ABSTRACT
Adaptation of bacterial pathogens to a host can lead to the selection and accumulation of specific mutations in their genomes with profound effects on the overall physiology and virulence of the organisms. The opportunistic pathogen Pseudomonas aeruginosa is capable of colonizing the respiratory tract of individuals with cystic fibrosis (CF), where it undergoes evolution to optimize survival as a persistent chronic human colonizer. The transcriptome of a host-adapted, alginate-overproducing isolate from a CF patient was determined following growth of the bacteria in the presence of human respiratory mucus. This stable mucoid strain responded to a number of regulatory inputs from the mucus, resulting in an unexpected repression of alginate production. Mucus in the medium also induced the production of catalases and additional peroxide-detoxifying enzymes and caused reorganization of pathways of energy generation. A specific antibacterial type VI secretion system was also induced in mucus-grown cells. Finally, a group of small regulatory RNAs was identified and a fraction of these were mucus regulated. This report provides a snapshot of responses in a pathogen adapted to a human host through assimilation of regulatory signals from tissues, optimizing its long-term survival potential.
IMPORTANCE
The basis for chronic colonization of patients with cystic fibrosis (CF) by the opportunistic pathogen Pseudomonas aeruginosa continues to represent a challenging problem for basic scientists and clinicians. In this study, the host-adapted, alginate-overproducing Pseudomonas aeruginosa 2192 strain was used to assess the changes in its transcript levels following growth in respiratory CF mucus. Several significant and unexpected discoveries were made: (i) although the alginate overproduction in strain 2192 was caused by a stable mutation, a mucus-derived signal caused reduction in the transcript levels of alginate biosynthetic genes; (ii) mucus activated the expression of the type VI secretion system, a mechanism for killing of other bacteria in a mixed population; (iii) expression of a number of genes involved in respiration was altered; and (iv) several small regulatory RNAs were identified, some being mucus regulated. This work highlights the strong influence of the host environment in shaping bacterial survival strategies.
Introduction
Introduction of bacteria into new environmental reservoirs, including tissues of infected human hosts, leads to large-scale reprogramming of their signal-transducing and regulatory networks to accommodate the conditions encountered in a particular niche. Pathogenic microorganisms similarly adjust their metabolism to favor effective utilization of available nutrients, often in competition with their hosts. Equally important is the activation or repression of expression of factors allowing pathogens to overcome the action of the various host defense mechanisms. Moreover, when an infection leads to persistent colonization, it is occasionally accompanied by microevolution and selection for variants with mutations in previously environmentally regulated genes, resulting in stable expression of physiological or virulence traits in the host. Mutations can be also selected to limit the production of potentially deleterious factors such as those targeted by the immune defenses. These changes occur without horizontal acquisition of genes, and they are referred to as pathogenicity-adaptive, or pathoadaptive, mutations (1, 2).
Among the most extensively studied examples of bacterial evolution within the infected host are those that arise from adaptive mutations during the chronic respiratory disease of individuals with CF (cystic fibrosis) (3). CF patients are colonized by a number of opportunistic bacterial pathogens, predominantly Pseudomonas aeruginosa, with specific strains coexisting in the respiratory tract for years or even decades. A number of genetic alterations leading to the loss of motility, quorum sensing, and production of lipopolysaccharide O-side chains and type IV pili have been identified in isolates from patients with chronic CF (4–6). The most widely recognized phenotype of P. aeruginosa isolates from CF individuals acquired through pathoadaptive mutations is the overproduction of the alginate exopolysaccharide, giving the colonies their characteristic mucoid appearance. The alginate capsule is a virulence factor protecting P. aeruginosa from killing by phagocytes in the respiratory tract (7, 8). The increased production of alginate in mucoid CF isolates is the result of mutations, most frequently in the gene encoding the anti-sigma factor MucA. In nonmucoid bacteria, MucA functions by limiting alginate production through its inhibitory interaction with the alternative sigma factor σ22 (also named AlgU or AlgT), which directs RNA polymerase to promoters of alginate biosynthetic genes. Loss of MucA inhibition leads to high-level expression of several AlgU-transcribed operons whose products are involved in polymerization, modification, and transport of alginate (9).
We have previously used DNA microarrays to identify genes in a nonmucoid laboratory strain that respond to the presence of human respiratory mucus in the growth medium (10). When the genes were exposed to mucus, an immediate repression of expression of flagellin was detected. The flagellin is not only the major protein component of the bacterial motility apparatus, but this protein is also a strong proinflammatory stimulant. This adaptive response of reducing the expression of a surface molecule involved in pathogen recognition very likely plays a role in the natural environment as well, where invertebrate predators of bacteria possess homologues of human Toll-like receptors (TLRs). It is therefore likely that mucus-induced suppression of flagellin synthesis represents a rapid response to avoidance of immune recognition by the flagellin receptor TLR5. A defect in flagellin synthesis is a common pathoadaptive mutation observed in strains isolated from chronically infected CF patients (11, 12). A number of mutations in various structural, regulatory, or assembly genes account for the loss of flagellar motility in such strains (4).
In the study described here, we built on our previous work on identifying mucus-responsive genes, this time in a stable mucoid, host-adapted CF isolate of P. aeruginosa. Analysis of the transcriptome of this strain provides an opportunity to study the consequences of environmental adaptation and pathoadaptive mutations for gene regulation in a manner not possible with genetically engineered mucoid isolates of common laboratory strains such as PAO1 or PA14. We used the method of deep sequencing-based transcriptome analysis (RNA-seq) to monitor the levels of all transcripts in bacteria grown in the presence and the absence of human respiratory mucus. We uncovered several physiological responses by this mucoid P. aeruginosa strain, including unexpected repression of alginate production in mucus with a concomitant activation of the antibacterial type VI secretion system (T6SS), that do not appear to be controlled by known regulatory mechanisms. Our results point toward a major contribution of the components of respiratory mucus in controlling the expression of virulence factors during chronic infection and thereby playing an important role in bacterial survival and its ecological fitness in the CF respiratory tract.
RESULTS AND DISCUSSION
Transcripome of P. aeruginosa 2192 determined by RNA-seq.
We used an isolate of P. aeruginosa (strain 2192) from a chronically infected CF patient (13) to probe its transcriptome during growth on media containing human respiratory mucus. This strain displayed a highly stable phenotype, with no loss of a mucoid colony appearance following inoculation of a single colony and growth for 24 h at 37°C in minimal (M63; see Materials and Methods) or rich (LB) liquid media and plating ca. 1,000 CFU on the same media solidified with agar (data not shown). Strain 2192 carried in its genome a mutation in the mucA gene with a substitution of a G for a T at base pair 538, replacing an aspartic acid codon with a stop codon. No other obvious mutations in the genome of strain 2192 that could have contributed to the maintenance of the mucoid phenotype were identified (14). To gain new insights into the regulatory networks that control gene expression in a mucoid P. aeruginosa strain adapted to chronic persistence within the CF lung, we used RNA-seq to compare transcriptomes of P. aeruginosa 2192 grown in minimal medium and in the same medium supplemented with pooled respiratory mucus collected from CF patients. The growth kinetics of the culture grown on minimal medium (referred to as the M-0 culture) showed that the mucus supplement (referred to as the M-50 culture) increased the growth rate in the exponential phase and the cell mass reached in the stationary phase only mildly (data not shown). Total RNA was isolated from P. aeruginosa 2192 grown to the late-exponential phase, and, following rRNA depletion, the remaining RNA was reverse transcribed into cDNA, fragmented, and sequenced using Single Molecule Sequencing (SMS) technology and a Helicos sequencing platform.
