In contrast, pdfr-1 males did not modulate their patterns of locomotion after mate deprivation (see below) (Fig. 2d). Thus, although nutritional status impacts the levels of reproductive drive, our data does not support a role for pdfr-1 in the signaling of overall nutritional state. Rather, our results are most consistent with a role for pdfr-1 in the signaling of reproductive drive.
PDF-1 signaling limits local search upon mate deprivation
C. elegans explores its environment in an optimal manner by modulating locomotion patterns in response to prior sensory experience. Hermaphrodites respond to loss of contact with a food source by increasing reversal frequency and turns, a behavior known as ARS (area restricted search) 21–23. ARS behavior tends to bring them back to the food. In hermaphrodites, ARS behavior depends on glutamate signaling from food-sensing chemosensory neurons and the glutamate receptor glr-1 21,24. After approximately 30 minutes, frequency of reversals and turns decreases, allowing dispersal in search of a new food source23.
We have shown previously that presence of mates affects the behavior of males at the food edge, inhibiting them from fully exiting the lawn. This modulation of exploration is an important component of the behavioral effect on males induced by contact experience with a mate 7. Here we examined whether loss of a source of food with or without mates induced the expression of ARS behavior in males. Second, we asked whether the pdf-1/pdfr-1 pathway modulated the response to loss of a source of food or loss of a source of mates.
Upon removal from a food source without mates, males expressed ARS behavior initially and began to disperse after 30 minutes off food (Fig. 2c). In contrast, glr-1 mutant males did not display ARS behavior upon removal from food and dispersed immediately showing that the response of males to loss of a food source is regulated similarly to that of hermaphrodites 21. Although pdfr-1 males displayed higher absolute levels of reversals compared to wild-type males, reversals were decreased two-fold after 30 minutes since last food exposure, as in wild-type males (Fig. 2c). This indicates that pdfr-1 does not function in the pathway that modulates reversal frequency in response to food experience, and that pdfr-1 mutants can reduce reversals.
Upon removal of a male from a food lawn with mates, the frequency of reversals was enhanced two-fold compared to removal from a lawn without mates (Fig 2d). Hence contact-experience with a mate enhanced ARS. The frequency of reversals decreased one hour after last experience with a mate and this decrease required the PDF-1/PDFR-1 pathway (Fig 2d). In contrast, reduction of ARS behavior upon mate-deprivation did not require neither the glutamate pathway (glr-1 background) nor the insulin signaling pathway (daf-2 background). Thus PDF-1 signaling functioned specifically to modulate ARS in response to mate experience, whereas components of the food response pathway were not required for this modulation.
We conclude that pdf-1/pdfr-1 and glr-1 act in parallel pathways to regulate ARS and dispersal in response to sensory experience: glr-1 promotes reversals upon recent food experience and pdfr-1 suppresses reversals upon mate deprivation. The relative contribution of each pathway to a distributed navigation circuit will determine the ability of males to leave food and explore in search of mates. Consistent with this interpretation, males with mutations in the food-searching pathway had a higher tendency to explore away from food than wild-type males and their levels of exploration were dependent on the function of pdfr-1 (Fig. 2e).
Mate-sensing neurons modulate the effects of PDF signaling
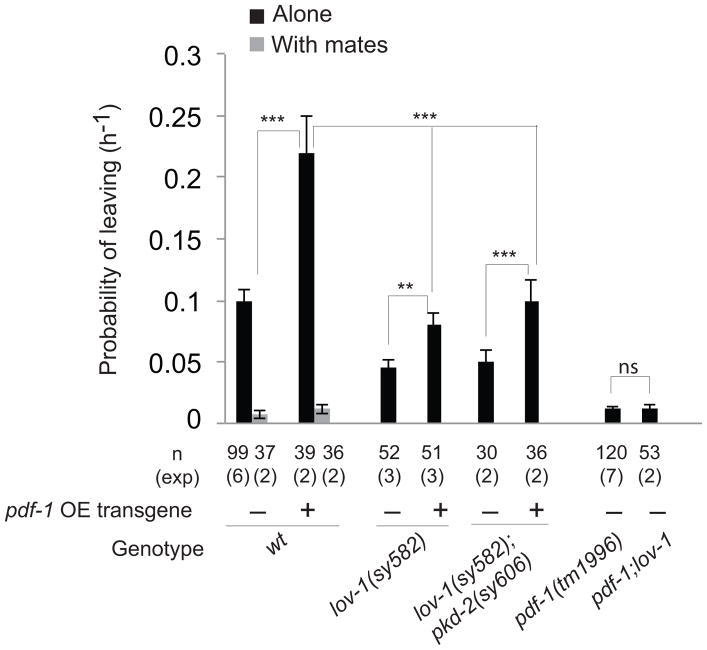
Wild-type males that have recently been in contact with a mate do not display mate-searching behavior 6,7 (Fig. 3). Introduction of a pdf-1 transgene into wild type males resulted in a two-fold increase in mate-searching activity that was completely suppressed by the presence of mates (Fig. 3). This indicates that previous experience with a mate completely inhibits the effects of pdf-1 signaling, and further supports the idea that pdf-1 acts within the circuit that signals reproductive drive after mate deprivation.
Figure 3. Mate sensation and ray activity modulate the effects of PDF-1 signaling.
The effects on mate-searching behavior (PL values) of pdf-1 overexpression (pdf-1 OE) in wild-type males with and without mates and in mutants with defects in B-type ray neuron function are shown. lov-1 and pkd-2 encode TRP polycystins required for ray neuron function. Error bars indicate SEM; n indicates total number of animals tested; (exp) indicates the number of independent population-based experiments. Maximum likelihood statistical analysis was used to compare PL values. ***p<0.001, **p<0.01, ns: no statistically significant difference (p≥0.05).
Mate-sensing male ray neurons stimulate mate-searching behavior by reducing reversals upon exiting food after mate deprivation 7. We asked whether ray neurons modulate the effects of pdf-1 signaling on mate-searching behavior. We first tested whether ray neurons were required for increased levels of mate-searching behavior produced by pdf-1 overexpression. To disrupt the function of a sub-set of ray neurons (B-type ray neurons) we used mutations in the TRP polycystin genes lov-1 and pkd-2 25,26. Disruption of B-type ray neurons reduced the levels of mate-searching behavior produced by the pdf-1 transgene although not to the levels of lov-1 and lov-1; pkd-2 mutants (Fig. 3). Thus, pdf-1 signaling has both, B-type ray neuron-dependent and independent effects on mate-searching behavior.
