Abstract
Two cDNA clones encoding endo-β-1,4-glucanases (EGases) were isolated from a radiata pine (Pinus radiata) cDNA library prepared from immature female strobili. The cDNAs PrCel1 (Pinus radiata cellulase 1) and PrCel2 encode proteins 509 and 515 amino acids in length, respectively, including putative signal peptides. Both proteins contain domains conserved in plant and bacterial EGases. The proteins PRCEL1 and PRCEL2 showed strong similarity to each other (76% amino acid identity), and higher similarity to TPP18 (73 and 67%, respectively), an EGase cloned from tomato (Lycopersicon esculentum) pistils, than to any other reported EGases. Northern-blot analyses indicated that both genes displayed a similar pattern of expression. The only significant difference was in the level of expression. In situ hybridizations were used to demonstrate that, within differentiating pine reproductive structures, PrCel1 expression was greatest in microsporangia in pollen strobili and near the developing ovule in the seed strobili. Expression was also found in vegetative tissues, especially in regions experiencing cell elongation, such as the elongating region of root tips. Both proteins have an ability to degrade carboxymethylcellulose in vitro. Genomic-blot analysis indicated the presence of a family of EGase genes in the radiata pine genome, and that PrCel1 and PrCel2 are transcribed from distinct one-copy genes.
The growth and development of floral organs involves many physiological processes, including modifications to the cell wall. EGases (cellulases) may play roles in cell wall loosening, which is required for expansion or major cell wall disruption. Cell expansion has been reviewed by Cosgrove (1993), who demonstrated that acid-induced extension of cell walls appears to require the activity of expansins. Wall-modifying enzymes such as endoxyloglucan transferase, and wall-degrading enzymes such as glucanases, are also likely to be involved, but there is no evidence that they can cause extension of isolated walls. Major cell wall disruption also occurs at several steps in the development of flower reproductive organs (del Campillo and Lewis, 1992). The callose wall that protects the meiotic cells is broken down during early pollen differentiation, releasing the microspores into the anther locule. Later, the tapetum begins to break down and the cytoplasm is released into the locule. Finally, the release of the mature pollen grains from the anthers is facilitated by the formation of a fissure, the stomium. Similarly, during pollen-stigma interactions, cell wall loosening of the papillary cells at the surface of the stigma has been reported. EGases have been shown to accumulate in anthers of beans and sweet peas in a developmentally regulated manner and may be involved in the cell wall disruption required for pollen differentiation.
Plant EGases typically lack the ability to degrade microcrystalline cellulose in vitro. Bacterial EGases, however, are able to degrade cellulose. Therefore, all EGases are sometimes referred to as cellulases. Genes encoding EGases have been isolated from many different plant species, including tomato (Lycopersicon esculentum) (Lashbrook et al., 1994; Milligan and Gasser, 1995), elder (Sambucus nigra) (Taylor et al., 1994), pea (Pisum sativum) (Wu et al., 1996), soybean (Glycine max) (Kemmerer and Tucker, 1994), Arabidopsis (Ferl, 1995), poplar (Populus alba) (Nakamura et al., 1995), kidney bean (Phaeseolus vulgaris) (Tucker and Milligan, 1991), and avocado (Persea americana) (Tucker et al., 1987). Some of these enzymes, including TomCel2 (Lashbrook et al., 1994), EGL1 (Wu et al., 1996), and AvoCel1 (Tucker et al., 1987), are primarily associated with fruit ripening. Another group, including BAC (Tucker and Milligan, 1991), SAC1 (Kemmerer and Tucker, 1994), TomCel1 (Lashbrook et al., 1994), and JET1 (Taylor et al., 1994), are associated with abscission. Yet another group of enzymes appears to be expressed predominantly in rapidly expanding tissues. Expression of TPP18 (Milligan and Gasser, 1995), which is identical to Cel4 (Brummell et al., 1997), occurs in growing pistils of tomato flowers, and to a lesser extent in stamens, but not in fully expanded flower parts. Expression is also high in the growing zones of etiolated hypocotyls and in expanding leaves. Here we report on the expression of two EGases cloned from reproductive structures of radiata (Monterey) pine (Pinus radiata).
As in angiosperms, the “flowering” of radiata pine starts with the transition of an undetermined axillary apex into a determinant reproductive apex, which develops into the strobili (cones). Reproductive buds are simple because they normally contain a single strobilus and no leaves. Mature male (pollen) cones are small (1–2 cm in length) and are made up of spirally arranged microsporophylls, each bearing two microsporangia (pollen sacs). The microspores develop into four-celled pollen grains. Female (seed) cones consist of an axis, which bears a specially arranged series of small appendages termed bracts. In the axil of each bract is a thick scale upon which two ovules are borne, attached to the adaxial surface of the cone scale near the base. Because the ovuliferous scales are lateral structures subtended by a bract, the entire cone is a “compound” strobilus, and may be compared in this respect with an inflorescence. Such female axes generally are located at the top of the adult tree, whereas male cones are located farther down the stem and contain only microsporophylls.
A few genes have previously been cloned from various parts or stages of developing radiata pine cones, including the cDNAs encoding genes preferentially expressed in immature female and male cone buds. Homologs of the angiosperm late-flowering, meristem-identity, and organ-identity genes regulate development of unisexual cones in the conifer radiata pine (Mouradov et al., 1996, 1997a, 1997b). MADS box genes have also been cloned from another conifer, Norway spruce (Picea abies) (Tandre et al., 1995). Two different cDNAs with homology to legumins have been isolated from fertilized ovules of white pine (Pinus strobus), but are not expressed in unfertilized ovules (Baker et al., 1996). Several cDNA clones encoding seed-storage proteins have also been isolated from Douglas fir and interior spruce megagametophytes (Newton et al., 1992; Leal and Misra, 1993). Mature megagametophytes have been used for many years to study isozyme variation, and are commonly used as sources of DNA for genome mapping because of their haploid condition.
