ABSTRACT
A novel human coronavirus (HCoV-EMC/2012) was isolated from a man with acute pneumonia and renal failure in June 2012. This report describes the complete genome sequence, genome organization, and expression strategy of HCoV-EMC/2012 and its relation with known coronaviruses. The genome contains 30,119 nucleotides and contains at least 10 predicted open reading frames, 9 of which are predicted to be expressed from a nested set of seven subgenomic mRNAs. Phylogenetic analysis of the replicase gene of coronaviruses with completely sequenced genomes showed that HCoV-EMC/2012 is most closely related to Tylonycteris bat coronavirus HKU4 (BtCoV-HKU4) and Pipistrellus bat coronavirus HKU5 (BtCoV-HKU5), which prototype two species in lineage C of the genus Betacoronavirus. In accordance with the guidelines of the International Committee on Taxonomy of Viruses, and in view of the 75% and 77% amino acid sequence identity in 7 conserved replicase domains with BtCoV-HKU4 and BtCoV-HKU5, respectively, we propose that HCoV-EMC/2012 prototypes a novel species in the genus Betacoronavirus. HCoV-EMC/2012 may be most closely related to a coronavirus detected in Pipistrellus pipistrellus in The Netherlands, but because only a short sequence from the most conserved part of the RNA-dependent RNA polymerase-encoding region of the genome was reported for this bat virus, its genetic distance from HCoV-EMC remains uncertain. HCoV-EMC/2012 is the sixth coronavirus known to infect humans and the first human virus within betacoronavirus lineage C.
IMPORTANCE
Coronaviruses are capable of infecting humans and many animal species. Most infections caused by human coronaviruses are relatively mild. However, the outbreak of severe acute respiratory syndrome (SARS) caused by SARS-CoV in 2002 to 2003 and the fatal infection of a human by HCoV-EMC/2012 in 2012 show that coronaviruses are able to cause severe, sometimes fatal disease in humans. We have determined the complete genome of HCoV-EMC/2012 using an unbiased virus discovery approach involving next-generation sequencing techniques, which enabled subsequent state-of-the-art bioinformatics, phylogenetics, and taxonomic analyses. By establishing its complete genome sequence, HCoV-EMC/2012 was characterized as a new genotype which is closely related to bat coronaviruses that are distant from SARS-CoV. We expect that this information will be vital to rapid advancement of both clinical and vital research on this emerging pathogen.
Introduction
Coronaviruses (CoVs) infect and cause disease in a wide variety of species, including bats, birds, cats, dogs, pigs, mice, horses, whales, and humans (1, 2). Recent studies suggest that bats act as a natural reservoir for coronaviruses (3–8). Coronaviruses may cause respiratory, enteric, hepatic, or neurological diseases with highly variable severity in their hosts. Until 2003, only two coronaviruses were known to infect humans. Human coronaviruses (HCoVs) HCoV-229E and HCoV-OC43 were identified in the 1960s as the causative agents of—generally mild—respiratory illnesses (9, 10). In 2002 to 2003, a previously unknown coronavirus—severe acute respiratory syndrome coronavirus (SARS-CoV)—caused a widespread outbreak of respiratory disease in humans, resulting in approximately 800 deaths and affecting around 30 countries (11–14). As a consequence of the renewed interest in coronaviruses after the SARS outbreak, two additional human coronaviruses were discovered after 2003: HCoV-NL63 in 2004 (15, 16) and HCoV-HKU1 in 2005 (17). A recent analysis of a large collection of human nasopharyngeal specimens using a Coronaviridae-wide primer set suggested that HCoV-229E, -OC43, -NL63, and -HKU1 are the only coronaviruses circulating in the human population (18).
Coronaviruses are enveloped single-stranded positive-sense RNA viruses with genomes of 25 to 32 kb, and the group includes the largest known genomes among the RNA viruses (1, 19). The coronaviruses form a subfamily (Coronavirinae) within the family Coronaviridae of the order Nidovirales. The International Committee on Taxonomy of Viruses (ICTV) has recognized four genera within the Coronavirinae subfamily: Alphacoronavirus, Betacoronavirus, and Gammacoronavirus, which were previously referred to as coronavirus groups 1, 2, and 3, and Deltacoronavirus (20). Coronaviruses are assigned to a genus on the basis of rooted phylogeny and calculation of pairwise evolutionary distances for seven highly conserved domains in the replicase polyprotein (1, 21) (C. Lauber and A. E. Gorbalenya, unpublished data). HCoV-229E and HCoV-NL63 are viruses belonging to the genus Alphacoronavirus (1). Four monophyletic lineages (A through D) with no formal taxonomic standing, some of them encompassing multiple virus species, are commonly recognized within the genus Betacoronavirus. Lineage A includes HCoV-OC43 and HCoV-HKU1 and lineage B SARS-CoV, all of which belong to different species. Lineages C and D include viruses detected only in bats, such as Rousettus bat coronavirus HKU9 (BtCoV-HKU9) (lineage D), Tylonycteris bat coronavirus HKU4 (BtCoV-HKU4), and Pipistrellus bat coronavirus HKU5 (BtCoV-HKU5) (both lineage C) (1). The genetic diversity of coronaviruses is likely facilitated by a high frequency of RNA recombination and the ability of their unusually large RNA genomes to both gain and lose domains (1, 22, 23). These factors are believed to have promoted the emergence of viruses with novel traits that are able to adapt to new hosts and ecological niches, sometimes causing zoonotic events.
For the present study, we report and analyze the complete genome sequence of the recently identified HCoV-EMC/2012, which was isolated from the sputum of a 60-year-old man who died in a hospital in Jeddah, Saudi Arabia, after developing acute respiratory distress syndrome (ARDS) and multiple organ dysfunction syndrome (MODS) in June 2012 (24). This virus appears to be closely related to the HCoV detected in a second patient who was transported from a hospital in Qatar to a hospital in London, 3 months after hospitalization of the first patient (25). These two cases of human infection with very similar or identical coronaviruses alarmed health authorities globally, as it was a reminder of the potential threat of coronaviruses to human health that was first highlighted by the SARS outbreak of 2003 (25). The sequence analysis of a small reverse transcription-PCR (RT-PCR) fragment that was first amplified from the HCoV-EMC/2012 genome revealed the highest similarity to two betacoronaviruses circulating in bats, BtCoV-HKU4 and -HKU5 (24). Here we present the complete genome sequence of the newly isolated HCoV-EMC/2012, accompanied by a detailed annotation of its genome organization and expression strategy. Furthermore, comparative genomic analysis and state-of-the-art classification and phylogenetic analyses were applied to determine the position of the novel agent with respect to previously characterized coronaviruses. We conclude that the HCoV-EMC/2012 genome organization and expression indeed most closely resemble those of BtCoV-HKU4 and -HKU5. However, based on our analysis and in line with the ICTV guidelines for the demarcation of coronavirus species, HCoV-EMC/2012 clearly qualifies to be recognized as the prototype of a novel species, which would thus constitute the first human coronavirus in lineage C of the genus Betacoronavirus.
