Abstract
Xanthine oxidoreductase (XOR) catalyzes the conversion of hypoxanthine to xanthine and xanthine to uric acid with concomitant reduction of either NAD+ or O2. The enzyme is a target of drugs to treat hyperuricemia, gout and reactive oxygen-related diseases. Human diseases associated with genetically determined dysfunction of XOR are termed xanthinuria, because of the excretion of xanthine in urine. Xanthinuria is classified into two subtypes, type I and type II. Type I xanthinuria involves XOR deficiency due to genetic defect of XOR, whereas type II xanthinuria involves dual deficiency of XOR and aldehyde oxidase (AO, a molybdoflavo enzyme similar to XOR) due to genetic defect in the molybdenum cofactor sulfurase. Molybdenum cofactor deficiency is associated with triple deficiency of XOR, AO and sulfite oxidase, due to defective synthesis of molybdopterin, which is a precursor of molybdenum cofactor for all three enzymes. The present review focuses on mutation or chemical modification studies of mammalian XOR, as well as on XOR mutations identified in humans, aimed at understanding the reaction mechanism of XOR and the relevance of mutated XORs as models to estimate the possible side effects of clinical application of XOR inhibitors.
Keywords: xanthine dehydrogenase, xanthine oxidase, xanthine oxidoreductase, xanthine oxidoreductase deficiency, flavoproteins, xanthinuria, hereditary xanthinuria, gout
1. Introduction
Xanthine oxidoreductase (XOR) catalyzes two hydroxylation steps in the metabolic pathway of purine degradation, i.e., hypoxanthine to xanthine and xanthine to uric acid, utilizing either NAD+ or O2[1–3] (Figure 1). In higher animals, XOR exists as a homodimer of 150 kDa subunits [4]. Each subunit contains one molybdenum center (molybdenum cofactor; Moco), one flavin adenine dinucleotide (FAD) cofactor and two distinct iron sulfur centers ([2Fe-2S] type) [1–3]. The purine hydroxylation reaction occurs at the molybdenum center. Electrons, which are transferred to molybdenum during the hydroxylation reaction, are further transferred to FAD via the two iron sulfur centers [5,6]. Finally, NAD+ or oxygen molecule, which is the final electron acceptor, is reduced at the FAD center.
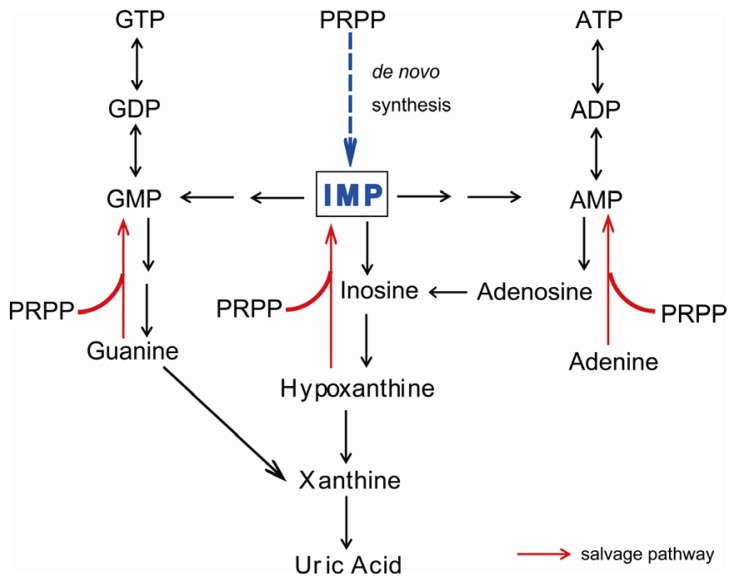
Figure 1.
Metabolic pathways of purine degradation in humans. Xanthine oxidoreductase (XOR) catalyzes the transformations of hypoxanthine to xanthine and xanthine to uric acid. XOR-deficient patients secrete xanthine, which is formed from guanine. Accumulated hypoxanthine is mostly converted to inosine monophosphate (IMP) via the salvage pathway using 5-phospho-α-D-ribosyl 1-pyrophosphate (PRPP) as a co-substrate.
XOR has two forms: xanthine dehydrogenase (XDH), which prefers NAD+ as the substrate and xanthine oxidase (XO), which prefers O2[1]. Historically, XDH and XO have been studied as distinct enzymes. XOR has been isolated only as the XO form from mammalian sources, whereas it has always been purified in the XDH form from other organisms [2]. It is becoming clear, however, that mammalian XORs exist in the XDH form under normal conditions in the cell, but are converted to the XO form during extraction or purification, either irreversibly by proteolysis or reversibly by oxidation of cysteine residues to disulfide bridges. In some particular cases, XDH can be converted to the XO form [2]. The mechanism of conversion from XDH to XO has been thoroughly elucidated in the past decade by means of a range of techniques, including X-ray crystal structure analysis of various mutants, and it has become clear that the protein environment influences the reactivity of the FAD cofactor towards different substrates through substantial conformational changes triggered by modifications located far from the cofactor [5,6].
The enzyme is a target of drugs to treat hyperuricemia, gout or reactive oxygen-related diseases [7,8]. It is distributed in various organs including liver, mammary gland and endothelial cells of vascular vessels [9,10]. The enzyme was proposed to be localized in peroxisomes of rat liver [11], but was found to be present in cytosol [12]. As XOR inhibitors significantly lower uric acid production and concentration in the blood, they can be used to treat gout. Allopurinol, which was introduced by Elion et al.[13], has been on the market for over 40 years [14]. In recent years, however, several companies have developed very effective inhibitors [14–16], of which one example is febuxostat [17]. Clinical trials indicate that febuxostat is superior to allopurinol in lowering uric acid production, although the mechanism of inhibition is different [18,19]. By means of enzymatic, spectroscopic and structural-biological analyses of the inhibition mechanism, it has been shown that these recently developed inhibitors bind tightly to both the oxidized and reduced forms of XOR in a highly structure-specific manner [15], whereas allopurinol, a substrate analogue, binds covalently to the reduced molybdenum atom (MoIV) after having been converted to the hydroxylated product (oxipurinol: alloxanthine) [20], mimicking the reaction intermediate formed during the hydroxylation reaction with xanthine as a substrate [21]. Although oxipurinol binds very tightly to the enzyme, it can be dissociated from the molybdenum (VI) by spontaneous reoxidation due to electron transfer to other centers with a half-time of 300 min at 25 °C [20]. Potent inhibition seems to be essential to lower the uric acid level in blood or tissue, since XOR is a house-keeping enzyme that exists abundantly in various organs [10]. However, it has been suggested that lowering uric acid levels may cause side effects in humans, since uric acid acts as a radical scavenger in the body [22,23]. Further, it is proposed that NO formed by XOR via reduction of NO2 (with any electron donor) may induce vasodilatation under ischemic conditions [24–26]. On the other hand, XOR has the potential to generate oxygen radical species (H2O2 and O2−) after conversion from XDH to XO [1–6]. O2− would rapidly react with NO to form ONOO−[27]. This reaction may serve to eliminate NO, at least in part, but the ONOO− produced is highly toxic [28]. As to the question of potential NO formation by XOR, the activity for NO formation from NO2 is extremely low, even under anaerobic conditions, although from a chemical point of view it is possible that the water-exchangeable hydroxyl group at OH-Mo(IV) can be replaced by NO2 to produce NO, since various compounds, such as uric acid (which reacts very slowly to form xanthine), can behave similarly, as discussed by Okamoto [29]. The reported kcat value of NO formation is 0.17 s−1 at 37 °C with NADH as an electron donor under anaerobic conditions [30], i.e., less than 1% of kcat for xanthine oxidizing activity (kcat value 15–20 s−1 at 25 °C) [31,32]. It is questionable whether such a weak activity can have any physiological significance, even under ischemic conditions. The present review focuses mainly on mutational studies of XOR and mutations associated with hereditary dysfunction of XOR in humans, since these are useful for understanding the enzyme reaction mechanism and also as models to estimate the possible side effects of using XOR inhibitors as drugs.
2. Symptoms of XOR Deficiency and Differential Diagnosis
Human diseases associated with genetic dysfunction of XOR are termed xanthinuria, because xanthine is excreted in the urine [33]. Although the enzyme catalyzes two steps of reaction, as described above, so that XOR dysfunction might be expected to be associated with tissue accumulation of hypoxanthine due to inhibition of the first step (conversion of hypoxanthine to xanthine), in fact hypoxanthine is not normally significantly excreted in urine [34,35]. Instead, hypoxanthine is converted to inosine monophosphate (IMP) owing to activation of the salvage pathway (Figure 1) [36]. Patients typically have low levels of uric acid (less than 1 mg/dL) in blood, so XOR-deficient patients are frequently identified based on measurement of uric acid in blood. Various diseases or disorders other than xanthinuria may lead to hypouricemia (Table 1). Renal hypouricemia, which can be caused by decreased re-absorption due to impaired function of urate transporter in the nephrons, is also clinically asymptomatic in most cases.
Table 1.
