ABSTRACT
Glycoprotein B (gB), gD, and gH/gL constitute the fusion machinery of herpes simplex virus (HSV). Prior studies indicated that fusion occurs in a stepwise fashion whereby the gD/receptor complex activates the entire process, while gH/gL regulates the fusion reaction carried out by gB. Trimeric gB is a class III fusion protein. Its ectodomain of 773 amino acids contains a membrane-proximal region (MPR) (residues 731 to 773) and two fusion loops (FLs) per protomer. We hypothesized that the highly hydrophobic MPR interacts with the FLs, thereby masking them on virions until fusion begins. To test this hypothesis, we made a series of deletion, truncation, and point mutants of the gB MPR. Although the full-length deletion mutants were expressed in transfected cells, they were not transported to the cell surface, suggesting that removal of even small stretches of the MPR was highly detrimental to gB folding. To circumvent this limitation, we used a baculovirus expression system to generate four soluble proteins, each lacking the transmembrane region and cytoplasmic tail. All retained the FLs and decreasing portions of the MPR [gB(773t) (gB truncated at amino acid 773), gB(759t), gB(749t), and gB(739t)]. Despite the presence of the FLs, all were compromised in their ability to bind liposomes compared to the control, gB(730t), which lacks the MPR. We conclude that residues 731 to 739 are sufficient to mask the FLs, thereby preventing liposome association. Importantly, mutation of two aromatic residues (F732 and F738) to alanine restored the ability of gB(739t) to bind liposomes. Our data suggest that the MPR is important for modulating the association of gB FLs with target membranes.
IMPORTANCE
To successfully cause disease, a virus must infect host cells. Viral infection is a highly regulated, multistep process. For herpesviruses, genetic material transfers from the virus to the target cell through fusion of the viral and host cell lipid membranes. Here, we provide evidence that the ability of the herpes simplex virus (HSV) glycoprotein B (gB) fusion protein to interact with the host membrane is regulated by its membrane-proximal region (MPR), which serves to cover or shield its lipid-associating moieties (fusion loops). This in turn prevents the premature binding of gB with host cells and provides a level of regulation to the fusion process. These findings provide important insight into the complex regulatory steps required for successful herpesvirus infection.
Introduction
Herpes simplex virus (HSV) has four envelope glycoproteins that are essential for virus entry into cells: glycoprotein D (gD), gH, gL, and gB. All herpesviruses use a combination of gB and the heterodimer gH/gL to carry out fusion (1–6); these three proteins are considered the core fusion machinery. For HSV, an additional protein, gD, is part of this machinery. Our current model of HSV fusion starts with the binding of gD to one of its receptors, likely transmitting a signal to gH/gL, which in turn acts upon gB to trigger fusion (7). HSV-1 gB is a 904-amino-acid type I membrane glycoprotein whose crystal structure identifies it as a class III fusion protein (8). Although there is no primary sequence conservation, HSV-1 gB shares a high degree of structural homology with other class III fusion proteins, including vesicular stomatitis virus (VSV) glycoprotein G (9), baculovirus gp64 (10), and Epstein-Barr virus (EBV) gB (11). According to ultrastructural data (8–11), these presumed postfusion conformations show that all are homotrimers with a long central coiled-coil structure reminiscent of class I fusion proteins. Yet, all have internal bipartite fusion loops (FLs) which are similar to the single internal FL of class II fusion proteins (5, 12–14). Single point mutations within either one of the gB fusion loops caused loss of cell-cell fusion and failure of soluble gB to associate with membranes (15, 16).
For VSV glycoprotein G, ultrastructural data are available for both pre- and postfusion forms (9, 17) and indicate that the FLs are situated near the transmembrane region and are close to the viral membrane in both forms. For the fusion loops to start and end in this position, it is presumed that an intermediate step occurs; in this intermediate step, the fusion loops reposition to the top of gB to interact with the cellular membrane. Then, as the conformation of gB changes to its postfusion form, it pulls the viral and cellular membranes close together to facilitate lipid mixing. The prevailing concept is that these hydrophobic loops would be masked on the virus surface prior to fusion activation to avoid premature or otherwise unwanted interactions that may be detrimental to virus infectivity.
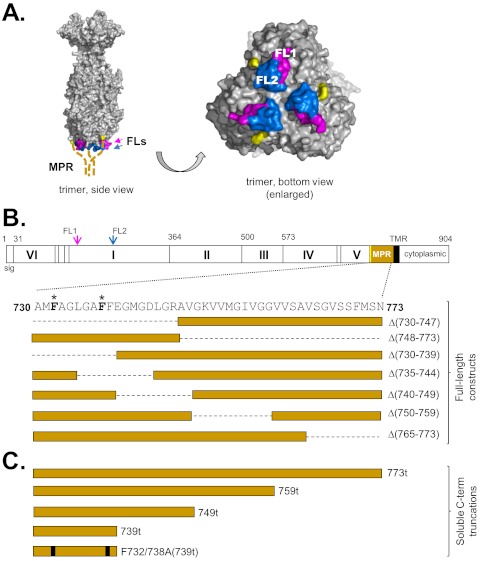
The form of HSV gB used for crystallization ended at amino acid 730, leaving the fusion loop region exposed (8). Residues downstream of amino acid 730 were initially excluded for crystallization trials due to their elevated hydrophobicity, which could impede crystal formation. These downstream residues (residues 731 to 773) constitute the gB membrane-proximal region (MPR) (8, 18–20). The structure of the MPR remains unknown for any of the class III fusion proteins, but it seems likely to be in close proximity to the FLs (Fig. 1A).
FIG 1.
(A) Surface representation of the gB trimer. Residues of FL1 (from all 3 monomers) are shown in pink, while residues of FL2 are shown in blue. The C termini of the solved gB monomers are highlighted (yellow). The unsolved MPRs (membrane-proximal regions) (amino acids 730 to 773) of each protomer are represented as dashed, thick tan lines emanating from the gB monomer C termini. FL1 and FL2 of one protomer are labeled in the bottom view of the trimer. (B) Schematic representation of a gB protomer, showing its structural domains (Roman numerals). FL1 and FL2 are indicated with arrows, colored as in panel A. Amino acid numbers are shown along the top. The MPR is shown in tan, and its residues are in expanded view below. Residues that were mutated in this study are highlighted in boldface type with an asterisk. Deletion mutants are designated boxes and aligned below the MPR amino acids, with a dashed line indicating deleted residues in the full-length protein. sig, signal sequence; TMR, transmembrane region. (C) Schematic representation of the soluble C-terminal (C-term) truncation mutants generated in this study. The point mutations F732A and F738A are indicated as black bars.
MPRs provide essential functions in different types of fusion proteins. Mutations within the MPRs of the fusion proteins for HIV (21), paramyxoviruses (22, 23), VSV (24), and baculovirus (25) abolish fusion but do not affect cell surface protein expression. MPRs are characteristically rich in aromatic residues (W, Y, and F) (26) that have been proposed to bridge the gap between the aqueous and hydrophobic environments present at the lipid interface (27). These aromatic residues could also act in concert (synergistically) with fusion peptides/loops to destabilize membranes and promote lipid mixing (26, 28). Indeed, aromatic residues are also vital in FL function; it is presumed that their side chains, along with their carbon backbones, are inserted into the target membrane (9, 29, 30). Interestingly, the MPR of baculovirus gp64 does not contain any aromatic residues, but its bulky leucine residues appear critical for cell-cell fusion (25). MPRs might also act as flexible tethers to help fusion proteins become properly positioned between two lipid bilayers, as described for VSV glycoprotein G (31).
Several studies of gB from different herpesviruses highlight the functional importance of its MPR. Most gB MPR mutants are defective in protein folding and cell surface expression (20, 32–34), making it difficult to assess its functional role. However, the few HSV gB MPR mutants that are expressed on the cell surface are impaired in cell-cell fusion or virus entry (32, 33).
