Abstract
Protein synthesis is highly regulated throughout nervous system development, plasticity, and regeneration. However, tracking the distributions of specific new protein species has not been possible in living neurons or at the ultrastructural level. Previously we created TimeSTAMP epitope tags, drug-controlled tags for immunohistochemical detection of specific new proteins synthesized at defined times. Here we extend TimeSTAMP to label new protein copies by fluorescence or photo-oxidation. Live microscopy of a fluorescent TimeSTAMP tag reveals that copies of the synaptic protein PSD95 are synthesized in response to local activation of growth factor and neurotransmitter receptors, and preferentially localize to stimulated synapses in rat neurons. Electron microscopy of a photo-oxidizing TimeSTAMP tag reveals new PSD95 at developing dendritic structures of immature neurons and at synapses in differentiated neurons. These results demonstrate the versatility of the TimeSTAMP approach for visualizing newly synthesized proteins in neurons.
INTRODUCTION
Spatiotemporal control of protein synthesis is essential for proper development, normal functioning, and adaptation of nervous systems. In embryonic neurons, proteins are synthesized in axonal growth cones during migration, and local inhibition of protein synthesis blocks growth cone responses to axon guidance cues1,2. Later in development, high levels of protein synthesis in dendrites and axons promote synapse formation3,4. In the mature nervous system, protein synthesis is induced by neuronal activity and required for memory consolidation in animals5,6. Persistence of long term potentiation (LTP) and long-term depression (LTD), activity-dependent changes in synaptic function believed to underlie learning, also requires new protein synthesis to persist beyond 1 hour7,8. The production and targeting of new proteins also appears critical, as inhibition of protein synthesis locally at stimulated synapses blocks late-phase LTP at those synapses9.
The intricate regulation of protein synthesis during differentiation and plasticity of subcellular structures such as axons and synapses suggests that those synthesized proteins are utilized in these structures. An attractive hypothesis for the function of activity-induced protein synthesis in memory formation is that new proteins incorporate into activated synapses, causing long-lasting changes in synaptic function10. However, which specific new protein species are locally incorporated during differentiation or plasticity and where they localize relative to subcellular structures undergoing change remains poorly understood10. An impediment to addressing these questions has been the lack of generalizable methods to visualize new copies of specific proteins in living cells and with subsynaptic spatial resolution.
We previously developed TimeSTAMP, a method for drug-controlled epitope tagging of newly synthesized proteins11. In this method, a cassette comprising the nonstructural protein 3 (NS3) protease domain of hepatitis C virus (HCV) flanked by cognate protease sites is fused between a protein and an epitope tag. The protease excises itself and the tag from proteins by default, but this can be blocked for proteins synthesized after a defined time by applying a cell-permeable HCV NS3 protease inhibitor. These epitope-based TimeSTAMP tags have been used to visualize distributions of new proteins of interest in cultured mammalian neurons and in fly brains using immunostaining of fixed samples11.
We now report an extension of the TimeSTAMP method to visualize new proteins in living cells by fluorescence microscopy and in fixed sections by high-resolution electron microscopy (EM). Using a new fluorescent tag to track new copies PSD95, we find that local dendritic stimulation of growth factor and neurotransmitter receptors induces local accumulation of new PSD95 in stimulated synapses and dendritic shafts. Using new fluorescent photo-oxidizing tags, we show by correlated light and electron microscopy that newly synthesized PSD95 molecules rapidly incorporate beneath the postsynaptic membrane at synapses. The ability of these new TimeSTAMP tags to visualize new proteins in living neurons and at an ultrastructural level will enable researchers to study the role of new protein synthesis and delivery in vitro and in vivo.
RESULTS
Development of fluorescent TimeSTAMPs
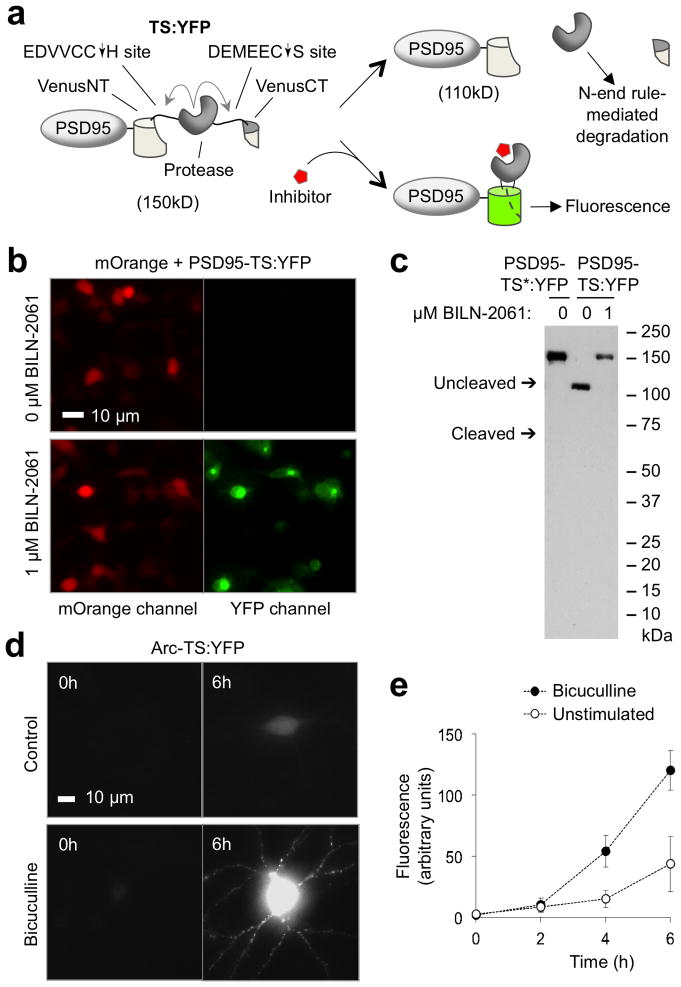
To track newly synthesized proteins of interest in living neurons, we first extended the TimeSTAMP technique to control maturation of fluorescent protein domains with drug. We reasoned that the NS3 protease placed within a fluorescent protein domain and allowed to remove itself in the absence of drug would generate fluorescent protein fragments that fail to associate and produce fluorescence, while protease inhibition by drug application would preserve linkage, allowing for fluorophore maturation (Fig. 1a). We first generated a single TimeSTAMP cassette to improve signal inducibility and to induce degradation of the protease following self-excision (Supplementary Fig. 1a). Cleavage efficiency and inhibitor binding were improved by fusing the 8-amino acid NS4A beta strand cofactor N-terminally to the NS3 domain. Breakthrough cleavage in the presence of drug was reduced by introducing the slow-cleaving T54A mutation into NS311. Inducible protease excision and elimination was achieved by introducing a new N-terminal cis-linked cleavage site with the P6-P1 sequence EDVVCC derived from the naturally preferred NS5A/5B substrate and with the N-end rule-inducing His at the P1′ position. The resulting cassette, TimeSTAMP2, showed improved control by NS3 protease inhibitors (complete inhibition at 1 μM BILN-2061 or ITMN-191 compared to 10 μM for original TimeSTAMP, Supplementary Fig. 1b,c). As desired, protease did not accumulate in the absence of drug (Supplementary Fig. 1d f). Similar to the original TimeSTAMP tags, TimeSTAMP2 allows drug-dependent epitope tagging in neurons and has no effect on synaptogenesis when targeted to synapses via PSD95 (Supplementary Fig. 1g,h).
Figure 1.
Development of TS:YFP, a TimeSTAMP tag with a YFP output. (a) Schematic of the TS:YFP cassette featuring protease-mediated fragmentation of a fluorescent protein, fused to the synaptic protein PSD95. (b) Imaging shows TS:YFP fluorescence is drug-dependent in HEK293 cells. BILN-2061 was continuously applied from 3 h after transfection to the time of imaging at 24 h after transfection. (c) Immunoblots confirm expected sizes for processed and full-length PSD95-TS:YFP fusion proteins in the absence or presence of drug, respectively (lanes 2 and 3). BILN-2061 was continuously applied from 3 h after transfection to the time of lysis at 24 h after transfection. In the left lane, PSD95-TS*:YFP is a protease-dead mutant of PSD95-TS:YFP in which the serine of the catalytic triad is mutated to alanine. (d) TS:YFP reveals that new Arc protein synthesis is induced by 20 μM bicuculline-stimulated synaptic activity in a 21-day in vitro (DIV) neuron. Images are of cells immediately before BILN-2061 addition and after 6 h in 1 μM BILN-2061. (e) Quantification of whole-cell Arc-TS:YFP fluorescence over time in unstimulated and stimulated neurons. Differences at 4 and 6 h are statistically significant (p ≤ 0.05 by unpaired two-sided t-test, n = 6 neurons per condition). Error bars represent standard error of the mean (SEM).
We next used TimeSTAMP2 to create autofluorescent drug-controlled labels of new protein copies. We screened insertions of the TimeSTAMP and TimeSTAMP2 modules into loops of fluorescent protein domains for drug-dependent fluorophore development. Among the constructs tested (Supplementary Fig. 2a), Venus yellow fluorescent protein (YFP) with the TimeSTAMP2 module inserted between amino acids 158 and 159 exhibited robust drug-dependent fluorescence (Fig. 1b). Immunoblotting confirmed that protease excision occurs efficiently in the absence of drug but is fully inhibited in 1 μM BILN-2061 (Fig. 1c). The fusion of this construct to PSD95 localized to synapses similarly to the previously characterized PSD95-CFP12 (Supplementary Fig. 2b), and it did not exert observable effects on synaptogenesis (Supplementary Fig. 2c). Therefore, this construct, TimeSTAMP:YFP (TS:YFP), can function as a drug-inducible continuous fluorescent label of newly synthesized proteins.