Causes of hypouricemia.
| Inherited disorders of purine metabolism |
|
|
| Genetic defects in the molybdoflavoprotein enzymes: |
| Xanthinuria type I (xanthine oxidoreductase deficiency) |
| Xanthinuria type II (molybdenum cofactor sulfurase deficiency: combined xanthine oxidoreductase and aldehyde oxidase deficiencies) |
| Molybdenum cofactor deficiency |
| Purine nucleoside phosphorylase deficiency |
| Phosphoribosylpyrophosphate synthetase deficiency |
|
|
| Secondary reduction in uric acid biosynthesis |
|
|
| Hepatic failure |
|
|
| Inherited renal hypouricemia (isolated renal tubule reabsorption defect) |
|
|
| Renal hypouricemia-1 [URAT1 (SLC22A12) deficiency] |
| Renal hypouricemia-2 [URAT9 (SLC22A9) deficiency] |
|
|
| Inherited causes of the Fanconi renotubular syndrome and its variants (the syndrome of multiple renal tubule reabsorption defects) |
|
|
| Fanconi renotubular syndrome 1 |
| Cystinosis (accumulation of intralysosomal cystine) |
| Galactosemia (galactose-1-phosphate uridylyltransferase deficiency) |
| Hereditary fructose intolerance (fructose 1-phosphate aldolase B deficiency) |
| Glycogen storage disease type 1 (glucose-6-phosphate deficiency) |
| Wilson’s disease [ATPase, Cu2+ transporting, beta polypeptide (ATP7B) deficiency] |
| Mitochondrial complex IV deficiency (cytochrome c oxidase deficiency) |
|
|
| Acquired causes of the Fanconi renotubular syndrome and its variants |
|
|
| Metal poisoning (e.g., Cd, Zn, Cu, Pb, Hg) |
| Multiple myeloma |
| Nephrotic syndrome |
| Malignant disease |
| Autoimmune disease (e.g., Sjogren’s syndrome) |
| Thermal burns |
| Primary hyperparathyroidism |
| Acute renal tubular necrosis |
| Renal transplant rejection |
|
|
| Drugs |
|
|
| Xanthine oxidoreductase inhibitor (e.g., allopurinol, febuxostat) |
| Drugs used either as uricosuric agents or to block other aspects of renal tubule excretion (e.g., sulfinpyrazone, probenecid, benzbromarone) |
| Non-steroidal anti-inflammatory drugs with uricosuric properties (e.g., phenylbutazone, azapropazone, high dose of aspirin) |
| Coumarin anticoagulants (e.g., warfarin) |
| Outdated tetracycline (5 alpha-6-anhydro-4-epitetracycline) |
|
|
| Nutritional deficiencies |
|
|
| Vitamines B12, C, D |
| Kwashiorkor |
Xanthinuria is classified into two subtypes, type I and type II (Table 1) [35]. The type I is due to a genetic defect of XOR, whereas the type II is due to a genetic defect in molybdenum cofactor sulfurase [37,38]. Aldehyde oxidase (AO), also a molybdoflavo enzyme, is similar to XOR. A terminal sulfide group is necessary as the third ligand in the active center of XOR and AO for enzymatic activation of these enzymes after biosynthesis of the molybdenum cofactor. Molybdenum cofactor sulfurase catalyzes this final maturation step by generating a protein-bound persulfide, which is the source of the terminal sulfur ligand of the molybdenum cofactor. Thus, lack of sulfurase results in type II xanthinuria. Type I and II xanthinuria are not clinically distinguishable. In order to differentiate them, allopurinol loading test and gene analysis are performed, because a measurement method for molybdenum cofactor sulfurase activity has not yet been established [39,40]. In the allopurinol loading test, oxipurinol is detected in serum and urine of type I xanthinuria patients after administration of allopurinol, as conversion of allopurinol to oxipurinol is catalyzed by XOR and AO, while oxipurinol is not detected in the case of type II xanthinuria. AO has broad substrate specificity, oxidizing different types of aldehydes and heterocyclic rings [41,42]. No clinical symptom or abnormal laboratory examination result due to lack of AO has yet been identified. However, it has recently been reported that AO plays an important role in the metabolism of numerous compounds. Thus, classification of type I and II xanthinuria might be indispensable for optimum medical treatment of patients with xanthinuria in the future.
In higher animals other than primates, xanthinuria is lethal due to kidney damage resulting from xanthine stones in the urinary tract [43–46]. Although primates have lost uricase during evolution and seem to have acquired tolerance to oxipurines, e.g., through downregulation of XOR gene expression, other animals convert uric acid to more soluble allantoin, catalyzed by peroxisomal uricase, and do not seem to have such tolerance [47]. Urolithiasis is sometimes accompanied with xanthinuria due to xanthine deposition, and rarely this may lead to acute renal failure [48–53]. In addition to its role in uric acid production, XOR has bactericidal activity via ROS generation under certain conditions, particularly in mammalian mammary gland [54]. In addition to the NO2 reduction as described in the previous section, XOR has been proposed to play a role in lactation, though the mechanism of the role in lactation remains unclear [55,56]. XOR has also been suggested to be implicated in hypertension, cardiovascular disorder, and adipogenesis [57,58]. Although the situation is not simple, clinical observations in xanthinuria patients with extremely low serum uric acid levels, who show no symptoms, suggest that administration of XOR inhibitors may not cause severe side effects if the inhibitor has no other effect than inhibition of XOR, except possibly in special cases, such as cancer, pregnant or breast-feeding patients. Many purine analogue cancer drugs, such as mercaptopurine, are known to be catabolized by XOR [59]. The third type of XOR deficiency, type III XOR deficiency, involves the molybdenum cofactor. Molybdenum cofactor deficiency involves triple deficiency of XOR, AO and SO (sulfite oxidase), due to a defect in the synthesis of molybdopterin, which is a precursor of molybdenum cofactor for all three enzymes. Symptoms of molybdenum cofactor deficiency include severe neurological disorder, lens dislocation and dysmorphism, and the outcome is poor [60].
3. Overall Structure of Human Xanthine Oxidoreductase (XOR)
The primary structure of human XOR was first reported by Ichida et al.[61], who isolated cDNA clones encoding human XOR by cross hybridization with rat cDNA, the structure of which was reported by Amaya in 1990 [62]. The XOR gene has 36 exons, and is located in chromosome 2p23.1 [63,64]. The primary structure of human XOR has 90% homology with the rat enzyme over the entire length. Although cloning of human XOR was subsequently reported by several groups [64–66], the sequences were all very similar, except for one reported by Wright et al that was later found to encode AO, not XOR. Although mammalian XORs and AOs from various sources have similar molecular weights and cofactors [2,6,67], their substrate specificities are different. AO exclusively utilizes O2 as the oxidizing substrate rather than NAD+, which is used by dehydrogenases. The specificities of the two enzymes for reducing substrates partially overlap, and each is capable of hydroxylating a distinct subset of a wide range of aldehydes and aromatic heterocycles. Purine bases are good substrates of XORs, but are not good substrates of AOs. The physiological substrates of mammalian AOs are not known, although the involvement of AOs in drug metabolism is well-established [42]. Structure-based sequence comparisons have identified residues in the vicinity of the active site molybdenum center of AO that differ from those in XOR, and these are most probably the determinants of the substrate preferences exhibited by the family members [68]. However, a glutamic acid residue, thought to represent an essential catalytic base, is strictly conserved in AOs and XORs, pointing to a common catalytic mechanism for all family members [69].
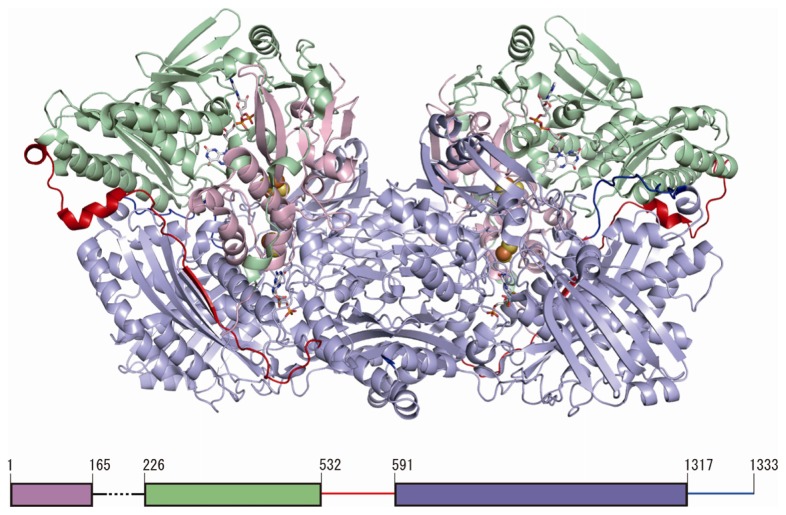
The crystal structures of human XOR from natural milk at 3.6 Å resolution (PDB: 2CKJ) and recombinantly produced XDH at 2.6 Å resolution (PDB: 2E1Q) [69] are available. Higher resolution structures of mammalian XORs are available for native bovine XDH and XO, as well as recombinantly produced rat XDH and XO, including various mutants [69,70]. The subunits in the crystal structures of all these mammalian XORs are arranged as identical dimers that display a distinct butterfly shape [4,69,70]. The dimensions of the whole enzyme molecule are about 155 Å × 90 Å × 70 Å (Figure 2). Each monomer is composed of three subdomains. The small N-terminal domain (residues 1 to 165 in the human enzyme) contains both of the iron-sulfur centers (Fe/S I and Fe/S II) and is connected to the second, FAD-containing domain (residues 226 to 532, colored light green in Figure 1) via a long, partially disordered segment consisting of residues 166 to 225. The FAD domain, in turn, is connected to the third, C-terminal domain via another extended segment (residues 533 to 590), which is also partially disordered. The third and largest domain (residues 591 to 1317, colored light blue in Figure 2) binds Moco close to the interface of the Fe/S- and FAD-binding domains, connected with a C-terminal loop (residues 1318–1333, colored blue in Figure 2) [4,69,70].
