Abstract
Most mammalian cells in nature are quiescent but actively transcribing mRNA for normal physiological processes; thus, it is important to investigate how endogenous and exogenous DNA damage compromises transcription in cells. Here we described a novel competitive transcription and adduct bypass (CTAB) assay to determine the effects of DNA lesions on the fidelity and efficiency of transcription. Using this strategy, we demonstrated that the oxidatively induced lesions 8,5′-cyclo-2′-deoxyadenosine (cdA) and 8,5′-cyclo-2′-deoxyguanosine (cdG), and methylglyoxal-induced N2-(1-carboxyethyl)-2′-deoxyguanosine (N2-CEdG) strongly inhibited transcription in vitro and in mammalian cells. In addition, cdA and cdG, but not N2-CEdG, induced transcriptional mutagenesis in vitro and in vivo. Furthermore, when located on the template DNA strand, all examined lesions were primarily repaired by transcription-coupled nucleotide excision repair (TC-NER) in mammalian cells. This newly developed CTAB assay should be generally applicable for quantitatively assessing how other DNA lesions impact DNA transcription in vitro and in cells.
The majority of mammalian cells are quiescent or slowly replicating in nature, whereas they must express numerous genes for normal physiological processes. Thus, it is important to protect these cells from engaging in aberrant transcription, which may be induced by various endogenous and exogenous DNA-damaging agents 1,2. In this context, some DNA lesions may partially or completely block transcription without inducing mutations, whereas others may allow for mutagenic lesion bypass by RNA polymerase (RNAP) through a process termed transcriptional mutagenesis 1,3. Stalling or blockage of a RNAP at a lesion site is a general signal for triggering TC-NER, a sub-pathway of NER that preferentially removes DNA lesions from the template strand of actively transcribed genes 3,4.
Many in vitro studies have been carried out to investigate transcriptional alterations induced by DNA lesions, using purified proteins and/or cell extracts necessary for prokaryotic or eukaryotic transcription. In these systems, a single structurely defined DNA lesion is placed site-specifically on the transcribed strand of DNA template and the resultant transcripts are subjected directly to polyacrylamide gel electrophoresis (PAGE) analysis. The extent of transcriptional lesion bypass can be determined by quantifying the relative amounts of full-length and truncated transcripts, and the nature of mutational events resulting from RNAP bypass of lesions can be assessed by utilizing reverse transcription (RT) and sequencing analysis of the resultant RT-PCR products 1,3,5. Several plasmid-based reporter assays have also been developed to examine the effects of DNA lesions on transcription in cells 1,3. Central to these in vivo assays is the use of a plasmid containing a site-specific lesion in the template strand of a reporter gene. Lesion-induced transcriptional alterations are determined by biochemical measurement of reporter gene activity. In addition, the influence of various DNA repair proteins on lesion-induced perturbation in transcription can be measured through the use of cells defective in these proteins, which provides important information about how the lesion is repaired in cells. Nevertheless, these reporter assays do not provide direct evidence for transcriptional alterations induced by DNA lesions. Moreover, these traditional methods require the use of DNA sequencing for determining the identities and frequencies of mutant transcripts and thus it is necessary to prepare and sequence a sufficient number of colonies for providing statistically robust information 1,3,6–12.
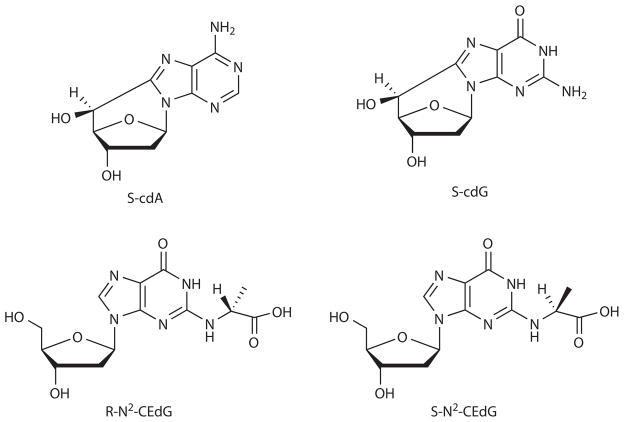
8,5′-Cyclopurine-2′-deoxynucleosides (cPu) represent a unique class of oxidatively induced DNA lesions in that they are considered substrates for NER 11,13,14. Both the (5′R) and (5′S) diastereomers of cdA and cdG (Fig. 1) have been detected in vitro and in vivo under various conditions, though the cellular levels of these lesions vary widely among different biological samples without exogenous oxidative stress 14–19. S-cdA strongly inhibits gene expression and affects transcriptional fidelity if located on the transcribed strand of an active gene in mammalian systems 11,12. Viewing the structural similarity of cdA and cdG, we reasoned that cdG may bear similar effects on transcription.
Figure 1.
Structures of S-cdA, S-cdG, R-N2-CEdG and S-N2-CEdG.
Methylglyoxal (MG) is a byproduct of the ubiquitous glycolysis pathway and high levels of MG are linked with diabetes and its complications 20. R- and S- diastereomers of N2-CEdG (Fig. 1) are the major stable MG-induced DNA adducts and may be widely distributed throughout the mammalian genome 21–25. It has been proposed that NER is involved in the repair of MG-induced mutations in E. coli cells 26. In addition, a recent study showed that MG-induced mutations occur more frequently in NER-deficient XP-G cells than in repair-proficient human cells, with a higher mutation frequency being associated with elevated levels of N2-CEdG 27. However, these observed mutations may also arise from other types of MG-induced DNA lesions since the lesion-bearing substrates were generated by directly treating plasmid DNA with MG 27. Furthermore, it remains unexplored how N2-CEdG impacts transcription.
