Abstract
To determine the evolutionary relationships among all members of the genus Atractylodes (Compositae, Cardueae), we conducted molecular phylogenetic analyses of one nuclear DNA (nrDNA) region (internal transcribed spacer, ITS) and one chloroplast DNA (cpDNA) region (intergenic spacer region of trnL-F). In ITS and ITS + trnL-F trees, all members of Atractylodes form a monophyletic clade. Atractylodes is a sister group of the Carlina and Atractylis branch. Atractylodes species were distributed among three clades: (1) A. carlinoides (located in the lowest base of the Atractylodes phylogenetic tree), (2) A. macrocephala, and (3) the A. lancea complex, including A. japonica, A. coreana, A. lancea, A. lancea subsp. luotianensis, and A. chinensis. The taxonomic controversy over the classification of species of Atractylodes is mainly concentrated in the A. lancea complex. In base on molecular results, the intraspecific division of Atractylodes lancea is not supported, and A. coreana should be treated as a synonym A. chinensis.
Keywords: Atractylodes, ITS, phylogenetics, plastid DNA, trnL-F
1. Introduciton
Atractylodes is an important medicinal genus in East Asia. With over 2000 years of medicinal history, all of its species have been used as medicine except for A. carlinoides. Their rhizomes are generally referred to as “Zhu” in ancient herbal literature and are clinically divided into “Cangzhu” and “Baizhu” in Traditional Chinese Medicine [1]. In China, Japan, South Korea and North Korea, Atractylodes is a traditional medicine for treating gastroduodenal diseases.
Although Atractylodes was established by Candolle in 1838 [2], the relationship between Atractylodes plants and other related genera should be clarified. This genus was considered by some taxonomists as Atractylis prior to its establishment [3–5]. After establishment of the genus, many taxonomists still placed it in Atractylis[3]. The sexuality of flowers is the main feature that distinguished Atractylodes from Atractylis. Atractylodes are dioecious and Atractylis are monoecious [5,6]. With further studies of numerous specimens and field observations, Atractylodes flowers were found to be either monoecious, as in A. macrocephala and A. carlinoides, or gynodioecious, as in A. lancea, A. japonica and A. coreana. The gynodioecism of Atractylodes can be attributed to the degeneration of stamens so that the monoecious flowers become pistillate. Sometimes the degeneration presents a continuous transition [7]. Based on morphologic observation, Atractylodes lancea and A. japonica are gynodioecious, whereas A. macrocephala is monoecious [7,8], similar to Atractylis. Based on its morphologic characteristics, Petit identified the closest relationship between Atractylis and Atractylodes and attributed two species (Atractylodes comosa and Atractylodes cuneata) in the Mediterranean area to Atractylodes[9]. Therefore, the relationship of Atractylodes with its related genera, Atractylis, should be examined.
On the other hand, the classification of interspecific and infraspecific taxa in Atractylodes is much debated [2–5]. As recorded in the “Flora Reipublicae Popularis Sinicae”, the genus has five species, namely, Atractylodes macrocephala, A. lancea, A. carlinoides, A. japonica and A. coreana[10–12]. However, as stipulated by the Chinese Pharmacopoeia, Cangzhu is derived from Atractylodes lancea and A. chinensis (a synonym of A. lancea in “Flora Reipublicae Popularis Sinicae”). In addition, the relationship between A. coreana and A. chinensis is controversial [13]. Hu et al.[14] published Atractylodes lancea subsp. luotianensis, whereas Zou et al.[13] suggested that A. lancea subsp. luotianensis should be included in A. lancea rather than be treated as a separate subspecies. However, as recorded in the “Flora of China” [15], Atractylodes comprises only four species: Atractylodes carlinoides, A. macrocephala, A. coreana and A. lancea. A. japonica was treated as a synonym of A. lancea. In fact, the rhizome of Atractylodes japonica is used as Cangzhu in China, and is used as Baizhu in Japanese Pharmacopoeia [16]. Controversies over the classification of species and infraspecific categories of Atractylodes affect the circulation and identification of medicinal materials. Hence, studies on the genetic relationships among Atractylodes species are conducive to understanding the relationship among different taxa, thereby confirming the classification of plants in this taxon.