A total of 35,130,649 (from M-0) and 25,010,555 (from M-50) reads were obtained for each cDNA library (see Table S1A in the supplemental material). Of these, 9,528,142 (from M-0) and 7,081,980 (from M-50) reads mapped uniquely to the genome of P. aeruginosa 2192, totaling 83,044,324 and 59,420,268 bases of sequenced P. aeruginosa 2192 cDNA, respectively. We utilized the rRNA depletion method, and 16S rRNA and 23S rRNA were largely removed, as assessed by the decreases of corresponding peaks in the Bioanalyzer analysis (data not shown). However, a high proportion of reads mapped to rRNA genes corresponding to 73.7% (M-0) and 75.2% (M-50) of the total number of mapped reads. The average numbers of reads per region other than rRNA were 213 (M-0) and 116 (M-50), with a coverage of 99.6% (M-0) and 98.3% (M-50) of the 6,197 coding sequences (CDSs) represented by at least a single read.
Differentially expressed genes in the absence or presence of CF mucus.
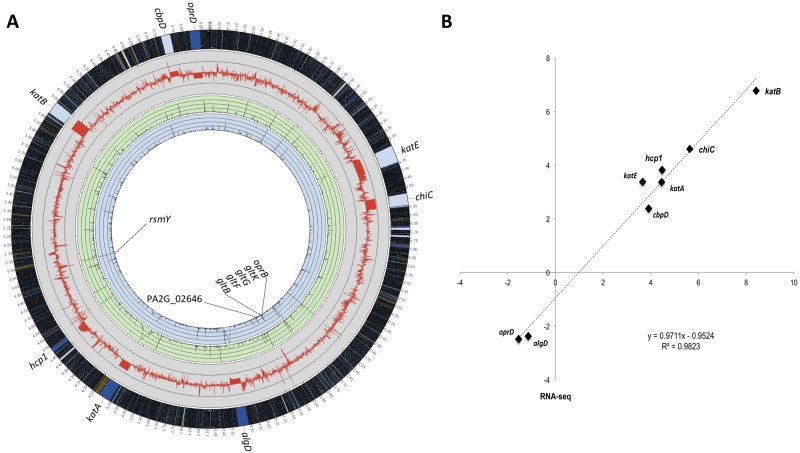
The absolute and relative distributions of reads in two media in the annotated genes of strain 2192 are shown in Fig. 1A (see also Table S1B in the supplemental material). The most highly expressed gene encoded the small regulatory RNA RsmY, accounting for ca. 15% of total reads. The genes PA2G_02646 (encoding a predicted membrane protein) and PA2G_02595 (gltK) were most strongly activated and repressed in mucus, respectively.
FIG 1.
Analysis and validation of RNA-seq experiments. (A) Global analysis of transcript levels in P. aeruginosa 2192 by RNA-seq. M-0 and M-50 refer to minimal media lacking and containing 50% respiratory mucus, respectively. The red line in the gray circle represents the baseline, and thin gray circular lines represent 64-fold changes (or log2 = 6) in expression (i.e., RPKM M-50/M-0 ratio values) of each gene. Green and blue circles correspond to the expression of each gene (represented as RPKM values) in bacteria grown under M-50 and M-0 conditions, respectively. In both of the colored circles, thin circular lines represent an RPKM value of 2,500, with a limit at 10,000. The outermost circle represents the full strain 2192 genome, with a ×50 magnification of the genes for which expression was confirmed by qRT-PCR (see Table S2 in the supplemental material). (B) Validation of RNA-seq results by qRT-PCR on selected genes. Mean log2 ratios of values determined in the qRT-PCR experiments are plotted against the mean log2 ratios of values determined in RNA-seq experiments (see Table S2 in the supplemental material).
To assess the reliability of RNA-seq in determining the relative abundances of individual transcripts in minimal and mucus-containing media, we used the identical total RNA samples and determined by real-time quantitative reverse transcription-PCR (qRT-PCR) the mRNA levels of six upregulated genes (katE, katA, hcp1, katB, chiC, and cbpD) and two downregulated genes (oprD and algD). The ratios of the transcripts from the M-0 and M-50 samples determined by RNA-seq to those obtained by qRT-PCR resulted in an excellent concordance, with a Pearson correlation value of 0.9823 obtained (Fig. 1B; see also Table S2 in the supplemental material). Therefore, RNA-seq appears to be a reliable method for analyzing steady-state levels of mRNA in total RNA samples.
We then organized the genes based on the ranking of normalized mRNA levels and grouped these into four categories: no, low, medium, and high expression (see Fig. S1 in the supplemental material). For the remainder of the article, we use this figure as a reference when commenting on relative transcript level of a specific gene compared to the rest of the transcriptome.
The analysis of RNA-seq data and comparisons identified 656 genes with altered transcript levels, with 378 genes showing an increase and 278 genes a decrease of at least 4-fold (i.e., log2 = 2) in mRNA concentrations in bacteria grown under M-50 conditions compared to M-0 conditions (see Table S3 in the supplemental material). Differentially expressed genes were classified into functional categories according to the PseudoCAP designations, occasionally requiring reannotation in strain 2192, based on information from other sequenced strains. Among these 656 genes, approximately 20% coded for hypothetical proteins of unknown function, equally represented in RNA samples obtained under both sets of conditions (see Fig. S2 and Table S3 in the supplemental material). In the presence of mucus, expression of genes coding for proteins involved in motility/attachment and the protein secretion/export apparatus and related to phage, transposons, or plasmids was induced whereas expression of genes coding for proteins involved in energy metabolism and transport of small molecules as well as genes coding for putative enzymes, secreted factors (toxins, enzymes, alginate), and two-component regulatory systems was repressed.
Repression of alginate production by mucus.
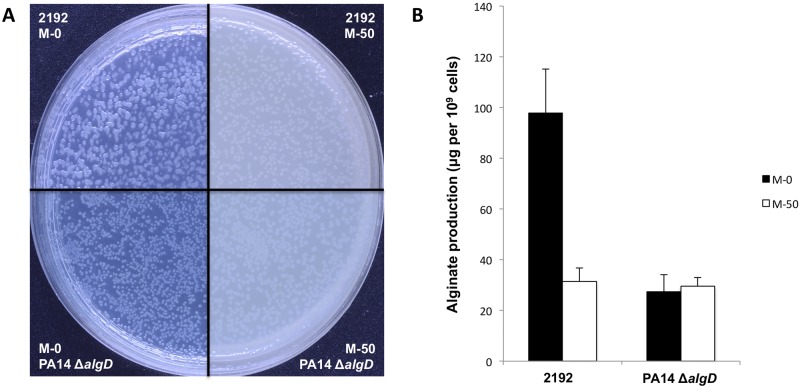
The overexpression of alginate in mucoid P. aeruginosa isolates from CF patients is due to mutations in regulatory genes, primarily in mucA, encoding the antagonist of the alternative sigma factor AlgU (15). Stable derepression of the alginate biosynthetic pathways provides a mechanism for producing copious amounts of this polysaccharide without environmental influences. The results of the RNA-seq analysis showed that the entire group of genes encoding proteins involved in the synthesis, modification, and export of alginate were highly expressed in the mucoid strain 2192 grown in minimal media but that the levels of the corresponding transcripts were reduced in media containing human mucus compared to media lacking this supplement. Specifically, the mRNA levels for each gene within the 12-gene cluster (algD, alg8, alg44, algK, algE, algG, algX, algL, algI, algJ, algF, and algA) were reduced, with fold changes between −4.8 and −9.4 (Fig. 2). No effect of the composition of media was observed for the moderately to highly expressed genes in the alginate regulatory operon (algU, mucABC, and D), and none of the other known regulatory genes (algQ, algP, algZ, algR, and algW) had a significant fold change in expression (see Table S4A in the supplemental material). Interestingly, bacteria grown on agar plates containing 50% mucus appeared less mucoid than those grown on media lacking mucus by visual inspection (Fig. 3A). Moreover, the effect of reduced transcription of genes encoding alginate biosynthetic enzymes was confirmed by quantitative determination of alginate production as measured by the amount of uronic acid produced by cells (Fig. 3B).