Herein, we developed a novel CTAB assay by employing non-replicating plasmids bearing a single, site-specifically inserted S-cdA, S-cdG, R-N2-CEdG or S-N2-CEdG on the transcribed strand and examined the impact of these lesions on transcription in vitro and in vivo. Moreover, we investigated the relative roles of four NER genes (XPA, XPC, ERCC1 and CSB) in the repair of these lesions in mammalian cells by interrogating how partial or complete loss of function(s) of one or more of these genes alter the yields of transcripts arising from the lesion-housing substrates.
RESULTS
Development of a CTAB assay
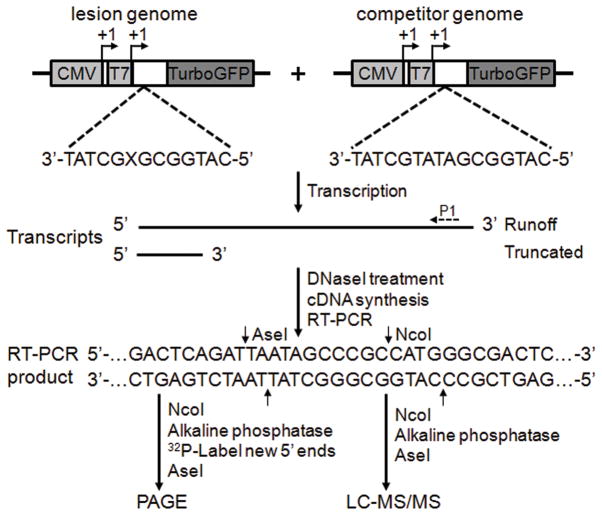
In this study, we designed a novel CTAB assay to investigate transcriptional alterations induced by DNAlesions in vitro and in cells (Fig. 2 and Supplementary Results, Supplementary Fig. 1). The development of this assay was inspired from the competitive replication and adduct bypass (CRAB) and restriction endonuclease and post-labeling (REAP) assays 28,29, which were developed previously for assessing how DNA lesions situated in a single-stranded M13 genome perturb the efficiency and fidelity of DNA replication in E. coli cells 28,29.
Figure 2. A schematic diagram depicts the CTAB assay system.
“X” indicates an S-cdA, S-cdG, dA or dG, which was located on the transcribed strand of TurboGFP gene downstream of the CMV and T7 promoters. The arrowheads indicate the +1 transcription start sites. Run-off RNA and truncated RNA resulting from transcription arrest at a lesion site are indicated. P1 represents a gene-specific primer used for reverse transcribing the run-off transcripts. Only the wild-type sequence of the RT-PCR products for the lesion-containing genome is shown. The RT-PCR products arising from the competitor genome are not depicted. The RT-PCR products were digested with two restriction enzymes (NcoI and AseI) and then subjected to PAGE or LC-MS/MS analyses. The cleavage sites of NcoI and AseI are designated with arrows.
For our purpose, we prepared non-replicating double-stranded vectors harboring a single, site-specifically inserted lesion (S-cdA, S-cdG, R-N2-CEdG or S-N2-CEdG), as well as the corresponding non-lesion control and competitor vectors. The competitor plasmids were designed to carry three additional nucleotides than the corresponding control plasmids situated between the two restriction sites used for CTAB assay. The cPu lesions were placed 58 and 39 nucleotides downstream of the transcription start site (TSS) of the cytomegalovirus (CMV) and T7 promoters, respectively, whereas the N2-CEdG lesions were situated 63 and 44 nucleotides downstream of the TSS of CMV and T7 promoters, respectively. This design allows for examining the effects of DNA lesions on transcriptional elongation by both single-subunit T7 RNA polymerase (T7 RNAP) and multi-subunit mammalian RNA polymerase II (RNAPII) with the same lesion-bearing plasmids. The single-subunit mitochondrial RNAPs are highly homologous to T7 RNAP 30. Thus, we chose T7 RNAP as a model for investigating the potential effect of these lesions on mitochondrial DNA transcription in mammalian cells.
In the CTAB assay, the lesion-bearing or undamaged control plasmids were pre-mixed with the competitor vector at specific molar ratios and used as templates for in vitro or in vivo transcription. The resulting transcripts were isolated and treated with RNase-free DNaseI to eliminate the contamination of residual DNA template. Although truncated transcripts may be generated when transcription arrests or pauses at or near a lesion site, only runoff transcripts were reverse transcribed to produce cDNA. The resultant RT-PCR products were digested with the appropriate restriction enzymes and subjected to PAGE and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses, which have been successfully employed to assess how a variety of DNA lesions affect DNA replication in E. coli and mammalian cells 25,31,32.
Using the CTAB assay, the degree to which a given DNA adduct inhibits transcription can be determined by the “relative bypass efficiency” (RBE). To determine this, the total amounts of restriction fragments arising from the transcription of the lesion-carrying plasmid are normalized to the corresponding fragment from the competitor genome generated in the same reaction, taking into account the initial molar ratio of the two genomes used for in vitro or in vivo transcription reactions. The RBE value is then calculated by dividing this ratio with that obtained from the control experiment where the corresponding lesion-free plasmid is co-transcribed with the competitor genome. In addition, the effect of a lesion on transcriptional fidelity can be quantified by the “relative mutation frequency” (RMF), that is, the percent of mutant restriction fragments, if detectable, in the total amounts of restriction fragments released from the lesion-carrying genome.