The findings in molecular systematics in recent years, especially genealogy trees built using gene sequences, have provided concrete evidence for the phylogeny of Compositae [17,18]. Some scholars have already established the genetic relationship of Atractylodes, which, however, failed to cover all the taxa of Atractylodes[13,19,20]. The limited number of individuals in each taxon limits the available experimental materials; hence, the genealogy of Atractylodes is incomplete. In this research, internal transcribed spacer (ITS) and trnL-F were used to determine the genealogy of Atractylodes because ITS and trnL-F sequences have provided satisfying results in other taxa of Compositae [21–23]. Samples of all Atractylodes species, including the controversial species and subspecies, were collected. The main purpose of the present study aims to establish a complete molecular systematic genetic relationship of Atractylodes (1) to ascertain the systematic relationship between Atractylodes and Atractylis (2) to determine the genetic relationship among the different taxa of Atractylodes.
2. Results and Discussion
2.1. The Features of Alignments in cpDNA (trnL-F) and nrDNA (ITS)
The newly sequenced ITS and chloroplast trnL-F from 49 Atractylodes samples are obtained (GenBank accession numbers are given in the Appendix Table 1). The sequences characteristics and phylogenetic information generated by the chloroplast trnL-F, nuclear ITS, and the combined ITS + trnL-F sequences is given in Table 1.
Table 1.
Comparison of Phylogenetic Information for Atractylodes Species from internal transcribed spacer (ITS), trnL-F, and combines data sets.
| Parameter | ITS | trnL-F | Combines |
|---|---|---|---|
| Number of accession | 49 | 49 | 49 |
| Range of sequence length (bp) | 642–643 | 799–801 | 1442–1443 |
| Characters of data matrix | 643 | 800 | 1443 |
| Number of constant sites (%) | 584 (90.82%) | 793 (99.13%) | 1377 (96.09%) |
| Number of parsimony-information sites (%) | 55 (8.55%) | 7 (0.88%) | 62 (4.32%) |
| Number of autopomorphic-information sites (%) | 4 (0.62%) | 0 (0.00%) | 4 (0.28%) |
| Consistency index (CI) | 0.892 | 0.969 | 0.891 |
| Retention index (RI) | 0.955 | 0.958 | 0.947 |
| Rescaled consistency index (RC) | 0.852 | 0.928 | 0.844 |
| Homoplasy index (HI) | 0.108 | 0.031 | 0.109 |
2.2. The Relationship between Atractylodes and Related Genera
The results of the partition homogeneity test (ILD) was p = 0.10, which indicates that the two datasets (ITS and trnL-F) show no significant incongruence. We applied Maximum parsimony (MP), Maximum likelihood (ML), and Bayesian Inference (BI) methods to conduct the phylogenetic analysis of the ITS and trnL-F sequences.
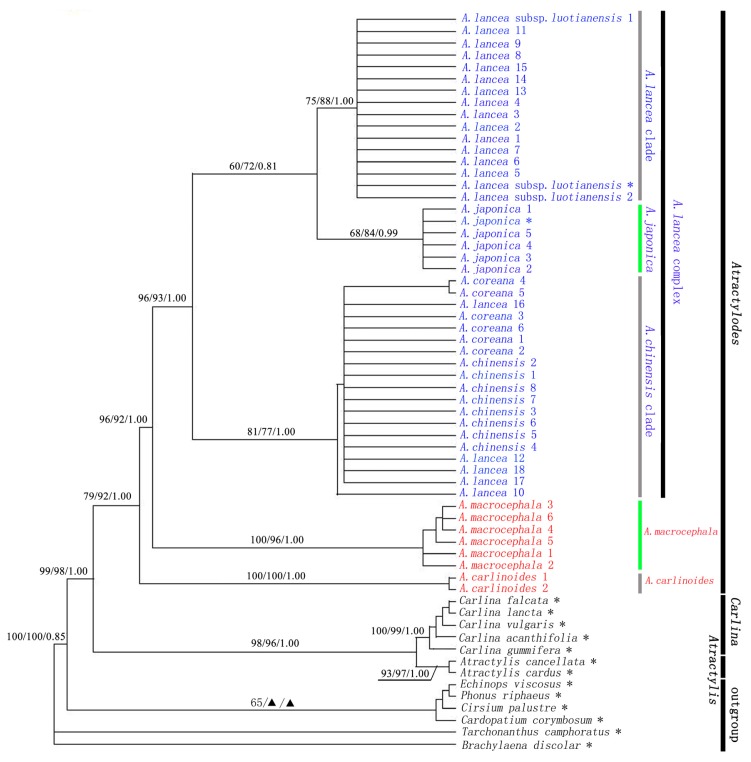
