Abstract
Gene essentiality changes are crucial for organismal evolution. However, it is unclear how essentiality of orthologs varies across species. We investigated the underlying mechanism of gene essentiality changes between yeast and mouse based on the framework of network evolution and comparative genomic analysis. We found that yeast nonessential genes become essential in mouse when their network connections rapidly increase through engagement in protein complexes. The increased interactions allowed the previously nonessential genes to become members of vital pathways. By accounting for changes in gene essentiality, we firmly reestablished the centrality-lethality rule, which proposed the relationship of essential genes and network hubs. Furthermore, we discovered that the number of connections associated with essential and non-essential genes depends on whether they were essential in ancestral species. Our study describes for the first time how network evolution occurs to change gene essentiality.
Gene essentiality varies across species and is one of the most dramatic phenotypic changes a gene can undergo1. For instance, deletion of MAP kinase kinase1 (Map2k1) did not affect the fitness of yeast, but its loss of function caused embryonic lethality in mouse2,3. In contrast, deletion of serine/threonine-protein kinase ICK caused lethality in yeast but had no apparent phenotypic effect in mouse4. Generally, orthologs are considered to deliver the same function in different species. Given that this is not always the case, why and how does essentiality of the same functional gene change between species?
The C-L rule explains that highly connected proteins in a network are more likely to be essential for cell viability5. However, a weak correlation between network connections and gene essentiality has led to controversies over the C-L rule6,7,8. A system-level understanding of how gene essentiality can change will give us a chance to understand the design principles of key biological processes and provide opportunity for predicting important gene functions.
Here, we investigated the mechanisms of gene essentiality changes in the framework of network expansion during evolution. We hypothesized that network rewiring has a significant effect on gene essentiality changes because rewiring of interactions enables genes to be integrated into new pathways9 and the new interactions can increase the probability of becoming involved in a vital biological process.
Results
Gene essentiality frequently changes during evolution
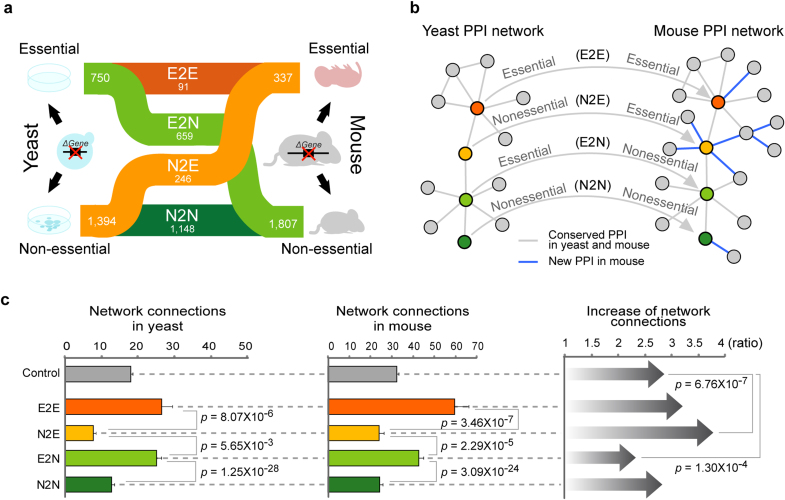
We found that a significant portion of 2,144 mouse genes with yeast orthologs changed their essentialities between mouse and yeast (Fig. 1a). We arranged the orthologous pairs of yeast and mouse genes into four phenotypic groups based on their changing essentiality patterns. We found 91 genes are essential in both yeast and mouse (E2E), 246 genes are nonessential in yeast but essential in mouse (N2E), 659 genes are essential in yeast but nonessential in mouse (E2N), and 1,149 genes are nonessential in both yeast and mouse (N2N). The list of yeast and mouse gene orthologs and their essentiality measurements can be accessed in Supplementary Table S1.
Figure 1. Increase in network connections and gene essentiality changes between yeast and mouse.