RESULTS
Sequencing of the HCoV-EMC/2012 genome.
Using a combination of approaches, including deep sequencing, cycle sequencing on a more traditional capillary sequencer, and determination of the genomic termini by rapid amplification of cDNA ends (RACE), the complete genome sequence of HCoV-EMC/2012 was determined from material that had been subjected to passage in cell culture 6 times. The data from the Roche 454 GS Junior deep-sequencing run yielded a total of 90,808 sequence reads, of which 87,256 were specific for HCoV-EMC/2012. Genome coverage ranged from 1 to 5,697 reads at single nucleotide positions, with an average of 1,006 reads in the deep-sequencing run. Based on the contigs assembled from these initial data, primers approximately 800 nucleotides (nt) apart were designed to amplify PCR fragments with 100-nt overlaps covering the entire virus genome (see Table S1 in the supplemental material for primer sequences). These amplicons were sequenced using Sanger sequencing, and a total of 104 sequence runs were assembled—along with the original 90,808 deep-sequencing reads—into a single contiguous sequence of 30,119 nt, including the first 12 nt of the 3′ poly(A) tail. Although 454 sequencing resulted in a higher single-read error rate than Sanger sequencing, the high coverage in the first data set largely corrected for these errors. Occasionally, the correct number of bases in homopolymer stretches was difficult to determine, which is a typical problem in 454 sequencing. Nevertheless, there was excellent agreement between the deep-sequencing data and the confirmatory Sanger sequencing. The final consensus sequence was submitted to GenBank (see below). This sequence contains only two ambiguous positions, nt 11623 and 27162. The variation at position 11623 translates into a Val or Gly uncertainty at amino acid (aa) 3782 of pp1a/pp1ab. Position 27162 was either a G or an A, with the A creating a premature stop codon for translation of open reading frame 5 (ORF5) (see Discussion). The verification of our consensus sequence awaits the availability of a second HCoV-EMC/2012 virus isolate or original specimen. The overall content of G and C residues in the HCoV-EMC/2012 genome was 41%, which is similar to values reported for other coronaviruses (37% to 42%) (14).
Genome organization and expression strategy.
Coronavirus genomes are polycistronic positive-stranded RNAs (Fig. 1A), of which the 5′-proximal three-fourths are occupied by the large replicase open reading frames ORF1a and ORF1b. These are translated from the genomic mRNA to produce polyproteins pp1a and, following −1 ribosomal frameshifting, pp1ab, which are subsequently cleaved into 15 or 16 nonstructural proteins (nsps) (19, 23, 26). The region downstream of ORF1b is characterized by containing a variable number of smaller genes, always including those encoding the spike (S), envelope (E), membrane (M), and nucleocapsid (N) structural proteins. These genes are translated from subgenomic (sg) mRNAs that form a 5′- and 3′-coterminal nested set with the viral genome. Subgenomic mRNAs are composed of a common 5′ leader sequence that is identical to the genomic 5′ region and a variable part of the 3′ quarter of the genome, with different sg mRNAs making different ORFs available for translation. The complement of the leader and “body” segments of the sg mRNAs are assumed to be joined during discontinuous negative-strand RNA synthesis. This step produces the templates for sg mRNA synthesis and is directed by a base-pairing interaction between conserved transcription-regulatory sequences (TRSs) (27–29). Such TRSs are found at the 3′ end of the leader sequence (leader TRS) and at different positions upstream of genes in the genomic 3′-proximal domain (body TRSs). The synthesis of subgenome-length negative-stranded RNAs is directed by the complement of a body TRS at the 3′ end of a nascent minus-strand base pairing with the leader TRS, with the extent of sequence complementarity being an important determinant of the level at which a given sg mRNA is produced.
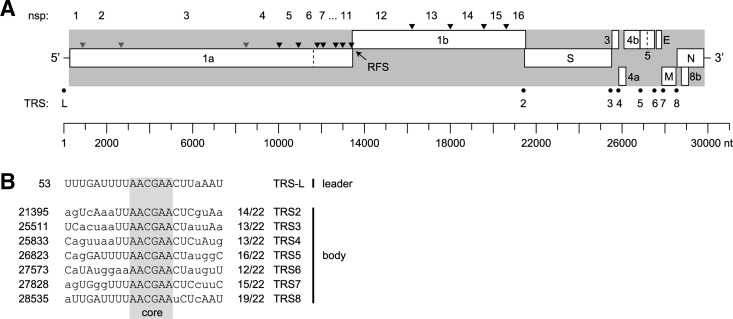
FIG 1.
Genome organization and expression of HCoV-EMC/2012. (A) The coding part of the genome and terminal untranslated regions are depicted, respectively, by a gray background and horizontal lines. Rectangles indicate ORFs and their locations in three reading frames. The dashed lines in ORF1a and ORF5 indicate base ambiguities observed during sequencing. Triangles represent sites in the replicase polyproteins pp1a and pp1ab that are predicted to be cleaved by papain-like proteinases (gray) or the 3C-like cysteine proteinase (black). Cleavage products are numbered nsp1 to nsp16, according to the convention established for other coronaviruses (23). The −1 ribosomal frameshift site (RFS) in the ORF1a/ORF1b overlap region is indicated. The location of the leader TRS (transcription-regulatory sequences) (L) and seven body TRSs (numbered) are highlighted by black dots. All coordinates correspond to the scale shown at the bottom. (B) Sequence comparison of leader TRS region and seven body TRSs. The fully conserved TRS core sequence AACGAA is highlighted. Nucleotides in the body TRSs are written in uppercase letters if the complementary nucleotide can base pair with the corresponding residue in the leader TRS region (including G-U base pairs). TRS starting coordinates in the HCoV-EMC/2012 genome are shown at the left; for the body TRSs, the numbers of (potential) base pairs with the leader TRS region are shown at the right.
Inspection of the genome sequence of HCoV-EMC/2012 revealed the two large, partially overlapping replicase open reading frames ORF1a and ORF1b, as well as (at least) nine downstream ORFs (Fig. 1A). The ORF1a sequence encodes the two protease domains conserved in all other coronaviruses, a papain-like protease (PL2pro) in nsp3 and a 3C-like protease (3CLpro; also known as the “main protease”) in nsp5. Sequence comparison with other coronaviruses allowed us to predict the putative pp1a/pp1ab cleavage sites and annotate the resulting nsp1 through -16 (Table 1). According to sequence conservation analyses performed with other coronaviruses, open reading frames ORF2, -6, -7, and -8a are predicted to encode the four canonical structural proteins of coronaviruses, the envelope proteins S, E, and M and the N protein, respectively (Fig. 1A; see also Fig. S1 in the supplemental material). A leader TRS and seven putative body TRSs could be readily identified, with the sequence 5′ AACGAA 3′ forming the conserved TRS core and potential TRS duplexes during leader-body joining ranging from 14 to 19 matches over a 22-nt window that includes the core of the leader TRS (Fig. 1B). From this analysis, it can be predicted that seven subgenomic mRNAs carrying a 67-nt common leader sequence would be produced in HCoV-EMC/2012-infected cells, with sizes ranging from ~4.7 kb for mRNA2 to ~1.7 kb for mRNA8. Experimental studies are needed to confirm the correct identification of the TRSs in the genomes of HCoV-EMC/2012 and related lineage C betacoronaviruses.