Previously, we suggested that the MPR could mask the hydrophobic FLs in the prefusion state (15) in a way that would be analogous to how the FLs of class II fusion proteins are masked at the dimer interface (35–37). Our goal here was to carry out a systematic analysis of the MPR. We first made a series of MPR deletion mutants in this region in full-length gB and expressed each mutant in mammalian cells. Unfortunately, none of these mutants were efficiently transported to the cell surface, a defect previously reported for other gB MPR deletions (20). We next constructed recombinant baculoviruses to express C-terminal gB truncation mutants, containing various lengths of the MPR. Despite the presence of the fusion loops, all of the proteins containing portions of the MPR were compromised in their ability to bind liposomes compared to the MPR-less gB(730t) (gB truncated at amino acid 730), including gB(739t), which contains only nine MPR residues. However, mutation of two aromatic residues in gB(739t) to alanine (F732A/F738A) restored the ability of this protein to bind liposomes. Thus, our data are consistent with a critical role for the MPR in regulating exposure of the gB fusion loops, thereby preventing premature association with lipid.
RESULTS
The crystal structure of HSV-1 gB (Fig. 1A) was solved using a soluble form of gB truncated at amino acid 730 (gB730t) (8) to avoid the hydrophobic membrane-proximal residues (residues 730 to 773) just upstream of the transmembrane region (TMR) that might impair crystal formation (18, 19). The form of HSV gB used for crystallization ended at amino acid 730, leaving the fusion loop region exposed (8). Residues downstream of amino acid 730 were initially excluded for crystallization trials due to their elevated hydrophobicity, which could impede crystal formation. We hypothesized that these 43 amino acids, constituting the MPR of gB, obscure the fusion loops, and prevent them from interacting with a target membrane until gB is “activated” for fusion (15). We assumed that these residues are in close proximity to the fusion loops based on the gB crystal structure (Fig. 1A, yellow).
MPR deletions impair HSV gB transport.
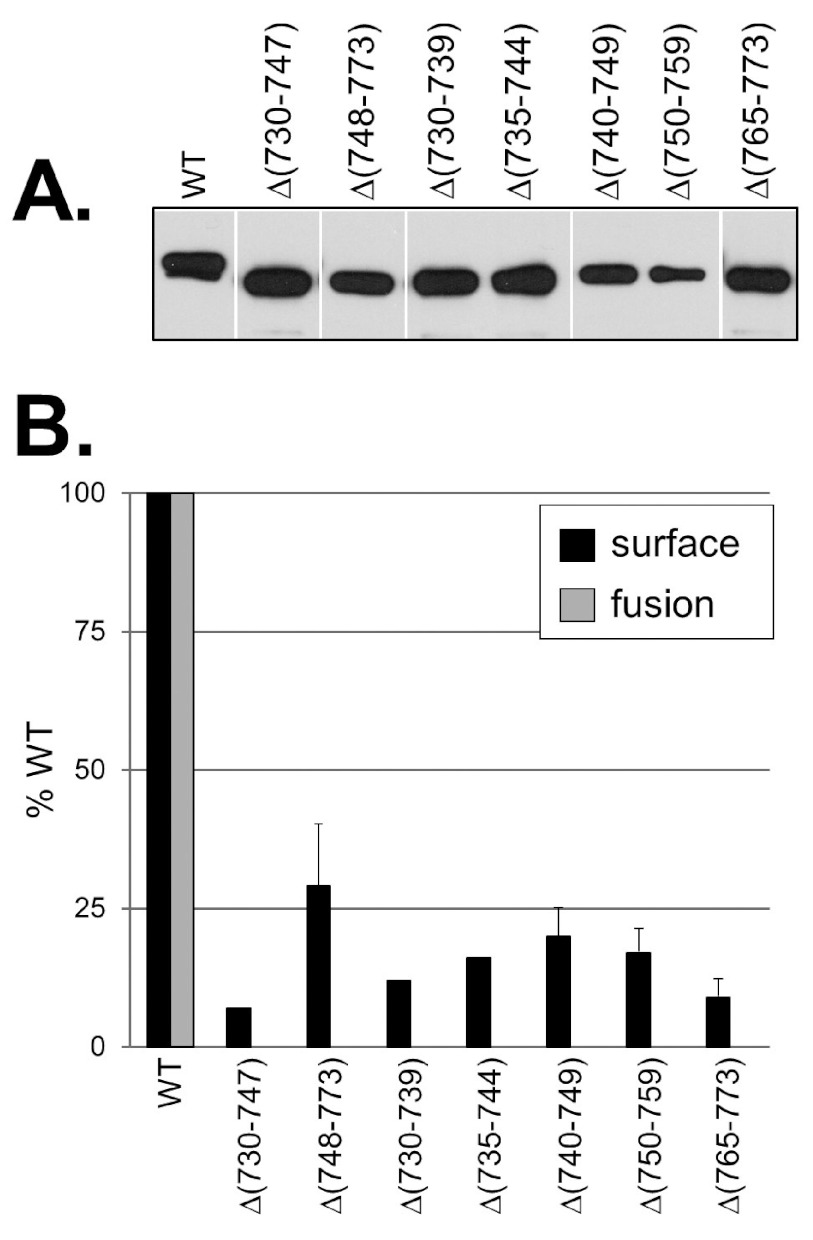
To determine whether the MPR regulated the ability of the fusion loops to associate with membranes, we first constructed seven MPR deletion mutants (within the context of full-length gB) (Fig. 1B). The N-terminal half of the MPR was deleted in gBΔ(730–747) (gB with residues 730 to 747 deleted), and the C-terminal half was deleted in gBΔ(748–773). The remaining mutants contained smaller deletions spanning the MPR: gBΔ(730–739), gBΔ(735–744), gBΔ(740–749), gBΔ(750–759), and gBΔ(765–773). All mutants were efficiently expressed in B78-C10 cells, as seen by Western blots of total cell lysates (Fig. 2A). However, when intact transfected cells were tested in a cell-based enzyme-linked immunosorbent assay (CELISA), all of the deletion mutants were poorly expressed (<30% that of the wild-type [WT] gB) on the cell surface, indicating that although the mutants were expressed, they were inefficiently transported to the cell surface (Fig. 2B, black bars). Consequently, none were able to function in a cell-cell fusion assay (Fig. 2B, gray bars). Thus, as seen in other studies (20, 34), deletion of even small sections of the gB MPR has a negative impact on proper protein folding and transport.
FIG 2.
Characterization of gB MPR deletion mutants. (A) Full-length MPR mutants were expressed in mammalian cells, and total cell extracts were analyzed by denaturing Western blotting. Blots were probed with the anti-gB PAb R68. (B) Protein surface expression as detected by CELISA (black bars). Transfected CHO-K1 cells were fixed with 3% paraformaldehyde, then incubated with the anti-gB PAb R69 and goat anti-mouse antibody conjugated to horseradish peroxidase (GAM-HRP). Cells transfected with empty vector DNA were used as a negative control, and this value was subtracted from the other experimental samples. Quantitative cell-cell fusion assay (gray bars) was performed with cocultivation of target CHO-K1 cells (expressing the luciferase protein and the HSV [herpes simplex virus] receptor HVEM) with effector CHO cells (expressing T7 polymerase, gD, gH, and gL, plus either WT gB, mutant gB, or empty vector DNA). Cell extracts were tested for light production 18 h later. Percent WT was calculated as follows: for CELISA = (sample absorbance/WT absorbance) × 100; for fusion assay = (RLU of test sample/RLU of WT) × 100 where RLU stands for relative light units.
Construction of soluble, MPR-containing gBs.