Effect of S-cdA and S-cdG on transcription in vitro
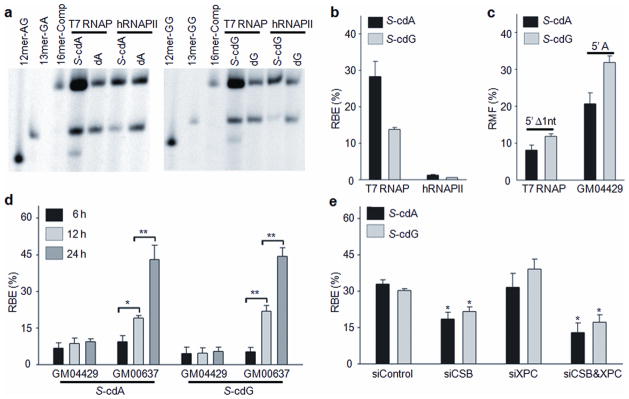
To examine the behavior of T7 RNAP and human RNAPII (hRNAPII) when they encounter an S-cdA or S-cdG, we allowed the lesion-bearing or control plasmid, along with the competitor plasmid, to undergo multiple rounds of in vitro transcription by T7 RNAP or HeLa cell nuclear extract, the latter of which supplies hRNAPII and its associated general transcription factors. PAGE analysis of restriction fragments of RT-PCR products showed that S-cdA and S-cdG substantially inhibited transcription by T7 RNAP, with the RBE values being ~ 28% and 14%, respectively. On the other hand, both lesions almost completely (99%) inhibited transcription by hRNAPII (Fig. 3a,b).
Figure 3. Transcriptional alterations induced by S-cdA and S-cdG.
(a) Representative gel images showing the restriction fragments released from the RT-PCR products in in vitro transcription systems using T7 RNAP or Hela nuclear extract (hRNAPII). The control/competitor genome ratio was 1:1 for all experiments while the ratios (lesion/competitor) for transcription by T7 RNAP and hRNAPII were 1:1 and 19:1, respectively. The restriction fragment arising from the competitor genome, i.e., d(CATGGCGATATGCTAT), is designated with ‘16mer-Comp’; ‘13mer-GA’, ‘13mer-GG’, ‘12mer-AG’, and ‘12mer-GG’ represent standard ODNs d(CATGGCGAGCTAT), d(CATGGCGGGCTAT), d(CATGGCAGCTAT), and d(CATGGCGGCTAT), respectively. The corresponding full gel is shown in Supplementary Fig. 16. (b) Relative bypass efficiencies (RBE) of S-cdA and S-cdG in in vitro transcription systems. (c) Relative mutation frequencies (RMF) of 5′Δ1nt mutation of S-cdA and S-cdG in in vitro transcription systems using T7 RNAP and RMF values of 5′ A mutation of S-cdA and S-cdG arising from in vivo transcription in NER-deficient XPA (GM04429) cells. (d) The RBE values of S-cdA and S-cdG based on in vivo transcription experiments using GM04429 and repair-proficient (GM00637) cells. (e) The RBE values of S-cdA and S-cdG in 293T cells treated with either or both CSB and XPC siRNAs. The data represent the mean and standard error of results from three independent experiments. ‘*’, P < 0.05; ‘**’, P < 0.01. The P values were calculated by using unpaired two-tailed Student’s t-test.
We also performed T7 RNAP-mediated transcription by including [α-32P]-CTP in the transcription reaction and analyzed the resulting transcripts directly by denaturing PAGE (Supplementary Fig. 2a–f). As expected, the RBE values of S-cdA and S-cdG determined by this method (Supplementary Fig. 2e) were comparable to that obtained by analysis of RT-PCR products (Fig. 3b). However, truncated transcripts resulting from transcription arrest at or near the S-cdA or S-cdG site were detected at relatively low frequencies of ~18% and 24%, respectively (Supplementary Fig. 2f). These results suggest that the presence of these lesions in template DNA strand may lead to a decreased transcription efficiency by arresting RNAPs at the lesion sites and by reducing the rate of transcription elongation at or near the lesion site. Although future studies are needed for understanding the mechanisms underlying the transcription impediment induced by these lesions, the inclusion of a lesion-free competitor template in the same transcription reaction mixture allowed the CTAB assay to accurately reveal the decreased transcription rate caused by a DNA lesion.
PAGE analysis also facilitated us to measure the frequencies of mutations induced by these lesions during in vitro transcription. We found no mutant transcripts generated from the hRNAPII-mediated in vitro transcription reaction of the S-cdA- or S-cdG-containing plasmid. However, T7 RNAP generated at least one type of mutant transcript, a single-nucleotide deletion immediately downstream of the lesion, which occurred at frequencies of 8% and 12% for S-cdA and S-cdG, respectively (Fig. 3a,c). We refer to this type of mutant transcripts as 5′Δ1nt, and confirmed their identities by LC-MS/MS analysis (Supplementary Fig. 3). We also attempted, but failed to detect other types of mutant transcripts by LC-MS/MS. These included single-base substitutions at the lesion site, single-nucleotide deletions opposite the lesion, and misincorporation of an adenosine (A) opposite the next nucleotide downstream of the lesion (5′A mutation). 5′A mutation was detected in a recent study of S-cdA-induced transcriptional mutagenesis in human cells 12.
Effect of S-cdA and S-cdG on transcription in cells
To assess how S-cdA and S-cdG affect DNA transcription in mammalian cells, the lesion-bearing or non-lesion control plasmids were mixed individually with the competitor genome at given molar ratios and co-transfected into repair-proficient (GM00637) and NER-deficient (GM04429, lacking XPA) human skin fibroblasts. At various time points following transfection, mRNA was isolated from the cells and the transcripts of interest were reverse transcribed, PCR-amplified and restriction digested for PAGE and LC-MS/MS analyses.
There was a time-dependent increase in RBE values for S-cdA and S-cdG in the NER-proficient cells, reaching approximately 45% by 24 h (Fig. 3d and Supplementary Fig. 4). In contrast, this increase was not observed in NER-deficient cells. The RBE values for S-cdA- and S-cdG-containing vectors in repair-competent cells were markedly greater than those in NER-deficient cells at 12 and 24 h after transfection. In addition, we found that only one type of mutant transcripts (5′A mutation) could be detected in the NER-deficient cells at frequencies of 21% and 32% for the S-cdA and S-cdG-containing vectors, respectively (Fig. 3c, Supplementary Fig. 5, and calibration curves are shown in Supplementary Fig. 6). Collectively, these data revealed that both S-cdA and S-cdG constitute strong impediments to transcription, and both induce transcriptional mutagenesis in human cells. Additionally, NER is required for the removal of these lesions in human cells.