To gain a greater understanding of the genes involved in the formation of pine reproductive structures, we constructed a cDNA library from immature female cones and differentially screened against vegetative buds. Here we report on the cloning, sequencing, and characterization of two of those genes, PrCel1 and PrCel2, that have very high homology to each other and to previously cloned EGases.
MATERIALS AND METHODS
Immature female, male, and vegetative shoots were collected from an adult radiata (Monterey) pine (Pinus radiata) tree, approximately 20 years of age and 30 m in height, in Victoria, Australia, commencing at the end of March until early April 1994. The cone-scale organization was visible at this stage of development, but no ovules and/or pollen grains were discernible. The PCBs and SCBs at this stage were approximately 1.5 mm long and weighed about 5 mg. Vegetative shoots at this stage were less than 2 mm long. SCBs and PCBs were collected at different stages of immature ovule and microsporangia development, from 0.5 to 5 mm long, throughout the winter until November 1994 (spring). Elongated needles were also collected during this period. Tissues were dissected and frozen in liquid nitrogen. Roots and stems were collected from seedlings grown for 3 to 4 weeks in growth cabinets.
Isolation of RNA
The various tissues were homogenized in buffer (5.5 m guanidinium thiocyanate, 25 mm sodium citrate, 0.5% sarcosyl, 0.2 m β-mercaptoethanol, and 4% [w/v] PVP [Mr 40,000]). The DNA was sheared by passing the homogenate through a 16-gauge needle, and the debris were pelleted. The RNA was pelleted through cesium trifluoroacetate (1.51 g/mL, Pharmacia Biotech) in a swinging bucket rotor, in a bench-top ultracentrifuge (Beckman TLX; 55,000 rpm for 3 h). The RNA pellets were resuspended in 10 mm Tris-HCl (pH 8.0), 1 mm EDTA, extracted with chloroform:butanol (4:1, v/v), and precipitated with potassium acetate and ethanol. Isolation of mRNA was performed using the Poly AT tract kit (Promega). For northern-blot hybridization experiments 20 μg of total RNA was run on a 1% formaldehyde/agarose gel, blotted onto nylon membranes, and hybridized to 32P-labeled PrCel1 and PrCel2 cDNA clones.
cDNA Library Construction and Screening
A cDNA library was constructed using the Pharmacia Time Saver cDNA synthesis kit, and mRNA was isolated from immature female cones weighing approximately 100 mg. These cones were about to emerge from the bud scales. Ten percent of the cDNA was cloned into λExCell arms (Pharmacia) and packaged into phage. This resulted in a cDNA library with approximately 86,000 clones. Duplicate plaque lifts were made, and each filter hybridized to the cDNA probe produced from either female cones or vegetative buds. Clones with much stronger hybridization to the cone probe than to the bud probe were plaque purified for further characterization.
Expression in Escherichia coli
For expression in E. coli, the coding regions of cDNA sequences encoding the mature protein sequences of PrCel1 and PrCel2 were amplified using Pfu DNA polymerase (Stratagene) and the following sets of forward and reverse primers. For the PrCel1 clone: primer 1, 5′-TAGGGCATGCAATTATACTATAGAGAGC-3′; primer 2, 5′-CGCAAGCTTTCAGGACATGGTAGAATG-3′. And for the PrCel2 clone: primer 3, 5′-ACATGCATGCCTTCCATAGAATTATACCTC-3′; primer 4, 5′-GGCAAGCTTCTAAG-CGAAACTGTGTGC-3′. Primers designed for 5′ end sequences (primers 1 and 2) contained SphI cloning sites and primers designed for the 3′ ends (primers 3 and 4) contained HindIII sites.
After amplification, the PCR fragments were digested by SphI and HindIII restriction enzymes and cloned into the E. coli expression vector pQE31 (Qiagen, Chatsworth, CA) digested by the same restriction enzymes. After expression of the recombinant proteins in E. coli, PRCEL1 and PRCEL2 proteins were purified using Ni-NTA resin (Qiagen) and run on gradient (4–20%) gels (Novex, San Diego, CA) using Tris-Gly SDS running buffer (Novex). Gels were stained with Coomassie blue G-250 stain (Colloidal Coomassie Staining Kit, Novex).
Assay of EGase Activity
Different amounts of Ni-NTA-purified protein (10, 100, and 500 ng) were loaded onto 1% agarose plates containing 0.2% CMC (Sigma) in 0.1 m phosphate buffer, pH 6.5, and incubated overnight at 37°C. Plates were stained with Congo red (1 mg/mL) for 10 min at room temperature and destained in 1 m NaCl for another 10 min.
Southern-Blot Hybridizations
Genomic DNA was isolated from needles using Genomic Tips 100 G (Qiagen). Twenty micrograms of genomic DNA was digested separately with HindIII and EcoRI restriction enzymes. DNA samples were run on 0.7% agarose gel, blotted, and hybridized to 32P-labeled PrCel1 and PrCel2 cDNA clones. Hybridization was performed at 65°C in 5× SSCP, 5× Denhardt's solution, and 0.5% SDS. Membranes were washed at 65°C in 2× SSC, 0.1% SDS (moderate stringency), and 0.1× SSC, 0.1% SDS (high stringency).
