Summary
Communication between pre- and post-synaptic cells is a key process in the development and modulation of synapses. Reciprocal induction between pre- and postsynaptic cells involves regulation of gene transcription, yet the underlying genetic program remains largely unknown. To investigate how innervation-dependent gene expression in postsynaptic cells supports synaptic differentiation, we performed comparative microarray analysis of Drosophila muscles before and after innervation, and of prospero mutants, which show a delay in motor axon outgrowth. We identified 84 candidate genes that are potentially up- or downregulated in response to innervation. By systematic functional analysis, we found that one of the downregulated genes, longitudinals lacking (lola), which encodes a BTB-Zn-finger transcription factor, is required for proper expression of glutamate receptors. When the function of lola was knocked down in muscles by RNAi, the abundance of glutamate receptors (GluRs), GluRIIA, GluRIIB and GluRIII, as well as that of p-21 activated kinase (PAK), was greatly reduced at the neuromuscular junctions (NMJs). Recordings of the synaptic response revealed a decrease in postsynaptic quantal size, consistent with the reduction in GluR levels. Lola appears to regulate the expression of GluRs and PAK at the level of transcription, because the amount of mRNAs encoding these molecules was also reduced in the mutants. The transcriptional level of lola, in turn, is downregulated by increased neural activity. We propose that Lola coordinates expression of multiple postsynaptic components by transcriptional regulation.
Keywords: Synapse formation, Transcriptional regulation, Neuromuscular junction, Drosophila, longitudinals lacking (lola), Glutamate receptor
Introduction
During initial synapse formation, reciprocal interaction between innervating neurons and their targets are essential for assembly of synaptic components, cytoskeletal organization and activation of gene expression (Goda and Davis, 2003; Li and Sheng, 2003; McAllister, 2007). Similarly, mutual trans-synaptic signaling is important for activity-dependent refinement of differentiated synapses (Kandel, 2001; Flavell and Greenberg, 2008). While short-term changes in synaptic properties can be induced by modulation of pre-existing proteins and/or mRNAs, such as trafficking, modification and local translation, long-term changes require transcriptional control of gene expression. During the formation of vertebrate neuromuscular junctions (NMJs), signals from presynaptic motor neurons are necessary for the regulation of gene expression of postsynaptic transmitter receptors, which are acetylcholine receptors (AChRs) (Sanes and Lichtman, 2001; Schaeffer et al., 2001; Kummer et al., 2006). Many studies have focused on the role of immediate-early genes (IEGs) and CREB-mediated transcriptional regulation in long-term synaptic plasticity and memory formation (Flavell and Greenberg, 2008; Cohen and Greenberg, 2008). Nonetheless, a large gap still exists in our knowledge about how multiple molecular pathways integrate and orchestrate the development and plasticity of synapses. In particular, despite extensive work on activity-induced genes, very few studies have established functional links between these activity-induced genes and the downstream target genes that ultimately regulate synaptic properties.
In this study, we used the Drosophila NMJ as a model to study gene expression changes in postsynaptic muscle cells in response to presynaptic innervation. The Drosophila NMJ is a glutamatergic synapse expressing ionotropic glutamate receptors (GluRs) and contains a number of synaptic components commonly found in mammalian synapses, such as the postsynaptic density protein Discs-Large/PSD-95 (Keshishian et al., 1996; Griffith and Budnik, 2006). Previous studies showed that immediate–early transcription factors such as CREB and AP-1 regulate the strength and/or morphology of this synapse (Davis et al., 1996; Sanyal et al., 2002) (reviewed in Sanyal and Ramaswami, 2006). Signaling pathways mediated by secreted factors, such as Wnts and Bmps, are known to regulate anterograde and/or retrograde interaction between the motor neurons and muscles that are important for synaptic development (McCabe et al., 2003; Ataman et al., 2008; Korkut et al., 2009) (reviewed in Griffith and Budnik, 2006). However, the final targets of these signaling cascades—the molecules that directly regulate the changes in synaptic structure and function—remain largely unknown.
In this study, we performed genome-wide microarray analyses of specific muscle cells and identified 84 candidate genes whose expression changed in response to innervation. By systematic functional analyses of the candidate genes, we found that longitudinals lacking (lola), a gene downregulated by innervation, plays a prominent role in the transcriptional control of a number of postsynaptic components. lola encodes a BTB-Zn-finger transcription factor with a number of different isoforms (Goeke et al., 2003; Horiuchi et al., 2003). This transcription factor, Lola, has been implicated in a wide range of developmental and cellular processes including axon guidance, neural specification and tumorigenesis (Madden et al., 1999; Crowner et al., 2002; Goeke et al., 2003; Ferres-Marco et al., 2006; Spletter et al., 2007). Previous studies suggest that Lola may execute its function by directly binding to DNA and regulating the expression of the target genes. Here we show that postsynaptic Lola transcriptionally regulates the expression level of the glutamate receptors GluRIIA, GluRIIB and GluRIII, as well as p-21 activated kinase (PAK). We also show that the transcriptional level of lola is downregulated by increased neural activity. We propose that postsynaptic Lola functions as a transcription factor that controls synapse formation and/or maturation by regulating the expression of multiple synaptic components.
Materials and Methods
Fly stocks
For microarray analysis, we used an allele of prospero (prosM4) (Broadie and Bate, 1993b) and y, w. Forced expression and RNA interference (RNAi) analyses were performed using the GAL4-UAS system (Brand and Perrimon, 1993). Elav-gal4 (Luo et al., 1994), 24B-Gal4 (Brand and Perrimon, 1993) or G14-Gal4 (Shishido et al., 1998), or 5053A-Gal4 (Ritzenthaler et al., 2000) were used to induce expression in all neurons, all muscles, or in M12, respectively. UAS-RNAi lines were obtained from the Vienna Drosophila RNAi Center (VDRC) and Fly Stocks of the National Institute of Genetics (NIG) (Dietzl et al., 2007). Lines and alleles used for the systematic functional analyses are listed in supplementary material Table S2. Animals were raised at 29°C for RNAi analyses and at 25°C for other analyses. Alleles of lola, lolaORC46 (Crowner et al., 2002) and lolaORE119 (Goeke et al., 2003), were used.
Microarray Analysis
The collection of embryonic somatic muscles was performed as previously described (Inaki et al., 2007) with the following modifications. For collection of muscles at 18 hr after egg laying (AEL), the preparation was treated with 1 mg / ml collagenase (Sigma, St. Louis, Missouri) for ∼30 sec to weaken intersegmental muscle-muscle attachments. For chip analysis, we prepared three samples for each developmental stage or genotype: one sample was prepared from 200 collected cells and two from 50 collected cells. We performed two or three rounds of cRNA amplification before biotin labeling for samples prepared from 200 cells or those prepared from 50 cells, respectively. Synthesis of biotin-labeled cRNA was performed with the IVT Labeling Kit (Affymetrix, Santa Clara, CA). Hybridization of the fragmented and labeled cRNA to Affymetrix Drosophila Genome 2.0 Genechip arrays were performed according to the Genechip Expression Analysis technical manual (Affymetrix). Gene expression data analyses were performed using Affymetrix GeneChip Operating Software 1.4 (Affymetrix). By comparing gene expression signals between the samples prepared from the same number of cells, we obtained the Change Call (I, increase; D, decrease; NC, no change). After three pairs of comparisons, we selected the genes that displayed Change Call ‘I’ in all three pairs. We carried out Gene Ontology analyses by batch query using NetAffx (Affymetrix).
Real-time reverse-transcription PCR
Real-time reverse-transcription PCR (RT-PCR) assays were performed with ABI Prism 7000 SDS or Applied Biosystems StepOne Real Time PCR System (Applied Biosystems, Foster City, CA) with SYBR Green fluorescence according to the manufacturer protocol. We used as templates the second-round amplified cDNAs of wild type muscles and pros mutant muscles, prepared as described for microarray analyses. The gene expression values were normalized using the Myosin heavy chain (Mhc) gene as a reference. To examine gene expression in larval muscles, total RNA was extracted from the body wall of wall-climbing third-instar larvae using Sepasol RNA I Super (Nacalai, Kyoto, Japan) according to the manufacturer instructions. Approximately 2 µg of total RNA was reverse-transcribed into cDNA using oligo-dT primer and SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA). The obtained values were normalized using the Gapdh1 gene as a reference.
Immunohistochemistry
Immunohistochemical staining of dissected larvae and embryos was performed as described previously (Inaki et al., 2007). The following primary antibodies were used at the indicated concentrations: goat anti-horse radish peroxidase (HRP; Jackson, West Grove, PA, 1∶4000); rabbit anti-Lola polyclonal antibodies (1∶150) (Giniger et al., 1994); mouse monoclonal anti-GluRIIA (8B4D2) and mouse monoclonal anti-Dlg (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA, 1∶10 and 1∶50, respectively); rabbit anti-GluRIIB and rabbit anti-GluRIII (1∶2500, and 1∶5000, respectively) (Marrus et al., 2004); mouse Nc82 (1∶100) (Wagh et al., 2006); rabbit anti-Pak antibody (1∶500) (Sone et al., 2000).
Quantification of NMJ morphology and immunofluorescence signals
We quantified the morphology of the NMJ with Imaris software (Bitplane, Zurich, Switzerland). The number of boutons and branches and terminal length were scored based on the morphology visualized with anti-HRP staining as previously described (Coyle et al., 2004). The terminal length was normalized to muscle area. The bouton and branch number were also normalized to muscle area in the analyses of pst RNAi mutants because there was a slight change in muscle area in the mutants (surface area, 84.3±2.8 [n = 10] in pst-RNAi-8588R-4 [p<0.05] and 66.4±2.7 [n = 17] in pst-RNAi-107243 [p<0.05] compared to 75.0±3.0 [n = 32] in control, × 103 µm2). To assess the content of particular molecules at synapses, confocal images were projected to create a two-dimensional image and analyzed with IPLab software (Scanalytics, Fairfax, VA, USA). To control for relative intensity, experimental and control samples were stained in the same dish and imaged under identical conditions. We defined the synaptic area as delimited by HRP immunoreactivity on the muscle surface and the synaptic intensity of GluR, PAK or BRP as the average immunoreactive signals within the synaptic area. The total area of GluRIIA-, PAK- and BRP-positive clusters were defined by the pixels with an intensity above a threshold set arbitrarily, and were normalized by the total area of muscles. The GluRIIA expression level of the embryonic NMJ was defined as the total immunoreactive signals within the area of GluRIIA-positive clusters. For analysis of Dlg levels, we measured net intensity of immunoreactive signals normalized by the stained area. The extra-junctional GluR expression level was defined as the average immunoreactive signal within arbitrarily chosen regions covering ∼1% of muscle surface area. All statistical analyses were done by Student's t-test. All quantitative data in the larvae were obtained from muscle 6/7 NMJs in abdominal segment 2.