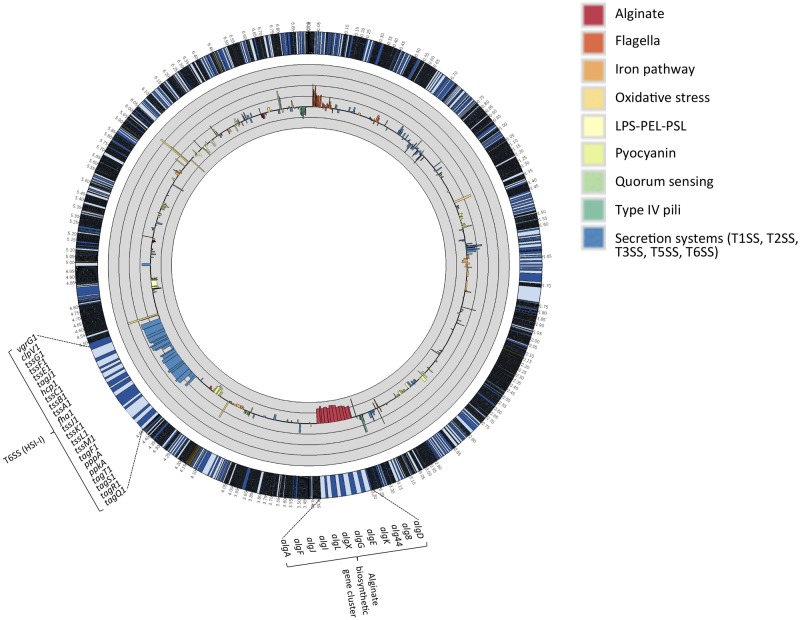
FIG 2.
Differential expression of selected virulence factor genes whose expression is influenced by mucus-containing media. Differences in the levels of expression of virulence genes measured under M-0 and M-50 conditions are presented. Each gene is represented by the log2 of the fold change (i.e., RPKM M-50/M-0 ratio values). The thick black circular line in the gray circle represents the baseline, and thin black circular lines represent 4-fold changes (i.e., a log2 scale). The outermost circle represents the full strain 2192 genome, with the regions corresponding to T6SS–HSI-I and alginate biosynthetic gene cluster magnified ×30.
FIG 3.
Production of alginate in the absence or the presence of mucus. (A) Morphological aspect of colonies of P. aeruginosa 2192 and PA14 ΔalgD (used as a negative control) on medium with no mucus (M-0) or 50% mucus (M-50) after 22 h of aerobic incubation at 37°C. (B) Quantitative estimation of alginate produced for two same strains in the same culture conditions using the uronic acid assay.
The finding that mucus represses alginate production in P. aeruginosa in a strain carrying a stable mutation in mucA, the negative regulator of the AlgU sigma factor, raises the possibility that the expression of this polysaccharide, even in mucoid strains, is controlled by the components of the CF lung by novel mechanisms. The transcription of both the biosynthetic and regulatory genes is initiated by the AlgU-containing RNA polymerase, and yet mucus influences the repression of only the biosynthetic gene cluster and not the operon containing the regulatory genes (15). Therefore, it is unlikely that AlgU functions as a regulatory element playing a role in transducing the signals derived from mucus toward the enhanced transcription of the algD-algA gene cluster. Such regulatory control could include any one of the uncharacterized environmentally responsive transcription factors or utilize posttranscriptional mechanisms that influence mRNA stability, such as small regulatory RNAs. Alginate is a polysaccharide assembled via energy-dependent polymerization processes, and, due to the copious amounts produced in a mucoid bacterium, its production represents a significant energy drain for the cells. The large amount of alginate produced on agar plates in vitro that gives colonies a mucoid appearance may represent an artifact of dysregulated synthesis in the absence of the signal provided by the respiratory mucus in vivo. Our findings suggest that during chronic respiratory tract colonization of CF patients, the levels of this polysaccharide are controlled by mucoid P. aeruginosa to provide amounts sufficient for its protective function during infection without imposing an energetically unsustainable metabolic burden on the bacterium.
Mucus regulates the expression of virulence factors.
The transcript levels of a number of genes specifying virulence factors (catalogued at http://www.mgc.ac.cn/VFs/main.htm) were also altered by growth in mucus (see Table S4A in the supplemental material). Among another large group of genes with increased levels of transcript in mucus-containing media were those encoding the determinants of the type VI secretion system (T6SS) (see Fig. 2; see also Table S4A in the supplemental material). This protein translocation apparatus delivers effectors to mammalian cells and to adjacent bacteria by a contact-dependent mechanism (16–18). The genome of P. aeruginosa encodes three complete T6SSs (HSI-I, HSI-II, and HSI-III), but only the components and not the regulators of the HSI-I gene cluster showed transcription in bacteria grown in respiratory mucus that was enhanced by magnitudes ranging from 1.3- to 14.1-fold (see Table S4A in the supplemental material). The levels of transcripts were not evenly distributed within the gene cluster, ranging from low to medium, with those for tssA1, tagQ1, tssB1, and hcp1 expressed at an elevated level in mucus-containing media. Interestingly, the segments of mRNAs corresponding to 5′ untranslated regions of a number of genes within the HSI-I cluster were present in an unusually high abundance in mucus-containing media (see Fig. S3 in the supplemental material). Mucus-induced expression of one of the T6SS proteins, Hcp1, was confirmed by Western immunoblotting (see Fig. S4A in the supplemental material).
