Abstract
Notch and Wnt are highly conserved signalling pathways that are used repeatedly throughout animal development to generate a diverse array of cell types. However, they often have opposing effects on cell-fate decisions with each pathway promoting an alternate outcome. Commonly, a cell receiving both signals exhibits only Wnt pathway activity. This suggests that Wnt inhibits Notch activity to promote a Wnt-ON/Notch-OFF output; but what might underpin this Notch regulation is not understood. Here, we show that Wnt acts via Dishevelled to inhibit Notch signalling, and that this crosstalk regulates cell-fate specification in vivo during Xenopus development. Mechanistically, Dishevelled binds and directly inhibits CSL transcription factors downstream of Notch receptors, reducing their activity. Furthermore, our data suggest that this crosstalk mechanism is conserved between vertebrate and invertebrate homologues. Thus, we identify a dual function for Dishevelled as an inhibitor of Notch signalling and an activator of the Wnt pathway that sharpens the distinction between opposing Wnt and Notch responses, allowing for robust cell-fate decisions.
Keywords: Dishevelled, Notch, Wnt, Signalling crosstalk, Xenopus
INTRODUCTION
The Notch and Wnt pathways are two of only a handful of highly conserved signalling pathways that control cell-fate decisions during the formation and maintenance of tissues in animal embryogenesis and adult homeostasis (Pires-daSilva and Sommer, 2003). However, they typically have distinct and opposing effects on cell-fate outcomes (Hayward et al., 2008). This antagonism is evident at multiple steps during cell-fate specification: from stem cell self-renewal and lineage commitment (Zhu and Watt, 1999; Lowell et al., 2000; Bouras et al., 2008; Zeng and Nusse, 2010) to the process of differentiation along a specific cell lineage, such as the intestinal epithelium (Fre et al., 2005; Stanger et al., 2005; van Es et al., 2005b; van Es et al., 2005a). Yet there are many examples where a cell receives both signals simultaneously. Within the mammary gland and the skin, stem cells are exposed to both Notch and Wnt pathway ligands (Lowell et al., 2000; Reddy et al., 2001; Brennan and Brown, 2004; Raafat et al., 2011). Similarly, both pathways can act on the same cell during sensory bristle development and wing margin formation in Drosophila (Axelrod et al., 1996; Rulifson et al., 1996; Brennan et al., 1999; Ramain et al., 2001). Despite receiving two signals that promote opposing fates, the cell is still able to resolve these inputs into a robust cell-fate decision. Commonly, this is resolved into a Wnt-ON/Notch-OFF response (Uyttendaele et al., 1998). However, how this occurs mechanistically is poorly understood. One possibility is direct inhibitory crosstalk with downstream Wnt signalling inhibiting the Notch pathway to stabilise cells in a Wnt-ON/Notch-OFF state (Hayward et al., 2008).
Notch receptors act as membrane-tethered transcription factors (Bray, 2006). Following binding of a DSL family ligand (Delta, Serrate, LAG-2 and Jagged), Notch proteins undergo γ-secretase-mediated cleavage and release the Notch intracellular domain (NICD). NICD then translocates to the nucleus and forms a transcriptional activator complex with CSL (CBF1, Suppressor of Hairless, LAG-1; termed RBPJκ in mice) factors and the co-activator Mastermind-like (MAML). A key mediator of Wnt/β-catenin signalling is the multi-domain protein Dishevelled (Dvl, Dsh in Drosophila) (MacDonald et al., 2009). Dishevelled acts as a scaffold to activate signalling by recruiting components of the β-catenin destruction complex to the Wnt receptors Frizzled-LRP5/6 (LDL receptor-related protein 5/6). This recruitment inactivates the destruction complex. Consequently, β-catenin accumulates and enters the nucleus, where it promotes expression of Wnt target genes.
Here, we show that Wnt signalling inhibits the Notch pathway and that this crosstalk occurs via Dishevelled. Furthermore, we demonstrate that Dvl2 inhibits Notch signalling in vivo to control cell-fate specification during Xenopus epidermal development. Investigation of the mechanism underlying Dishevelled-Notch crosstalk reveals that Dishevelled limits signalling by all four vertebrate Notch paralogues. This occurs through inhibition of the NICD transcriptional activator complex, by binding and reducing the level of the CSL transcription factor within the nuclear pool of active transcription factors. Our data also indicate that this crosstalk mechanism is conserved between vertebrates and invertebrates. These findings reveal that, in response to Wnt signalling, Dishevelled inhibits CSL transcription factors to regulate Notch signalling and cell-fate decisions in vivo.
