ABSTRACT
Horizontal gene transfer (HGT) is largely responsible for increasing the incidence of antibiotic-resistant infections worldwide. While studies have focused on HGT in vivo, this work investigates whether the ability of pathogens to persist in the environment, particularly on touch surfaces, may also play an important role. Escherichia coli, virulent clone ST131, and Klebsiella pneumoniae harboring extended-spectrum-β-lactamase (ESBL) blaCTX-M-15 and metallo-β-lactamase blaNDM-1, respectively, exhibited prolonged survival on stainless steel, with approximately 104 viable cells remaining from an inoculum of 107 CFU per cm2 after 1 month at 21°C. HGT of bla to an antibiotic-sensitive but azide-resistant recipient E. coli strain occurred on stainless steel dry touch surfaces and in suspension but not on dry copper. The conjugation frequency was approximately 10 to 50 times greater and occurred immediately, and resulting transconjugants were more stable with ESBL E. coli as the donor cell than with K. pneumoniae, but blaNDM-1 transfer increased with time. Transconjugants also exhibited the same resistance profile as the donor, suggesting multiple gene transfer. Rapid death, inhibition of respiration, and destruction of genomic and plasmid DNA of both pathogens occurred on copper alloys accompanied by a reduction in bla copy number. Naked E. coli DNA degraded on copper at 21°C and 37°C but slowly at 4°C, suggesting a direct role for the metal. Persistence of viable pathogenic bacteria on touch surfaces may not only increase the risk of infection transmission but may also contribute to the spread of antibiotic resistance by HGT. The use of copper alloys as antimicrobial touch surfaces may help reduce infection and HGT.
IMPORTANCE
Horizontal gene transfer (HGT) conferring resistance to many classes of antimicrobials has resulted in a worldwide epidemic of nosocomial and community infections caused by multidrug-resistant microorganisms, leading to suggestions that we are in effect returning to the preantibiotic era. While studies have focused on HGT in vivo, this work investigates whether the ability of pathogens to persist in the environment, particularly on touch surfaces, may also play an important role. Here we show prolonged (several-week) survival of multidrug-resistant Escherichia coli and Klebsiella pneumoniae on stainless steel surfaces. Plasmid-mediated HGT of β-lactamase genes to an azide-resistant recipient E. coli strain occurred when the donor and recipient cells were mixed together on stainless steel and in suspension but not on copper surfaces. In addition, rapid death of both antibiotic-resistant strains and destruction of plasmid and genomic DNA were observed on copper and copper alloy surfaces, which could be useful in the prevention of infection spread and gene transfer.
Introduction
The discovery in 1928 of an antimicrobial compound containing a β-lactam ring that exerted its effect by disrupting bacterial cell wall synthesis and construction revolutionized the treatment of infectious diseases. Continuous use, overuse, and misuse of many antimicrobials in human and animal health applications and food production and the lack of effective regimens to prevent the spread of infection over the last 50 years has led to the evolution of many classes of β-lactamase enzymes and the subsequent development of many generations of β-lactams to try to overcome this (reviewed in reference 1). The most worrying trend is the horizontal transmission of antibiotic resistance genes via mobile elements such as plasmids and transposons, especially in Gram-negative pathogens (1–3).
Over the last 15 years, β-lactamase enzymes that have an extended spectrum of activity (ESBL) against the majority of β-lactams, including cephalosporins but not carbapenemases, have evolved. One of these, CTX-M-15, initially found in Escherichia coli but now found in other members of Enterobacteriaceae and frequently associated with a specific lineage, uropathogenic clone ST131 (1, 2), has spread worldwide. It is often located on highly mobile IncFII plasmids (4, 5) and associated with mobile genetic element IS26 (1). The risk of infection is particularly high in individuals in association with prolonged hospitalization, catheterization, nursing home residency, previous antibiotic treatment, underlying renal or liver pathology, and travel to high-risk areas (4). This has generated a high selection pressure, particularly because the use of carbapenemases has been necessary to treat ESBL infections and now β-lactamases have evolved that are effective against carbapenems as well. There has been no choice for treatment, because delay in the use of carbapenems increases mortality in ESBL patients (6). Klebsiella pneumoniae carbapenemases (KPC) have now spread worldwide, and it has been estimated that up to 40% of intensive care unit (ICU) patients in New York harbor carbapenem-resistant Klebsiella strains, with a 14-day mortality rate of 47% (reviewed in reference 4). There are several metallo-β-lactamases, including VIM and NDM-1, with 1 or 2 zinc ions in the active site, and these are more prevalent in Europe and Asia (1, 4, 7).
NDM-1 was isolated in 2008 from a patient who had visited India and contracted a urinary tract infection (UTI) (8). One significance of this important discovery was that the gene was not only present in different species in the same patient but was also located on a plasmid associated with genes conferring resistance not only to all β-lactams but also to other antibiotic classes and those affecting virulence and pathogenesis. The subsequent increase in nosocomial infections caused by bacteria harboring NDM-1 has been linked to travel to India, Pakistan, and the Balkans (1, 7, 9), especially for surgical procedures, including transplantation and cosmetic surgery often described as “medical tourism” (1, 7, 9). A worrying development is the spread to the community, resulting in infections primarily from orofecal transmission via direct contact with contaminated humans, animals, or the environment as well as via the ingestion of contaminated food and water (4, 10–12). The discovery that in certain areas there is widespread environmental contamination with NDM-1, lack of access to adequate sanitation, and extensive environmental contamination with fecal matter (4) and that the gene is increasingly found in E. coli, a major cause of community diarrhea, is a huge cause for concern; in Pakistan, approximately 20% of the population carry the gene (4, 7). In addition, initial isolates were not associated with clonal spread; however, the recent discovery of blaNDM-1 in the ubiquitous E. coli ST131 lineage and the recent discovery of the gene on the IncFII plasmid which has been primarily responsible for the rapid dissemination of blaCTX-M15 is adding to concerns of a similar pandemic spread (4, 7, 13). In addition, as for methicillin-resistant Staphylococcus aureus (MRSA), asymptomatic colonization, which may precede infection, may mask the size of the reservoir (4). NDM enzymes, such as NDM-4, are now evolving that are more catalytically efficient (14). A clonal outbreak observed in the United Kingdom, when 9 patients were contaminated, was traced to a failure of disinfection of a single endoscope camera. blaNDM-1 is now found in a large number of Enterobacteriaceae and opportunistic environmental species, although carriage is not as stable in vitro in some nonfermenters, including Acinetobacter baumannii and Pseudomonas aeruginosa, although NDM-1 has been isolated from these species (4, 7, 15).
This is such a serious problem it has been likened to the development of acquired immunodeficiency syndrome in the 1980s and the current epidemic of drug-resistant tuberculosis (16). The main concern is that the lack of effective antimicrobials and the delay in the development of new ones may return us to the preantibiotic era such that minor infections may escalate into serious, systemic life-threatening pathologies (1, 4, 16).
Within clinical and domestic environments, touch surfaces such as stainless steel are employed primarily for their resilience, cost, and ability to be cleaned regularly. However, several pathogens have been shown to exhibit prolonged survival on stainless steel and other dry surfaces, and microscopic striations on apparently smooth surfaces can harbor viable microorganisms that evade cleaning regimens (17–19). Guet-Revillet et al. (20) observed contamination by fecal carriage of ESBL-producing Enterobacteriaceae, especially Klebsiella, in clinical environments frequented by patients and also saw no difference in levels of environmental contamination before and after routine cleaning procedures.
Consequently, we investigated the survival of E. coli (clone ST131) and K. pneumoniae containing blaCTX-M-15 and blaNDM-1, respectively, on abiotic surfaces. It has been shown that these genes are transferable in suspension in vitro and even in situ in the gut to other species (8, 15, 21). We investigated if horizontal gene transfer could occur on stainless steel surfaces, which may have implications for the rapid spread of these genes to potentially hardier and more virulent bacteria. We also investigated the efficacy of antimicrobial copper alloy surfaces which could be employed in high-risk areas in clinical and community settings.