TS:YFP fluorescence arising during drug treatment persisted after drug washout (Supplementary Fig. 2d), consistent with previous findings that fluorescent protein fragment assembly is irreversible13. Thus, TS:YFP should function for pulse labeling by irreversibly labeling proteins synthesized in a drug-defined pulse period followed by production of unlabeled protein after washout. This would be conceptually analogous to pulse-chase labeling with radioactive metabolic precursors, without requiring a “chase” with excess nonradioactive precursors to dilute out radioactive precursors. To test optical pulse labeling, we expressed in primary rat embryonic hippocampal neurons the transmembrane synaptic adhesion molecule Neuroligin3 (NLGN3) tagged with TS:YFP. At 14 days in vitro (DIV), we induced fluorescence on new proteins by drug incubation for 18 h followed by washout. Time-lapse microscopy showed fluorescence appearing during the pulse period, initially in perinuclear structures consistent with endoplasmic reticulum and Golgi and then at the cellular membrane and in synapses (Supplementary Fig. 2e). After washout, fluorescence decreased from all locations with an approximate half-life of 24 h, consistent with half-life measurements performed by isotopic labeling14. These results confirm that TS:YFP can be used for optical pulse labeling.
To test the ability of TS:YFP to report on activity-dependent protein synthesis, we tracked the production of Arc, which is translationally induced by synaptic activity8, in basal and stimulated conditions. In this reporter, we fused TS:YFP C-terminally to Arc coding sequence and included the complete pre-mRNA sequence with the 3′ untranslated region (UTR) and introns, which are involved in Arc mRNA localization and translational induction by neuronal activity15. In 21-DIV hippocampal neurons expressing this Arc-TS:YFP reporter, no fluorescence was detectable from the transfected neurons prior to BILN-2061 treatment, while fluorescence increased over time in 1 μM BILN-2061 (Fig. 1d). Bicuculline, which promotes action potential generation in excitatory glutamatergic neurons by antagonizing GABA receptors, increased the rate of fluorescence development (Fig. 1e), consistent with previous observations16, thereby confirming that TS:YFP can report activity-dependent protein translation.
To simultaneously visualize newly synthesized proteins from two species, we also created an orange fluorescent TimeSTAMP (TS:OFP) using an alternative design based on cleavage-induced N-end rule degradation of a fully intact mKO2 orange fluorescent protein (Supplementary Fig. 3a). In the absence of drug, the released protease-mKO2 fusion was degraded, as assessed by direct fluorescence (Supplementary Fig. 3b) and immunoblotting (Supplementary Fig. 3c), whereas EGFP, mCherry, or Venus in place of mKO2 were resistant to degradation (Supplementary Fig. 3d). This TS:OFP tag may be useful together with TS:YFP for tracking new copies of two proteins simultaneously. However, one clear advantage of TS:YFP is that the fluorescent protein remains intact after drug washout for monitoring protein turnover by fluorescence.
Stimulus-dependent PSD95 translation revealed by TimeSTAMP
We next used fluorescent TimeSTAMP to tested whether synaptic proteins synthesized after plasticity induction incorporate into stimulated synapses. It has been suggested that activity-induced proteins are delivered to stimulated synapses to promote plasticity persistence. PSD95 functions to build a postsynaptic density (PSD) by cross-linking receptors and cytoskeletal elements17, and its synthesis is induced in synaptoneurosomes by mGluR stimulation18. However, whether local stimulation induces local synaptic accumulation of new PSD95 has not been investigated.
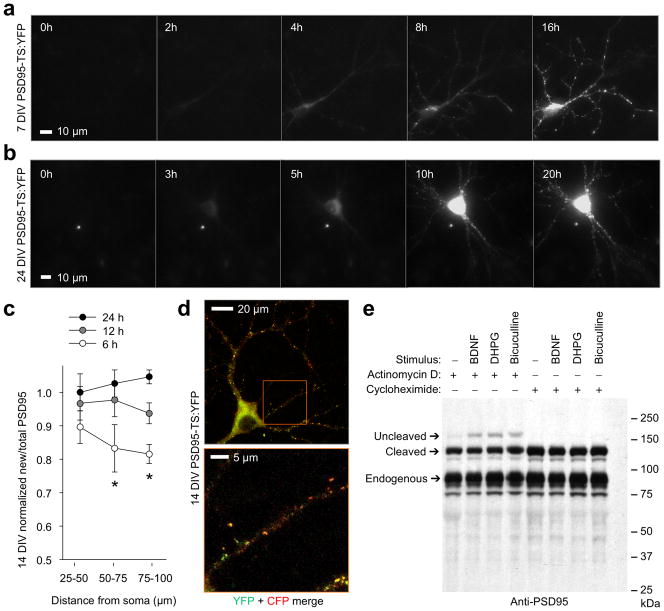
To visualize new PSD95 in living neurons, we fused PSD95 coding sequence to TS:YFP and encoded the complete native 3′ UTR, required for mRNA stabilization and translational control18–20. Neurons expressing this PSD95-TS:YFP reporter exhibited no fluorescence in the absence of BILN-2061, but fluorescence accumulated over time in its presence (Fig. 2a). Transfected neurons undergoing synaptogenesis at 7 DIV showed drug-dependent fluorescence in dendritic puncta that appeared simultaneously with dendrite arborization (Fig. 2a), suggesting incorporation of new PSD95-TS:YFP into nascent synapses as previously observed with TimeSTAMP tags11. At 24 DIV, drug-dependent fluorescence arose gradually over 24 h in puncta throughout the dendritic tree, consistent with accumulation at pre-existing synapses (Fig. 2b). Similarly, ratios of new/total PSD95 in neurons at 14 DIV revealed slower synaptic accumulation of newly synthesized PSD95-TS:YFP further from the cell body, as indicated by ratios of PSD95-TS:YFP/PSD95-CFP fluorescence (Fig. 2c,d), similar to earlier observations 11. These results confirm that TS:YFP can track new copies of PSD95 and that under basal conditions new PSD95 expression does not occur preferentially in dendrites.
Figure 2.
Tracking of basal and activity-induced PSD95 production with TimeSTAMP. (a) New PSD95 accumulates in dendritic puncta during arborization in a 7-DIV neuron. New puncta appear on distal branches simultaneously with their extension. Times in 1 μM BILN-2061 are shown. (b) At 21 DIV, new PSD95 accumulates in puncta throughout the dendritic tree. (c) Distally located synapses incorporate less new PSD95 than proximal ones in 14-DIV neurons. PSD95-TS:YFP fluorescence in segments of the primary dendrite was divided by cotransfected PSD95-CFP fluorescence for each time, then normalized to the initial value in the 25-μm segment. Mean differences between times were significant for 50–75 μm and 75–100 μm segments by ANOVA (p = 0.018 and 0.020 respectively, n = 7 segments each). Differences between 24 h and 6 h were significant by post-hoc Tukey’s tests (p = 0.017 and 0.016 for segments at 50–75 μm and 75–100 μm, respectively). Error bars represent SEM. (d) Upper image, a representative neuron from (c) showing that PSD95 synthesized for 20 h in 1 μM BILN-2061 under basal conditions exists in a gradient from the cell body. PSD95-TS:YFP protein is in green while PSD95-CFP, a marker of total PSD95, is in red. Lower image, enlarged view of boxed region in upper image. (e) TimeSTAMPa revealed induction of PSD95 synthesis by stimuli. 14-DIV neurons expressing PSD95-GFP-TimeSTAMPa were treated for 3 h with 10 μM BILN-2061 and stimuli along with actinomycin D to block transcriptional upregulation or cycloheximide to block protein synthesis. Anti-PSD95 immunoblotting reveals proteins synthesized during BILN-2061 incubation migrating at the uncleaved size. 100 ng/mL BDNF, 50 μM DHPG, and 20 μM bicuculline each induce PSD95 synthesis above baseline independent of transcription. Endogenous PSD95 serves as a loading control.
We next investigated the regulation of PSD95 translation in experimental models of synaptic plasticity. The TrkB receptor tyrosine kinase and type 1 metabotropic glutamate receptors (mGluRs) regulate protein synthesis in synaptic plasticity, and TrkB activation by brain-derived neurotrophic factor (BDNF) is both necessary for the late phase of electrically induced LTP21 and sufficient to induce long-lasting increases in synaptic currents that resemble electrically induced LTP22. Activation of type 1 mGluRs by pharmacological agonists CHPG and DHPG can either induce LTD or decrease stimulation thresholds for LTP, depending on context23. Both receptor classes activate the PI3K-mTOR pathway24,25, which can promote PSD95 translation26. To determine if TrkB or type 1 mGluR pathways regulate PSD95 translation, we performed immunoblotting experiments with PSD95-GFP-TimeSTAMPa, composed of PSD95 fused to GFP and the TimeSTAMPa cassette and encoding the complete PSD95 3′ UTR11. Bath stimulation of 14-DIV neurons with BDNF or the type I mGluR agonist DHPG increased the amount of new PSD95-GFP, detected as a slower migrating species in the presence of BILN-2061 (Fig. 2e). This induction was blocked by cycloheximide but insensitive to actinomycin D, indicating dependence on protein synthesis but not transcription. Bicuculline also enhanced new PSD95-GFP protein production. These results indicate that PSD95 synthesis is regulated by neurotrophin and neurotransmitter receptors and by synaptic activity.
We next hypothesized that TrkB or type 1 mGluR activation can induce local accumulation of new PSD95 at stimulated dendritic regions and synapses. To perform local stimulations, we microfabricated polydimethylsiloxane devices in which two culture compartments are separated by a 50-μm thick barrier traversed by 7-μm wide microchannels connecting the compartments (Supplementary Fig. 4a). This device is based on earlier devices for axonal isolation27, but the barrier is thinner to permit dendrites to traverse. Compounds applied to one compartment diffuse down the microchannels, but compartment concentrations do not detectably change over 12 h even without active microfluidic control due to the small volumes of the microchannels compared to the compartment (Supplementary Fig. 4b d). When neurons are transfected and plated in both compartments, some extend their dendrites through the microchannels to the opposite compartment, allowing their distal dendrites to be selectively stimulated (Supplementary Fig. 4e).