The main function of the HSI-I T6SS identified to date is bactericidal activity directed at other microorganisms following the translocation of its three secreted substrates, TseI, TseII, and TseIII, into the target bacterial cells (19). Increased expression of a killing mechanism may provide a competitive advantage to P. aeruginosa against other microorganisms that cohabitate the CF respiratory tract (20–24). The HSI-I T6SS locus, together with other genes involved in biofilm formation, is controlled by the GacS/GacA two-component system (and its antagonist RetS), via a regulatory mechanism involving two small regulatory RNAs, RsmZ and RsmY, and its cognate RNA-binding protein, RsmA (25, 26). The availability of the comprehensive transcriptome of strain 2192 allowed us to compare the levels of expression of the genes within the HSI-I locus and to compare those to the levels of expression of the genes within the GacS/GacA regulon. In a previous study (27), the most significant GacS/GacA-dependent regulation coordinated with HSI-I was the activation of genes encoding the Pel and Psl polysaccharide. RNA-seq analysis of P. aeruginosa 2192 clearly showed that the biosynthetic genes for the Psl (PA2G_01334 to PA2G_01348) were not activated by mucus and that the mRNA levels of the majority of the genes were unaffected, with the exception of pslN, whose expression was activated 3.3-fold (see Table S4A in the supplemental material). The transcripts for genes for the Pel polysaccharide (PA2G_02472 to PA2G_02478) showed similar fluctuations, ranging from 2.2-fold activation to −2.4-fold repression (see Table S4A in the supplemental material). Similarly, the expression of the genes for cyanide production (hcnA, hcnB, hcnC) is GacS/GacA dependent (26) and yet their mRNA levels show virtually no effect of mucus in the medium (see Table S4A in the supplemental material). Another group of genes repressed by the GacA/GacS pathway are the genes specifying the type III secretion system. However, the results of RNA-seq analyses of P. aeruginosa 2192 showed that the transcript levels of these 36 genes (regulators, components of the secretion machinery, and effectors) were not significantly regulated by mucus, with individual genes both modestly activated or repressed (see Table S4A in the supplemental material). Genes approaching significance in terms of mRNA levels were pscP, exsB, and pscL, which were repressed −3.8, −3.3, and −4.9-fold, respectively, in cells grown in mucus. The levels of exoS, exoT, and exoY were also not significantly influenced by the presence of mucus in the medium. We therefore conclude that the signals provided by the mucus in regulating the genes for the HSI-1 T6SS do not utilize the GacS/GacA pathway.
Genes for two functionally related proteins, chitinase- and chitin-binding protein (ChiC and CbpD, respectively), were also upregulated in P. aeruginosa 2192 grown in the presence of human respiratory mucus, increasing from medium- to high-level expression (24.3-fold for chiC and 5.2-fold for cbpD) (see Table S3A in the supplemental material). The increase of chitinolytic activity was phenotypically confirmed by using by a chitinase assay (see Fig. S4B in the supplemental material). Chitinases catalyze the hydrolysis of chitin, a linear polymer of N-acetylglucosamine residues linked in β-1,4 glycosidic bonds. Several microbial chitinases have been described previously and are believed to function primarily in nutrient acquisition in their natural habitats, since mammalian tissues lack chitin. However, chitinase-deficient mutants of Listeria monocytogenes are attenuated in virulence and are unable to spread into mouse liver (28), while an analogous mutant of Legionella pneumophila has a reduced ability to persist in the mouse lung (29). Bacterial chitin-binding proteins, often coregulated with chitinases, serve often as bacterial adhesins (30). To date, there is no evidence that P. aeruginosa Chic and CbpD proteins have a function in P. aeruginosa infections, and high concentrations of chiC and cbpD mRNA in cells grown in the presence of mucus may simply reflect the composition of the media, where a polysaccharide mimic of chitin may induce expression of these genes.
Mucus activates genes involved in protection of bacteria against oxidative stress.
Chronic colonizers of the CF respiratory tract are contiguously exposed to oxidative stress created by the action of influxing of neutrophils. When experiencing oxidative stress, P. aeruginosa activates the expression of a group of genes via the transcriptional regulator OxyR, which responds to signals that include reactive oxygen species. The regulon controlled by OxyR consists of a number of oxidative stress-responsive genes encoding two catalases (KatA and KatB), alkyl hydroperoxide reductase B (AhpB), alkyl hydroperoxide reductase CF (AhpCF), and an ankrin-like protein (AnkB) (31, 32). Another putative catalase, KatE, is encoded in the genome of P. aeruginosa, but its role in protecting the cells against oxidative stress is not known. Transcripts for all of these OxyR-regulated genes were expressed at medium levels in minimal medium lacking mucus, but their expression was induced from 10- to 100-fold by growth in mucus-containing media (see Table S4A in the supplemental material), indicating that mucus directly or indirectly controls the activities of this global transcriptional regulator. Alternatively, pro-oxidants released by neutrophils could indirectly control the expression of a number of genes involved in the observed oxidative stress response. The increase of catalase activity was phenotypically confirmed by using a catalase assay (see Fig. S4C in the supplemental material).
Mucus influences several metabolic pathways in P. aeruginosa 2192.
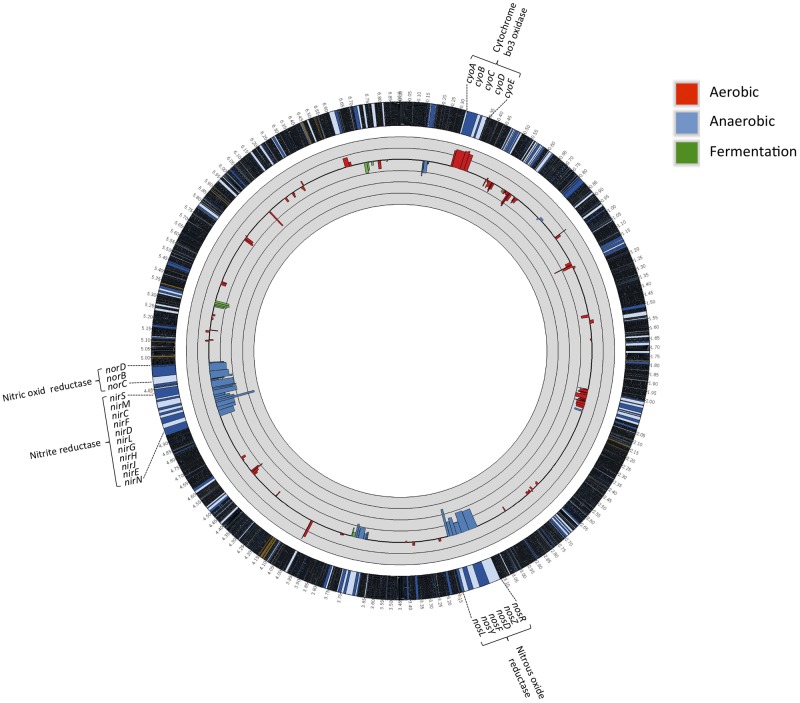
The ability of P. aeruginosa to occupy diverse niches, including the human respiratory tract, requires it to be able to carry out highly versatile energy metabolism (33). Its branched respiratory chain terminates with multiple terminal oxidases and denitrification enzymes. To support aerobic respiration, five terminal oxidases can be expressed in P. aeruginosa. Denitrification involves conversion of nitrate to molecular nitrogen, allowing P. aeruginosa to grow anaerobically. In addition to the denitrification, arginine catabolism can provide cellular ATP to P. aeruginosa growing anaerobically. Expression of genes involved in catabolism, as assessed by RNA-seq analysis, provides an insight into the pathways favored by P. aeruginosa for optimal energy generation within the milieu of respiratory secretions and specific physical conditions. Strong induction of the cyoA-cyoE operon was observed in P. aeruginosa growing on mucus (Fig. 4; see also Table S4B in the supplemental material). This operon encodes one of the two quinol oxidases, an enzyme of relatively low affinity for oxygen, indicating enhanced access of these cells to oxygen. Interestingly, P. aeruginosa can express two operons encoding Cbb3 oxidase type 1 (Cbb3-1) and Cbb3-2, and while the former was expressed in both media, expression of the gene cluster for Cbb3-1 (ccoN2-ccoO-ccoQ-ccoP2) was repressed (2.6- to 6.2-fold) in the media containing respiratory mucus (see Table S1B and Table S4B in the supplemental material). It is likely that the different terminal oxidases function under unique sets of environmental conditions, and it is apparent that mucus provides a signal for activation of one (Cyo) and repression of another (Cbb3-2). Multiple terminal oxidases of such different characteristics and the differential use of them under different conditions must contribute to adjust its metabolism for optimal energy generation.