RESULTS AND DISCUSSION
ESBL-producing E. coli and NDM-1-producing K. pneumoniae exhibit prolonged survival on stainless steel surfaces.
Previous work in our laboratory had shown that both Gram-positive and Gram-negative pathogens survive for long periods on stainless steel surfaces (18, 22). In this new work, we observed the same for ESBL-producing E. coli and NDM-1-producing K. pneumoniae: with a starting inoculum of approximately 107 CFU per cm2 of metal surface, there was a 2-log reduction in viable cell numbers for both species over the first 10 days, gradually declining to a 3- to 4-log reduction at 4 weeks. Viable E. coli bacteria were detected at 100 days (Fig. 1). In addition, there was no significant reduction in viability when cells were exposed to other common surfaces, including glass, ceramic tiles, and acrylic, for 5 h (not shown). The results suggest that the presence of antibiotic resistance genes that confer an advantage to the organism in vivo does not affect fitness in the environment. In addition, the prolonged environmental survival can increase the risk of infection spread (23).
FIG 1.
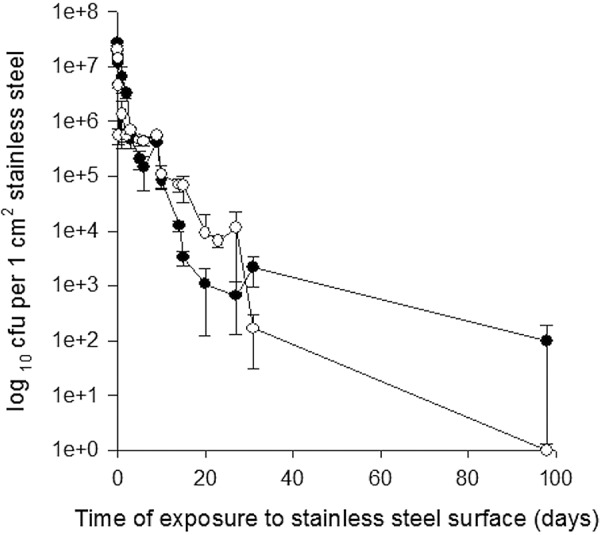
Survival of E. coli (NCTC 13441) (●) and K. pneumoniae (NCTC 13443) (○) containing blaCTX-M-15 and blaNDM-1, respectively, on stainless steel at 22°C. Approximately 107 CFU in 20 µl were inoculated onto 1-cm2 metal coupons in bacteriological medium. Cells were removed and assessed for culturability as described in the text. For both species, a 2-log reduction in viable cell numbers was observed over the first 10 days, gradually declining to a 3- to 4-log reduction over the following month. Viable E. coli bacteria were detected at 100 days. Error bars represent ± SD (standard deviations), and data are from multiple independent experiments.
Horizontal transfer of β-lactamase genes occurs in suspension and on stainless steel surfaces.
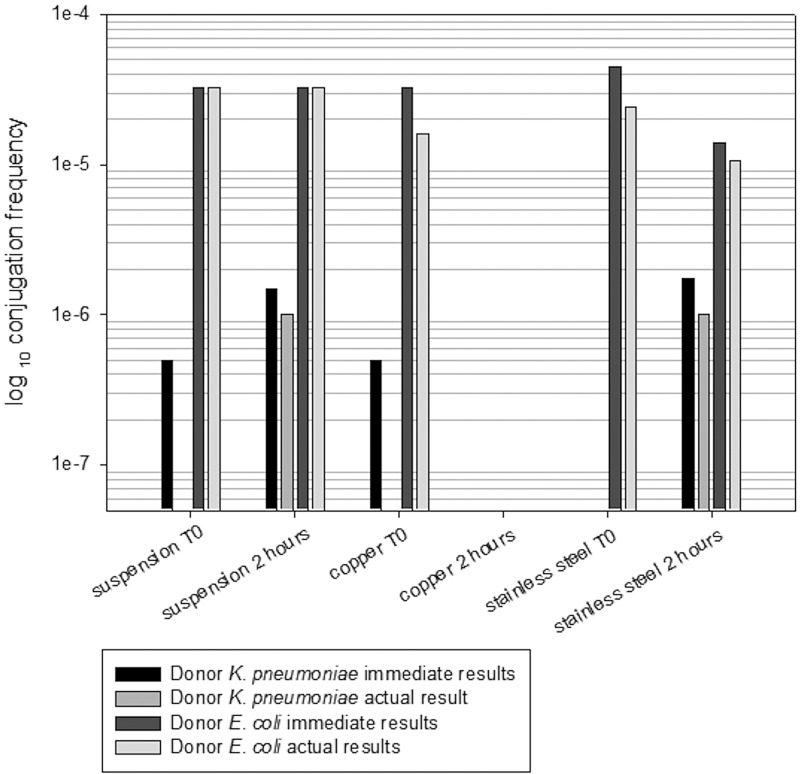
Clinical isolates are often screened for the ability to transfer genes to recipient strains in suspension. Using an azide-resistant recipient strain of E. coli, we selected for transconjugants produced in suspension or on surfaces at room temperature (21°C ± 2°C) using culture medium containing antibiotic and sodium azide concentrations that together would kill donor and recipient cells. The frequency of conjugation was over 10 times greater and more stable with E. coli containing blaCTX-M-15 as the donor than with K. pneumoniae containing blaNDM-1 (Fig. 2).
FIG 2.
Comparison of the frequencies of conjugation of donor E. coli (NCTC 13441) and K. pneumoniae (NCTC 13443) containing blaCTX-M-15 and blaNDM-1, respectively, versus the recipient E. coli strain (Azr [F− met pro], sensitive to β-lactam antibiotics) in suspension and on surfaces. Conjugation frequency is expressed as the number of transconjugants per donor cell as described in the text and is shown as the immediate result (initial isolation) or as the actual result following subculture (i.e., stable transconjugants that retain the gene). Transfer of blaCTX-M15 occurred immediately in the suspension at a frequency of approximately 3 × 10−5 and did not increase after 2 h. Similar results were observed on stainless steel, but no transconjugants were recovered from copper after 2 h of contact. Transfer of blaNDM-1 from K. pneumoniae occurred at a lower frequency of approximately 3 × 10−7, and although transfer did occur immediately when the cells were mixed in suspension and on copper, the transconjugants were not stable and died on subculturing. However, after 2 h, stable transconjugants were produced on stainless steel and in suspension.
Gene transfer from ESBL E. coli to strain J53 in suspension occurred in the few minutes required to process t0 samples, i.e., as soon as the cells were mixed together. After 2 h, there was no change in the conjugation frequency (approximately 3 × 10−5), and all transconjugants were stable upon subculture. On stainless steel, at t0, the conjugation frequency was twice that of cells in suspension, but the actual number of transconjugants which were stable was less than in suspension. After 2 h of contact, there was no increase in gene transfer, suggesting that the significant gene transfer occurred as soon as cells were mixed at t0.
The transfer of carbapenemase genes occurred at a significantly lower frequency (~4 × 10−7). In suspension, unstable transconjugants were produced at t0; however, over 2 h, the number of transconjugants increased (~2 × 10−6) and the majority of these were then stable.
Horizontal transmission of carbapenemase genes is often accompanied by transmission of flanking genes, including those conferring resistance to other antimicrobials and affecting expression, including multiple promoters and novel replicase genes (reviewed in reference 4). In our work, transconjugants generated from both species had the same multidrug resistance profile as the donor strain, demonstrating multiple gene transfer and the potential for rapid dissemination of multiple antibiotic resistance genes to the same and other species. E. coli serotype O157: H7 has evolved as a significant pathogen by the acquisition of genes from 53 different species, including pathogenicity islands and virulence factors responsible for attachment, colonization, and production of toxins, and gene transfer on towels and chopping boards has been observed (24). The advent of ESBL- and metallo-β-lactamase-producing organisms has rendered virtually all β-lactams obsolete and has severely limited treatment options, and a huge concern is that these pathogens may also acquire other genes, increasing virulence. Increased mortality caused by severe infections with Pseudomonas metallo-β-lactamase producers has already been observed, but there is a wide variation in the results of infections by Enterobacteriaceae (1). The lack of development of new antimicrobials (3) and the emergence of resistance to resurrected “savage” older treatments (25) to counteract this exacerbates the problem. A few potential treatments, including new applications for existing drugs such as the angiotensin converting enzyme (ACE) inhibitor d-captopril, and new treatments, including a non-β-lactam inhibitor of metallo-β-lactamases, NXL104 (1).