ITS and ITS + trnL-F phylogenetic trees, constructed based on three commonly used phylogenetic analysis methods (MP, ML, and BI) were very similar in terms of topologic structure, and only differed in bootstrap support, as shown in Figure 1,S1. The phylogenetic trees show that Atractylis and Carlina gathered into a clade, with higher bootstrap support rates (ITS + trnL-F: MP-BS/ML-BS/BI-PP = 98/96/1.00, Figure 1; ITS: MP-BS/ML-BS/BI-PP = 82/92/1.00, Figure S1; trnL-F: MP-BS/ML-BS/BI-PP = 83/95/1.00, Figure S2). Atractylodes is a sister group of the Atractylis and Carlina clade (ITS + trnL-F: MP-BS/ML-BS/BI-PP = 99/98/1.00, Figure 1; ITS: MP-BS/ML-BS/BI-PP = 85/96/1.00, Figure S1; trnL-F: MP-BS/ML-BS/BI-PP = 90/93/1.00, Figure S2). Although Atractylodes presents a relatively close genetic relationship with Atractylis in terms of morphologic characteristics [9,10]. However, in terms of biotypes and chemical composition, Atractylodes has obvious differences from Atractylis. Atractylodes is distributed in the Korean Peninsula, Japan, Russia, and other areas in Central China, in East Asia. Atractylodes is an endemic genus in the East Asia. Atractylis is geographically distributed in the Mediterranean area. The Carlina genus is distributed in the Canary Islands, the Mediterranean, and in temperate areas of Europe and Asia. Carlina is distributed in the Altay Area in Xinjiang Province, China, and is only represented by one species, Carlina biebersteinii. Carlina and Atractylis share a relatively close genetic relationship which is consistent with their geographic distribution.
Figure 1.
The 50% majority rule consensus tree from Bayesian Inference (BI) analysis of the combined ITS and trnL-F dataset; Maximum parsimony and ML bootstrap values and Bayesian posterior probabilities are shown above branches (MP/ML/BI). ▲ indicates ≤ 50% support. Taxa with * were downloaded from GenBank. Among the Atractylodes, the taxon with red color represent red flower, and the blue colored have white flower. Taxa with green color have obvious petiole, and gray colored have no petiole.
2.3. Atractylodes is a Monophyletic Group
According to our phylogenetic analyses, all taxon of Atractylodes formed a well-supported clade, with high support rates (ITS + trnL-F: MP-BS/ML-BS/BI-PP = 79/92/1.00, Figure 1; ITS: MP-BS/ML-BS/BI-PP = 83/95/1.00, Figure S1; trnL-F: ML-BS = 0.96, Figure S2). It was shown that Atractylodes is a monophyletic group.
2.4. Atractylodes carlinoides is Sister Species of the Rest of Atractylodes Species
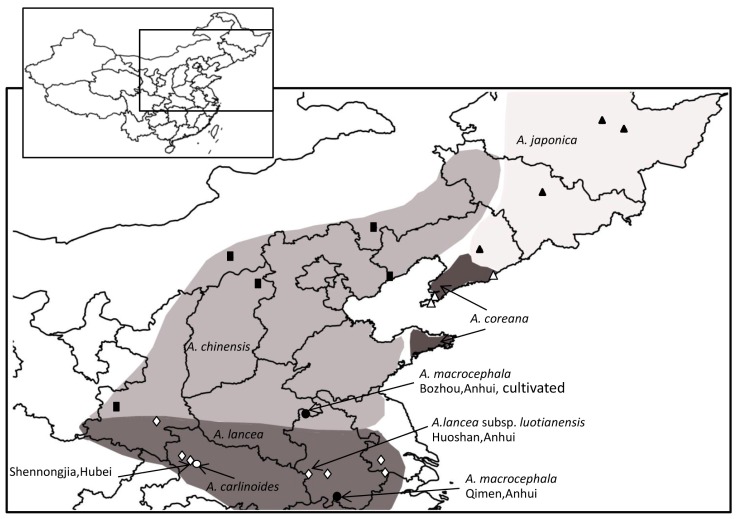
Atractylodes carlinoides is endemic to Shennongjia and its vicinities (Figure 2). Previous studies were often lacking research due to the narrow distribution of Atractylodes carlinoides[19,20,24]. Morphologically, Atractylodes carlinoides presents significant differences from other Atractylodes species such as the presence of basal leaves in mature plants, the uniramous aerial stem, and the underdeveloped rhizome [6]. According to ITS and ITS + trnL-F phylogenetic trees, Atractylodes can be divided into three branches: A. carlinoides, A. macrocephala, and A. lancea complex. Among them, Atractylodes carlinoides is sister species of the rest of Atractylodes species, thus obtaining higher bootstrap support rates (ITS + trnL-F and ITS: MP-BS/ML-BS/BI-PP = 100/100/1.00, Figure 1,S1).