Reducing ray neuron function in pdf-1 mutants, by introducing the lov-1 mutation, did not enhance the defects in mate-searching behavior (Fig. 3). Together, these results are consistent with the interpretation that PDF-1/PDFR-1 signaling sets the level of reproductive drive for mate-searching behavior, which is modulated by the activity of ray neurons and mate-sensation. In a context where the drive to explore is low, such as in pdf-1 or pdfr-1 mutants, ray function becomes irrelevant.
PDFR-1 acts in gender-shared sensory neurons
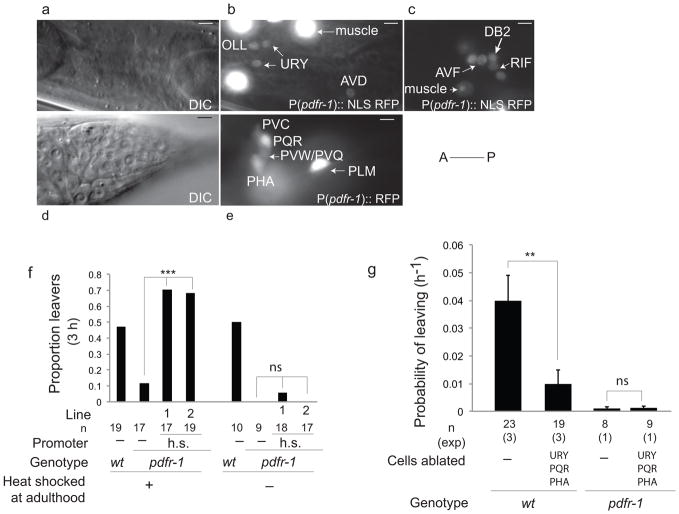
To determine the site of action of PDFR-1 we generated a variety of transcriptional reporters and rescue constructs. Transcriptional reporters containing 3kb of the endogenous promoter upstream of the translational start site show expression in neurons and body wall muscle (Fig. 4a–e) and show similar expression in both males and hermaphrodites. Co-localization with other reporters and anatomical criteria enabled identification of the expressing neurons as the ciliated sensory neurons OLL, PHA and PQR; the non-ciliated sensory neurons URY and URX; the touch receptor neurons (ALM, PLM, AVM and PVM); interneurons in the retro-vesicular ganglion RIF and AVF; the command interneurons AVD and PVC; the ring motor neurons RMED and RMEV and two other neurons tentatively identified as PVQ or PVW and DB2 (Fig. 4a–e). No expression was observed in amphid or male-specific neurons.
Figure 4. pdfr-1 is required in sensory neurons URY, PQR and PHA to produce mate-searching behavior.
(a–e) Expression of P(pdfr-1)::RFP in males at the L4 stage. (a and d) DIC images, (b, c and e) fluorescence images. Neurons in the head (a, b) and the retro-vesicular ganglion (c) labeled with a pdfr-1::NLS RFP reporter. (d and e), neurons in the tail labeled with a pdfr-1::RFP reporter. Anterior is to the left in all images. (a, b, d and e) lateral views; (c) ventral view. Scale bars, 5 μm
(f) Rescue of mate-searching behavior in pdfr-1(bx142) mutants with a heat-shock inducible promoter driving isoforms b and d. Lines indicate independent arrays of the transgene. Graphs show the proportion of males that left food after 3 hours of leaving assay. n indicates number of animals tested; χ2 test was used for statistical analysis, ***p<0.001, ns: no statistically significant difference (p≥0.05).
(g) Effects of laser ablation of URY, PQR and PHA on mate-searching behavior in wild type and pdfr-1(bx142) males. The rate of leaving in non-ablated control wild-type males was slightly reduced by the presence of the transgenes used to identify the target neurons. Error bars indicate SEM; n indicates total number of animals tested; (exp) indicates the number of independent population-based experiments. Maximum likelihood statistical analysis was used to compare PL values. **p<0.01, ns: no statistically significant difference (p≥0.05).
The defects in mate-searching behavior were fully rescued with constructs that express pdfr-1 cDNAs under the control of the 3kb pdfr-1 promoter. Three different isoforms (a, b and c) encoded by the pdfr-1 locus have been identified 9,10. From a mixed stage and mixed gender mRNA pool we isolated five isoforms: a, b, c and two new isoforms d and e. The five isoforms differ in their extracellular and intracellular domains (Supplementary Fig. 2a). Simultaneous expression of both isoforms b and d under the 3kb pdfr-1 promoter fully rescued the defects in mate searching behavior of pdfr-1 mutants (Table 1 and Supplementary Fig. 2b). Having obtained full rescue with isoforms b and d together, we did not test any other isoform combinations and all subsequent rescue experiments were performed with both pdfr-1 b and d isoforms.
Table 1.
Rescue and expression of transgenes driving pdfr-1 isoforms b and d under different promoters. Black and white circles indicate positive or negative expression in that cell respectively; a mixed circle indicates variable expression.