Walsh et al. (7) observed that the highest frequency of conjugation between cells in suspension occurred at 30°C compared to 25°C and 37°C and suggested that gene transfer may be more frequent in areas of the world with a higher ambient temperature. This may be extrapolated to the gene transfer on surfaces that we have observed. The combination of the ability to survive for long periods in the environment and the propensity to acquire genes increasing virulence has resulted in the development of significant pathogens. Guet-Revillet et al. (20) identified contamination of surfaces in clinical facilities that had been occupied by ESBL-colonized or -infected children and observed that the contamination was more common with K. pneumoniae than E. coli. Our results suggest that transfer of blaCTX-M-15 to recipient cells can occur immediately and that transfer of blaNDM-1 from Klebsiella can occur on surfaces with increasing numbers over time at room temperature. This suggests that immediate effective decontamination of surfaces exposed to patients is required; otherwise, gene transfer to other pathogens in the vicinity could occur. Prolonged pathogen survival and horizontal gene transfer (HGT) pose a significance risk not only for exacerbations of multiple antibiotic-resistant health care-associated infections but also for widespread dissemination in public transportation systems where hand contact is unavoidable, particularly in societies where personal hygiene compliance is poor.
Horizontal transfer of β-lactamase genes does not occur on antimicrobial copper surfaces.
A reduction in the microbial burden and infection rate has previously been observed with dry copper alloy surfaces for other bacterial, viral, and fungal pathogens in laboratory studies and clinical trials incorporating copper alloys into a range of fitments, including door handles and push plates, toilet seats, bed rails, and intravenous poles in high-risk clinical and community environments throughout the world (18, 19, 22, 26–35). Previous work with enterococci has suggested that dry copper surfaces evoke death by release of copper ions and effects on growth and respiration; genomic and plasmid DNA is a primary target (18, 22). DNA is also degraded in Gram-negative cells exposed to copper but not as rapidly; the cell membrane is a primary target because depolarization occurs immediately on contact and peroxidation of membrane lipids occurs (32, 36). After we had observed HGT on stainless steel, we investigated whether it would occur on dry copper surfaces.
On copper surfaces, the transconjugation frequency of blaCTX-M-15 was the same as that seen in cells in suspension at t0 but a small, significant (P < 0.001) number of transconjugants were unstable (Fig. 2). No transconjugants were evident after 2 h of contact, presumably due to the effects of antimicrobial copper. On copper at t0, no stable blaNDM-1-containing transconjugants were produced (the same frequency as cells in suspension), and none were evident after 2 h of contact. The results suggest that copper surfaces may prevent HGT.
Under conditions of wet fomite or dry touch surface contamination with E. coli and K. pneumoniae carrying blaCTX-M-15 and blaNDM-1, respectively, the pathogens die rapidly on antimicrobial copper surfaces at room temperature (21°C ± 2°C).
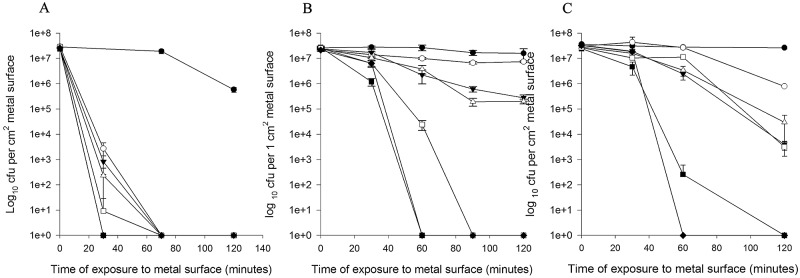
Approximately 107 CFU of ESBL-producing E. coli in phosphate-buffered saline (PBS) were spread over 1-cm2 coupons of copper and alloys containing between 95% and 60% copper (Table 1) in 20 µl or 1 µl to mimic wet fomite and dry touch surface contamination, respectively. Results for E. coli fomite contamination are shown in Fig. 3. Cells inoculated in PBS (panel A) were dead on copper and copper nickel after 30 min of contact. Death occurred on all other alloys by 90 min, although the rate of killing over the first 30 min was proportional to the percentage of copper, i.e., the rate was higher for phosphor bronze than Muntz metal containing 95% and 60% copper, respectively. When cells were inoculated in complex matrices of carbohydrates, proteins, and lipids to represent naturally occurring soils (bacteriological media tryptone soy broth [TSB] [panel B] and brain heart infusion broth [BHIB] [panel C]), death on copper and copper alloys was delayed.
TABLE 1.
Composition of metal coupons
| Metal type | UNSa no. | % composition |
|||||
|---|---|---|---|---|---|---|---|
| Cu | Zn | Sn | Ni | Fe | Cr | ||
| Copper | C11000 | 100 | |||||
| Phosphor bronze (contains ~0.26% P) | C51000 | 95 | 5 | ||||
| Copper nickel | C70600 | 89 | 10 | 1 | |||
| Cartridge brass | C26000 | 70 | 30 | ||||
| Nickel silver | C75200 | 65 | 17 | 18 | |||
| Muntz metal | C28000 | 60 | 40 | ||||
| Stainless steel | S30400 | 8 | 74 | 18 | |||
UNS, unified numbering system.
FIG 3.
Survival of a wet fomite inoculum of extended-spectrum-β-lactamase-producing E. coli (NCTC 13441) containing blaCTX-M-15 on copper and copper alloys at 22°C inoculated in a range of matrices. Approximately 107 CFU in 20 µl were inoculated onto the following 1-cm2 metal coupons in PBS (A), tryptone soy broth (TSB) (B), or brain heart infusion broth (BHIB) (C) (see Table 1 for constituents of coupons): S30400 (●), C28000 (○), C75200 (▾), C26000 (Δ), C70600 (▪), C51000 (□), and C11000 (♦). Cells were removed and assessed for culturability as described in the text. Prolonged survival was observed on stainless steel with no significant reduction in cell viability for all matrices. Cells in PBS died very rapidly, but death was delayed in the other two matrices. However, there was a significant reduction in numbers of viable cells at 2 h for all alloys and matrices (P < 0.05) except cells inoculated in TSB onto C28000, the alloy with the lowest copper content. Error bars represent ± SD, and data are from multiple independent experiments.
In TSB, death occurred by 60 min on copper and copper nickel and 90 min on phosphor bronze, and a significant reduction (P < 0.05) occurred on cartridge brass and nickel silver following 2 h of contact. On Muntz metal, which has the lowest copper content tested, the reduction in viable cells was not significant at 2 h (P = 0.243).
In BHIB, which has high lipid content, death occurred on copper and copper nickel within 2 h and there was a significant reduction in viable cells on all other alloys (a 3- to 4-log reduction on phosphor bronze, cartridge brass, and nickel silver).
This delay may have been due either to constituents in the matrices chelating copper ions and therefore preventing access to the cells or possibly to interactions with the Gram-negative outer membrane. Previous experiments with Gram-positive enterococci indicate the latter, because there was no significant delay between kill times in complex matrices on copper surfaces.