Specificity of new PSD95 for stimulated synapses
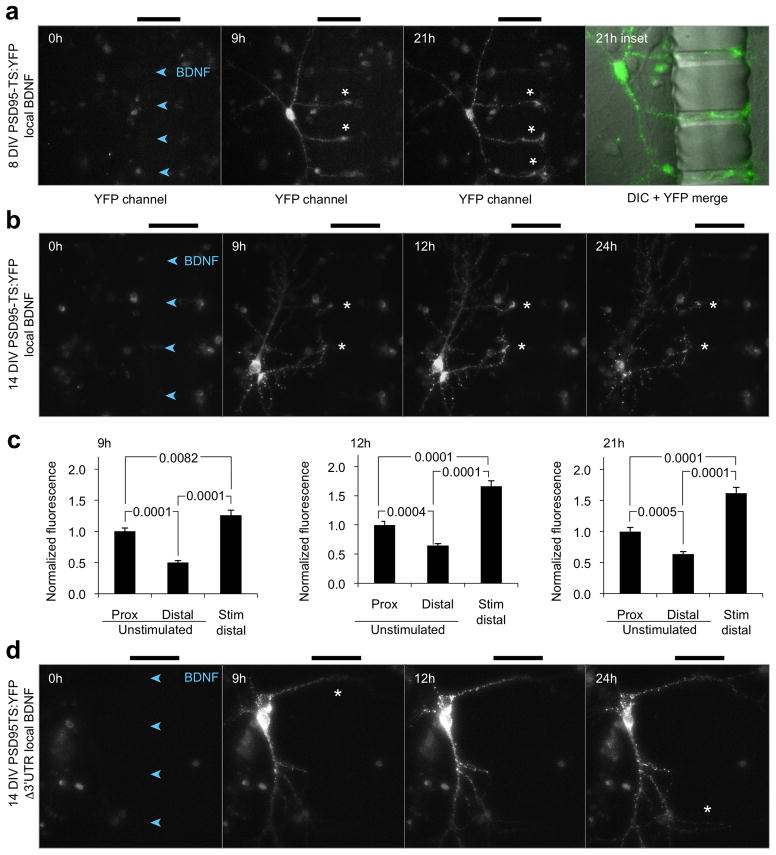
To determine whether distributions of new PSD95 copies can be spatially regulated in developing neurons, we stimulated distal dendrites of 8-DIV neurons expressing PSD95-TS:YFP with BDNF and added BILN-2061 to both chambers to monitor new PSD95 anywhere. New PSD95 copies were enriched at tips of neurites extending toward the BDNF-stimulated compartment in the microchannels (Fig. 3a). These structures are likely developing dendrites rather than axons due to their multiple number, thickness, and presence of PSD95. Fluorescence intensity per dendritic length was higher in BDNF-stimulated distal segments than in proximal segments of the same dendrite and higher in stimulated dendritic segments than in segments from matched unstimulated dendrites equally far from the cell body (Fig. 3a). To determine if new PSD95 protein can preferentially accumulate in stimulated synapses in response to BDNF, performed this experiment in 14-DIV neurons. Local BDNF stimulation induced intense accumulations of new PSD95 in shafts and synapses of dendrites entering the microchannels beginning at 9 h and persisting to 24 h (Fig. 3b). At 9, 12, and 21 h, synaptic intensities of new PSD95 (adjusted for synapse size) in stimulated distal regions were 26–62% higher than in unstimulated proximal regions of the same dendrite and 121–153% higher than in unstimulated distal regions of dendrites from the same cell that were similarly distant from the cell body (Fig. 3c). These results reveal that new PSD95 copies can be preferentially expressed at BDNF-stimulated dendritic regions and synapses.
Figure 3.
BDNF-dependent local accumulation of new PSD95 in neurons. neurons expressing PSD95-TS:YFP were treated with 100 ng/mL BDNF in the right chamber at time 0 while 1 μM BILN-2061 was applied to both chambers, also at time 0. Arrowheads within the channels point in the direction of BDNF diffusion, and bars above the images mark the location of the 50-μm barrier. (a) New PSD95 accumulates at BDNF-stimulated growth cones in an 8-DIV neuron expressing PSD95-TS:YFP. Asterisks mark growth cones with new PSD95. The fourth panel is an inset of the 21-h field showing merged TS:YFP fluorescence (green) and differential interference contrast (DIC, grayscale) images. (b) New PSD95 is enriched at BDNF-stimulated regions in two 14-DIV neurons (asterisks) expressing PSD95-TS:YFP. (c) New PSD95 mean concentrations are higher in synapses of stimulated regions (within 10 μm of the microchannels, n = 31) versus synapses located more proximally on the same dendritic branch (n = 40 synapses) as well as versus synapses equidistant from the soma on an unstimulated branch (n = 36 synapses). Differences were significant by one-way ANOVA at all time points (p = 0.0001). p values of posthoc unpaired two-tailed t-tests with Tukey’s correction are indicated on the charts. Error bars represent SEM. (d) New PSD95 is not preferentially detected at BDNF-stimulated synapses when the 3′ UTR is removed.
The reporter constructs used in the above experiments included the complete PSD95 3′ UTR, which mediates localization of the mRNA to dendrites and translational regulation by FMRP18,20. Removal of the 3′ UTR would thus be expected to prevent dendritic localization of the mRNA and FMRP-dependent translational induction, and this truncation would test whether local BDNF-induced signals recruit PSD95 proteins synthesized in an FMRP-independent manner in the cell body. In neurons expressing PSD95-TS:YFP lacking the 3′ UTR and dendritically stimulated with BDNF, new PSD95 was primarily expressed in the cell body with no specific enrichment in stimulated regions at any time (Fig. 3d). These results indicate that dendritic translation of PSD95 and/or FMRP involvement is required for localized dendritic accumulation of new PSD95 at stimulated synapses, and they demonstrate that TS:YFP can be used to further study 3′ UTR function in mediating activity-dependent local accumulation of new synaptic proteins.
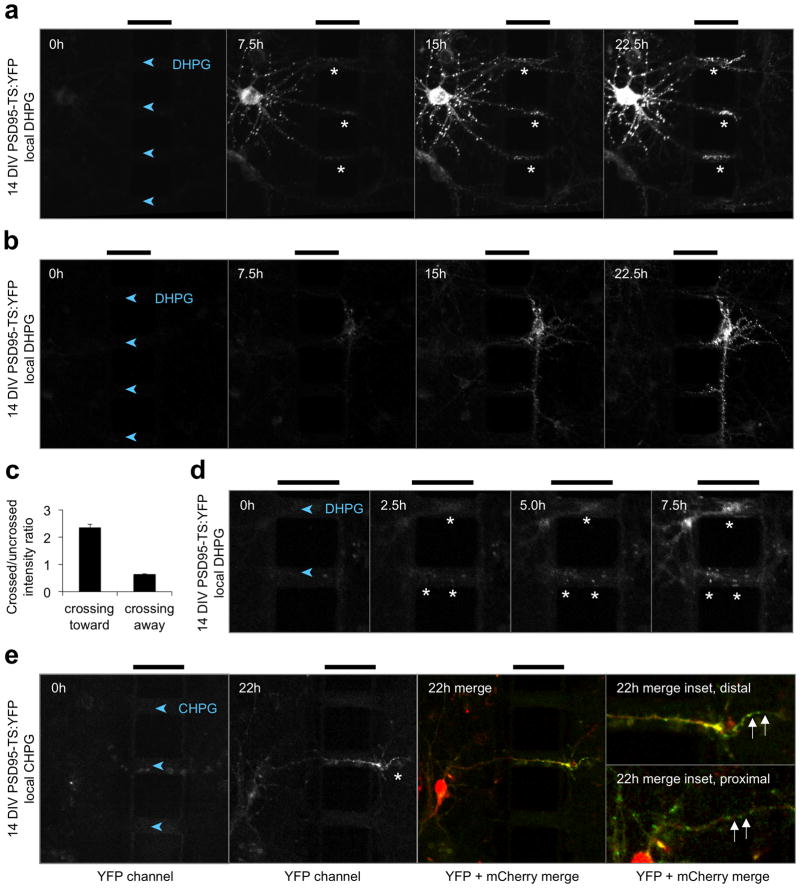
To determine if new PSD95 protein are also recruited to synapses after mGluR stimulation, we stimulated distal dendrites of 14-DIV neurons expressing PSD95-TS:YFP with DHPG. We again observed synaptic accumulation of new PSD95 in stimulated regions (Fig. 4a and Supplementary Videos 1,2). New PSD95 levels were enriched 2.4-fold in stimulated segments compared to unstimulated segments equidistant from the cell body at 22.5 h (Fig. 4c), the magnitude of which is similar to increased levels of translationally regulated protein that were previously observed with neurotransmitter receptor modulation28,29 and cannot be accounted for solely by the 20% increase in PSD size previously observed in response to BDNF30. By contrast, dendrites extended toward the unstimulated side showed less new PSD95 (Fig. 4b,c), ruling out that the effect is a non-specific result of crossing through a microchannel. In some neurons, new PSD95 enrichment was first visible in stimulated synapses 2.5 h after stimulation began (Fig. 4d) and persisted for the duration of the experiment. Similar results were obtained with CHPG, a specific agonist of mGluR5, the predominant type 1 mGluR at hippocampal synapses 31 (Fig. 4e). Close examination revealed that new PSD95 was present in both synapses and dendritic shafts within the stimulated region (Fig. 4a,e). These findings show that neurotransmitter receptor activation in a dendritic region can induce accumulation of new copies of PSD95 in the stimulated region, including at synapses. These results further demonstrate the ability of TS:YFP to visualize new copies of a synaptic protein during local dendritic stimulations.
