Abstract
Generally, most miRNAs that were up-regulated during differentiation promoted adipogenesis, but our research indicated that up-regulation of miR-145 in porcine preadipocytes did not promote but inhibit adipogenesis. In this study, miR-145 was significantly up-regulated during porcine dedifferentiated fat (DFAT) cells differentiation. In miR-145 overexpressed DFAT cells, adipogenesis was inhibited and triglycerides accumulation was decreased after hormone stimulation (P<0.05). Furthermore, up-regulation of miR-145 expression repressed induction of mRNA levels of adipogenic markers, such as CCAAT/enhancer-binding protein α (C/EBPα), and peroxisome proliferator-activated receptor γ2 (PPARγ2). These effects caused by miR-145 overexpression were mediated by Insulin receptor substrate 1 (IRS1) as a mechanism. These data suggested that induced miR-145 expression during differentiation could inhibit adipogenesis by targeting IRS1, and miR-145 may be novel agent for adipose tissue engineering.
Keywords: miR-145, IRS1, inhibit, dedifferentiated fat cells, adipogenesis.
Introduction
MicroRNAs (miRNAs), 18-26nt endogenously expressed post-transcriptional modifiers, play crucial roles in diverse biological processes by degrading mRNA and/or inhibiting translation. Some of them involved in maturation of adipocytes, such as miR-1431, 2, miR-369-5p3, miR-27a/b4, 5. However, it is far from understanding their functions and mechanisms completely. Making such research is of great interest for uncovering the molecular mechanism of adipose tissue developmemt. Up till now most miRNAs up-regulated during adipogenesis promoted adipocyte differentiation, but there were few miRNA induced during adipogenesis inhibited the process. For example, miR-31 which was up-regulated during murine mesenchymal stem cell line C3H10T1/2 adipogenesis, inhibited adipogenesis6. In vitro primary cell culture system could mock in vivo environment well, porcine preadipocytes has been suggested as a substitute for human preadipocytes and novel model systems for studying the mechanisms of adipocyte differentiation 7. Therefore, our recent study screened miRNAs profiles during porcine intramuscular and subcutaneous preadipocytes differentiation to understand the role of miRNAs in adipocytes differentiation better. In both cell lines, miR-145 was up-regulated significantly during adipogenesis, and it was expressed notably higher in fat tissues than in some other tissues. Recent studies indicated that miR-145 played roles in inhibiting cancer cells proliferation 8, 9, repressing pluripotency and chondrogenic differentiation 10, 11, and inducing differentiation of multipotent neural crest stem cells 12, 13. In these biological processes, miR-145 functioned by targeting different mRNAs. Most target mRNAs of miR-145 played roles in aidpocytes differentiation, such as KLF4 and KLF5 14, 15. However, to our knowledge, no report has been conducted to link miR-145 and any of its target genes to adipogenesis.
Therefore, our investigation focused on the role of miR-145 in adipogenesis. During dedifferentiated fat (DFAT) cells differentiation, miR-145 was up-regulated. Lentivirus overexpression of miR-145 significantly inhibited DFAT cells differentiation. This inhibiting function was conducted by targeting Insulin receptor substrate 1 (IRS1) that is essential for adipogenesis. Therefore, the inhibiting function of miR-145 in adipogenesis may contribute to adipose tissue engineering.
Materials and methods
Pig and Primary DFAT cells. All animal procedures were in strict accordance with the guidelines of the Institutional Animal Care and Use Committee, and all animal work was approved by the Animal Care and Use Committee of Guangdong Province, China. Cells were isolated from no more than 24 h old male landrace and induced to differentiation using the methods established by Nobusue and Kano 7. Differentiated DFAT cells were stained with oil red O as decribed previously 16. Intracellular oil red O was extracted in 100% isopropanol and quantified by the absorbance at 510 nm 17.
Triglycerides assay. Cells were lysed and triglycerides contents were determined using a Triglyceride Kit (Biosino Bio-Technology and Science Inc., Beijing, China) according to the manufacturer's protocol. Total protein in the cell lysate was quantified with the Bradford Protein Assay Kit (Beyotime,Shanghai, China).
RNA extraction and qRT-PCR: Total RNA was extracted using TRIzol reagent (Invitrogen, California, USA) according to the manufacturer's instructions. First-strand cDNA of mRNAs and miRNAs were obtained using the Reverse Transcription System (Promega, Madison, WI, USA) and All-in OneTM miRNA First-Stand cDNA Synthesis Kit (GeneCopoeia, Rockville, MD, USA), respectively. QRT-PCR was performed using SYBR Premier Dimer EraserTM (TaKaRa, Osaka, Japan) on a LightCycler480 (Roche Basel, Switzerland), and relative quantification (△△Ct) method was used to analyze the data. Endogenous GAPDH mRNA was used as reference, as it was expressed stable during DFAT adipogenesis 18, 19. The primers for mRNA and miRNA were as follows: GAPDH, F: ACAGTCAAGGCGGAGAACG,
R: GGCAGAAGGGGCAGAGAT, 204bp; C/EBPα,
F: TGGACAAGAACAGCAACGAG, R: ACCTTCTGTTGAGTCTCCACG, 109bp; C/EBPβ, F: GGTGGACAAGCACAGCGA, R: TGCTGCGTCTCCAGGTTG, 105bp; PPARγ2, F: TTGATTTCTCCAGCATTTCC, R: GGCTCCACTTTGATGGCACT, 126bp; ap2/FABP4, F: GGAAACTTGTCTCCAGTGAAAAC,
R: TGGTGCTCTTGACTTTCCTGT, 228bp; ssc-miR-145,
F: GTCCAGTTTTCCCAGGAATCCCTTA; ssc-miR-145,
R: TGCTGTCAACGATACGCTACG, 84bp.