Figure 2.
Structure of human XOR. The structure illustrated is that of a human mutant dimeric XDH [69] (PDB: 2E1Q). The Fe/S, FAD, and molybdopterin domains are colored light pink, light green and light blue, respectively. The interdomain loop (residues 533–590) is colored red. C-terminal is colored blue. A schematic representation of the domain structure in relation to the primary sequence is shown at the bottom.
4. Residues Crucial for Enzyme Function: Experimental Studies
In order to elucidate the mechanisms of hydroxylation at the molybdenum center, electron transfer within the redox centers and reoxidation of the reduced FAD by the natural substrate, NAD+ or molecular oxygen, various chemical modification and mutation studies have been performed during the last two decades. The amino acid residues of human XOR corresponding to those that have so far been found to be crucial for enzyme function, either by chemical modification or by mutagenesis studies with bovine or rat XORs, are summarized in Table 2.
Table 2.
Residues crucial for enzyme function revealed by experimental studies.
| Corresponding human residue No. | Residue in experimental animal | Function | Experiments |
|---|---|---|---|
| The Fe/S domain | |||
|
| |||
| Cys43 | rat Cys43 | Fe/S II ligand | mutation to Ser [71] |
| Cys51 | rat Cys51 | Fe/S II ligand | mutation to Ser or Ala [71] |
| Cys116 | rat Cys115 | Fe/S I ligand | mutation to Ser [71] |
| Lys185 | rat Lys184 | interdomain | Trypsin [62] |
|
| |||
| The FAD domain | |||
|
| |||
| Arg427 | bovine Arg427 | A member of the cluster XDH/XO conversion | mutation to Gln [72] |
| Arg335 | bovine Arg335 | A member of the cluster XDH/XO conversion | mutation to Ala [72] |
| Trp336 | bovine Trp336 & rat Trp335 | A member of the cluster XDH/XO conversion | mutation to Ala [72] |
| Phe337 | rat Phe336 | redox potential of FAD | mutation to Leu (to be published) |
| Tyr393 | chicken Tyr419 | NAD+ binding | chemical modification with FSBA [73] |
| Asp429 | rat Asp428 | redox potential of FAD | mutation (to be published) |
| Cys536 | rat Cys535 | disulfide formation with Cys992 XDH/XO conversion | mutation to Ala [70] & chemical modification with FDNB [74] |
| Lys552 | rat Lys551 | Interdomain trypsin XDH/XO | Trypsin [62] |
|
| |||
| The Moco domain | |||
|
| |||
| Lys755 | bovine Lys754 | kcat slower | chemical modification with FDNB [74,75] |
| Lys772 | bovine Lys771 | kcat slower | chemical modification with FDNB [74,75] |
| Glu803 | human | purine binding | mutation to Val [69] |
| Arg881 | human | purine binding | mutation to Met [69] |
| Cys993 | rat Cys992 | disulfide with Cys535 XDH/XO conversion | mutation to Arg [70] & chemical modification with FDNB [74] |
| Glu1262 | human | mutation to Ala [69,76] | |
| Cys1318 | rat Cys1316 | disulfie with Cys1324? | mutation to Ser [70] |
| Cys1326 | rat Cys1324 | disulfide with Cys1316? | mutation to Ser [70] & chemical modification with FDNB [74] |
4.1. The N-Terminal Fe/S Domain
This domain contains a cluster of two distinct [2Fe-2S] types, having different EPR signals and redox potentials, and these are named the Fe/S I and Fe/S II centers [77–79]. The Fe/S I signal displays g-values of g1,2,3 = 2.022, 1.932, 1.894, with line-widths and relaxation properties typical of a [2Fe-2S] cluster, while Fe/S II has g-values of g1,2,3 = 2.110, 1.991, 1.902, with unusually broad line widths and relaxation properties. The latter signals can only be observed below 25 K [80]. Site-directed mutagenesis studies employing heterologously expressed rat XOR have allowed assignment of the two distinct types of EPRsignals to the respective clusters [71], with Fe/S I being located in the unusual-113Cys-Xaa2-116Cys-//-148Cys-Xaa1-15 Cys-motif in the α-helical domain and Fe/S II in the N-terminal-43Cys-X-48Cys-X-51Cys-//-73Cys-motif in the ferredoxin-like domain. This establishes the sequence of electron transfer within the enzyme molecule as Mo → Fe/S I → Fe/S II → FAD. It was noted that the mutation at Cys43Ser or Cys51Ala, which are both components of Fe/S I, resulted in the appearance of insoluble or monomeric proteins, suggesting the importance of the Fe/S I cluster for protein conformation and/or folding [71].
4.2. The Intermediate FAD Domain
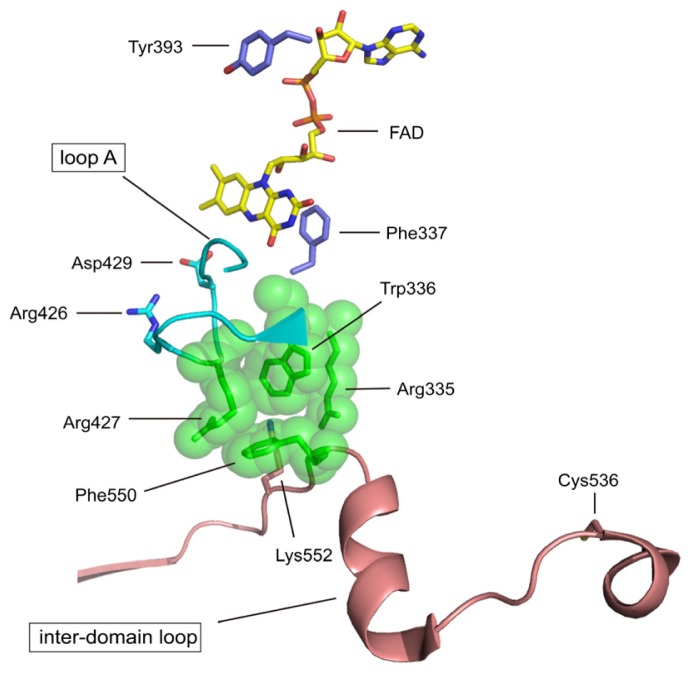
The domain binds its cofactor FAD in a deep cleft; in the NAD-free form, the si-face of the isoalloxazine ring is exposed to solvent (Figure 3). The same space allows the substrate NAD access to the flavin, and the two ring systems stack on top of each other [81]. Modification of the chicken XDH residue corresponding to Tyr419 (human Tyr393) with fluorosulfonylbenzoyl adenosine (FSBA) resulted in loss of activity towards NAD+[73], suggesting that this tyrosine residue is indeed involved in NADH binding, as indicated by the crystal structure of the rat XDH-NADH complex (Tomoko Nishino, K. Okamoto, E.F. Pai and Takeshi Nishino, unpublished data). In contrast to the open si-side, the re-side of the flavin ring is in tight contact with residues of the protein chain, e.g., the side chain of Phe336 (human Phe337) lies parallel to the isoalloxazine ring. Mutation study indicated that this phenyl-flavin pair may serve to tune the cofactor’s FAD redox potential (Tomoko Nishino, K. Okamoto, E.F. Pai and Takeshi Nishino, unpublished data). In the crystal structures of XDH and XO, the location of so-called loop A (residues 423–433 in human XOR) is very different in rat and bovine XORs. In rat and bovine XDH, the side chain of Asp428 (rat sequence, corresponding to human & bovine Asp429) in the loop is close to C6 of the flavin. This residue must be a major contributor to the strong negative charge at the flavin-binding site [4,81]. Mutation of this residue with rat XOR changes the reactivity of FAD by changing its redox potential (Y. Kawaguchi et al. unpublished). In XO conformation, Asp428 moves away from the flavin ring and the guanidinium group of Arg425 replaces it, approaching the nearest atom of the isoalloxazine ring to within 6.3 Å. This reversal of the electrostatic potential surrounding the redox-active part of the FAD cofactor matches predictions based on biochemical and biophysical studies of the XDH and XO forms [82–84]. Bovine Arg427, Arg335, Trp336 and Phe549 (human Arg335, Trp336, Arg427 and Phe550) are components of a unique cluster of four amino acids [72], which are held together mostly via π-cation interactions in the XDH form. Phe549 (rat Phe549, human Phe550) is located in the long linker between the intermediate FAD and C-terminal Moco domains. In the XO form, however, this cluster is disrupted (Figure 3) [5]. An equivalent effect can be achieved by mutating one of these residues with rat XOR [72]. Proteolysis at Lys551 (human Lys552) [62], leading to drastically increased mobility of the linker peptide between the intermediate FAD and C-terminal Moco domains, or disulfide formation between Cys535 and Cys992 [70,74], causing conformational strain, breaks Phe549 out of this tight arrangement. Disruption of the cluster is accompanied with movement of the active site loop A. Recent studies suggest that the conversion from XDH to XO is in equilibrium [85]; the highly packed amino acid cluster, binding of NAD+/NADH and insertion of the C-terminal peptide shift the equilibrium towards the XDH form, while disulfide formation between Cys535 and Cys992 (human Cys536 and Cys993) or proteolysis in the linker between the FAD and the Moco domains disrupts the amino acid cluster and moves the active site loop A. In rat enzyme extrusion of the C-terminal peptide, by formation of a disulfide bond between Cys1316 and Cys1324, shifts the equilibrium partially to the XO form.