Plasmid DNA Sequencing
Sequencing reactions were carried out using the Dye Terminator Cycle DNA Sequencing Kit (Perkin-Elmer Cetus) and products were separated on an automated sequenator (model 377, Applied Biosystems). Sequence data were analyzed using the GCG sequence-analysis program (Genetics Computer Group, Madison, WI).
In Situ Hybridization
The antisense probe used in this study was a single-stranded, digoxigenin-labeled RNA probe derived from the original PrCel1 cDNA clone in a pExCell vector (Pharmacia). The plasmid was linearized by digestion with HindIII (in the polylinker) and used as a template for synthesizing RNA using SP6 polymerase. As a control, a single-stranded, digoxigenin-labeled sense probe was synthesized using T7 polymerase from the same plasmid linearized with SfiI (in the polylinker). Plant material was fixed in 3.7% formaldehyde, dehydrated, cleared, and embedded in wax, and sections were prepared for in situ hybridization according to Jack et al. (1992), with minor modifications; Histoclear (National Diagnostics, Buckinghamshire, UK) was used instead of xylene throughout the procedure. Larger plant material was presectioned to about 1 mm in thickness to aid infiltration of the fixative. Tissues were gently agitated in fixative for a total period of 3 h. Sections were baked onto slides coated with 3-aminopropyltriethoxysilane at 65°C for 18 h. The RNA probe was subjected to mild alkali hydrolysis by heating at 60°C for 1 h in 100 mm carbonate buffer (pH 10.2), and approximately 8% of each labeling reaction was added to 120 μL of hybridization buffer (Jack et al., 1992) and coated onto each slide. Slides were incubated overnight at 42°C, then washed by the method of Jack et al. (1992). After the RNase A incubation and low- and high-stringency washes, the slides were rinsed twice in PBS, then stored in this buffer at 4°C for 5 min to overnight. Sections were mounted in Histomount (National Diagnostics) and examined under bright-field and dark-field illumination. For each experiment, similar sections were hybridized with the sense and antisense probes in parallel.
Construction of Phylogenic Tree
Alignment of conceptual amino acid sequences and the phylogenic-tree analysis were performed with the GCG program using the Phylogeny Interface Package. The signal peptide sequences were omitted in these analyses.
RESULTS
Organogenic Sequence of Seed Cone and Pollen Cone Differentiation in Radiata Pine
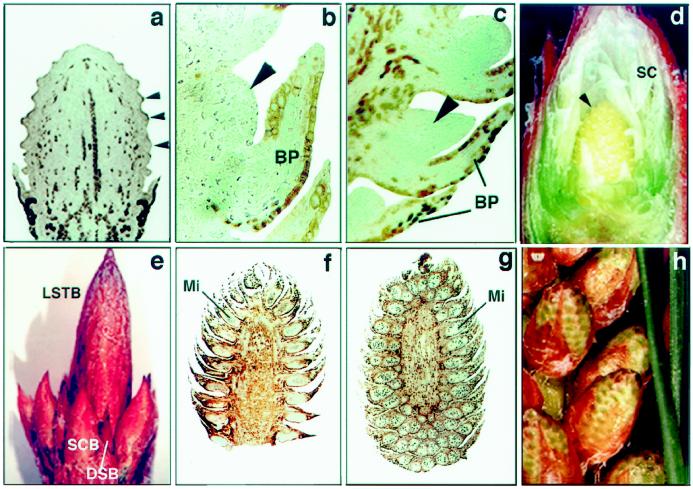
In radiata pine lateral branches are terminated by a LSTB. The LSTB could be classified as vegetative, containing only vegetative DSBs; as male, containing both PCBs and DSBs; or as female, containing both SCBs and DSBs. The SCBs became anatomically differentiated with initiation of bracts, which were first apparent as numerous conspicuous and regularly spaced pockets of randomly dividing cells in the peripheral zone of the SCB apex (Fig. 1a, 5-mg cone). Ovuliferous scale primordia were initiated from hypodermal cells on the adaxial base of bracts (not shown). As the bract and ovuliferous scale developed, the scale became displaced from the cone-bud axis and a fused bract-ovuliferous scale complex resulted (Fig. 1b, 50-mg cone). Figure 1, c and d, illustrates more developed (100 mg) SCBs with more defined ovules. A female LSTB at a later stage of SCB development (300–500 mg) is shown in Figure 1e. Differentiation of PCBs is associated with the development of the sporogenous pollen mother cells within microsporophylls initiating on the peripheral zone of the PCB apex (Fig. 1f). Figure 1g shows the PCB after the completion of microsporophyll initiation. A male LSTB with more developed (300 mg) PCB is shown in Figure 1h.
Figure 1.
Organogenic sequence of SCBs and PCBs during differentiation in radiata pine. a, Longitudinal section of an SCB with initiated bracts (arrowheads) (5 mg), ×28.5. b, Longitudinal section of a 50-mg SCB showing fused bract-ovuliferous scale primordia complex. An arrowhead indicates the ovuliferous scale primordia, ×180. c and d, Longitudinal section of a 100-mg SCB with developing bracts and ovuliferous scales (arrowheads), ×170 and ×5, respectively. e, Female LSTB with developing SCB (100–300 mg) and DSB, ×1.5. f, Five-milligram PCB with developing microsporophylls (Mi), ×15. g, Longitudinal section of a developing 100-mg PCB after the completion of microsporophyll (Mi) initiation, ×10. h, Male LSTB with developing PCB (300 mg), ×1.7. BP, Bract primordia; SC, sterile cataphylls.