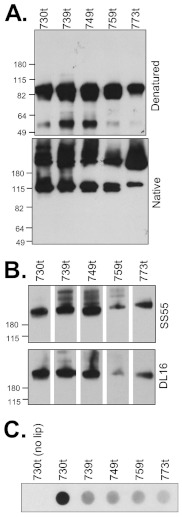
To characterize the biochemical properties of the gB MPR, we created and purified a series of soluble gB ectodomains (no transmembrane region [TMR] or endodomain) that lack portions of the MPR. We previously characterized the gB ectodomain gB(730t), lacking all of the MPR, and found that it remains oligomeric, retains all of its major epitopes, and associates with liposomes (8, 16, 38). Here, we compared it with both soluble forms of gB bearing partial MPRs [gB(759t), gB(749t), and gB(739t)] and a complete ectodomain containing the full MPR, gB(773t) (Fig. 1C). All were expressed and secreted into the supernatant of baculovirus-infected cells (Fig. 3A). When the proteins were examined on a nondenaturing gel, all contained both monomeric and trimeric forms of gB (Fig. 3A, bottom panel). Although all of the truncated proteins were recognized by monoclonal antibodies (MAbs) with trimer-specific, conformation-dependent epitopes (DL16 and SS55), indicative of proper protein folding, gB(759t) appeared to react less well (Fig. 3B). All MPR-containing gBs also retained their ability to react with MAbs with epitopes in each functional region (data not shown).
FIG 3.
Soluble gB MPR-containing proteins are expressed and folded correctly. (A) Four gB mutants (739t, 749t, 759t, and 773t) were cloned and expressed in a baculovirus expression system as secreted forms with each protein truncated after the indicated amino acid. Proteins were detected with the PAb R69 and visualized by denaturing or “native” Western blots. (B) Reactivity of gB MPR-containing proteins with the conformation-dependent MAbs SS55 or DL16 via “native” Western blotting. The positions of molecular mass standards (in kilodaltons) are shown to the left of the blots in panels A and B. (C) Liposome flotation assay. Purified soluble glycoproteins were incubated with liposomes for 1 h at 37°C. Samples were adjusted to 1 M KCl, incubated for an additional 15 min, layered beneath a discontinuous 5 to 40% sucrose gradient, centrifuged for 3 h, and then fractionated. The top, liposome-containing fraction was analyzed by dot blotting with the PAb R68. lip, liposomes.
The presence of the MPR affects binding of soluble gB to liposomes.
(i) Analysis by liposome flotation.
We previously observed that the MPR-less soluble gB(730t) interacts in vitro with liposomes when the two are mixed (16, 38), presumably recapitulating the interaction of the gB fusion loops with the cell membrane during virus-cell fusion. Initially, we used the liposome flotation assay to assess whether the MPR, or portions of it, interfered with this interaction.
Purified soluble proteins were individually incubated with liposomes (phosphatidylcholine-cholesterol) at 37°C. Each protein-liposome mixture was then adjusted to 40% sucrose, layered beneath a sucrose step gradient, and centrifuged to allow the liposomes and any associated proteins to float to the top, while proteins that failed to bind liposomes would remain at the bottom of the gradient. The liposome-containing (top) fractions were analyzed by dot blotting using a polyclonal anti-gB antibody for visualization (Fig. 3C). As previously observed (16), gB(730t) was not detected in the upper fractions in the absence of liposomes but floated to the top of the gradient in their presence. In the case of the MPR-containing gBs, the ability to coassociate with liposomes decreased markedly. The data shown in Fig. 3C suggest that there might be a small difference in binding between gB(739t) and gB(773t), but this type of analysis is not quantitative. Therefore, we turned to surface plasmon resonance (SPR) (biosensor) to obtain a more accurate measurement of liposome binding (39).
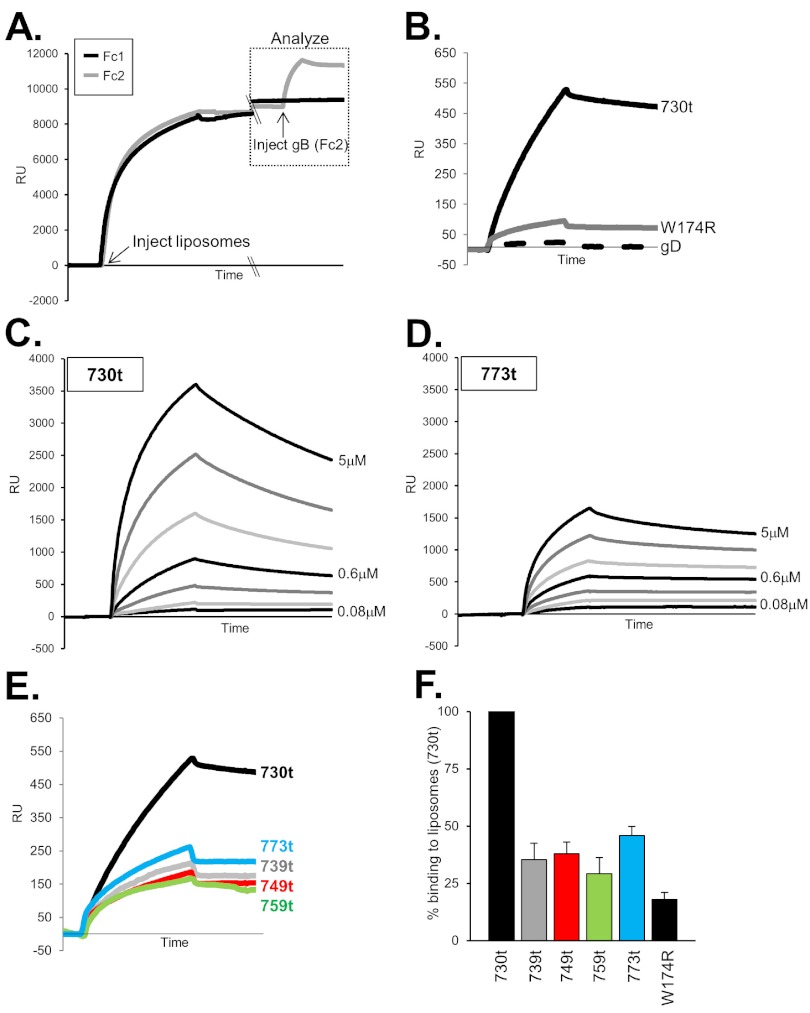
(ii) Analysis by SPR/biosensor.
Liposomes are able to bind to the lipophilic L1 chip of the BIAcore instrument, which detects mass changes by SPR (40). In this assay, liposomes were first injected and allowed to flow across an L1 chip surface, and liposome binding, i.e., the mass change due to binding, was detected as an increase in response units (RU) (Fig. 4A). Approximately 8,500 RU of liposomes was sufficient to saturate each flow cell of the chip, and this amount was used in each experiment. Next, each gB protein was added and allowed to flow across the chip for 240 s. An increase in RU was indicative of protein binding to the immobilized liposomes. It is this second response curve (gB-liposome binding [Fig. 4A, boxed]) that is shown in Fig. 4B to E. To set up the appropriate binding conditions for our proteins, we first compared gB(730t), known to bind liposomes in a flotation assay, to both the fusion loop mutant gB-W174R(730t) and to gD(306t), two proteins that do not bind liposomes (16). As expected, gB(730t) readily bound to the immobilized liposomes, as indicated by the increase in RU, while gB-W174R(730t) and gD(306t) did not (Fig. 4B). Furthermore, gB(730t) bound to liposomes in a dose-dependent manner, showing an increase in RU from 0.08 µM to 5 µM protein (Fig. 4C). Thus, we demonstrated that biosensor analysis yields reproducible and quantitative data on membrane binding by soluble glycoproteins.
FIG 4.