Figure 2.
Geographic distribution of Atractylodes and sample collection location in China (▲: A.japonica; ■: A.chinensis; δ: A. coreana; ⋄: A. lancea or A.lancea subsp luotianensis; •: A. macrocephala;○: A. carlinoides).
2.5. Genetic Relationship of the Atractylodes lancea Complex
According to ITS and ITS + trnL-F phylogenetic trees, Atractylodes macrocephala independently formed a branch as the sister group of the A. lancea complex, with high bootstrap support rates (ITS + trnL-F: MP-BS/ML-BS/BI-PP = 96/92/1.00, Figure 1; ITS: MP-BS/ML-BS/BI-PP = 90/88/0.98, Figure S1). Atractylodes macrocephala and A. carlinoides share some common aspects, including a large inflorescence diameter, red flowers, and monoecism. However, only Atractylodes macrocephala has pinnately divided leaves and plump, developed rhizomes. The common features of the Atractylodes lancea complex include smaller inflorescence diameters, white flower, and gynodioecism. In the phylogenetic tree, Atractylodes macrocephala is the sister group of the A. lancea complex, which is consistent with the findings of Zhou et al. and Mizukami et al.[13,19].
The taxonomic controversy over the classification of species and infraspecific taxa of Atractylodes is mainly concentrated in the A. lancea complex. The classification of the Atractylodes lancea complex is mainly based on leaf traits, such as petiole, divided leaves, and the degree of the division. Variation in these traits is related to geographical distribution and ecological environment, which leads to different classifications by scholars in terms of macrospecies or microspecies classification that ultimately generated masses of taxonomic controversy [6,25,26]. In our styudy, the Atractylodes lancea complex was divided into three subclades: A. lancea clade, A. chinensis clade, and A. japonica (Figure 1,S1).
Atractylodes lancea clade is mainly distributed in South China (such as in Anhui, Jiangsu, and Hubei Provinces), including A. lancea subsp. luotianensis and A. lancea. A. lancea subsp. luotianensis is a new geographic subspecies established by Hu [14], and it is found at altitudes above 600 m in the Dabie Mountain Area in Anhui and Hubei Province. The main difference of this subspecies from Atractylodes lancea is the larger diameter of its inflorescence. Continuous survey at different elevations of the Dabie Mountain area revealed that the traits of A. lancea continuously change with altitude [7]. In the present research, ITS analysis and the joint analysis of ITS and trnL-F demonstrate that Atractylodes lancea subsp. luotianensis shares a short genetic distance with A. lancea, which is consistent with the findings of Zou et al.[13]. Hence, the intraspecific division of Atractylodes lancea is not supported by our study.
Atractylodes chinensis clade includes A. coreana, A. chinensis, and a part of A. lancea. Whether Atractylodes chinensis should be identified as a species, a variant (A. lancea var. chinensis), or a synonym of A. lancea has been a controversial issue because A. chinensis is hard to distinguish from A. lancea in terms of biomorphology [6,10,25]. Shi [6,10] identified Atractylodes lancea as a polytypic species based on morphologic studies, and distinguishing A. chinensis from A. lancea is difficult based on traits such as leaf shape. Shi [6,10] also believed that Atractylodes chinensis should be a synonym of A. lancea. Scholars thought that A. lancea was in the south side of Qinlin Mountains, A. chinensis was in the north side, and there were some transitional types between the two species (Figure 2). Shiba et al.[24] attributed the continuous variation to hybrids of Atractylodes lancea and A. chinensis, which suggests that A. lancea and A. chinensis should be two independent species. However, in the phylogenetic tree, the Atractylodes lancea in Taibai and Zhen’an at the South Slope of Qinlin Mountains in Shaanxi Province are clustered with all A. chinensis and A. coreana. In the population from Shennongjia, two strains are clustered with Atractylodes lancea clade and one strain is clustered with A. chinensis clade. This finding indicates the inconsistent morphologic features and genetic differentiation between Atractylodes lancea and A. chinensis.