PRCEL1 and PRCEL2 Are Similar to Angiosperm and Bacterial EGases
Approximately 60,000 cDNA clones from an immature female cone library were differentially screened using probes from immature cones and vegetative buds. Thirty-eight clones, representing mRNAs preferentially expressed in cones, were isolated. The clones were assigned to 13 groups based on cross-hybridization and patterns of expression. Some groups contained only one clone, whereas others contained several.
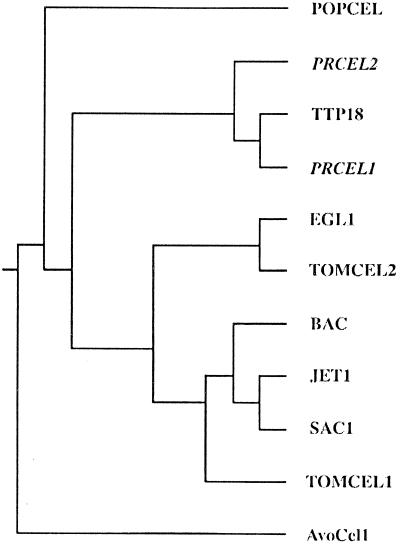
A few clones, representing genes with much greater levels of expression in cones than vegetative buds, were sequenced and characterized. Clones within two groups, PrCel1 and PrCel2, have deduced amino acid sequences very similar to that of plant EGases. PrCel1 and PrCel2 encode proteins of 509 and 515 amino acids, respectively. The difference in lengths is attributable to sequences at the N termini. Both proteins have hydrophobic N-terminal sequences comprising, respectively, the first 24 and 29 amino acids of PRCEL1 and PRCEL2. The mature proteins were predicted to have a molecular mass of approximately 54 kD, with estimated pIs of 8.06 (PRCEL1) and 6.96 (PRCEL2). Comparison between PRCEL1 and PRCEL2 reveals 71% amino acid identity between immature protein sequences, and 76% identity between mature protein sequences. Both proteins contain N-glycosylation sites within the mature peptides, at positions 25 to 28 for PRCEL1 and at positions 277 to 280 for PRCEL2. Amino acids, or blocks of amino acids generally conserved among other EGases, are also found in the pine proteins. Both PRCEL1 and PRCEL2 are very similar to TPP18 (CEL4), an EGase cloned from tomato (Lycopersicon esculentum) pistils (Milligan and Gasser, 1995; Brummell et al., 1997), with 73 and 67% identity, respectively. They are less similar (60 and 65% identity, respectively) to POPCEL, an EGase from poplar (Populus alba) (Nakamura et al., 1995), and EGL1 from pea (Pisum sativum) (60 and 59%, respectively) (Wu et al., 1996). A lower level of homology (approximately 50–55%) was found with EGases associated with ripening in tomato (TomCel2; Lashbrook et al., 1994) and avocado (Persea americana) (AvoCel1; Tucker et al., 1987). The amino acid sequences of PRCEL1 and PRCEL2 did not show strong homology with abscission zone-associated EGases (Fig. 2). The proteins TomCel1 (Lashbrook et al., 1994), BAC (Tucker and Milligan, 1991), SAC1 (Kemmerer and Tucker, 1994), and JET1 (Taylor et al., 1994) have 48 to 50% identity to the pine proteins. There was also some homology to cellulases from bacterial species. For example, starting at PRCEL1 residue 45, there is a 77-amino-acid-long sequence that is 43 to 56% identical to homologous sequence regions in EGases from Thermomonospora fusca, Clostridium thermocellum, Clostridium cellulolyticum, Clostridium stercocarium, and Cellulomonas fimi (not shown).
Figure 2.
Phylogenic tree showing the relationships between the plant EGases: TOMCEL1 (GenBank accession no. U13054); TOMCEL2 (GenBank accession no. U13055); EGL1 (GenBank accession no. L41046); POPCEL (GenBank accession no. D32166); TPP18 (GenBank accession no. U20590); PRCEL1 (GenBank accession no. U76725); PRCEL2 (GenBank accession no. U76756); AvoCel1 (GenBank accession no. M17634); SAC1 (GenBank accession no. U00730); JET1 (GenBank accession no. X74290); and BAC (GenBank accession no. P22503).
PrCel1 and PrCel2 Are Members of Multigene Families
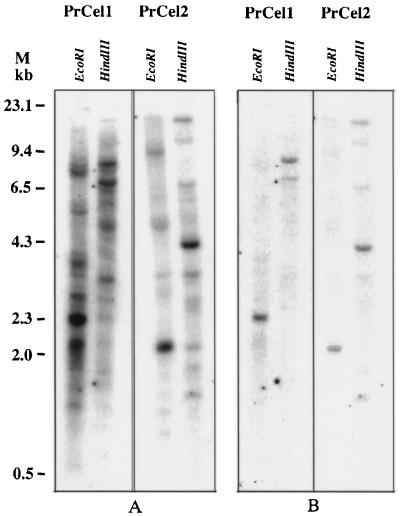
Southern-blot hybridizations were performed to determine the genomic organization of the radiata pine EGase gene family. In this experiment, only restriction enzymes that do not cut the two clones were used to digest genomic DNA samples. The DNA fragments that constitute the coding regions of the PrCel1 and PrCel2 clones were hybridized to genomic DNA digested with HindIII and EcoRI restriction enzymes. The 1287-bp PstI fragment of the pSPT18-412 (PrCel1) and the 1259-bp XbaI fragment of the pSPT18-69 (PrCel2) clone contain no EcoRI or HindIII sites. Hybridizations performed using moderate stringency revealed multiple hybridizing bands for both PrCel1 and PrCel2 genes (Fig. 3A). All shared bands disappeared after hybridizations at high-stringency conditions (0.1× SSC, 0.1% SDS, 65°C), which revealed only one main band for each of two restriction digests (Fig. 3B). This result indicates the presence of a family of EGase genes in the radiata pine genome, and that PrCel1 and PrCel2 are transcribed from distinct single genes.