Immunoblot analysis
Two hundred first-instar larvae were ground in RIPA buffer containing protease inhibitors (Complete mini; Roche Diagnostics, Mannheim, Germany) as described by Thomas et al. (Thomas et al., 1997) and loaded onto 4%–12% SDS-PAGE gels (TEFCO, Tokyo, Japan). Immunoblotting was performed using mouse monoclonal anti-Tubulin (12G10; Developmental Studies Hybridoma Bank, 1∶2000), anti-GluRIIA (MAb 8B4D2 described above, 1∶50), goat anti-mouse IgG HRP conjugate (Jackson, 1∶1000) and donkey anti-goat IgG IRDye 680(LI-COR Biosciences, Lincoln, NE, USA, 1∶2500) and imaged and quantified by Odyssey infrared imaging system (LI-OCR). To control for loading, the level of GluRIIA was normalized to the level of Tubulin.
Electrophysiology
Intracellular recordings were made from muscle 6 in abdominal segments 3 and 4, and excitatory junctional potentials (EJPs) and miniature EJPs were recorded as described previously (Morimoto et al., 2010). HL3-containing medium at varying concentrations of extracellular Ca2+ were used (0.375 mM for mEJP recordings and 1.5 mM for EJP recordings). The peak amplitudes of ten EJPs were measured and averaged in each recording. All statistical analyses were done by Student's t-test.
Manipulation of neural activity with dTRPA1
Embryos of elav-Gal4/UAS-dTRPA1 (Pulver et al., 2009) were raised at 22°C, collected at 18 hr AEL and transferred to a PCR tube containing 25 µl of Halocarbon 700 oil (Sigma). Repetitive, spaced stimuli, consisting of 2-min intervals at a higher temperature (27°C) spaced by 5 min of rest (22°C) were applied in a PCR machine for 12 hrs. The larvae were then transferred to an agar plate and reared at 22°C until third instar. Control embryos (elav-Gal4/+) underwent the same temperature cycling.
Results
Gene expression changes in innervated muscle cells
In the Drosophila NMJ, synaptogenesis starts at ∼13 hr AEL, when motor neurons contact the target muscles for the first time (Broadie and Bate, 1993a). During the process of intimate interaction between the presynaptic motor neurons and the postsynaptic muscles, the synapses gradually mature and become fully functional by 17 hr AEL (Broadie and Bate, 1993a; Kohsaka et al., 2007). Using Affymetrix DNA microarrays, we performed two series of comparative gene expression analysis of muscles (Fig. 1A). First, we compared gene expression profiling of muscles before (12.75 hr AEL) and after (18 hr AEL) innervation (“before versus after” comparison). Second, we compared gene expression in muscles of wild-type versus prospero (pros) mutants; ∼80% of the muscles in pros mutants lack innervation but otherwise differentiate normally (“with versus without” comparison, at 18 hr AEL) (Broadie and Bate, 1993b). Genes that are differentially expressed in the first comparison would include genes that are up- or downregulated by innervation. However, these would also include genes that are involved in other aspects of muscle differentiation and whose expression is regulated at this stage, such as genes involved in the maturation of the muscle contraction apparatus. On the other hand, the second comparison would identify genes that are specifically regulated by innervation because the differentiation of muscle itself is normal in pros mutants (Broadie and Bate, 1993b). By conducting comparative analyses based on these two categories, we aimed to identify genes that function in synapse formation and whose expression is regulated by innervation.
Fig. 1. Microarray analyses of genes whose expression is regulated by innervation during synaptogenesis.
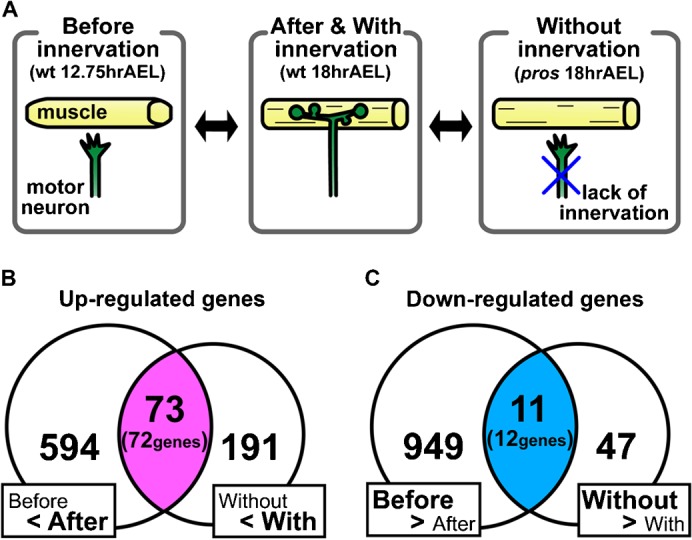
(A) Schematic drawings of the experimental design. (B,C) Venn diagrams showing the number of probe sets that displayed similar or differential regulation in two series of gene expression comparative analyses. For the probe sets that displayed similar regulation in the two comparisons, the number of the corresponding genes is also shown in parenthesis.
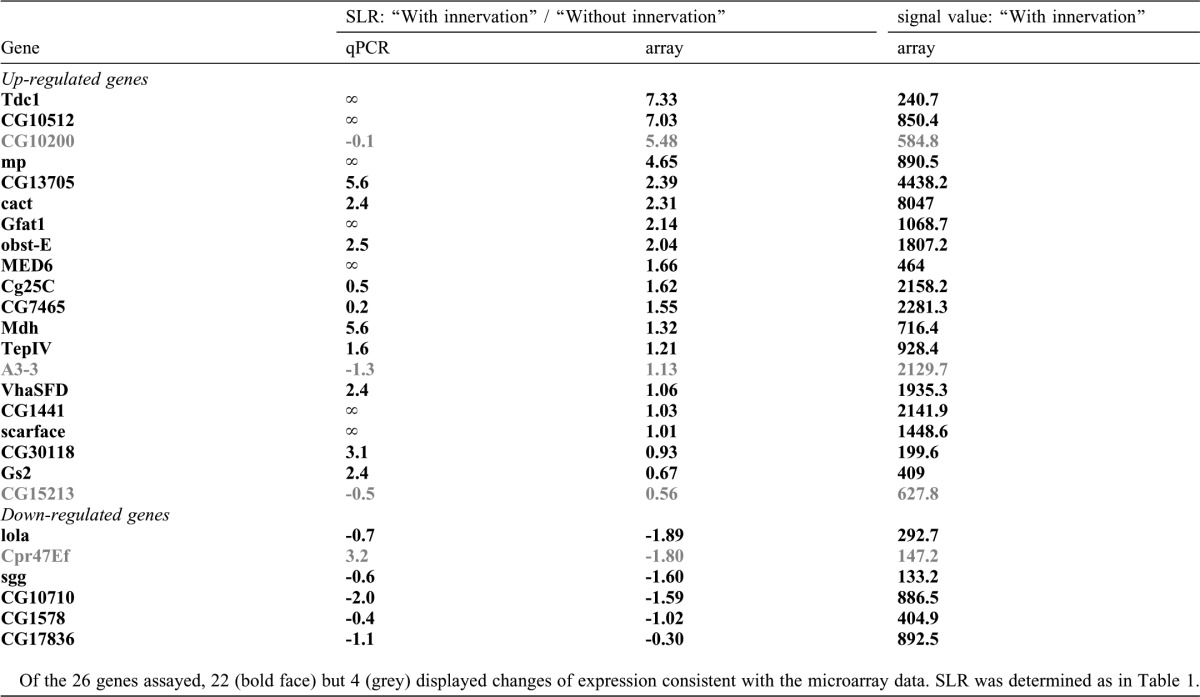
Using the single-cell collection technique that we developed previously (Inaki et al., 2007), we performed microarray analyses of four neighboring ventral muscles, M6, M7, M12 and M13, using Affymetrix Drosophila Genome chips (see Materials and Methods for detail). Because these muscles show similar morphology, run in parallel and attach to adjacent muscle insertion sites, they likely share common genetic programs for synapse formation. We therefore used a combination of the four muscles as a source for the microarray analyses. We prepared three independent samples each for the two comparative expression analyses described above (“before versus after” and “with versus without” innervation) and focused on genes that displayed differential expression reproducibly. We identified 72 genes that showed increased signals in muscles “after” and “with” innervation compared respectively to muscles “before” and “without” innervation. We defined these as “genes upregulated by innervation” (Fig. 1B; Table 1). We also identified 12 genes that showed decreased signals in the same comparisons and defined them as “genes downregulated by innervation” (Fig. 1C; Table 1). To validate the microarray results, we analyzed the expression of randomly-chosen subsets of the identified genes by quantitative RT-PCR (qPCR). Differential expression (“with vs. without”) was confirmed for 22 of the 26 genes assayed, indicating 85% concordance (Table 2).
Table 1. Genes whose expression is predicted to be regulated by nerve innervation.