Figure 3.
Structure of the active site cavity of FAD in human XOR. FAD is shown as a yellow colored stick model. The amino acid residues experimentally studied with various systems are listed in Table 2. The unique amino acid cluster consisting of the side chains of Arg427, Arg335, Trp336 and Phe550, is shown as a space-filling model in green (PDB: 2E1Q).
4.3. The C-Terminal Moco Domain
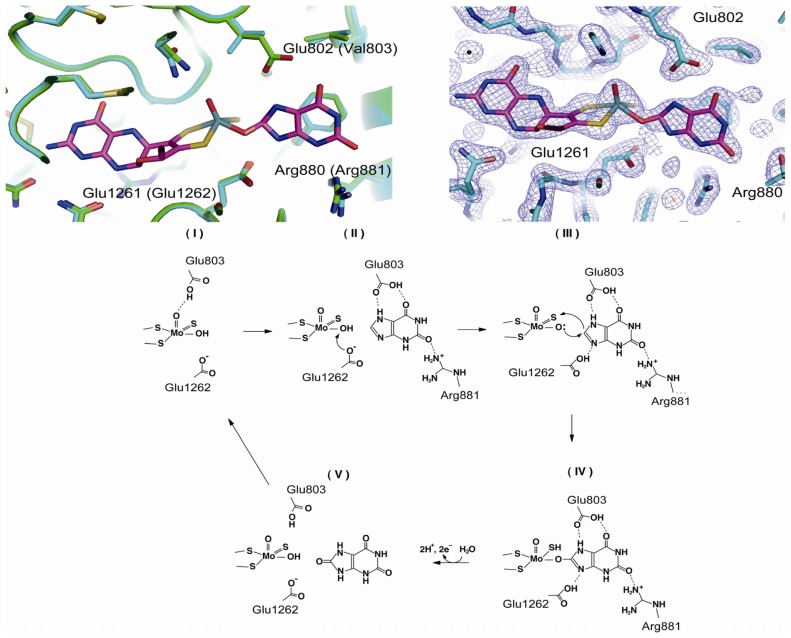
The large third domain (residues 591 to 1317, colored light blue in Figure 2) sequesters Moco close to the interface of the Fe/S- and FAD-binding domains. However, recombinantly expressed proteins, including human and rat enzymes, lack Moco, likely due to overloading of the Moco synthesis and insertion enzymes in the expression system [69,70]. High-resolution crystal structure analysis of a mutant of rat XDH (1.7 Å) indicated that the conformation of the polypeptide chain surrounding Moco is very similar to that found in the native bovine milk enzyme [86]. Although the amino acid residues in the active site do not differ greatly in their positions and orientations, crystallographic information regarding the interactions of amino acid residues with substrates and inhibitors is based only on data for native fully active bovine XOR, and the mechanism of hydroxylation has been well understood only in the last decade. The amino acid residues directly involved in substrate binding and catalysis are Glu803, Arg881 and Glu1262 (human sequence) [69] (Figure 4). In the oxidized form of XORs, the Mo ion is in the +VI oxidation state, surrounded by an oxo– (=O) at the apical position, and one hydroxo (–OH) and one sulfido (=S) ligand in the equatorial plane [16], in addition to the two vicinal sulfur ligands contributed by the pterin group (Figure 4). It is now generally accepted that XOR transfers the –OH to the substrate [6,76] (Figure 4). Proton transfer occurs upon substrate binding from Mo-OH to Glu1262, and the protonated Glu1262 forms a hydrogen bond with substrate nitrogen atom, facilitating nucleophilic attack on the adjoining carbon by the oxygen atom, which has become a base (Mo-O−) [6,76]. When the residue corresponding to Glu1262 was mutated, the enzyme was completely inactivated [69,87]. Regarding the activating role of the charged residues of the active center, it was found that purine hydroxylation activity is significantly decreased by mutation of two residues, Glu803 and Arg881, in the active site cavity of human XOR into the corresponding residues in the amino acid sequence of AO, Val803 and Met881, respectively [69]. However, the mutants exhibited significant AO activity. Proposed binding modes of substrates hypoxanthine and xanthine (Figure 4) have been proposed based on kinetic analysis of mutants, as illustrated in Figure 4. Those binding modes suggest that the activation mechanism facilitates nucleophilic reaction through hydrogen bond formation between the substrate and amino acid residues (Figure 4 bottom). The interaction of the 2-position keto group (C=O) and Arg881 is crucial for the efficacy of hydroxylation of the 8-position. These mechanisms are consistent with the metabolic sequence that hydroxylation of the 2-position of hypoxanthine precedes that at the 8-position [88–90]. X-Ray crystallography of the urate-bound reduced bovine XDH having full activity is consistent with this binding mode [86], as are the results of QM/MM studies with bovine XOR [91]. It was reported that two lysine residues were modified with fluorodinitrobenzene (FDNB) at pH 8.5, resulting in a decrease of activity due to slower release of the product, urate [75]. These residues were identified as Lys754 and Lys771 with rat XOR [74], both of which are located near the surface of the Moco domain, which may explain their accessibility to this chemical reagent. One of the nitro groups of DNB incorporated into a lysine residue of the enzyme was reported to be converted to an amino group due to reduction by substrate xanthine; this residue is most likely Lys771, which is rather close to the active site of the molybdenum center. Possible mechanisms will be discussed below.
Figure 4.
Binding modes of the substrate xanthine and mechanism of its hydroxylation. Upper left, superposition of the two crystal structures around Moco of human E803V mutant XDH (cyan) and reduced native bovine XDH in the urate-bound form (green) [86]. Upper right, electron-density map of reduced native bovine XDH with bound urate [86] (PDB: 3AMZ). Lower, proposed hydroxylation mechanism based on the crystal structure of the urate-bound form and the results of mutation studies [69].
5. Mutations Causing Type I Xanthinuria
Although inherited XOR deficiency was first reported in 1954 [33], detailed analysis of mutation sites of XOR was first reported in 1997 [37], and subsequently there have been several reports on XOR protein mutations associated with xanthinuria, as summarized in Table 3, including recent work on SNPs not necessarily associated with xanthinuria. The incidence of XOR deficiency, including type II, has been reported to be 1/69,000, but SNP analysis suggested a higher frequency of mutation in XOR, possibly because most mutations not cause dysfunction, being asymptomatic or merely producing a lower level of uric acid in blood.
Table 3.
Mutants causing type I xanthinuria.
| Codon change | Amino acid change | Codon number | Phenotype | Reference |
|---|---|---|---|---|
| c. 140_141insG (c. 140dupG) | p.Cys48LeufsX12 | 47 | Xanthinuria, type 1 | [92] |
| c. 445C > T | p.Arg149Cys | 149 | Xanthinuria, type 1 | [93] |
| c. 641delC | p.Pro214GlnfsX4 | 214 | Xanthinuria, type 1 | [94,95] |
| c. 682C > T | p.Arg228X | 228 | Xanthinuria, type 1 | [37] |
| c. 1664_1665insC (c.1664dupC) | p.Ala556SerfsX15 | 555 | Xanthinuria, type 1 | [96] |
| c. 1663C > T | p.Pro555Ser | 555 | Decreased activity | [97] |
| c. 1820G > A | p.Arg607Gln | 607 | Decreased activity | [97] |
| c. 1868C > T | p.Thr623Ile | 623 | Decreased activity | [97] |
| c. 2107A > G | p.Ile703Val | 703 | Increased activity | [97] |
| c. 2164A > T | p.Lys722X | 722 | Xanthinuria, type 1 | [98] |
| c. 2473C > T | p.Arg825X | 825 | Xanthinuria, type 1 | [95] |
| c. 2567delC | p.Thr856LysfsX73 | 856 | Xanthinuria, type 1 | [37,96] |
| c. 2641C > T | p.Arg881X | 881 | Xanthinuria, type 1 | [95] |
| c. 2727C > A | p.Asn909Lys | 909 | Decreased activity | [97] |
| c. 2729C > A | p.Thr910Lys | 910 | XDH deficiency | [97] |
| c. 2729C > T | p.Thr910Met | 910 | Xanthinuria, type 1 | [52,92] |
| c. 3449C > G | p.Pro1150Arg | 1150 | Decreased activity | [97] |
| c. 3662A > G | p.His1221Arg | 1221 | Increased activity | [97] |
| c. 3953G > A | p.Cys1318Tyr | 1318 | Decreased activity | [97] |
Any mutation that causes nonsense substitution [92,94–96,98,99] can be expected to cause loss of activity, since the active site of xanthine hydroxylation lies in the C-terminal domain and therefore truncated proteins should be inactive for hydroxylation. Arg881X is the longest peptide among the reported mutants having a stop codon (Table 3), and as the stop codon site is just at the active site region, as described above, it seems very likely that an active site cavity cannot be formed.
The mutation of Arg149Cys at the Fe/S I cluster motif [93] may influence the formation of the cluster, resulting in loss of electron transfer, even if the protein is completely processed and folded. Thr910 is located at a distance of 7.3 Å from Mo=S in the molybdenum center. Mutation of this residue to a bulky methionine or lysine residue seems likely to result in the loss of Moco or its sulfur atom, which is essential for the activity. Alternatively, insertion of the lysine residue may change the electrostatic environment in the active center cavity.