TABLE 1 .
Cleavage products of the replicase polyproteins of HCoV-EMC/2012
| Cleavage product | Position in polyprotein pp1a/pp1aba |
Protein size (no. of amino acids) |
Putative functional domain(s)b |
|---|---|---|---|
| nsp1 | 1Met-Gly193 | 193 | |
| nsp2 | 194Asp-Gly853 | 660 | |
| nsp3 | 854Ala-Gly2740 | 1887 | ADRP, PL2pro, TM1 |
| nsp4 | 2741Ala-Gln3247 | 507 | TM-2 |
| nsp5 | 3248Ser-Gln3553 | 306 | 3CLpro |
| nsp6 | 3554Ser-Gln3845 | 292 | TM-3 |
| nsp7 | 3846Ser-Gln3928 | 83 | |
| nsp8 | 3929Ala-Gln4127 | 199 | Putative primase |
| nsp9 | 4128Asn-Gln4237 | 110 | |
| nsp10 | 4238Ala-Gln4377 | 140 | |
| nsp11 | 4378Ser-Leu4391 | 14 | |
| nsp12 | 4378Ser-Gln5310 | 933 | RdRp |
| nsp13 | 5311Ala-Gln5908 | 598 | ZD, HEL1 |
| nsp14 | 5909Ser-Gln6432 | 524 | ExoN, NMT |
| nsp15 | 6433Gly-Gln6775 | 343 | NendoU |
| nsp16 | 6776Ala-Arg7078 | 303 | OMT |
Amino acids of the replicase proteins pp1a and pp1ab were numbered with the assumption that a −1 ribosomal frameshift occurs to express ORF1b, as in other coronaviruses (see text); the use of the slippery sequence UUUAAAC is predicted to result in a peptide bond between Asn4385 and Arg4386 in pp1ab.
The major transmembrane domains and a selection of the most conserved domains with enzymatic activities that have been characterized functionally and/or structurally in coronaviruses are listed. Abbreviations: PL2pro, papain-like proteinase 2; ADRP, ADP-ribose 1″-phosphatase; TM, transmembrane domain; 3CLpro, 3C-like cysteine proteinase; RdRp, RNA-dependent RNA polymerase; ZD, putative zinc-binding domain; HEL1, superfamily 1 helicase; ExoN, 3′-to-5′ exonuclease; NMT, N7-methyltransferase; NendoU, nidoviral endoribonuclease specific for U; OMT, S-adenosylmethionine-dependent ribose 2′-O-methyltransferase.
Furthermore, mRNA4 and -8 are predicted to be functionally bicistronic, with ribosomal leaky scanning being the likely translation initiation mechanism for both ORF4b and ORF8b. The ORF4b AUG codon is not preceded by a separate body TRS, and the 241-nt sequence separating the 5′ ends of ORF4a and ORF4b is entirely devoid of AUG codons. The AUG codon of the current ORF8b, an internal ORF that is overlapped by the N protein gene (ORF8a) and is present in all betacoronaviruses, is the third AUG codon on mRNA8, but sequence analysis and comparison with the BtCov-HKU4 and -HKU5 sequences (8) suggests that the 5′ end of ORF8b may have become truncated relatively recently (see Discussion).
Twenty-two additional putative ORFs of 150 to 432 nt in length were detected throughout the genome of HCoV-EMC/2012, overlapping the major ORFs. In contrast to the ORFs shown in Fig. 1A, these 22 additional ORFs are not positioned (immediately) downstream of a body TRS, and hence it is unlikely that they are expressed. The synthesis of the replicase pp1ab polyprotein of HCoV-EMC/2012 involves −1 programmed ribosomal frameshifting, with nt 13427 to 13433 predicted to form the conserved “slippery sequence” (5′ UUUAAAC 3′) in the ORF1a/ORF1b overlap region that is typical for coronaviruses (30). The frameshift region is followed by a predicted RNA hairpin, formed by nucleotides at positions 13439 to 13450 base pairing with those at 13462 to 13473, with potential RNA pseudoknot formation occurring by base pairing of the loop of the hairpin (nt 13452 to 13460) with a downstream complementary sequence (nt 13506 to 13514). As is common in coronavirus genomes, nontranslated sequences are found only at the genomic termini, with the 5′ and 3′ untranslated regions (278 and 300 nt, respectively) having sizes similar to those found in other family members. The only other apparently untranslated region in the genome that is larger than 50 nucleotides concerns the intergenic region between ORF5 and -6 (nt 27515 to 27589). This region appears to be conserved between HCoV-EMC/2012, BtCoV-HKU4, and BtCoV-HKU5, with sequence identities ranging from 63% to 84%, but we have no explanation for this observation thus far.
Phylogenetic relations and taxonomic position of HCoV-EMC/2012.
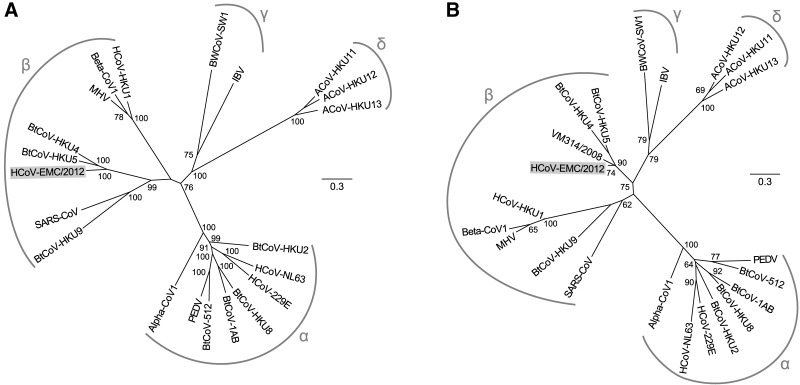
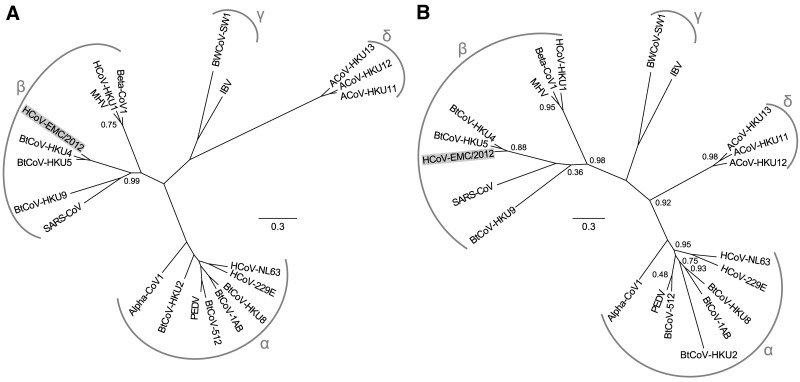
Phylogenetic trees were inferred using nucleotide sequences for ORF1ab (Fig. 2A) and a 332-nt fragment from ORF1b (Fig. 2B) encoding the most conserved part of the RNA-dependent RNA polymerase (RdRp) domain, which is commonly targeted in virus discovery studies. The first tree was produced for a representative set of coronaviruses for which complete genome sequences are available. In the second tree, we also included coronaviruses for which only partial genome sequences are known, particularly that of P.pipi/VM314/2008/NLD (31) which produced the best match with HCoV-EMC/2012. In both trees, HCoV-EMC/2012 clearly groups within lineage C of the genus Betacoronavirus, relatively close to BtCoV-HKU4 and BtCoV-HKU5. However, based on the 332-nt fragment from ORF1b, HCoV-EMC/2012 is more closely related to bat-derived isolate VM314/2008 (GenBank accession number GQ259977), which was isolated from Pipistrellus bats in The Netherlands 4 years ago. Phylogenetic trees were also constructed based on amino acid sequences, using coronavirus-wide conserved domains of replicative proteins in pp1ab (Fig. 3A) as well as using conserved parts of structural proteins (Fig. 3B). In both trees, HCoV-EMC/2012 clusters with betacoronaviruses, supporting its classification as a member of the genus Betacoronavirus.