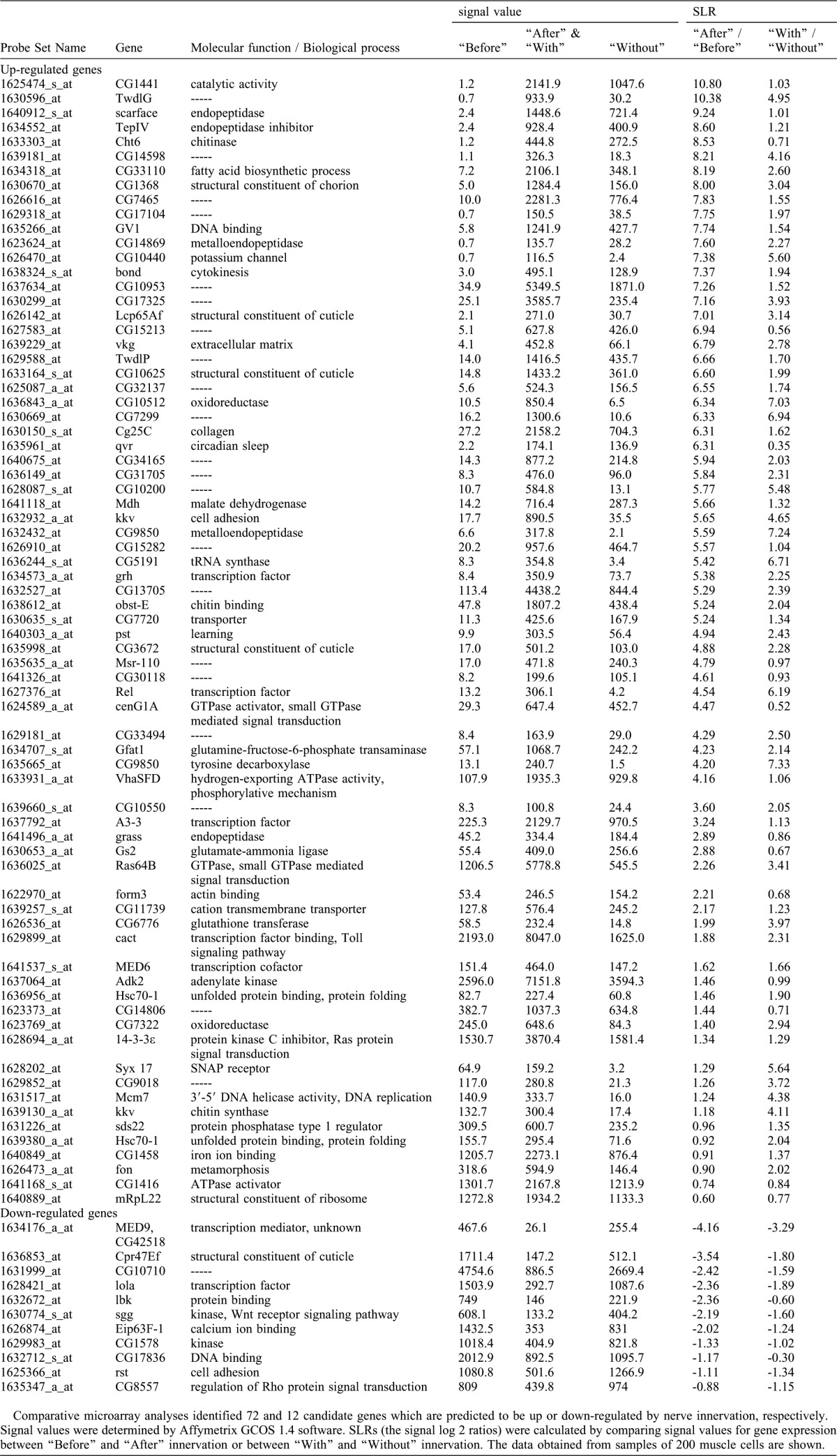
Table 2. Vertification of microarray data by qPCR.
pastrel is involved in synaptic growth
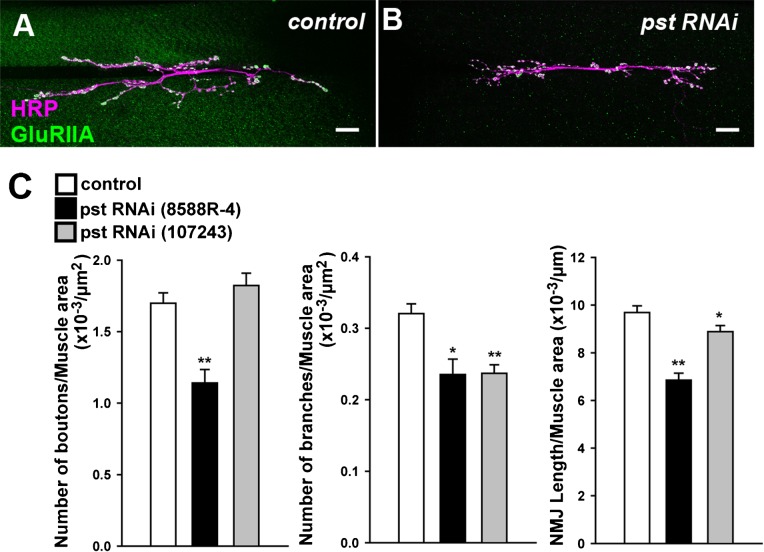
The 72 upregulated and 12 downregulated candidate genes described above were categorized into multiple functional classes of proteins (supplementary material Table S1). Among the upregulated genes was cactus (cact), one of the few genes known to function in muscles to regulate synaptic development and/or function (Cantera et al., 1999; Heckscher et al., 2007). To study whether other candidate genes also have roles in synaptic development, we performed loss-of-function (LOF) analyses on 37 of the candidate genes whose predicted molecular structure and/or function are related to synaptogenesis (supplementary material Table S2), using existing LOF mutant alleles and RNA interference (RNAi)-mediated gene knock-down. We visualized the NMJ by staining the presynaptic membrane (anti-HRP) and postsynaptic GluRIIA, and looked for changes in the terminal morphology and/or receptor clustering in the mutants. This screening revealed a function for pastrel (pst) in proper synaptic growth and lola in proper GluRIIA expression. No dramatic phenotypes were observed in the LOF mutants of the remaining genes examined.
RNAi knock-down of pastrel (pst) with two independent constructs that target different portions of the mRNA resulted in decreased bouton number, branch number and NMJ length (Fig. 2) (number of boutons per muscle area, 1.1±0.094 in pst-RNAi-8588R-4 [p<0.0005] versus 1.7±0.072 in control, × 10−3/µm2; number of branches per muscle area, 0.24±0.021 in pst-RNAi-8588R-4 [p<0.05] and 0.24±0.012 in pst-RNAi-107243 [p<0.0005] versus 0.32±0.014 in control, × 10−3/µm2; NMJ length per muscle area, 6.9±0.3 in pst-RNAi-8588R-4 [p<0.0005] and 8.9±0.3 in pst-RNAi-107243 [p<0.05] versus 9.7±0.3 in control, × 10−3 µm−1; n = 10, 17, 32).
Fig. 2. The role of pastrel (pst) in regulating NMJ outgrowth.
(A,B) Third-instar larval 6/7 NMJs immunostained with anti-HRP (magenta) and anti-GluRIIA (green) antibodies. (A) control, (B) pst RNAi. Scale bar = 20 µm. (C) Quantification of the phenotypes (*p<0.05, ** p<0.0005). Quantitative data presented in this and the following figures are presented as the mean value (± s.e.m.).
pst has been implicated in the processes of learning and memory (Dubnau et al., 2003). The expression of pst appears to be upregulated with innervation. Therefore, pst may be part of the genetic program that is induced by innervation to promote synaptic growth.
lola, downregulated by innervation, controls GluRIIA expression
The systematic functional analyses described above revealed that lola plays a critical role in the regulation of postsynaptic GluRIIA (Fig. 3). lola encodes a BTB-Zn-finger transcription factor with at least 20 unique protein isoforms, some of which are expressed in somatic muscles (Goeke et al., 2003). In our microarray screen, lola was identified as a gene downregulated by innervation; the expression level was 5-fold lower in 18-hr AEL compared to 12.75 hr AEL and 4-fold higher in the pros mutants compared to wild type (Table 1). qPCR analyses further confirmed differential expression: lola expression was 1.6-fold higher in pros mutants compared to the wild type (Table 2). Thus, lola expression appears to be downregulated by innervation. Lola protein was expressed in the nuclei of muscles in 12.75 hr and 18 hr embryos and in third instar larvae (supplementary material Fig. S1); see also Giniger et al. (Giniger et al., 1994).
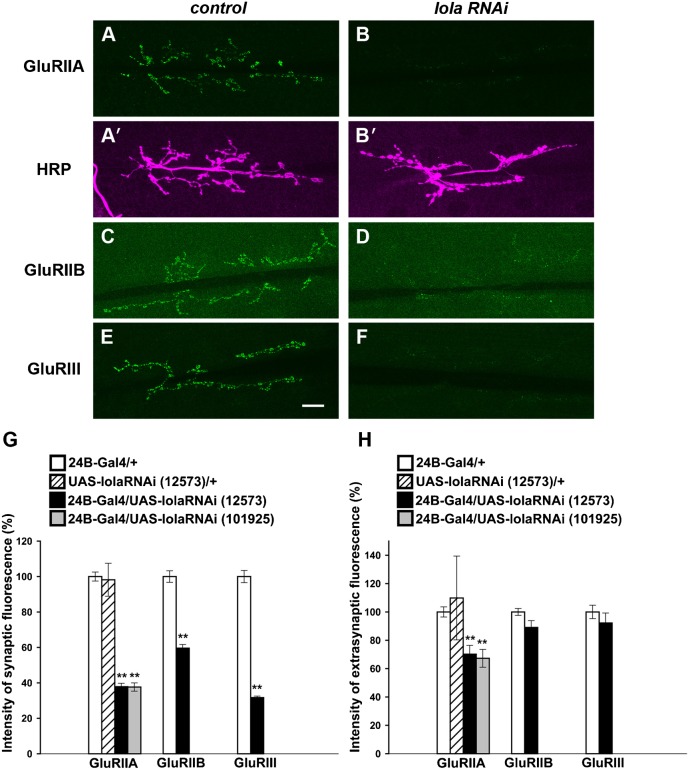
Fig. 3. Synaptic GluR abundance is decreased at the larval NMJ in lola RNAi mutants.