SNP analysis suggests that mutations of XOR may be quite frequent [97]. Although the conditions of activity determination, such as XDH/XO ratio and content of the desulfo-form of each mutant may have varied, it was reported that mutation of some residues not directly involved in the catalysis may result in partial loss of activity, possibly through effects on the protein conformation. It is intriguing to note that mutants Ile703Val and His1221Arg show increased activity due to an increase of Vmax. Those residues are located not in the active site cavity, but rather at the surface of the C-terminal Moco domain. As stopped-flow studies with XDH showed that the rate-limiting step of the overall reaction is release of urate, such mutation might increase the rate of release of urate. It has been reported that the kcat value of bacterial XDH is 10 times higher [87] and the enzyme inhibition pattern is very different from that of mammalian enzyme, i.e., bacterial XDH was not efficiently inhibited by febuxostat, a potent inhibitor of the mammalian enzyme [100]. Molecular dynamic simulation indicated that the bacterial enzyme molecule is much more mobile due to different mobility of surface amino acid residues, suggesting that the release rate of urate may be slower than that of the mammalian enzyme. This may be consistent with the finding that the modification of surface amino acid residues with FDNB caused slower release of urate, as described in the previous section.
6. Type II Xanthinuria Is the Consequence of Mutation of Human Moco Sulfurase Gene
As described above, the molybdenum atom of XOR and AO is coordinated by 5 atoms, of which one is a sulfide atom (Mo=S). During hydroxylation, two electrons from the substrate are transferred as hydride to Mo=S to form Mo-SH. The natural preparation is known to contain a significant amount of inactive form in which the sulfide atom (S) is replaced by an oxygen atom (O) [101]. The ratio Mo=O/Mo=S varies from batch to batch. The enzyme can be inactivated spontaneously by loss of sulfide, and the sulfide can also be removed by CN treatment to give SCN [101,102]. Fully active enzyme (Mo=S) can be separated using affinity chromatography [103,104]. The amount of desulfo-form seems to be regulated by the sulfur-donating activity in various organisms, including fly [102] or chicken [105,106], suggesting the existence of a sulfur-donating enzyme, Moco sulfurase. In Drosophila melanogaster, some mutations at maroon-like locus (ma-l) are known to cause inactivation of both XDH and XO [107], and combined deficiency of XOR and AO in humans was reported [108]. In 1995, it was proposed that type II xanthinuria might be due to a defect in sulfur donation, resulting in combined deficiency of XOR and AO [35]. Subsequently the ma-l gene, bovine Moco sulfurase gene and finally human Moco sulfurase gene were cloned and sequenced; all of them are members of a superfamily having a NifS-like domain in the N-terminal followed by a possible Moco-binding domain with a total of 888 amino acids [38,109,110]. Two independent xanthinuria patients were found to having a mutation that converts codon 419 to a nonsense codon [38]. Subsequently, other mutants, Ala156 to Pro [111] and Arg776 to Cys [112], were reported to cause type II xanthinuria. As human Moco sulfurase has not yet been successfully expressed as a soluble protein and its three-dimensional structure is not available, we can only speculate that the mutations cause some conformational change or folding error that affects Moco binding. Further studies can be expected on this interesting protein and on the mechanism of sulfur incorporation, including the question of whether the sulfur atom is incorporated before or after Moco is incorporated into XOR or AO protein.
Acknowledgments
This work was supported by Grant-in Aids (T.N. 24659144, K. O. 24590393, K. I. 23591205) for scientific research from the Japanese Ministry of Education, Science, Sports and Culture and the Gout Research Foundation of Japan.
Footnotes
Conflict of Interest
Authors declare no conflict of interests.
References
- 1.Nishino T. The conversion of xanthine dehydrogenase to xanthine oxidase and the role of the enzyme in reperfusion injury. J. Biochem. 1994;116:1–6. doi: 10.1093/oxfordjournals.jbchem.a124480. [DOI] [PubMed] [Google Scholar]
- 2.Hille R., Nishino T. Flavoprotein structure and mechanism. 4. Xanthine oxidase and xanthine dehydrogenase. Faseb J. 1995;9:995–1003. [PubMed] [Google Scholar]
- 3.Hille R. The Mononuclear Molybdenum Enzymes. Chem. Rev. 1996;96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- 4.Enroth C., Eger B.T., Okamoto K., Nishino T., Nishino T., Pai E.F. Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: Structure-based mechanism of conversion. Proc. Natl. Acad. Sci. USA. 2000;97:10723–10728. doi: 10.1073/pnas.97.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishino T., Okamoto K., Eger B.T., Pai E.F., Nishino T. Mammalian xanthine oxidoreductase mechanism of transition from xanthine dehydrogenase to xanthine oxidase. FEBS J. 2008;275:3278–3289. doi: 10.1111/j.1742-4658.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- 6.Hille R., Nishino T., Bittner F. Molybdenum enzymes in higher organisms. Coord. Chem. Rev. 2011;255:1179–1205. doi: 10.1016/j.ccr.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elion G.B. Enzymatic and metabolic studies with allopurinol. Ann. Rheum. Dis. 1966;25:608–614. doi: 10.1136/ard.25.suppl_6.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry C.E., Hare J.M. Xanthine oxidoreductase and cardiovascular disease: Molecular mechanisms and pathophysiological implications. J. Physiol. 2004;555:589–606. doi: 10.1113/jphysiol.2003.055913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellsten-Westing Y. Immunohistochemical localization of xanthine oxidase in human cardiac and skeletal muscle. Histochemistry. 1993;100:215–222. doi: 10.1007/BF00269094. [DOI] [PubMed] [Google Scholar]
- 10.Linder N., Rapola J., Raivio K.O. Cellular expression of xanthine oxidoreductase protein in normal human tissues. Lab Investig. 1999;79:967–974. [PubMed] [Google Scholar]
- 11.Angermuller S., Bruder G., Volkl A., Wesch H., Fahimi H.D. Localization of xanthine oxidase in crystalline cores of peroxisomes. A cytochemical and biochemical study. Eur. J. Cell. Biol. 1987;45:137–144. [PubMed] [Google Scholar]
- 12.Ichikawa M., Nishino T., Nishino T., Ichikawa A. Subcellular localization of xanthine oxidase in rat hepatocytes: High-resolution immunoelectron microscopic study combined with biochemical analysis. J. Histochem. Cytochem. 1992;40:1097–1103. doi: 10.1177/40.8.1619276. [DOI] [PubMed] [Google Scholar]
- 13.Elion G.B., Kovensky A., Hitchings G.H. Metabolic studies of allopurinol, an inhibitor of xanthine oxidase. Biochem. Pharmacol. 1966;15:863–880. doi: 10.1016/0006-2952(66)90163-8. [DOI] [PubMed] [Google Scholar]
- 14.Pacher P., Nivorozhkin A., Szabo C. Therapeutic effects of xanthine oxidase inhibitors: renaissance half a century after the discovery of allopurinol. Pharmacol. Rev. 2006;58:87–114. doi: 10.1124/pr.58.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto K., Eger B.T., Nishino T., Kondo S., Pai E.F., Nishino T. An extremely potent inhibitor of xanthine oxidoreductase. Crystal structure of the enzyme-inhibitor complex and mechanism of inhibition. J. Biol. Chem. 2003;278:1848–1855. doi: 10.1074/jbc.M208307200. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto K., Matsumoto K., Hille R., Eger B.T., Pai E.F., Nishino T. The crystal structure of xanthine oxidoreductase during catalysis: Implications for reaction mechanism and enzyme inhibition. Proc. Natl. Acad. Sci. USA. 2004;101:7931–7936. doi: 10.1073/pnas.0400973101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terkeltaub R. Update on gout: New therapeutic strategies and options. Nat. Rev. Rheumatol. 2010;6:30–38. doi: 10.1038/nrrheum.2009.236. [DOI] [PubMed] [Google Scholar]
- 18.Becker M.A., Schumacher H.R., Jr, Wortmann R.L., MacDonald P.A., Eustace D., Palo W.A., Streit J., Joseph-Ridge N. Febuxostat compared with allopurinol in patients with hyperuricemia and gout. N. Engl. J. Med. 2005;353:2450–2461. doi: 10.1056/NEJMoa050373. [DOI] [PubMed] [Google Scholar]