Figure 4.
Local new PSD95 accumulation is induced by local metabotropic glutamate receptor activation. (a) New PSD95 is enriched in DHPG-stimulated regions in a 14-DIV neuron (asterisks). Time-lapse imaging was performed in the presence of 1 μM BILN-2061 in both chambers and 100 μM DHPG in the right chamber for the times shown. (b) Representative neuron exhibiting no enrichment of new PSD95 in distal dendrites crossing from the DHPG-stimulated compartment into the unstimulated compartment. (c) The mean ratio of new PSD95 in synapses within a 20-μm dendritic segment crossing the barrier versus in a 20-μm non-crossing dendritic segment from the same neuron was quantified at 22.5 h. This ratio was greater than 1 in neurons with cell bodies in the unstimulated compartment extending dendrites into the stimulated compartment, but less than 1 for neurons extending dendrites in the opposite direction (p = 0.009 by unpaired two-tailed t-test, n = 8 and 4 segments for crossing-toward and crossing-away neurons, respectively). Error bars represent SEM. (d) Enrichment of new PSD95 in synapses and dendrites encountering DHPG (asterisks) can be seen as early as 2.5 h. (e) Similar enrichment of new PSD95 in stimulated dendrites was observed in response to 300 μM of the mGluR5-selective agonist CHPG. Asterisks mark new PSD95 at stimulated synapses. The third panel shows merged TS:YFP (green) and mCherry (red). In the magnified insets, new PSD95 is seen in the dendritic shaft in the stimulated region (top) but not in the unstimulated region (bottom). The intensity gain of the TS:YFP signal in the bottom inset is twice that of the other images. In all images, arrowheads within the channels point in the direction of DHPG or CHPG diffusion, and bars above the images mark the 50-μm barrier.
Development of photo-oxidizing TimeSTAMPs
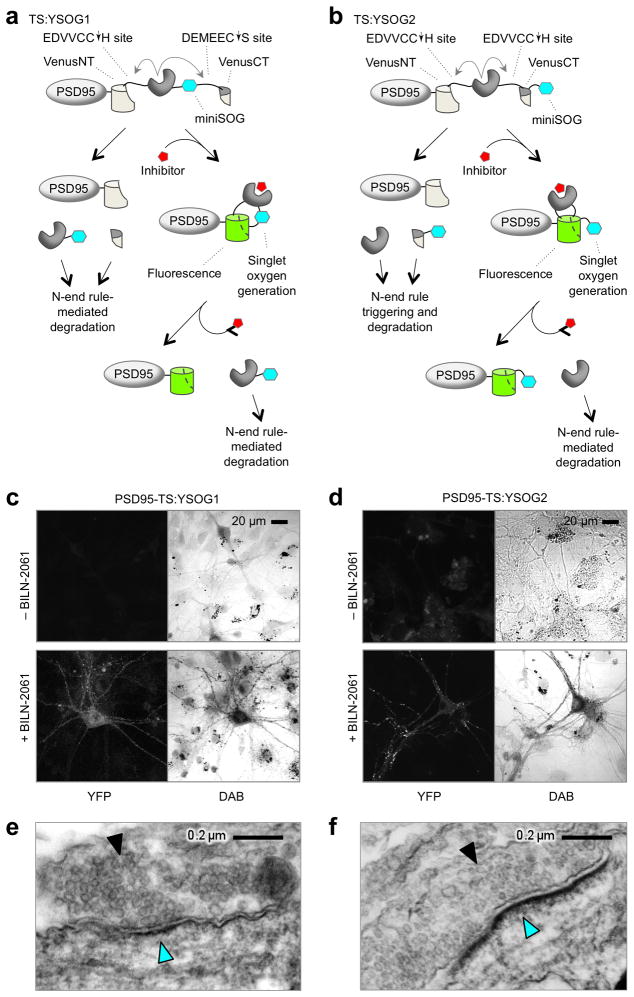
We next sought to visualize new proteins of interest in neurons with ultrastructural resolution by EM. Because two epitope tags in TS:YFP are drug-dependent (Supplementary Fig. 5a e), we first tested peroxidase-conjugated antibodies to deposit diaminobenzidine (DAB) for EM imaging. However, the permeabilization steps required for antibody penetration resulted in poor ultrastructural detail (Supplementary Fig. 5f). Therefore, we modified TS:YFP to add the 12-kD genetically encoded singlet oxygen generator miniSOG, which can mediate photo-oxidation of DAB for EM visualization32. Photo-oxidation with miniSOG removes concerns about non-specific antibody binding and does not require sample permeabilization, allowing for better ultrastructural preservation32. We created two designs incorporating miniSOG (Fig. 5a,b). In TimeSTAMP:YFPminiSOG 1 (TS:YSOG1), miniSOG was fused to the protease domain in TS:YFP for degradation by the N-end rule in the absence of drug but preservation in its presence (Fig. 5a). In TS:YSOG2, miniSOG was fused after the C-terminal YFP fragment, and the C-terminal cleavage sequence was changed to allow for cleavage-induced N-end rule-mediated degradation of miniSOG in the absence of drug but preservation in its presence (Fig. 5b). Although the second version is limited by fusion to the C-terminus of proteins of interest because it degrades its C-terminal cleavage product, its advantage over the first is its applicability for pulse labeling. Because miniSOG in TS:YSOG2 is covalently linked to the C-terminal YFP fragment it will remain bound to the protein of interest during drug pulse and washout (Fig. 5b), whereas miniSOG in TS:YSOG1 would be cleaved away and degraded with the protease during drug washout (Fig. 5a).
Figure 5.
Design and development of photo-oxidizing TimeSTAMPs. Schematics of the TS:YSOG1 (a) and TS:YSOG2 (b) cassettes with drug-dependent singlet oxygen generation ability encoded by the miniSOG domain (aqua), fused to the synaptic protein PSD95. PSD95-TS:YSOG1 (c) and PSD95-TS:YSOG2 (d) produce correlated drug-dependent fluorescence and DAB images in neurons. PSD95-TS:YSOG1 (e) and PSD95-TS:YSOG2 (f) DAB signal is localized to PSDs of neurons incubated with drug. Solid arrowhead marks abundant 30- to 40-nm vesicles indicative of a presynaptic bouton. Cyan arrowhead indicates DAB signal at and beneath the apposing postsynaptic membrane. The excess black dots present in transmitted light images in (c) and (d) are non-specific miniSOG-independent DAB precipitates that commonly form when photo-oxidizing cultured neurons. Because this signal is mainly extracellular and does not match YFP fluorescence, it was easily distinguishable from true miniSOG signal at transmitted light and EM levels.
In lysates from PSD95-TS:YSOG1 or PSD95-TS:YSOG2-expressing cells grown without BILN-2061, blotting for the AU1 or HA epitope, respectively, in the miniSOG fragment detected no protein (Supplementary Fig. 6a,b), confirming efficient cleavage and N-end rule-mediated degradation. In the continual presence of BILN-2061, only uncleaved PSD95-TS:YSOG1 or PSD95-TS:YSOG2, respectively, were detected, as expected. Both constructs exhibited fluorescence induction by BILN-2061 similar to TS:YFP (Supplementary Fig. 6c). Like PSD95-TS:YFP, PSD95-TS:YSOG1 and PSD95-TS:YSOG2 both localized to synaptic sites on dendritic spines (Supplementary Fig. 6d,e) without disrupting synaptogenesis (Supplementary Fig. 6f).
We tested the performance of photo-oxidizing TimeSTAMP tags in correlated light and electron microscopy. In 12-DIV neurons expressing PSD95-TS:YSOG1, fluorescence was negligible in the absence of drug, but appeared throughout cells after 48 h in drug (Fig. 5c). We fixed these neurons without permeabilization and performed miniSOG-dependent photo-oxidation of DAB with blue light. The resulting DAB precipitate overlapped well with the YFP signal by light microscopy, indicating specificity for the reporter protein (Fig. 5c). By EM, fine subcellular structures such as microtubules and synaptic vesicles were clearly visible in photo-oxidized samples (Fig. 5e), demonstrating excellent ultrastructure preservation and fine resolution of synaptic elements similar to conventional label-free samples33. The majority of DAB label localized to submembrane locations across from synaptic vesicles, consistent with PSD95 localization, and DAB signal was dependent on drug (Supplementary Fig. 7a). Similar results were obtained with PSD95-TS:YSOG2 (Fig. 5d,f and Supplementary Fig. 7b).
In early experiments, miniSOG-independent DAB signal appeared in neuronal mitochondria after photo-oxidation (Supplementary Fig. 8a). Endogenous mitochondrial iron-sulfur complexes can photosensitize oxygen at the wavelengths used with miniSOG, but mersalyl acid abolishes this effect.28 Indeed, mersalyl acid treatment (5 mM, 30 minutes) before photo-oxidation reduced mitochondrial DAB signal 8-fold (Supplementary Fig. 8b,c). With this step included, EM of DAB-positive puncta in neurons expressing PSD95 tagged with either TS:YSOG1 or TS:YSOG2 reliably revealed synaptic structures with excellent ultrastructural preservation and with the highest electron density underneath the postsynaptic membrane, as expected (Supplementary Fig. 8d,e). These results confirm that photo-oxidizing TimeSTAMP tags allow high-resolution EM imaging.