To determine the influence of various NER sub-pathways on transcriptional alterations induced by S-cdA and S-cdG, we employed small interfering RNAs (siRNAs) to knock-down the expression of either or both XPC and CSB, which are specifically required for global genome repair (GG-NER) and TC-NER, respectively 4. We chose 293T cells for the siRNA experiments because the transfection efficiency is high and the cells are considered to have normal capacity for NER. Real-time PCR and Western blot results showed that the knock-down was highly efficient for both XPC and CSB genes (Supplementary Fig. 7). The RBE values for both S-cdA and S-cdG were significantly lower in CSB knockdown cells than in control siRNA-treated cells, whereas there was no siginificant diference between the XPC knockdown cells than the control (Fig. 3e and Supplementary Fig. 8). Relative to knockdown of CSB alone, simultaneous knockdown of CSB and XPC conferred a slight but statistically insignificant decrease in transcription bypass efficiency for S-cdA and S-cdG; the RBE values dropped from 19% to 13% and from 22% to 17% for S-cdA and S-cdG, respectively (Fig. 3e and Supplementary Fig. 8). These results indicated that S-cdA and S-cdG in the template strand are primarily repaired by TC-NER.
Effect of N2-CEdG on transcription in vitro
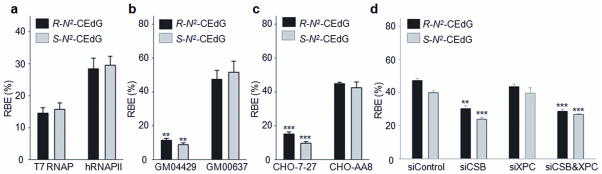
Having established a rigorous quantitative method for assessing the effects of cdA and cdG on transcription, we decided to employ the same method to examine how the two diastereomers of N2-CEdG alter the efficiency and fidelity of transcription. To this end, we first investigated the behavior of T7 RNAP and hRNAPII when they encounter an R- or S-N2-CEdG. PAGE analysis of restriction fragments of RT-PCR products showed that both diastereomers of N2-CEdG considerablely inhibited transcription by T7 RNAP and hRNAPII, with the RBE values being ~ 15% and 29%, respectively (Fig. 4a and Supplementary Fig. 9a).
Figure 4. Transcriptional alterations induced by R- and S-N2-CEdG.
(a) Relative bypass efficiencies (RBE) of R- and S-N2-CEdG in in vitro transcription systems using T7 RNAP or HeLa nuclear extract (hRNAPII). (b) The RBE values of R- and S-N2-CEdG in in vivo transcription systems with NER-deficient XP-A (GM04429) and wild-type (GM00637) human fibroblast cells. (c) The RBE values of R- and S-N2-CEdG in ERCC1-deficient (CHO-7-27) and wild-type (CHO-AA8) CHO cells. (d) The RBE values of R- and S-N2-CEdG in 293T cells treated with either or both CSB and XPC siRNAs. The data represent the mean and standard error of results from three independent experiments. ‘**’, P < 0.01; ‘***’, P < 0.001. The P values were calculated by using unpaired two-tailed Student’s t-test.
PAGE analysis showed that the 10 mer product (10mer-CT) with the wild-type sequence can be clearly distinguished from the corresponding 10-mers carrying a single-base substitution at the lesion site; however, no such mutant transcripts were detectable in the restriction mixtures (Supplementary Fig. 9a). We further interrogated the fragments in the same restriction digestion mixtures using LC-MS and MS/MS analyses. We monitored the fragmentation of the [M-3H]3− ions of the complementary 14-mer fragments [d(GCAAAPCTTGAGCT), where “P” designates A, T, C, or G]. It turned out that only the wild-type sequence [d(GCAAAGCTTGAGCT)] could be detected in the mixture (Supplementary Fig. 10). With the LC-MS/MS method, we also attempted, but failed to observe other types of mutant transcripts, including the 5′A mutation and the single-nucleotide deletion opposite the lesion (Supplementary Figs. 10 and 11).
Effect of N2-CEdG on transcription in cells
To assess how N2-CEdG lesions affect DNA transcription in vivo, we performed the CTAB assay using the repair-competent GM00637 and XPA-deficient GM04429 cells for in vivo transcription. The RBE values for R- and S-N2-CEdG in GM00637 cells were ~ 48% and 52%, respectively, and the RBE values for the two lesions were markedly lower in XPA-deficient GM04429 cells at 24 h after transfection (~ 11% and 9%, Fig. 4b and Supplementary Fig. 9b). These results indicated that NER is required for repairing these two minor-groove N2-dG lesions.
To confirm and extend the data acquired for the human cells, we examined the effect of N2-CEdG on DNA transcription in two types of Chinese hamster ovary (CHO) cells, i.e., AA8 (wild-type) and CHO-7-27 (NER-deficient Ercc1 mutant cells) 33. The RBE values for both diastereomers of N2-CEdG were significantly lower in CHO-7-27 cells than in AA8 cells at 24 h following transfection (Fig. 4c and Supplementary Fig. 9c), lending further evidence to support the important role of NER in the removal of N2-CEdG lesions in mammalian cells.
In keeping with what we observed from in vitro transcription reaction, these in vivo results unveiled that N2-CEdG is able to substantially inhibit DNA transcription in mammalian cells. In addition, the results from PAGE and LC-MS/MS analyses showed that these lesions are not mutagenic in mammalian systems (Supplementary Figs. 9b,c, 12 and 13).