Western blotting. Cells were lysed in cell lysis buffer (Beyotime,Shanghai, China), and protein content was quantified as mentioned above. 40 μg total cellular protein was fractionated by 10% (w/v) SDS/PAGE and electronically transferred to 0.45 μm PVDF membrane (Roche Basel, Switzerland). The membrane was rinsed with TBS-Tween20 (TBST), blocked for 2 h in TBST containing 5% (w/v) skimmed milk, and incubated with primary antibody for 1 h. The membrane was washed with TBST and incubated for 1 h with secondary antibody conjugated to HRP. Blots were visualized using an ECL detection kit (Thermo Scientific, massachusetts, USA). The primary antibody for IRS1 was monoclonal anti-IRS1 (#05-1085, millipore, Bedford, MA, USA). GAPDH protein was used as reference (sc-59540, Santa Cruz Biotechnology, California, USA), and the secondary antibody of both IRS1 and GAPDH was HRP-conjugated goat anti-mouse IgG.
Conservation analysis. MiR-145 sequences of different species in miRBase (18.0) were downloaded. Then the phylogenetic analysis was performed using software MEGA5 according to the manufacturer's instructions.
Lentivirus production and infection. Ssc-miR-145 with BamHI and EcoRI restriction enzyme digestion sites at 5' and 3' end was cloned into a BamHI-EcoRI-treated vector pLVX-ShRNA2 (Clontech). Empty vector or expression plasmids were co-transfected into 70%-80% confluent 293T cells with lentivirus packaging vectors. Virus supernatant was collected twice at day 1 and day 2 after transfection. Primary DFAT cells were cultured to 50%-60% confluence and infected with lentivirus at 20 multiplicity of infection (MOI), and the cells were re-infected with the same amount of lentivirus on the next day. The infection efficiency was judged by GFP expression observation on the third day.
Target prediction. An online database mirecords (http://mirecords.biolead.org/) was used to predict the targets of miR-145 based on sequence homology. In 6 databases integrated in mirecords, IRS1 was predicted simultaneously. A database (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) was used to predict hybridation of miR-145 to IRS1 3' UTR.
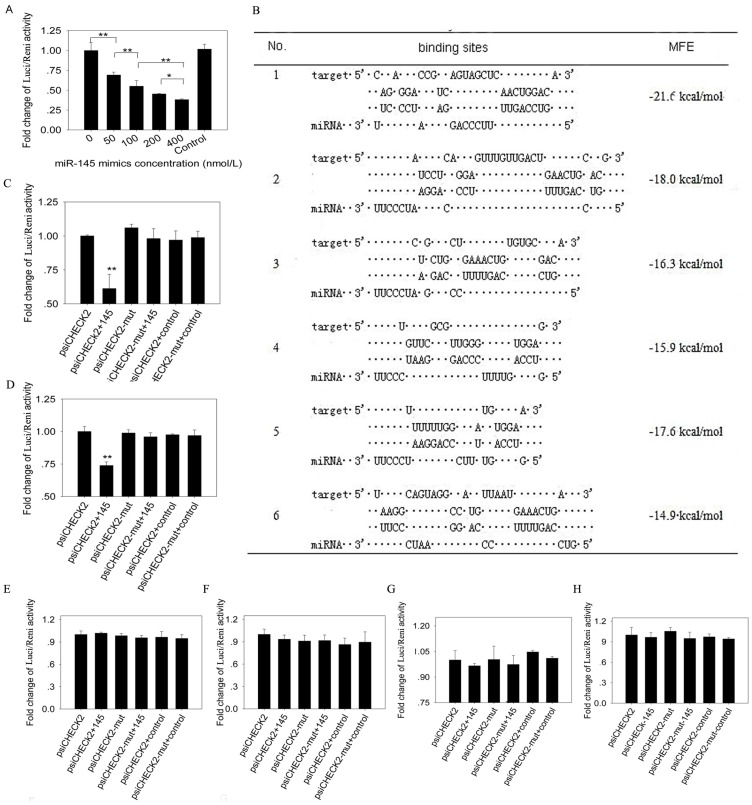
Luciferase reporter assay. 293T cells were plated in 48-well plates (Corning, Sanford, ME, USA) at a density of 6×104 cells per well. 50 ng psiCHECK2 vector (Promega, Madison, WI, USA) containing IRS1 3' UTR were co-transfected into the cells with different concentration of miR-145 mimics (final concentrations were 0, 50, 100, 200 and 400nmol/l) using Lipofectamine 2000 (Invitrogen, California, USA), and mimics control with 400 nmol/l was used. 50 ng psiCHECK2 vector containing different binding sites or mutant binding sites were co-transfected with miR-145 mimics or a mimics control (final concentration was 400 nmol/l). Four replicates were made for each transfection. Firefly and Renilla luciferase activities were measured with the Dual-Glo luciferase assay system (Promega, Madison, WI, USA) at 48 h after transfection. The following primers were used to amplify porcine IRS1 3' UTR from total RNA. The product was 1094 bp in length covering the entire IRS1 3'UTR. XhoI-3'UTR primer: CCGCTCGAGCTCAACTGGACATCACAGCAG, NotI-3'UTR primer: TTGCGGCCGCTAAAGATCAACAGTCTCTAGTTTA. We predicted 6 potential binding sites in porcine IRS1 3' UTR. The 6 sites and their scrambled sequences were obtained by annealing sense and antisense strand, and then the annealed sequences were inserted into XhoI-NotI treated psiCHECK2 vector.