Liposome binding assay and biosensor analysis. (A) Liposomes are injected and allowed to flow across flow cell 1 (Fc1) and Fc2, with binding at saturation (~8,500 response units [RU]). Soluble gB(730t) is then injected across Fc2 at 5 µl/min for 240 s, and binding to liposomes is measured as an increase in RU (response units); this portion of the graph (boxed) is shown and analyzed in subsequent figures. A double-slash denotes a break in the x axis (no injections were performed during that time period). (B) Response curve showing binding of control proteins gB(730t) (positive control), fusion loop mutant gB-W174R(730t) (negative control), and soluble gD (negative control). (C and D) Twofold serial dilutions of gB(730t) (C) or gB(773t) (D) were injected across the liposome-coated flow cell at 10 µl/min for 2 min to evaluate ligand-liposome association. After each injection, the surface preparation protocol was performed to remove protein and liposomes from the chip, regenerating the surface to the RU baseline. (E) Each soluble protein (156 nM) was allowed to flow across an L1 chip containing immobilized liposomes as described above for panel A. The flow rate was 5 µl/min. (F) Bar graph representation of gB-liposome binding via biosensor. Values are averages of at least 3 separate experiments, with samples presented as percent binding of gB(730t) (set at 100%). Error bars represent standard errors of the means.
The liposome binding capacity of the MPR-containing gB forms was then analyzed within the same range of protein concentrations as used for gB(730t). We found that gB(773t), which contains the complete MPR, bound to liposomes in a dose-dependent manner, albeit at a much lower level than seen for gB(730t) (compare Fig. 4C and D), but curve fitting for kinetic analysis (40) was not possible with our data sets, possibly due to complications associated with liposome binding. None of the MPR-containing gB truncations bound to liposomes as well as gB(730t), although all exhibited a low level of measurable binding (Fig. 4E), especially compared to the two negative controls (Fig. 4B). Of importance, we detected no significant differences in gB-liposome binding between proteins containing portions of MPR or the complete MPR [gB(739t), gB(749t), gB(759t), or gB(773t)]. Upon averaging the maximum binding from several experiments (i.e., after 2 min of injection of 0.16 µM gB), we determined that gB(739t) bound to liposomes at a level that was 35% of that of gB(730t), 38% of gB(749t), 29% of gB(759t), and 46% of gB(773t) (Fig. 4F). In comparison, the negative-control gB-W174R(730t) bound at 18% of that of gB(730t). The data show that the presence of the first 9 amino acids of the MPR (residues 731 to 739) are sufficient to significantly diminish the association of gB with liposomes.
The decrease in liposome binding is not due to aggregation of the MPR mutants.
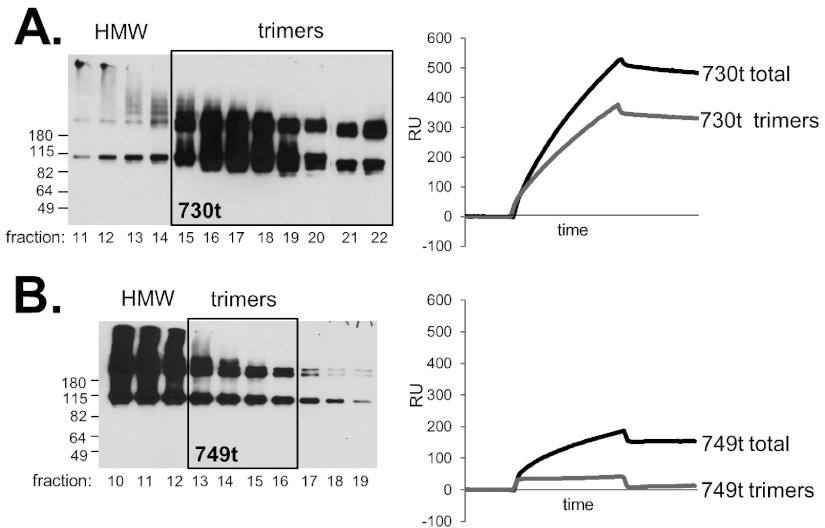
Our data suggest that the MPR specifically obscures the fusion loops and prevents their interaction with lipids; however, another possibility is that the hydrophobic character of the MPR promotes gB aggregation, possibly obscuring the fusion loops nonspecifically and preventing binding to membranes/lipids. To address this issue, we used gel filtration to separate out any high-molecular-weight (HMW) (i.e., aggregated) gB species from those fractions predominantly containing gB trimers (Fig. 5). We then tested the pooled trimeric fractions for liposome binding using SPR (Fig. 5, right panels). Each protein was chromatographed by size exclusion on a Sepharose column as previously described (41). Multiple fractions were obtained and evaluated by Western blotting, and a representative group of data are shown in Fig. 5. The bulk of gB(730t) eluted in the trimeric fractions (boxed in Fig. 5A), and much less was found in the earlier, HMW fractions. In contrast, the bulk of the MPR-containing gBs [represented by gB(749t) in Fig. 5B] was in the HMW fractions (Fig. 5B and data not shown). For each protein, the trimeric fractions were pooled, and equal concentrations of protein were injected over the liposome-L1 chip. As expected, the trimeric pool of gB(730t) bound well to liposomes (Fig. 5A, right panel). Importantly, the trimeric pools of MPR-containing gBs were unable to bind liposomes, as exemplified by gB(749t) (Fig. 5B). Indeed, the modest binding of unfractionated gB(749t) was substantially reduced by removal of the HMW material, suggesting that its binding was due to nonspecific association of aggregated material. Together, our results indicate that aggregation of MPR-containing gB does not prevent liposome binding but instead indicate that MPR-containing gB trimers are specifically compromised in their liposome-binding ability compared to gB(730t), presumably due to the FLs being obscured by the MPR.
FIG 5.
Gel filtration/size exclusion chromatography of gB(730t) (A) and gB(773t) (B). Fractions containing mostly trimeric species (boxed in the Western blot) were tested via biosensor for liposome binding at 156 nM gB for 240 s. Fractions containing high-molecular-weight (HMW) species, samples to the right of the box on the Western blot) were excluded from study. “Total” refers to the gB sample before it underwent gel filtration.
A double point mutation within the MPR restores gB-liposome binding.