| Promoter |
pdfr-1 positive cells
|
other neurons | Rescue | Lines | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| muscle | OLL | URY | RMED/V | URX | AVD | TRN | RIF | AVF | DB2 | PVC | PQR | PHA | PVW/PVQ | ||||
|
| |||||||||||||||||
| pdfr-1 (3kb) | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ○ | +++ | 4/4 |
| rab-3 | ○ | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | +++ | 2/4 |
| pdfr-1 (2kb proximal) | ● | ○ | ● | ◑ | ◑ | ○ | ● | ○ | ● | ○ | ○ | ◑ | ◑ | ◑ | ○ | ++ | 2/7 |
| glr-4 | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ? | ○ | ● | ○ | ○ | ● | +++ | 4/4 |
| npr-1 | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ● | +++ | 3/4 |
| pdfr-1 (1kb prox) + osm-6 | ● | ● | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ● | ++ | 2/2 |
| osm-6 | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ○ | ● | + | 1/5 |
| pdfr-1 (1kb proximal) | ● | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | − | 5/5 |
| pdfr-1 (1kb medial) | ○ | ● | ○ | ● | ◑ | ● | ● | ○ | ● | ○ | ● | ○ | ○ | ● | ○ | − | 4/4 |
| pdfr-1 (1kb distal) | ○ | ● | ○ | ○ | ● | ● | ● | ● | ○ | ● | ○ | ○ | ○ | ○ | ○ | − | 5/5 |
| gcy-32 | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ● | − | 5/5 |
| pdfr-1 (1kb prox) + gcy-32 | ● | ○ | ● | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ● | − | 4/4 |
| glr-1 | ○ | ○ | ● | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ? | ● | − | 4/4 |
| eat-4 | ○ | ● | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ? | ● | − | 4/4 |
| tol-1 | ○ | ○ | ● | ○ | ○ | ○ | ● | ? | ? | ○ | ○ | ○ | ○ | ○ | ? | − | 3/3 |
| mec-17 | ○ | ○ | ○ | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | − | 4/4 |
| unc-7 + unc-4 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ● | ● | ? | ○ | ○ | ○ | ○ | ● | − | 4/4 |
The number of + indicates the levels of rescue: +++ indicates mate-searching/probability of leaving (PL)not significantly different from wild type (PL=0.1 and p≥0.05); ++ indicates 0.1>PL>0.045; + indicates PL=0.025; − indicates PL<0.025.
The proportion of lines that displayed the levels of PL assigned is indicated.
To test whether PDFR-1 is required during development or in adulthood, we drove expression of pdfr-1 cDNA under a heat-shock-inducible promoter 27. Induction of the heat-shock promoter at adulthood fully rescued the mate searching defects in both lines tested, indicating that PDFR-1 is not required during development for male exploratory behavior (Fig. 4f). The rescuing effects produced by heat-shock induction were extinguished after five hours, indicating that PDFR-1 needs to be regularly produced in the adult to stimulate mate-searching behavior.
Expression of pdfr-1 pan-neuronally (with a rab-3 promoter) restored mate-searching behavior in pdfr-1 mutants (Table 1 and Supplementary Fig. 2b). In contrast, expression in muscle (with a 1kb proximal pdfr-1 promoter region) did not (Table 1 and Supplementary Fig. 2b). Together, these results indicate that PDFR-1 functions in neurons in the adult male to produce mate-searching behavior.
To determine which of the pdfr-1-expressing neurons are responsible for the production of mate searching we performed a series of rescue experiments with smaller regions of the pdfr-1 promoter and with heterologous promoters (Table 1 and Supplementary Fig. 2b). This analysis suggested that pdfr-1 is required in three classes of neurons, URY (4 neurons), PQR (1 neuron) and PHA (2 neurons) to elicit reproductive drive (Table 1 and Supplementary Fig. 2b). To confirm this hypothesis, we restored pdfr-1 expression in all 3 classes of neurons by crossing together two arrays that individually produced a minimal rescue of mate-searching behavior: the pdfr-1 1kb minimal promoter (URY) and the osm-6 promoter (PQR, PHA, OLL and other pdfr-1-negative ciliated neurons). This experiment significantly rescued mate-searching behavior in pdfr-1 mutants (Table 1 and Supplementary Fig. 2b). These results implicate the sensory neurons URY, PQR, which sense the internal environment of the animal, and PHA, which is exposed to the outside, as part of the circuit where pdfr-1 functions for the production of mate-searching behavior.
Laser ablation of all three classes of neurons, URY, PQR and PHA, together, significantly decreased mate-searching behavior in wild-type males (Fig. 4g). These results indicate that URY, PQR and PHA are necessary for the production of wild-type levels of mate-searching behavior and suggest that pdfr-1 stimulates mate-searching behavior through positive regulation of the activity of these neurons.
The interneuron pair AIM is a source of PDF-1
To determine the relevant source of the ligand PDF-1 for mate searching, we expressed a pdf-1 genomic fragment or a pdf-1 cDNA under an endogenous promoter containing 3.4 kb upstream of the translational start site. Both of these constructs fully rescued the mate searching defects of pdf-1 mutants (Supplementary Fig. 3). This 3.4 kb promoter region drives expression in both sexes in the head interneurons SAAV, SAAD, SIAV, AVB, AIM, RMG and two other unidentified ciliated neurons (possibly ASK and AFD) (Fig. 5a–b). In the tail, pdf-1 was consistently expressed in interneurons PVN, LUA, PVT and the pair PVS and PVU (called PVP left and right in the hermaphrodite) (Fig. 5c–d). We also saw expression in male specific neurons of the ventral cord CP7, CP8, in PDC and in one unidentified interneuron, possibly PVX, in the pre-anal ganglion (Fig. 5c–d).
pdf-1 acts instructively in males, but not in hermaphrodites, to produce mate-searching behavior (Fig. 1e). Ablation of all pdf-1-positive male specific neurons in wild-type males with a laser micro-beam did not cause any significant reduction in mate-searching behavior (Fig. 5e). Therefore, male-specific neurons are not a necessary source of neuropeptide secretion for the stimulation of mate-searching behavior. In contrast, ablation of the AIM pair caused a significant reduction in mate searching, indicating that AIM interneurons are a component of the mate-searching circuit (Fig. 5e). Expression of pdf-1 in AIM and PVS/U (and other pdf-1 negative neurons) with the mbr-1 promoter 28 or in AIM and PVT (and other pdf-1 negative neurons) with the zig-3 promoter 29 restored mate-searching behavior in pdf-1 mutants (Fig. 5e and Supplementary Fig. 3). These results indicate that AIM is an important source of PDF-1 release for the stimulation of mate-searching behavior.
Taken together, our results demonstrate that the PDF-1/PDFR-1 pathway acts in a sexually dimorphic-manner, yet, functions in neurons that are present in both males and hermaphrodites. This indicates that the PDF-1/PDFR-1 circuit is dimorphic in the functional properties of the neurons and/or in their connectivity.