(A–F) Confocal images of NMJs stained for GluRIIA (A,B), GluRIIB (C,D), GluRIII (E,F) and HRP (A′,B′) in control (A,A′,C,E) or in lola RNAi mutants (B,B′,D,F). Scale bar = 20 µm. (G) Quantification of fluorescence intensity for GluRIIA, GluRIIB and GluRIII staining. (H) Extrasynaptic GluR levels in lola RNAi mutants. Data represent % of control fluorescence intensity. **p<0.0005.
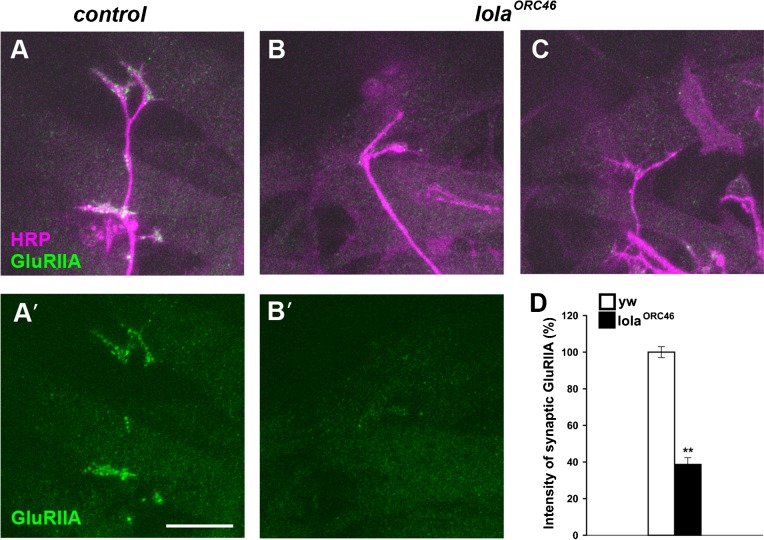
We knocked down lola expression in muscles using RNAi constructs. The level of lola mRNA in the body wall was reduced to 37±6.5% (n = 9) of control levels in the RNAi mutants, as assessed by qPCR analysis. We observed a dramatic decrease in GluRIIA abundance in NMJs in the mutants (Fig. 3A,B). This phenotype was seen when two independent RNAi constructs that target different regions of the mRNA were used. In contrast, there were no abnormalities in the morphology of the NMJs as assessed by anti-HRP staining (Fig. 3A′,B′, Fig. 6H). Thus, lola knock-down specifically alters GluRIIA abundance without affecting the area of the NMJs. We quantified the change in the abundance of GluRIIA by measuring the fluorescence intensity of GluRIIA at the NMJ as described previously (Albin and Davis, 2004). The abundance of GluRIIA decreased to ∼40% of control levels (Fig. 3G) (38±1.9% in lola-RNAi-12573 [n = 15] and 38±2.3% in lola-RNAi-101925 [n = 17] compared to 100±2.6% in 24B-Gal4/+ [n = 27] and 98±9.3% in UAS-lola-RNAi-12573/+ [n = 8]). Similar changes in GluRIIA abundance in the RNAi mutants were also observed at the late embryonic NMJ (20 hr AEL) (supplementary material Fig. S2). Furthermore, even more dramatic changes in GluRIIA abundance were observed in the lolaORC46 embryos (Fig. 4). lolaORC46 is a null allele and is embryonic lethal. In lolaORC46 embryos, motor axon projection and muscle patterning are partly deranged with some muscles entirely lacking nerve innervation (Madden et al., 1999). We therefore focused on muscles that received normally-sized nerve terminals (as visualized with a presynaptic marker, anti-HRP) and found a dramatic decrease in the density of synaptic GluRIIA (Fig. 4) (39±3.7 in lolaORC46 [n = 27] compared to 100±3.0% in yw [n = 101], p<0.0005). These data indicate that Lola is necessary for normal GluRIIA expression at the NMJ.
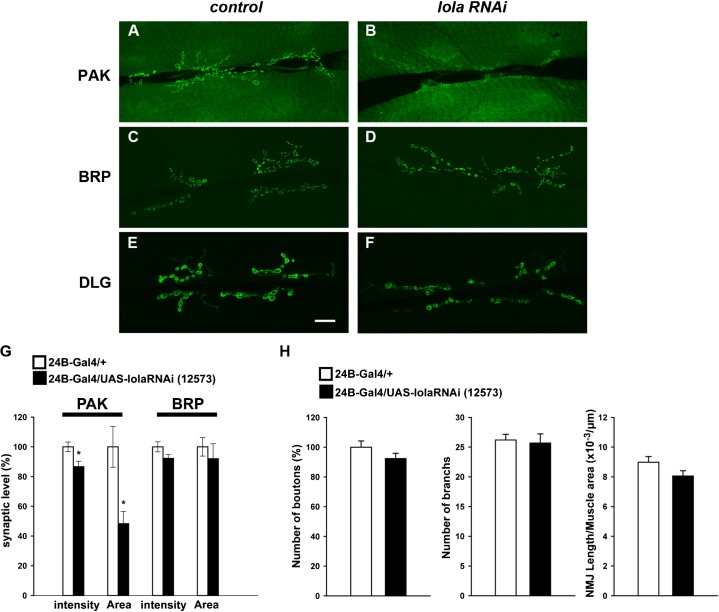
Fig. 6. Postsynaptic PAK and DLG abundance, but not presynaptic BRP abundance, are altered in lola RNAi mutants.
(A–F) Confocal images of NMJs stained for PAK (A,B) and BRP (C,D) and DLG (E,F) in control (A,C,E) and in lola RNAi mutants (B,D,F). Scale bar = 20 µm. (G) Quantification of the fluorescence intensity of PAK and BRP staining (*p<0.05). Data represent % of average fluorescence intensity among control animals. (H) Quantitative analysis of presynaptic morphology in lola RNAi mutants. The number of boutons represent % of the control value, which was 110.6±4.7.
Fig. 4. Synaptic GluRIIA abundance is decreased at the embryonic NMJs in lola mutants.
(A–C) Confocal images of NMJs in dorsal muscles (20 hr AEL) stained for GluRIIA (green) and HRP (magenta) in control (A,A′) and lolaORC46 (B,B′,C) embryos. Scale bar = 10 µm. (D) Quantification of the GluRIIA intensity within the HRP-positive NMJs. **p<0.0005.
The decrease in GluRIIA abundance at the NMJ could be due to the decrease in total GluRIIA expression in muscles or to defects in proper accumulation of GluRIIA at the NMJs. If the latter is the case, then an increase in the level of GluRIIA in the extra-junctional region would be expected. However, we did not detect any such increase in the RNAi mutants. Instead, fluorescence intensity of GluRIIA in the extra-junctional region was significantly decreased in lola RNAi mutants (Fig. 3H) (70±6.3 in 24B-Gal4/UAS-lola-RNAi-12573 [n = 15] and 67±6.3 in 24B-Gal4/UAS-lola-RNAi-101925 [n = 17] compared to 100±3.6% in 24B-Gal4/+ [n = 27] and 110±30 in UAS-lola-RNAi-12573/+ [n = 8]). Thus, postsynaptic lola knock-down appears to impair GluRIIA levels at the NMJ without altering the overall distribution of the receptors. We also examined the total amount of GluRIIA expression by immunoblotting. The total amount of GluRIIA protein in lola RNAi larvae was significantly reduced compared to control (Fig. 5) (expression in lola-RNAi [24B-Gal4/UAS-lola-RNAi-101925] was reduced to 35±10% of the control [24B-Gal4/+] level [n = 2]). These results strongly suggest that Lola is required for the control of total GluRIIA expression in muscles rather than its proper accumulation at the NMJs. Overexpression of lola in muscles driven by 24B-Gal4 did not induce obvious changes in NMJ morphology and level of GluRIIA expression (supplementary material Table S2).
Fig. 5. Total amount of GluRIIA protein expression is decreased in lola RNAi mutants.
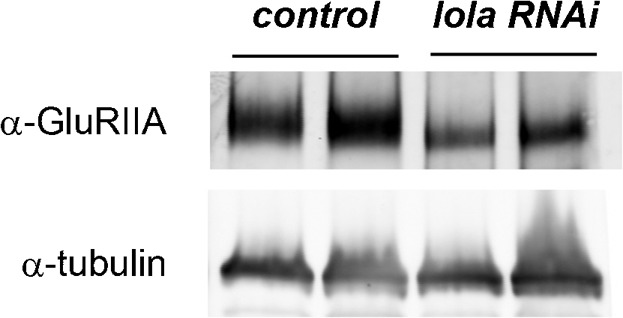
Two independent lysates from first instar larvae of control and lola RNAi mutants were loaded and analyzed by immunoblotting for GluRIIA and tubulin expression.
Lola is required for proper expression of GluRIIB, GluRIII and PAK
We next asked if Lola is required for proper expression of other GluR subunits, GluRIIB and GluRIII (Featherstone et al., 2005; Marrus et al., 2004; Petersen et al., 1997; Qin et al., 2005). As is the case for GluRIIA, the fluorescence intensity of GluRIIB and GluRIII at the NMJ was dramatically decreased in the mutants (Fig. 3C–G) (GluRIIB, 60±2.1[n = 15] compared to 100±3.3 [n = 12], p<0.0005; GluRIII, 32±0.8 [n = 12] compared to 100±3.4 [n = 12], p<0.0005; data represent % value in lola-RNAi-12573 compared to 24B-Gal4/+). The abundance of GluRIIB and GluRIII at extra-junctional muscle areas was not also increased (Fig. 3H) (GluRIIB, 89±4.8 [n = 15] compared to 100±2.5 [n = 12]; GluRIII, 92±7.0 [n = 12] compared to 100±4.7 [n = 12]). Thus, Lola appears to be required for proper expression of at least three GluR subunits—GluRIIA, GluRIIB and GlulRIII—at the NMJs.