(a) Gene essentiality changes between yeast and mouse. The numbers of essential and nonessential genes of yeast (left) and mouse (right) are presented. (b) A network evolution model describing gene essentiality changes. (c) Network connections of the four phenotypic classes. The first two panels display the average number of network connections in yeast and mouse respectively. Error bars indicate the standard error. The last panel shows the fold increase in the average number of connections in mouse relative to yeast.
Increase of network connections explains gene essentiality changes
We hypothesized that the frequent gene essentiality changes we observed are related to interaction rewiring, which allows genes to integrate into, or separate from, important biological pathways10,11,12,13. To test this hypothesis, we examined the increase of network connections between yeast and mouse protein-protein interaction (PPI) networks (Fig. 1b). It has been suggested that the number of protein interactions are highly correlated with the complexity of the organism14,15. Protein interactions were measured by experiments from yeast and mouse separately and the network connections between yeast and mouse were compared by ortholog mapping (see Materials and Methods). We found that all the four classes of essentiality changes increased the average network connections in mouse relative to yeast, but the amount of increase was quite different in the four classes. In particular, N2E genes have the highest increase in network connections, whereas E2N genes have the smallest increase among the four phenotypic groups. The increase in connectivity was most significant in N2E genes compared to all genes (p = 6.76 × 10−7; Fig. 1c), whereas the increase for E2N genes was significantly smaller than the average (p = 1.30 × 10−4).
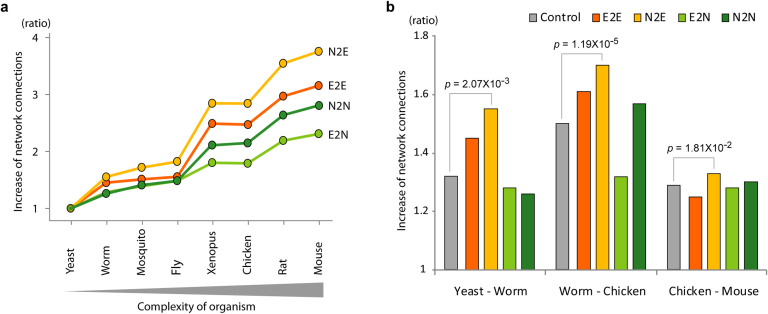
Because of a large evolutionary distance between yeast and mouse, we investigated more species pairs that diverged enough but closer than the distance between yeast and mouse. We found that all genes gradually increased their network connections in the course of evolution (Fig. 2a) but N2E genes increased network connections fastest among all phenotypic groups from the comparison of closer species (Fig. 2b). These results suggest that essential genes in unicellular organisms that become nonessential in multicellular organisms, fail to rapidly expand their network connections in the course of evolution.
Figure 2. Comparison of network connections in various species.
(a) Increase in network connections by the complexity of organisms. The fold increase in the number of connections relative to yeast is plotted. (b) Increase of network connections between various species pairs. The fold increases of network connections in worm over yeast, chicken over worm, and mouse over chicken were presented.