Figure 3.
Genomic Southern-blot analyses. Total radiata pine genomic DNA (20 μg per lane) was digested with EcoRI and HindIII and hybridized with 32P-labeled PrCel1 and PrCel2 probes. Blots were hybridized and washed at low-stringency (A) and high-stringency conditions (B). M indicates the markers in kilobases.
Expression
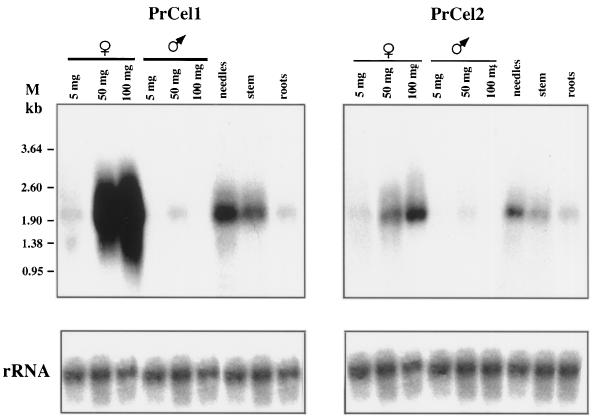
Northern-blot and in situ hybridizations were used to determine the temporal and spatial expression of the PrCel1 and PrCel2 genes. Expression of both genes increased markedly in PCBs and SCBs during their development, associated with growth from 5 to 100 mg in weight. Strong expression of the PrCel1 gene was detected in immature 50- and 100-mg SCBs (Fig. 4). A significantly lower level of expression was found in small (5-mg) SCBs. In the PCBs a low but still detectable level of expression was found in 50-mg cones. Low-level expression was also observed in 5- and 100-mg cones, but only after a longer exposure of 1 week (not shown). The PrCel2 gene displayed a similar pattern of expression to PrCel1 in male and female reproductive organs, but at a much lower level than PrCel1. In vegetative organs both genes showed a similar pattern of expression, with stronger levels for PrCel1. Expression was strong in developing needles and low in stems and roots.
Figure 4.
Expression of PrCel1 and PrCel2 genes in different radiata pine tissues. Total RNA (20 μg per lane) isolated from 5, 50, and 100 mg of SCB and PCB, needles, stems, and roots was hybridized with 32P-labeled PrCel1 and PrCel2 probes. The loading of equivalent amounts of RNA was confirmed by hybridization with a 32P-labeled rRNA clone. M, Markers (in kb).
In situ hybridization was used to investigate the potential involvement of PrCel1 in initiation and development of reproductive organs, SCBs, PCBs, vegetative organs, DSBs, seedlings, and root tips. The pattern of PrCel1 mRNA localization was determined by in situ hybridization of digoxigenin-labeled RNA probes to longitudinal and transverse sections of developing female and male cone and vegetative buds. Some of the most informative examples of expression of PrCel1 at different stages of SCB, PCB, and DSB development are shown in Figure 5. PrCel1 transcript was first detected in differentiated SCBs, within a group of cells initiating ovuliferous scale primordia at the adaxial part of the bract primordia. At this stage, the bract primordia were differentiated but the ovuliferous scale primordia had not emerged (Fig. 5a, lower bract). Expression of PrCel1 at a later stage of ovuliferous scale primordia development in the differentiated SCB is also shown in Figure 5a (upper bract). At this stage, signals were highly localized in emergent ovuliferous scale primordia and not detected in sterile bract primordia (Fig. 5, a and b). At the later stages of SCB development (50 and 100 mg), PrCel1 transcripts accumulated almost uniformly within ovuliferous scale primordia (Fig. 5, c and d). No expression was found at these stages in bract primordia or sterile cataphylls.
Figure 5.
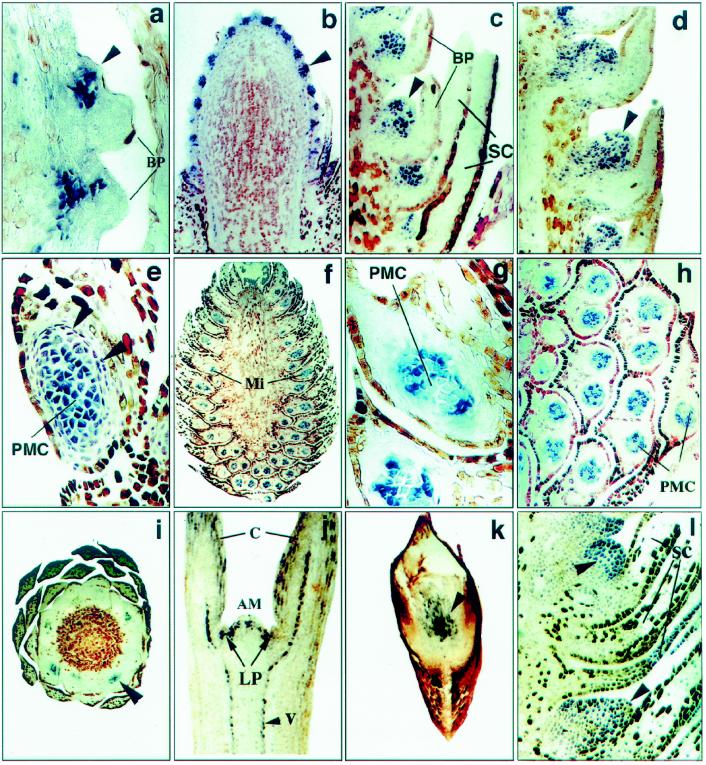
In situ localization of PrCel1 transcripts in the developing SCB, PCB, and DSB. SCB (position of ovuliferous primordia is shown by arrowheads): a and b, 5-mg SCB, ×50 and ×18, respectively; c, 50-mg SCB, ×30; d,100-mg SCB, ×22.5. PCB: e, Early stages of microsporophyll development in 5-mg PCB. Arrowheads show two layers of tapetal cells, ×45; f, 100-mg PCB, ×8.3; g, microsporophylls from the upper part of a 100-mg PCB, ×45; h, microsporophylls from the bottom part of a 100-mg PCB, ×45; i, cross-section of a 100-mg PCB (pollen mother cells are shown by the arrowhead), ×8.6; j, 7-d-old seedling, ×13.8; k, root tip (scattered cells are shown by the arrowhead), ×36.6; and l, vegetative LSTB with differentiated DSB (arrowhead), ×21.6. AM, Apical meristem; BP, bract primordia; C, cotyledons; LP, leaf primordia; PMC, pollen mother cells; SC, sterile cataphylls; V, vascular cells. All sections except i and k are longitudinal.