FIG 2.
Phylogenetic trees for HCoV-EMC/2012 and selected other coronaviruses. Unrooted maximum likelihood phylogenies inferred from the nucleotide sequences of full-length ORF1ab (A) or a 332-nt fragment from the RdRp-encoding domain of ORF1b (B) are shown. HCoV-EMC/2012 and 20 viruses representing the recognized species diversity of coronaviruses were included, with bat-derived isolate VM314/2008 also included in the analysis presented in panel B (31). The viruses and corresponding species used are Alphacoronavirus 1 (Alpha-CoV1), Human coronavirus 229E (HCoV-229E), Human coronavirus NL63 (HCoV-NL63), Miniopterus bat coronavirus 1 (BtCoV-1AB), Miniopterus bat coronavirus HKU8 (BtCoV-HKU8), Porcine epidemic diarrhea virus (PED), Rhinolophus bat coronavirus HKU2 (BtCoV-HKU2), Scotophilus bat coronavirus 512 (BtCoV-512), Betacoronavirus 1 (Beta-CoV1), Human coronavirus HKU1 (HCoV-HKU1), Murine coronavirus (MHV), Tylonycteris bat coronavirus HKU4 (BtCoV-HKU4), Pipistrellus bat coronavirus HKU5 (BtCoV-HKU5), Rousettus bat coronavirus HKU9 (BtCoV-HKU9), Severe acute respiratory syndrome-related coronavirus (SARS-CoV), Avian coronavirus (IBV), Beluga whale coronavirus SW1 (BWCoV-SW1), Bulbul coronavirus HKU11 (ACoV-HKU11), Thrush coronavirus HKU12 (ACoV-HKU12), and Munia coronavirus HKU13 (ACoV-HKU13). Bootstrap values above 50 are shown. Arcs and symbols indicate the four coronavirus genera. The scale bar represents the number of nucleotide substitutions per site.
FIG 3.
Phylogenetic trees for HCoV-EMC/2012 and selected other coronaviruses. Unrooted maximum likelihood phylogenies based on coronavirus-wide conserved protein domains in replicase pp1ab (A) or on the conserved parts of structural proteins S2, E, M, and N (B) for HCoV-EMC/2012 and 20 viruses representing the recognized species diversity of coronaviruses are shown (see Fig. 2 legend for names and abbreviations). Branch support values are based on the Shimodaira-Hasegawa-like procedure and are in the range of zero to one; only nonoptimal values smaller than one are shown. Arcs and symbols indicate the four coronavirus genera. The scale bars represent average numbers of substitutions per amino acid position.
ICTV assigns newly identified members of the family Coronaviridae to a subfamily and genus on the basis of rooted phylogeny and calculation of pairwise evolutionary distances for seven replicase domains (1, 21). To establish whether HCoV-EMC/2012 indeed prototypes a new species, amino acid sequence alignments were generated for each of these domains and concatenated, after which the sequence identity of HCoV-EMC/2012 with closely related strains was calculated. For this purpose, the full genomes of 9 strains, derived from 3 species, belonging to Betacoronavirus lineage C were available (Table 2). Amino acid sequence identity between conserved replicase domains of HCoV-EMC/2012 and those of other lineage C viruses ranged from 57% (ADP-ribose 1″-phosphatase [ADRP]) to 94% (helicase [Hel]). Overall amino acid sequence identities to BtCoV-HKU4 and BtCoV-HKU5 strains across the conserved domains were around 75% and 76.7%, respectively. These percentages are well below the threshold of 90% amino acid sequence identity that is used for coronavirus species identification by the ICTV. The distance between HCoV-EMC/2012 and members of these two species is as large as that observed upon interspecies comparison of other species pairs, for example, Murine coronavirus versus Human coronavirus HKU1 or Porcine epidemic diarrhea virus versus Scotophilus bat coronavirus 512 (Fig. 3). Consequently, we propose that HCoV-EMC/2012 prototypes a novel species of lineage C of the genus Betacoronavirus.
TABLE 2 .
Percent amino acid sequence identity between conserved domains of the replicase polyprotein of HCoV-EMC/2012 and established betacoronavirusesa
| Virus strain | % amino acid sequence identity with conserved domain of the indicated HCoV-EMC/2012 replicase polyproteinb |
|||||||
|---|---|---|---|---|---|---|---|---|
| ADRP | 3CLpro | RdRp | Hel | ExoN | NendoU | O-MT | All domains | |
| BtCoV-HKU4.1 | 57.4 | 81.0 | 90.0 | 92.1 | 85.4 | 72.6 | 83.4 | 75.1 |
| BtCoV-HKU4.2 | 57.5 | 81.0 | 90.0 | 92.1 | 85.4 | 72.6 | 83.4 | 75.1 |
| BtCoV-HKU4.3 | 57.4 | 81.0 | 90.0 | 92.1 | 85.4 | 72.6 | 83.4 | 75.1 |
| BtCoV-HKU4.4 | 57.5 | 81.0 | 89.9 | 92.1 | 85.4 | 72.6 | 83.4 | 74.9 |
| BtCoV/133/2005 | 57.6 | 80.7 | 89.9 | 91.6 | 86.4 | 72.0 | 83.4 | 74.9 |
| BtCoV-HKU5.1 | 57.6 | 82.6 | 92.1 | 93.8 | 91.7 | 79.7 | 85.3 | 76.7 |
| BtCoV-HKU5.2 | 57.6 | 82.0 | 92.2 | 93.8 | 91.7 | 80.0 | 85.3 | 76.7 |
| BtCoV-HKU5.3 | 57.2 | 82.0 | 92.2 | 93.8 | 91.7 | 80.0 | 85.3 | 76.7 |
| BtCoV-HKU5.5 | 57.3 | 82.0 | 92.2 | 93.8 | 91.7 | 80.0 | 85.3 | 76.7 |
Genome similarities between coronavirus HCoV-EMC/2012 and coronaviruses BtCoV-HKU4 and BtCoV-HKU5.