N2E genes have integrated into vital biological pathways
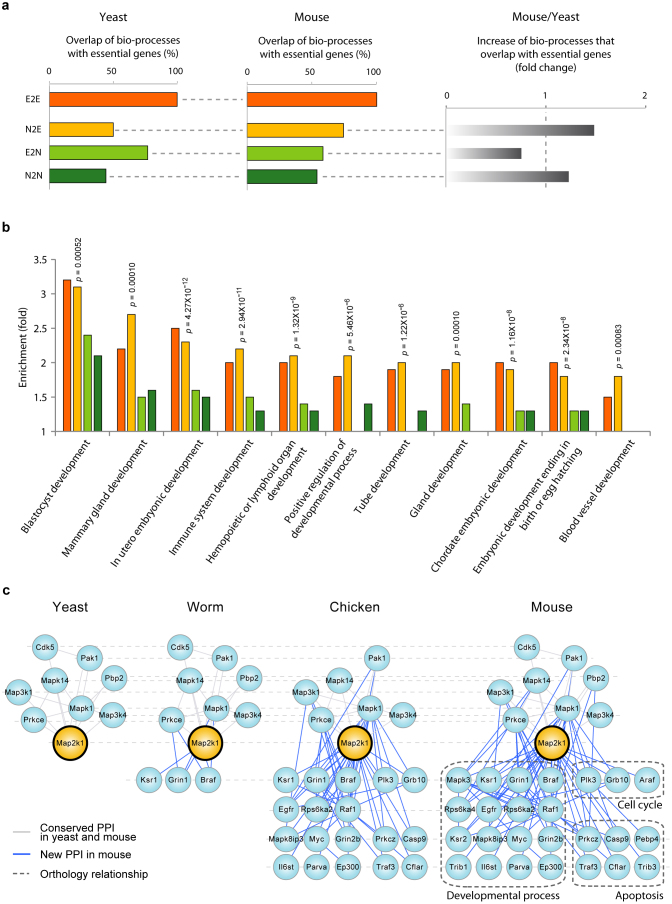
Next we asked whether the increased connections create new connections to core biological functions and thereby increased essentiality. It has been suggested that genes may become essential by participating in core pathways9, but evidence for this hypothesis has heretofore been lacking. We find that new interactions gained from network expansion do tend to cause integration of N2E genes into vital pathways of essential genes (Fig. 3a). Functional enrichment analysis of gene ontology of biological processes (BPs) was carried out for interactions formed by N2E, E2N, N2N, and E2E genes in yeast and mouse (Supplementary Table S2). The analysis reveals that interactions of N2E genes gained from network expansion have dramatically increased their participation in essential BPs of E2E genes. Specifically, in yeast, interactions of N2E genes share 50% of BPs with E2E genes, but in mouse, the fraction sharply increases to 74%. Whereas interactions of E2N genes share 77% of BPs with E2E genes in yeast, the fraction decreases to 59% in mouse.
Figure 3. Functional enrichment analysis of essentiality changing genes.
(a) Comparison Biological processes of N2E, E2N, and N2N genes with those of E2E genes in yeast and mouse. (b) Enrichment of biological processes in the four phenotypic groups. Gene ontology terms that are significantly enriched (p < 0.001) in N2E genes are presented. (c) Network connections of Map2k1 in yeast, worm, chicken, and mouse. Interaction partners Map2k1 in yeast (left) and mouse (right) are depicted with the orthologs connected by dotted lines.
Many N2E genes become integrated into BPs that are vital for the development of multicellular organisms (Fig. 3b and Table 1). Interactions of N2E proteins are highly enriched in developmental processes where a single misregulation could cause embryonic lethality. For example, the expanded network connections of Map2k1, an N2E gene, are involved in key pathways in multicellular organisms (Supplementary Table S3). Map2k1 participates in placenta development in mouse via newly evolved interactions. It has eight interaction partners in the yeast PPI network, but its network connections increased to 23 in the mouse PPI network (Fig. 3c). Consequently, the deletion of Map2k1 is not lethal in yeast, but causes embryonic lethality in mice2,3. Among the interaction partners of Map2k1 is epidermal growth factor receptor, EGFR, which regulates the epidermal growth factor pathway that is crucial for cell growth and morphogenesis16.
Table 1. Developmental processes of N2E genes in mouse.