We also examined the expression of other postsynaptic components. Pak kinase (PAK) colocalizes with GluRIIA at the post-synaptic density (PSD) and is involved in the PSD assembly (Albin and Davis, 2004; Parnas et al., 2001; Rasse et al., 2005). In lola RNAi mutants, the level of synaptic PAK was significantly reduced (Fig. 6A,B,G) (synaptic intensity, 87±3.7 [n = 17] compared to 100±3.2 [n = 16], p<0.05; expression field area, 48±8.2 [n = 17] compared to 100±13 [n = 16], p<0.05). We also found a significant reduction in synaptic levels of Dlg, a scaffolding protein highly enriched at the perisynaptic muscle membrane (Fig. 6E,F) (postsynaptic intensity, 85±2.0 [n = 22] compared to 100±2.4 [n = 24], p<0.0005). These data indicate that postsynaptic Lola is required for proper expression of a number of postsynaptic components.
Presynaptic terminals and active zones are normal in lola mutants
The lola RNAi mutants developed into normally sized third-instar larvae with no obvious behavioral abnormalities. No apparent abnormality was found in the morphology or size of muscles in the mutants (surface area, 82.7±2.9 [n = 19] in UAS-lola-RNAi compared to 86.8±2.9 [n = 22] in control, × 103 µm2 [p>0.05]). The morphology of the presynaptic terminals also appeared normal (Fig. 3A′,B′, Fig. 6H). There was no significant change in the bouton number, branch number or NMJ length (bouton number, 92±3.6 [n = 17] compared to 100±4.2% [n = 17]; branch number, 26±1.5 [n = 19] compared to 26±1.0 [n = 22]; NMJ length per muscle area, 8.1±0.4 [n = 19] compared to 9.0±0.4 [n = 22], × 10−3 µm−1; data represent values in 24B-Gal4/UAS-lola-RNAi-12573 compared to control [24B-Gal4/+]). We also examined the expression and distribution of an active zone marker Bruchpilot (BRP) (Wagh et al., 2006), and found no abnormality (Fig. 6C,D,G) (synaptic intensity, 92±2.6 [n = 10] compared to 100±3.4% [n = 16]; expression field area, 92±10 [n = 10] compared to 100±6.2% [n = 16]). Thus, despite the severe reduction in the expression of a number of postsynaptic components, the morphology and molecular expression of the apposing active zones appeared normal.
lola inhibition alters postsynaptic quantal size but not evoked response
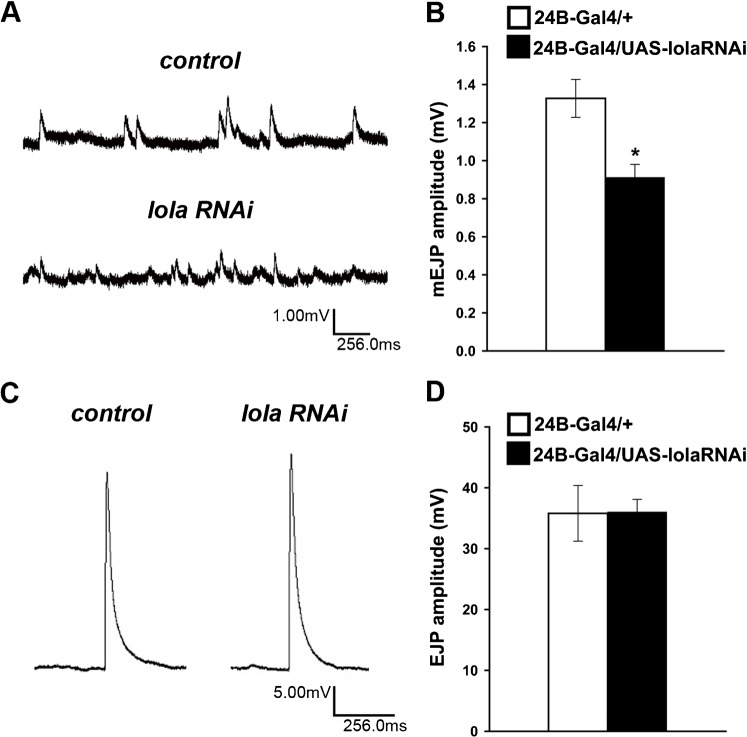
To investigate the physiological effects of the postsynaptic lola knock-down, we performed intracellular recordings from muscle 6 in the third-instar larvae. We tested the postsynaptic response to both spontaneous and evoked transmitter release. The mean amplitude of spontaneous miniature excitatory junctional potentials (mEJPs), or quantal size, is a measure of postsynaptic sensitivity to transmitters. A significant reduction in quantal size was observed in lola RNAi mutants (Fig. 7A,B) (0.91±0.068 mV in 24B-GAL4/UAS-lola-RNAi-12573 [n = 11] compared to 1.33±0.10 mV in 24B-Gal4/+ [n = 8], p<0.05). Previous studies suggested that the density of channels is an important determinant of the quantal size (Petersen et al., 1997). Therefore, our data are consistent with the decreased GluR expression observed in the imaging analysis. However, there was no significant change in the amplitude of excitatory junctional potential (EJP) in lola RNAi mutants, despite the decrease in the quantal size (Fig. 7C,D) (36.0±2.07 mV in lola-RNAi [n = 11] compared to 35.8±4.58 mV in control [n = 8]). Similar compensation for the decrease in quantal size has been reported for several mutants, including GluRIIA (Davis et al., 1998; Paradis et al., 2001; Petersen et al., 1997). These previous studies suggested that retrograde signals from the postsynaptic cells increased the presynaptic transmitter release to compensate for postsynaptic defects. The mechanism for the compensation in lola mutants remains to be determined.
Fig. 7. Quantal size of NMJs is decreased in lola RNAi mutants.
(A) Representative traces of spontaneous neurotransmitter release in 0.375 mM calcium in control (upper trace) and in lola RNAi mutants (lower trace). (B) Quantification of quantal size. *p<0.05. (C) Representative traces of evoked transmitter release in 1.5 mM calcium in control (left) and lola RNAi mutants (right). (D) Quantification of the amplitude of EJPs.
Lola regulates transcription level of GluRs and PAK
The imaging and physiological analyses described above show that postsynaptic Lola is necessary for normal expression of GluRs and other synaptic components. Since lola encodes a BTB-Zn-finger transcription factor (Goeke et al., 2003; Cavarec et al., 1997), Lola might regulate the transcriptional level of GluRs. To assess this possibility, we examined the level of GluR mRNAs in the mutants. We extracted total RNA from the larval body wall and performed real-time quantitative PCR. We found that the levels of GluRIIA and GluRIIB mRNAs in lola RNAi mutants are dramatically reduced (Fig. 8) (GluRIIA, 42±7.9 [n = 9] compared to 100±2.9 [n = 9], p<0.0005; GluRIIB, 56±14.4 [n = 9] compared to 100±6.4 [n = 9], p<0.05; data represent % values in lola-RNAi-12573 compared to control [24B-Gal4/+]). We also found a smaller but significant decrease in the level of GluRIII (75±8.7 [n = 9] compared to 100±3.5 [n = 9], p<0.05). Furthermore, we found a significant reduction of PAK mRNAs (59±6.4 [n = 9] compared to 100±2.9 [n = 9], p<0.0005). On the other hand, the mRNA transcript level of Dlg and Myosin heavy chain (Mhc), a protein involved in muscle contraction, was not affected (Dlg, 87±11.3 [n = 9] compared to 100±9.6 [n = 9]; Mhc,116±8.4 [n = 9] compared to 100±4.5 [n = 9]). These data indicate that Lola positively regulates the transcription level of glutamate receptors and PAK but not Dlg.
Fig. 8. The transcript levels of GluRs and PAK decreased in lola RNAi mutants.
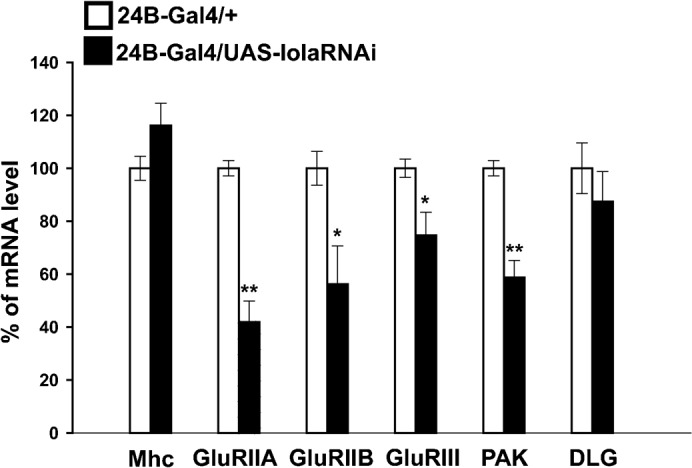
Real-time quantitative PCR analysis of the levels of GluRIIA, GluRIIB, GluRIII, PAK, DLG and Mhc mRNA in lola RNAi mutants. The transcript level of GluRIIA, GluRIIB, GluRIII and PAK, but not DLG and Mhc, were significantly reduced (n = 9, *p<0.05, **p<0.0005).