Tracking newly synthesized PSD95 proteins by EM
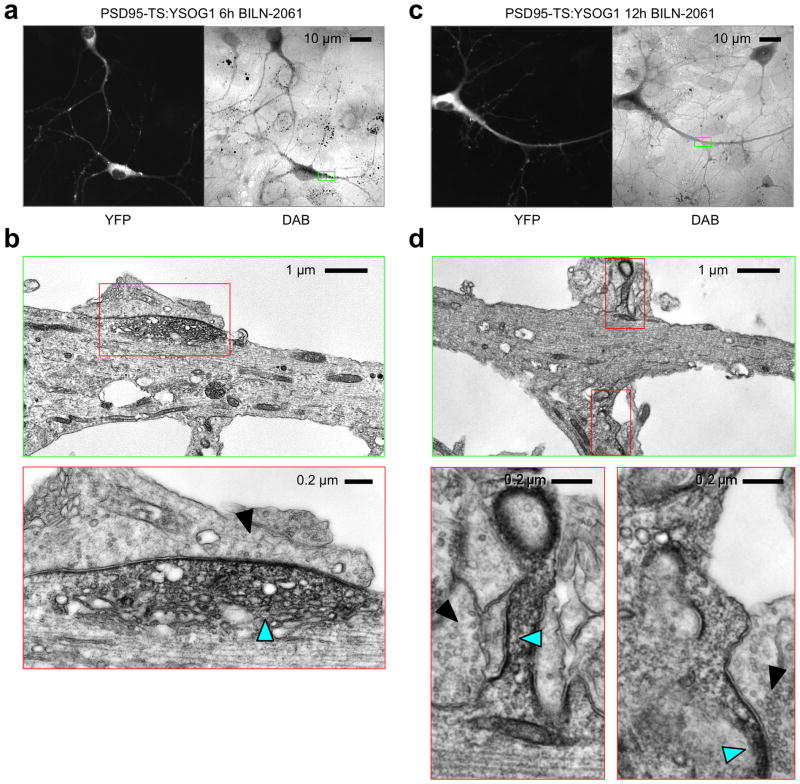
We examined distributions of freshly synthesized PSD95 using TS:YSOG1 by treating neurons expressing PSD95-TS:YSOG1 at 12 DIV, a time of synaptogenesis, with 1 μM BILN-2061 for 6 h before fixation. By fluorescence, YFP signal appeared diffusely throughout the dendrite except for a few large puncta (Fig 6a). These puncta appeared larger than typical PSDs, but the resolution of fluorescence microscopy was insufficient to determine if they contain synapses. We then performed photo-oxidation and obtained a pattern of DAB deposition similar to YFP fluorescence. EM visualization of one dendritic PSD95 accumulation located at a junction with an axon in the fluorescence image (Fig. 6a) revealed it to be within a dendritic protrusion and containing diffuse DAB signal (Fig. 6b). There was little signal in the rest of the dendrite, suggesting that new PSD95 was preferentially localized to these diffuse accumulations (Fig. 6b). The dimensions of this accumulation was 2 μm wide and 0.6 μm deep, larger in each axis than a mature PSD33. The presence of PSD95 that is not membrane associated implies that some PSD95 protein may not be palmitoylated34. In addition, the microtubule network appeared disorganized in this protrusion, suggesting that this was a dynamic structure in the cell35. This protrusion was apposed to an axonal segment with a low density of vesicles (fewer than 4 within 100 nm), a criterion used to describe immature synapses under EM36. We speculate that this structure may be a newly forming synapse, as new synapses often arise at sites with diffuse accumulations of PSD9511,37.
Figure 6.
Tracking new PSD95 populations using TS:YSOG1. (a) After 6 h in BILN-2061, new PSD95 protein was visible throughout shafts with enrichment in 2 μm-wide accumulations. (b) EM imaging of one accumulation after photo-oxidation revealed a diffuse accumulation of membrane-bound and cytoplasmic PSD95 (cyan arrowhead) in contact with an axonal region with a low density of vesicles (solid arrowhead). (c) After 12 h in BILN-2061, new PSD95 protein was visible throughout shafts and in numerous puncta less than 1 μm wide. (d) EM imaging of two small puncta revealed membranous and submembranous PSD95 (cyan arrowhead) in contact with an axonal region with high vesicular density indicative of a mature synapse (solid arrowhead).
In PSD95-TS:YSOG1-expressing neurons treated with BILN-2061 for 12 h, we observed labeling of new PSD95 throughout dendrites by both fluorescence and DAB (Fig. 6c). In one region we examined by EM (Fig. 6c), the highest signal was observed within spines (Fig. 6d), and new PSD95 preferentially accumulated on membranes contacting apposing mature presynaptic structures (Fig. 6d). These accumulations measured 500 nm wide and less than 30 nm deep, dimensions typical for a mature PSD33. Thus, new PSD95 populations appear at mature synapses within 12 h.
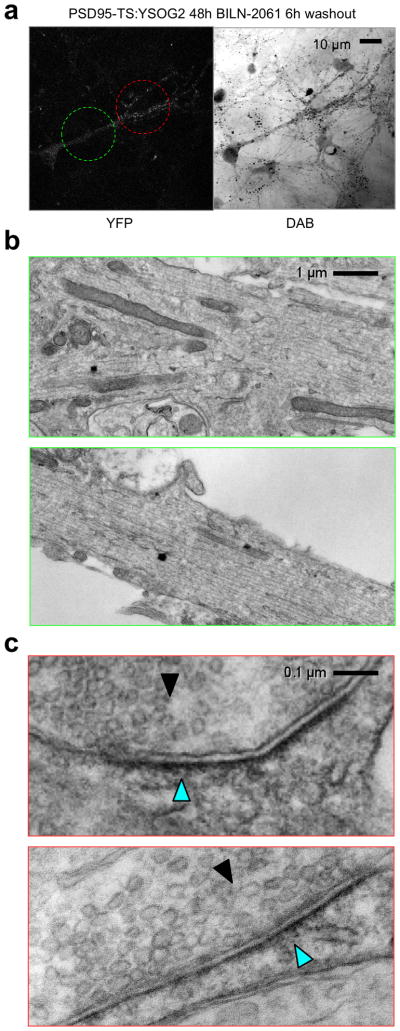
To pulse-label PSD95 synthesized in a specific time window, we incubated 12-DIV neurons expressing PSD95-TS:YSOG2 with 1 μM BILN-2061 for 48 h and then removed drug for 6 h. By fluorescence, we observed YFP signal in small and bright puncta in distal neuronal processes (Fig. 7a). We also noticed a decrease in diffuse YFP signal in soma and proximal processes compared to neurons maintained continuously in drug (Fig. 7a, 6b), suggesting that proteins labeled in the pulse period have moved out of these structures. After photo-oxidation, we visualized by EM two dendritic areas within one cell (Fig. 7a). A more proximal area that had only dim diffuse signal by fluorescence microscopy exhibited no specific DAB signal by EM, suggesting clearance of PSD95 proteins from the cytosol 6 h after synthesis (Fig. 7b). A more distal area containing multiple fluorescent puncta smaller than 1 μm in diameter exhibited PSD95 proteins localized to mature synapses, as indicated by submembranous staining apposing presynaptic structures with abundant vesicles (Fig. 7c). Taken together, these results show that new PSD95 proteins localize to synapses by 6 h after synthesis in 14-DIV neurons.
Figure 7.
Pulse labeling of older PSD95 populations using TS:YSOG2. (a) After 48 h in BILN-2061 followed by 6 h washout, pulse-labeled PSD95 protein was visible at low levels in shafts with enrichment in numerous puncta less than 1 μm wide. (b) EM imaging in a dendritic shaft area without fluorescent puncta revealed no accumulations of pulse-labeled PSD95. (c) EM imaging in an area with fluorescent puncta revealed pulse-labeled PSD95 at multiple synapses, detected as membrane-bound aggregations of 500 nm width (cyan arrowheads) facing a presynaptic zone with a large number of synaptic vesicles (solid arrowheads).
DISCUSSION
Comparison of TimeSTAMPs with other translation reporters
We developed fluorescent and photo-oxidizing TimeSTAMP tags to visualize new copies of proteins of interest in living neurons and by EM. These new TimeSTAMP tags exhibit a unique combination of features suitable for studying the fates of newly synthesized proteins during nervous system development and plasticity. First, they are genetically encoded, so they can be appended to specific proteins of interest. Second, they are drug-controllable, which both provides an easy means to control the time period of new protein labeling and allows easy control in large tissue volumes. Lastly, they are fluorescent and photo-oxidizing, allowing visualization of new protein copies in living cells and by EM.
As protein tags, TimeSTAMPs can report on all steps that control protein expression and localization, including transcriptional, translational, and post-translational regulatory events and protein-protein interactions. This is especially critical for studying synaptic proteins, which are dynamically regulated by multiple mechanisms. For instance, proteins of the PSD are held together by a network of protein-protein interactions, which are regulated by post-translational modifications such as phosphorylation38, palmitoylation39, proteolysis40, and ubiquitination41. In contrast, destabilized GFP reporters cannot be used as fusion tags because of their destabilizing nature, and while capable of reporting transcriptional and translational control28,29, do not report on subsequent steps of protein localization, interactions, or modification.
Monomeric fluorescent protein timers and photoconvertible fluorescent proteins42 can in theory be used as fluorescent tags that report on protein age in living cells. However, a population of timer molecules changes color gradually over many hours due to an autocatalytic conversion reaction occurring at different times in each molecule, and thus fluorescent protein timers cannot provide the same temporal resolution or control as TimeSTAMP42. Monomeric photoconvertible fluorescent proteins can be fused to a protein of interest and converted from one emission wavelength to another using violet light42, after which only new proteins will fluoresce at the unconverted wavelength. However, detecting low concentrations of new proteins requires complete conversion throughout the cell, and the prolonged exposures and high intensities required often cause phototoxicity43. Extending the use of photoconvertible proteins to a large volume in vivo would also be difficult, whereas drugs are routinely perfused throughout a slice or injected into a brain region.