| Developmental process | N2E genes |
|---|---|
| blastocyst development | Cul3, Smarcb1, Ada, Sp3, Junb |
| mammary gland development | Phb2, Atp7b |
| in utero embryonic development | Prmt1, Sin3a, Cul3, Slc30a1, Ccnb2, Smarcb1, Msh2, Ube2a, Mapk1, Myo1e, Mecom, Sp3, Ccnb1, Plcg1, Junb, Lig4, Fgfr1, Ada, Hsf1, Map2k1 |
| immune system development | Exo1, Msh2, Maea, Ung, Sp3, Rps19, Slc11a2, Xrcc6, Lig4, Blm, Sgpl1, Msh6, G6pdx, Ccnb2, Mlh1, Myo1e, Tcea1, Ada, Hells, Sod2, Dnaja3 |
| hemopoietic or lymphoid organ development | G6pdx, Ccnb2, Msh2, Myo1e, Maea, Sp3, Rps19, Slc11a2, Lig4, Blm, Sgpl1, Tcea1, Ada, Hells, Sod2, Dnaja3 |
| positive regulation of developmental process | Hmgb1, Junb, Lig4, Xrcc6, Fgfr1, Ada, Map2k1, Mapk14 |
| tube development | Phb2, Hmgb1, Timeless, Sp3, Ppp3r1, Ptges3, Fgfr1, Ada |
| gland development | Phb2, Atp7b, Fgfr1 |
| chordate embryonic development | Msh2, Ube2a, Phgdh, Sp3, Junb, Lig4, Fgfr1, Map2k1, Prmt1, Sin3a, Cul3, Slc30a1, Ccnb2, Smarcb1, Mapk1, Myo1e, Mecom, Ccnb1, Plcg1, Ada, Hsf1, Atm |
| embryonic development ending in birth or egg hatching | Msh2, Ube2a, Phgdh, Sp3, Junb, Lig4, Fgfr1, Map2k1, Prmt1, Sin3a, Cul3, Slc30a1, Ccnb2, Smarcb1, Mapk1, Myo1e, Mecom, Ccnb1, Plcg1, Ada, Hsf1, Atm |
| blood vessel development | Myo1e, Mapk1, Sphk2, Vezf1, Junb, Ppap2b, Sgpl1, Atg5, Fgfr1, Map2k1, Mapk14 |
Gene essentiality change is related with protein complex membership
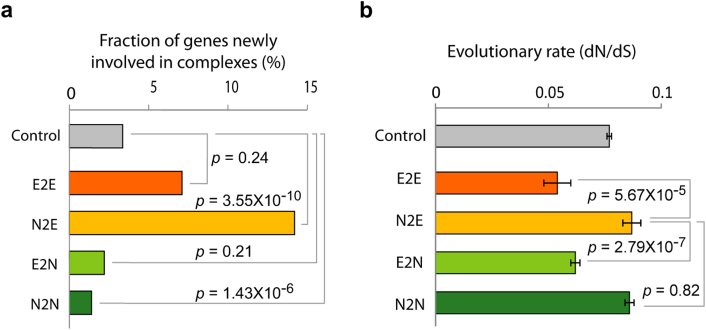
We next asked how N2E genes have quickly increased their network connections at the molecular level. We examined the membership changes of protein complexes between yeast and mouse, and found that N2E genes showed the highest rate of engaging in protein complexes among the four groups (p = 3.55 × 10−10; Fig. 4a). For example, Map2k1 is not a member of a protein complex in yeast, but becomes a member of the Ksr1 scaffold protein complex in multicellular organisms17. This suggests that protein complex membership may be an important mechanism for expanding network connections that can affect gene essentiality changes18,19.
Figure 4. Protein complex membership and evolution of gene essentiality changes.
(a) Fraction of genes newly involved in protein complexes are compared in each phenotypic group. (b) Evolutionary rates (dN/dS) of each phenotypic group in yeast. The evolutionary rates (dN/dS) were calculated from nucleotide sequences for 3,392 orthologous open reading frames (ORFs) in four Saccharomyces species including S. cerevisiae, S. paradoxus, S. mikatae, and S. bayanus.