MATERIALS AND METHODS
Cell culture
CHO-K1 cells (John Gallagher, Paterson Institute for Cancer Research, Manchester, UK) were cultured in Ham's F12 medium with Glutamax (Invitrogen, Carlsbad, USA), supplemented with 10% FBS [Biowest, Nuaillé, France], 1% non-essential amino acids, 50 U/ml penicillin and 50 μg/ml streptomycin (Lonza, Basel, Switzerland). HEK 293T and NIH3T3 cells (Anthony Brown, Weill Medical College, Cornell University, New York, NY, USA) and SHEP neuroblastoma cells (Patrick Mehlen, Centre Léon Bérard, Lyon, France), were cultured in DMEM (Lonza) supplemented as above. SHEP/RBPJκ-Luc cells are a stable cell line carrying the p10XRBPJκ-Luc reporter vector. Cells were maintained at 37°C in 5% CO2 in a humidified incubator. To inhibit GSK3β, cells were cultured overnight with 20 mM LiCl or 10 μM SB216763 (Ascent Scientific, Bristol, UK) using 20 mM KCl or DMSO as controls. To inhibit γ-secretase, cells were cultured overnight with 5 μM DAPT (Merck Chemicals, Nottingham, UK) using DMSO as a control. Control conditioned medium and conditioned medium containing Wnt1 were recovered from SHEP cells and Wnt1-expressing SHEP cells cultured in serum-free medium for 24 hours, respectively.
Plasmids, expression constructs and transcriptional reporters
The following plasmids were generous gifts: mWnt1/pLNCX (Anthony Brown, Weill Medical College, Cornell University, New York, USA); RBPJκ/pCMX and VP16-RBPJκ/pCMX (Tasuko Honjo, Kyoto University, Japan); p10xRBPJκ-Luc and p10xRBPJκ-lacZ (Grahame MacKenzie, Lorantis, Cambridge, UK); ΔN-hN1-3/pcDNA4 His-Max C (Anne-Marie Buckle, University of Manchester, UK); mN1/pcDNA3 (Jeff Nye, Northwestern University Medical School, Chicago, USA); mN1ICD/pEGFP-N1 (Vincent Zecchini, University of Cambridge, UK); TOPflash (Louise Howe, Weill Medical College, Cornell University, New York, USA); NRE Su(H)-reporter construct, and the Su(H)-VP16/pUAST and Drosophila Dsh/pMT expression constructs (Sarah Bray, University of Cambridge, UK); GSK3β K85R (Trevor Dale, Cardiff University, UK); XDvl2- and Ds1-myc and -GFP expression constructs (Sergei Sokol, Mount Sinai Medical Center, New York, USA); and hGR-XSu(H)-ANK/pCS2 and XNICD/pCS2 (Nancy Papalopulu, University of Manchester, UK). mDvl2, mβ-catenin, hNotch4 and MAML1 cDNAs were obtained from Geneservice Cambridge, UK (IMAGE clones 6402000, 5709247, 9021650 and 6407060, respectively). pRL-CMV and pGL3-basic were obtained from Promega, pEGFP-N1 and pGBKT7 were from Clontech (Mountain View, USA), and pcDNA3.1(+) and pcDNA6V5-his from Invitrogen. All primer sequences are shown in supplementary material Table S1. The following plasmids were generated in our laboratory.
ΔN-mN1/pSecTagNC
The sequence encoding the extracellular juxtamembrane, transmembrane and intracellular domains of mN1 were cloned as HindIII/BclI-digested PCR product (amplified using the primers N1 5210F and N1 5589R from the mN1/pcDNA3 template) and a BclI/EcoRI restriction fragment of mN1/pcDNA3. These two fragments were ligated into pSecTagNC (Brennan et al., 2004) digested with HindIII/EcoRI.
ΔN-mN1-GFP/pSecTagNC
ΔN-mN1-GFP was generated by cloning an NheI/SacI fragment from ΔN-mN1/pSecTagNC and a SacI/NotI fragment from mN1ICD/pEGFP-N1 into pSecTagNC cut NheI/NotI.
ΔN-mN1-ΔNCR/pSecTagNC
ΔN-mN1ΔNCR was cloned from the ΔN-mN1/pSecTagNC template as an XhoI/EcoRI restriction fragment ligated with a BspEI/XhoI-digested PCR product (amplified using the primers N1 5970F and N1 6400R) into ΔN-mN1/pSecTagNC digested with BspEI/EcoRI.
ΔN-mN1-ΔC238/pSecTagNC
ΔN-mN1/pSecTagNC was digested with BspEI/HindIII (blunted) and ligated into ΔN-mN1/pSecTagNC BspEI/EcoRI (blunted)
ΔN-mN1-ΔC351/pSecTagNC
ΔN-mN1/pSecTagNC was digested to yield two restriction fragments: (1) BspEI/XhoI (blunted) and (2) BspHI (blunted)/EcoRI. These two fragments were ligated into ΔN-mN1/pSecTagNC (BspEI/EcoRI).
ΔN-mN1-ΔC425/pSecTagNC
ΔN-mN1/pSecTagNC was digested with HindIII (blunted)/EcoRI and ligated with a ΔN-mN1-ΔNCR/pSecTagNC BspEI/XhoI(blunted) restriction fragment into ΔN-mN1/pSecTagNC (BspEI/EcoRI).