Discussion
Our work reveals an evolutionarily conserved neuropeptide system as a major regulator of reproductive drive in C. elegans. The risky decision to explore away from a food source in search for mates depends on the balance of two competing drives, feeding and reproduction. Mates and food are sensed through two independent sensory circuits with input to a distributed network for exploration. Our results indicate that the lack of mate-searching behavior in pdfr-1 mutants reflects a reduction in the reproductive drive and corresponding increase in the relative contribution of the food–searching circuit to the network for exploration. pdfr-1 does not appear to function within the food-searching circuit. First, pdfr-1 mutants do not act as though they are nutritionally deprived, and, like wild-type animals, reach satiety in nutritionally high food (Figure 2b). Second, pdfr-1 mutants modulate their locomotion in response to previous food experience (Figure 2c). Third, pdfr-1 expression in the food-sensing circuit with the glr-1 or osm-6 promoters is not sufficient to restore mate-searching behavior (Table 1). pdfr-1 is required in sensory neurons exposed to the internal environment of the animal, URY and PQR and a ciliated sensory neuron exposed to the outside, PHA. Thus the pdf-1/pdfr-1 system appears to modulate a circuit that senses the internal state of the male and antagonizes the food-sensing circuit. This is consistent with a role for neuropeptides as molecular indicators of internal states and modulators of circuit properties and contributions 30.
Gender may be considered another dimension of internal state that shapes behavioral decision-making. Unlike their hermaphrodite counterparts, C. elegans males must find a mate in order to reproduce. Like other physiological needs, reproduction is subject to homeostatic regulation, the drive to reproduce increasing after deprivation and decreasing after experience 31. The pdf-1/pdfr-1 pathway is an important component of the drive for mate-searching behavior after mate deprivation. pdf-1 and pdfr-1 mutants do not produce mate-searching behavior and are unable to modulate their locomotion to disperse after mate-deprivation (Figure 1b and 2d). The effects of PDF-1 signaling are regulated by the activity of mate-sensing ray neurons and are completely suppressed by previous experience with a mate (Figure 3). Our results indicate that the neural circuit for mate-searching behavior that conveys the reproductive drive to explore is under the regulation of internal state-sensing neurons, modulated by the pdf-1/pdfr-1 pathway, and mate-sensing ray neurons. Mate-sensation may block the effects of PDF-1 signaling by regulating the secretion of the neuropeptide ligand or by modulating the activity of the receptor-expressing neurons or downstream targets. Determining the exact mechanism awaits further studies.
The pdf-1/pdfr-1 pathway acts in non sex-specific neurons to produce a male-specific behavior. The sexual dimorphism may lie in the intrinsic functional characteristics of the pdf-1/pdfr-1 neurons or in their connectivity. Indeed, there is evidence for dimorphism in both, molecular expression and connectivity within circuits shared by both males and hermaphrodites 32,33. The wiring diagram of connectivity of the C. elegans male indicates that dimorphic synaptic connections are extensive throughout the shared nervous system and suggests possible sites for integration of the pdfr-1 circuit within the navigation network 33. Both, PHA and PQR synapse directly to AVA, the command interneuron responsible for backward movement. Another possible site of integration are the male-specific EF interneurons, which are post-synaptic to PQR, PHA and ray neurons and are required for the suppression of mate-searching behavior by the presence of mates 7. The connectivity of URY and AIM in the male is not known.
Our work opens new avenues for the study of the secretin family of neuropeptides in circuits regulating appetitive reproductive behaviors. We have shown that the PDF-1/PDFR-1 pathway is an important component of the reproductive drive for mate searching. A role for pigment dispersing factor in the regulation of internal drives appears to be phylogenetically conserved. PDF regulates male sex drive in Drosophila 34. In the monarch butterfly, the circadian pacemaker system, which is regulated by PDF, is involved in the induction of the migratory state 3. In humans, members of the secretin family of neuropeptide receptors, orthologs of pdfr-1 35, have been associated with disorders affecting motivation such as bipolar disorder 36 and post-traumatic stress syndrome 37. Determining how the PDF-1/PDFR-1 system modulates neuronal physiology and circuit properties will reveal the mechanisms by which neuropeptides shape drives and behavioral decision-making.
Materials and methods
Strains used
CB1490, him-5(e1490) was used as the wild-type strain for males; Bristol N2 was used as wild-type hermaphrodites and unc-51(e369) hermaphrodites were used for retention experiments;
EM938, pdfr-1(bx142);him-5(e1490)
PT2248, pdf-1(tm1996);him-5(e1490)
PT2159, pdf-1(tm1996);pdf-2(tm4393);him-5(1490)
PT2177, pdf-2(tm4393);him-5(e1490)
PT2547 pdfr-1(tm4457);him-5(e1490)
PT2367,myEx721[P(pdf-1)3kb::pdf-1genomic + P(unc-122)::gfp]
PT2422, myEx721[P(pdf-1)3kb::pdf-1genomic + P(unc-122)::gfp];him-5(e1490)