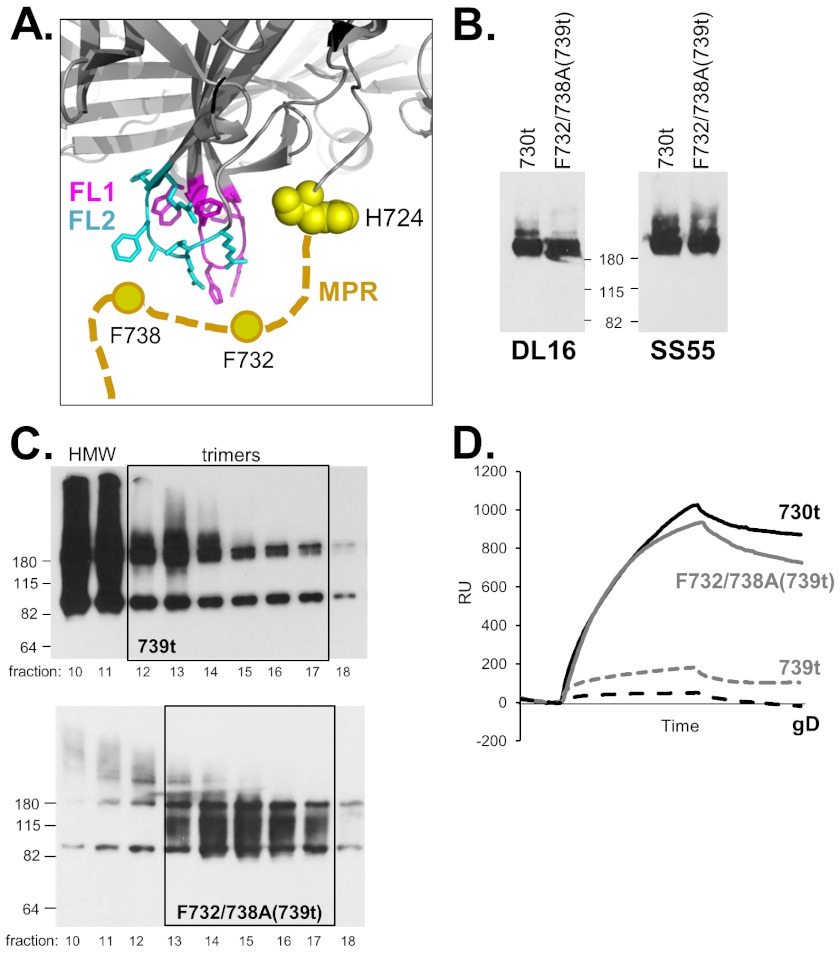
Because mutants such as gB-W174R(730t) do not bind liposomes (Fig. 4B), it is clear that association of gB(730t) with lipid membranes occurs via its fusion loops (16). Here we have shown that the first nine residues of the MPR are sufficient to reduce this binding (Fig. 4E and F). Among residues 731 to 739 (MFAGLGAFF) there are four hydrophobic amino acids, F732, L735, F738, and F739. These residues could stabilize the MPR and through their side chains possibly form contacts with the fusion loops or surrounding residues. To address the roles of these nine residues, we selected F732 and F738, as based on our model, they were most likely to form such contacts (Fig. 6A) and mutated them to alanine, creating a double point mutant within the context of gB(739t), gB-F732/738A(739t) (Fig. 1C). We predicted that this mutant would prevent putative interactions between the MPR and the underlying residues of the fusion loops (modeled in Fig. 6A) and therefore might improve gB-liposome binding in the context of gB(739t). The mutant protein was recognized by MAbs against conformation-dependent epitopes, indicating that it was properly folded (Fig. 6B). Unlike its parent protein gB(739t), very few HMW species were observed when gB-F732/738A(739t) was analyzed by gel filtration (Fig. 6C). Interestingly, biosensor analysis revealed that gB-F732/738A(739t) bound liposomes much better than gB(739t) did and nearly as well as gB(730t) did (Fig. 6D). These data suggest that the phenylalanine mutations compromise the ability of the MPR to shield the fusion loops, permitting their exposure and subsequent association with lipid membranes.
FIG 6.
(A) Model of a possible interaction between the gB fusion loops and MPR. The gB trimer is rendered in cartoon form in gray, focusing on the fusion loops (FL1 shown in pink and FL2 shown in blue) and C terminus of one of the protomers (H724, yellow spheres). The side chains of both fusion loops are shown. The MPR is depicted as a thick tan dashed line, with the aromatic residues F732 and F738 represented as circles. (B) Native Western blot of soluble gB proteins, probed with the indicated MAbs. The positions of molecular mass standards (in kilodaltons) are shown between the two blots. (C) Native Western blot of gel filtration fractions as described in the legend to Fig. 5. (D) gB-liposome binding assay (biosensor analysis) as described in the legend to Fig. 4A. Soluble gB(730t) served as the positive control and soluble gD as the negative control.
DISCUSSION
Both class I and class II viral fusion proteins have their fusion peptides/loops buried inside the molecule in the prefusion form (42). Here, we suggest that for the class III fusion protein HSV gB, the MPR serves this role. Soluble gB ectodomains containing various lengths of the MPR were all reduced in liposome binding compared to the MPR-less gB(730t); this included gB(739t), which contains only nine MPR residues. Mutation of two phenylalanine residues to alanine (F732A/F738A) in the gB(739t) MPR sequence restored the ability of this protein to bind liposomes. We believe that substitution of alanine residues for F732 and F738 disrupts the association of the MPR with the fusion loops, thereby allowing the fusion loops to be unimpeded and now able to bind lipid membranes. Our data are consistent with a role for the MPR in regulating the lipid association capability of the gB fusion loops.
Use of biosensor to evaluate gB-liposome binding.
To study gB-liposome interactions, we used SPR/biosensor in addition to our previously described liposome flotation assay (16, 38). SPR has been used in the HIV field to dissect binding of antibody, MPR (of the viral fusion protein gp41), and liposome (reviewed in reference 43). Here we have applied it to the HSV gB fusion protein. Since liposomes remain as individual vesicles when bound to an L1 sensor chip (44), as they do with liposome flotation, the two assays should be in agreement to measure protein-liposome interactions. Measuring liposome binding by SPR has several advantages over the standard flotation assay. First, it is far easier and faster to measure binding, as multiple samples can be analyzed on a single chip. Moreover, SPR allows for real-time measurement of gB-liposome binding, whereas the flotation assay relies on a secondary observation (antibody binding on a dot or Western blot) taking place approximately 6 h after the proteins and liposomes are coincubated. The quantitative nature of the biosensor also allowed us to compare percent binding of each experimental sample to binding of the wild type [in this case, MPR-containing gBs, gB fusion loop mutants, or gD to gB(730t)].
The HSV gB MPR is sensitive to mutation.
Unlike the MPRs of other fusion proteins (21–25), the MPR of gB is particularly sensitive to deletion (20) (Fig. 2). While our data point to MPR residues 731 to 739 as having a role in modulating fusion, the remainder of the MPR remains essential at the very least for protein folding and transport; this may be due to hydrophobic residues in the FLs or the MPR being recognized by endoplasmic reticulum (ER) chaperones, leading to ER retention. While deletions within the gB MPR are detrimental in the context of full-length proteins, deletion of the MPR in soluble gB ectodomains does not affect protein expression in our baculovirus system (nor does inclusion of the MPR). Although the soluble gB ectodomain is believed to be in a postfusion form, we are hypothesizing that the MPR functions as a fusion loop “mask” even when gB is in a prefusion form prior to fusion. In fact, a prefusion structure of VSV glycoprotein G (17) and model of EBV gB (11) suggest that structural domain I, which contains the fusion loops, remains in the same relative conformation both pre- and postfusion, suggesting that this strategy is sound. In further support, the FLs remain in close proximity to the ectodomain C terminus (and therefore MPR) in both pre- and postfusion forms.
Previously characterized MPR point mutants in full-length HSV-1 gB were found to be complementation defective for gB-null virus and exhibited a lower rate of entry, although two of the four mutants characterized were also defective in protein trafficking (33). None of these point mutations were within residues 731 to 739, which we have defined as having an effect on liposome binding. In additional experiments, we found that separate mutation of F732A or F738A (in the context of full-length gB) had no effect on cell surface expression, yet each had a negative effect on cell-cell fusion (data not shown). This result is in accordance with our data from the soluble gB-F732/738A double mutant. We predict that the decrease in cell-cell fusion would be explained by premature “triggering” of the FL, due to disruption of important MPR contacts required to maintain gB in a prefusion form. It is of note that gB residue F738 is conserved in all alphaherpesviruses (8, 33).
Roles of the MPR in other viruses.
MPR mutations in both VSV glycoprotein G (24) and baculovirus gp64 (25) abolish fusion but do not affect cell surface protein expression, indicative of a role for the MPR in the function of these proteins. In VSV glycoprotein G, the amino acids upstream of the TMR in the linear sequence are modeled as “stretching” from the top of the protein structure (crown) down to the transmembrane region in the postfusion structure (9), and this region is hypothesized to act as a flexible tether for positioning of the fusion protein between the viral and cellular lipid bilayers (31). Unlike the gB fusion loops, those of baculovirus gp64 are hypothesized to have a second role, in virus-cell attachment (10, 45). Therefore, one would expect that the triggering of gp64 fusion loops would occur earlier in the pathway (to facilitate virus-cell attachment) than for gB (to initiate virus-cell fusion), leaving the MPRs of gp64 and gB to perhaps serve different functions.
How might the gB MPR and FLs work together?