Statistical analysis. One-Way ANOVA and Bivariate Correlations analyses were conducted using SPSS16.0, and a probability less than 0.05 was chosen as significant.
Results
Dynamic changes of miR-145 during DFAT cells differentiation
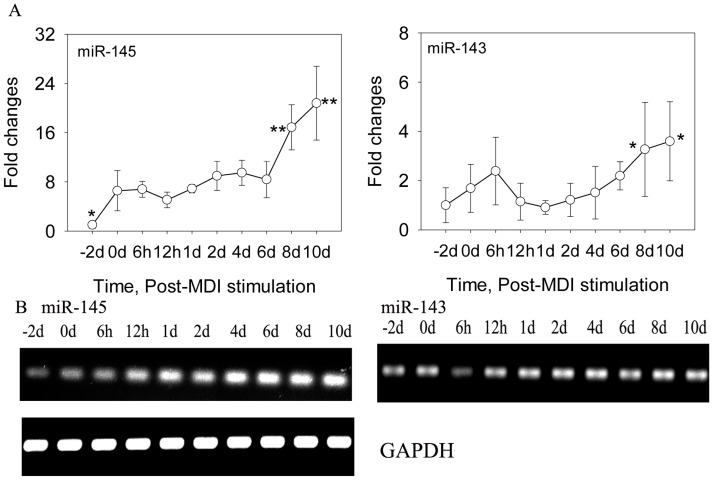
DFAT cells were isolated using the ceiling culture method established previously 7, and the DFAT cells could be obtained in 24 days (Figure 1). Then the cells were induced to differentiate with high efficiency in differentiation medium (0.5 mM 3-isobutyl-1-methylxanthine, 0.25 μM dexamethasone, and 5 mg/l insulin, MDI) (Figure 2). To determine the changes of miR-145 during DFAT cells differentiation, cells were collected at different time points, including -2 day (-2 d), 0 d, 6 h,12 h,1 d, 2 d, 4 d, 6 d, 8 d and 10 d. QRT-PCR and semi-RT-PCR results showed that miR-145 was significantly up-regulated during DFAT cells differentiation especially at 8 d and 10 d, which was similar with miR-143 (Figure 3A and B). Expression levels of the two miRNAs showed high correlation coefficients (Pearson, r=0.785, P<0.01; Sperman, r=0.711, P<0.01). The endogenous mRNA GAPDH reference expressed stably during the whole adipogenesis process (Figure 3B).
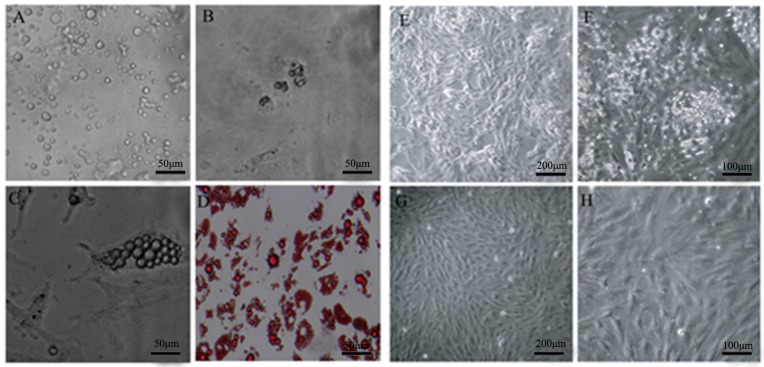
Fig 1.
Isolation of DFAT cells using the ceiling culture method. (A) Morphology of purified porcine mature adipocytes. (B) Morphology of cells at 2 d after seeding. (C) Morphology of cells at 4 d after seeding. (D) Morphology of cells at 7 d after seeding, and the cells were stained with oil red O and viewed under microscope. (E) and (F) Morphology of cells at 14 d after seeding. (G) and (H) Morphology of cells at 24 d after seeding.
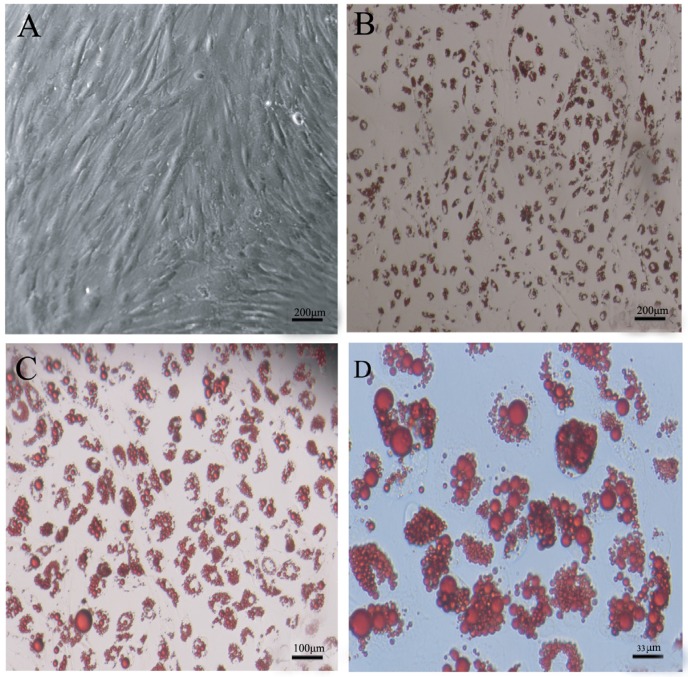
Fig 2.