In “Flora Reipublicae Popularis Sinicae” and “Flora of China”, Atractylodes coreana was regarded as a separate species. A. coreana was only distributed in Liaodong and Shadong Peninsulas (Figure 2), and it have different characteristics such as lower and middle cauline leaves narrowly elliptic to ovate-lanceolate, undivided. However, in ITS and ITS + trnL-F phylogenetic trees, Atractylodes coreana and A. chinensis are clustered into one clade. Shiba et al.[24] indicated that all samples of Atractylodes chinensis and A. coreana have the same ITS genotype. This study show that A. coreana should be treated as a synonym
Atractylodes japonica was regarded as a separate species in “Flora Reipublicae Popularis Sinicae”. However, it was a synonym of Atractylodes lancea in “Flora of China”. According to ITS and ITS + trnL-F phylogenetic trees, Atractylodes japonica is clustered into its own branch among the A. lancea complex, and it is closely related to A. lancea clade. Atractylodes japonica have different morphological characters from the other taxa in A. lancea complex: the leaves have long petioles and are generally divided or completely divided into 3–5 lobes. The trnK phylogenetic tree revealed that Atractylodes japonica and A. lancea are most closely related [19]. Guo also indicated that Atractylodes japonica is most closely related to the A. lancea in Jiangsu Province while not that closely related to A. chinensis in Hebei, Beijing, and Shaanxi [27].
The phylogenetic trees show that Atractylodes lancea complex formed a monophyletic group, with higher support rate (ITS + trnL-F: MP-BS/ML-BS/BI-PP = 96/93/1.00, Figure1; ITS: MP-BS/ ML-BS/BI-PP = 88/90/1.00, Figure S1). However, three branches in the Atractylodes lancea complex (A. lancea clade, A. chinensis clade, and A. japonica) were lack of high support rate. Geographically, Atractylodes lancea and A. chinensis was distributed in the south and north side of Qinlin-Huaihe River each other. Morphologically, some specimens of Atractylodes chinensis or A. lancea in transitional areas are hard to clarify. In the phylogenetic tree, some samples of the A. lancea are clustered in A. chinensis clade. In the population from Shennongjia, two strains are clustered with Atractylodes lancea clade and one strain is clustered with A. chinensis clade. This finding indicates that geographical distribution, morphologic features and genetic differentiation of Atractylodes lancea clade and A. chinensis clade are not entirely consistent. For a profound understanding of the relationship between the genetic differentiation of Atractylodes and its geographic distribution, an in-depth investigation into the subject with molecular phylogeography is needed.
3. Experimental Section
3.1. Selection of Taxa and Outgroups
Samples were collected from related producing areas according to voucher specimens or literature based on the view of microspecies, because the classification of species and infraspecific taxa of Atractylodes is much debated (The areas where the samples are produced are shown in Figure 2 and Table S1) [5,6,10,26]. The tested materials were fresh leaves. After collection, the samples were immediately dried in silica gel for DNA extraction and sequence analysis. After identification by the first author, the samples were preserved and deposited with the Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences.
Atractlis, Carlina, and Atractylodes belong to subtribe Carlininae [28]. To study the relationship of Atractylodes with Atractlis, and Carlina, the genera Brachylaena, Cardopatium, Cirsium, Echinops, Phonus and Tarchonanthus were used as outgroups in the phylogenetic analyses based on the studies of Garcia-Jacas et al.[28] and Susanna et al.[17]. Their ITS and trnL-F sequences were downloaded from GenBank (GenBank accession numbers are shown in Table S2).