There are several hypotheses on what triggers gB activation, thereby exposing the fusion loops and promoting fusion of the virus and cell membranes (42). A low-pH, “histidine switch” model has been proposed for gp64 (46) and was also tested for HSV gB by Stampfer and colleagues (47), who found that only one of the two fusion loops (fusion loop 2 [FL2], containing H263) changed conformation at low pH; the bulk of the soluble gB(730t) remained unchanged in structure. Although this work was done on soluble gB (thought to be in the postfusion conformation), it fits with a prefusion model of EBV gB which predicts that domain I (containing the FLs) retains its fold between pre- and postfusion forms (11). Therefore, we suggest that the change in FL2 in response to low pH is sufficient to disengage the FLs from the MPR and expose them to lipids. Indeed, when gB(739t) was incubated at pH 5, its ability to bind liposomes increased to 68% that of gB(730t) (data not shown). However, we need to keep in mind that HSV gB also functions at neutral pH in certain cell types (48–51) and activation may occur through gHgL-gB interactions (7, 52), and perhaps even the gB cytoplasmic tail (53). Regardless of these possibilities, our data highlight the importance of the MPR in regulating exposure of the gB fusion loops and suggest that they play a critical role in maintaining gB in an inactive form, i.e., unable to insert its fusion loops into a target membrane until it is triggered to execute virus-cell fusion.
MATERIALS AND METHODS
Cells.
Mouse B78H1 melanoma cells engineered to express the gD receptor nectin-1 (C10 cells) were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 5% fetal bovine serum (FBS) and 500 µg/ml of G418. Chinese hamster ovary (CHO-K1) cells were grown in Ham’s F-12 medium containing 10% FBS. CHO-K1 and C10 cells were kindly provided by P. G. Spear. Sf9 (Spodoptera frugiperda) cells were grown in Sf900II serum-free medium (Gibco).
Antibodies.
Rabbit polyclonal antibodies (PAbs) R68 and R69 were raised against full-length gB1 purified from infected cells as previously described (54). gB-specific monoclonal antibodies (MAbs) SS55 and DL16 were characterized previously and recognize discontinuous (conformation-dependent) epitopes and are trimer specific (55).
Plasmids.
Full-length gB constructs containing deletions within the MPR, pSS1000 [gBΔ(730–747)], pSS1001 [gBΔ(748–773)] pSS1002 [gBΔ(730–739)], pSS1003 [gBΔ(735–744)], pSS1004 [gBΔ(740–749)], pSS1005 [gBΔ(750–759)], and pSS1008 [gBΔ(765–773)], were created using the QuikChange XL site-directed mutagenesis kit (Stratagene Cloning Systems) as described previously (56). QuikChange primers were designed to “loop out” unwanted residues during amplification of template pPEP98 (containing the full-length gB gene from KOS, a gift of P. Spear) (2) and were as follows (deletion between underlined nucleotides, only forward primers shown): pSS1000, 5′-CATCCACGCCGACGCCAACGCCGCGGTCGGCAAGGTGGTGATGGGC; pSS1001, 5′-CGAGGGGATGGGCGACCTGGGGCGCCCCTTTGGGGCGCTGGCCGTGGG; pSS1002, 5′-CACGCCGACGCCAACGCCGAGGGGATGGGCGACCTG; pSS1003, 5′-GCCGCCATGTTCGCGGGCCTGGGGCGCGCGGTCGGC; pSS1004, 5′-GGCCTGGGCGCGTTCTTCGGCAAGGTGGTGATGGGC; pSS1005, 5′-GACCTGGGGCGCGCGGTCGTGGTATCGGCCGTGTCG; and pSS1008, 5′-GGCGGCGTGGTATCGGCCGTGCCCTTTGGGGCGCTGGCC. All deletions were confirmed by sequencing the gB gene. Plasmids for expression of full-length HSV glycoproteins (pPEP99, pPEP100, and pPEP101) and those needed in the cell-cell fusion assay (pCAGGS/MCS, pT7EMCLuc [Luc stands for luciferase], and pCAGT7) were gifts of P. G. Spear (2, 57).
Production and purification of soluble HSV glycoproteins.
To construct a baculovirus vector expressing the MPR-containing gB ectodomain, we PCR amplified residues 526 to 773 of gB1 from template pKBXX (containing the gB gene from KOS, a gift of S. Person) using primers 5′-CGGCTGCAGTTTACGTACAA (PstI site underlined) and 5′-CGCGAGTTCAATTGGACATGAAGGAGGACAC (EcoRI site underlined). This fragment was cloned into PstI-EcoRI-digested pCW289 (58) to generate pLH633 [gB(773t)]. Other truncations of gB1 were generated from template pLH633 by QuikChange mutagenesis, generating stop codons (underlined) at residues 740 (primer 5′-GGCCTGGGCGCGTTCTTCTGAATTCGGTACCGACTCTGC), 750 (primer 5′-GACCTGGGGCGCGCGGTCTGAATTCGGTACCGACTCTGC), and 760 (primer 5′-GATGGGCATCGTGGGCGGCTGAATTCGGTACCGACTCTGC). The resulting constructs were named pSS1010 [gB(739t)], pSS1013 [gB(749t)], and pSS1015 [gB(759t)], respectively. A double point mutant, gB-F732/738A(739t) (pSS1023), was generated from template pSS1010 by QuikChange using primer 5′-GCCAACGCCGCCATGGCCGCGGGCCTGGGC and 5′-GCGGGCCTGGGCGCGGCCTTCGAGGGGATGGGC (substitutions underlined) to sequentially add first F732A and then F738A to the coding sequence. All mutations were verified by sequencing the entire gB gene. Recombinant baculoviruses were generated as previously described (59). The resulting soluble gB proteins were purified with a DL16 immunosorbent column (58). Soluble gB-W174R(730t) was described previously (16). Soluble gD(306t) was purified from baculovirus-infected insect cells as detailed elsewhere (59, 60).
Western blotting.
Purified, soluble proteins were mixed with an equal volume of 2× sample buffer containing either no reducing agent and 0.2% SDS (“native” conditions) or 200 mM dithiothreitol and 2% SDS (“denaturing” conditions) (61). Proteins from denatured samples were also boiled for 5 min before electrophoresis. For full-length proteins, C10 cells were grown on 6-well plates and transfected with the desired plasmids according to the GenePORTER protocol (Gene Therapy Systems, Inc.). At 24 to 48 h posttransfection, cells were lysed in 200 µl of lysis buffer (10 mM Tris [pH 8], 150 mM NaCl, 10 mM EDTA, 1% NP-40, 0.5% deoxycholic acid), 5 µl of which was used for electrophoresis (denaturing conditions). All proteins were resolved by polyacrylamide gel electrophoresis and transferred to nitrocellulose for Western blotting.
Fusion assay.
To detect cell-cell fusion, we used a previously described luciferase reporter assay (2, 57, 62). Briefly, CHO-K1 cells were grown in 24-well plates and transfected with plasmids carrying genes encoding T7 RNA polymerase (pCAGT7), gD (pPEP99), gH (pPEP100), and gL (pPEP101) and either wild-type gB (pPEP98), mutant gB, or empty vector (pCAGGS/MCS). To prepare receptor-expressing cells, CHO-K1 cells growing in six-well plates were transfected with a plasmid encoding the firefly luciferase gene under control of the T7 promoter (pT7EMCLuc) and a plasmid encoding HVEM (herpesvirus entry mediator) (pSC386). Six hours posttransfection, receptor-expressing cells were trypsinized and added to the glycoprotein-expressing cells. At 18 h after cocultivation, the cells were washed with phosphate-buffered saline (PBS), lysed in reporter lysis buffer (Promega), and frozen. Finally, the extracts were thawed and mixed with 100 µl of luciferase substrate (Promega) and immediately assayed for light output by luminometry.
CELISA.