Neural activity induces changes in lola expression
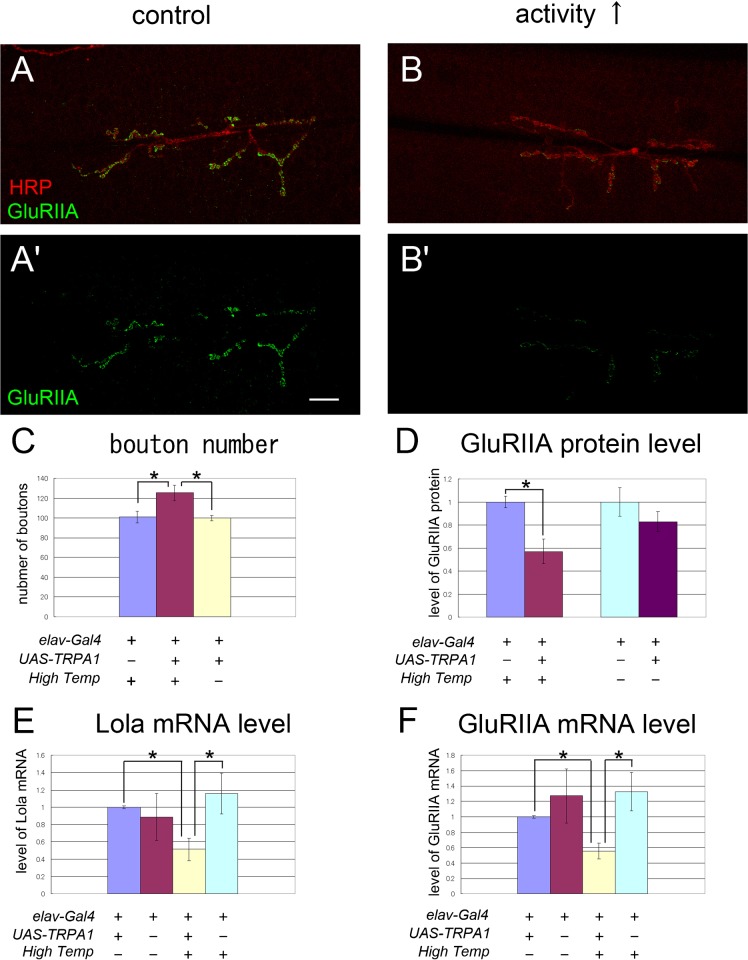
Having shown that Lola is involved in the transcriptional regulation of a number of synaptic components, we next asked how the expression of lola itself is regulated. Our microarray analyses showed that the level of lola transcript is changed by innervation. Therefore, one possibility is that neural activity modulates lola expression. To address this, we used the Drosophila warmth-activated cation channel TRPA1 (dTRPA1) (Pulver et al., 2009) to activate all neurons during late embryonic and early 1st instar larval stages and studied the effect on the expression of lola in the third instar larvae. We applied repetitive 2-min intervals at a higher temperature (27°C) spaced by 5 min of rest (22°C), for 12 hrs from AEL18. Such spaced induction of increased activity with Channelrhodopsin or high K+ has previously been shown to induce changes in synaptic structure and function in larval NMJs (Ataman et al., 2008). The spaced stimuli with dTRPA1 increased the number of boutons (Fig. 9A–C), indicating that our stimulation protocol successfully induced plastic changes in the NMJs. The stimulation protocol also decreased the level of lola mRNAs (Fig. 9E), indicating that neural activity negatively regulates lola expression. The levels of GluRIIA mRNA and protein were also decreased upon the activity elevation (Fig. 9D,F). These results suggest that increased neural activity downregulates the transcription level of lola and downstream GluRIIA.
Fig. 9. The transcript levels of lola and GluRIIA decreased after increased activity.
(A–B′) Confocal images of NMJs stained for HRP (A,B) and GluRIIA (A–B′) in control (A,A′) or in a larva stimulated with dTRPA (B,B′). Scale bar = 10 μm. (C,D) Quantification of the number of boutons and relative fluorescence intensity for GluRIIA staining. The number of boutons was increased and the level of GluRIIA was decreased upon dTRPA stimulation with pulses of high temperature shift (High Temp, *p<0.05). The temperature shift itself had no significant effect on these values in the control animals. (E,F) Real-time quantitative PCR analysis of the level of lola (E) and GluRIIA (F) mRNA. The transcript level of Lola and GluRIIA were significantly reduced by increased activity (*p<0.05). Data represent % of control values in C-F.
Discussion
Microarray analysis of genes regulated by innervation
We used the highly accessible and well-characterized Drosophila NMJ to study gene expression changes in the postsynaptic cells upon presynaptic innervation. We performed comparative genome-wide gene expression profiling of muscles by individually picking up the cells at specific developmental stages (before and after innervation) and in pros mutants that lack nerve innervation. Through two series of comparative expression profiling (“before versus after” and “with versus without” innervation), we identified genes whose expression is potentially up- or downregulated upon innervation (72 and 12 genes, respectively). Subsequent systematic functional analyses of the candidate genes identified two genes, pst and lola, that play roles in synaptic development. Muscle-specific knock-down of pst decreased the number of boutons and branches. Since manipulating the expression of pst in muscles affected the growth of the presynaptic terminals, some retrograde signals derived from postsynaptic muscles are likely to be involved in causing the phenotypes. Previous studies describe several molecules that act as muscle-secreted retrograde signals (e.g., the BMP ortholog Gbb) and coordinate synaptic growth (McCabe et al., 2003; McCabe et al., 2004). pst may function in the molecular pathway(s) that regulate the activity of such retrograde signals.
Lola as a regulator of multiple postsynaptic components
We found that another candidate gene identified by the microarray analyses, lola, plays a prominent role in the regulation of GluR abundance. lola encodes a transcriptional factor with a BTB-Zn-finger motif. Previous studies on the Drosophila NMJ identified several molecular pathways that regulate GluR abundance through post-transcriptional mechanisms. Subsynaptic protein synthesis, mediated by aggregates of protein synthesis machinery, affects the level of postsynaptic GluRIIA (Sigrist et al., 2000; Sigrist et al., 2003). NF-κB/Dorsal, IκB/Cactus and IRAK/Pelle colocalize postsynaptically around GluR clusters and regulate postsynaptic GluR density through unknown post-transcriptional mechanism(s) (Cantera et al., 1999; Heckscher et al., 2007). A serine threonine kinase, PAK, which colocalizes with GluRs, regulates GluR abundance via its interaction with the adaptor protein Dreadlocks (Dock) (Albin and Davis, 2004; Parnas et al., 2001). Unlike these molecules, Lola regulates the abundance of GluRs at the level of mRNA. When the function of lola was knocked down in muscles, the mRNA levels of both of the specific subunits, GluRIIA and the GluRIIB, as well as the common subunit GluRIII were decreased. The abundance of GluR proteins at the synapses was also decreased.
In addition to the three GluR genes, lola also regulates the transcriptional level of another postsynaptic component, PAK. Thus, lola is a potential candidate gene that may control synaptic function by coordinating transcription of multiple post-synaptic components. A similar role has been proposed for Lola during axon guidance. During midline crossing of commissural axons, Lola regulates the expression of the guidance cue Slit and its receptor Robo (Crowner et al., 2002). We also observed that synaptic accumulation of Dlg protein but not the level of its mRNA is abnormal in lola mutants. This is likely a secondary consequence of the change in the expression level of other synaptic regulators whose mRNA level is directly controlled by Lola. A strong candidate is PAK since PAK is known to regulate synaptic abundance of Dlg (Albin and Davis, 2004). Thus, Lola appears to regulate the expression of synaptic components both directly and indirectly.
It remains to be determined how Lola controls the transcriptional level of GluRs. Lola is a transcription factor that can activate gene expression by binding to specific DNA sequences in cultured cells (Cavarec et al., 1997). Thus, Lola may directly modulate the transcription level of GluRs by binding to the regulatory region of the genes. Alternatively, Lola may indirectly modulate GluR levels by regulating the transcription and/or activity of other molecules. lola is a highly complex genetic locus and encodes at least 20 different splice isoforms (Goeke et al., 2003; Horiuchi et al., 2003). Most isoforms have four common exons that contain a BTB or POZ domain and distinct exons that encode one or a pair of zinc-finger domains. The lola sequence that was used in the present RNAi analyses targets all Lola isoforms. While some RNAi lines that target specific Lola isoforms are available, none of them targets the isoforms that are expressed in muscles. Isoform-specific conventional mutants are available for two Lola isoforms (lolaORE4 and lolaORE50 for isoform K, and lolaORE119 for isoform L) (Goeke et al., 2003). Of these, isoform L is expressed in muscle cells, so we tested whether lolaORE119 mutants have defects in GluR abundance but found no abnormality. It is thus currently difficult to determine isoform-specific functions of Lola at the NMJ.
Possible role of Lola in activity-dependent changes in postsynaptic gene expression
The results of the microarray and qPCR analyses showed that the expression level of lola is downregulated by innervation. lola expression was downregulated after innervation and upregulated in pros mutants, which lack innervation. Similarly, increasing neural activity decreased the transcriptional level of lola. Thus, lola mRNA expression appears to be negatively regulated by innervation and neural activity. Since the expression of GluRs and other synaptic components are in turn regulated by lola, lola may link changes in neural activity to changes in postsynaptic gene expression. Consistent with this view, increased neural activity decreased the expression of the GluRIIA transcript and protein, concomitant with the decrease in lola expression. It is however currently unknown whether lola is solely responsible for the changes in GluRIIA expression in the activity misregulated NMJs. We also could not detect obvious changes in the expression of Lola protein in the aneural muscles (in pros mutants) or in activity-misregulated muscles (in dTRPA1 expressing larvae). Role of lola in coordinating activity-dependent changes in postsynaptic components remains to be explored in future studies.
How lola expression is regulated by neural innervation and/or activity also remains to be determined. Signaling mediated by PKA and CaMKII has been previously implicated in activity-dependent regulation of GluRIIA expression (Davis et al., 1998; Haghighi et al., 2003; Kazama et al., 2003; Morimoto-Tanifuji et al., 2004). Whether lola is regulated by these or other activity-dependent signaling mechanisms will be an interesting question to address in the future.
The negative regulation of lola by innervation and increased activity is somewhat counterintuitive in light of the fact that lola positively regulates the level of GluR and other postsynaptic components. The synaptic expression level of GluRs is known to increase after innervation (Broadie and Bate, 1993c; Chen and Featherstone, 2005). Some protocols that increase larval activity are known to induce an increase in GluRIIA expression (Schuster, 2006). Increasing motoneuron activity also increases their terminal size (Ataman et al., 2008; Budnik et al., 1990). One possibility is that Lola is part of the homeostatic mechanism(s) that maintain a proper level of synaptic strength. It has been shown that a decrease in postsynaptic function is compensated by an increase in presynaptic synaptic release at the Drosophila NMJs (Davis, 2006; Dickman and Davis, 2009; Frank et al., 2009). Similarly, Lola may control the appropriate level of postsynaptic components according to the level of presynaptic activity. For example, when presynaptic activity is too strong, expression of lola may be downregulated to reduce the level of GluRs at the postsynaptic cells. Conversely, when presynaptic innervation drops to an insufficient level in pros mutants, lola may be upregulated to compensate for the decreased transmission by increasing the level of GluRs.