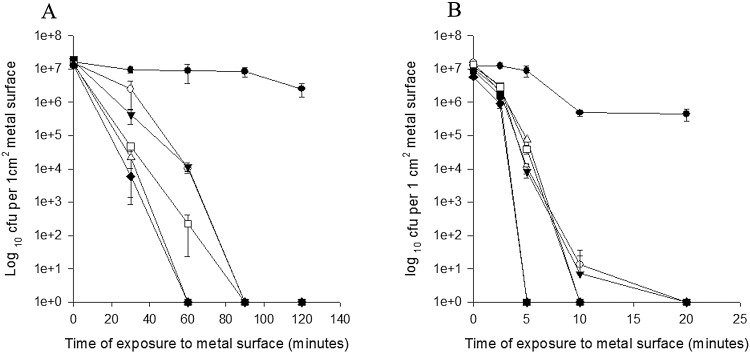
Similar results were observed for K. pneumoniae, with slightly longer times to death: all cells had died on copper, copper nickel, and cartridge brass by 60 min or 90 min for the remaining alloys (Fig. 4A). Death occurred considerably more quickly with dry touch surface contamination, a phenomenon we have observed previously (32): death occurred on copper and copper nickel by 5 min and by 20 min for the remaining alloys (Fig. 4B).
FIG 4.
Survival of a wet fomite (A) and dry touch surface (B) inoculum of K. pneumoniae (NCTC 13443) containing blaNDM-1 on copper and copper alloys at 22°C. Approximately 107 CFU in 20 µl (dries in 30 min) or 1 µl (dries in seconds) were inoculated onto the following 1-cm2 metal coupons in PBS: S30400 (●), C28000 (○), C75200 (▾), C26000 (Δ), C70600 (▪), C51000 (□), and C11000 (♦). Cells were removed and assessed for culturability as described in the text. Prolonged survival was observed on stainless steel, but rapid death occurred on copper and copper alloys in proportion to the percentage of copper, especially for the dry touch surface contamination, where all cells were dead on copper and copper nickel after 5 min of contact. Similar results were obtained for extended-spectrum-β-lactamase-producing E. coli (not shown). Error bars represent ± SD, and data are from multiple independent experiments.
Therefore, the rate of killing on copper and copper alloys is determined not only by the aqueous content of the inoculating matrix but also by the constituents of the matrix itself. Death was most rapid in PBS and slightly slower in complex matrices. Similar results were observed by Noyce et al. (28), who observed a delay in killing of E. coli O157 on copper surfaces in the presence of blood and fat in meat juices.
As expected, in light of the long-term survival noted previously, no significant reduction in viable cells on stainless steel in all three matrices was observed after 2 h of exposure at room temperature. The high inoculum used for these experiments represents a worst-case scenario, because, in reality, the naturally occurring bioburden of surfaces is much less (31), and previous experiments have shown that death occurs even more rapidly with a reduced inoculum size (18).
The results do highlight the fact that antimicrobial copper and copper alloy can provide a constant killing surface, providing that suitable cleaning protocols are employed that use nonchelating substances to ensure continuous copper ion release.
Exposure to dry copper surfaces inhibits the respiration of β-lactamase producers.
Bacteria may exist in a viable-but-nonculturable (VBNC) state under stressful environmental conditions with the potential to cause infection if conditions change to those more optimal for growth. Bacteria in a VBNC state can be identified by the presence of intact, respiring cells that do not grow in culture. CTC (5-cyano-2,3-ditolyl tetrazolium chloride) is a nonfluorescent redox dye that can be used to detect active respiration in bacteria by acting as an alternative electron acceptor. When respiration is occurring, the dye is reduced to a red, fluorescent non-water-soluble formazan product.
Although culture results suggested that no viable cells could be recovered from copper and cartridge brass surfaces following 2 h of contact at room temperature (Fig. 3A and 4A), cells were stained with CTC to determine if any VBNC cells were present (results are shown for ESBL-producing E. coli in Fig. S1 in the supplemental material). Actively respiring cells (stained red) were evident on stainless steel (panel B) after 2 h of contact but not on copper (panel F) or brass (panel D) surfaces (see Fig. S1 in the supplemental material), suggesting the absence of VBNC cells. The same preparations were also stained with the cell-permeable, low-toxicity, green fluorescent stain SYTO 9, which exhibits brighter fluorescence when bound to double-stranded DNA of live and dead cells. On stainless steel (panel A), the cells stained brightly, suggesting intact nucleic acid. However, on copper (panel E) and brass (panel C), the staining intensity was diminished, suggesting that DNA breakdown had occurred and that the dye was unable to bind. We have observed similar results in enterococci and other E. coli species (22, 32).
Therefore, the use of antimicrobial surfaces in conjunction with efficient cleaning and disinfection routines could help in the prevention of infection spread.
Degradation of plasmid DNA occurs in β-lactamase-producing E. coli and K. pneumoniae exposed to copper surfaces accompanied by reduced copy numbers of bla genes.
We have observed the prevention of HGT on copper surfaces. We extracted the entire plasmid DNA of β-lactamase producers exposed to copper surfaces to ascertain the extent of DNA damage.
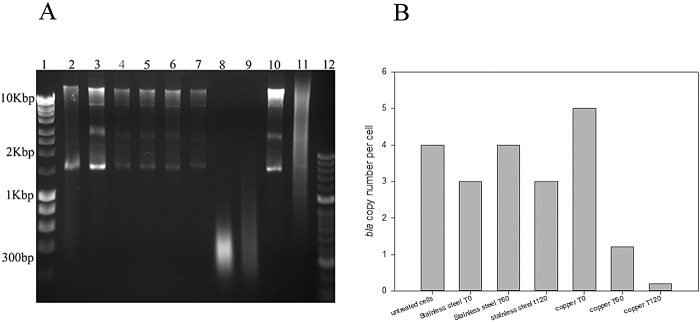
For E. coli containing blaCTX-M-15, the plasmid profiles of untreated cells (Fig. 5A, lanes 3 and 10) and cells exposed to stainless steel for 0, 60, and 120 min (Fig. 5A, lanes 4, 5, and 6) are similar, with several plasmids of multiple sizes greater than 1.5 kbp. However, on copper there was a progressive degradation of the plasmid DNA with time so that after 120 min a smear of small fragments with an average size of 300 bp was present (Fig. 5A, lanes 7, 8, and 9). The concentration of the bla gene determined by quantitative PCR (qPCR) in the same plasmid samples indicates that, although the copy number was reduced following contact with copper surfaces (after 60 min, the copy number was less than 1, suggesting that many cells did not contain a plasmid), copies were still detectable. However, the qPCR amplifies only a small fraction of the gene, and we have not yet investigated the expression and functionality of the β-lactamase itself. Interestingly, the cells that had been removed from copper immediately (t0) had a greater copy number than control cells (Fig. 5B).
FIG 5.
Degradation of plasmid DNA and a reduction of copy numbers of blaCTX-M-15 occur in extended-spectrum-β-lactamase-producing E. coli exposed to copper but not stainless steel surfaces; wet fomite inoculum. (A) The plasmid DNAs of untreated cells (lanes 3 and 10), heat-killed cells (lane 11), or cells exposed to metal surfaces (stainless steel, 0, 60, and 120 min in lanes 4, 5, and 6, respectively; copper, 0, 60, and 120 min in lanes 7, 8, and 9, respectively) were purified and separated by agarose gel electrophoresis as described in the text. The DNAs of untreated cells and those exposed to stainless steel demonstrate the same plasmid bands, indicating that no degradation of DNA had occurred. The reduction of fragment size and smearing of plasmid DNA from cells exposed to copper suggest that extensive degradation was occurring which increased with time. Plasmid DNA from K. pneumoniae is represented in lane 2. Control lanes represent Bioline Hyperladder I (lane 1) and Hyperladder II (lane 12). (B) The same plasmid preparations were assessed for concentrations of blaCTX-M-15 with gene-specific qPCR as described in the text. Approximately 3 to 4 copies of the gene were present in untreated cells and in those exposed to stainless steel. The copy number increased slightly on cells exposed to copper and immediately removed but then diminished to <1 upon longer exposure; i.e., many cells did not contain the gene.