We next employed siRNA knock-down of XPC and CSB to assess how defects in GG-NER and TC-NER sub-pathways, respectively, affect transcription bypass efficiencies of N2-CEdG in 293T cells. We found that knockdown of CSB significantly reduced the transcription bypass efficiency, with the RBE values for R- or S-N2-CEdG decreasing from 47% and 40% in control siRNA-treated cells to 30% and 24% in CSB knockdown cells, respectively (Fig. 4d and Supplementary Fig. 14). In contrast, knockdown of XPC had no effect. Moreover, simultaneous knockdown of CSB and XPC did not confer a statistically significant change in the RBE values for R- or S-N2-CEdG, relative to knockdown of CSB alone. Taken together, these results indicated that TC-NER, but not GG-NER, is required for the removal of N2-CEdG lesions from the template strand of an actively transcribed gene in mammalian cells.
DISCUSSION
In this study, we designed a novel transcription assay (i.e., the CTAB assay) to investigate quantitatively how site-specifically inserted DNA lesions affect the efficiency and fidelity of DNA transcription in vitro and in vivo. In the CTAB assay, the entire population of RNA transcripts is used for the assessment of transcriptional bypass and mutagenesis. Thus, the method provides statistically robust and quantitative conclusions about how DNA lesions inhibit transcription and invoke transcriptional mutagenesis in vitro and in vivo. The method is also efficient because it does not require phenotypic selection and colony picking. In addition, the incorporation of LC-MS/MS into the assay facilitates the unambiguous identification of mutant transcripts, thereby providing accurate assessment of transcriptional mutagenesis occurring at or near the lesion site. It is also of note that we recently developed a high-throughput method, which employs a barcoding strategy and next-generation sequencing, for the quantitative and simultaneous assessment of how multiple DNA lesions perturb the efficiency and accuracy of DNA replication in cells 34. It can be envisaged that, in combination with next-generation sequencing, the CTAB assay is amenable for simultaneously examining the effects of multiple DNA lesions on transcription. Furthermore, because the effect of the DNA damage on transcription is modulated by the repair capacity of cells 6,35,36, the CTAB assay can be used for examining the relative roles of various proteins in the repair of structurally defined DNA lesions via manipulating the expression of one or more genes, as demonstrated in the present study.
Our results from the CTAB assay revealed that S-cdG and, to a lesser degree, S-cdA in the template DNA strand act as significant inhibitions to transcription by T7 RNAP and hRNAPII. In this vein, it was reported recently that S-cdG and, to a lower extent, S-cdA strongly block DNA replication in E. coli cells 34,37. In addition, both diastereomers of cdA were found to substantially block primer extension mediated by human DNA polymerase η in vitro 38, though it remains unexplored how these lesions compromise DNA replication in mammalian cells.
We also found that transcriptional bypass of S-cdA and S-cdG can give rise to mutations. We observed 5′A mutations for S-cdA in NER-deficient human cells, as previously reported 12, albeit in a different sequence context. More importantly, we showed for the first time that bypass of S-cdG also generates 5′A mutations in the transcript, suggesting that this type of mutation may be considered a signature mutation induced by cPu lesions during transcription. With T7 RNAP, we found a new type of aberrant transcript, where the polymerase skips a nucleotide rather than incorporating an AMP opposite the next nucleotide downstream (i.e., on the 5′ side) of the lesion. Differences in the behavior of T7 RNAP and mammalian RNAPII are observed with other lesions 39, and the structure and active site conformation between these two classes of RNA polymerases are drastically different 40,41. Thus, it is not surprising to observe different types of transcriptional mutations induced by T7 RNAP and hRNAPII. Different from the mutations induced during transcription, replicative bypass of S-cdA and S-cdG in E. coli cells mainly gave rise to nucleobase substitutions at the lesion site, with S-cdA and S-cdG yielding A→T and G→A mutations at frequencies of 11% and 20%, respectively 34.
Consistent with previous reports 11,13,14,16,42, our results demonstrate that the RBE values for S-cdA- and S-cdG are lower in NER-deficient cells than in NER-competent cells. Moreover, we found that XPC-mediated GG-NER is dispensable, whereas CSB-mediated TC-NER plays an important role in the repair of S-cdA and S-cdG in the DNA template strand.
The minor-groove N2 position of guanine is a major site for DNA modification induced by various carcinogens 43. Several studies have shown that N2-dG lesions can be bypassed accurately and efficiently during DNA replication in vitro and in vivo, in particular by specialized translesion synthesis polymerases 25,31,44,45. Owing to the lack of effect of N2-dG lesions on DNA replication, this type of lesions have been suggested to be “stealth lesions” 46. The results from the present study, along with previous in vitro studies 46,47, showed that these minor-groove N2-dG lesions are able to strongly inhibit DNA transcription when present on the DNA template strand. N2-ethyl-2′-deoxyguanosine (N2-EtdG) was found, from primer extension assays, to be a strong block to both single-subunit T7 RNAP and multi-subunit RNAPs in vitro 47. In addition, mammalian RNAPII was found to exclusively incorporate the correct nucleotide opposite N2-EtdG 47. Likewise, N2-furfuryl-dG, a structural mimic of the principal nitrofurazone-induced dG lesions, absolutely blocks transcription by E. coli RNAP when located in the template DNA strand 46. To our knowledge, the present study is the first to demonstrate that N2-dG lesions are strong impediments to transcription in mammalian cells, although neither diastereomer of N2-CEdG leads to detectable transcriptional mutagenesis.
We also found that CSB-mediated TC-NER plays a significant role in the repair of N2-CEdG in the DNA template strand in mammalian cells. This is in keeping with a previous proposal that NusA-dependent TC-NER pathway plays an important role in removing a class of DNA lesions typified by the N2-furfuryl-dG in E. coli 46. N2-CEdG is produced endogenously, and its levels in mammalian cells is enhanced by elevated concentrations of glycolytic metabolites 23,25. Thus, N2-CEdG lesions, if left unrepaired, may lead to detrimental biological consequences and contribute to the etiology of diabetic complications and other human diseases by blocking DNA transcription.