In PCBs expression of the PrCel1 gene was also detected at the early stage of microsporophyll development. Accumulation of the transcripts at this stage was detected in pollen mother cells (at the stage when microsporophylls just emerge from the inflorescence apex, 5-mg cones) (Fig. 5e), and persisted through the later stages of development (100-mg PCB) (Fig. 5, f–i). At early stages of PCB development (5 mg), PrCel1 transcripts were also found in two layers of tapetal cells surrounding the pollen mother cells (Fig. 5, e and g). At the later stages of PCB development, when initiation of microsporophylls was already completed (100 mg), hybridization signals were detected only within pollen mother cells, not in tapetal cells (Fig. 5, g and h). Cross-sectional analysis of 100-mg PCBs showed that expression of PrCel1 was restricted to a group of cells within the reproductive tissues (microsporophylls) (Fig. 5i). In 7-d-old seedlings (Fig. 5j), the PrCel1 gene was expressed in dividing scattered cells in the apical meristem, in the primary leaf primordia, and in the vascular tissue forming in the cotyledons and the hypocotyl. Scattered cells in the elongating region of root tips also demonstrated high levels of expression (Fig. 5k). In vegetative LSTBs expression of PrCel1 was detected in differentiated DSBs (Fig. 5l). These data suggest that PrCel1 may be expressed in cells undergoing elongation associated with cell division. PrCel2 showed a very low hybridization signal that was difficult to distinguish from the background signal.
The Products of PrCel1 and PrCel2 Have EGase Activity in Vitro
To analyze whether PRCEL1 and PRCEL2 proteins show some EGase activity in vitro, the coding regions of PrCel1 and PrCel2 clones encoding the mature protein sequences were amplified by PCR using the primers P1-P2 and P3-P4 (see Methods). To check the PCR fragments, they were cloned into the Bluescript SKII vector. These plasmids were linearized and used as a template for a transcription/translation reaction (TNT-Coupled Wheat Germ Extract System, Promega). Resulting products were electrophoresed on gradient Tris-Gly gels (4–20%), dried, and exposed to x-ray film (Kodak). Both reactions revealed peptides of the predicted size (approximately 55 kD, not shown).
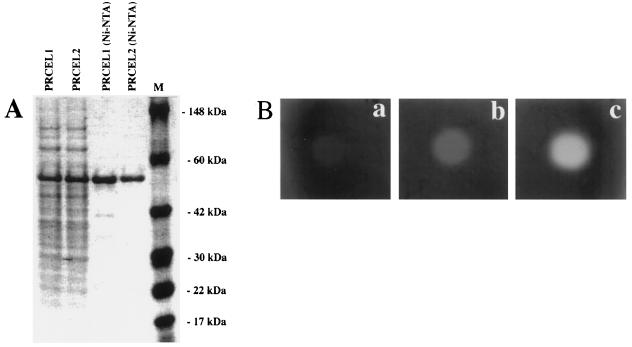
The PCR fragments constituting the sequences encoding the mature PRCEL1 and PRCEL2 proteins were cloned into the E. coli expression vector pQE31 (Qiagen), which contains a 6xHis tag upstream from the multiple cloning site (see Methods). PAGE analysis of the samples isolated from the E. coli expression culture induced with 2 mm isopropylthio-β-galactoside showed strong bands around 55 kD (Fig. 6a). The PRCEL1 and PRCEL2 proteins were purified from the total E. coli protein mixture using Ni-NTA resin.
Figure 6.
A, SDS-PAGE of total and Ni-NTA-purified proteins extracted from E. coli carrying pQE30-PrCel1 (PRCEL1) and pQE30-PrCel2 (PRCEL2) vectors after induction with isopropylthio-β-galactoside; B, EGase assay of PRCEL1. Different amounts of Ni-NTA-purified protein (a, 10 ng; b,100 ng; and c, 500 ng) were dotted onto 1% agarose plates containing 0.2% CMC and stained with Congo red (1 mg/mL) (see Methods).
To determine EGase activity, protein extracts were serially diluted and loaded onto agarose plates containing CMC (0.2%) in phosphate buffer (0.1 m, pH 6.5) and incubated overnight at 37°C. Enzyme activity was visualized by staining with Congo red (Fig. 6b). Both proteins displayed the same pattern of EGase activity, which was increased with larger amounts of protein. To determine whether PRCEL1 and PRCEL2 were stable at elevated temperatures, equal amounts of proteins were exposed to different temperatures (4, 37, and 65°C for 2 h) and then tested for enzymatic activity. Neither protein showed EGase activity at 65°C (not shown).