3.2. DNA Extraction, Amplification, and Sequencing
The cetyltrimethylammonium bromide method [29] was used to extract genomic DNA from the silica gel-dried leaves. The chloroplast DNA sequences were amplified using trnL as the forward primer and trnF as the reverse primer [30]. The nuclear ribosomal sequence was amplified using ITS5 as the forward primer and ITS4 as the reverse primer [31]. All sequences used 20 μL PCR reaction contained 14.7 μL of ddH2O, 2 μL of 10× buffer (TaKaRa Biotech, Dalian, Beijing, China), 1.6 μL of dNTP (TaKaRa Biotech, Dalian, Beijing, China), 0.5 μL of each primer, 0.5 μL of template DNA (1 μg), and 0.2 μL of Ex Taq DNA polymerase (TaKaRa Biotech, Dalian, Beijing, China). The PCR conditions were as follows: initial denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C for 1 min, 54 °C (ITS) or 58 °C (trnL-F) for 30 s, 72 °C for 1 min 40 s, and final extension at 72 °C for 5 min. The PCR products were run on 1.0% agarose gel in 1.0× Tris-borate-EDTA buffer, purified using a Tiangen Midi Purification Kit (Tiangen Biotech, Beijing, China), and then sequenced using a Bigdye Terminator Cycles Sequencing Ready Reaction Kit and Applied Biosystems ABI3730 DNA Sequencer.
3.3. Sequence Alignment and Phylogenetic Analysis
Susanna et al.[17] used ITS, trnL-F, and matK to study the phylogenetic relationship of Cardueae. ITS, trnL-F, and matK sequence and analysis was also applied in the present study. However, our studies show that the substitution sites of matK in Atractylodes are very limited; thus, ITS and trnL-F were reported.
The raw sequences were assembled and edited using the BioEdit software ver.5.0.9 [32] and adjusted manually as necessary. The novel sequences generated from Atractylodes and the sequences downloaded from GenBank were subjected to multiple alignment using the CLUSTALW 1.83 software package with default settings [33]. ITS and trnL-F + ITS datasets were constructed, and all contained 49 individuals from 7 taxa.
To examine the extent of conflict between the ITS and trnL-F datasets, the partition homogeneity test (also known as the incongruence length difference, ILD; [34]) was carried out using PAUP*. The text was implemented with 1000 partition homogeneity test replicates, using a heuristic search option with simple addition of taxa, TBR branch swapping and MaxTrees ser to 1000. The results suggest that both data sets are congruent (p-value > 0.05) and can be combined.
Parsimony analysis. Maximum parsimony (MP) analyses were carried out using PAUP* v.4.0b10 [35]. For each analysis, maximum parsimony trees were sought using the heuristic search strategies of PAUP* (with 1000 replicate analyses, random stepwise addition of taxa, tree bisection and reconnection (TBR) branch swapping, and setting the maximum number of trees to 10,000). Bootstrap values were calculated from 1,000,000 replicate analyses using fast stepwise addition of taxa, and only those values compatible with the majority-rule consensus tree were recorded. All most parsimonious trees (MPTs) were saved and PAUP* was used to compute a strict consensus. Tree lengths, consistency index (CI) and retention index (RI) were calculated excluding uninformative characters.
Bayesian Inference (BI) analysis. Datasets were conducted using MrBayes version 3.1.2 [36]. Prior to these analyses, the best-fit nucleotide substitution models of each partition were selected by AIC implemented in MrModeltest v.2.3 [37] (GTR + G for the ITS datasets, GTR for the cpDNA datasets, GTR + I + G for the cpDNA +nrDNA datasets). Four simultaneous Markov chain Monte Carlo (MCMC) chains were run using MrBayes version 3.1.2 [38], there heated and one cold, with the temperature adjusted to 0.5 in order to keep an appropriate heat range for the four chains. From a random starting tree, the Bayesian analysis was run for 20 million generations, and the trees were saved to a file every 1000 generations. Branch lengths of the trees were saved. Each analysis reached stationarity (the average standard deviation of split frequencies between runs ≤0.01) well before the end of the run. Burn-in (=5,000) trees were discarded, and the remaining trees and their parameters were saved. The 50% majority rule consensus tree was constructed. The results of the Bayesian analysis are reported as posterior probabilities [38], which are equal to the percentage of trees sampled when a given clade is resolved. Only PP scores exceeding 50% are shown.
Maximum likelihood (ML) analysis. ML analyses were performed using PHYML version 2.4 [39] using the same model of substitution with 1000 bootstrap replicates.