To increase network connections rapidly, N2E genes may have acquired new interaction sites through fast adaptive evolution. To test this possibility, we examined the evolutionary rates of E2E, N2E, E2N, and N2N genes in various yeast species, and discovered that N2E genes have rapidly evolved. Evolutionary rates of yeast genes were calculated as the ratio of nonsynonymous substitutions (dN) to synonymous substitutions (dS) from the four complete genomes of Saccharomyces species20. As shown in Fig. 4b, N2E genes show a rapid evolutionary rate compared to E2E (p = 5.67 × 10−5) and E2N genes (p = 2.79 × 10−7). Interestingly, the evolutionary rates of N2E and N2N genes were similar (p = 0.82). The rapid evolutionary rate of N2N genes is probably due to low selective pressure on nonessential genes.
Discussion
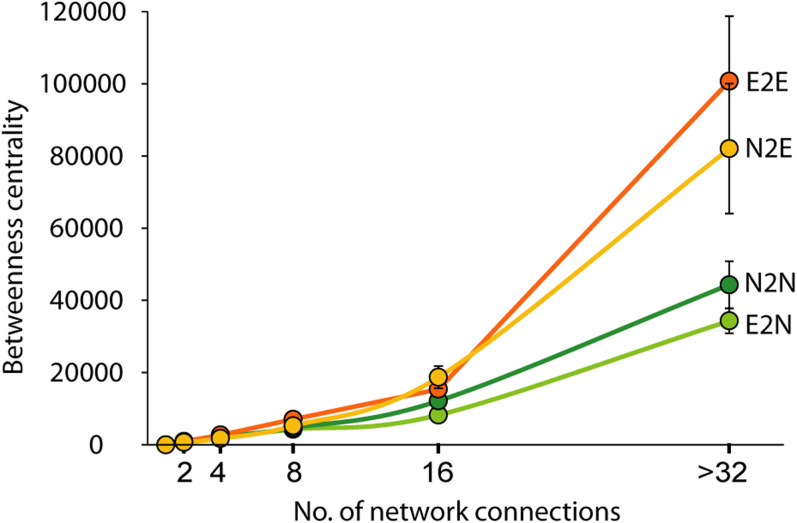
Having confirmed that network evolution influences gene essentiality changes, we asked how interaction rewiring has impacted the information flow of biological networks. Betweenness centrality is a measure of a node's centrality in a network equal to the number of shortest paths between all pairs of nodes that pass through that node. Proteins with high betweenness centrality tend to interact with many different functional groups21 and are important for controlling information flow in the network22,23. We discovered that the betweenness centrality of N2E genes is higher than those of N2N and E2N genes when they have same number of network connections (Fig. 5). Of the four groups, E2E genes have the highest betweenness centrality due to their importance in information flow in PPI network. However, N2E genes showed a dramatic increase in betweenness centrality if they were highly connected (>16 network connections). The increased betweenness centrality affects the functional role of N2E genes by reforming the modular architecture of the PPI network. Although both N2E and N2N genes were nonessential in yeast, the extensive rewiring of network connections for N2E genes in more complex organisms enables them to connect with various functional modules, thereby controlling information flow around newly evolved essential genes.
Figure 5. Information flow about essentiality changing genes.
Betweenness centrality of four phenotypic groups was presented by the number of network connection. Circles correspond to the mean betweenness of data points with interval by log scale. Error bars indicate the standard error.