DISCUSSION
We have described the cloning and characterization of two EGases from reproductive tissues of radiata pine. Both PRCEL1 and PRCEL2 are similar to previously cloned plant EGases. Although some overlap exists, many of the previously cloned EGases are preferentially associated with abscission or ripening. EGases from different species with possible similar functions (i.e. abscission or ripening) are generally more similar to each other than EGases from the same species with different functions. The likely function(s) of PRCEL1 and PRCEL2 are not suggested by homology to other plant EGases, since neither pine EGase showed distinct homology to EGases associated with ripening or abscission. Both pine proteins, however, show slightly higher similarity to EGases associated with fruit ripening than with abscission (Fig. 2).
PRCEL1 and PRCEL2 are very similar to each other, and both are more similar to TPP18 (CEL4), an EGase from tomato pistils (Milligan and Gasser, 1995; Brummell et al., 1997), than to any other reported EGases. Tomato TPP18 (CEL4) has stronger homology to PRCEL1 and PRCEL2 than to other angiosperm EGases, including other tomato EGases. Although the function of TPP18 (CEL4) is unknown, it is preferentially expressed in elongating tomato pistil tissue and rapidly expanding vegetative tissues. PrCel1 and PrCel2 mRNA were also found in both reproductive and vegetative tissues.
It is suggested that EGases may play multiple roles during the development of angiosperm reproductive tissues. They may be involved in loosening the cell wall for expansion, or in the major disruptions of the cell wall required during the development of anthers. They may also be involved in the dissolution of the cell walls of the stigma surface cells to facilitate pollen tube penetration (del Campillo and Lewis, 1992). Although the reproductive structures of pine are very different from those of most angiosperms, the EGases may have similar functions. Both PrCel1 and PrCel2 were isolated from rapidly growing female reproductive structures. In situ hybridizations showed that PrCel1 expression in parts of the cones, in vegetative buds, and in root tips is primarily localized to cells in rapidly growing regions. Therefore, PRCEL1 and PRCEL2 may play a role in cell wall loosening associated with cell expansion. Alternatively, they may play a role in the degradation events during development of the vascular system. In male cones PrCel1 expression is high in the regions containing the microsporangia, and may be involved in the major cell wall disruption associated with the production of mature pollen.
Another possible role for EGases in cell expansion is the generation of xyloglucan oligosaccharides. Oligosaccharides produced by a fungal cellulase have been shown to promote elongation of pea stem segments (McDougall and Fry, 1990). Xyloglucan fragments generated by EGase during auxin-stimulated cell elongation may serve a regulatory role during cell elongation. At low concentrations, the xyloglucans have an anti-auxin activity and may prevent excessive growth. At higher concentrations, there appears to be a second growth-restoring activity, which may mimic auxin. The gene EGL1, an EGase gene cloned from elongating pea epicotyls, has been shown to be induced by auxin (Wu et al., 1996).
Many plant EGases are members of small multigene families. Pines have very large genomes and enormous amounts of repetitive DNA. Some cloned genes, such as one encoding alcohol dehydrogenase in radiata pine, are members of very large multigene families (Harry et al., 1989). Other genes, however, such as one encoding phenylalanine ammonia-lyase, which are encoded by small multigene families in angiosperms, are reported to be single-copy genes in pine (Whetten and Sederoff, 1992). Genomic-blot analysis has indicated the presence of a family of EGase genes in the radiata pine genome, and that PrCel1 and PrCel2 are transcribed from distinct one-copy genes.
Almost all plant cellulases belong to the cellulase family E (Henrissat et al., 1989) or to the glycosyl hydrolase family 9 (Henrissat, 1991; Henrissat and Bairoch, 1996). The PRCEL1 protein has a region (positions 476–494) that is homologous to glycosyl hydrolase family 9 active-site consensus pattern 2. The PRCEL2 protein has two active sites (positions 420–436 and 482–501) with homology to consensus pattern 1. Comparison of the pine proteins with the three-dimensional structure of endonuclease D of C. thermocellum has revealed putative catalytic residues at positions Asp-100 (base) and Glu-487 (proton donor) in PRCEL1, and Asp-106 (base) and Glu-494 (proton donor) in PrCEL2.
The fact that plant EGases have no cellulase binding domain and belong to the same cellulase family E, or the glycosyl hydrolases family 9, is probably not without significance. Perhaps the function of these enzymes is not to achieve extensive degradation of crystalline cellulose. The activity exhibited with CMC does not imply activity with crystalline cellulose; there is indeed no evidence that plant “cellulases” have a significant activity with crystalline cellulose. Identification of the real substrate of plant cellulases would help in understanding their role.
ACKNOWLEDGMENTS
We thank Dr. Bernard Henrissat from Centre de Recherches sur les Macromolécules Végétales-Centre National de la Recherche Scientifique for help and technical advice concerning analysis of the cellulose-binding and catalytic domains of PRCEL1 and PRCEL2. We also thank Dr. Tim Sawbridge for help with the sequence alignment and construction of the phylogenetic tree, and Dr. Keith Mitchelson and Corinna Lange for their critical reading of the manuscript.