DFAT cells could differentiate into mature adipocytes. (A) Morphology of DFAT cells at 0 d before differentiation. (B), (C), (D) Differentiated DFAT cells were fixed and stained with oil red O at 8 d.
Fig 3.
MiR-145 and miR-143 was up-regulated during DFAT cells differentiation. (A) QRT-PCR results of miR-145 and miR-143 during DFAT cells differentiation, and the results were first normalized to endogenous mRNA GAPDH and fold changes were calculated to preadipocytes (-2 d). Triplicates were made and data were shown as mean ± SD. One-Way ANOVA analysis was conducted, P<0.05 was shown in one star, and P<0.01 was shown in two star. (B) Semi-RT-PCR results of miR-145, miR-143 and GAPDH reference during DFAT cells differentiation.
Conservation analysis of miR-145
To determine whether miR-145 is conservative among species, the sequences of all miR-145 in miRBase (18.0) were phylogenetic analyzed. After phylogenetic analysis, miR-145 was conserved among these species, and ssc-miR-145 had the closest distance with human, mouse and rat (Supplementary Material: Figure S1).
Up-regulaion of miR-145 inhibited DFAT cells differentiation
To obtain more cells expressing miR-145 stably, the DFAT cells were infected twice with lentivirus. The infection efficiencies of both miR-145 and mock vector control were more than 90% as judged by the expression of GFP two days after the first infection (Supplementary Material: Figure S2A). After induction, miR-145 expression levels were quantified at 0 d, 4 d and 8 d. In this assay, besides the DFAT cells infected with miR-145 (DFAT-miR-145) and mock vector lentivirus (DFAT-mock) were induced to differentiation, DFAT cells in differentiation medium (DFAT-MDI) was also included as control. As expected, miR-145 showed significantly higher expression levels in DFAT-miR-145 than the two controls at 0 d, 4 d and 8 d after hormone stimulation (P<0.01, Supplementary Material: Figure S2B).
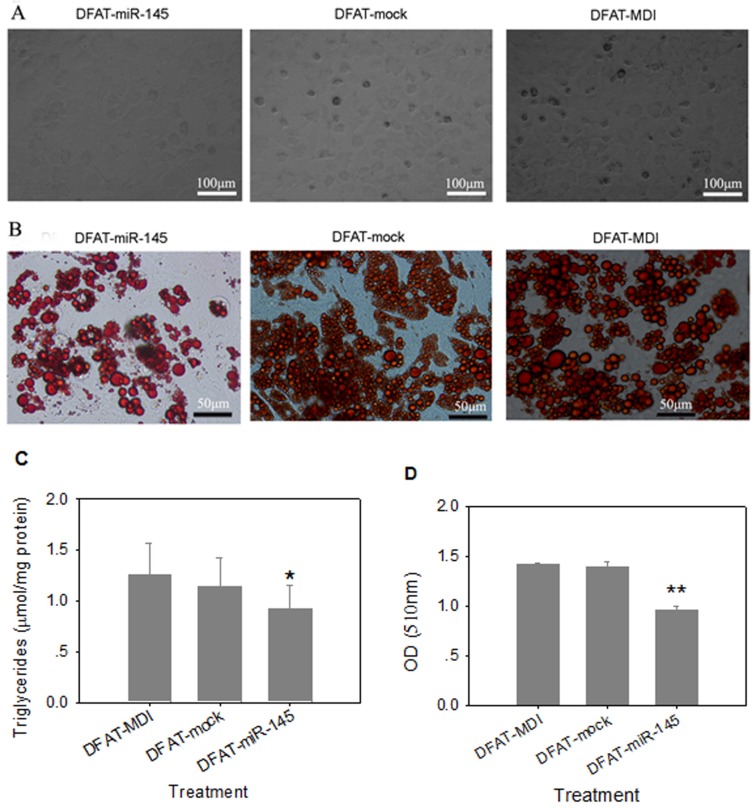
Three days after differentiation, cells infected with miR-145 lentivirus were round in appearance without lipid droplets. However, lipid droplets were accumulated in DFAT-mock and DFAT-MDI controls after 3 days differentiation (Figure 4A). At 8 d of differention, DFAT-miR-145 accumulated less triglycerides than DFAT-mock and DFAT-MDI (P<0.05, Figure 4B and Figure 4C). Intracellular oil red O quantification results were similar with triglyceride contents determination (Figure 4D). These results indicated that miR-145 inhibited DFAT cells differentiation.
Fig 4.
Up-regulation of miR-145 with lentivirus inhibited DFAT cells differentiation. DFAT cells were infected with miR-145 and mock vector lentivirus twice, and the cells were induced to differention with MDI. DFAT-cells induced directly was used as control. (A) Morphology of DFAT cells at 3 d after MDI stimulation. (B) DFAT cells were fixed and stained with oil red O at 8 d after MDI stimulation. (C)Triglycerides contents in DFAT cells under different conditions were measured at 8 d of differentiation, triplicate of cells were made, and data were mean ± SD. (D) Intracellular oil red O in (B) cells was extracted in 100% isopropanol, then the absorbance was measured at 510 nm, the data based on triplicate of cells, data were mean ± SD.
Luciferase reporter assay
Luciferase reporter assay showed that firefly luciferase activity reduced significantly as the increasing concentration of miR-145 mimics (Figure 5A). To further determine the binding sites of miR-145 to IRS1 3' UTR, we predicted 6 binding sites (Figure 5B) and inserted them into psiCHECK2 vector, respectively. MiR-145 mimics co-transfected with site1 or site2 psiCHECK2 constructs could significantly decrease firefly luciferase activity (P<0.01), while the above effects were abolished by site1 and site2 mutant (Figure 5C and D). The other 4 sites displayed no changes in firefly luciferase activity (Figure 5E -H). These results indicated that miR-145 could directly bind to the 3' UTR of IRS1 and the binding was in dose dependent manner.