- 19.Kamatani N., Fujimori S., Hada T., Hosoya T., Kohri K., Nakamura T., Ueda T., Yamamoto T., Yamanaka H., Matsuzawa Y. An allopurinol-controlled, randomized, double-dummy, double-blind, parallel between-group, comparative study of febuxostat (TMX-67), a non-purine-selective inhibitor of xanthine oxidase, in patients with hyperuricemia including those with gout in Japan: phase 3 clinical study. J. Clin. Rheumatol. 2011;17:S13–S18. doi: 10.1097/RHU.0b013e31821d36cc. [DOI] [PubMed] [Google Scholar]
- 20.Massey V., Komai H., Palmer G., Elion G.B. On the mechanism of inactivation of xanthine oxidase by allopurinol and other pyrazolo [3,4-d]pyrimidines. J. Biol. Chem. 1970;245:2837–2844. [PubMed] [Google Scholar]
- 21.Okamoto K., Eger B.T., Nishino T., Pai E.F., Nishino T. Mechanism of inhibition of xanthine oxidoreductase by allopurinol: Crystal structure of reduced bovine milk xanthine oxidoreductase bound with oxipurinol. Nucleos. Nucleot. Nucleic Acids. 2008;27:888–893. doi: 10.1080/15257770802146577. [DOI] [PubMed] [Google Scholar]
- 22.Ames B.N., Cathcart R., Schwiers E., Hochstein P. Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: A hypothesis. Proc. Natl. Acad. Sci. USA. 1981;78:6858–6862. doi: 10.1073/pnas.78.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glantzounis G.K., Tsimoyiannis E.C., Kappas A.M., Galaris D.A. Uric acid and oxidative stress. Curr. Pharm. Des. 2005;11:4145–4151. doi: 10.2174/138161205774913255. [DOI] [PubMed] [Google Scholar]
- 24.Palmer R.M., Ferrige A.G., Moncada S. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature. 1987;327:524–526. doi: 10.1038/327524a0. [DOI] [PubMed] [Google Scholar]
- 25.Jones S.P., Bolli R. The ubiquitous role of nitric oxide in cardioprotection. J. Mol. Cell Cardiol. 2006;40:16–23. doi: 10.1016/j.yjmcc.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Lundberg J.O., Weitzberg E., Gladwin M.T. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 2008;7:156–167. doi: 10.1038/nrd2466. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi K., Miki M., Tagawa S. Pulse-radiolysis study of the reaction of nitric oxide with superoxide. J. Chem. Soc. Dalton. Trans. 1995:2885–2889. doi: 10.1039/DT9950002885. [DOI] [Google Scholar]
- 28.Nishino T., Nakanishi S., Okamoto K., Mizushima J., Hori H., Iwasaki T., Nishino T., Ichimori K., Nakazawa H. Conversion of xanthine dehydrogenase into oxidase and its role in reperfusion injury. Biochem. Soc. Trans. 1997;25:783–786. doi: 10.1042/bst0250783. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto K., Kusano T., Nishino T. Chemical Nature and Reaction Mechanisms of the Molybdenum Cofactor of Xanthine Oxidoreductase. Curr. Pharmaceut. Des. 2012 doi: 10.2174/1381612811319140010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Samouilov A., Liu X., Zweier J.L. Characterization of the magnitude and kinetics of xanthine oxidase-catalyzed nitrite reduction. Evaluation of its role in nitric oxide generation in anoxic tissues. J. Biol. Chem. 2001;276:24482–24489. doi: 10.1074/jbc.M011648200. [DOI] [PubMed] [Google Scholar]
- 31.Massey V., Brumby P.E., Komai H. Studies on milk xanthine oxidase. Some spectral and kinetic properties. J. Biol. Chem. 1969;244:1682–1691. [PubMed] [Google Scholar]
- 32.Saito T., Nishino T. Differences in redox and kinetic properties between NAD-dependent and O2-dependent types of rat liver xanthine dehydrogenase. J. Biol. Chem. 1989;264:10015–10022. [PubMed] [Google Scholar]
- 33.Dent C.E., Philpot G.R. Xanthinuria, an inborn error (or deviation) of metabolism. Lancet. 1954;266:182–185. doi: 10.1016/s0140-6736(54)91257-x. [DOI] [PubMed] [Google Scholar]
- 34.Kojima T., Nishina T., Kitamura M., Hosoya T., Nishioka K. Biochemical studies on the purine metabolism of four cases with hereditary xanthinuria. Clin. Chim. Acta. 1984;137:189–198. doi: 10.1016/0009-8981(84)90179-7. [DOI] [PubMed] [Google Scholar]
- 35.Simmonds H.A., Reiter S., Nishino T. Hereditary xanthinuria. In: Scriver C.R., editor. The Metabolic and Molecular Bases of Inherited Disease. 7th ed. McGraw-Hill Health Professions Division; New York, NY, USA: 1995. pp. 1781–1797. [Google Scholar]
- 36.Mateos F.A., Puig J.G., Jimenez M.L., Fox I.H. Hereditary xanthinuria. Evidence for enhanced hypoxanthine salvage. J. Clin. Invest. 1987;79:847–852. doi: 10.1172/JCI112893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ichida K., Amaya Y., Kamatani N., Nishino T., Hosoya T., Sakai O. Identification of two mutations in human xanthine dehydrogenase gene responsible for classical type I xanthinuria. J. Clin. Invest. 1997;99:2391–2397. doi: 10.1172/JCI119421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ichida K., Matsumura T., Sakuma R., Hosoya T., Nishino T. Mutation of human molybdenum cofactor sulfurase gene is responsible for classical xanthinuria type II. Biochem. Biophys. Res. Commun. 2001;282:1194–1200. doi: 10.1006/bbrc.2001.4719. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto T., Higashino K., Kono N., Kawachi M., Nanahoshi M., Takahashi S., Suda M., Hada T. Metabolism of pyrazinamide and allopurinol in hereditary xanthine oxidase deficiency. Clin. Chim. Acta. 1989;180:169–175. doi: 10.1016/0009-8981(89)90348-3. [DOI] [PubMed] [Google Scholar]
- 40.Ichida K., Yoshida M., Sakuma R., Hosoya T. Two siblings with classical xanthinuria type 1: Significance of allopurinol loading test. Intern. Med. 1998;37:77–82. doi: 10.2169/internalmedicine.37.77. [DOI] [PubMed] [Google Scholar]
- 41.Pryde D.C., Dalvie D., Hu Q., Jones P., Obach R.S., Tran T.D. Aldehyde oxidase: An enzyme of emerging importance in drug discovery. J. Med. Chem. 2010;53:8441–8460. doi: 10.1021/jm100888d. [DOI] [PubMed] [Google Scholar]
- 42.Garattini E., Terao M. The role of aldehyde oxidase in drug metabolism. Expert Opin. Drug Metab. Toxicol. 2012;8:487–503. doi: 10.1517/17425255.2012.663352. [DOI] [PubMed] [Google Scholar]
- 43.Kucera J., Bulkova T., Rychla R., Jahn P. Bilateral xanthine nephrolithiasis in a dog. J. Small Anim. Pract. 1997;38:302–305. doi: 10.1111/j.1748-5827.1997.tb03471.x. [DOI] [PubMed] [Google Scholar]
- 44.Van Zuilen C.D., Nickel R.F., van Dijk T.H., Reijngoud D.J. Xanthinuria in a family of Cavalier King Charles spaniels. Vet. Q. 1997;19:172–174. doi: 10.1080/01652176.1997.9694766. [DOI] [PubMed] [Google Scholar]
- 45.Tsuchida S., Kagi A., Koyama H., Tagawa M. Xanthine urolithiasis in a cat: A case report and evaluation of a candidate gene for xanthine dehydrogenase. J. Feline Med. Surg. 2007;9:503–508. doi: 10.1016/j.jfms.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miranda M., Rigueira L., Suarez M.L., Carbajales P., Moure P., Fidalgo L.E., Failde D., Vazquez S. Xanthine nephrolithiasis in a galician blond beef calf. J. Vet. Med. Sci. 2010;72:921–923. doi: 10.1292/jvms.09-0494. [DOI] [PubMed] [Google Scholar]
- 47.Oda M., Satta Y., Takenaka O., Takahata N. Loss of urate oxidase activity in hominoids and its evolutionary implications. Mol. Biol. Evol. 2002;19:640–653. doi: 10.1093/oxfordjournals.molbev.a004123. [DOI] [PubMed] [Google Scholar]
- 48.Bradbury M.G., Henderson M., Brocklebank J.T., Simmonds H.A. Acute renal failure due to xanthine stones. Pediatr. Nephrol. 1995;9:476–477. doi: 10.1007/BF00866732. [DOI] [PubMed] [Google Scholar]
- 49.Thomas N., Stephen D.C., Abraham B., Kekre N., Seshadri M.S. Xanthinuria—An unusual cause for renal stone disease. J. Assoc. Physicians. India. 1996;44:203–204. [PubMed] [Google Scholar]
- 50.Kiss A., Barenyi M., Csontai A. Xanthine stone in the urinary bladder of a male child. Urol. Int. 1999;63:242–244. doi: 10.1159/000030458. [DOI] [PubMed] [Google Scholar]
- 51.Al-Eisa A.A., Al-Hunayyan A., Gupta R. Pediatric urolithiasis in Kuwait. Int. Urol. Nephrol. 2002;33:3–6. doi: 10.1023/a:1014419830292. [DOI] [PubMed] [Google Scholar]
- 52.Arikyants N., Sarkissian A., Hesse A., Eggermann T., Leumann E., Steinmann B. Xanthinuria type I: A rare cause of urolithiasis. Pediatr. Nephrol. 2007;22:310–314. doi: 10.1007/s00467-006-0267-3. [DOI] [PubMed] [Google Scholar]