BtCoV-HKU4 and BtCoV-HKU5 (8) are the closest relatives of HCoV-EMC/2012 for which full-length genome sequences are available (see above). Accordingly, comparison of the genomes of these three viruses revealed important similarities, including the organization of the “accessory protein genes,” ORF3a through ORF5, residing between the S protein gene and those encoding the E, M, and N proteins. Upon annotating this region of the BtCoV-HKU4 and BtCoV-HKU5 genomes, Woo et al. (8) identified the body TRSs for sg mRNA3, mRNA4, and mRNA5 but unfortunately did not follow standard coronavirus nomenclature, naming the downstream open reading frames ORF3a through ORF3d (encoding ns3a through ns3d) rather than ORF3, ORF4a, ORF4b, and ORF5 (Fig. 1A). The similarities of all ORFs and proteins of HCoV-EMC/2012, BtCoV-HKU4, and BtCoV-HKU5 were calculated, and percentages of sequence identity are summarized in Table 3. The lowest percentages of sequence identity to BtCoV-HKU4 and BtCoV-HKU5 were observed for ORF3 at the nucleotide level (46.4% and 46.0%, respectively) and for ORF4b at the amino acid level (23.5% and 25.9%, respectively). The highest percentages of sequence identity to BtCoV-HKU4 and BtCoV-HKU5 were observed for the E ORF at the nucleotide level (74.6% and 75.1%, respectively) and for the M ORF at the amino acid level (82.6% and 82.2%, respectively). These data further supported the characterization of HCoV-EMC/2012 as a close relative of BtCoV-HKU4 and BtCoV-HKU5.
TABLE 3 .
Percent identity between open reading frames of coronavirus HCoV-EMC/2012 and coronaviruses BtCoV-HKU4 and BtCoV-HKU5 at the nucleotide and amino acid levels
| Annotation in HCoV-EMC/2012 |
Annotation in BtCoV-HKU4 and BtCoV-HKU5a |
% identity to BtCoV-HKU4b |
% identity to BtCoV-HKU5b |
||
|---|---|---|---|---|---|
| nt | aa | nt | aa | ||
| ORF1ab | ORF1ab | 70.6 | 72.2 | 70.7 | 73.8 |
| S | S | 66.3 | 66.1 | 63.8 | 63.5 |
| ORF3 | NS3a | 46.4 | 34.9 | 46.0 | 31.4 |
| ORF4a | NS3b | 51.5 | 37.5 | 47.8 | 38.0 |
| ORF4b | NS3c | 35.1 | 23.5 | 45.2 | 25.9 |
| ORF5 | NS3d | 56.6 | 46.9 | 58.1 | 54.2 |
| E | E | 74.6 | 69.5 | 75.1 | 68.2 |
| M | M | 72.8 | 82.6 | 73.0 | 82.2 |
| N | N | 67.2 | 71.8 | 66.7 | 67.8 |
| ORF8b | Undescribed | 45.3 | 32.1 | 48.0 | 33.8 |
DISCUSSION
Coronaviruses have been known for quite some time as viruses that cause a variety of diseases in humans and animals (32, 33). The discovery of a coronavirus as the causative agent of SARS revived the interest in coronaviruses and resulted in a rapid increase of the number of identified coronaviruses, as well as of the number of full coronavirus genome sequences. Until this study, lineage C of the genus Betacoronavirus (formerly known as subgroup 2c) included virus isolates from bats. Here, we determined and analyzed the complete genome sequence of a previously unknown lineage C betacoronavirus that was isolated from the sputum of a 60-year-old male suffering from acute pneumonia and renal failure in the Kingdom of Saudi Arabia whose death was probably a consequence of this infection (24).
The sequencing of the full HCoV-EMC/2012 genome was greatly facilitated by the advent of high-throughput techniques. Using an optimized random amplification deep-sequencing approach, approximately 90% of the virus genome was covered with high accuracy in a single run. Using the data from this first run, primers could be designed to perform conventional Sanger sequencing for confirmation. This combination of techniques allowed the determination of the complete virus genome within a few days, without a requirement for prior knowledge of the virus genome under investigation. The error rate in 454 deep sequencing was generally higher than in Sanger sequencing, but the high coverage across the HCoV-EMC/2012 virus genome (up to 5,697 reads per nucleotide position) corrected for most of the incorrect base callings. The sequence obtained using the 454 platform aligned almost perfectly with that obtained by Sanger sequencing, with the exception of two nucleotide positions. The deep-sequencing data revealed variation at position 11623 (U or G), with G occurring in 44% of the reads, suggesting that ORF1a-encoded residue 3782 can be either valine (codon GUC) or glycine (codon GGC). The valine codon was the more abundant codon at this position in HCoV-EMC/2012, and valine is also present in most other betacoronaviruses. At position 27162, both G and A were detected in different runs, with an A in 45% of the reads. This G-to-A substitution introduces a premature stop codon (UGG to UAG) in ORF5. The virus stock that we sequenced was derived from passage of the virus from a sputum specimen six times in Vero cell culture. Hence, the observed sequence variants may reflect either natural heterogeneity or emerging genetic changes that occurred during virus passage in cell culture. Additional HCoV-EMC/2012 virus isolates or patient materials are currently not available to verify these genome sequence ambiguities at positions 11623 and 27162.
Adaptation to cell culture leading to a loss of functionality of genes, and in particular in relation to the so-called “accessory protein genes,” has previously been described for a variety of coronaviruses, including SARS-CoV (2, 34). These genes, like ORF3 through ORF5 of HCoV-EMC/2012, are dispersed between the structural protein genes (35) and in some cases may even overlap such a gene, as in the case of the ORF overlapping the N protein gene in betacoronaviruses (Fig. 1A) (23, 36). The origin of most accessory protein genes remains unclear, although for some, acquisition by recombination with cellular or heterologous viral sequences seems plausible (37, 38). Accessory gene functions have been probed by reverse genetics (knockout mutants) for a variety of coronaviruses, including SARS coronavirus (39), which established that they are not essential for replication in cell culture systems. In animal models, on the other hand, profound effects on pathogenesis after the inactivation (or transfer to a heterologous coronavirus) of accessory protein genes have been previously described (40–42). In some cases, accessory gene products have been implicated in immune evasion, e.g., by interfering with cellular innate immune signaling (43).