Fig 5.
MiR-145 binds to IRS1 3' UTR region and there were 2 binding sites. (A) 50 ng psiCHECK2 vector containing IRS13'UTR were co-transfected with different concentrations of miR-145 mimics (final concentrations were 0, 50, 100, 200 and 400 nmol/l) into 293T cells. 400 nmol/l non-targeting mimics control was also co-transfected with psiCHECK2 vector containing IRS13'UTR. Firefly luciferase activity was normalized to Renilla luciferase. Then fold changes were normalized to 0 concentration. Four replicates were made for each transfection, and the data were shown as mean ± SD. (B) Six putative binding sites of miR-145 in IRS1 3' UTR region were predicted. (C) -(H) Determination of miR-145 binding sites in the 3' UTR of IRS1. 50 ng psiCHECK2 vector containing binding sites or binding sites mutant at seed region were co-transfected with miR-145 mimics or non-targeting mimics control (final concentration was 400 nmol/l) into 293T cells. Firefly luciferase activity was normalized to Renilla luciferase. Then fold changes were normalized to psiCHECK2. Replicates and data analysis were the same as A. (C) Site 1, (D) Site 2, (E) Site 3, (F) Site 4, (G) Site 5, (H) Site 6.
Effects of up-regulation of miR-145 on adipogenic markers and IRS1
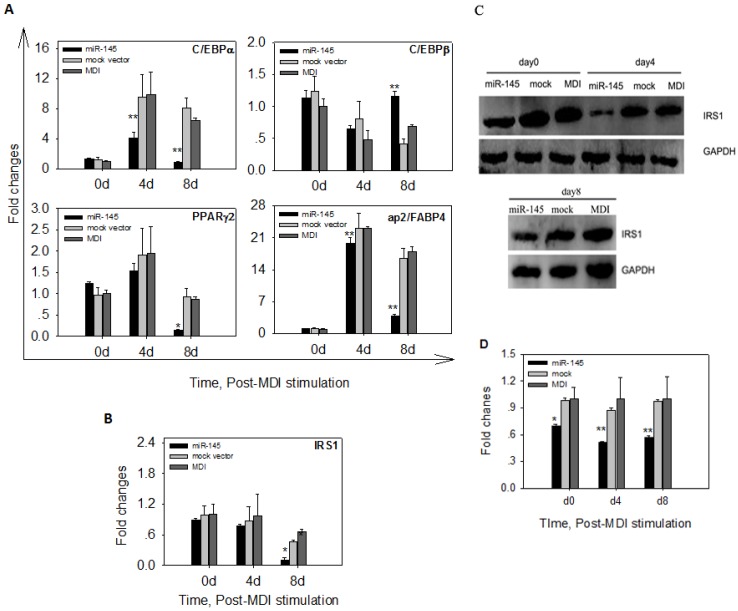
To determine whether overexpression of miR-145 affected mRNA levels of adipogenic markers, we quantified mRNA levels of C/EBPα, C/EBPβ, PPARγ2 and ap2/FABP4 in DFAT cells at 0 d, 4 d and 8 d after MDI stimulation. The early marker C/EBPβ displayed similar expression levels among DFAT-miR-145, DFAT-mock and DFAT-MDI at 4d of differentiation. At 8 d, C/EBPβ mRNA expression levels in DFAT-miR-145 was significantly higher than DFAT-mock and DFAT-MDI (P<0.01, Figure 6A). C/EBPα and ap2/FABP4 expression levels in DFAT-miR-145 were lower than DFAT-mock and DFAT-MDI (P<0.01) at 4 d of adipogenesis. At 8 d, the two markers were expressed markedly lower in DFAT-miR-145 than DFAT-mock and DFAT-MDI (P<0.01, Figure 6A). PPARγ2 was expressed similar among DFAT-miR-145, DFAT-mock and DFAT-MDI at 4 d after stimulation (P<0.01). At 8 d, PPARγ2 expression levels in DFAT-miR-145 were lower than DFAT-mock and DFAT-MDI (P<0.01, Figure 6A).
Fig 6.
Up-regulation of miR-145 affected adipogenic markers and IRS1. (A) The expression levels of 4 adipogenic markers (C/EBPα, C/EBPβ, PPARγ2 and ap2/FABP4) in cells infected with miR-145 and mock vector control at 0 d, 4 d and 8 d were analyzed. The data were first normalized to endogenous mRNA GAPDH and fold changes were calculated to DFAT-MDI preadipocytes (0 d). Triplicates were made and data were shown as mean ± SD. One-Way ANOVA analysis was conducted, P<0.05 was shown in one star, and P<0.01 was shown in two star. (B) IRS1 mRNA changes after up-regulation miR-145 expression. Data were analyzed as A. (C) IRS1 protein expression was down-regulated by up-regulation of miR-145. (D) IRS1 and GAPDH protein in (C) were quantified. Statistical analysis was the same as (A).
The mRNA and protein expressions of target gene IRS1 were also detected after MDI stimulation. No statistical differences were found at mRNA levels among DFAT-miR-145, DFAT-mock and DFAT-MDI at 0 d and 4 d after stimulation(P>0.05). At 8 d, the mRNA expression of IRS1 in DFAT-miR-145 was lower than DFAT-mock and DFAT-MDI (P<0.01, Figure 6B). The protein levels of IRS1 at 0 d, 4 d and 8 d after stimulation were significantly lower in DFAT-miR-145 than DFAT-mock and DFAT-MDI (Figure 6C and 6D).