- 53.Gargah T., Essid A., Labassi A., Hamzaoui M., Lakhoua M.R. Xanthine urolithiasis. Saudi J. Kidney Dis. Transpl. 2010;21:328–331. [PubMed] [Google Scholar]
- 54.Martin H.M., Hancock J.T., Salisbury V., Harrison R. Role of xanthine oxidoreductase as an antimicrobial agent. Infect. Immun. 2004;72:4933–4939. doi: 10.1128/IAI.72.9.4933-4939.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vorbach C., Scriven A., Capecchi M.R. The housekeeping gene xanthine oxidoreductase is necessary for milk fat droplet enveloping and secretion: Gene sharing in the lactating mammary gland. Gene Dev. 2002;16:3223–3235. doi: 10.1101/gad.1032702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McManaman J.L., Palmer C.A., Wright R.M., Neville M.C. Functional regulation of xanthine oxidoreductase expression and localization in the mouse mammary gland: Evidence of a role in lipid secretion. J. Physiol. 2002;545:567–579. doi: 10.1113/jphysiol.2002.027185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakazono K., Watanabe N., Matsuno K., Sasaki J., Sato T., Inoue M. Does superoxide underlie the pathogenesis of hypertension? Proc. Natl. Acad. Sci. USA. 1991;88:10045–10048. doi: 10.1073/pnas.88.22.10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson R.J., Kang D.H., Feig D., Kivlighn S., Kanellis J., Watanabe S., Tuttle K.R., Rodriguez-Iturbe B., Herrera-Acosta J., Mazzali M. Is there a pathogenetic role for uric acid in hypertension and cardiovascular and renal disease? Hypertension. 2003;41:1183–1190. doi: 10.1161/01.HYP.0000069700.62727.C5. [DOI] [PubMed] [Google Scholar]
- 59.Elion G.B. The purine path to chemotherapy. Science. 1989;244:41–47. doi: 10.1126/science.2649979. [DOI] [PubMed] [Google Scholar]
- 60.Johnson J.L., Duran M. Molybdenum Cofactor Deficiency and Isolated Sulfite Oxidase Deficiency. In: Scriver C.R., Childs B., Kinzler K.W., Vogelstein B., editors. The Metabolic & Molecular Bases of Inherited Disease. 8th ed. McGraw-Hill; New York, NY, USA: 2001. pp. 3163–3177. [Google Scholar]
- 61.Ichida K., Amaya Y., Noda K., Minoshima S., Hosoya T., Sakai O., Shimizu N., Nishino T. Cloning of the cDNA encoding human xanthine dehydrogenase (oxidase): Structural analysis of the protein and chromosomal location of the gene. Gene. 1993;133:279–284. doi: 10.1016/0378-1119(93)90652-j. [DOI] [PubMed] [Google Scholar]
- 62.Amaya Y., Yamazaki K., Sato M., Noda K., Nishino T., Nishino T. Proteolytic conversion of xanthine dehydrogenase from the NAD-dependent type to the O2-dependent type. Amino acid sequence of rat liver xanthine dehydrogenase and identification of the cleavage sites of the enzyme protein during irreversible conversion by trypsin. J. Biol. Chem. 1990;265:14170–14175. [PubMed] [Google Scholar]
- 63.Minoshima S., Wang Y., Ichida K., Nishino T., Shimizu N. Mapping of the gene for human xanthine dehydrogenase (oxidase) (XDH) to band p23 of chromosome 2. Cytogenet. Cell Genet. 1995;68:52–53. doi: 10.1159/000133887. [DOI] [PubMed] [Google Scholar]
- 64.Xu P., Huecksteadt T.P., Hoidal J.R. Molecular cloning and characterization of the human xanthine dehydrogenase gene (XDH) Genomics. 1996;34:173–180. doi: 10.1006/geno.1996.0262. [DOI] [PubMed] [Google Scholar]
- 65.Wright R.M., Vaitaitis G.M., Wilson C.M., Repine T.B., Terada L.S., Repine J.E. cDNA cloning, characterization, and tissue-specific expression of human xanthine dehydrogenase/xanthine oxidase. Proc. Natl. Acad. Sci. USA. 1993;90:10690–10694. doi: 10.1073/pnas.90.22.10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saksela M., Raivio K.O. Cloning and expression in vitro of human xanthine dehydrogenase/oxidase. Biochem. J. 1996;315:235–239. doi: 10.1042/bj3150235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garattini E., Fratelli M., Terao M. Mammalian aldehyde oxidases: Genetics, evolution and biochemistry. Cell Mol. Life Sci. 2008;65:1019–1048. doi: 10.1007/s00018-007-7398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boer D.R., Thapper A., Brondino C.D., Romao M.J., Moura J.J. X-ray crystal structure and EPR spectra of “arsenite-inhibited” Desulfovibriogigas aldehyde dehydrogenase: A member of the xanthine oxidase family. J. Am. Chem. Soc. 2004;126:8614–8615. doi: 10.1021/ja0490222. [DOI] [PubMed] [Google Scholar]
- 69.Yamaguchi Y., Matsumura T., Ichida K., Okamoto K., Nishino T. Human xanthine oxidase changes its substrate specificity to aldehyde oxidase type upon mutation of amino acid residues in the active site: roles of active site residues in binding and activation of purine substrate. J. Biochem. 2007;141:513–524. doi: 10.1093/jb/mvm053. [DOI] [PubMed] [Google Scholar]
- 70.Nishino T., Okamoto K., Kawaguchi Y., Hori H., Matsumura T., Eger B.T., Pai E.F., Nishino T. Mechanism of the conversion of xanthine dehydrogenase to xanthine oxidase: Identification of the two cysteine disulfide bonds and crystal structure of a non-convertible rat liver xanthine dehydrogenase mutant. J. Biol. Chem. 2005;280:24888–24894. doi: 10.1074/jbc.M501830200. [DOI] [PubMed] [Google Scholar]
- 71.Iwasaki T., Okamoto K., Nishino T., Mizushima J., Hori H. Sequence motif-specific assignment of two [2Fe-2S] clusters in rat xanthine oxidoreductase studied by site-directed mutagenesis. J. Biochem. 2000;127:771–778. doi: 10.1093/oxfordjournals.jbchem.a022669. [DOI] [PubMed] [Google Scholar]
- 72.Kuwabara Y., Nishino T., Okamoto K., Matsumura T., Eger B.T., Pai E.F., Nishino T. Unique amino acids cluster for switching from the dehydrogenase to oxidase form of xanthine oxidoreductase. Proc. Natl. Acad. Sci. USA. 2003;100:8170–8175. doi: 10.1073/pnas.1431485100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nishino T. The nicotinamide adenine dinucleotide-binding site of chicken liver xanthine dehydrogenase. Evidence for alteration of the redox potential of the flavin by NAD binding or modification of the NAD-binding site and isolation of a modified peptide. J. Biol. Chem. 1989;264:5468–5473. [PubMed] [Google Scholar]
- 74.Nishino T. The conversion from the dehydrogenase type to the oxidase type of rat liver xanthine dehydrogenase by modification of cysteine residues with fluorodinitrobenzene. J. Biol. Chem. 1997;272:29859–29864. doi: 10.1074/jbc.272.47.29859. [DOI] [PubMed] [Google Scholar]
- 75.Nishino T., Tsushima K., Hille R., Massey V. Inhibition of milk xanthine oxidase by fluorodinitrobenzene. J. Biol. Chem. 1982;257:7348–7353. [PubMed] [Google Scholar]
- 76.Huber R., Hof P., Duarte R.O., Moura J.J., Moura I., Liu M.Y., LeGall J., Hille R., Archer M., Romao M.J. A structure-based catalytic mechanism for the xanthine oxidase family of molybdenum enzymes. Proc. Natl. Acad. Sci. USA. 1996;93:8846–8851. doi: 10.1073/pnas.93.17.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Palmer G., Massey V. Electron paramagnetic resonance and circular dichroism studies on milk xanthine oxidase. J. Biol. Chem. 1969;244:2614–2620. [PubMed] [Google Scholar]
- 78.Porras A.G., Palmer G. The room temperature potentiometry of xanthine oxidase. Ph-dependent redox behavior of the flavin, molybdenum, and iron-sulfur centers. J. Biol. Chem. 1982;257:11617–11626. [PubMed] [Google Scholar]
- 79.Caldeira J., Belle V., Asso M., Guigliarelli B., Moura I., Moura J.J., Bertrand P. Analysis of the electron paramagnetic resonance properties of the [2Fe-2S]1+ centers in molybdenum enzymes of the xanthine oxidase family: Assignment of signals I and II. Biochemistry. 2000;39:2700–2707. doi: 10.1021/bi9921485. [DOI] [PubMed] [Google Scholar]
- 80.Hille R., Hagen W.R., Dunham W.R. Spectroscopic studies on the iron-sulfur centers of milk xanthine oxidase. J. Biol. Chem. 1985;260:10569–10575. [PubMed] [Google Scholar]
- 81.Ishikita H., Eger B.T., Okamoto K., Nishino T., Pai E.F. Protein conformational gating of enzymatic activity in xanthine oxidoreductase. J. Am. Chem. Soc. 2012;134:999–1009. doi: 10.1021/ja207173p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Massey V., Schopfer L.M., Nishino T., Nishino T. Differences in protein structure of xanthine dehydrogenase and xanthine oxidase revealed by reconstitution with flavin active site probes. J. Biol. Chem. 1989;264:10567–10573. [PubMed] [Google Scholar]