The apparent absence of selection pressure on coronavirus accessory protein genes during cell culture passage may explain the relatively high frequency with which loss of functionality appears to occur. The detection of an internal termination codon in part of the HCoV-EMC/2012 ORF5 sequences (45% of the reads) may constitute another example of such an event, which would lead to the truncation of the ORF5 protein after 107 amino acids. This would resemble a 29-nt deletion that occurred in the SARS-CoV genome, which resulted in the truncation of ORF8 (34, 44), and a 45-nt in-frame deletion in ORF7b of the same virus that emerged upon cell culture passage (23).
Our analysis identified a potential ORF underlying the N protein gene (ORF8a), which is a common feature in betacoronaviruses. This ORF was not previously described for BtCoV-HKU4 and BtCoV-HKU5 (8) but is conserved in the genome sequences of both viruses (see Fig. S1J in the supplemental material). Remarkably, in HCoV-EMC/2012, both the 5′ and 3′ parts of the ORF appear to have been truncated. In BtCoV-HKU4 and BtCoV-HKU5, the ORF8b AUG codon would be the second AUG on sg mRNA8, making leaky ribosomal scanning a likely mechanism for translation initiation. In HCoV-EMC/2012, however, this AUG codon (positions 28606 to 28608) seems to have been mutated to AUA. Conservation of the sequence immediately downstream of this position, which is now formally upstream of ORF8b in HCoV-EMC/2012, was observed with BtCoV-HKU4 and BtCoV-HKU5, suggesting that the putative loss of this AUG codon may also have been a relatively recent event. In the 3′ part of ORF8b, sequence alignment of HCoV-EMC/2012 with BtCoV-HKU4 and BtCoV-HKU5 suggests that the former acquired a premature termination codon at positions 29099 to 29101 (UAA). Although we cannot at present assess the timing of these events in HCoV-EMC/2012 evolution, due to the lack of alternative samples for this species, the presumed loss of ORF8b functionality may also be a consequence of virus passage in cell culture.
To classify newly identified coronaviruses as the prototype of a novel virus species, it is required that the amino acid sequence identity in the conserved replicase domains in all intervirus pairwise comparisons is below the 90% threshold (1). Here, we propose HCoV-EMC/2012 to represent a novel species of the betacoronavirus genus, since the amino acid sequence identities between HCoV-EMC/2012 and its closest relatives BtCoV-HKU4 and BtCoV-HKU5 in the seven conserved domains of ORF1ab were 75% and 77%, respectively. These viruses were originally detected in Asia in lesser bamboo bats (Tylonycteris pachypus) and Japanese house bats (Pipistrellus abramus), respectively (8). This proposed classification will remain provisional until approved by ICTV.
The ICTV guidelines for coronavirus species demarcation require the availability of a (nearly) complete genome sequence prior to virus classification. However, there is considerable correlation between the results based on full-genome sequence analysis and those determined using the most conserved part of the ORF1b-encoded RdRp domain, which is commonly used in screening for new coronaviruses. In 2010, this partial sequence was reported for a betacoronavirus (VM314/2008) that was isolated 2 years earlier from a Pipistrellus pipistrellus bat in The Netherlands. This virus was provisionally classified a betacoronavirus based on a 332-nt fragment from the RdRp-encoding domain of ORF1b (31), which shares 88% nucleotide sequence identity with HCoV-EMC/2012, the highest identity with any coronavirus sequence available in the public domain. Although this high similarity is not sufficient to resolve the taxonomic relation between HCoV-EMC/2012 and isolate VM314/2008, it suggests that they may both belong to the same coronavirus species. Establishing the genome sequence of VM314/2008, or closely related viruses, is urgently required to verify this hypothesis. Based on the genetic relation between HCoV-EMC/2012 and bat coronaviruses, it is tempting to speculate that HCoV-EMC/2012 emerged from bats—either directly or via an intermediate animal host, possibly Pipistrellus bats. This bat species is known to be present in the Kingdom of Saudi Arabia and neighboring countries.
Although most infections of human coronaviruses are relatively mild, the infection by HCoV-EMC/2012 with fatal outcome, and a similar severe case of an infection with a closely related coronavirus in London (25), is a reminder that certain coronaviruses may cause severe and sometimes fatal infections in humans. It is important to develop an animal model that can be used to fulfill Koch’s postulates for the novel virus, by demonstrating that the isolated virus can indeed cause the observed disease. The availability of the HCoV-EMC/2012 genome sequence will facilitate the development of a variety of diagnostic assays that can be used to study the prevalence and clinical impact of HCoV-EMC/2012 infections in humans. The first generation of assays for this purpose has recently been described (45). We anticipate that the availability of this full-length virus genome sequence will be valuable for the development of additional applied and fundamental research.
MATERIALS AND METHODS
Virus propagation.
Patient material had been subjected to passage in Vero cells four times in the Dr. Soliman Fakeeh Hospital, Jeddah, Saudi Arabia. Subsequently, in the Erasmus Medical Center, Rotterdam, The Netherlands, LLC-MK2 cells were inoculated with HCoV-EMC/2012 in minimal essential medium (MEM-Eagle) with Earle’s salts (BioWhittaker, Verviers, Belgium), supplemented with 2% serum, 100 U/ml penicillin, 100 mg/ml streptomycin, and 2 mM glutamine. Vero cells were inoculated with virus in Dulbecco’s modified Eagle medium (BioWhittaker) supplemented with 1% serum, 100 U/ml penicillin, 100 mg/ml streptomycin, and 2 mM glutamine. After inoculation, the cultures were incubated at 37°C in a CO2 incubator and checked daily for cytopathic changes. Three days after inoculation, supernatant from Vero cells was collected and used for virus genome characterization.
Arbitrarily primed PCR and virus genome sequencing.