METHODS
Transcription template construction
Construction of the non-replicating vectors bearing a single lesion (S-cdA, S-cdG, R-N2-CEdG or S-N2-CEdG), as well as the corresponding non-lesion control and competitor vectors were described in the Supplementary Methods
In vitro transcription assay
The lesion-bearing or the corresponding non-lesion control plasmids were seperately mixed with the competitor genome at indicated molar ratios and then digested with NotI to yield linear DNA templates. The T7 RNAP-mediated reactions contained 50 ng of DNA template, 0.5 mM each of the four non-radioactive ribonucleotides (ATP, CTP, GTP and UTP) and 20 U T7 RNAP (Promega) in a final volume of 20 μL. The reaction mixture was incubated at 37°C for 60 min, following the manufacturer’s recommended procedures. HeLa nuclear extract (Promega) was used as the source for the hRNAPII machinery. The reactions contained 50 ng of DNA template, 0.4 mM each of all four non-radioactive ribonucleotides and 8 U HeLa nuclear extract in a final volume of 25 μL. The reaction was incubated at 30°C for 60 min, following the vendor’s recommended conditions.
In vivo transcription assay
The lesion-bearing or the corresponding non-lesion control plasmids were mixed with the competitor genome at indicated molar ratios and then used for transfection experiments. CHO (CHO-7-27 and AA8) and human fibroblast (GM00637 and GM04429) cells (1×105) were seeded in 24-well plates and cultured overnight. The cells were then co-transfected with 50 ng mixed genome and 450 ng carrier DNA (self-ligated pGEM-T vector, Promega) using Lipofectamine 2000 (Invitrogen) following the vendor’s instructions. The cells were harvested for RNA extraction at indicated time points after transfection. For siRNA experiments, all siRNAs were purchased from Dharmacon: XPC ON-TARGETplus SMARTpool (L-016040), CSB ON-TARGETplus SMARTpool (L-004888) and siGENOME Non-Targeting siRNA (D-001210). The 293T cells were seeded in 24-well plates at 40–60% confluence level and transfected with 25 pmol siRNAs for each gene using Lipofectamine 2000 (Invitrogen). After a 48 h incubation, 50 ng of mixed genomes was transfected into the cells together with another aliquot of siRNA and 450 ng carrier DNA. The cells were harvested for RNA extraction 24 h after transfection.
RNA extraction and RT-PCR
RNA transcripts arising from in vitro or in vivo transcription were extracted using the RNeasy Mini Kit (QIAGEN). Isolated RNA was treated twice with the DNA-free kit (Ambion). Subsequently, cDNA synthesis was performed by employing M-MLV reverse transcriptase (Promega) with a mixture of an oligo(dT)16 primer and a gene-specific primer (5′-TCGGTGTTGCTGTGAT-3′). Approximately 5% of the resulting cDNA was used as a template for RT-PCR amplification with Phusion high-fidelity DNA polymerase (New England Biolabs). The primers were 5′-GCTAGCGCTACCGGACTCAG-3′ and 5′-TGCTGCGGATGATCTTGTCG-3′, and the RT-PCR amplification started at 98°C for 30 s, followed with 35 cycles at 98°C for 10 s, 60°C for 30 s, and 72°C for 15 s, and a final extension at 72°C for 5 min. The RT-PCR products were purified by QIAquick PCR Purification Kit (Qiagen) and stored at −20°C until use. The same PCR conditions were used for confirm the elimination of DNA contamination by the absence of PCR product when RNA was employed directly as template (Supplementary Fig. 15).
PAGE analysis
In order to analyze the transcription products of S-cdAand S-cdG using PAGE, a portion of the above RT-PCR fragments was treated with 10 U NcoI and 1 U shrimp alkaline phosphatase in 10 μL NEB buffer 3 at 37°C for 30 min, followed by heating at 80°C for 20 min to deactivate the phosphatase. The mixture was then treated in a 15 μL NEB buffer 3 with 5 mM DTT, ATP (50 pmol cold, premixed with 1.66 pmol [γ-32P]ATP) and T4 polynucleotide kinase. The reaction was continued at 37°C for 30 min, followed by heating at 65°C for 20 min to deactivate the polynucleotide kinase. To the reaction mixture was subsequently added 10 U AseI, and the solution was incubated at 37°C for 30 min, followed by quenching with 15 μL formamide gel loading buffer containing xylene cyanol FF and bromophenol blue dyes. The mixture was loaded onto a 30% polyacrylamide gel (acrylamide:bis-acrylamide=19:1) with 7 M urea and products were quantified by phosphorimager analysis with a Typhoon 9410 Variable Mode Imager and ImageQuant software (GE Healthcare). A similar method was used to PAGE analysis of the transcription products of R- or S-N2-CEdG except that SacI and FspI were the restriction enzymes employed for the PAGE analysis.
LC-MS/MS analysis
In order to identify the transcription products of S-cdA and S-cdG using LC-MS/MS, RT-PCR products were treated with 50 U NcoI and 20 U shrimp alkaline phosphatase in 250 μL NEB buffer 3 at 37°C for 2 h, followed by heating at 80°C for 20 min. To the resulting solution was added 50 U of AseI, and the reaction mixture was incubated at 37°C for 1 h, followed by extraction once with phenol/chloroform/isoamyl alcohol (25:24:1, v/v). The aqueous portion was dried with Speed-vac, desalted with HPLC and dissolved in 12 μL water. The ODN mixture was subjected to LC-MS/MS analysis, where the LTQ linear ion trap mass spectrometer was set up for monitoring the fragmentation of the [M-3H]3- ions of the 13mer [d(CATGGCGPGCTAT), where “P” designates A, T, C, or G, d(CATGGCTGGCTAT), d(CATGGCTAGCTAT)] and 12mer [d(CATGGCGGCTAT) and d(CATGGCAGCTAT)] ODNs, and the [M-4H]4- ion of the 16mer [i.e., d(CATGGCGATATGCTAT)] ODN. LC-MS/MS was performed as described in the Supplementary Methods. A similar method was used to identify the transcription products of Rand S-N2-CEdG-containing substrates, where the mass spectrometer was set up for monitoring the fragmentation of the[M-3H]3- ions of the 14-mer [d(GCAAAPCTTGAGCT), where “P” designates A, T, C, or G, and d(GCAATGCTTGAGCT)] and 13-mer [d(CATCAAGCTTTGC) and d(GCAAACTTGAGCT)] ODNs.