PT2364, npr-1(n1353);him-5(e1490)
DA508, npr-1(n1353)
PT2247, glr-1(ky176);him-5(e1490)
PT2176, dpy-17 (e164) pdfr-1(bx142); him-5(e1490)
PT2246, dpy-17(e164)pdfr-1(bx142)glr-1(ky176);him-5(e1490)
PT2260, him-5(e1490);akIs9[Pglr-1:glr-1(A687T)]
PT2548, daf-16(mu86);him-5(e1490),
PT2549, daf-16(mu86);pdfr-1(bx142);
EM814, daf-2(e1370);him-5(e1490);
PT2501, daf-2(e1370);myEx721[P(pdf-1)3kb::pdf-1genomic + P(unc-122)::gfp];him-5(e1490)
PT2350, egl-4(n479);him-5(e1490)
PT839, osm-9(ky10);him-5(e1490)
PT2300, osm-9(ky10);pdfr-1(bx142);him-5(e1490)
PT2193, eat-4(n2474);him-5(e1490)
PT2195, dpy-17(e164)eat-4(n2474)pdfr-1(bx142);him-5(e1490)
PT2479, pkd-2(sy606);lov-1(sy582);him-5(e1490);myEx721[P(pdf-1)3kb::pdf-1genomic + P(unc-122)::gfp]
PT2480, pdf-1(tm1996);lov-1(sy582);him-5(e1490)
PT2481, lov-1(sy582);him-5(e1490);myEx721[P(pdf-1)3kb::pdf-1genomic + P(unc-122)::gfp]
PT2417, pdfr-1(bx142);him-5(e1490);myEx768[P(hsp-16.2)::pdfr-1cDNA isof b+P(hsp-16.2)::pdfr-1cDNA isof d + unc-122::gfp]
PT2315, him-5(e1490); myEx731[P(pdfr-1)3kb::rfp + P(unc-122)::gfp]
PT2351, him-5(e1490);myEx741[P(pdfr-1)3kb::nls rfp + P(unc-122)::gfp]
PT2294, pdf-1(tm1996);him-5(e1490);myEx721[P(pdf-1)3kb::pdf-1genomic + P(unc-122)::gfp]
PT2249, him-5(e1490);myEx696[P(pdf-1)3kb::rfp + P(unc-122)::gfp]
PT2565, IqIs3(osm-6::GFP); him-5(e1490)
PT2493, pdf-1(tm1996);him-5(e1490);myEx783[P(flp-10)::pdf-1cDNA) + P(unc-122)::rfp]
PT2504, pdf-1(tm1996);him-5(e1490);myEx786[P(pdf-1)3kb::pdf-1cDNA) + P(unc-122)::rfp]
PT2282, pdfr-1(bx142);him-5(e1490);myEx709[P(pdfr-1)3kb::pdfr-1 cDNA isof b + P(pdfr-1)3kb::pdfr-1 cDNA isof d + P(unc-122)::gfp]
PT2427, pdfr-1(bx142);him-5(e1490);myEx769[P(pdfr-1)2kb::pdfr-1 cDNA isof b + P(pdfr-1)2kb::pdfr-1 cDNA isof d +P(unc-122)::gfp]
PT2459, pdfr-1(bx142);him-5(e1490);myEx774[P(pdfr-1)1kb distal::pdfr-1cDNA isof b + P(pdfr-1)1kb distal::pdfr-1cDNA isof d + unc-122::gfp]
PT2460, him-5(e1490);myEx775[P(pdfr-1)1kb distal::nls rfp tag + unc-122::gfp]
PT2471, pdfr-1(bx142);him-5(e1490);myEx776[P(pdfr-1)1kb medial::pdfr-1cDNA isof b + P(pdfr-1)1kb medial::pdfr-1cDNA isof d + unc-122::gfp]
PT2472, him-5(e1490);myEx777[P(pdfr-1)1kb medial::nls rfp + unc-122::gfp]
PT2473, pdfr-1(bx142);him-5(e1490);myEx778[P(pdfr-1)1kb proximal::pdfr-1cDNA isof b + P(pdfr-1)1kb proximal::pdfr-1cDNA isof d + unc-122::rfp]
PT2474, him-5(e1490);myEx779[P(pdfr-1)1kb proximal::nls rfp + unc-122::gfp]
PT2356, pdfr-1(bx142);him-5(e1490);myEx745[P(osm-6)::pdfr-1cDNA isof b + P(osm-6)::pdfr-1cDNA isof d + unc-122::gfp]
PT2141, pdfr-1(bx142); him-5(e1490)V;myEx694[P(rab-3)::pdfr-1cDNA isof b + P(rab-3)::pdfr-1cDNA isof d + P(unc-122)::gfp]
PT2346, pdfr-1(bx142);him-5(e1490);myEx739[P(mec-17)::pdfr-1cDNA isof b + P(mec-17)::pdfr-1cDNA isof d + P(unc-122)::gfp]
PT2358, pdfr-1(bx142);him-5(e1490);myEx747[P(eat-4)::pdfr-1cDNA isof b + P(eat-4)::pdfr-1cDNA isof d + P(unc-122)::gfp]
PT2373, pdfr-1(bx142);him-5(e1490);myEx751[P(glr-1)::pdfr-1 cDNA isof b + Pglr-1::isof d + P(unc-122)::gfp]
PT2416, pdfr-1(bx142);him-5(e1490);myEx767[P(unc-4)::pdfr-1cDNA isof b + P(unc-4)::pdfr-1cDNA isof d + P(unc-7b)::pdfr-1cDNA b + P(unc-7b)::pdfr-1cDNA isof d + P(unc-122)::gfp]
PT2475, pdfr-1(bx142);him-5(e1490);myEx780[P(gcy-32)::pdfr-1cDNA isof b + P(gcy-32)::pdfr-1cDNA isof d + P(unc-122)::gfp]
PT2494, pdfr-1(bx142);him-5(e1490);myEx785[P(tol-1)5kb::pdfr-1cDNA isof b + P(tol-1)5kb::pdfr-1cDNA isof d + P(unc-122)::rfp]
Mapping
The bx142 mutation was placed on chromosome III between dpy-17 and unc-32 using single nucleotide polymorphisms 11 and two point genetic mapping. The recessive bx142 mutation was crossed to deficiency strains covering the region between dpy-17 and unc-32. The F1 offspring were tested in the leaving assay. The proportion of animals with the Las(bx142) phenotype was scored. A 1/2 proportion of Las animals in the F1 offspring indicated no complementation. Deficiency strains used:
BC4638, dpy-17(e164) sDF127(s2428)unc-32(e189)III;sDp3(III,f)
BC4634, dpy-17(e164) sDF125(s2424)unc-32(e189)III;sDp3(III,f)
CX2914, nDf16/dpy-17(e164)unc-32(e189)III
A pool of fosmids containing the genomic region between the two genes with known physical positions deleted in sDF125 (left limit, F23F12.3) and nDf16 (right limit, odr-4) was injected into bx142 mutants (3 of 5 lines rescued). Complementation tests with available mutants of genes within that region (acr-5, oig-1, acr-21) and sequencing (srxa-5, srh-40, rbf-1, pdfr-1) revealed a mutation in the coding region of pdfr-1. The bx142 was subsequently rescued by injection of the genomic fragment containing the pdfr-1 locus.
DNA constructs
cDNAs were isolated by RT-PCR from a mixed gender and mixed stage mRNA pool and gene-specific primers:
pdfr-1 cDNAs: 5′ ctgactcatatcatggcggatg 3′ and 5′ tgacacggtgggtttgacaac 3′ or 5′ ttatggagattttgtggagcgattgg3′. The 3′UTR of the unc-54 gene was used in the pdfr-1 cDNA constructs.
pdf-1 cDNA: 5′ atgaacagattcatcatttc3′ and 5′ aacatttgttatgctgaac 3′. The pdf-1 endogenous 3′UTR was used in the constructs. We used a starting coding site 279 nucleotides downstream from the one previously annotated in Wormbase. The pdf-1 genomic region was amplified with primers:
5′ aaaaggttgctttaccgagacgtg 3′ and 5′ gccacttttcacgcttcagca 3′
Rescuing and expression constructs were constructed by PCR fusion 48 and injected at 10 or 15 ng/μl except for P(mbr-1)::pdf-1 and P(zig-3)::pdf-1, which were injected at 50 ng/μl.