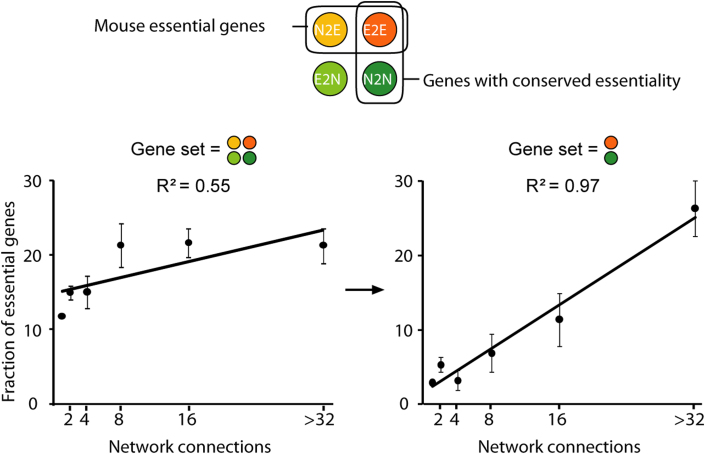
Our findings on the evolution of networks allow us to firmly reestablish the C-L rule by showing that highly connected genes in a network are indeed more essential when network rewiring is properly considered. The C-L rule has been debated because of an apparent weak correlation between network connection and gene essentiality6,7,8. We suspected that the poor correlation may have occurred because the evolution of gene essentiality was not considered previously (Fig. 6). According to the C-L rule, essential genes in yeast will have a relatively high connectivity. If rewiring leads it to become nonessential in mouse (E2N), connections will decrease relative to essential mouse genes (see above), but not enough evolutionary time may have occurred to descend to the level of a nonessential gene that was already nonessential in yeast (N2N). Similarly, if a nonessential gene becomes essential in mouse (N2E), then connections are generally added rapidly (see above), but insufficient evolutionary time may have occurred to achieve the connection level of a gene that was already essential in yeast and remained essential in mouse. As shown in Fig. 6, when we only consider genes with conserved essentiality in both yeast and mouse, the correlation between connectivity and essentiality becomes extremely high (R2 = 0.97). In other words, when we set a common starting point in the connectivity race, essential genes do acquire more connections than non-essential genes. Thus, the C-L rule does explain the relationship between gene essentiality and network connection. It also suggests that interaction rewiring should be properly considered for predicting gene essentiality on a genome-wide scale through the mapping of orthologs24.
Figure 6. The influence of evolutionary history on the C-L rule.
When all genes are considered regardless of evolutionary history (left panel), the correlation between connectivity and essentiality is relatively weak. If only genes with conserved essentiality are considered (right panel), the correlation is dramatically improved. Error bars indicate the standard error.
The relationship between gene essentiality changes and the increase of network connections is also true for relatively young genes that are found from either yeast or mouse. Among mouse genes that do not have yeast orthologs, 2,189 were found to be essential (X2E) and 12,207 were nonessential (X2N). We found that X2E has significantly more network connections than X2N in the mouse PPI network (p = 2.16 × 10−72). Meanwhile, of yeast genes without mouse orthologs, 427 were found to be essential (E2X) and 3,983 were nonessential (N2X). Similarly, E2X were found to have significantly more network connections than N2X (p = 5.33 × 10−21). These biases of network connections in young genes suggest that genes engaging in more interactions are likely to be essential. When young genes first arose, they are likely to be nonessential because their ancestral species survived without them and they share network connections with their parental genes9. As they underwent interaction rewiring, those that gained more interactions became essential and had more chances to be a member of vital pathways.
To our knowledge, this study highlights for the first time that interaction rewiring is a key to the evolution of gene essentiality. Relating network rewiring with phenotypic changes will improve our understanding of the functional evolution of genes.
Methods
Essential and nonessential genes of yeast and mouse
Phenotype data of mouse gene deletions were obtained from Mouse Genome Informatics (www.informatics.jax.org/). These phenotypes were identified from random gene disruption, gene trap mutagenesis, and targeted deletion25. Genes annotated as essential phenotypes, such as embryonic lethality (MP: 0002080), prenatal lethality (MP: 0002081), survival postnatal lethality (MP: 0002082), abnormal reproductive system morphology (MP: 0002160), or abnormal reproductive system physiology (MP: 0001919) were classified as essential genes. All other mouse genes were classified as nonessential genes. This process identified 2,071 essential 12,928 nonessential mouse genes.
Gene essentiality data of yeast were manually compiled from the Comprehensive Yeast Genome Database (http://mips.helmholtz-muenchen.de/genre/proj/yeast/) and large-scale experiments26. The dataset contained 1,178 essential and 4,904 nonessential yeast genes.