Abbreviations:
- CMC
carboxymethylcellulose
- DSB
dwarf shoot bud
- EGase
endo-β-1,4-glucanase
- LSTB
long-shoot terminal bud
- PCB
pollen-cone bud
- SCB
seed-cone bud
Footnotes
LITERATURE CITED
- Baker SS, Rugh CL, Whitmore FW, Kamalay JC. Genes encoding 11 S globulin-like proteins are expressed in the megagametophyte soon after fertilization in eastern white pine (Pinus strobusL.) Int J Plant Sci. 1996;157:453–461. [Google Scholar]
- Brummell DA, Bird CR, Schuch W, Bennett AB. An endo-1,4-β-glucanase expressed at high levels in rapidly expanding tissues. Plant Mol Biol. 1997;33:87–95. doi: 10.1023/a:1005733213856. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. How do plant cell walls extend? Plant Physiol. 1993;102:1–6. doi: 10.1104/pp.102.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campillo E, Lewis LN. Occurrence of 9.5 cellulase and other hydrolases in flower reproductive organs undergoing major cell wall disruption. Plant Physiol. 1992;99:1015–1020. doi: 10.1104/pp.99.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferl RJ. An ArabidopsiscDNA encoding beta glucanase. Plant Mol Biol. 1995;29:883–889. [Google Scholar]
- Harry DE, Mordecai KS, Kinlaw CS, Loopstra CA, Sederoff RR (1989) DNA sequence diversity in alcohol dehydrogenase genes from pines. In Proceedings of the 20th Southern Forest Tree Improvement Conference, Charleston, SC. The National Technical Information Services, U.S. Department of Commerce, Springfield, VA, pp 373–380
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Claeyssens M, Tomme P, Lemesle L, Mornon JP. Cellulase families revealed by hydrophobic cluster analysis. Gene. 1989;81:83–95. doi: 10.1016/0378-1119(89)90339-9. [DOI] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thalianaencodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Kemmerer EC, Tucker ML. Comparative study of cellulases associated with adventitious root initiation, apical buds, and leaf, flower, and pod abscission zones in soybean. Plant Physiol. 1994;104:557–562. doi: 10.1104/pp.104.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook CC, Gonzalez-Bosch C, Bennett AB. Two divergent endo-β-1,4-glucanase genes exhibit overlapping expression in ripening fruit and abscising flowers. Plant Cell. 1994;6:1485–1493. doi: 10.1105/tpc.6.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal I, Misra S. Molecular cloning and characterization of a legumin-like storage protein cDNA of Douglas fir seeds. Plant Mol Biol. 1993;21:709–715. doi: 10.1007/BF00014555. [DOI] [PubMed] [Google Scholar]
- McDougall GJ, Fry SC. Xyloglucan oligosaccharides promote growth and activate cellulase. Evidence for a role of cellulase in cell expansion. Plant Physiol. 1990;93:1042–1048. doi: 10.1104/pp.93.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan SB, Gasser CS. Nature and regulation of pistil-expressed genes in tomato. Plant Mol Biol. 1995;28:691–711. doi: 10.1007/BF00021194. [DOI] [PubMed] [Google Scholar]
- Mouradov A, Glassick T, Teasdale R. Isolation and characterization of a FLORICAULA/LEAFY-like cDNA from Pinus radiata (PGR 97-026) Plant Physiol. 1997a;113:664. [Google Scholar]
- Mouradov A, Glassick T, Teasdale R. Isolation and characterization of a new MADS-box cDNA from Pinus radiata (PGR 97-027) Plant Physiol. 1997b;113:665. [Google Scholar]
- Mouradov A, Glassick T, Vivian-Smith A, Teasdale R. Isolation of a MADS box gene family from Pinus radiata (PGR 96-002) Plant Physiol. 1996;110:1047. [Google Scholar]
- Nakamura S, Mori H, Sakal F, Hayashi T. Cloning and sequencing of a cDNA for poplar endo-β-1,4-glucanase. Plant Cell Physiol. 1995;36:1229–1235. [PubMed] [Google Scholar]
- Newton CH, Flinn BS, Sutton BCS. Vicilin-like seed storage proteins in the gymnosperm interior spruce (Picea glaucalengelmanii) Plant Mol Biol. 1992;20:315–322. doi: 10.1007/BF00014500. [DOI] [PubMed] [Google Scholar]
- Tandre K, Albert V, Sundas A, Engstrom P. Conifer homologues to genes that control floral development in angiosperms. Plant Mol Biol. 1995;27:6978. doi: 10.1007/BF00019179. [DOI] [PubMed] [Google Scholar]
- Taylor JE, Coupe SA, Ploton S, Roberts JA. Characterization and accumulation pattern of an mRNA encoding an abscission-related endo-β-1,4-glucanase from leaflets of Sambucus nigra. Plant Mol Biol. 1994;24:961–964. doi: 10.1007/BF00014449. [DOI] [PubMed] [Google Scholar]
- Tucker ML, Durbin ML, Clegg MT, Lewis LN. Avocado cellulase: nucleotide sequence of a putative full-length cDNA clone and evidence for a small gene family. Plant Mol Biol. 1987;9:197–203. doi: 10.1007/BF00166456. [DOI] [PubMed] [Google Scholar]
- Tucker ML, Milligan SB. Sequence analysis and comparison of avocado fruit and bean abscission cellulases. Plant Physiol. 1991;95:928–933. doi: 10.1104/pp.95.3.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetten R, Sederoff R. Phenylalanine ammonia-lyase from loblolly pine. Purification of the enzyme and isolation of complementary DNA clones. Plant Physiol. 1992;98:380–386. doi: 10.1104/pp.98.1.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Blumer JM, Darvill AG, Albersheim P. Characterization of an endo-β-1,4-glucanase gene induced by auxin in elongating pea epicotyls. Plant Physiol. 1996;10:163–170. doi: 10.1104/pp.110.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]