A powerful non-genetically encoded approach to labeling new proteins is to metabolically incorporate physically or chemically distinct amino acids. Classically this was performed with radioactive amino acids for detection by autoradiography. Recently the approach has been extended to amino acids with nonradioactive isotopes for detection by mass spectroscopy and, in the BONCAT technique, to unnatural amino acids bearing reactive chemical groups that can then be conjugated to fluorophores or affinity tags44. These metabolic labeling approaches differ in applicability from TimeSTAMP in two ways. First, they result in the labeling of all new proteins proportional to abundance rather than specifically labeling proteins of interest. Second, they currently do not allow live visualization of new proteins. For visualization with autoradiography, samples must be fixed and then exposed to film. For visualization with BONCAT, cells must be starved of natural amino acids prior to unnatural amino acid addition, then after the labeling period, fixed and reacted in non-physiological conditions. Metabolic labeling approaches, however, enable de novo identification of synthesized proteins, either by mass spectrometry for heavy isotopes, or by reaction to affinity groups and purification for chemically reactive amino acids. Metabolic labeling methods such as BONCAT can therefore be used to screen for previously unknown locally translated or activity-induced proteins. In contrast, TimeSTAMPs are intended to visualize specific proteins of interest, and thus are well suited for further investigation of any proteins identified in screens by mass spectroscopy or BONCAT.
Preferential incorporation of PSD95 at stimulated synapses
The precise function of activity-induced protein synthesis in synaptic plasticity is still poorly understood. An attractive hypothesis is that proteins synthesized in response to synaptic activity function at activated synapses to promote long-lasting structural changes6,45. This hypothesis predicts that new copies of specific proteins synthesized after synaptic activation will localize to activated synapses. However, an alternative hypothesis is that new protein synthesis replenishes pre-existing proteins that are rapidly consumed during synaptic plasticity45,46. If copies of a particular protein synthesized in response to activity are used primarily to replenish global stocks rather than to remodel activated synapses, old copies would be utilized by the stimulated synapses and new proteins would resupply a dendritic pool to be utilized later by all synapses. These hypotheses thus make different predictions about the fates of newly synthesized proteins.
Our results demonstrate that copies of PSD95 synthesized after localized stimulation preferentially accumulate in stimulated dendritic regions and synapses. This is the first direct observation that copies of a synaptic structural protein synthesized after local dendritic stimulation can preferentially incorporate into stimulated synapses. Because new protein copies are preferentially incorporated at stimulated synapses, our findings support the hypothesis that new synthesis of PSD95 has synapse-autonomous functions and is not solely functioning in global replenishment. Multiple mechanisms have been described that can contribute to the localization of new PSD95 molecules to activated synapses, including local translation18, activity-induced palmitoylation34, and phosphorylation34,38. Interestingly, PSD95 molecules within PSDs are immobile during baseline activity but exhibit increased exchange between postsynaptic and cytoplasmic locations after LTP or LTD induction39,47. Preferential accumulation of new copies of synaptic proteins such as PSD95 within stimulated dendritic regions could thus provide synapses undergoing plasticity with proteins to rebuild PSDs or supply fresh copies of proteins that have not yet experienced irreversible post-translational modifications such as proteolysis by calpain40. Our finding of high PSD95 levels in stimulated dendritic regions both in synapses and dendritic shafts is consistent with this possibility.
Visualization of new protein copies in synaptic plasticity had previously only been possible for the protein Arc, which is expressed at low levels basally and is strongly transcriptionally and translationally induced after synaptic activity such that induced protein increases can be attributed to new proteins. In the dentate gyrus of rats, Arc protein levels increase rapidly and specifically in dendritic regions undergoing LTP48. However, whether new Arc proteins function to maintain LTP persistence in a synapse-autonomous manner is not yet clear. Arc has a well-characterized function in promoting AMPA-type glutamate receptor removal from synapses in LTD16, and its requirement in LTP persistence may be due in part to homeostatic removal of glutamate receptors from non-potentiated synapses. Unlike Arc, which is trafficked with endocytic vesicles away from synapses and is degraded within hours16,39, PSD95 has a half-life of days and a subpopulation is stably bound in the synapse with a persistence of many hours11,39. Once incorporated into specific synapses, new PSD95 could thus conceivably exert long-lasting effects on synaptic function.
Other studies have inferred synaptic incorporation of activity-induced proteins but have not directly demonstrated it. A Dendra2 message tagged with the sensorin 3′ UTR is translated near activated synapses in Aplysia neurons, suggesting that sensorin itself may be synthesized and utilized at activated synapses43, but this has not yet been directly confirmed. Preexisting Homer-1a protein, marked by photoactivation of a PA-GFP tag 4 h prior to synaptic activation, can be recruited specifically to activated synapses in mammalian neurons49. However, whether copies of Homer-1a synthesized after synaptic activation target activated synapses has not been studied, and whether Homer-1a synthesis could contribute to the protein synthesis dependence of LTP persistence remains unclear. Fluorescent TimeSTAMP tagging of sensorin or Homer-1a could be used to determine if synapse-specific utilization of new copies follows their activity-dependent synthesis.
A previous study using local subcellular fluorescence recovery after photobleaching (FRAP) demonstrated that local TrkB stimulation using bead-immobilized BDNF non-specifically enhanced PSD95-GFP movement throughout the entire dendritic tree without preferential enhancement at the stimulation site30. Local FRAP measurements report replacement rates of total PSD95 in the photobleached location from all sources outside that location. In contrast, our study specifically investigated the fates of newly synthesized PSD95 copies. If total protein concentrations are higher than new protein concentrations, then increased accumulation of new proteins may not be apparent when visualizing the total protein population. Furthermore, increased retention of PSD95 at stimulated synapses, for instance by palmitoylation or phosphorylation34,38, would not increase FRAP and indeed could decrease it if the area of FRAP resides within a larger area of increased retention. The previous study also did not include the native 3′ UTR in the PSD95 constructs. Because the 3′ UTR mediates mRNA stabilization, dendritic localization, and translational induction18,20, its influence on these events would not have been detected.
Ultrastructural pulse labeling of PSD95
Our results demonstrate that visualizing new and old PSD95 proteins by correlated light and electron microscopy with photo-oxidizing TimeSTAMP tags TS:YSOG1 and TS:YSOG2 provides a more complete picture of their localization and environment. By fusing these reporters to PSD95, we found that new and old PSD95 populations exhibited distinct localization patterns both at the cellular and synaptic level. PSD95 copies less than 6 h old were found in dendritic protrusions opposite immature presynaptic termini and less abundant in PSDs of mature spines, whereas older PSD95 populations were preferentially observed at mature synapses. This indicates that PSD95 turns over more slowly at PSDs of mature synapses, consistent with previous reports from optical microcopy39. New PSD95 populations also exhibited unique synaptic accumulation at each of these structures. New copies that were less than 6 h old were observed to accumulate diffusely in the cytoplasm in a protrusion apposing a vesicle-sparse presynaptic terminus. By contrast, copies of PSD95 more than 6 h old were not observed in the cytoplasm and accumulated in dense membrane-bound structures apposing vesicle-rich presynaptic membranes. This could suggest that recruitment of PSD95 to mature postsynaptic sites may require palmitoylation, which has been suggested previously37. Therefore, younger PSD95 molecules do not simply replace older PSD95 populations, but display distinct localization and may have distinct functions at growing areas of the neurons.
Without EM, it would be difficult to distinguish PSD95 labeling at immature versus mature synapses because of the limited contextual information of light microscopy. Super-resolution optical microscopy allows resolution of single targets below the diffraction limit but cannot provide the same contextual information as photo-oxidizing TimeSTAMPs because it is limited by the number of distinct fluorophores that can be imaged simultaneously. It would be difficult to optically label a protein of interest such as PSD95 together with markers required for context, such as membranes, vesicles, and microtubules. EM inherently includes such context. The only alternative technique for EM imaging of new vs. old copies of a genetically defined protein involves sequential FlAsH and ReAsH labeling of tetracysteine motifs50. However, the modest singlet oxygen quantum yield of ReAsH makes it much less sensitive than miniSOG. The biarsenical dyes also suffer from higher nonspecific background labeling, potential toxicity, and difficulty in application to intact tissues and organisms. Photo-oxidizing TimeSTAMPs offer a relatively simple method for ultrastructural resolution of either newer or older copies of a genetically selected protein.
METHODS
Reagents
Plasmids encoding PSD95 or Arc fused to TimeSTAMP cassettes11 were modified by standard molecular biology techniques including polymerase chain reaction, restriction enzyme digestion, and ligation to create new TimeSTAMP variants. New linker sequences were introduced with synthetic oligonucleotides. All subcloned fragments were sequenced in their entirety to confirm successful construction. Full sequences of all plasmids in this study are available upon request.
The following compounds were obtained from Sigma: bicuculline, CHPG, DHPG, actinomycin D, and cycloheximide. Alexa Fluor 647 carboxylic acid succinimidyl ester was obtained from Invitrogen. BILN-2061 and ITMN-191 were synthesized by a contract synthesis company (Acme). BDNF was obtained from Chemicon, Primary antibodies used were mouse monoclonal anti-PSD95 (Neuromab), mouse monoclonal anti-AU1 (Covance MMS-130R), rat monoclonal anti-HA (Roche), and rabbit anti-synapsin (Chemicon). For immunoblotting, primary antibodies were used at 0.1–0.4 μg/mL and HRP-conjugated goat secondary antibodies (Zymed) at 0.1 μg/mL. For immunofluorescence, primary antibodies were used at 0.5–1 μg/mL and Alexa Fluor 568- and 647-conjugated goat secondary antibodies (Invitrogen) at 0.5 μg/mL.