The results were similar for the “dry” inoculum (see Fig. S2 in the supplemental material), where there was minimal effect on plasmid DNA from cells exposed to stainless steel for 0, 10, 30, and 60 min (see Fig. S2A, lanes 2 to 5, in the supplemental material) but rapid degradation on copper at the same time points (see Fig. S2A, lanes 6 to 9). It is interesting that some degradation occurred immediately on contact and that DNA fragments had massively degraded to 300-bp fragments by 10 min. qPCR detection of blaCTX-M-15 (see Fig. S2B in the supplemental material) also showed a reduction in copy numbers in cells exposed to copper, but there was greater variation in copy numbers in all cells.
Naked DNA degrades on copper surfaces and in a temperature-dependent manner.
We have previously shown that enterococcal genomic and plasmid DNA is actively degraded on copper and copper alloy surfaces (18, 22) as a result of copper ion release and generation of reactive oxygen species. We have also observed the effect of copper on Gram-negative bacteria and observed an immediate effect on the outer membrane and a much slower degradation of the bacterial DNA (32). There is some dispute with regard to whether the DNA degradation observed is the cause or effect of release of lytic enzymes and compounds post-cell death. Naked plasmid DNA (60 ng) was applied to the surface of 1-cm2 stainless steel or copper coupons in 1- or 10-µl volumes (dry within few seconds or by 10 min, respectively) for 20 min at room temperature. On stainless steel, the DNA remained intact in large fragments (several bands are visible due to the presence of several plasmid polymeric, relaxed, and supercoiled isoforms) (Fig. 6A, lanes 4 and 5). On copper, the DNA completely degraded in the rapidly dried application but traces remained in the “wet” application (Fig. 6A, lanes 6 and 7).
FIG 6.
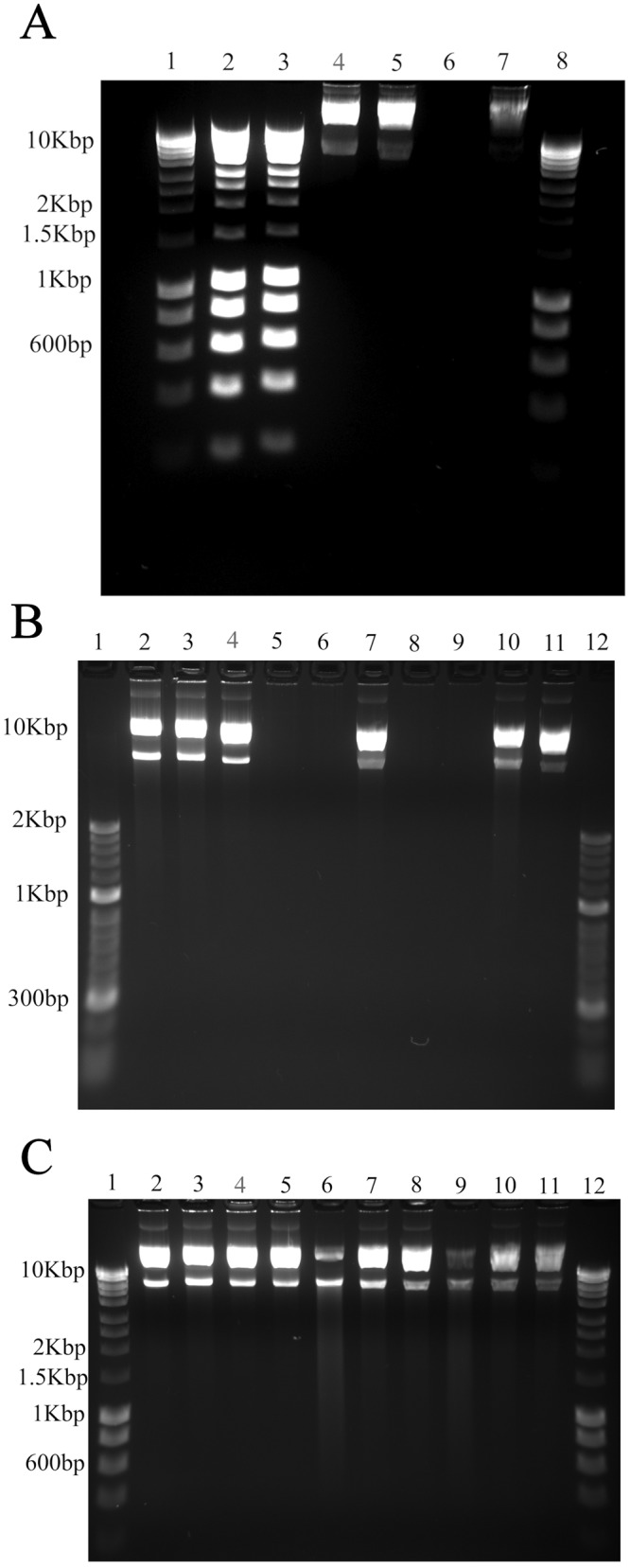
Degradation of naked plasmid DNA (pBR322) on copper surfaces is dependent on temperature and aqueous content. (A) Sixty nanograms of naked plasmid DNA was applied to metal surfaces for 20 min at room temperature in 1 µl, which dried in seconds (“dry”), or in 10 µl (“wet”). The DNA was removed by pipetting and integrity determined by agarose electrophoresis, as described in the text. For the dry inoculum, plasmid DNA recovered from stainless steel (lane 4) produced two multimer bands, the same as plasmid DNA that had not been exposed to metal (B, lane 11), but the DNA completely degraded on copper (lane 6). The wet inoculum was the same on stainless steel (lane 5), and although extensive degradation occurred on copper (lane 7), traces of DNA were still visible. Control lanes (lanes 1 and 8) represent Bioline Hyperladder I. The ladder DNA was applied directly to stainless steel (lane 2) and copper (lane 3) surfaces. No breakdown of the DNA was evident because the chelator, EDTA, and glycerol components used to stabilize DNA for long-term storage protected the DNA from damaging copper ions released from the surface. (B) Plasmid DNA (“dry” inoculum) was applied directly to stainless steel (lanes 2 to 4), cartridge brass (lanes 5 to 7), or copper (lanes 8 to 10) surfaces at 37°C (lanes 2, 5, and 8), 22°C (lanes 3, 6, and 9), or 4°C (lanes 4, 7, and 10) for 20 min. On stainless steel, DNA remained intact at all temperatures. However, on brass and copper, the DNA degraded completely at 37°C and 22°C, but very little degradation occurred at 4°C. (C) Plasmid DNA (“wet” inoculum) was also applied to stainless steel (lanes 3 to 5), cartridge brass (lanes 6 to 8), or copper (lanes 9 to 11) surfaces at 37°C (lanes 3, 6, and 9), 22°C (lanes 4, 7, and 10), or 4°C (lanes 5, 8, and 11) for 45 min. The DNA remained intact on stainless steel the same as it did on the dry inoculum. On brass and copper, results were similar to those seen with dry inoculation, i.e., most DNA broke down at the highest temperature of 37°C, but the effect was not as extensive. The amount of DNA breakdown was proportional to the copper content. Control lanes (lanes 1 and 12) represent Bioline Hyperladder I, and lane 2 is the untreated plasmid.
When the DNA was applied to copper (Fig. 6B, lanes 8 to 10) or cartridge brass (Fig. 6B, lanes 5 to 7) (dry inoculum) at a range of temperatures (4°C, 22°C, and 37°C, respectively), the DNA totally fragmented after 20 min at 37°C and 22°C but not at 4°C. The inoculum was not completely dry at this temperature.
Figure 6C shows the results seen with the DNA applied in 20 µl at 37°C (lanes 3, 6, and 9), 22°C (lanes 4, 7, and 10), or 4°C (lanes 5, 8, and 11): the extent of degradation after 45 min was greater on copper (lanes 9 to 11) than on cartridge brass (lanes 6 to 8), presumably due to reduced copper content in the latter, and became greater as the temperature increased. No change in DNA exposed to stainless steel (lanes 3 to 5) was observed under any of the 3 temperature conditions.