The pdfr-1 distal and medial promoter regions (which do not include the proximal region of the pdfr-1 promoter) were built to include the minimal promoter found in pPD95.75 (indicated as capital letters in the primers).
3kb: 5′ cactactttcagcgacctttacttgctc 3′ and 5′ gatatgagtcagtgatgaa 3′
2kb: 5′ caccctccaaatgtcttgaaactga 3′ and 5′ gatatgagtcagtgatgaa 3′
1kb distal: 5′ cactactttcagcgacctttacttgctc 3′ and 5′ TTTGGGTCCTTTGGCCAATCtcagtttcaagacatttgga 3″
1kb medial: 5′ ctaattacttcggacgggtctagacgtt 3′, and 5′ TTTGGGTCCTTTGGCCAATCaccagctttattgcgtgac 3′
1kb proximal: 5′ cacgcaataaagctggttagggaag 3′ and 5′ gatatgagtcagtgatgaa 3′
Primers for heterologous promoters driving the pdfr-1 cDNAs, amplified from a fosmid library:
rab-3: 5′ gcgagttttgactggctttc 3′, 5′ cctgatttcaggcggcggaa 3′
unc-4: 5′ cgacgtcgtccgagattaaagattc 3′, 5′ tttcactttttggaagaag 3′
eat-4: 5′ ggagttcgaaactgcagcaatga 3′, 5′ ggtttctgaaaatgatgatg 3′
glr-4: 5′ caaaaaggcacacttgagctg 3′, 5′ gctgtgtaaaagtttagctc 3′
npr-1: 5′ acgatctgtgtcgcgttttgattt 3′, 5′ ttggcctatgtctgaaattt 3′
gcy-32: 5′ gccatggtgttaatacgtcaagcaa 3′, 5′ tctataatacaatcgtgat 3′
glr-1: 5′ actacgtggagtcgagggctca 3′, 5′ tgtgaatgtgtcagattgg 3′
osm-6: 5′ cgcagttttcattacatttcatgggtatt 3′, 5′ agatgtatactaatgaagg 3′
unc-7b: 5′ aagaagggggaaagaaagcactgaa 3′, 5′ cggttcaggattgctggag 3′
tol-1: 5′ tatgtaggttgcctgtgtgcctgt 3′, 5′ tttcgttgtgtcatgtgatc 3′
mec-17: 5′ cttgagaggtccggcaacttgtttg 3′, 5′ cggtgaagcagcttctc 3′
heat-shock (from pPD.49.78): 5′ gctatgaccatgattacgccaagc 3′, 5′ cggggatccagtgagatg 3′
Primers to amplify the pdf-1 promoter for expression constructs:
Reporter 1: 5′ aaaaggttgctttaccgagacgtg 3′,5′ caacgccgagcttatcaac 3′
Reporter 2: 5′ aaaaggttgctttaccgagacgtg 3′, 5′ ctcattaagattatcccacta 3′
Reporter 3: 5′ aaaaggttgctttaccgagacgtg 3′, 5′ cttcactcaacgtcaacatg 3′
Reporter 4: 5′ gtgacgtcgacatgattcagaca 3′, 5′ cttcactcaacgtcaacatg 3′
Primers to amplify heterologous promoters driving pdf-1 cDNA:
pdf-1prom2: 5′ aaaaggttgctttaccgagacgtg 3′, 5′ cttcactcaacgtcaacatg 3′
rab-3: 5′ gcgagttttgactggctttc 3′, 5′ cctgatttcaggcggcggaa 3′
mbr-1: 5′ aataattgcgaaaagggcgattagc 3′, 5′ attgtattttcgtaggtaca 3′
zig-3: 5′ gtatggatgtccaacgatgagcttg 3′, 5′ ctatattttgcatttggaaaa 3′
Behavioral assays
Leaving assay
As previously described in 6,7. A population of between 10 and 20 worms (picked the night before at the L4 stage) was assayed by placing each worm individually in a Petri plate (9 cm diameter and 10 ml agar) in the center of a 20 μl (10 mm diameter) patch of food (E. coli OP50 seeded the night before). To assay retention, each individual test male was placed together with 2 paralyzed unc-51(e369) hermaphrodites. Plates were kept at 20°C during the 24 hrs assay and at 4 time points the proportion of males that had left the food was scored blindly. A worm was considered a leaver and left censored if at the scoring time point it had reached a distance 1 cm away from the edge of the plate (3 cm away from the food edge).
Hermaphrodite leaving assay
As previously described in 14, with some modifications. 30 L4 hermaphrodites were picked the night before of the assay onto a fresh plate. Conditioning plates (with a 70 μl food lawn) and assay plates (with a 10 μl food lawn) were seeded with a fresh culture of E. coli OP50 diluted in LB to A600=2.0. 90 minutes after seeding the plates, 10 young adult hermaphrodites were placed in the conditioning plate for 30 minutes. 7 hermaphrodites were then transferred from the conditioning plate to the assay plate and left for 1 hour before recording the assay for 20 minutes. Leaving events were scored manually by examining the video recordings. A leaving event was scored if the whole body of the worm was outside of the bacterial lawn and did not reverse immediately to return to the food. The rate of leaving events was calculated by dividing the number of leaving events per worm per minute of assay. Experiments of each genotype were performed at least 5 times. Statistical analysis was performed with Kruskal Wallis test.
Locomotion assays
Single worm assays were performed on 5 cm plates with or without food. Worms at the L4 stage were isolated from mates the night before on a plate with food. Body bends on food were scored by eye during 3 minutes observation periods under a dissecting microscope.
Locomotion in response to food: reversals and omega turns for each worm were scored during 5 minutes on food. The worm was then transferred to a transition plate without food and immediately to a scoring plate without food where it was left for 1 minute to settle. Reversals and omega turns were scored for 5 minutes and again 30 minutes later.