Construction of yeast and mouse PPI networks
We constructed yeast and mouse PPI networks by integrating 22 protein interaction databases10: the Bio-molecular Interaction Network Database (BIND), the Human Protein Reference Database (HPRD), the Molecular Interaction database (MINT), DIP, IntAct, BioGRID, Reactome, the Protein-Protein Interaction Database (PPID), BioVerse, CCS-HI1, the comprehensive resource of mammalian protein complexes (CORUM), IntNetDB, the Mammalian Protein-Protein Interaction Database (MIPS), the Online Predicted Human Interaction Database (OPHID), Ottowa, PC/Ataxia, Sager, Transcriptome, Complexex, Unilever, protein-protein interaction database for PDZ-domains (PDZBase), and a protein interaction dataset from the literature. We removed low-confidence interactions that were not supported by direct experimental evidence. The resulting integrated PPI network comprises 101,777 interactions between 11,043 proteins. Based on the integrated PPI network, we then constructed yeast and mouse PPI networks by ortholog mapping. The interactions were transferred to yeast and mouse when both orthologs in an interacting pair were present. Orthologous gene pairs were obtained from the Inparanoid database (http://inparanoid.sbc.su.se). Only the 100% confidence orthologous pair in each ortholog group was used in the analysis. The final yeast PPI network comprises 14,024 interactions between 1,367 yeast proteins; the mouse PPI network comprises 78,582 interactions between 9,210 mouse proteins.
Gene ontology analysis
To investigate bio-processes mediated by the interactions of E2E, N2E, E2N, and N2N genes, we analyzed the GO annotations of direct network neighbors. We used DAVID27 for gene set enrichment analysis. Statistically overrepresented bio-process terms of each group were analyzed and the fold enrichment was calculated by comparing the frequencies of genes with a GO annotation between a gene group and a genome. The analyses were conducted for yeast and mouse, separately. Only bio-processes that were overrepresented with p-value lower than 0.001 were employed.
Protein complex data
We obtained yeast protein complex data from a curated consensus set which catalogs 518 protein complexes through a combination of various high-throughput data28. Mouse protein complex data were obtained from CORUM database which lists 454 manually curated mouse complexes29.
Calculation of evolutionary rate (dN/dS)
The evolutionary rates (dN/dS) of the genes in Saccharomyces species were computed by using nucleotide sequences for 3,392 orthologous open reading frames (ORF) in S. cerevisiae, S. paradoxus, S. mikatae, and S. bayanus20. A maximum likelihood phylogeny was constructed for each ORF using PHYLIP30. Then, the number of synonymous nucleotide substitutions per synonymous site (dS) and the number of nonsynonymous substitutions per nonsynonymous site (dN) were calculated by using PAML program31.
Author Contributions
J.K. and S.K. designed the study. J.K., J.B., S.K. wrote the paper. I.K, S.K.H. analyzed data.
Supplementary Material
supplementary table 1
supplementary table 2
supplementary table 3
Acknowledgments
This work was supported in part by Korean National Research Foundation grants (2012002568, 20110027840, and R312011000101 of the World Class University program).