4. Conclusions
In Atractylodes, the ITS region from the nuclear genome was more variable than the trnL-trnF region from the plastid genome. All members of Atractylodes formed a well-supported clade, which is a sister group of the Carlina and Atractylis. Atractylodes species were distributed among three clades: A. carlinoides, A. macrocephala and the A. lancea complex (including A. japonica, A. coreana, A. lancea, A. lancea subsp. luotianensis, and A. chinensis). Atractylodes carlinoides is a sister species of the rest of the Atractylodes species. The taxonomic controversy over the classification of species of Atractylodes is mainly concentrated on the A. lancea complex. A. lancea subsp. luotianensis shares a short genetic distance with A. lancea; therefore, it should be treated as a synonym of A. lancea. Based on molecular results, Atractylodes coreana should be treated as a synonym of A. chinensis.
Acknowledgements
This research was supported by National Natural Science Foundation of China (NSFC:81130070, 81072989) and National Key Technology Research and Development Program of the Ministry of Science and Technology of China (2012BAI29B02).
References
- 1.Peng H.S., Wang D.Q. The history and differentiation of raw Atractylodes commodity in successive ages. Chin. J. Med. Hist. 2007;37:15–18. [PubMed] [Google Scholar]
- 2.De Candolle A.P. Prodromus Systematis Naturalis Regni Vegetabilis, Pars VI. Treuttel & Würtz; Paris, France: 1838. [Google Scholar]
- 3.Ling Y. Les Composees Chinoises De L’herbier De L’academie De Peiping. Contr. Inst. Bot. Nat. Acad. Peip. 1935;III:132–134. [Google Scholar]
- 4.Hu S.Y. The Compositaeof China (II) Quart J. Taiwan. Mus. 1965;XVIII:233–334. [Google Scholar]
- 5.Fu S.M., Fang H.J., Liu G.S., Xiao P.G. A study on the medicinal plants of the genus Atractylodes. Acta Phytotaxon. Sin. 1981;19:195–201. [Google Scholar]
- 6.Shi Z. Flora Reipublicae Popularis Sinicae. Vol. 78 Science Press; Beijing, China: 1987. [Google Scholar]
- 7.Peng H.S., Wang D.Q. Studies on population biology of transitional types of genus Atractylodes in Anhui Province. Chin. J.Chin. Mater. Med. 2007;32:793–797. [PubMed] [Google Scholar]
- 8.Xu G.F., Yu Y.L., Liu J., Wang L.X. Observations on the morphological charcter of Atractylodes japonica Koidz. Et Kitam. J. Jiamusi Med. Coll. 1993;16:15–17. [Google Scholar]
- 9.Petit D.P. Generic interrelationships of theCardueae (Compositae): A cladistic analysis of morphological data. Plant Syst. Evol. 1997;207:173–203. [Google Scholar]
- 10.Shi Z. On the nomenclature of Chinese drug “Cangzhu”. Acta Phytotaxon. Sin. 1981;19:318–322. [Google Scholar]
- 11.Kunio I., Takasi Y., David E.B., Hideaki O. Flora of Japan. III Kodansha; Tokyo, Japan: 1997. [Google Scholar]
- 12.Bobrov E., Bochantsev V., Iljin M., Linchevskii I., Lipshits S., Yu S.E.V., Cherepanov S., Cherneva O., Yuzepchuk S. Flora of the USSR. Vol. 27 Bishen Singh Mahendrapal Singh; Dehradun, Moscow, Russia: 1997. [Google Scholar]
- 13.Zou X.X., Huang L.Q., Cui G.H., Yuan Q.J., Peng Y., Liu Y., Xiao P.G. Genetic relationship of Atractylodes plants. Acta. Pharm. Sin. 2009;44:680–686. [PubMed] [Google Scholar]
- 14.Hu S.L., Feng X.F., Wang J., Ge X.G. A new subspecies of Atractylodes from China. Acta Phytotaxon. Sin. 2001;39:84–86. [Google Scholar]
- 15.Wu Z.Y. Flora of China. Vol. 20 Science Press; Beijing, China: 2011. [Google Scholar]
- 16.Committee J.P.E. The Japanese Pharmacopoeia. 16th ed. Hirohawa Press; Tokyo, Japan: 2011. [Google Scholar]
- 17.Susanna A., Garcia-Jacas N., Hidalgo O., Vilatersana R., Garnatje T. The Cardueae (Compositae) revisited: Insights from ITS, trnL-trnF, and matK nuclear and chloroplast DNA analysis. Ann. Mo. Bot. Gard. 2006;93:150–171. [Google Scholar]