Locomotion in response to mates: 10 males (picked at the L4 stage the night before) were placed in a plate with 20 mates on a 1 cm patch of food for 3 hours. A male that was observed to have initiated the response and backing steps of mating was transferred to a transition plate without food and immediately to a scoring plate without food. The male was left to settle for 1 minute and reversals were scored for 5 minutes. The male was subsequently transferred to a plate with food in isolation for 1 hour and then, transferred to a transition and scoring plate with no food where reversals were scored again for 5 minutes.
Assays for response to food experience and mate experience were performed in different days. Wild type and mutant worms were assayed in parallel. Mann-Whitney test was used for statistical analysis.
Quiescence assay
As previously described in 20 with some modifications. Worms were grown in E. coli HB101 for several generations. Prior to the assay about 20 worms (picked at the L4 stage the night before) were transferred to a fresh plate for 5 hours before scoring lack of movement and pharyngeal pumping under a dissecting microscope. χ2 test was used for statistical analysis.
Response efficiency assays
Males were picked at the L4 stage the night before. Single males were then placed with 30 mates (hermaphrodites) in a 20 mm food lawn. The number of contacts with a mate until the male responded was scored for a maximum of 3 minutes. A good response was scored if the male placed its tail ventral down on the mate body and backed to make a turn. A bad response was scored if the male lost contact during backing to the opposite end of the mate body. Mann-Whitney test was used for statistical analysis.
Statistical analysis
Calculation of leaving rates and statistical analysis: As previously demonstrated in 6 male leaving behavior was modeled with the exponential function: N(t)/N(0)=exp(-λt). N(0) is equal to the number of worms at time zero, N(t) is the number of worms at time t (in hours). λ is the PL value or probability of leaving, per worm, per hour. PL values for each genotype and condition were calculated using R (http://www.R-project.org) to fit the censored data with an exponential parametric survival model, using maximum likelihood. The hazard values obtained were reported as the PL values. To estimate the PL values, S.E.M. and the 95% confidence intervals, worms from each experimental treatment were then pooled across replicas and contrasted against controls using maximum likelihood.
Values in graphs represent means and error bars represent standard error of the mean (SEM). Non-parametric statistical tests were used for locomotion and hermaphrodite leaving assays because the data did not always follow a normal distribution. Kruskal-Wallis test was used to compare multiple groups of independent data (genotypes) and Mann-Whitney test was used to compare 2 groups of independent data (conditions) against each other.
Laser ablations
The standard protocol was used 49. L4 or adult animals were mounted on a glass slide on a 5% agarose pad with 20mM NaAzide as anesthetic. Animals were left to recover for 1 day and then assayed. The IqIs3(osm-6::GFP) and myEx741[P(pdfr-1)3kb::nls rfp+P(unc-122)::gfp] transgenes were used to identify URY, PQR and PHA neurons. The myEx696[P(pdf-1)3kb::rfp] transgene was used to identify AIM, AVB and male-specific pdf-1 positive cells.
Heat-shock induction
Control and test worms were heat-shocked for 45 minutes at 37 °C, left to recover for 90 minutes at 20 °C and then tested in the leaving assay. Proportion of leavers was scored 3 hours later.
Supplementary Material
Acknowledgments
We thank the Maricq, Rongo and Hobert laboratories for strains and reagents; the Mitani laboratory and Japan’s National BioResource Project for mutants; additional strains were obtained from the CGC. We thank R. Poole and members of the Barr laboratory for advice and many helpful discussions on the manuscript; L. Vaynerchuk for experimental aid. This research was supported by NIH grant 2R01DK059418 to M.M.B.
Footnotes
Author contributions: A.B. designed and performed the experiments and co-wrote the manuscript; R.G. performed the genetic screen; C.F. contributed to mapping process; S.W.E. and M.M.B. co-wrote and discussed the manuscript with A.B.
References
- 1.Pfaff DW. The Physiological mechanisms of motivation. Springer Verlag; 1982. [Google Scholar]
- 2.von Frisch O. Animal migration. HarperCollins; 1969. [Google Scholar]
- 3.Reppert SM. A Colorful Model of the Circadian Clock. Cell. 2006;124:233–236. doi: 10.1016/j.cell.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Mowrey WR, Portman DS. Sex differences in behavioral decision-making and the modulation of shared neural circuits. Biol Sex Differ. 2012;3:8. doi: 10.1186/2042-6410-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendesky A, Bargmann CI. Genetic contributions to behavioural diversity at the gene-environment interface. Nat Rev Genet. 2011;12:809–820. doi: 10.1038/nrg3065. [DOI] [PubMed] [Google Scholar]
- 6.Lipton J, Kleemann G, Ghosh R, Lints R, Emmons SW. Mate searching in Caenorhabditis elegans: a genetic model for sex drive in a simple invertebrate. J Neurosci. 2004;24:7427–7434. doi: 10.1523/JNEUROSCI.1746-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrios A, Nurrish S, Emmons SW. Sensory regulation of C. elegans male mate-searching behavior. Curr Biol. 2008;18:1865–1871. doi: 10.1016/j.cub.2008.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleemann G, Jia L, Emmons SW. Regulation of Caenorhabditis elegans male mate searching behavior by the nuclear receptor DAF-12. Genetics. 2008;180:2111–2122. doi: 10.1534/genetics.108.093773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janssen T, et al. Functional characterization of three G protein-coupled receptors for pigment dispersing factors in Caenorhabditis elegans. J Biol Chem. 2008;283:15241–15249. doi: 10.1074/jbc.M709060200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janssen T, et al. Discovery and characterization of a conserved pigment dispersing factor-like neuropeptide pathway in Caenorhabditis elegans. Journal of Neurochemistry. 2009;111:228–241. doi: 10.1111/j.1471-4159.2009.06323.x. [DOI] [PubMed] [Google Scholar]
- 11.Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- 12.Janke DL, et al. Interpreting a sequenced genome: toward a cosmid transgenic library of Caenorhabditis elegans. Genome Res. 1997;7:974–985. doi: 10.1101/gr.7.10.974. [DOI] [PubMed] [Google Scholar]
- 13.Stewart HI, et al. Lethal mutations defining 112 complementation groups in a 4.5 Mb sequenced region of Caenorhabditis elegans chromosome III. Mol Gen Genet. 1998;260:280–288. doi: 10.1007/pl00013816. [DOI] [PubMed] [Google Scholar]