Discussion
Emerging researches revealed that miR-145 was involved in several key cellular functions, including cancer cells proliferation 8, 9, human embryonic stem cells differentiation 10, 11, and smooth muscle fate 12, 13. However, no reports implied that miR-145 possesed a function in adipogenesis. Here our results demonstrated that up-regulated miR-145 expression inhibited adipogenesis, which was different from most miRNAs, including miR-21020 and miR-103/10721. During porcine adipocytes differentiation, miR-145 was mainly expressed at the late stage, and loss of proliferative potential is generally a characteristic of terminal differentiation of adipocytes 22. These suggested that miR-145 expressed at higher level in non-proliferating cells, which had been confirmed by Rocca et al.(2011) 23. In addition, similar dynamic patterns in miR-145 and miR-143 expression levels during the DFAT cells adipogenesis were observed, and they both belonged to the same cluster. MiR-143 was a well-known marker of adipogenesis, and it promoted adipogenesis by targeting ERK5 1, 24. Mir-145 and miR-143 had totally opposite roles in adipogenesis, which due to their different seed sequences. Because miRNAs functioned mainly by through binding of seed region (at position 2-8 from the 5′ end) to target 3′ UTRs 25 . These findings enlarged our understanding that miRNAs in the same cluster may have opposite roles in adipogenesis. According to previous studies, up-regulated miRNAs during aidpogenesis may promote preadipocytes differentiation, including miR-21 26 and the miR-17-92 cluster 16. However, miR-31 was up-regulated during murine mesenchymal stem cell adipogenesis, and it inhibited adipogenesis 6. In addition, miR-378 was down-regulated in thicker backfat of pig and bovine with more mature adipocytes 27, 28, it increased the size of lipid droplets 29. These findings enriched our understanding that miRNAs induced during differentiation may also inhibit adipogenesis.
We further studied a mechanism for the inhibition function of miR-145 in adipogenesis. In the present study, miR-145 is a translational repressor of its IRS1 target gene during DFAT cells differentiation. IRS1 is a major member of the insulin receptor substrate family, and its activation is essential for lipogenesis stimulated by insulin 30, 31. Decreased expression of IRS-1 in embryonic fibroblast cells severely decreased the expression of C/EBPα and PPARγ 30. Similarly, up-regulation of miR-145 in DFAT cells reduced IRS1 protein expression dramatically, which may decrease expression of adipogenic marker genes in the present study. The down-regulation in 3'UTR and binding site1/2 luciferase assays indicated that miR-145 targeted IRS1 by binding directly to 3' UTR, since mutations of the miR-145 seed sites abolish this down-regulation. However, at mRNA level, IRS1 was not negatively regulated by miR-145, though its expression was significantly lower in DFAT-miR-145 cells than DFAT-mock and DFAT-MDI cells at 8 d of differentiation. One explanation may be that DFAT-miR-145 cells had significantly lower differentiation potential than DFAT-mock and DFAT-MDI cells. In additon, IRS1 mRNA expression levels were also significantly lower in DFAT-GM (growth medium) cells at 4 d and 8 d, and few DFAT-GM cells differentiated into mature adipocytes (data not shown). In accordance with this study, miR-145 also did not negatively regulate IRS1 at mRNA levels in human colon cancer cells 9. These results confirmed the hypothesis that miR-145 is presumably acting on the translation of IRS1 9. IRS proteins are conserved during evolution 9, so we did not validate the function of IRS1 during porcine preadipocytes differentiation. Though this study discovered that miR-145 inhibited adipogenesis by down-regulated IRS1 gene expression, and IRS1 further affected adipogenic markers expression, which may finally inhibited adipogensis of porcine preadipocytes. This may be a mechanism for miR-145, but additional studies will need to investigate the complete regulatory mechanism of miR-145 in adipogenesis. Intensive studies of miR-145 targets revealed that many targets of miR-145 in adipocytes differentiation may exist. Among these targets genes are Krüppel-like factor 4 (KLF4), KLF5 and insulin-like growth factor receptor (IGF-IR) were proved as targets of has-miR-145 10, 12, 13, 32. KLF4 and KLF5 are essential zinc-finger transcription factors promoting adipocyte differentiation 14, 15. IGF-1R was responsible for the induction of adipogenesis, and IGF-1R deletion in male mice decreased body fat accumulation and increased energy expenditure 33. However, the effects of up-regulated miR-145 expression on IGF-1R was not determined in the present study, because IGF-1R was validated as another target of ssc-miR-145(data not shown).
Pig may be a better biomedical model organism for study human obesity and related diseases, because the pig is physiologically more similar and possibly evolutionarily more closely related to human than rodents 34. What's more, miRNAs have conserved function across species 2, and porcine preadipocytes has been suggested as a substitute for human preadipocytes 7. Therefore, miR-145 may be a novel agent for therapy of obesity and related metabolic syndrome, which was studied in cancer cells 35 and tumor tissues 36. Further studies shoud be done on human to apply the function of miR-145.
In conclusion, our present study suggested that miR-145 was up-regulated during porcine adipocytes differentiation, and it inhibited adipocytes differentiation by targeting insulin signaling pathway member IRS1 at translation level. These results demonstrated that miR-145 was involved in fat tissue development, and miR-145 may be new agent for adipose tissue engineering.
Supplementary Material
Fig. S1: MiR-145 was conservative in evolution. Fig. S2: Overexpression of miR-145.