FIG 4.
Differential expression of respiration genes in P. aeruginosa 2192 in M-0 and M-50 media. Each gene is represented by the log2 of the fold change (i.e., RPKM M-50/M-0 ratio values). The thick black circular line in the gray circle represents the baseline, and thin black circular lines represent 4-fold changes (i.e., a log2 scale). The outermost circle represents the full strain 2192 genome, with a ×30 magnification of the regions corresponding to cyo, nir, nor, and nos gene clusters.
Denitrification consists of four sequential steps catalyzed by specific metalloenzymes: reduction of nitrate to nitrite by nitrate reductase (NarGHI), NirS-catalyzed reduction of nitrite to nitric oxide (NO), and reduction of NO to N2O catalyzed by NO reductase NorCB, terminating with the conversion of N2O to N2 by nitrous oxide reductase NosZ. Our RNA-seq data show a substantial reduction in transcript levels in mucus-grown P. aeruginosa for genes encoding enzymes involved in denitrification and clustered in the nir, nos, and nor operons (Fig. 4; see also Table S4B in the supplemental material). Moreover, the other anaerobic energy-generating pathways involving arginine catabolism were also repressed in media containing mucus. These pathways include the genes for the arginine/ornithine antiporter (arcD), arginine deaminase (arcA), catabolic ornithine carboyltransferase (arcB), and carbamate kinase (arcC) (see Table S4B in the supplemental material). Total RNA used in the RNA-seq analysis was isolated from the bacteria grown without any specific attempts to create an atmosphere of reduced oxygen, and P. aeruginosa in the respiratory tract has been reported to very likely grow under anaerobic or microaerophilic conditions (34, 35). Therefore, we cannot exclude the possibility that the conditions used in this work do not exactly reflect those found in the CF respiratory tract. Moreover, the utilization of specific energy-generating pathways may be influenced by other factors. For example, overproduction of the alginate in mucoid strains may create anaerobic conditions for individual cells, as it has been shown that the restriction of oxygen diffusion imposed by alginate is not confined to mucoid isolates but that the same effect is seen when alginate is added to media on an isogenic nonmucoid revertant, leading to an increase of the expression of denitrification genes (36). However, as shown above, growth in mucus has a repressive effect on the expression of alginate biosynthetic genes, which can lead to an increase in access of oxygen to cells and a reduction in the expression of genes whose products are involved in anaerobic energy generation.
Examination of the mRNA concentrations in cells grown in minimal medium and medium supplemented with mucus revealed unexpected patterns of expression of several metabolic genes, indicating that host adaption selected for stable genetic changes affecting signal transduction and regulation of expression of regulatory or signal transduction pathways. For example, we have observed unexpected repression of several metabolic genes involved in degradation of unusual natural products (see Table S3B in the supplemental material). Specifically, the dit genes that were previously identified within the large 112-kb genomic island have been found among P. aeruginosa isolates only in strain PA2192 (14). These genes encode enzymes of abietane diterpenoid metabolism in Burkholderia xenovorans, and they were shown to be positively regulated when bacteria were grown in minimal medium supplemented with dehydroabietic acid (37). Since the compounds have so far been identified only in plants and wood pulp, it is unclear why the genes whose products are involved in their catabolism are even expressed in minimal medium and repressed in the same medium containing 50% respiratory mucus.
Similarly, we detected high-level expression of the glcDEFG operon, encoding the glyoxylate oxidase subunits GlcDEF and GlcG, a protein of unknown function, in nominal medium and its repression (from −69.6- to −27.8-fold change) in medium with the mucus supplement (see Table S3B in the supplemental material). The unlinked glcB gene (PA2G_04399) was highly expressed in both types of media (see Table S1B in the supplemental material). In Escherichia coli, the glc operon is regulated by the GlcC transcriptional activator located upstream of the glcDEFG operon and is induced by growth on glycolate. The glyoxylate oxidase catalyzes the formation of glyoxylate from glycolate, and malate synthase (GlcB) converts glyoxylate to malate (38). The basis of mucus repression of the genes encoding the dit and glc genes remains unclear.
We detected an interesting example of environmental adaptation of P. aeruginosa 2192 in the regulation of the pathways for utilization of acyclic monoterpenes. A limited number of bacterial species, including P. aeruginosa, carry the atuR-atuABCDEFGH gene cluster, encoding the regulator and the enzymes for utilization of the acyclic terpene citronellol family as the sole carbon source (39, 40). The pathway feeds into the leucine/isovalerate utilization pathway (referred to as either liuR-ABCDE or gnyR-DBHAL), leading to the formation of acetyl-coenzyme A (CoA). The expression of atu genes is regulated, and they are expressed only in the presence of acyclic monopteroids and not in the presence of glucose or leucine/isovalerate substrates of the Liu pathway (41). AtuR is a repressor, binding as a dimer to two 13-bp inverted-repeat sequences in the atuA promoter region (42). In our RNA-seq analysis, several of the genes involved in the metabolism of acyclic terpenes, atuBCDEFG, showed higher (between 2.2- and 8.7-fold) expression in minimal medium than in the same medium containing human respiratory mucus (see Table S1B and Table S3B in the supplemental material). The mRNA level for the AtuR repressor gene was not influenced by mucus; neither was the level of atuA and atuH transcripts. In contrast, the liu (gny) genes were expressed poorly in minimal medium but were activated by mucus (see Table S3A in the supplemental material). An increase in the expression of the liu genes induced by leucine or isovalerate very likely present in mucus and simultaneous repression of atu genes could explain why the levels of these two gene clusters were reciprocal in cells grown in the presence and absence of mucus. However, the basis for high-level expression of the atu genes in media containing glucose and lacking the substrate inducer is less clear. The protein sequences of the 198-amino-acid AtuR proteins in sequenced P. aeruginosa genomes are nearly identical, with a single difference in the 2192 sequence (D197N) from that of PAO1. The 13-bp inverted DNA sequences of the operator regions in PAO1 and 2192 are also identical. However, three nucleotide substitutions can be found in the 40-bp regions separating these inverted repeats. Although these differences could account for the constitutive expression of the atuABCDEFGH cluster in the absence of substrate, the genes are still repressed in mucus, suggesting that additional regulatory mechanisms may control their expression in strain 2192 and possibly other strains adapted to chronic persistence in the CF respiratory tract.
Regulation of the genes of flagellar motility.