To detect gB cell surface expression, we used a modification of a cell-based enzyme-linked immunosorbent assay (CELISA). CHO-K1 cells growing in 96-well plates were transfected with pCAGT7, pPEP99, pPEP100, pPEP101, and a plasmid encoding either wild-type gB (pPEP98), mutant gB, or empty vector (pCAGGS/MCS). Forty nanograms of each plasmid/well and 0.5 µl of Lipofectamine 2000 (Invitrogen) were used. The cells were exposed to the DNA-Lipofectamine 2000 mixture for 5 h, after which the mixture was replaced with growth medium. The cells were grown overnight, fixed in 3% paraformaldehyde, and rinsed with PBS. The cells were then incubated for 1 h with PAb R68 diluted in PBS with 3% bovine serum albumin (3% bovine serum albumin-PBS) and incubated for 30 min with goat anti-rabbit antibodies coupled to horseradish peroxidase. The cells were rinsed with 20 mM citrate buffer (pH 4.5) and incubated with peroxidase substrate (Moss, Inc.), and the absorbance at 405 nm was recorded using a microtiter plate reader. The absorbance values of cells transfected with the empty vector pCAGGS/MCS were subtracted, and data were normalized to WT gB.
Liposome flotation assay.
Liposome flotation assay conditions were adapted from previously described methods (16, 63, 64). Liposomes were purchased from Encapsula Nanosciences (Nashville, TN) at a size of 400 nm, containing a 1.7:1 molar ratio of soy-phosphatidylcholine to cholesterol. Briefly, purified soluble gB (1 µg), liposomes (25 µg), 1× protease inhibitor cocktail (Roche), and 15 µl of 200 mM sodium citrate were combined, and the final reaction mixture volume was adjusted to 50 µl with PBS. Protein-liposome mixtures were then incubated at 37°C for 1 h. To eliminate unwanted electrostatic protein-lipid associations, mixtures were incubated with 1 M KCl for 15 min at 37°C before being loaded at the bottom of a sucrose gradient. Mixtures were adjusted to 40% sucrose in a final volume of 500 µl and overlaid with 4 ml of PBS with 25% sucrose (25% sucrose-PBS) and 500 µl of 5% sucrose-PBS. The gradients were centrifuged for 3 h in a Beckman SW55Ti rotor at 246,000 × g at 4°C. Seven equal fractions (approximately 700 µl each) were collected, starting from the top of the gradient. Dot blotting onto nitrocellulose was performed using 225 µl per fraction and probed with the anti-gB PAb R68.
Surface plasmon resonance (biosensor) experiments to detect gB-liposome binding.
Surface plasmon resonance (SPR) experiments were performed using a BIAcore 3000 or BIAcore X100 optical biosensor (GE Healthcare, BIAcore Life Sciences) at 25°C. Filtered and degassed HBS-N buffer (10 mM HEPES [pH 7.4], 150 mM NaCl) was used in all liposome association experiments. We used an L1 sensor chip (BIAcore), as it has a hydrophobic surface capable of binding liposomes. Liposomes were purchased from Encapsula Nanosciences (Nashville, TN) at a size of 400 nm, containing a 1.7:1 molar ratio of soy-phosphatidylcholine to cholesterol. To prepare the chip surface for liposome binding, it was washed sequentially with 20 µl of 1% octyl-β-d-glucopyranoside, 20 µl of 0.5% SDS, 10 µl of 1% octyl-β-d-glucopyranoside, and 10 µl of 30% ethanol. Liposomes (1 mM, diluted in HBS-N buffer) were injected until the chip was saturated, giving a signal of approximately 8,500 RU. Once bound, the liposomes remained on the chip, and there was no appreciable dissociation (no measurable off-rate). Purified soluble proteins diluted in HBS-N buffer were then individually injected at various concentrations (achieved by dilution in HBS-N buffer) at a flow rate of 5 µl/min. After injection of the soluble protein was complete (typically 20 µl for 240 s), the RU was recorded and used to determine the level of protein binding. After each protein injection, the surface preparation protocol was performed to remove protein and liposomes from the chip, regenerating the surface to the RU baseline.
Gel filtration/size exclusion chromatography.
Size exclusion chromatography of gB (41) was performed on an AKTA purifier equipped with a Superdex S200 (24-ml) column (GE Healthcare) equilibrated with phosphate-buffered saline. The Superdex column was calibrated by using thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), RNase A (14 kDa), and aprotinin (6 kDa); blue dextran was used to determine the void volume (GE Healthcare). For each sample, 5 µl of each fraction was separated by SDS-PAGE and the high-molecular-weight, trimeric, and monomeric forms of gB were visualized by Western blotting with an anti-gB PAb (R68). Fractions that contained mostly trimers/monomers were pooled and tested by SPR for liposome binding.
ACKNOWLEDGMENTS
Funding for this project was through NIH grants R01-AI056045 (R.J.E.), R01-AI076231 (R.J.E.), and R37-A8289 (G.H.C.) from the National Institute of Allergy and Infectious Diseases. S.S.S. was funded in part by the VMD/PhD program of the School of Veterinary Medicine at the University of Pennsylvania. We thank P. Spear and S. Person for reagents, Lauren Hirao for constructing pLH633, and Brian Hannah for help with gB(773t).
We are grateful to our labmates, especially John Gallagher, for thoughtful discussions regarding the manuscript. Special thanks to Leslie King of the School of Veterinary Medicine for careful editing of the manuscript.
Footnotes
Citation Shelly SS, et al. 2012. The membrane-proximal region (MPR) of herpes simplex virus gB regulates association of the fusion loops with lipid membranes. mBio 3(6):e00429-12. doi:10.1128/mBio.00429-12.