Cell culture
All cell culture reagents were obtained from Invitrogen unless otherwise indicated. HEK293A cells (Invitrogen) were cultured in DMEM medium with 10% v/v FBS, 50 U/mL penicillin, and 50 μg/mL streptomycin and transfected with Lipofectamine 2000. Hippocampal neurons were dissociated by papain from embryonic day 18 (E18) or postnatal day 0 (P0) Sprague Dawley rats, transfected by Amaxa electroporation (Lonza AG), cultured in Neurobasal with B27 supplement, 2 mM GlutaMAX, 50 U/mL penicillin, and 50 μg/mL streptomycin as previously described11. All animal procedures were approved by Institutional Animal Care and Use Committee of the University of California, San Diego or of Stanford University.
Compartmentalized neuronal culture chambers
Silicon wafer masters were made by soft photolithography on two layers of photoresist with patterning provided by two transparency masks created in CAD software and printed on a 20,000 dots-per-inch printer. Polydimethylsiloxane (PDMS) prepolymer and catalyst (Dow Corning) were mixed at a 10:1 ratio and allowed to polymerize on the masters at 70°C overnight. Blocks were cut out, sterilized, and adhered to washed coverglasses, then the channels were coated with poly-D-lysine. More detailed procedures have been previously published27.
Immunoblotting
For analysis of TimeSTAMP cleavage by immunoblotting, HEK293A cells were transfected with Lipofectamine for 3 h, then transferred to fresh medium without or with BILN-2061. 1–2 days later, cells were rinsed quickly in Hank’s Buffered Saline Solution (HBSS) and immediately lysed in 0.1% boiling sodium dodecyl sulfate (SDS) loading buffer. Lysates were sonicated to shear DNA and then run on NuPage 4–12% Novex Bis-Tris SDS polyacrylamide gels (Invitrogen). Proteins were transferred onto polyvinylidene fluoride membrane (Millipore) by electroblotting, which were then blocked with 10% nonfat dried milk in tris-buffered saline with 0.1% Tween-20 (TBST), incubated in primary antibody overnight in 5% bovine serum albumin in TBST at 4 °C, washed in TBST to remove excess primary antibody, incubated in HRP-conjugated secondary antibody in 10% nonfat dried milk in TBST for 45 min at room temperature, and finally rinsed 3 times for 10 min each in TBST. Proteins were visualized by chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate, Thermo) and film (Amersham Hyperfilm Blue). For analysis of stimulus-dependent PSD95 synthesis in neurons by immunoblotting, transfected dissociated P0 rat hippocampal neurons were plated and maintained in 6-well dishes. Following treatment by the desired pharmacological agents, cells were rinsed quickly with HBSS and immediately lysed in boiling SDS, then immunoblotting was performed as above.
Immunofluorescence
To quantify effects of TimeSTAMP expression on synaptic density, hippocampal neurons were dissociated, electroporated, and plated on washed coverslips coated with poly-D-lysine. Neurons were cultured in the absence of BILN-2061 or in its presence from 4 to 14 DIV. Neurons were fixed by the addition of one culture volume of 8% paraformaldehyde for 15 minutes at room temperature, then washed in phosphate-buffered saline (PBS), blocked in PBS with 5% non-immune goat serum, and probed for synapsin and HA according to standard procedures. Specificity of secondary antibodies was confirmed in control samples without primary antibody. Coverslips were mounted in Vectashield (Chemicon) and sealed with nail polish. Conditions were randomized and blinded, then images of 7–10 neurons per condition were obtained on a Zeiss Axiovert 200M with a LSM 5 Live confocal scanner, using a 40× water-immersion lens with numerical aperture (NA) 1.2 and one of the following filter sets: 495/10 nm excitation, 515 nm dichroic, 535/25nm emission (YFP); 460/20 nm excitation, 515 nm dichroic, 535/25 nm emission (miniSOG); or 540/25 nm excitation, 560 nm dichroic, 595/50 nm emission (mOrange, mKO2, or mCherry). A stack of optical sections with 1024 × 1024 resolution at 1-μm intervals through each neuron was obtained and then flattened in a maximum intensity projection. Blinded quantitation of synaptic density was performed as previously described11. Analysis was performed on an Apple Macintosh notebook computer using the public domain NIH ImageJ program (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image).
Time-lapse microscopy
To follow fluorescence development of TS:YFP tags in transfected HEK293A cells, cells were cultured and transfected on glass-bottom dishes in DMEM with 10% fetal bovine serum. One day after transfection, DMEM was exchanged for HBSS with B27. Cells were imaged on a Zeiss Axiovert 200M equipped with a xenon arc lamp, a 20×0.7 NA lens, a Photometrics Cascade II 1024 camera, and a stage-top environmental chamber with temperature set to 35°C and humidity to 100%. Images were acquired before and at various times after BILN-2061 addition with exposure times selected to avoid sensor saturation. To plot normalized fluorescence versus time, total background-subtracted fluorescence from individual cells at each time point was measured using ImageJ software and normalized to the maximum value for each cell.
To visualize new synaptic protein synthesis in neurons, dissociated P0 hippocampal neurons were transfected with TimeSTAMP reporters and a mCherry marker by electroporation. They were then cultured in Neurobasal with B27 in glass-bottom dishes or PDMS chambers. Cultures were maintained at 37°C in 5% carbon dioxide and 100% humidity. Half of the medium was replaced every 2 days. On the day of imaging, medium was exchanged for HBSS with B27. Cells were imaged by epifluorescence on the Zeiss Axiovert 200M as above. They were also imaged on an Olympus IX81 using a 20× 0.7 NA lens and a FV1000 confocal scanning system. For confocal imaging, the following settings were used: excitation with a 488-nm argon-ion laser line at 10% power and a 559-nm laser diode at 10% power, pinhole 200 μm, scan resolution 1024 × 1024 pixels, scanning speed 1 μs per pixel, photomultiplier voltage 700 V, digital gain 1. In either imaging system, fields including transfected neurons identified by mCherry fluorescence were imaged before and at various times after BILN-2061 addition. A stack of optical sections at 1-μm intervals through each neuron was obtained at each position and time point and then flattened in a maximum intensity projection.
Immunocytochemistry and photo-oxidation for EM
For conventional immuno-EM labeling, cells were fixed in 4% paraformaldehyde and 0.1% glutaraldehyde in PBS (0.1 M, pH 7.4) for 30 min, rinsed several times in chilled buffer, and incubated in permeabilizing buffer (0.1% Triton X-100 in PBS) for 5 min. Then cells were incubated in primary antibody (monoclonal antibody to HA 1:1000, Roche clone 12CA5) in 0.01% Triton X-100 and 5% normal goat serum (NGS) in PBS at 4°C for 3 h to overnight, secondary conjugated to biotin (Goat anti mouse, 1:1000) in 5% NGS in PBS at 4°C for 1–2 h, and streptavidin-HRP (1:100) in 5% NGS in PBS at 4 °C for 1 hour, allowing for sufficient washing in 5% NGS in PBS on ice after each incubation. Fluorescent cells were identified and registered by the grid on the cover glass of these dishes. Confocal images of these cells were taken with minimum exposure using a MRC-1024 inverted confocal microscope (BioRad) to identify transfected cells and for correlative light microscopic imaging. Then the buffer was replaced with a solution of 1 mg/mL DAB and 0.001% H2O2 in PBS. Cells in the registered grids were monitored under a transmission microscope until a light brown reaction product became visible. The dish was then removed from the microscope and washed in chilled buffer (5 times for 2 min) and post-fixed in 2% glutaraldehyde in 0.1 M PBS on ice for 20 min and then in 1% osmium tetroxide (Electron Microscopy Sciences) in 0.1 M PBS on ice for 30 min. Cells were then washed in chilled buffer twice and rinsed in distilled water and stained with 2% aqueous uranyl acetate (Ted Pella Inc.) for 1 h to overnight at 4°C. The samples were then dehydrated in a cold graded ethanol series (20%, 50%, 70%, 90%, 100%, 100%, 100%) 2 min each, rinsed once in room temperature anhydrous ethanol, and infiltrated in Durcupan ACM resin (Electron Microscopy Sciences) using 1:1 anhydrous ethanol and resin for 30 min, then 100% resin twice for 1 h each, then into fresh resin and polymerized in a vacuum oven at 60°C for 48 h. Sample preparation for photo-oxidation was performed as described previously.24 Incubation with mersalyl acid (5 mM in cacodylate buffer for 30 min on ice) followed by several rinse steps in chilled buffer before photo-oxidation was added in some experiments to reduce DAB staining of mitochondria.
Electron Microscopy
Labeled and imaged areas of embedded cultured cells were identified by transmitted light microscopy. Areas of interest were sawed out using a jeweler’s saw and mounted on dummy acrylic blocks with cyanoacrylic adhesive. The coverslip was carefully removed, ultrathin sections were cut using an ultramicrotome, and electron micrographs were imaged using a 1200 TEM (JEOL) operating at 80 keV.
Statistical Analysis
Statistical comparisons between two groups for a measure of interest were performed with two-tailed t-tests assuming unpaired samples with significance level set at α = 0.05. Comparisons between more than two groups for a measure of interest were performed by one-factor ANOVA followed by pairwise Tukey’s tests if ANOVA revealed unequal means at a significance level of α = 0.05. Prior to t-tests and Tukey’s tests, F-tests were first used to reject a null hypothesis of equal variances for a two-tailed distribution with significance level set at α = 0.05. If the null hypothesis of equal variances was accepted then Tukey’s test (for ANOVA post-hoc analysis) or a t-test for samples with equal variances was performed; otherwise Dunnett’s T3 test (for ANOVA post-hoc analysis) or a t-test for samples with unequal variances was performed. For analysis of dendritic segment TS:YFP fluorescence or synaptic density, sample numbers were determined by the number of healthy transfected neurons in each condition. For analysis of synaptic TS:YFP fluorescence in chambers, sample numbers were determined by the number of puncta in each segment. Statistical tests were performed in Excel (Microsoft) and Prism (Graphpad).