We have previously reported that in PAK, a nonmucoid P. aeruginosa strain, the transcription of the flagellin gene (fliC) is repressed in the presence of mucus whereas the rest of the apparatus of flagellar biogenesis is unaffected (10). This effect likely involves recognition of the host environment by the bacterial pathogen, leading to repression of expression of the flagellin, a major proinflammatory ligand of P. aeruginosa, recognized by the TLR5 receptor (43). In strain 2192, we observed a significant increase in transcript levels of the genes involved in the assembly of flagella, while expression of its fliC gene was activated 1.9-fold (see Table S1B and Table S4A in the supplemental material). It is difficult to interpret these results in light of the observation that this strain is nonmotile and lacks assembled flagella. One possible explanation is that the shutoff of flagellin synthesis in a nonmucoid strain requires the presence of the assembled hook (FlgE) (44), which is not present in the absence of a flagellum. Thus, the flagellar shutoff in nonmucoid strains is essential for avoiding early recognition by the innate host defenses, and this regulatory mechanism is eliminated in this strain, which is adapted to survival in the chronically infected hosts. The genetic basis of this observation was not apparent from the analysis of the sequence polymorphisms present in the sequences of the structural and regulatory flagellar loci of strain 2192 (14).
Detection of sRNAs.
The strand-specific sequencing platform of the Helicos technology can be used to identify small RNAs (sRNAs). Thirty-four candidate sRNAs, including 10 novel small RNAs, were identified in strain 2192 (see Table S5 in the supplemental material) (45). Of these, two were differentially expressed under the conditions used here. One (PA2G_03487.1) was highly expressed in minimal medium, but expression was dramatically repressed in the presence of mucus (fold change, −117.3), while expression of the second (PA2G_05393.1) was significantly induced in the presence of mucus (fold change, 8.0) (see Table S5 in the supplemental material). The existence of both of these RNA species was validated by Northern blot analysis (not shown). Therefore, it is conceivable that these two sRNAs have a regulatory function in P. aeruginosa in the CF mucus environment. Their mRNA targets are currently being investigated using RNA-seq of wild-type P. aeruginosa 2192 and its mutants lacking the sRNA genes.
In conclusion, we used RNA-seq to identify genes in a mucoid CF isolate of P. aeruginosa whose transcript levels are influenced by growth in human respiratory mucus. We uncovered unexpected changes in the expression of genes involved in immune avoidance (alginate), survival under oxidative stress (catalases), and bacterial niche competition (the T6SS) and in a number of metabolic functions. In certain instances, the control of expression of these genes appeared to function without utilization of previously described regulatory mechanisms. Our findings suggest the existence of signal transduction and regulatory pathways in P. aeruginosa, and likely in other human pathogens, that can be detected only when bacteria are exposed to specialized environments that closely mimic the contents of human tissues. Application of molecular techniques should provide a wealth of information about novel mechanisms that control bacterial adaptation to host environments during infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
For RNA-seq, P. aeruginosa 2192 (13) was grown at 37°C in two different media. Expectorated sputum (5 to 10 ml) (referred to here as mucus) from 25 CF patients was retrieved from the Hospital Microbiology Laboratory at the University of Florida Medical Center and kept frozen at −70°C until needed. This material was used with supplemented 50% (vol/vol) or not solid (1.5% agar) M63 minimal medium [15 mM (NH4)2SO4, 22 mM KH2PO4, 40 mM K2HPO4, 1 mM MgSO4, 25 µM FeCl2, 10 mM d-glucose, 9 mM l-lactic acid]. Prior to use, mucus was sterilized by UV irradiation for 30 min in Stratagene Stratalinker 1800 (Agilent Genomics). Some mutants from the P. aeruginosa PA14 transposon insertion mutant library (46) were used for confirmatory phenotypic assays.
Oligonucleotides, PCR, and DNA sequencing.
All synthesized oligonucleotides were obtained from Invitrogen (see Table S6 in the supplemental material), and confirmatory DNA sequencing was performed at Genewiz Inc. (Cambridge, MA). PCR techniques were performed in accordance with standard protocols, and DNA fragments were purified with a QIAquick PCR purification or QIAquick gel extraction kit (Qiagen).
RNA isolation.
Bacteria were cultured (for 22 h at 37°C) to the late-exponential-growth phase on solid M63 agar plates, and total RNA was extracted using the hot phenol method. Residual chromosomal DNA was removed by treating samples with a Turbo DNA-free kit (Ambion). DNase-treated RNA samples were quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific), and the integrity of the results (RNA integrity number [RIN] > 8) was assessed using an Agilent 2100 bioanalyzer.
For RNA-seq analysis, a MICROBExpress kit (Ambion) was used according to the manufacturer’s recommendations to remove the 23S and 16S rRNA from the total RNA samples. To evaluate the degree of rRNA depletion, the samples were analyzed using an Agilent 2100 bioanalyzer.
Quantitative RT-PCR.
qRT-PCR experiments were performed using a Kapa SYBR Fast One-Step qRT-PCR kit (Kapa Biosystems) and an Eppendorf Mastercycler ep realplex instrument (Eppendorf) with the following program: 42°C for 5 min, 95°C for 5 min, and 40 cycles of 95°C for 3 s and 60°C for 20 s. Transcript levels in each sample (without rRNA depletion) were determined by absolute quantification using serial dilutions of PCR products (47). Expression of the rpsL gene as a housekeeping control gene was measured. Each experiment was performed in duplicate.
cDNA synthesis and preparation of the library for high-throughput sequencing.
cDNA was prepared and modified for Helicos sequencing as previously described (48). Briefly, first-strand cDNA was synthesized from 800 ng of RNA, using 1.2 µg of random hexamer primers and 1,000 U of Superscript III reverse transcriptase (Invitrogen). RNA was removed using RNase H and RNase A, and cDNA was sheared at 4°C by sonication using a Misonix 4000 sonicator (Qsonica) with the amplitude set at 60% for 20 min (20 s on/off pulses), generating fragments of 60 to 200 nt. Sheared cDNA was treated with terminal transferase (NEB) and dATP, generating the 100-to-200-nt 3′ poly(A) tail that is necessary for Helicos sequencing. The samples were purified with a MinElute kit (Qiagen) and then quantified spectrophotometrically using a NanoDrop 1000 spectrophotometer (Thermo Scientific). The samples were sequenced using a HeliScope single-molecule sequencer (Helicos Biosciences) at the Molecular Biology Core Facilities of the Dana-Farber Cancer Institute (Boston, MA).
RNA-seq data analysis.
The poly(T) tail of the raw reads was removed by filtering using the Helisphere program (Helicos Biosciences) and then mapped against the genomic sequence of P. aeruginosa 2192 (GenBank accession no. CH482384.1) using CLC Genomics Workbench software v5.1 (CLC bio). The number of reads uniquely aligned to each genomic position (coding sequence, putative small RNA) was determined using BEDTools software v2.16.2 (49). Since the genome sequence of P. aeruginosa 2192 is incomplete, the number of reads aligning to rRNA regions and some sRNAs (e.g., Prrf1 and Prrf2) was evaluated by mapping reads against the genomic sequence of P. aeruginosa PAO1 (GenBank accession no. AE004091.2). To normalize the expression of genes in different RNA-seq samples, values corresponding to the number of reads per kilobase per million mapped (RPKM) were calculated as follows: (number of reads for the gene × 109)/(total number of reads × size of the gene). Differentially expressed genes were identified using a log2 absolute fold change value greater than 2. Products of differentially expressed genes were classified by functional category according to PseudoCAP function classes (http://www.pseudomonas.com/). RKPM values for each gene were plotted and visualized as a circle using the Circos program (50). The sequence coverage per base was plotted and visualized using the genome browser Artemis v13.0 (51). Identification of putative small RNAs was performed using a program created in-house, and the results were compared to the list generated previously (45).
Uronic acid assay.