No degradation of DNA occurred on stainless steel (wet or dry) at any of the temperatures tested.
There is controversy in the literature, with some researchers believing that the DNA degradation observed is entirely a result of post-cell-death breakdown and toxicity from released products (37). Our experiments performed with naked DNA suggest that active DNA breakdown does occur on copper and brass surfaces that is more extensive in rapidly drying samples at temperatures greater than or equal to room temperature. However, at 4°C, DNA degradation is very slow but, given that the optimal temperature for conjugation is 30°C, the chances of conjugation may be minimal. Perhaps the DNA denaturation we have observed in Gram-negative species here represents a combination of active processes and lysis of dead cells releasing deleterious compounds.
CONCLUSION
It is estimated that up to 10% of patients admitted to modern hospitals acquire one or more infections and that the proportion rises to 25% in some developing countries. In Europe and the United States, this is equivalent to 4 and 2 million cases, leading to approximately 37,000 and 99,000 deaths per year, respectively, with a combined resultant cost of over 13 billion dollars (3). The meteoric rise in ESBL-producing E. coli and Klebsiella spp., which have been the most common Enterobacteriaceae responsible for the nosocomial spread in ESBL, may now be followed by carbapenemase producers. Nordmann et al. (4) warn of an increase in E. coli NDM-1 and OXA-48 community-acquired infections and Klebsiella species KPC, IMP, NDM, and OXA-48 infections in high-risk hospitalized individuals. It is very difficult to estimate the size of the reservoir of multidrug resistance genes surviving in resilient bacteria over the long term in the environment along with increased colonization of humans and animals. This study demonstrated that HGT readily occurs on dry touch surfaces such as stainless steel, providing a potentially important route for multidrug resistance emergence and dissemination in public buildings and transportation systems if surfaces are not regularly and efficiently cleaned. Although previous studies in our laboratory have demonstrated that plasmid and genomic DNA degradation of antibiotic-resistant enterococci and E. coli O157 occurs on dry copper surfaces (18, 32), the next step is to investigate whether HGT in these and other pathogens, including the transfer of specific genes responsible for enhanced virulence and toxin production as well as resistance to antimicrobial therapies, is prevented. The effect of various environmental conditions, contact times, and bacterial densities on the rate of HGT also has to be determined. The use of copper alloys in clinical and community settings could help reduce infection spread and also reduce the incidence of horizontal transmission genes conferring drug resistance, virulence, and pathogenesis and expression efficiency. Considerable concern has been expressed and blame has been assigned in describing the nature, origin, and nomenclature of the newly evolving resistance genes, but more accessible global travel has made the world a much smaller place, and the evolution of potentially untreatable infectious diseases will eventually and inevitably affect us all.
MATERIALS AND METHODS
Bacterial strains.
Klebsiella pneumoniae NCTC 13443 (encodes blaNDM-1 metallo-beta-lactamase) and Escherichia coli NCTC 13441 (encodes blaCTX-M-15 on plasmid pEK499) (5) were supplied by Health Protection Agency, Porton Down, United Kingdom. Sodium azide-resistant E. coli J53 (J53 Azr [F− met pro]), used as the recipient strain in conjugation experiments, was kindly supplied by George Jacoby (Lahey Clinic, Burlington, United States) (38) and James Anson (University of Liverpool, United Kingdom).
Preparation of sample surfaces.
Metal coupons (10 by 10 by 0.5 mm) were degreased in acetone, stored in absolute ethanol, and flamed prior to use as described previously (18). The constituents of each metal tested are detailed in Table 1, and all were supplied by the Copper Development Association. Other surfaces were glass microscope slides (Fisher; Thermo Scientific), ceramic tiles, and acrylic (polymethylmethacrylate [PMMA]). These surfaces, and stainless steel as a method comparison control, were degreased in mild detergent solution and sterilized by immersion in ethanol for 10 min. Samples were dried under a sterile airflow prior to use (results described in text).
Culture preparation.
Bacterial stocks were prepared on Protect Cryo beads (Microbiological Supply Company, United Kingdom) and stored at −80°C. Cultures were prepared by inoculation of 1 bead into 15 ml sterile brain heart infusion broth (BHIB) or TSB (Oxoid, United Kingdom) and incubated aerobically at 37°C for 16 ± 2 h.
Metal coupon inoculation (vortex/bead method) and assessment of viable cells by culture to simulate wet fomite or dry touch surface contamination.
The surfaces of coupons were inoculated with approximately 107 CFU in either bacteriological medium or PBS. The volume was 20 µl or 1 µl for the same number of cells, with the former representing a wet fomite inoculum that dries in 30 to 40 min at 22°C and the latter drying in seconds in a manner equivalent to a dry touch surface contamination. Drying time was included in the exposure time, and this method has been described previously by us for enterococci (18). Bacteria were removed from the coupons at the required time point using a vortex procedure and 5 ml phosphate-buffered saline (PBS) containing EDTA (20 mM) to chelate and neutralize free copper ions and 2-mm-diameter glass beads and subjected to a vortex procedure for 30 s. A range of dilutions was prepared immediately, and aliquots were plated onto nutrient agar and tryptone soy agar (TSA) for K. pneumoniae and E. coli, respectively. When dry, the plates were inverted and incubated aerobically at 37°C for up to 48 h to quantify viable cells. Duplicate coupons were analyzed for each time point.
Inoculation of glass, ceramic, acrylic, and stainless steel surfaces and assessment of viable cells by culture (wet fomite contamination only).
These surfaces were inoculated as described for metal coupons, but the cells were removed by addition of 100 µl PBS which was vigorously pipetted up and down and the cells transferred to a sterile tube. This was repeated 5 more times; the cells were then pooled, and the volume was increased to 5 ml with the addition of sterile PBS. Dilutions were prepared and aliquots spread over nutrient agar (NA) and TSA plates as described for metal surfaces.
Detection of respiring cells on metal surfaces in situ using the redox dye CTC (5-cyano-2,3-ditolyl tetrazolium chloride): “wet” inoculum only.
Actively respiring bacteria can reduce the redox dye, CTC, to red fluorescent formazan which can be visualized by epifluorescence microscopy. Copper, cartridge brass, and stainless steel coupons were inoculated with ESBL-producing E. coli in petri dishes as described. When the coupons had dried (45 min), 50 µl of 5 mM CTC was added to each coupon, which was covered and incubated for a further 65 min at 37°C. A further 50 µl of the double-stranded DNA stain SYTO 9 (5 µM) was added to each coupon, which was incubated at room temperature for 10 min. Coupons were observed using epifluorescence microscopy with a metal halide light source and a long-working-distance 100× objective as described previously (18, 22, 39). A positive control of inoculum was stained in suspension at the same time.
Detection of changes in the integrity of plasmid DNA from CTX-M-15- and NDM-1-producing E. coli and K. pneumoniae, respectively.
Cells were inoculated onto 1-cm2 metal coupons as described for culture assessment (10 coupons per time point). Cells were removed and pooled, and the plasmid DNA was extracted using a Qiaprep Spin miniprep kit (Qiagen, United Kingdom) according to the manufacturer’s instructions. Fragments were separated on 0.9% (wt/vol) agarose gel using a GelRed nucleic acid prestaining kit (Biotium, United Kingdom) and Hyperladder I and II size markers (Bioline, United Kingdom). Gels were observed in a Syngene UV light box and photographed using GeneSnap software.
Quantification of blaCTX-M-15 and blaNDM-1 using quantitative PCR of purified plasmid DNA.
Primers were designed to amplify 80- and 118-bp fragments of blaNDM-1 and blaCTX-M15 as follows: for accession number FN396876, plasmid pKpANDM-1, sense, CCGCCATCCCTGACGATC (position 2969), antisense, GTCTGGCAGCACACTTCCT (position 3048); and for accession number *000046, plasmid pEK499_p079, sense, TGAGGCTGGGTGAAGTAAGTG (position 68), antisense, CCTGGGTTGTGGGGGATAAAA (position 185) (PrimerDesign Ltd., Southampton, United Kingdom). Amplification was performed on a Bio-Rad iQ5 cycler, and standard curves were prepared from known copy number standards to determine copy numbers in test samples.