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China (863 Program) (No. 2011AA100304), Natural Science Foundation of Guangdong (S2011010001380) and the Key Projects in the Science & Technology Pillar Program of Guangzhou (2008Z1-E121).
References
- 1.Takanabe R, Ono K, Abe Y, Takaya T, Horie T, Wada H. et al. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochemical and Biophysical Research Communications. 2008;376:728–32. doi: 10.1016/j.bbrc.2008.09.050. doi:10.1016/j.bbrc.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 2.Esau C, Kang XL, Peralta E, Hanson E, Marcusson EG, Ravichandran LV. et al. MicroRNA-143 regulates adipocyte differentiation. Journal of Biological Chemistry. 2004;279:52361–5. doi: 10.1074/jbc.C400438200. doi:DOI 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 3.Bork S, Horn P, Castoldi M, Hellwig I, Ho AD, Wagner W. Adipogenic Differentiation of Human Mesenchymal Stromal Cells Is Down-Regulated by microRNA-369-5p and Up-Regulated by microRNA-371. Journal of Cellular Physiology. 2011;226:2226–34. doi: 10.1002/jcp.22557. doi:Doi 10.1002/Jcp.22557. [DOI] [PubMed] [Google Scholar]
- 4.Yoo W, Lee J, Park S, Kim Y-S, Lim C, Yoon E. et al. Albumin expression is required for adipocyte differentiation of 3T3-L1 cells. Biochemical and Biophysical Research Communications. 2010;397:170–5. doi: 10.1016/j.bbrc.2010.05.067. doi:10.1016/j.bbrc.2010.05.067. [DOI] [PubMed] [Google Scholar]
- 5.Karbiener M, Fischer C, Nowitsch S, Opriessnig P, Papak C, Ailhaud G. et al. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochemical and Biophysical Research Communications. 2009;390:247–51. doi: 10.1016/j.bbrc.2009.09.098. doi:10.1016/j.bbrc.2009.09.098. [DOI] [PubMed] [Google Scholar]
- 6.Sun F, Wang J, Pan Q, Yu Y, Zhang Y, Wan Y. et al. Characterization of function and regulation of miR-24-1 and miR-31. Biochemical and Biophysical Research Communications. 2009;380:660–5. doi: 10.1016/j.bbrc.2009.01.161. doi:10.1016/j.bbrc.2009.01.161. [DOI] [PubMed] [Google Scholar]
- 7.Nobusue H, Kano K. Establishment and characteristics of porcine preadipocyte cell lines derived from mature adipocytes. Journal of Cellular Biochemistry. 2010;109:542–52. doi: 10.1002/jcb.22431. doi:10.1002/jcb.22431. [DOI] [PubMed] [Google Scholar]
- 8.Sachdeva M, Mo YY. MicroRNA-145 Suppresses Cell Invasion and Metastasis by Directly Targeting Mucin 1. Cancer Research. 2010;70:378–87. doi: 10.1158/0008-5472.CAN-09-2021. doi:Doi 10.1158/0008-5472.Can-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi B, Sepp-Lorenzino L, Prisco M, Linsley P, deAngelis T, Baserga R. Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. the Journal of Biological Chemistry. 2007;282:32582–90. doi: 10.1074/jbc.M702806200. doi:M702806200 [pii] 10.1074/jbc.M702806200. [DOI] [PubMed] [Google Scholar]
- 10.Xu N, Papagiannakopoulos T, Pan GJ, Thomson JA, Kosik KS. MicroRNA-145 Regulates OCT4, SOX2, and KLF4 and Represses Pluripotency in Human Embryonic Stem Cells. Cell. 2009;137:647–58. doi: 10.1016/j.cell.2009.02.038. doi:DOI 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 11.Yang B, Guo HF, Zhang YL, Chen L, Ying DJ, Dong SW. MicroRNA-145 Regulates Chondrogenic Differentiation of Mesenchymal Stem Cells by Targeting Sox9. Plos One. 2011;6:e21679.. doi: 10.1371/journal.pone.0021679. doi:ARTN e21679 DOI 10.1371/journal.pone.0021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN. et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–U80. doi: 10.1038/nature08195. doi:Doi 10.1038/Nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xin M, Small EM, Sutherland LB, Qi XX, McAnally J, Plato CF. et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes & Development. 2009;23:2166–78. doi: 10.1101/gad.1842409. doi:Doi 10.1101/Gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K. et al. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metabolism. 2005;1:27–39. doi: 10.1016/j.cmet.2004.11.005. doi:DOI 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metabolism. 2008;7:339–47. doi: 10.1016/j.cmet.2008.02.001. doi:10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ. et al. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proceedings of the National Academy of Sciences. 2008;105:2889–94. doi: 10.1073/pnas.0800178105. doi:10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Q, Lee YJ, Yun Z. Differentiation arrest by hypoxia. Journal of Biological Chemistry. 2006;281:30678–83. doi: 10.1074/jbc.C600120200. doi:C600120200 [pii] 10.1074/jbc.C600120200. [DOI] [PubMed] [Google Scholar]
- 18.Nicholls PK, Harrison CA, Walton KL, McLachlan RI, O'Donnell L, Stanton PG. Hormonal Regulation of Sertoli Cell Micro-RNAs at Spermiation. Endocrinology. 2011;152:1670–83. doi: 10.1210/en.2010-1341. doi:Doi 10.1210/En.2010-1341. [DOI] [PubMed] [Google Scholar]