Alginate production was quantified as previously described (52). Briefly, P. aeruginosa 2192 cells grown on M63 minimal medium plates for 22 h at 37°C were collected and washed twice in phosphate-buffered saline (PBS). The alginate in the supernatants was precipitated with 1 vol of ice-cold isopropanol. After centrifugation, the pellet was resuspended in water and treated with DNase I and RNase A for 1 h at 37°C followed by proteinase K digestion for an additional hour at 37°C. The uronic acid concentrations were determined by the uronic acid assay (53). Uronic acid concentrations were determined using a standard curve with brown alga alginate (Sigma). The amount of alginate in each sample was normalized to the number of cells per sample. Each experiment was performed in duplicate.
Western blot analysis.
Production of Hcp1 protein was evaluated by Western blot analysis as previously described using specific anti-Hcp1 antibodies (54). P. aeruginosa 2192 cells were grown on M63 plates for 20 h, 22 h, and 30 h at 37°C. Total proteins were extracted using BugBuster protein extraction reagent (Novagen) according to the manufacturer’s recommendations, and 1 µg of proteins was loaded per lane. P. aeruginosa PA14 Δhcp1 was used as a negative control. Each experiment was performed in duplicate.
Chitinase and catalase assays.
The chitinase activity was determined using a chitinase assay kit, fluorometric (Sigma), whereas the catalase activity was estimated using an Amplex Red catalase assay kit (Invitrogen). For both assays, P. aeruginosa 2192 cells were grown on M63 plates for 22 h at 37°C, and total proteins were extracted using BugBuster protein extraction reagent (Novagen) according to the manufacturer’s recommendations. PA14 ΔchiC and PA14 ΔkatA were used as negative controls in the chitinase and catalase assays, respectively. Both activities in each sample were normalized to the number of cells per sample. Each experiment was performed in duplicate.
SUPPLEMENTAL MATERIAL
Distribution of the number of genes according to the RKPM values in the absence (condition M-0) or the presence (condition M-50) of CF (cystic fibrosis) mucus. The expression of the genes was categorized as follows: not expressed (RPKM = 0); low expression (RPKM = >0 to 10); medium expression (RPKM = >10 to 500); high expression (RPKM > 500). See Materials and Methods for the definition of RPKM. Download Figure S1, TIF file, 0.4 MB.
PseudoCAP functional categories of the 656 genes induced or repressed by 50% mucus. Percentages of induced and repressed genes with a change in expression level greater than 2 log2-fold are represented in black and white bars, respectively. Statistically significant differences (using the Fisher exact test) are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Download Figure S2, TIF file, 0.5 MB.
Expression of genes of the HSI-I locus. RNA-seq data were visualized using Artemis software (51). Gene coordinates of P. aeruginosa 2192 are indicated horizontally. The base count, indicated vertically, is representative of the number of times each base was mapped by a single read. Green and red lines represent base reads under conditions without mucus (M-0) and with 50% mucus (M-50), respectively. Predicted transcriptional start sites are also reported. Download Figure S3, TIF file, 0.5 MB.
Confirmation of RNA-seq data by phenotypic approaches. (A) Production of Hcp-1 in the absence or the presence of mucus. Samples were taken at different time points: 20 h (mid-exponential phase), 22 h (late exponential phase), and 30 h (stationary phase). Western immunoblot analysis was performed by using polyclonal anti-Hcp1 antibodies and 1 µg total protein extract per lane. The PA14 Δhcp1 strain served as a negative control. (B) Chitinase activity in the absence or the presence of mucus. PA14 ΔchiC was used as a negative control. (C) Catalase activity in the absence or the presence of mucus. PA14 ΔkatA was used as a negative control. Download Figure S4, TIF file, 0.5 MB.
Global and detailed data of RNA-seq experiments. (a) Summary of coverage from RNA-seq. (b) Total reads from RNA-seq and relative ratios of transcript levels.
qRT-PCR-validation of selected genes.
Genes differentially expressed under M-0 and M-50 conditions (i.e., fold change ≥ 4). (a) Genes whose transcript levels increased during growth in mucus compared to minimal media. (b) Genes whose transcript levels decreased during growth in mucus compared to minimal media.
Effect of mucus on selected genes. (a) Putative virulence genes. The list of virulence genes was compared to those listed on http://www.mgc.ac.cn/VFs/main.htm. (b) Genes involved in respiration.
Candidate small RNAs found in P. aeruginosa 2192.
Primers used for qRT-PCR.
ACKNOWLEDGMENTS
We warmly thank Bryan W. Davies and William P. Robins for their useful help and Marek Basler for providing us anti-Hcp antibodies.
The work in G.N.’s laboratory was partially supported by FDOH grant 09KW-10. R.R. and J.J. were supported by the NIH (grant 1R01AI078770-01). Work in S.L.’s laboratory was supported by grant R37 AI021451 from the NIH.
Footnotes
Citation Cattoir V, et al. 2012. Transcriptional response of mucoid Pseudomonas aeruginosa to human respiratory mucus. mBio 3(6):e00410-12. doi:10.1128/mBio.00410-12.
REFERENCES
- 1. Sokurenko EV, Hasty DL, Dykhuizen DE. 1999. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 7:191–195 [DOI] [PubMed] [Google Scholar]
- 2. Maurelli AT. 2007. Black holes, antivirulence genes, and gene inactivation in the evolution of bacterial pathogens. FEMS Microbiol. Lett. 267:1–8 [DOI] [PubMed] [Google Scholar]
- 3. Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith EE, et al. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cramer N, Wiehlmann L, Tümmler B. 2010. Clonal epidemiology of Pseudomonas aeruginosa in cystic fibrosis. Int. J. Med. Microbiol. 300:526–533 [DOI] [PubMed] [Google Scholar]
- 6. Hogardt M, Heesemann J. 2010. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int. J. Med. Microbiol. 300:557–562 [DOI] [PubMed] [Google Scholar]
- 7. Schwarzmann S, Boring JR. 1971. Antiphagocytic effect of slime from a mucoid strain of Pseudomonas aeruginosa. Infect. Immun. 3:762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leid JG, et al. 2005. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 175:7512–7518 [DOI] [PubMed] [Google Scholar]
- 9. Jain S, Ohman DE. 2004. Alginate biosynthesis, p 53–81 In Ramos JL, Pseudomonas, vol 3: biosynthesis of macromolecules and molecular metabolism. Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- 10. Wolfgang MC, Jyot J, Goodman AL, Ramphal R, Lory S. 2004. Pseudomonas aeruginosa regulates flagellin expression as part of a global response to airway fluid from cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 101:6664–6668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luzar MA, Thomassen MJ, Montie TC. 1985. Flagella and motility alterations in Pseudomonas aeruginosa strains from patients with cystic fibrosis: relationship to patient clinical condition. Infect. Immun. 50:577–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mahenthiralingam E, Campbell ME, Speert DP. 1994. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect. Immun. 62:596–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pier GB, Matthews WJ, Jr, Eardley DD. 1983. Immunochemical characterization of the mucoid exopolysaccharide of Pseudomonas aeruginosa. J. Infect. Dis. 147:494–503 [DOI] [PubMed] [Google Scholar]
- 14. Mathee K, et al. 2008. Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. U. S. A. 105:3100–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ramsey DM, Wozniak DJ. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56:309–322 [DOI] [PubMed] [Google Scholar]
- 16. Filloux A, Hachani A, Bleves S. 2008. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154:1570–1583 [DOI] [PubMed] [Google Scholar]
- 17. Pukatzki S, McAuley SB, Miyata ST. 2009. The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12:11–17 [DOI] [PubMed] [Google Scholar]