Horizontal transfer of blaCTX-M-15 and blaNDM-1 from donor cells to recipient E. coli J53 Azr (F− met pro) cells on surfaces.
Cultures of blaCTX-M-15- and blaNDM-1-containing E. coli and K. pneumoniae, respectively (donor cells), and recipient E. coli J53 Azr cells were prepared in BHIB. The cells were pelleted and mixed together at a 10:1 ratio of donor to recipient in the same volume of Mueller-Hinton broth (Oxoid, United Kingdom); 20 µl mixed cells (total cell concentration, 5 × 108 CFU/ml) was applied immediately to the surface of copper or stainless steel coupons. Cells were removed by gentle pipetting in 2× 100 µl sterile PBS and spread over medium selecting for transconjugants, i.e., TSA containing 100 µg/ml sodium azide and 2 µg/ml cefotaxime (for the selection of J53 that had received blaCTX-M-15) or 2 µg/ml meropenem (for the selection of J53 that had received blaNDM-1). Prior to the experiment, donor cells were found to be sensitive to sodium azide and recipient cells sensitive to the antibiotic concentrations that were used. This method differs from that described for culture, so efficiency of recovery was determined. All of the donor cells and between 99.71% and 99.94% of the recipient J53 cells were recovered.
Transconjugants were subcultured, plasmids were prepared, and DNA integrity was checked by gel electrophoresis; qPCR confirmed the presence and copy numbers of blaCTX-M-15 and blaNDM-1. Those transconjugants that were isolated initially on selective media but did not survive subculture and/or lost blaCTX-M-15 and blaNDM-1 when retested by gene-specific qPCR were considered to be unstable.
The conjugation frequency was calculated from the number of transconjugants per coupon/number of donor cells per coupon (and from equivalent numbers of cells in suspension). blaCTX-M-15 transconjugants were also resistant to the same macrolide, fluoroquinolone, aminoglycoside, and glycopeptide antibiotics as the donor strain, as shown by testing with a disc diffusion assay (Oxoid, United Kingdom). The same was true of blaNDM-1 transconjugants.
Assessment of the integrity of naked plasmid DNA on metal surfaces at a range of temperatures.
Sixty nanograms of purified E. coli plasmid BR322 DNA (Sigma-Aldrich, United Kingdom) was spread over 1-cm2 copper, cartridge brass, or stainless steel surfaces in a final volume of 1 or 10 µl. After 20 or 45 minutes of contact at 4, 22, or 37°C, the DNA was removed by addition of 10 µl distilled deionized water to the surface, gentle pipetting, and transfer to a DNase-free tube. The integrity of the DNA samples was investigated by agarose gel electrophoresis as described previously.
It should, however, be noted that this is a commercial plasmid preparation lyophilized in a solution containing 1 mM EDTA to retain stability. The sample was diluted to give a final concentration of <5 µM EDTA, which is significantly less than the 20 mM concentration found to be protective on copper surfaces in our previous work.
Statistical analysis.
Data are expressed as means ± standard deviations (SD) and are the result of multiple independent experiments. Differences between duplicate samples were assessed using the Mann-Whitney rank t test. Group comparisons were analyzed using the Mann-Whitney U test where statistical significance was expressed as P < 0.05. Statistical analyses were performed using Sigma Stat version 3.5, and graphical representations were performed using Sigma Plot version 11.
SUPPLEMENTAL MATERIAL
Inhibition of respiration of extended-spectrum-β-lactamase-producing E. coli occurs on cartridge brass (C and D) and copper (E and F) but not stainless steel (A and B) surfaces following 2 h of contact at 22°C. Approximately 107 CFU in 20 µl were inoculated onto 1-cm2 metal coupons in PBS. Once dry, the cells were stained in situ with the redox dye, CTC (which is reduced to a red fluorescent product in actively respiring cells) (B, D, and F), and with the nonvital stain, SYTO 9 (which fluoresces green when intercalated into double-stranded DNA) (A, C, and E), as described in the text. Actively respiring cells were present on stainless steel, and bright SYTO 9 staining suggests that the DNA of live and dead cells was intact. On copper and brass surfaces, no respiring cells were present, which, together with the culture results, suggests the absence of VBNC (viable-but-nonculturable) cells on this surface. In addition, the reduction in SYTO 9 staining suggests that DNA had broken down and that the dye was unable to bind. Bar, 10 µm. Download Figure S1, TIF file, 3.1 MB.
Degradation of plasmid DNA and a reduction of copy numbers of blaCTX-M-15 occur in extended-spectrum-β-lactamase-producing E. coli exposed to copper but not stainless steel surfaces; dry touch surface contamination (107 CFU in 1 µl per cm2 of metal surface). (A) The plasmid DNAs of untreated cells (lane 12) or cells exposed to metal surfaces (stainless steel, 0, 10, 30, and 60 min in lanes 2, 3, 4, and 5, respectively; copper, 0, 10, 30, and 60 min in lanes 6, 7, 8, and 9, respectively) were purified and separated by agarose gel electrophoresis as described in the text. The DNAs of untreated cells and those exposed to stainless steel demonstrate the same plasmid bands, indicating that no degradation of DNA had occurred. The reduction of fragment size and smearing of plasmid DNA from cells exposed to copper suggest that extensive degradation had occurred by 10 min. Plasmid DNA from E. coli is represented in lane 10. Control lanes represent Bioline Hyperladder I (lane 1) and Hyperladder II (lane 13). (B) The same plasmid preparations were assessed for concentrations of blaNDM-1 with gene-specific qPCR as described in the text. Approximately 2 copies of the gene were present in untreated cells and in those exposed to stainless steel. The copy number increased slightly on cells exposed to copper and immediately removed but then diminished to <1 upon longer exposure, i.e., many cells did not contain the gene. Download Figure S2, TIF file, 0.8 MB.
ACKNOWLEDGMENTS
This research was supported by the Copper Development Association, New York, NY.
Footnotes
Citation Warnes SL, Highmore CJ, Keevil CW. 2012. Horizontal transfer of antibiotic resistance genes on abiotic touch surfaces: implications for public health. mBio 3(6):e00489-12. doi:10.1128/mBio.00489-12.