Another possibility is involvement in the pruning of ectopic synapses. During the formation of neuromuscular connectivity in the embryos, reduction in electric activity in motor neurons results in an increase in ectopic synapses on non-target muscles (Jarecki and Keshishian, 1995; Carrillo et al., 2010). While involvement of presynaptic mechanisms has been shown by a recent study (Carrillo et al., 2010), postsynaptic Lola may also contribute to the regulation of ectopic synapses. For example, lola expression may be downregulated after innervation by proper motor neurons, to prevent the formation of ectopic synapses. Possible roles of lola in these processes will be determined in future studies.
Several studies suggest that Lola can regulate the chromatin structure and function in an epigenetic manner (Ferres-Marco et al., 2006; Zhang et al., 2003). Thus, Lola may be involved in the epigenetic control of synaptic plasticity, as shown in other systems (Guan et al., 2004; Levenson and Sweatt, 2005). Identification of Lola as a regulator of multiple postsynaptic components will provide a unique entry point to the understanding of transcriptional regulation of synapse formation and plasticity at the Drosophila NMJ.
Supplementary Material
Acknowledgments
We are grateful to E. Giniger, T. Aigaki, A. DiAntonio, M. Saitoe, A. Hofbauer, C. Hama, Bloomington and Kyoto Stock Center, Vienna Drosophila RNAi Center (VDRC), Fly Stocks of the National Institute of Genetics (NIG), The Exelixis Collection at the Harvard Medical School, and the Developmental Studies Hybridoma Bank for fly stocks and reagents. We acknowledge H. Miyakawa and members of the Nose lab for helpful discussion, H. Kohsaka for help in staining of the embryos, Y. Fujioka, H. Kato and M. Sotodate for technical assistance. This study was supported by a Grant-in-Aid for Scientific Research B and for Scientific Research on Priority Areas-Molecular Brain Science-from the MEXT to A.N.
Footnotes
Competing interests: We declare that there are no competing interests.
References
- Albin S. D., Davis G. W. (2004). Coordinating structural and functional synapse development: postsynaptic p21-activated kinase independently specifies glutamate receptor abundance and postsynaptic morphology. J. Neurosci. 24, 6871–6879 10.1523/JNEUROSCI.1538-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataman B., Ashley J., Gorczyca M., Ramachandran P., Fouquet W., Sigrist S., Budnik V. (2008). Rapid activity-dependent modifications in synaptic structure and function require bidirectional Wnt signaling. Neuron 57, 705–718 10.1016/j.neuron.2008.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Broadie K. S., Bate M. (1993a). Development of the embryonic neuromuscular synapse of Drosophila melanogaster. J. Neurosci. 13, 144–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadie K., Bate M. (1993b). Muscle development is independent of innervation during Drosophila embryogenesis. Development 119, 533–543. [DOI] [PubMed] [Google Scholar]
- Broadie K., Bate M. (1993c). Innervation directs receptor synthesis and localization in Drosophila embryo synaptogenesis. Nature 361, 350–353 10.1038/361350a0 [DOI] [PubMed] [Google Scholar]
- Budnik V., Zhong Y., Wu C. F. (1990). Morphological plasticity of motor axons in Drosophila mutants with altered excitability. J. Neurosci. 10, 3754–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantera R., Kozlova T., Barillas-Mury C., Kafatos F. C. (1999). Muscle structure and innervation are affected by loss of Dorsal in the fruit fly, Drosophila melanogaster. Mol. Cell. Neurosci. 13, 131–141 10.1006/mcne.1999.0739 [DOI] [PubMed] [Google Scholar]
- Carrillo R. A., Olsen D. P., Yoon K. S., Keshishian H. (2010). Presynaptic activity and CaMKII modulate retrograde semaphorin signaling and synaptic refinement. Neuron 68, 32–44 10.1016/j.neuron.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavarec L., Jensen S., Casella J. F., Cristescu S. A., Heidmann T., Chen K. (1997). Molecular cloning and characterization of a transcription factor for the copia retrotransposon with homology to the BTB-containing lola neurogenic factor. Mol. Cell. Biol. 17, 482–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Featherstone D. E. (2005). Discs-large (DLG) is clustered by presynaptic innervation and regulates postsynaptic glutamate receptor subunit composition in Drosophila. BMC Biol. 3, 1 10.1186/1741-7007-3-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S., Greenberg M. E. (2008). Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annu. Rev. Cell Dev. Biol. 24, 183–209 10.1146/annurev.cellbio.24.110707.175235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle I. P., Koh Y. H., Lee W. C., Slind J., Fergestad T., Littleton J. T., Ganetzky B. (2004). Nervous wreck, an SH3 adaptor protein that interacts with Wsp, regulates synaptic growth in Drosophila. Neuron 41, 521–534 10.1016/S0896-6273(04)00016-9 [DOI] [PubMed] [Google Scholar]
- Crowner D., Madden K., Goeke S., Giniger E. (2002). Lola regulates midline crossing of CNS axons in Drosophila. Development 129, 1317–1325. [DOI] [PubMed] [Google Scholar]
- Davis G. W. (2006). Homeostatic control of neural activity: from phenomenology to molecular design. Annu. Rev. Neurosci. 29, 307–323 10.1146/annurev.neuro.28.061604.135751 [DOI] [PubMed] [Google Scholar]
- Davis G. W., Schuster C. M., Goodman C. S. (1996). Genetic dissection of structural and functional components of synaptic plasticity. III. CREB is necessary for presynaptic functional plasticity. Neuron 17, 669–679 10.1016/S0896-6273(00)80199-3 [DOI] [PubMed] [Google Scholar]
- Davis G. W., DiAntonio A., Petersen S. A., Goodman C. S. (1998). Postsynaptic PKA controls quantal size and reveals a retrograde signal that regulates presynaptic transmitter release in Drosophila. Neuron 20, 305–315 10.1016/S0896-6273(00)80458-4 [DOI] [PubMed] [Google Scholar]
- Dickman D. K., Davis G. W. (2009). The schizophrenia susceptibility gene dysbindin controls synaptic homeostasis. Science 326, 1127–1130 10.1126/science.1179685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Dubnau J., Chiang A. S., Grady L., Barditch J., Gossweiler S., McNeil J., Smith P., Buldoc F., Scott R., Certa U. et al. (2003). The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 13, 286–296 10.1016/S0960-9822(03)00064-2 [DOI] [PubMed] [Google Scholar]
- Featherstone D. E., Rushton E., Rohrbough J., Liebl F., Karr J., Sheng Q., Rodesch C. K., Broadie K. (2005). An essential Drosophila glutamate receptor subunit that functions in both central neuropil and neuromuscular junction. J. Neurosci. 25, 3199–3208 10.1523/JNEUROSCI.4201-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferres-Marco D., Gutierrez-Garcia I., Vallejo D. M., Bolivar J., Gutierrez-Aviño F. J., Dominguez M. (2006). Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature 439, 430–436 10.1038/nature04376 [DOI] [PubMed] [Google Scholar]
- Flavell S. W., Greenberg M. E. (2008). Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu. Rev. Neurosci. 31, 563–590 10.1146/annurev.neuro.31.060407.125631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank C. A., Pielage J., Davis G. W. (2009). A presynaptic homeostatic signaling system composed of the Eph receptor, ephexin, Cdc42, and CaV2.1 calcium channels. Neuron 61, 556–569 10.1016/j.neuron.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Tietje K., Jan L. Y., Jan Y. N. (1994). lola encodes a putative transcription factor required for axon growth and guidance in Drosophila. Development 120, 1385–1398. [DOI] [PubMed] [Google Scholar]
- Goda Y., Davis G. W. (2003). Mechanisms of synapse assembly and disassembly. Neuron 40, 243–264 10.1016/S0896-6273(03)00608-1 [DOI] [PubMed] [Google Scholar]
- Goeke S., Greene E. A., Grant P. K., Gates M. A., Crowner D., Aigaki T., Giniger E. (2003). Alternative splicing of lola generates 19 transcription factors controlling axon guidance in Drosophila. Nat. Neurosci. 6, 917–924 10.1038/nn1105 [DOI] [PubMed] [Google Scholar]
- Griffith L., Budnik V. (2006). Plasticity and second messengers during synapse development. Int. Rev. Neurobiol. 75, 237–265 10.1016/S0074-7742(06)75011-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J. S., Haggarty S. J., Giacometti E., Dannenberg J. H., Joseph N., Gao J., Nieland T. J., Levenson J. M., O'Riordan K. J., Brown K. D. et al. (2004). Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 279, 40545–40559 10.1074/jbc.M404920200 [DOI] [PubMed] [Google Scholar]
- Haghighi A. P., McCabe B. D., Fetter R. D., Palmer J. E., Hom S., Goodman C. S. (2003). Retrograde control of synaptic transmission by postsynaptic CaMKII at the Drosophila neuromuscular junction. Neuron 39, 255–267 10.1016/S0896-6273(03)00427-6 [DOI] [PubMed] [Google Scholar]
- Heckscher E. S., Fetter R. D., Marek K. W., Albin S. D., Davis G. W. (2007). NF-kappaB, IkappaB, and IRAK control glutamate receptor density at the Drosophila NMJ. Neuron 55, 859–873 10.1016/j.neuron.2007.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T., Giniger E., Aigaki T. (2003). Alternative trans-splicing of constant and variable exons of a Drosophila axon guidance gene, lola. Genes Dev. 17, 2496–2501 10.1101/gad.1137303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaki M., Yoshikawa S., Thomas J. B., Aburatani H., Nose A. (2007). Wnt4 is a local repulsive cue that determines synaptic target specificity. Curr. Biol. 17, 1574–1579 10.1016/j.cub.2007.08.013 [DOI] [PubMed] [Google Scholar]