- 18. Silverman JM, Brunet YR, Cascales E, Mougous JD. 2012. Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 66:453–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Russell AB, et al. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harris JK, et al. 2007. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc. Natl. Acad. Sci. U. S. A. 104:20529–20533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sibley CD, et al. 2008. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 105:15070–15075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klepac-Ceraj V, et al. 2010. Relationship between cystic fibrosis respiratory tract bacterial communities and age, genotype, antibiotics and Pseudomonas aeruginosa. Environ. Microbiol. 12:1293–1303 [DOI] [PubMed] [Google Scholar]
- 23. Guss AM, et al. 2011. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J. 5:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Madan JC, et al. 2012. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. mBio 3:e00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mougous JD, et al. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brencic A, et al. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73:434–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goodman AL, et al. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745–754 [DOI] [PubMed] [Google Scholar]
- 28. Chaudhuri S, et al. 2010. Contribution of chitinases to Listeria monocytogenes pathogenesis. Appl. Environ. Microbiol. 76:7302–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DebRoy S, Dao J, Söderberg M, Rossier O, Cianciotto NP. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc. Natl. Acad. Sci. U. S. A. 103:19146–19151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jude BA, Martinez RM, Skorupski K, Taylor RK. 2009. Levels of the secreted Vibrio cholerae attachment factor GbpA are modulated by quorum-sensing-induced proteolysis. J. Bacteriol. 191:6911–6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heo YJ, et al. 2010. The major catalase gene (katA) of Pseudomonas aeruginosa PA14 is under both positive and negative control of the global transactivator OxyR in response to hydrogen peroxide. J. Bacteriol. 192:381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ochsner UA, Vasil ML, Alsabbagh E, Parvatiyar K, Hassett DJ. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182:4533–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arai H. 2011. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front. Microbiol. 2:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alvarez-Ortega C, Harwood CS. 2007. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65:153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Worlitzsch D, et al. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hassett DJ. 1996. Anaerobic production of alginate by Pseudomonas aeruginosa: alginate restricts diffusion of oxygen. J. Bacteriol. 178:7322–7325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smith DJ, Park J, Tiedje JM, Mohn WW. 2007. A large gene cluster in Burkholderia xenovorans encoding abietane diterpenoid catabolism. J. Bacteriol. 189:6195–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pellicer MT, Badía J, Aguilar J, Baldomà L. 1996. glc locus of Escherichia coli: characterization of genes encoding the subunits of glycolate oxidase and the glc regulator protein. J. Bacteriol. 178:2051–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Díaz-Pérez AL, Zavala-Hernández AN, Cervantes C, Campos-García J. 2004. The gnyRDBHAL cluster is involved in acyclic isoprenoid degradation in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 70:5102–5110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Höschle B, Gnau V, Jendrossek D. 2005. Methylcrotonyl-CoA and geranyl-CoA carboxylases are involved in leucine/isovalerate utilization (Liu) and acyclic terpene utilization (Atu), and are encoded by liuB/liuD and atuC/atuF, in Pseudomonas aeruginosa. Microbiology 151:3649–3656 [DOI] [PubMed] [Google Scholar]
- 41. Förster-Fromme K, et al. 2006. Identification of genes and proteins necessary for catabolism of acyclic terpenes and leucine/isovalerate in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 72:4819–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Förster-Fromme K, Jendrossek D. 2010. AtuR is a repressor of acyclic terpene utilization (Atu) gene cluster expression and specifically binds to two 13 bp inverted repeat sequences of the atuA-atuR intergenic region. FEMS Microbiol. Lett. 308:166–174 [DOI] [PubMed] [Google Scholar]
- 43. Ramphal R, et al. 2008. Control of Pseudomonas aeruginosa in the lung requires the recognition of either lipopolysaccharide or flagellin. J. Immunol. 181:586–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jyot J, Sonawane A, Wu W, Ramphal R. 2007. Genetic mechanisms involved in the repression of flagellar assembly by Pseudomonas aeruginosa in human mucus. Mol. Microbiol. 63:1026–1038 [DOI] [PubMed] [Google Scholar]
- 45. Wurtzel O, et al. 2012. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown at body temperature. PLoS Pathog. 8:e1002945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liberati NT, et al. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yoder-Himes DR, et al. 2009. Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc. Natl. Acad. Sci. U. S. A. 106:3976–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mandlik A, et al. 2011. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10:165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quinlan AR, Hall IM. 2010. Bedtools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rutherford K, et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 52. Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65:876–895 [DOI] [PubMed] [Google Scholar]
- 53. Knutson CA, Jeanes A. 1968. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal. Biochem. 24:470–481 [DOI] [PubMed] [Google Scholar]
- 54. Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of the number of genes according to the RKPM values in the absence (condition M-0) or the presence (condition M-50) of CF (cystic fibrosis) mucus. The expression of the genes was categorized as follows: not expressed (RPKM = 0); low expression (RPKM = >0 to 10); medium expression (RPKM = >10 to 500); high expression (RPKM > 500). See Materials and Methods for the definition of RPKM. Download Figure S1, TIF file, 0.4 MB.
PseudoCAP functional categories of the 656 genes induced or repressed by 50% mucus. Percentages of induced and repressed genes with a change in expression level greater than 2 log2-fold are represented in black and white bars, respectively. Statistically significant differences (using the Fisher exact test) are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Download Figure S2, TIF file, 0.5 MB.
Expression of genes of the HSI-I locus. RNA-seq data were visualized using Artemis software (51). Gene coordinates of P. aeruginosa 2192 are indicated horizontally. The base count, indicated vertically, is representative of the number of times each base was mapped by a single read. Green and red lines represent base reads under conditions without mucus (M-0) and with 50% mucus (M-50), respectively. Predicted transcriptional start sites are also reported. Download Figure S3, TIF file, 0.5 MB.
Confirmation of RNA-seq data by phenotypic approaches. (A) Production of Hcp-1 in the absence or the presence of mucus. Samples were taken at different time points: 20 h (mid-exponential phase), 22 h (late exponential phase), and 30 h (stationary phase). Western immunoblot analysis was performed by using polyclonal anti-Hcp1 antibodies and 1 µg total protein extract per lane. The PA14 Δhcp1 strain served as a negative control. (B) Chitinase activity in the absence or the presence of mucus. PA14 ΔchiC was used as a negative control. (C) Catalase activity in the absence or the presence of mucus. PA14 ΔkatA was used as a negative control. Download Figure S4, TIF file, 0.5 MB.
Global and detailed data of RNA-seq experiments. (a) Summary of coverage from RNA-seq. (b) Total reads from RNA-seq and relative ratios of transcript levels.
qRT-PCR-validation of selected genes.
Genes differentially expressed under M-0 and M-50 conditions (i.e., fold change ≥ 4). (a) Genes whose transcript levels increased during growth in mucus compared to minimal media. (b) Genes whose transcript levels decreased during growth in mucus compared to minimal media.
Effect of mucus on selected genes. (a) Putative virulence genes. The list of virulence genes was compared to those listed on http://www.mgc.ac.cn/VFs/main.htm. (b) Genes involved in respiration.
Candidate small RNAs found in P. aeruginosa 2192.
Primers used for qRT-PCR.