- 14.Bendesky A, Tsunozaki M, Rockman MV, Kruglyak L, Bargmann CI. Catecholamine receptor polymorphisms affect decision-making in C. elegans. Nature. 2011;472:313–318. doi: 10.1038/nature09821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milward K, Busch KE, Murphy RJ, de Bono M, Olofsson B. Neuronal and molecular substrates for optimal foraging in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1106134109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gloria-Soria A, Azevedo RBR. npr-1 Regulates foraging and dispersal strategies in Caenorhabditis elegans. Curr Biol. 2008;18:1694–1699. doi: 10.1016/j.cub.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 17.Dickson L, Finlayson K. VPAC and PAC receptors: From ligands to function. Pharmacology & Therapeutics. 2009;121:294–316. doi: 10.1016/j.pharmthera.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 19.Ogg S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 20.You YJ, Kim J, Raizen DM, Avery L. Insulin, cGMP, and TGF-β Signals Regulate Food Intake and Quiescence in C. elegans: A Model for Satiety. Cell Metabolism. 2008;7:249–257. doi: 10.1016/j.cmet.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hills T, Brockie PJ, Maricq AV. Dopamine and glutamate control area-restricted search behavior in Caenorhabditis elegans. J Neurosci. 2004;24:1217–1225. doi: 10.1523/JNEUROSCI.1569-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wakabayashi T, Kitagawa I, Shingai R. Neurons regulating the duration of forward locomotion in Caenorhabditis elegans. Neurosci Res. 2004;50:103–111. doi: 10.1016/j.neures.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Gray JM, Hill JJ, Bargmann CI. A circuit for navigation in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102:3184–3191. doi: 10.1073/pnas.0409009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chalasani SH, et al. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature. 2007;450:63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- 25.Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- 26.Barr MM, et al. The Caenorhabditis elegans autosomal dominant polycystic kidney disease gene homologs lov-1 and pkd-2 act in the same pathway. Curr Biol. 2001;11:1341–1346. doi: 10.1016/s0960-9822(01)00423-7. [DOI] [PubMed] [Google Scholar]
- 27.Flowers EB, et al. The Groucho ortholog UNC-37 interacts with the short Groucho-like protein LSY-22 to control developmental decisions in C. elegans. Development. 2010;137:1799–1805. doi: 10.1242/dev.046219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kage E, et al. MBR-1, a Novel Helix-Turn-Helix Transcription Factor, Is Required for Pruning Excessive Neurites in Caenorhabditis elegans. Current Biology. 2005;15:1554–1559. doi: 10.1016/j.cub.2005.07.057. [DOI] [PubMed] [Google Scholar]
- 29.Aurelio O. Immunoglobulin-Domain Proteins Required for Maintenance of Ventral Nerve Cord Organization. Science. 2002;295:686–690. doi: 10.1126/science.1066642. [DOI] [PubMed] [Google Scholar]
- 30.Bargmann CI. Beyond the connectome: How neuromodulators shape neural circuits. Bioessays. 2012:n/a–n/a. doi: 10.1002/bies.201100185. [DOI] [PubMed] [Google Scholar]
- 31.Davidson J. Biological bases of sexual behavior. 1974 [Google Scholar]
- 32.Portman DS. Genetic control of sex differences in C. elegans neurobiology and behavior. Adv Genet. 2007;59:1–37. doi: 10.1016/S0065-2660(07)59001-2. [DOI] [PubMed] [Google Scholar]
- 33.Jarrell TA, et al. The Connectome of a Decision-Making Neural Network. Science. 2012;337:437–444. doi: 10.1126/science.1221762. [DOI] [PubMed] [Google Scholar]
- 34.Fujii S, Amrein H. Ventral lateral and DN1 clock neurons mediate distinct properties of male sex drive rhythm in Drosophila. Proc Natl Acad Sci USA. 2010;107:10590–10595. doi: 10.1073/pnas.0912457107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joao CR, Cardoso, et al. Evolution of secretin family GPCR members in the metazoa. BMC Evolutionary Biology. 2006;11:99, 6, 108. doi: 10.1186/1471-2148-6-108. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soria Virginia, et al. Differential Association of Circadian Genes with Mood Disorders: CRY1 and NPAS2 are Associated with Unipolar Major Depression and CLOCK and VIP with Bipolar Disorder. Neuropsychopharmacology. 2010;35:1279. doi: 10.1038/npp.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ressler KJ, et al. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470:492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science. 2011;331:304–308. doi: 10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coates JC, de Bono M. Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans. Nature. 2002;419:925–929. doi: 10.1038/nature01170. [DOI] [PubMed] [Google Scholar]
- 40.Morsci NS, Barr MM. Kinesin-3 KLP-6 regulates intraflagellar transport in male-specific cilia of Caenorhabditis elegans. Curr Biol. 2011;21:1239–1244. doi: 10.1016/j.cub.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu S, Avery L, Baude E, Garbers DL. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc Natl Acad Sci USA. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maricq AV, Peckol E, Driscoll M, Bargmann CI. Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature. 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- 43.Lee RY, Sawin ER, Chalfie M, Horvitz HR, Avery L. EAT-4, a homolog of a mammalian sodium-dependent inorganic phosphate cotransporter, is necessary for glutamatergic neurotransmission in caenorhabditis elegans. J Neurosci. 1999;19:159–167. doi: 10.1523/JNEUROSCI.19-01-00159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pujol N, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, et al. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
- 46.Zeynep F, Altun BCZ-WW, Hall DH. High Resolution Map of Caenorhabditis elegans Gap Junction Proteins. Developmental dynamics: an official publication of the American Association of Anatomists. 2009;238:1936. doi: 10.1002/dvdy.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lickteig KM, et al. Regulation of neurotransmitter vesicles by the homeodomain protein UNC-4 and its transcriptional corepressor UNC-37/groucho in Caenorhabditis elegans cholinergic motor neurons. J Neurosci. 2001;21:2001–2014. doi: 10.1523/JNEUROSCI.21-06-02001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boulin T, Etchberger JF, Hobert O. Reporter gene fusions. WormBook. 2006:1–23. doi: 10.1895/wormbook.1.106.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bargmann CI, Avery L. Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 1995;48:225–250. doi: 10.1016/s0091-679x(08)61390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.