Other methods
Materials and additional methods are described in Supplementary Methods.
Supplementary Material
Acknowledgments
We thank Profs. Timothy R. O’Connor, Gerd P. Pfeifer, and Michael Seidman for providing cell lines and plasmid. This work was supported by the National Institutes of Health (R01 DK082779, R01 ES019873 and R01 CA101864 to Y. W., and R01 ES016114 to L. J. N.).
Footnotes
Author contributions
C.Y., X.D., B.Y., and Y.W. designed research; C.Y., X.D., B.Y., and J.(Jianshuang) W. performed research; C.Y., X.D., and Y.W. analyzed data; C.Y., J.(Jin) W., P.J.B., L.J.N. and Y.W. wrote the paper; Y.W. conceived and supervised the study.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html. Correspondence and requests for materials should be addressed to Y.W.
References
- 1.Bregeon D, Doetsch PW. Transcriptional mutagenesis: causes and involvement in tumour development. Nat Rev Cancer. 2011;11:218–27. doi: 10.1038/nrc3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 3.Saxowsky TT, Doetsch PW. RNA polymerase encounters with DNA damage: transcription-coupled repair or transcriptional mutagenesis? Chem Rev. 2006;106:474–88. doi: 10.1021/cr040466q. [DOI] [PubMed] [Google Scholar]
- 4.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–70. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 5.Kuraoka I, Tanaka K. Assays for transcription elongation by RNA polymerase II using oligo(dC)-tailed template with single DNA damage. Methods Enzymol. 2006;408:214–23. doi: 10.1016/S0076-6879(06)08013-X. [DOI] [PubMed] [Google Scholar]
- 6.Saxowsky TT, Meadows KL, Klungland A, Doetsch PW. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:18877–82. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bregeon D, Peignon PA, Sarasin A. Transcriptional mutagenesis induced by 8-oxoguanine in mammalian cells. PLoS Genet. 2009;5:e1000577. doi: 10.1371/journal.pgen.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns JA, Dreij K, Cartularo L, Scicchitano DA. O6-methylguanine induces altered proteins at the level of transcription in human cells. Nucleic Acids Res. 2010;38:8178–87. doi: 10.1093/nar/gkq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bregeon D, Doddridge ZA, You HJ, Weiss B, Doetsch PW. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli. Mol Cell. 2003;12:959–70. doi: 10.1016/s1097-2765(03)00360-5. [DOI] [PubMed] [Google Scholar]
- 10.Viswanathan A, You HJ, Doetsch PW. Phenotypic change caused by transcriptional bypass of uracil in nondividing cells. Science. 1999;284:159–62. doi: 10.1126/science.284.5411.159. [DOI] [PubMed] [Google Scholar]
- 11.Brooks PJ, et al. The oxidative DNA lesion 8,5′-(S)-cyclo-2′-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J Biol Chem. 2000;275:22355–62. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 12.Marietta C, Brooks PJ. Transcriptional bypass of bulky DNA lesions causes new mutant RNA transcripts in human cells. EMBO Rep. 2007;8:388–93. doi: 10.1038/sj.embor.7400932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuraoka I, et al. Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc Natl Acad Sci U S A. 2000;97:3832–7. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaruga P, Dizdaroglu M. 8,5′-Cyclopurine-2′-deoxynucleosides in DNA: mechanisms of formation, measurement, repair and biological effects. DNA Repair (Amst) 2008;7:1413–25. doi: 10.1016/j.dnarep.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Belmadoui N, et al. Radiation-induced formation of purine 5′,8-cyclonucleosides in isolated and cellular DNA: high stereospecificity and modulating effect of oxygen. Org Biomol Chem. 2010;8:3211–9. doi: 10.1039/c004531d. [DOI] [PubMed] [Google Scholar]
- 16.Chatgilialoglu C, Ferreri C, Terzidis MA. Purine 5′,8-cyclonucleoside lesions: chemistry and biology. Chem Soc Rev. 2011;40:1368–82. doi: 10.1039/c0cs00061b. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, et al. Quantification of oxidative DNA lesions in tissues of Long-Evans Cinnamon rats by capillary high-performance liquid chromatography-tandem mass spectrometry coupled with stable isotope-dilution method. Anal Chem. 2011;83:2201–9. doi: 10.1021/ac103099s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Errico M, et al. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. 2006;25:4305–15. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez H, et al. Lymphoblasts of women with BRCA1 mutations are deficient in cellular repair of 8,5′-Cyclopurine-2′-deoxynucleosides and 8-hydroxy-2′-deoxyguanosine. Biochemistry. 2007;46:2488–96. doi: 10.1021/bi062022p. [DOI] [PubMed] [Google Scholar]
- 20.Thornalley PJ. Pharmacology of methylglyoxal: formation, modification of proteins and nucleic acids, and enzymatic detoxification--a role in pathogenesis and antiproliferative chemotherapy. Gen Pharmacol. 1996;27:565–73. doi: 10.1016/0306-3623(95)02054-3. [DOI] [PubMed] [Google Scholar]
- 21.Frischmann M, Bidmon C, Angerer J, Pischetsrieder M. Identification of DNA adducts of methylglyoxal. Chem Res Toxicol. 2005;18:1586–92. doi: 10.1021/tx0501278. [DOI] [PubMed] [Google Scholar]
- 22.Schneider M, et al. Determination of glycated nucleobases in human urine by a new monoclonal antibody specific for N2-carboxyethyl-2′-deoxyguanosine. Chem Res Toxicol. 2004;17:1385–90. doi: 10.1021/tx049929d. [DOI] [PubMed] [Google Scholar]