To characterize the viral genome, we used a random amplification deep-sequencing approach as the first step. Virus-containing supernatant was centrifuged for 10 min at 3,000 rpm to remove cellular debris. This supernatant was then filtered through a 0.45-µm-pore-size centrifugal filter unit (Millipore, Amsterdam, The Netherlands) to minimize bacterial contamination. Omnicleave endonuclease (Epicenter Biotechnologies, Madison, WI) was used to remove any free DNA and RNA, according to the manufacturer’s protocol. Subsequently, viral RNA was extracted from the purified, infected cell culture supernatant using a High Pure RNA isolation kit (Roche Diagnostics, Almere, The Netherlands). To remove contaminating mammalian rRNA, a Ribo-Zero RZH110424 rRNA removal kit (Epicenter Biotechnologies, Madison, WI) was used according to the manufacturer’s protocol. RNA was reverse transcribed using circular permuted primers (46) that were extended with random hexamer sequences, namely, CCCACCACCAGAGAGAAAN(6), ACCAGAGAGAAACCCACCN(6), GAGAAACCCACCACCAGAN(6), GGAGGCAAGCGAACGCAAN(6), AAGCGAACGCAAGGAGGCN(6), and ACGCAAGGAGGCAAGCGAN(6). Per reaction, reverse transcription mixtures contained 6 µl RNA, 1 µl primer (20 pmol), 0.5 µl (20 U) RNase inhibitor (Promega, Leiden, The Netherlands), 1 µl (10 mM each) deoxynucleoside triphosphates (Roche), and 5 µl water. After a 5 min incubation at 65°C for optimal primer hybridization to the template, 4 µl (10×) first-strand buffer, 1 µl (200 U/µl) SuperScript III reverse transcriptase (Invitrogen, Bleiswijk, The Netherlands), 1 µl (0.1 M) dithiothreitol (DTT), and 0.5 µl (20 U) RNase inhibitor (Promega) were added to the mixture in a 20-µl volume. To obtain cDNA, the reverse transcription mixture was sequentially incubated at 25°C for 5 min and at 42°C for 1 h. After 3 min at 95°C and 2 min on ice, 1 µl Klenow DNA polymerase (5 U) (New England BioLabs Inc., Ipswich, MA) was added and the mixture was sequentially incubated at 25°C for 5 min, 37°C for 1 h, and 75°C for 20 min to obtain double-stranded cDNA. The cDNA was purified using a MinElute PCR purification kit (Qiagen, Venlo, The Netherlands) according to the instructions of the manufacturer. To amplify the purified cDNA, a PCR with the individual circular permuted primers without the random hexamer was performed. The PCR mixture contained 2 µl primer (40 pmol), 2 µl purified cDNA, 1.25 µl (10 mM each) deoxynucleoside triphosphate (Roche), 5 µl (10×) PfuUltra II Rxn buffer, and 1 µl (2.5 U) PfuUltra II DNA polymerase (Stratagene, Amsterdam, The Netherlands). Water was added to reach a final volume of 50 µl. The PCR mixture was incubated at 95°C for 2 min and then for 40 cycles of 95°C for 20 s, 56°C for 1 min, and 72°C for 2 min, followed by a final extension at 72°C for 10 min. Fragments were purified using a MinElute PCR purification kit (Qiagen) according to the instructions of the manufacturer.
Amplified fragments were sequenced using a 454/Roche GS Junior sequencing platform. A fragment library was created according to the manufacturer’s protocol without DNA fragmentation (GS FLX Titanium rapid library preparation; Roche), selecting for fragments larger than 100 bp. The emulsion-based PCR (emPCR) (amplification method Lib-L) and GS Junior sequencing run were performed according to the instructions of the manufacturer (Roche). The sequence reads were trimmed at 30 nt from the 3′ and 5′ ends to remove all primer sequences. Sequence reads were assembled into contigs using CLC Genomics 5.5.1 software (CLC Bio, Aarhus, Denmark). Using this deep-sequencing approach, approximately 90% of the virus genome sequence was obtained.
As a second step, specific primers were designed to amplify overlapping fragments of approximately 800 bp by RT-PCR. These PCR products were purified from agarose gels and sequenced using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands) and a 3130XL genetic analyzer (Applied Biosystems), according to the instructions of the manufacturers. The genomic 5′- and 3′-terminal sequences were determined using a FirstChoice RLM-RACE kit (Ambion, Bleiswijk, The Netherlands).
Virus classification.
Newly identified members of the family Coronaviridae are generally assigned by the ICTV to a subfamily and genus on the basis of rooted phylogeny and calculation of pairwise evolutionary distances for seven replicase polyprotein domains (1): the ADP-ribose 1″-phosphatase (ADRP) in nsp3, the coronavirus 3C-like (3 CL) protease (3CLpro, or “main protease”) in nsp5, the RNA-dependent RNA polymerase (RdRp) in nsp12, the helicase (Hel) in nsp13, the exoribonuclease (ExoN) in nsp14, the nidoviral endoribonuclease specific for U (NendoU) in nsp15, and the ribose-2′-O-methyltransferase (O-MT) in nsp16. Amino acid sequence alignments were generated for each of these domains using ClustalW within the BioEdit (version 7.0.5.3) (47) program and concatenated, after which the sequence identity of HCoV-EMC/2012 with closely related strains was calculated. For this purpose, the full genomes of 9 strains, derived from 3 species, belonging to Betacoronavirus lineage C were available.
To support virus classification, protein-based phylogenetic trees were generated. Multiple amino acid alignments, including sequences of HCoV-EMC/2012 and one representative of each of the 20 recognized species of the subfamily Coronavirinae, were produced for the following proteins, using the Viralis platform (48) followed by manual correction: ADRP, the N-terminal part of PLP2, TM1, Y domain, nsp4 to nsp16, and the C-terminal part of the spike (S) protein (S2), envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein. From each protein alignment, the most informative blocks (49) were extracted using the BAGG program (50), and only these strongly conserved alignment regions were used for further analyses. Two concatenated alignments were used. The first included replicase pp1ab protein regions (4,110 aa positions, gap content of 0.9%), and the second included regions in the C-terminal domain of the S protein (S2) and the E, M, and N proteins (1,127 aa positions, gap content of 3.9%). ProtTest version 3.2 (51) was used to select the best-fitting model of protein evolution. For both datasets, the LG model with rate heterogeneity (4 categories) ranked top among 112 models tested, with a relative weight of 0.98 under the Bayesian information criterion (BIC) and 0.74 under the corrected Akaike information criterion (AICc) for the first data set and 0.97/0.75 (BIC/AICc) for the second data set. Hence, this model was applied for maximum likelihood phylogeny reconstruction using PhyML version 3.0 (52).
Phylogenetic reconstruction.
Nucleotide sequences were aligned using the ClustalW software running within the BioEdit (version 7.0.5.3) (47) program and MAFFT version 6 (53). Maximum likelihood phylogenetic trees with 100 bootstrap replicates were estimated under the general time-reversible model (GTR) + I + Γ4 and the transversion model (TVM) + I + Γ4 (determined by ModelTest [54]), using PhyML 3.0 software (52). For both the 332-nt ORF1ab alignment that included isolate VM314/2008 and the alignment of the complete ORF1ab, the GTR + I + Γ4 model ranked top among 65 models tested, with relative weights of 0.8185 and 1.000 under AIC, respectively.
Nucleotide sequence accession number.
The final HCoV-EMC/2012 consensus sequence was submitted to GenBank under accession number JX869059.
SUPPLEMENTAL MATERIAL
Alignments of (poly)proteins encoded by the genome of HCoV-EMC/2012 and its two closest relatives, BtCoV-HKU4 and BtCoV-HKU5. Expert-curated multiple alignments for pp1ab (A), S protein (B), ORF3 protein (C), ORF4a protein (D), ORF4b protein (E), ORF5 protein (F), E protein (G), M protein (H), N protein (I), and the ORF8b product (J) are shown. (A) Amino acid residues in pp1ab between which cleavage by viral proteinases is predicted to occur are highlighted in gray. Download Figure S1, DOC file, 0.1 MB.
Primers designed to amplify ~800-nucleotide PCR fragments with 100-nucleotide overlaps covering the entire HCoV-EMC/2012 genome.
ACKNOWLEDGMENTS
We thank Igor Sidorov, Dmitry Samborskiy, and Alexander Kravchenko (partially supported through the MoBiLe program) for help and administration of the Viralis.