- 19.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proceddings of the National Academy of Sciences of the United States of America. 2006;103:2422–7. doi: 10.1073/pnas.0511041103. doi:DOI 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie H, Lim B, Lodish HF. MicroRNAs Induced During Adipogenesis that Accelerate Fat Cell Development Are Downregulated in Obesity. Diabetes. 2009;58:1050–7. doi: 10.2337/db08-1299. doi:10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin L, Chen Y, Niu Y, Chen W, Wang Q, Xiao S. et al. A deep investigation into the adipogenesis mechanism: Profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/β-catenin signaling pathway. BMC Genomics. 2010;11:320.. doi: 10.1186/1471-2164-11-320. doi:10.1186/1471-2164-11-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth MJ, Sparks RL, Wharton W. Proadipocyte cell lines: models of cellular proliferation and differentiation. Journal of Cell Science. 1993;106( Pt 1):1–9. doi: 10.1242/jcs.106.1.1. [DOI] [PubMed] [Google Scholar]
- 23.La Rocca G, Shi B, Audia A, Ferrari-Amorotti G, Mellert HS, Calabretta B. et al. Regulation of microRNA-145 by growth arrest and differentiation. Experimental Cell Research. 2011;317:488–95. doi: 10.1016/j.yexcr.2010.11.010. doi:DOI 10.1016/j.yexcr.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV. et al. MicroRNA-143 Regulates Adipocyte Differentiation. Journal of Biological Chemistry. 2004;279:52361–5. doi: 10.1074/jbc.C400438200. doi:10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 25.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L. et al. Widespread siRNA "off-target" transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–87. doi: 10.1261/rna.25706. doi:10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YJ, Hwang SJ, Bae YC, Jung JS. MiR-21 Regulates Adipogenic Differentiation through the Modulation of TGF-beta Signaling in Mesenchymal Stem Cells Derived from Human Adipose Tissue. Stem Cells. 2009;27:3093–102. doi: 10.1002/stem.235. doi:Doi 10.1002/Stem.235. [DOI] [PubMed] [Google Scholar]
- 27.Li G, Li Y, Li X, Ning X, Li M, Yang G. MicroRNA identity and abundance in developing swine adipose tissue as determined by solexa sequencing. Journal of Cellular Biochemistry. 2011;112:1318–28. doi: 10.1002/jcb.23045. doi:10.1002/jcb.23045. [DOI] [PubMed] [Google Scholar]
- 28.Jin W, Dodson MV, Moore SS, Basarab JA, Guan LL. Characterization of microRNA expression in bovine adipose tissues: a potential regulatory mechanism of subcutaneous adipose tissue development. BMC Molecular Biology. 2010;11:29.. doi: 10.1186/1471-2199-11-29. doi:1471-2199-11-29 [pii] 10.1186/1471-2199-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacDougald OA, Gerin I, Bommer GT, McCoin CS, Sousa KM, Krishnan V. Roles for miRNA-378/378*in adipocyte gene expression and lipogenesis. American Journal of Physiology-Endocrinology and Metabolism. 2010;299:E198–E206. doi: 10.1152/ajpendo.00179.2010. doi:10.1152/ajpendo.00179.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miki H, Yamauchi T, Suzuki R, Komeda K, Tsuchida A, Kubota N. et al. Essential role of insulin receptor substrate 1 (IRS-1) and IRS-2 in adipocyte differentiation. Molecular and Cellular Biology. 2001;21:2521–32. doi: 10.1128/MCB.21.7.2521-2532.2001. doi:10.1128/MCB.21.7.2521-2532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angela M. Valverde CRK, and Manuel Benito. Insulin Signaling in Insulin Receptor Substrate ( I R S ) -1-Deficient Brown Adipocytes. Diabetes. 1999;48:2122–31. doi: 10.2337/diabetes.48.11.2122. doi: 10.2337/diabetes.48.11.2122. [DOI] [PubMed] [Google Scholar]
- 32.La Rocca G, Shi B, Badin M, De Angelis T, Sepp-Lorenzino L, Baserga R. Growth inhibition by microRNAs that target the insulin receptor substrate-1. Cell Cycle. 2009;8:2255–9. doi: 10.4161/cc.8.14.9026. doi.org/10.4161/cc.8.14.9026. [DOI] [PubMed] [Google Scholar]
- 33.Freude S, Schilbach K, Hettich MM, Bronneke HS, Zemva J, Krone W. et al. Neuron-specific deletion of a single copy of the insulin-like growth factor-1 receptor gene reduces fat accumulation during aging. Hormone and Metabolic Research. 2012;44:99–104. doi: 10.1055/s-0031-1298018. doi:10.1055/s-0031-1298018. [DOI] [PubMed] [Google Scholar]
- 34.Jorgensen FG, Hobolth A, Hornshoj H, Bendixen C, Fredholm M, Schierup MH. Comparative analysis of protein coding sequences from human, mouse and the domesticated pig. BMC biology. 2005;3:2.. doi: 10.1186/1741-7007-3-2. doi:10.1186/1741-7007-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Guo H, Zhang H, Wang H, Qian G, Fan X. et al. Putative tumor suppressor miR-145 inhibits colon cancer cell growth by targeting oncogene friend leukemia virus integration 1 gene. Cancer. 2011;117:86–95. doi: 10.1002/cncr.25522. doi: 10.1002/cncr.25522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ugras S, Brill E, Jacobsen A, Hafner M, Socci ND, Decarolis PL. et al. Small RNA sequencing and functional characterization reveals MicroRNA-143 tumor suppressor activity in liposarcoma. Cancer Research. 2011;71:5659–69. doi: 10.1158/0008-5472.CAN-11-0890. doi:10.1158/0008-5472.CAN-11-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: MiR-145 was conservative in evolution. Fig. S2: Overexpression of miR-145.