References
- Chen W. H., Minguez P., Lercher M. J. & Bork P. OGEE: an online gene essentiality database. Nucleic Acids Res 40, D901–906 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissonauth V., Roy S., Gravel M., Guillemette S. & Charron J. Requirement for Map2k1 (Mek1) in extra-embryonic ectoderm during placentogenesis. Development 133, 3429–3440 (2006). [DOI] [PubMed] [Google Scholar]
- Nadeau V. et al. Map2k1 and Map2k2 genes contribute to the normal development of syncytiotrophoblasts during placentation. Development 136, 1363–1374 (2009). [DOI] [PubMed] [Google Scholar]
- Anderson S. J. & Perlmutter R. M. A signaling pathway governing early thymocyte maturation. Immunol Today 16, 99–105 (1995). [DOI] [PubMed] [Google Scholar]
- Jeong H., Mason S. P., Barabasi A. L. & Oltvai Z. N. Lethality and centrality in protein networks. Nature 411, 41–42 (2001). [DOI] [PubMed] [Google Scholar]
- Coulomb S., Bauer M., Bernard D. & Marsolier-Kergoat M.-C. Gene essentiality and the topology of protein interaction networks. Proceedings. Biological sciences/The Royal Society 272, 1721–1725 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi T. K. B. et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nature Genetics 38, 285–293 (2006). [DOI] [PubMed] [Google Scholar]
- Batada N. N., Hurst L. D. & Tyers M. Evolutionary and physiological importance of hub proteins. PLoS computational biology 2, e88 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhang Y. E. & Long M. New Genes in Drosophila Quickly Become Essential. Science 330, 1682–1685 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. et al. Rewiring of PDZ domain-ligand interaction network contributed to eukaryotic evolution. PLoS Genet 8, e1002510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M. G., Sikora M., Costanzo M., Boone C. & Kim P. M. Network evolution: rewiring and signatures of conservation in signaling. PLoS Comput Biol 8, e1002411 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. et al. Reshaping of global gene expression networks and sex-biased gene expression by integration of a young gene. EMBO J 31, 2798–2809 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. et al. Frequent recent origination of brain genes shaped the evolution of foraging behavior in Drosophila. Cell reports 1, 118–132 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C. & Chothia C. Protein family expansions and biological complexity. PLoS Comput Biol 2, e48 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K., Fu Z., Hou L. & Han J. D. Impacts of protein-protein interaction domains on organism and network complexity. Genome Res 18, 1500–1508 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M. C. & Rosenblum N. D. Genetic regulation of branching morphogenesis: lessons learned from loss-of-function phenotypes. Pediatr Res 54, 433–438 (2003). [DOI] [PubMed] [Google Scholar]
- McKay M. M., Ritt D. A. & Morrison D. K. Signaling dynamics of the KSR1 scaffold complex. Proc Natl Acad Sci U S A 106, 11022–11027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam T. J. & Snel B. Protein complex evolution does not involve extensive network rewiring. PLoS Comput Biol 4, e1000132 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L. J., Jensen T. S., de Lichtenberg U., Brunak S. & Bork P. Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature 443, 594–597 (2006). [DOI] [PubMed] [Google Scholar]
- Hirsh A. E., Fraser H. B. & Wall D. P. Adjusting for selection on synonymous sites in estimates of evolutionary distance. Mol Biol Evol 22, 174–177 (2005). [DOI] [PubMed] [Google Scholar]
- Dunn R., Dudbridge F. & Sanderson C. M. The use of edge-betweenness clustering to investigate biological function in protein interaction networks. BMC bioinformatics 6, 39 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. & Fell D. A. The small world inside large metabolic networks. Proc Biol Sci 268, 1803–1810 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Tombor B., Albert R., Oltvai Z. N. & Barabasi A. L. The large-scale organization of metabolic networks. Nature 407, 651–654 (2000). [DOI] [PubMed] [Google Scholar]
- Deng J. et al. Investigating the predictability of essential genes across distantly related organisms using an integrative approach. Nucleic Acids Res 39, 795–807 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B.-Y. & Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proceedings of the National Academy of Sciences of the United States of America 105, 6987–6992 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener U. et al. CYGD: the Comprehensive Yeast Genome Database. Nucleic Acids Res 33, D364–368 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T. & Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- Benschop J. J. et al. A consensus of core protein complex compositions for Saccharomyces cerevisiae. Mol Cell 38, 916–928 (2010). [DOI] [PubMed] [Google Scholar]
- Ruepp A. et al. CORUM: the comprehensive resource of mammalian protein complexes--2009. Nucleic Acids Res 38, D497–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (phylogeny and inference package). Version. 3.6. Distributed by the author, Department of Genetics, University of Washington, Seattle. (2003).
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13, 555–556 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary table 1
supplementary table 2
supplementary table 3