The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7) under EMPERIE grant agreement no. 223498 and SILVER grant agreement no. 260644.
Footnotes
Citation van Boheemen S, et al. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3(6):e00473-12. doi:10.1128/mBio.00473-12.
REFERENCES
- 1. de Groot RJ, et al. 2012. Family Coronaviridae, p. 806–828 In King AMQ, Adams MJ, Cartens EB, Lefkowitz EJ, Virus taxonomy, the 9th report of the international committee on taxonomy of viruses. Academic Press, San Diego, CA. [Google Scholar]
- 2. Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7:439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gloza-Rausch F, et al. 2008. Detection and prevalence patterns of group I coronaviruses in bats, northern Germany. Emerg. Infect. Dis. 14:626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lau SK, et al. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 102:14040–14045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li W, et al. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679 [DOI] [PubMed] [Google Scholar]
- 6. Pfefferle S, et al. 2009. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg. Infect. Dis. 15:1377–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vijaykrishna D, et al. 2007. Evolutionary insights into the ecology of coronaviruses. J. Virol. 81:4012–4020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Woo PC, et al. 2007. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 81:1574–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hamre D, Procknow JJ. 1966. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 121:190–193 [DOI] [PubMed] [Google Scholar]
- 10. McIntosh K, Dees JH, Becker WB, Kapikian AZ, Chanock RM. 1967. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. U. S. A. 57:933–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drosten C, et al. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J. Med. 348:1967–1976 [DOI] [PubMed] [Google Scholar]
- 12. Marra MA, et al. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399–1404 [DOI] [PubMed] [Google Scholar]
- 13. Peiris JS, Guan Y, Yuen KY. 2004. Severe acute respiratory syndrome. Nat. Med. 10:S88–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rota PA, et al. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399 [DOI] [PubMed] [Google Scholar]
- 15. Fouchier RA, et al. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. U. S. A. 101:6212–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van der Hoek L, et al. 2004. Identification of a new human coronavirus. Nat. Med. 10:368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Woo PC, et al. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zlateva KT, et al. 2012. No novel coronaviruses identified in a large collection of human nasopharyngeal specimens using family-wide CODEHOP-based primers. Arch. Virol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adams MJ, Carstens EB. 2012. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2012). Arch. Virol. 157:1411–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lauber C, Gorbalenya AE. 2012. Partitioning the genetic diversity of a virus family: approach and evaluation through a case study of picornaviruses. J. Virol. 86:3890–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Masters PS. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66:193–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Snijder EJ, et al. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zaki AM, et al. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J. Med., in press. [DOI] [PubMed] [Google Scholar]
- 25. Bermingham A, et al. 2012. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 17:pii=10290 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20290 [PubMed] [Google Scholar]
- 26. Ziebuhr J, Snijder EJ, Gorbalenya AE. 2000. Virus-encoded proteinases and proteolytic processing in the nidovirales. J. Gen. Virol. 81:853–879 [DOI] [PubMed] [Google Scholar]
- 27. Pasternak AO, Spaan WJ, Snijder EJ. 2006. Nidovirus transcription: how to make sense…? J. Gen. Virol. 87:1403–1421 [DOI] [PubMed] [Google Scholar]
- 28. Sawicki SG, Sawicki DL, Siddell SG. 2007. A contemporary view of coronavirus transcription. J. Virol. 81:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sola I, Mateos-Gomez PA, Almazan F, Zuñiga S, Enjuanes L. 2011. RNA-RNA and RNA-protein interactions in coronavirus replication and transcription. RNA Biol. 8:237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Firth AE, Brierley I. 2012. Non-canonical translation in RNA viruses. J. Gen. Virol. 93:1385–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reusken CB, et al. 2010. Circulation of group 2 coronaviruses in a bat species common to urban areas in Western Europe. Vector Borne Zoonotic Dis. 10:785–791 [DOI] [PubMed] [Google Scholar]
- 32. Holmes KV, Lai MC. 1996. Coronaviridae: the viruses and their replication, p. 1075–1093 In Fields BN, Knipe P, Howley PM, Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- 33. McIntosh K. 1996. Coronaviruses, p. 1095–1103 In Fields BN, Knipe P, Howley PM, Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- 34. Keng CT, et al. 2011. SARS coronavirus 8b reduces viral replication by down-regulating E via an ubiquitin-independent proteasome pathway. Microbes Infect. 13:179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Narayanan K, Huang C, Makino S. 2008. Coronavirus accessory proteins, p. 235–244 In Perlman S, Gallagher T, Snijder EJ, Nidoviruses. ASM Press, Washington, DC [Google Scholar]
- 36. Senanayake SD, Brian DA. 1997. Bovine coronavirus I protein synthesis follows ribosomal scanning on the bicistronic N mRNA. Virus Res. 48:101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mazumder R, Iyer LM, Vasudevan S, Aravind L. 2002. Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res. 30:5229–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Snijder EJ, den Boon JA, Horzinek MC, Spaan WJ. 1991. Comparison of the genome organization of toro- and coronaviruses: evidence for two nonhomologous RNA recombination events during Berne virus evolution. Virology 180:448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yount B, et al. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 79:14909–14922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cruz JL, et al. 2011. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog. 7:e1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Haan CA, Masters PS, Shen X, Weiss S, Rottier PJ. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pewe L, et al. 2005. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 79:11335–11342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Frieman M, et al. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 81:9812–9824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oostra M, de Haan CA, Rottier PJ. 2007. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J. Virol. 81:13876–13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cornman VM, et al. 2012. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 17:pii=20285 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20285 [DOI] [PubMed] [Google Scholar]
- 46. Welsh J, McClelland M. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 18:7213–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment Editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 48. Gorbalenya AE, et al. 2010. Practical application of bioinformatics by the multidisciplinary VIZIER consortium. Antiviral. Res. 87:95–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552 [DOI] [PubMed] [Google Scholar]
- 50. Antonov IV, Leontovich AM, Gorbalenya AE. 2008. BAGG—Blocks accepting gaps generator, version 1.0 http://www.genebee.msu.su/~antonov/bagg/cgi/bagg.cgi
- 51. Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 53. Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Posada D, Crandall KA. 1998. ModelTest: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignments of (poly)proteins encoded by the genome of HCoV-EMC/2012 and its two closest relatives, BtCoV-HKU4 and BtCoV-HKU5. Expert-curated multiple alignments for pp1ab (A), S protein (B), ORF3 protein (C), ORF4a protein (D), ORF4b protein (E), ORF5 protein (F), E protein (G), M protein (H), N protein (I), and the ORF8b product (J) are shown. (A) Amino acid residues in pp1ab between which cleavage by viral proteinases is predicted to occur are highlighted in gray. Download Figure S1, DOC file, 0.1 MB.
Primers designed to amplify ~800-nucleotide PCR fragments with 100-nucleotide overlaps covering the entire HCoV-EMC/2012 genome.