- 18.Panero J.L., Funk V.A. The value of sampling anomalous taxa in phylogenetic studies: Major clades of the Asteraceae revealed. Mol. Phylogenet. Evol. 2008;47:757–782. doi: 10.1016/j.ympev.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 19.Mizukami H., Shimizu R., Kohjyouma M., Kohda H., Kawanishi F., Hiraoka N. Phylogenetic analysis of Atractylodes plants based on chloroplast trnK sequence. Biol. Pharm. Bull. 1998;21:474–478. doi: 10.1248/bpb.21.474. [DOI] [PubMed] [Google Scholar]
- 20.Ge Y.F., Hang Y.Y., Xia B., Wei Y.L. Sequencing of trnL-F and analysis of interspecific genetic relationship of five medicinal species in Atractylodes DC. J. Plant Resour. Environ. 2007;16:12–16. [Google Scholar]
- 21.Guo Y.P., Ehrendorfer F., Samuel R. Phylogeny and systematics of Achillea (Asteraceae-Anthemideae) inferred from nrITS and plastid trnL-F DNA sequences. Taxon. 2004;53:657–672. [Google Scholar]
- 22.Fernández I.Á., Aguilar J.F., Panero J.L., Feliner G.N. A phylogenetic analysis of Doronicum (Asteraceae, Senecioneae) based on morphological, nuclear ribosomal (ITS), and chloroplast (trnL-F) evidence. Mol. Phylogenet. Evol. 2001;20:41–64. doi: 10.1006/mpev.2001.0954. [DOI] [PubMed] [Google Scholar]
- 23.Sánchez-Jiménez I., Lazkov G.A., Hidalgo O., Garnatje T. Molecular systematics of Echinops L.(Asteraceae, Cynareae): A phylogeny based on ITS and trnL-trnF sequences with emphasis on sectional delimitation. Taxon. 2010;59:698–708. [Google Scholar]
- 24.Shiba M., Kondo K., Miki E., Yamaji H., Morota T., Terabayashi S., Takeda S., Sasaki H., Miyamoto K., Aburada M. Identification of medicinal Atractylodes based on ITS sequences of nrDNA. Biol. Pharm. Bull. 2006;29:315–320. doi: 10.1248/bpb.29.315. [DOI] [PubMed] [Google Scholar]
- 25.Liu S.E. Claves Plantarum Chinae Boreali-Olientalis. Science Press; Beijing, China: 1959. [Google Scholar]
- 26.Guo Y.Z., Chen F.K., Zhao J.F., Zhou Y., Yao N.X., Zhang Y.Y. Studies on Cangzhu. In: Lou Z.Q., Qin B., editors. Species Systematization and Quality Evaluation of Commonly Used Chinese Traditional Drugs. Vol. 3. Peking University Medical Press; Beijing, China: 2003. pp. 743–778. [Google Scholar]
- 27.Guo L.P., Huang L.Q., Wang M., Feng X.F., Fu G.F., Yan Y.N. A preliminary study on relationship between Atractylodes lancea and A. chinensis as analyzed by RAPD. Zhongguo Zhong Yao Za Zhi. 2001;26:156–158. [PubMed] [Google Scholar]
- 28.Garcia-Jacas N., Garnatje T., Susanna A., Vilatersana R. Tribal and subtribal delimitation and phylogeny of the Cardueae (Asteraceae): A combined nuclear and chloroplast DNA analysis. Mol. Phylogenet. Evol. 2002;22:51–64. doi: 10.1006/mpev.2001.1038. [DOI] [PubMed] [Google Scholar]
- 29.Doyle J.J., Doyle J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 30.Taberlet P., Gielly L., Pautou G., Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- 31.White T.J., Bruns T., Lee S., Taylor J. PCR Protocols A Guide to Methods and Applications. Academic Press; New York, NY, USA: 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics; pp. 315–322. [Google Scholar]
- 32.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 33.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farris J.S., Källersjö M., Kluge A.G., Bult C. Testing significance of incongruence. Cladistics. 1994;10:315–319. [Google Scholar]
- 35.Swofford D.L. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates; Sunderland, MA, USA: p. 2000. [Google Scholar]
- 36.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 37.Nylander J. Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University; Uppsala, Sweden: 2004. MrModeltest v2. [Google Scholar]
- 38.Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 39.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]