REFERENCES
- 1. Bush K, Fisher JF. 2011. Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from gram-negative bacteria. Annu. Rev. Microbiol. 65:455–478 [DOI] [PubMed] [Google Scholar]
- 2. Woodford N, Turton JF, Livermore DM. 2011. Multiresistant gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35:736–755 [DOI] [PubMed] [Google Scholar]
- 3. World Health Organization 2011. Report on the burden of endemic health care-associated infection worldwide. World Health Organization, Geneva, Switzerland: ISBN 978 92 4 150150 7 [Google Scholar]
- 4. Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol. 19:588–595 [DOI] [PubMed] [Google Scholar]
- 5. Woodford N, et al. 2009. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 53:4472–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tuon FF, Kruger M, Terreri M, Penteado-Filho SR, Gortz L. 2011. Klebsiella ESBL bacteremia-mortality and risk factors. Braz. J. Infect. Dis. 15:594–598 [DOI] [PubMed] [Google Scholar]
- 7. Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11:355–362 [DOI] [PubMed] [Google Scholar]
- 8. Yong D, et al. 2009. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumarasamy KK, et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cohen Stuart J, et al. 2012. Comparison of ESBL contamination in organic and conventional retail chicken meat. Int. J. Food Microbiol. 154:212–214 [DOI] [PubMed] [Google Scholar]
- 11. O’Keefe A, Hutton TA, Schifferli DM, Rankin SC. 2010. First detection of CTX-M and SHV extended-spectrum beta-lactamases in Escherichia coli urinary tract isolates from dogs and cats in the United States. Antimicrob. Agents Chemother. 54:3489–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Overdevest I, et al. 2011. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg. Infect. Dis. 17:1216–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bonnin RA, Poirel L, Carattoli A, Nordmann P. 2012. Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS One 7:e34752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nordmann P, Boulanger AE, Poirel L. 2012. NDM-4 metallo-β-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob. Agents Chemother. 56:2184–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Potron A, Poirel L, Nordmann P. 2011. Plasmid-mediated transfer of the bla(NDM-1) gene in Gram-negative rods. FEMS Microbiol. Lett. 324:111–116 [DOI] [PubMed] [Google Scholar]
- 16. Bonomo RA. 2011. New Delhi metallo-β-lactamase and multidrug resistance: a global SOS? Clin. Infect. Dis. 52:485–487 [DOI] [PubMed] [Google Scholar]
- 17. Kramer A, Schwebke I, Kampf G. 2006. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 6:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Warnes SL, Green SM, Michels HT, Keevil CW. 2010. Biocidal efficacy of copper alloys against pathogenic enterococci involves degradation of genomic and plasmid DNAs. Appl. Environ. Microbiol. 76:5390–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilks SA, Michels H, Keevil CW. 2005. The survival of Escherichia coli O157 on a range of metal surfaces. Int. J. Food Microbiol. 105:445–454 [DOI] [PubMed] [Google Scholar]
- 20. Guet-Revillet H, et al. 2012. Environmental contamination with extended-spectrum beta-lactamases: is there any difference between Escherichia coli and Klebsiella spp? Am. J. Infect. Control 40:845–848 [DOI] [PubMed] [Google Scholar]
- 21. Goren MG, et al. 2010. Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerg. Infect. Dis. 16:1014–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Warnes SL, Keevil CW. 2011. Mechanism of copper surface toxicity in vancomycin-resistant enterococci following wet or dry surface contact. Appl. Environ. Microbiol. 77:6049–6059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boyce JM. 2007. Environmental contamination makes an important contribution to hospital infection. J. Hosp. Infect. 65(Suppl 2):50–54 [DOI] [PubMed] [Google Scholar]
- 24. Kruse H, Sørum H. 1994. Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl. Environ. Microbiol. 60:4015–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McGann P, et al. 2012. Complete sequence of a novel 178-kilobase plasmid carrying bla(NDM-1) in a Providencia stuartii strain isolated in Afghanistan. Antimicrob. Agents Chemother. 56:1673–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mehtar S, Wiid I, Todorov SD. 2008. The antimicrobial activity of copper and copper alloys against nosocomial pathogens and Mycobacterium tuberculosis isolated from healthcare facilities in the Western Cape: an in-vitro study. J. Hosp. Infect. 68:45–51 [DOI] [PubMed] [Google Scholar]
- 27. Noyce JO, Michels H, Keevil CW. 2006. Potential use of copper surfaces to reduce survival of epidemic meticillin-resistant Staphylococcus aureus in the healthcare environment. J. Hosp. Infect. 63:289–297 [DOI] [PubMed] [Google Scholar]
- 28. Noyce JO, Michels H, Keevil CW. 2006. Use of copper cast alloys to control Escherichia coli O157 cross-contamination during food processing. Appl. Environ. Microbiol. 72:4239–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noyce JO, Michels H, Keevil CW. 2007. Inactivation of influenza A virus on copper versus stainless steel surfaces. Appl. Environ. Microbiol. 73:2748–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmidt MG. 2011. Copper surfaces in the ICU reduced the relative risk of acquiring an infection while hospitalised. BMC Proc. 5(Suppl 6):O53 [Google Scholar]
- 31. Schmidt MG, et al. 2012. Sustained reduction of microbial burden on common hospital surfaces through introduction of copper. J. Clin. Microbiol. 50:2217–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Warnes SL, Caves V, Keevil CW. 2012. Mechanism of copper surface toxicity in Escherichia coli O157:H7 and salmonella involves immediate membrane depolarization followed by slower rate of DNA destruction which differs from that observed for Gram-positive bacteria. Environ. Microbiol. 14:1730–1743 [DOI] [PubMed] [Google Scholar]
- 33. Weaver L, Michels HT, Keevil CW. 2008. Survival of Clostridium difficile on copper and steel: futuristic options for hospital hygiene. J. Hosp. Infect. 68:145–151 [DOI] [PubMed] [Google Scholar]
- 34. Weaver L, Michels HT, Keevil CW. 2010. Potential for preventing spread of fungi in air-conditioning systems constructed using copper instead of aluminium. Lett. Appl. Microbiol. 50:18–23 [DOI] [PubMed] [Google Scholar]
- 35. Wilks SA, Michels HT, Keevil CW. 2006. Survival of Listeria monocytogenes Scott A on metal surfaces: implications for cross-contamination. Int. J. Food Microbiol. 111:93–98 [DOI] [PubMed] [Google Scholar]
- 36. Hong R, Kang TY, Michels CA, Gadura N. 2012. Membrane lipid peroxidation in copper alloy-mediated contact killing of Escherichia coli. Appl. Environ. Microbiol. 78:1776–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Espírito Santo C, et al. 2011. Bacterial killing by dry metallic copper surfaces. Appl. Environ. Microbiol. 77:794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jacoby GA. 1998. Epidemiology of extended-spectrum beta-lactamases. Clin. Infect. Dis. 27:81–83 [DOI] [PubMed] [Google Scholar]
- 39. Keevil CW. 2003. Rapid detection of biofilms and adherent pathogens using scanning confocal laser microscopy and episcopic differential interference contrast microscopy. Water Sci. Technol. 47:105–116 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Inhibition of respiration of extended-spectrum-β-lactamase-producing E. coli occurs on cartridge brass (C and D) and copper (E and F) but not stainless steel (A and B) surfaces following 2 h of contact at 22°C. Approximately 107 CFU in 20 µl were inoculated onto 1-cm2 metal coupons in PBS. Once dry, the cells were stained in situ with the redox dye, CTC (which is reduced to a red fluorescent product in actively respiring cells) (B, D, and F), and with the nonvital stain, SYTO 9 (which fluoresces green when intercalated into double-stranded DNA) (A, C, and E), as described in the text. Actively respiring cells were present on stainless steel, and bright SYTO 9 staining suggests that the DNA of live and dead cells was intact. On copper and brass surfaces, no respiring cells were present, which, together with the culture results, suggests the absence of VBNC (viable-but-nonculturable) cells on this surface. In addition, the reduction in SYTO 9 staining suggests that DNA had broken down and that the dye was unable to bind. Bar, 10 µm. Download Figure S1, TIF file, 3.1 MB.
Degradation of plasmid DNA and a reduction of copy numbers of blaCTX-M-15 occur in extended-spectrum-β-lactamase-producing E. coli exposed to copper but not stainless steel surfaces; dry touch surface contamination (107 CFU in 1 µl per cm2 of metal surface). (A) The plasmid DNAs of untreated cells (lane 12) or cells exposed to metal surfaces (stainless steel, 0, 10, 30, and 60 min in lanes 2, 3, 4, and 5, respectively; copper, 0, 10, 30, and 60 min in lanes 6, 7, 8, and 9, respectively) were purified and separated by agarose gel electrophoresis as described in the text. The DNAs of untreated cells and those exposed to stainless steel demonstrate the same plasmid bands, indicating that no degradation of DNA had occurred. The reduction of fragment size and smearing of plasmid DNA from cells exposed to copper suggest that extensive degradation had occurred by 10 min. Plasmid DNA from E. coli is represented in lane 10. Control lanes represent Bioline Hyperladder I (lane 1) and Hyperladder II (lane 13). (B) The same plasmid preparations were assessed for concentrations of blaNDM-1 with gene-specific qPCR as described in the text. Approximately 2 copies of the gene were present in untreated cells and in those exposed to stainless steel. The copy number increased slightly on cells exposed to copper and immediately removed but then diminished to <1 upon longer exposure, i.e., many cells did not contain the gene. Download Figure S2, TIF file, 0.8 MB.