- Jarecki J., Keshishian H. (1995). Role of neural activity during synaptogenesis in Drosophila. J. Neurosci. 15, 8177–8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel E. R. (2001). The molecular biology of memory storage: a dialogue between genes and synapses. Science 294, 1030–1038 10.1126/science.1067020 [DOI] [PubMed] [Google Scholar]
- Kazama H., Morimoto-Tanifuji T., Nose A. (2003). Postsynaptic activation of calcium/calmodulin-dependent protein kinase II promotes coordinated pre- and postsynaptic maturation of Drosophila neuromuscular junctions. Neuroscience 117, 615–625 10.1016/S0306-4522(02)00923-5 [DOI] [PubMed] [Google Scholar]
- Keshishian H., Broadie K., Chiba A., Bate M. (1996). The drosophila neuromuscular junction: a model system for studying synaptic development and function. Annu. Rev. Neurosci. 19, 545–575 10.1146/annurev.ne.19.030196.002553 [DOI] [PubMed] [Google Scholar]
- Kohsaka H., Takasu E., Nose A. (2007). In vivo induction of postsynaptic molecular assembly by the cell adhesion molecule Fasciclin2. J. Cell Biol. 179, 1289–1300 10.1083/jcb.200705154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkut C., Ataman B., Ramachandran P., Ashley J., Barria R., Gherbesi N., Budnik V. (2009). Trans-Synaptic Transmission of Vesicular Wnt Signals through Evi/Wntless. Cell 139, 393–404 10.1016/j.cell.2009.07.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer T. T., Misgeld T., Sanes J. R. (2006). Assembly of the postsynaptic membrane at the neuromuscular junction: paradigm lost. Curr. Opin. Neurobiol. 16, 74–82 10.1016/j.conb.2005.12.003 [DOI] [PubMed] [Google Scholar]
- Levenson J. M., Sweatt J. D. (2005). Epigenetic mechanisms in memory formation. Nat. Rev. Neurosci. 6, 108–118 10.1038/nrn1604 [DOI] [PubMed] [Google Scholar]
- Li Z., Sheng M. (2003). Some assembly required: the development of neuronal synapses. Nat. Rev. Mol. Cell Biol. 4, 833–841 10.1038/nrm1242 [DOI] [PubMed] [Google Scholar]
- Luo L., Liao Y. J., Jan L. Y., Jan Y. N. (1994). Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8, 1787–1802 10.1101/gad.8.15.1787 [DOI] [PubMed] [Google Scholar]
- Madden K., Crowner D., Giniger E. (1999). LOLA has the properties of a master regulator of axon-target interaction for SNb motor axons of Drosophila. Dev. Biol. 213, 301–313 10.1006/dbio.1999.9399 [DOI] [PubMed] [Google Scholar]
- Marrus S. B., Portman S. L., Allen M. J., Moffat K. G., DiAntonio A. (2004). Differential localization of glutamate receptor subunits at the Drosophila neuromuscular junction. J. Neurosci. 24, 1406–1415 10.1523/JNEUROSCI.1575-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister A. K. (2007). Dynamic aspects of CNS synapse formation. Annu. Rev. Neurosci. 30, 425–450 10.1146/annurev.neuro.29.051605.112830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe B. D., Marqués G., Haghighi A. P., Fetter R. D., Crotty M. L., Haerry T. E., Goodman C. S., O'Connor M. B. (2003). The BMP homolog Gbb provides a retrograde signal that regulates synaptic growth at the Drosophila neuromuscular junction. Neuron 39, 241–254 10.1016/S0896-6273(03)00426-4 [DOI] [PubMed] [Google Scholar]
- McCabe B. D., Hom S., Aberle H., Fetter R. D., Marques G., Haerry T. E., Wan H., O'Connor M. B., Goodman C. S., Haghighi A. P. et al. (2004). Highwire regulates presynaptic BMP signaling essential for synaptic growth. Neuron 41, 891–905 10.1016/S0896-6273(04)00073-X [DOI] [PubMed] [Google Scholar]
- Morimoto-Tanifuji T., Kazama H., Nose A. (2004). Developmental stage-dependent modulation of synapses by postsynaptic expression of activated calcium/calmodulin-dependent protein kinase II. Neuroscience 128, 797–806 10.1016/j.neuroscience.2004.07.018 [DOI] [PubMed] [Google Scholar]
- Morimoto T., Nobechi M., Komatsu A., Miyakawa H., Nose A. (2010). Subunit-specific and homeostatic regulation of glutamate receptor localization by CaMKII in Drosophila neuromuscular junctions. Neuroscience 65, 1284–1292 10.1016/j.neuroscience.2009.11.059 [DOI] [PubMed] [Google Scholar]
- Paradis S., Sweeney S. T., Davis G. W. (2001). Homeostatic control of presynaptic release is triggered by postsynaptic membrane depolarization. Neuron 30, 737–749 10.1016/S0896-6273(01)00326-9 [DOI] [PubMed] [Google Scholar]
- Parnas D., Haghighi A. P., Fetter R. D., Kim S. W., Goodman C. S. (2001). Regulation of postsynaptic structure and protein localization by the Rho-type guanine nucleotide exchange factor dPix. Neuron 32, 415–424 10.1016/S0896-6273(01)00485-8 [DOI] [PubMed] [Google Scholar]
- Petersen S. A., Fetter R. D., Noordermeer J. N., Goodman C. S., DiAntonio A. (1997). Genetic analysis of glutamate receptors in Drosophila reveals a retrograde signal regulating presynaptic transmitter release. Neuron 19, 1237–1248 10.1016/S0896-6273(00)80415-8 [DOI] [PubMed] [Google Scholar]
- Pulver S. R., Pashkovski S. L., Hornstein N. J., Garrity P. A., Griffith L. C. (2009). Temporal dynamics of neuronal activation by Channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. J. Neurophysiol. 101, 3075–3088 10.1152/jn.00071.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Schwarz T., Kittel R. J., Schmid A., Rasse T. M., Kappei D., Ponimaskin E., Heckmann M., Sigrist S. J. (2005). Four different subunits are essential for expressing the synaptic glutamate receptor at neuromuscular junctions of Drosophila. J. Neurosci. 25, 3209–3218 10.1523/JNEUROSCI.4194-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasse T. M., Fouquet W., Schmid A., Kittel R. J., Mertel S., Sigrist C. B., Schmidt M., Guzman A., Merino C., Qin G. et al. (2005). Glutamate receptor dynamics organizing synapse formation in vivo. Nat. Neurosci. 8, 898–905. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler S., Suzuki E., Chiba A. (2000). Postsynaptic filopodia in muscle cells interact with innervating motoneuron axons. Nat. Neurosci. 3, 1012–1017 10.1038/79833 [DOI] [PubMed] [Google Scholar]
- Sanes J. R., Lichtman J. W. (2001). Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat. Rev. Neurosci. 2, 791–805 10.1038/35097557 [DOI] [PubMed] [Google Scholar]
- Sanyal S., Ramaswami M. (2006). Activity-dependent regulation of transcription during development of synapses. Int. Rev. Neurobiol. 75, 287–305 10.1016/S0074-7742(06)75013-9 [DOI] [PubMed] [Google Scholar]
- Sanyal S., Sandstrom D. J., Hoeffer C. A., Ramaswami M. (2002). AP-1 functions upstream of CREB to control synaptic plasticity in Drosophila. Nature 416, 870–874 10.1038/416870a [DOI] [PubMed] [Google Scholar]
- Schaeffer L., de Kerchove d'Exaerde A., Changeux J. P. (2001). Targeting transcription to the neuromuscular synapse. Neuron 31, 15–22 10.1016/S0896-6273(01)00353-1 [DOI] [PubMed] [Google Scholar]
- Schuster C. M. (2006). Experience-dependent potentiation of larval neuromuscular synapses. Int. Rev. Neurobiol. 75, 307–322 10.1016/S0074-7742(06)75014-0 [DOI] [PubMed] [Google Scholar]
- Shishido E., Takeichi M., Nose A. (1998). Drosophila synapse formation: regulation by transmembrane protein with Leu-rich repeats, CAPRICIOUS. Science 280, 2118–2121. [DOI] [PubMed] [Google Scholar]
- Sigrist S. J., Thiel P. R., Reiff D. F., Lachance P. E., Lasko P., Schuster C. M. (2000). Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature 405, 1062–1065 10.1038/35016598 [DOI] [PubMed] [Google Scholar]
- Sigrist S. J., Reiff D. F., Thiel P. R., Steinert J. R., Schuster C. M. (2003). Experience-dependent strengthening of Drosophila neuromuscular junctions. J. Neurosci. 23, 6546–6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M., Suzuki E., Hoshino M., Hou D., Kuromi H., Fukata M., Kuroda S., Kaibuchi K., Nabeshima Y., Hama C. (2000). Synaptic development is controlled in the periactive zones of Drosophila synapses. Development 127, 4157–4168. [DOI] [PubMed] [Google Scholar]
- Spletter M. L., Liu J., Liu J., Su H., Giniger E., Komiyama T., Quake S., Luo L. (2007). Lola regulates Drosophila olfactory projection neuron identity and targeting specificity. Neural. Dev. 2, 14 10.1186/1749-8104-2-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas U., Kim E., Kuhlendahl S., Koh Y. H., Gundelfinger E. D., Sheng M., Garner C. C., Budnik V. (1997). Synaptic clustering of the cell adhesion molecule fasciclin II by discs-large and its role in the regulation of presynaptic structure. Neuron. 4, 787–799 10.1016/S0896-6273(00)80961-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh D. A., Rasse T. M., Asan E., Hofbauer A., Schwenkert I., Dürrbeck H., Buchner S., Dabauvalle M. C., Schmidt M., Qin G. et al. (2006). Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49, 833–844 10.1016/j.neuron.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Zhang W., Wang Y., Long J., Girton J., Johansen J., Johansen K. M. (2003). A developmentally regulated splice variant from the complex lola locus encoding multiple different zinc finger domain proteins interacts with the chromosomal kinase JIL-1. J. Biol. Chem. 278, 11696–11704 10.1074/jbc.M213269200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.