- 23.Li H, et al. N2-carboxyethyl-2′-deoxyguanosine, a DNA glycation marker, in kidneys and aortas of diabetic and uremic patients. Kidney Int. 2006;69:388–92. doi: 10.1038/sj.ki.5000064. [DOI] [PubMed] [Google Scholar]
- 24.Synold T, et al. Advanced glycation end products of DNA: quantification of N2-(1-Carboxyethyl)-2′-deoxyguanosine in biological samples by liquid chromatography electrospray ionization tandem mass spectrometry. Chem Res Toxicol. 2008;21:2148–55. doi: 10.1021/tx800224y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan B, Cao H, Jiang Y, Hong H, Wang Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2′-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc Natl Acad Sci U S A. 2008;105:8679–84. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murata-Kamiya N, Kaji H, Kasai H. Deficient nucleotide excision repair increases base-pair substitutions but decreases TGGC frameshifts induced by methylglyoxal in Escherichia coli. Mutat Res. 1999;442:19–28. doi: 10.1016/s1383-5718(99)00054-6. [DOI] [PubMed] [Google Scholar]
- 27.Tamae D, Lim P, Wuenschell GE, Termini J. Mutagenesis and repair induced by the DNA advanced glycation end product N2-1-(carboxyethyl)-2′-deoxyguanosine in human cells. Biochemistry. 2011;50:2321–9. doi: 10.1021/bi101933p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci U S A. 2004;101:14051–6. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaney JC, Essigmann JM. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 2006;408:1–15. doi: 10.1016/S0076-6879(06)08001-3. [DOI] [PubMed] [Google Scholar]
- 30.Masters BS, Stohl LL, Clayton DA. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 31.Yuan B, et al. The roles of DNA polymerases kappa and iota in the error-free bypass of N2-carboxyalkyl-2′-deoxyguanosine lesions in mammalian cells. J Biol Chem. 2011;286:17503–11. doi: 10.1074/jbc.M111.232835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong H, Cao H, Wang Y. Formation and genotoxicity of a guanine-cytosine intrastrand cross-link lesion in vivo. Nucleic Acids Res. 2007;35:7118–27. doi: 10.1093/nar/gkm851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rolig RL, et al. Survival, mutagenesis, and host cell reactivation in a Chinese hamster ovary cell ERCC1 knock-out mutant. Mutagenesis. 1997;12:277–83. doi: 10.1093/mutage/12.4.277. [DOI] [PubMed] [Google Scholar]
- 34.Yuan B, Wang J, Cao H, Sun R, Wang Y. High-throughput analysis of the mutagenic and cytotoxic properties of DNA lesions by next-generation sequencing. Nucleic Acids Res. 2011;39:5945–5954. doi: 10.1093/nar/gkr159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clauson CL, Oestreich KJ, Austin JW, Doetsch PW. Abasic sites and strand breaks in DNA cause transcriptional mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:3657–62. doi: 10.1073/pnas.0913191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clauson CL, Saxowsky TT, Doetsch PW. Dynamic flexibility of DNA repair pathways in growth arrested Escherichia coli. DNA Repair (Amst) 2010;9:842–7. doi: 10.1016/j.dnarep.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jasti VP, et al. (5′S)-8,5′-Cyclo-2′-deoxyguanosine is a strong block to replication, a potent pol V-dependent mutagenic lesion, and is inefficiently repaired in Escherichia coli. Biochemistry. 2011;50:3862–5. doi: 10.1021/bi2004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuraoka I, et al. Oxygen free radical damage to DNA. Translesion synthesis by human DNA polymerase eta and resistance to exonuclease action at cyclopurine deoxynucleoside residues. J Biol Chem. 2001;276:49283–8. doi: 10.1074/jbc.M107779200. [DOI] [PubMed] [Google Scholar]
- 39.Tornaletti S. Transcription arrest at DNA damage sites. Mutat Res. 2005;577:131–45. doi: 10.1016/j.mrfmmm.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Steitz TA. The structural changes of T7 RNA polymerase from transcription initiation to elongation. Curr Opin Struct Biol. 2009;19:683–90. doi: 10.1016/j.sbi.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kornberg R. The molecular basis of eukaryotic transcription (Nobel Lecture) Angew Chem Int Ed Engl. 2007;46:6956–65. doi: 10.1002/anie.200701832. [DOI] [PubMed] [Google Scholar]
- 42.Brooks PJ. The 8,5′-cyclopurine-2′-deoxynucleosides: candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair. DNA Repair (Amst) 2008;7:1168–79. doi: 10.1016/j.dnarep.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi JY, Guengerich FP. Kinetic evidence for inefficient and error-prone bypass across bulky N2-guanine DNA adducts by human DNA polymerase iota. J Biol Chem. 2006;281:12315–24. doi: 10.1074/jbc.M600112200. [DOI] [PubMed] [Google Scholar]
- 44.Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase kappa. J Biol Chem. 2006;281:21062–72. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 45.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–8. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 46.Cohen SE, et al. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:15517–22. doi: 10.1073/pnas.1005203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng TF, Hu X, Gnatt A, Brooks PJ. Differential blocking effects of the acetaldehyde-derived DNA lesion N2-ethyl-2′ deoxyguanosine on transcription by multisubunit and single subunit RNA polymerases. J Biol Chem. 2008;283:27820–8. doi: 10.1074/jbc.M804086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.