- 83.Saito T., Nishino T., Massey V. Differences in environment of FAD between NAD-dependent and O2-dependent types of rat liver xanthine dehydrogenase shown by active site probe study. J. Biol. Chem. 1989;264:15930–15935. [PubMed] [Google Scholar]
- 84.Hunt J., Massey V. Purification and properties of milk xanthine dehydrogenase. J. Biol. Chem. 1992;267:21479–21485. [PubMed] [Google Scholar]
- 85.Tsujii A., Nishino T. Mechanism of transition from xanthine dehydrogenase to xanthine oxidase: Effect of guanidine-HCL or urea on the activity. Nucleos. Nucleot. Nucleic Acids. 2008;27:881–887. doi: 10.1080/15257770802146569. [DOI] [PubMed] [Google Scholar]
- 86.Okamoto K., Kawaguchi Y., Eger B.T., Pai E.F., Nishino T. Crystal Structures of Urate Bound Form of Xanthine Oxidoreductase: Substrate Orientation and Structure of the Key Reaction Intermediate. J. Am. Chem. Soc. 2010;132:17080–17083. doi: 10.1021/ja1077574. [DOI] [PubMed] [Google Scholar]
- 87.Leimkuhler S., Stockert A.L., Igarashi K., Nishino T., Hille R. The role of active site glutamate residues in catalysis of Rhodobacter capsulatus xanthine dehydrogenase. J. Biol. Chem. 2004;279:40437–40444. doi: 10.1074/jbc.M405778200. [DOI] [PubMed] [Google Scholar]
- 88.Bergmann F., Dikstein S. Studies on uric acid and related compounds III. Observations on the specificity of mammalian xanthine oxidases. J. Biol. Chem. 1956;223:765–780. [PubMed] [Google Scholar]
- 89.Cao H., Pauff J.M., Hille R. Substrate orientation and catalytic specificity in the action of xanthine oxidase: The sequential hydroxylation of hypoxanthine to uric acid. J. Biol. Chem. 2010;285:28044–28053. doi: 10.1074/jbc.M110.128561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jezewska M.M. Xanthine accumulation during hypoxanthine oxidation by milk xanthine oxidase. Eur. J. Biochem. 1973;36:385–390. doi: 10.1111/j.1432-1033.1973.tb02923.x. [DOI] [PubMed] [Google Scholar]
- 91.Metz S., Thiel W. A combined QM/MM study on the reductive half-reaction of xanthine oxidase: substrate orientation and mechanism. J. Am. Chem. Soc. 2009;131:14885–14902. doi: 10.1021/ja9045394. [DOI] [PubMed] [Google Scholar]
- 92.Nakamura M., Yuichiro Y., Jorn Oliver S., Tomohiro M., Schwab K.O., Takeshi N., Tatsuo H., Ichida K. Identification of a xanthinuria type I case with mutations of xanthine dehydrogenase in an Afghan child. Clin. Chim. Acta. 2012;414:158–160. doi: 10.1016/j.cca.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 93.Sakamoto N., Yamamoto T., Moriwaki Y., Teranishi T., Toyoda M., Onishi Y., Kuroda S., Sakaguchi K., Fujisawa T., Maeda M., et al. Identification of a new point mutation in the human xanthine dehydrogenase gene responsible for a case of classical type I xanthinuria. Hum. Genet. 2001;108:279–283. doi: 10.1007/s004390100477. [DOI] [PubMed] [Google Scholar]
- 94.Jurecka A., Stiburkova B., Krijt J., Gradowska W., Tylki-Szymanska A. Xanthine dehydrogenase deficiency with novel sequence variations presenting as rheumatoid arthritis in a 78-year-old patient. J. Inherit. Metab. Dis. 2010 doi: 10.1007/s10545-009-9011-z. [DOI] [PubMed] [Google Scholar]
- 95.Stiburkova B., Krijt J., Vyletal P., Bartl J., Gerhatova E., Korinek M., Sebesta I. Novel mutations in xanthine dehydrogenase/oxidase cause severe hypouricemia: Biochemical and molecular genetic analysis in two Czech families with xanthinuria type I. Clin. Chim. Acta. 2012;413:93–99. doi: 10.1016/j.cca.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 96.Levartovsky D., Lagziel A., Sperling O., Liberman U., Yaron M., Hosoya T., Ichida K., Peretz H. XDH gene mutation is the underlying cause of classical xanthinuria: A second report. Kidney Int. 2000;57:2215–2220. doi: 10.1046/j.1523-1755.2000.00082.x. [DOI] [PubMed] [Google Scholar]
- 97.Kudo M., Moteki T., Sasaki T., Konno Y., Ujiie S., Onose A., Mizugaki M., Ishikawa M., Hiratsuka M. Functional characterization of human xanthine oxidase allelic variants. Pharmacogenetics Genom. 2008;18:243–251. doi: 10.1097/FPC.0b013e3282f55e2e. [DOI] [PubMed] [Google Scholar]
- 98.Gok F., Ichida K., Topaloglu R. Mutational analysis of the xanthine dehydrogenase gene in a Turkish family with autosomal recessive classical xanthinuria. Nephrol. Dial. Transplant. 2003;18:2278–2283. doi: 10.1093/ndt/gfg385. [DOI] [PubMed] [Google Scholar]
- 99.Fujiwara Y., Kawakami Y., Shinohara Y., Ichida K. A case of hereditary xanthinuria type 1 accompanied by bilateral renal calculi. Intern. Med. 2012;51:1879–1884. doi: 10.2169/internalmedicine.51.6891. [DOI] [PubMed] [Google Scholar]
- 100.Kikuchi H., Fujisaki H., Furuta T., Okamoto K., Leimkuhler S., Nishino T. Different inhibitory potency of febuxostat towards mammalian and bacterial xanthine oxidoreductases: Insight from molecular dynamics. Sci. Rep. 2012;2:331. doi: 10.1038/srep00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Massey V., Edmondson D. On the mechanism of inactivation of xanthine oxidase by cyanide. J. Biol. Chem. 1970;245:6595–6598. [PubMed] [Google Scholar]
- 102.Wahl R.C., Rajagopalan K.V. Evidence for the inorganic nature of the cyanolyzable sulfur of molybdenum hydroxylases. J. Biol. Chem. 1982;257:1354–1359. [PubMed] [Google Scholar]
- 103.Nishino T., Nishino T., Tsushima K. Purification of highly active milk xanthine oxidase by affinity chromatography on Sepharose 4B/folate gel. FEBS Lett. 1981;131:369–372. doi: 10.1016/0014-5793(81)80406-1. [DOI] [PubMed] [Google Scholar]
- 104.Ikegami T., Nishino T. The presence of desulfo xanthine dehydrogenase in purified and crude enzyme preparations from rat liver. Arch. Biochem. Biophys. 1986;247:254–260. doi: 10.1016/0003-9861(86)90582-5. [DOI] [PubMed] [Google Scholar]
- 105.Nishino T. Purification of hepatic xanthine dehydrogenase from chicken fed a high-protein diet. Biochim. Biophys. Acta. 1974;341:93–98. doi: 10.1016/0005-2744(74)90069-2. [DOI] [PubMed] [Google Scholar]
- 106.Itoh R., Nishino T., Usami C., Tsushima K. An immunochemical study of the changes in chicken liver xanthine dehydrogenase activity during dietary adaptation. J. Biochem. 1978;84:19–26. doi: 10.1093/oxfordjournals.jbchem.a132107. [DOI] [PubMed] [Google Scholar]
- 107.Wahl R.C., Warner C.K., Finnerty V., Rajagopalan K.V. Drosophila melanogaster ma-l mutants are defective in the sulfuration of desulfo Mo hydroxylases. J. Biol. Chem. 1982;257:3958–3962. [PubMed] [Google Scholar]
- 108.Reiter S., Simmonds H.A., Zollner N., Braun S.L., Knedel M. Demonstration of a combined deficiency of xanthine oxidase and aldehyde oxidase in xanthinuric patients not forming oxipurinol. Clin. Chim. Acta. 1990;187:221–234. doi: 10.1016/0009-8981(90)90107-4. [DOI] [PubMed] [Google Scholar]
- 109.Amrani L., Primus J., Glatigny A., Arcangeli L., Scazzocchio C., Finnerty V. Comparison of the sequences of the Aspergillus nidulans hxB and Drosophila melanogaster ma-l genes with nifS from Azotobacter vinelandii suggests a mechanism for the insertion of the terminal sulphur atom in the molybdopterin cofactor. Mol. Microbiol. 2000;38:114–125. doi: 10.1046/j.1365-2958.2000.02119.x. [DOI] [PubMed] [Google Scholar]
- 110.Watanabe T., Ihara N., Itoh T., Fujita T., Sugimoto Y. Deletion mutation in Drosophila ma-l homologous, putative molybdopterin cofactor sulfurase gene is associated with bovine xanthinuria type II. J. Biol. Chem. 2000;275:21789–21792. doi: 10.1074/jbc.C000230200. [DOI] [PubMed] [Google Scholar]
- 111.Yamamoto T., Moriwaki Y., Takahashi S., Tsutsumi Z., Tuneyoshi K., Matsui K., Cheng J., Hada T. Identification of a new point mutation in the human molybdenum cofactor sulferase gene that is responsible for xanthinuria type II. Metabolism. 2003;52:1501–1504. doi: 10.1016/s0026-0495(03)00272-5. [DOI] [PubMed] [Google Scholar]
- 112.Peretz H., Naamati M.S., Levartovsky D., Lagziel A., Shani E., Horn I., Shalev H., Landau D. Identification and characterization of the first mutation (Arg776Cys) in the C-terminal domain of the Human Molybdenum Cofactor Sulfurase (HMCS) associated with type II classical xanthinuria. Mol. Genet. Metabol. 2007;91:23–29. doi: 10.1016/j.ymgme.2007.02.005. [DOI] [PubMed] [Google Scholar]