Supplementary Material
Acknowledgments
We would like to thank the members of the Tsien and Ellisman laboratories for helpful discussion, especially Varda Lev-Ram, Stephen Adams, and Thomas Deerinck. This work was supported by a NIH Pharmacology Training grant (M.T.B), the NSF Graduate Research Fellowships Program (M.T.B), the Lucille Packard Children’s Hospital Pediatric Research Fund (Y.G.), World Class University Program grant R31-2008-000-10083-0 from the Korea Research Foundation of the South Korea Ministry of Education, Science and Technology (N.L.J.), the Howard Hughes Medical Institute and NIH grant 4R37NS027177-23 (R.Y.T.), NIH grant P41GM103412-24 (M.H.E.), NIH grant R01NS076860 (M.Z.L.), and the Burroughs Wellcome Fund (M.Z.L.). M.Z.L. is a Rita Allen Foundation Scholar.
Footnotes
AUTHOR CONTRIBUTIONS
M.T.B. conceived, designed and performed electron microscopy experiments and co-wrote the manuscript. J.Y. designed and validated constructs. Y.G. designed and performed dendritic stimulation experiments. Y.J.K. and N.L.J. designed and fabricated microfluidic chambers. X.S. assisted with the study design. M.R.M. assisted with electron microscopy sample preparation and imaging. M.H.E. and R.Y.T. supervised the project and provided advice. M.Z.L. conceived, designed, and performed dendritic stimulation experiments, co-wrote the manuscript, and provided supervision and advice.
References
- 1.Jung H, O’Hare CM, Holt CE. Translational regulation in growth cones. Curr Opin Genet Dev. 2011;21:458–464. doi: 10.1016/j.gde.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welshhans K, Bassell GJ. Netrin-1-induced local beta-actin synthesis and growth cone guidance requires zipcode binding protein 1. J Neurosci. 2011;31:9800–9813. doi: 10.1523/JNEUROSCI.0166-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillips LL, Pollack AE, Steward O. Protein synthesis in the neuropil of the rat dentate gyrus during synapse development. J Neurosci Res. 1990;26:474–482. doi: 10.1002/jnr.490260410. [DOI] [PubMed] [Google Scholar]
- 4.Sebeo J, et al. Requirement for protein synthesis at developing synapses. J Neurosci. 2009;29:9778–9793. doi: 10.1523/JNEUROSCI.2613-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Schuman EM, Dynes JL, Steward O. Synaptic regulation of translation of dendritic mRNAs. J Neurosci. 2006;26:7143–7146. doi: 10.1523/JNEUROSCI.1796-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bekinschtein P, et al. Persistence of long-term memory storage requires a late protein synthesis-and BDNF- dependent phase in the hippocampus. Neuron. 2007;53:261–277. doi: 10.1016/j.neuron.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Bourtchouladze R, et al. Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learn Mem. 1998;5:365–374. [PMC free article] [PubMed] [Google Scholar]
- 8.Waung MW, Huber KM. Protein translation in synaptic plasticity: mGluR-LTD, Fragile X. Curr Opin Neurobiol. 2009;19:319–326. doi: 10.1016/j.conb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradshaw KD, Emptage NJ, Bliss TV. A role for dendritic protein synthesis in hippocampal late LTP. Eur J Neurosci. 2003;18:3150–3152. doi: 10.1111/j.1460-9568.2003.03054.x. [DOI] [PubMed] [Google Scholar]
- 10.Abraham WC, Williams JM. LTP maintenance and its protein synthesis-dependence. Neurobiol Learn Mem. 2008;89:260–268. doi: 10.1016/j.nlm.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Lin MZ, Glenn JS, Tsien RY. A drug-controllable tag for visualizing newly synthesized proteins in cells and whole animals. Proc Natl Acad Sci U S A. 2008;105:7744–7749. doi: 10.1073/pnas.0803060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magliery TJ, et al. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J Am Chem Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 14.Zhang C, et al. A neuroligin-4 missense mutation associated with autism impairs neuroligin-4 folding and endoplasmic reticulum export. J Neurosci. 2009;29:10843–10854. doi: 10.1523/JNEUROSCI.1248-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bramham CR, Worley PF, Moore MJ, Guzowski JF. The immediate early gene arc/arg3.1: regulation, mechanisms, and function. J Neurosci. 2008;28:11760–11767. doi: 10.1523/JNEUROSCI.3864-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shepherd JD, et al. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrlich I, Malinow R. Postsynaptic density 95 controls AMPA receptor incorporation during long-term potentiation and experience-driven synaptic plasticity. J Neurosci. 2004;24:916–927. doi: 10.1523/JNEUROSCI.4733-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Todd PK, Mack KJ, Malter JS. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci U S A. 2003;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zalfa F, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waterhouse EG, Xu B. New insights into the role of brain-derived neurotrophic factor in synaptic plasticity. Mol Cell Neurosci. 2009;42:81–89. doi: 10.1016/j.mcn.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 23.Auerbach BD, Bear MF. Loss of the fragile X mental retardation protein decouples metabotropic glutamate receptor dependent priming of long-term potentiation from protein synthesis. J Neurophysiol. 2010;104:1047–1051. doi: 10.1152/jn.00449.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ronesi JA, Huber KM. Homer interactions are necessary for metabotropic glutamate receptor-induced long-term depression and translational activation. J Neurosci. 2008;28:543–547. doi: 10.1523/JNEUROSCI.5019-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci. 2004;24:7366–7377. doi: 10.1523/JNEUROSCI.1739-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CC, Huang CC, Wu MY, Hsu KS. Insulin stimulates postsynaptic density-95 protein translation via the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway. J Biol Chem. 2005;280:18543–18550. doi: 10.1074/jbc.M414112200. [DOI] [PubMed] [Google Scholar]
- 27.Taylor AM, et al. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith WB, Starck SR, Roberts RW, Schuman EM. Dopaminergic stimulation of local protein synthesis enhances surface expression of GluR1 and synaptic transmission in hippocampal neurons. Neuron. 2005;45:765–779. doi: 10.1016/j.neuron.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Wang KH, et al. In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell. 2006;126:389–402. doi: 10.1016/j.cell.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 30.Yoshii A, Constantine-Paton M. BDNF induces transport of PSD-95 to dendrites through PI3K-AKT signaling after NMDA receptor activation. Nat Neurosci. 2007;10:702–711. doi: 10.1038/nn1903. [DOI] [PubMed] [Google Scholar]
- 31.Benarroch EE. Metabotropic glutamate receptors: synaptic modulators and therapeutic targets for neurologic disease. Neurology. 2008;70:964–968. doi: 10.1212/01.wnl.0000306315.03021.2a. [DOI] [PubMed] [Google Scholar]
- 32.Shu X, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X, et al. PSD-95 is required to sustain the molecular organization of the postsynaptic density. J Neurosci. 2011;31:6329–6338. doi: 10.1523/JNEUROSCI.5968-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noritake J, et al. Mobile DHHC palmitoylating enzyme mediates activity-sensitive synaptic targeting of PSD-95. J Cell Biol. 2009;186:147–160. doi: 10.1083/jcb.200903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goellner B, Aberle H. The synaptic cytoskeleton in development and disease. Dev Neurobiol. 2012;72:111–125. doi: 10.1002/dneu.20892. [DOI] [PubMed] [Google Scholar]
- 36.Toni N, et al. Synapse formation on neurons born in the adult hippocampus. Nat Neurosci. 2007;10:727–734. doi: 10.1038/nn1908. [DOI] [PubMed] [Google Scholar]
- 37.Bresler T, et al. The dynamics of SAP90/PSD-95 recruitment to new synaptic junctions. Mol Cell Neurosci. 2001;18:149–167. doi: 10.1006/mcne.2001.1012. [DOI] [PubMed] [Google Scholar]
- 38.Kim MJ, et al. Synaptic accumulation of PSD-95 and synaptic function regulated by phosphorylation of serine-295 of PSD-95. Neuron. 2007;56:488–502. doi: 10.1016/j.neuron.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Sturgill JF, Steiner P, Czervionke BL, Sabatini BL. Distinct domains within PSD-95 mediate synaptic incorporation, stabilization, and activity-dependent trafficking. J Neurosci. 2009;29:12845–12854. doi: 10.1523/JNEUROSCI.1841-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zadran S, Bi X, Baudry M. Regulation of calpain-2 in neurons: implications for synaptic plasticity. Mol Neurobiol. 2010;42:143–150. doi: 10.1007/s12035-010-8145-1. [DOI] [PubMed] [Google Scholar]
- 41.Bingol B, Sheng M. Deconstruction for reconstruction: the role of proteolysis in neural plasticity and disease. Neuron. 2011;69:22–32. doi: 10.1016/j.neuron.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Stepanenko OV, et al. Modern fluorescent proteins: from chromophore formation to novel intracellular applications. Biotechniques. :51, 313–4, 316, 318. doi: 10.2144/000113765. passim (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang DO, et al. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324:1536–1540. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dieterich DC, et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernandez PJ, Abel T. The role of protein synthesis in memory consolidation: progress amid decades of debate. Neurobiol Learn Mem. 2008;89:293–311. doi: 10.1016/j.nlm.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gold PE. Protein synthesis inhibition and memory: formation vs amnesia. Neurobiol Learn Mem. 2008;89:201–211. doi: 10.1016/j.nlm.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanpied TA, Kerr JM, Ehlers MD. Structural plasticity with preserved topology in the postsynaptic protein network. Proc Natl Acad Sci U S A. 2008;105:12587–12592. doi: 10.1073/pnas.0711669105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 49.Okada D, Ozawa F, Inokuchi K. Input-specific spine entry of soma-derived Vesl-1S protein conforms to synaptic tagging. Science. 2009;324:904–909. doi: 10.1126/science.1171498. [DOI] [PubMed] [Google Scholar]
- 50.Gaietta G, et al. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.