REFERENCES
- 1. Browne H, Bruun B, Minson T. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J. Gen. Virol. 82:1419–1422 [DOI] [PubMed] [Google Scholar]
- 2. Pertel PE, Fridberg A, Parish ML, Spear PG. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313–324 [DOI] [PubMed] [Google Scholar]
- 3. Pertel PE. 2002. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J. Virol. 76:4390–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eisenberg RJ, et al. 2012. Herpes virus fusion and entry: a story with many characters. Viruses 4:800–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. 2011. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 9:369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kinzler ER, Compton T. 2005. Characterization of human cytomegalovirus glycoprotein-induced cell-cell fusion. J. Virol. 79:7827–7837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. 2010. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J. Virol. 84:12292–12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heldwein EE, et al. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220 [DOI] [PubMed] [Google Scholar]
- 9. Roche S, Bressanelli S, Rey FA, Gaudin Y. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187–191 [DOI] [PubMed] [Google Scholar]
- 10. Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM. 2008. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 15:1024–1030 [DOI] [PubMed] [Google Scholar]
- 11. Backovic M, Longnecker R, Jardetzky TS. 2009. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 106:2880–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kielian M, Rey FA. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4:67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lamb RA, Paterson RG, Jardetzky TS. 2006. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology 344:30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Steven AC, Spear PG. 2006. Viral glycoproteins and an evolutionary conundrum. Science 313:177–178 [DOI] [PubMed] [Google Scholar]
- 15. Hannah BP, Heldwein EE, Bender FC, Cohen GH, Eisenberg RJ. 2007. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J. Virol. 81:4858–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hannah BP, et al. 2009. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J. Virol. 83:6825–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roche S, Rey FA, Gaudin Y, Bressanelli S. 2007. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 315:843–848 [DOI] [PubMed] [Google Scholar]
- 18. Pellett PE, Kousoulas KG, Pereira L, Roizman B. 1985. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J. Virol. 53:243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bzik DJ, Fox BA, DeLuca NA, Person S. 1984. Nucleotide sequence specifying the glycoprotein gene, gB, of herpes simplex virus type 1. Virology 133:301–314 [DOI] [PubMed] [Google Scholar]
- 20. Rasile L, Ghosh K, Raviprakash K, Ghosh HP. 1993. Effects of deletions in the carboxy-terminal hydrophobic region of herpes simplex virus glycoprotein gB on intracellular transport and membrane anchoring. J. Virol. 67:4856–4866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Salzwedel K, West JT, Hunter E. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tong S, et al. 2001. Three membrane-proximal amino acids in the human parainfluenza type 2 (HPIV 2) F protein are critical for fusogenic activity. Virology 280:52–61 [DOI] [PubMed] [Google Scholar]
- 23. Zhou J, Dutch RE, Lamb RA. 1997. Proper spacing between heptad repeat B and the transmembrane domain boundary of the paramyxovirus SV5 F protein is critical for biological activity. Virology 239:327–339 [DOI] [PubMed] [Google Scholar]
- 24. Jeetendra E, et al. 2003. The membrane-proximal region of vesicular stomatitis virus glycoprotein G ectodomain is critical for fusion and virus infectivity. J. Virol. 77:12807–12818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Z, Blissard GW. 2009. The pretransmembrane domain of the Autographa californica multicapsid nucleopolyhedrovirus gp64 protein is critical for membrane fusion and virus infectivity. J. Virol. 83:10993–11004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Suárez T, Gallaher WR, Agirre A, Goñi FM, Nieva JL. 2000. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 74:8038–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Killian JA, von Heijne G. 2000. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 25:429–434 [DOI] [PubMed] [Google Scholar]
- 28. Jeetendra E, Robison CS, Albritton LM, Whitt MA. 2002. The membrane-proximal domain of vesicular stomatitis virus G protein functions as a membrane fusion potentiator and can induce hemifusion. J. Virol. 76:12300–12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Modis Y, Ogata S, Clements D, Harrison SC. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313–319 [DOI] [PubMed] [Google Scholar]
- 30. Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375:291–298 [DOI] [PubMed] [Google Scholar]
- 31. Roche S, Albertini AA, Lepault J, Bressanelli S, Gaudin Y. 2008. Structures of vesicular stomatitis virus glycoprotein: membrane fusion revisited. Cell. Mol. Life Sci. 65:1716–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin E, Spear PG. 2007. Random linker-insertion mutagenesis to identify functional domains of herpes simplex virus type 1 glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 104:13140–13145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wanas E, Efler S, Ghosh K, Ghosh HP. 1999. Mutations in the conserved carboxy-terminal hydrophobic region of glycoprotein gB affect infectivity of herpes simplex virus. J. Gen. Virol. 80:3189–3198 [DOI] [PubMed] [Google Scholar]
- 34. Zheng Z, Maidji E, Tugizov S, Pereira L. 1996. Mutations in the carboxyl-terminal hydrophobic sequence of human cytomegalovirus glycoprotein B alter transport and protein chaperone binding. J. Virol. 70:8029–8040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gibbons DL, et al. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427:320–325 [DOI] [PubMed] [Google Scholar]
- 36. Roussel A, et al. 2006. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure 14:75–86 [DOI] [PubMed] [Google Scholar]
- 37. Zhang Y, et al. 2004. Conformational changes of the flavivirus E glycoprotein. Structure 12:1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cairns TM, et al. 2011. Capturing the herpes simplex virus core fusion complex (gB-gH/gL) in an acidic environment. J. Virol. 85:6175–6184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Besenicar M, Macek P, Lakey JH, Anderluh G. 2006. Surface plasmon resonance in protein-membrane interactions. Chem. Phys. Lipids 141:169–178 [DOI] [PubMed] [Google Scholar]
- 40. General Electric Company May 2007. Biacore X100 handbook, edition AC GE Healthcare Bio-Sciences AB, Uppsala, Sweden [Google Scholar]
- 41. Silverman JL, Sharma S, Cairns TM, Heldwein EE. 2010. Fusion-deficient insertion mutants of herpes simplex virus type 1 glycoprotein B adopt the trimeric postfusion conformation. J. Virol. 84:2001–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hearty S, Conroy PJ, Ayyar BV, Byrne B, O’Kennedy R. 2010. Surface plasmon resonance for vaccine design and efficacy studies: recent applications and future trends. Expert Rev. Vaccines 9:645–664 [DOI] [PubMed] [Google Scholar]
- 44. Anderluh G, Besenicar M, Kladnik A, Lakey JH, Macek P. 2005. Properties of nonfused liposomes immobilized on an L1 Biacore chip and their permeabilization by a eukaryotic pore-forming toxin. Anal. Biochem. 344:43–52 [DOI] [PubMed] [Google Scholar]
- 45. Zhou J, Blissard GW. 2008. Identification of a GP64 subdomain involved in receptor binding by budded virions of the baculovirus Autographica californica multicapsid nucleopolyhedrovirus. J. Virol. 82:4449–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Z, Blissard GW. 2011. Autographa californica multiple nucleopolyhedrovirus gp64 protein: roles of histidine residues in triggering membrane fusion and fusion pore expansion. J. Virol. 85:12492–12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stampfer SD, Lou H, Cohen GH, Eisenberg RJ, Heldwein EE. 2010. Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1. J. Virol. 84:12924–12933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fuller AO, Spear PG. 1987. Anti-glycoprotein D antibodies that permit adsorption but block infection by herpes simplex virus 1 prevent virion-cell fusion at the cell surface. Proc. Natl. Acad. Sci. U. S. A. 84:5454–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nicola AV, McEvoy AM, Straus SE. 2003. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J. Virol. 77:5324–5332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wittels M, Spear PG. 1991. Penetration of cells by herpes simplex virus does not require a low pH-dependent endocytic pathway. Virus Res. 18:271–290 [DOI] [PubMed] [Google Scholar]
- 51. Milne RS, Nicola AV, Whitbeck JC, Eisenberg RJ, Cohen GH. 2005. Glycoprotein D receptor-dependent, low-pH-independent endocytic entry of herpes simplex virus type 1. J. Virol. 79:6655–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Atanasiu D, et al. 2010. Bimolecular complementation defines functional regions of herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J. Virol. 84:3825–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Silverman JL, et al. 2012. Membrane requirement for the folding of the HSV-1 gB cytodomain suggests a unique mechanism of fusion regulation. J. Virol. 86:8171–8184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Handler CG, Eisenberg RJ, Cohen GH. 1996. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J. Virol. 70:6067–6070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bender FC, et al. 2007. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J. Virol. 81:3827–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cairns TM, et al. 2007. N-terminal mutants of herpes simplex virus type 2 gH are transported without gL but require gL for function. J. Virol. 81:5102–5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Okuma K, Nakamura M, Nakano S, Niho Y, Matsuura Y. 1999. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254:235–244 [DOI] [PubMed] [Google Scholar]
- 58. Bender FC, et al. 2003. Specific association of glycoprotein B with lipid rafts during herpes simplex virus entry. J. Virol. 77:9542–9552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sisk WP, et al. 1994. High-level expression and purification of secreted forms of herpes simplex virus type 1 glycoprotein gD synthesized by baculovirus-infected insect cells. J. Virol. 68:766–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rux AH, et al. 1998. Functional region IV of glycoprotein D from herpes simplex virus modulates glycoprotein binding to the herpesvirus entry mediator. J. Virol. 72:7091–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cohen GH, Isola VJ, Kuhns J, Berman PW, Eisenberg RJ. 1986. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J. Virol. 60:157–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lazear E, et al. 2008. Engineered disulfide bonds in herpes simplex virus type 1 gD separate receptor binding from fusion initiation and viral entry. J. Virol. 82:700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Doms RW, Helenius A, White J. 1985. Membrane fusion activity of the influenza virus hemagglutinin. The low pH-induced conformational change. J. Biol. Chem. 260:2973–2981 [PubMed] [Google Scholar]
- 64. Kielian M, Klimjack MR, Ghosh S, Duffus WA. 1996. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J. Cell Biol. 134:863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]