ΔN-hN4/pSecTagNC
The sequence encoding the extracellular juxtamembrane, transmembrane and intracellular domains of hN4 were cloned as HindIII/SacI-digested PCR product (hNotch4 template and the primers hN4 4357 F and hN4 4563 R) and a SacI/BamHI restriction fragment of the hNotch4 cDNA ligated together into HindIII/BamHI-digested pSecTagNC.
mDvl2/pcDNA6V5-his
mDvl2 cDNA was cloned from pYx-Asc (Geneservice) into pcDNA6V5-his as an EcoRI/SalI restriction fragment and a SalI/XbaI-digested PCR fragment generated using mDvl2 1834F and mDvl2 2331R primers.
ΔN-mDvl2/pcDNA6V5-his
A HindIII/SfoI-digested PCR fragment generated using T7 and mDvl2 132R was cloned with a NaeI/XbaI-digested PCR fragment generated using mDvl2 1654F and BGHR into pcDNA6V5-his digested with HindIII and XbaI.
ΔC-mDvl2/ pcDNA6V5-his
The EcoRI/NotI restriction fragment from mDvl2-V5-his was cloned with A NotI/XhoI-digested PCR fragment generated using mDvl2 793F and mDvl2 1668R into pcDNA6V5-his digested with EcoRI/XhoI.
Dsh/pcDNA3.1(+)
A cDNA encoding Dsh was recovered from Dsh/pMT as an ApoI restriction fragment and cloned into pcDNA3.1(+) digested with EcoRI.
mβ-catenin/pcDNA3.1myc-hisA
mβ-catenin cDNA was cloned from pYx-Asc (Geneservice) into pcDNA3.1myc-hisA (digested KpnI/XhoI) as a KpnI/SacI restriction fragment and a SacI/XhoI-digested PCR fragment using mβ-cat 1914F and mβ-cat 2555R primers.
RBPJκ/pEGFP-N1
Two fragments encoding RBPJκ were ligated into pEGFP-N1 (Clontech) digested with EcoRI/XmaI (blunted): (1) RBPJκ/pCMX digested with EcoRI/KpnI and (2) a PCR fragment using RBPJκ BglIIF and RBPJκ BglIIR digested with KpnI/BglII (blunted).
VP16-RBPJκ-GFP/pcDNA3.1
A KpnI/NotI fragment from RBPJκ/pEGFP-N1 was cloned into VP16-RBPJκ/pcDNA3.1 digested KpnI/NotI.
XSu(H)-ANK/pCS2
The XSu(H)-ANK encoding sequence was removed from hGR-XSu(H)-ANK using ClaI and XbaI sites and inserted into ClaI/XbaI-digested pCS2.
Su(H)-VP16/pcDNA3.1(+)
A cDNA encoding the Su(H)-VP16 fusion protein was recovered from Su(H)-VP16/pUAST as a BglII/XbaI restriction fragment and cloned into pcDNA3.1(+) digested with BamHI/XbaI.
MAML-GAL4/pcDNA3.1(+)
A cDNA encoding the GAL4 DNA-binding domain was recovered from pGBKT7 (Clontech) as a HindIII/BamHI fragment and cloned into to pcDNA3.1(+)-digested HindIII/BamHI to generate a GAL4/pcDNA3.1 vector. The MAML1 cDNA was introduced into this intermediate vector (digested SfiI/EcoRI) as a SfiI/AvrII-digested PCR fragment generated using mMAML1 176F and mMAML1 548R and an AvrII/ApoI restriction fragment from the mMAML1/pYx-Asc IMAGE clone (Geneservice).
GAL4-VP16/pcDNA3.1(+)
A cDNA encoding the GAL4 DNA-binding domain was recovered from GAL4/pcDNA3.1(+) as a HindIII/NcoI fragment and cloned with a cDNA encoding the VP16 transactivation domain [recovered from VP16-RBPJκ/pCMX as a NcoI/HindIII (blunted) fragment] into pcDNA3.1(+)-digested HindIII/XbaI (blunted).
UAS-Luc
A DNA fragment containing the five GAL4 DNA-binding sites and the hsp70 promoter from pUAST was cloned into pGL3-basic as a SacI/BglII fragment.
Site-directed mutagenesis
All point mutations were generated using the QuikChange site-directed mutagenesis kit (Agilent, La Jolla, USA). See supplementary material Table S1 for mutagenic primer sequences.
Transfections
Cells were transfected with plasmids using Lipofectamine and Plus reagents (Invitrogen) and with siRNA using Lipofectamine 2000 (Invitrogen). For luciferase assays, each well of a 24-well plate was transfected 48 hours before lysis with a total of 250 ng DNA, including 50 ng of p10xRBPJκ-Luc, NRE or TOPflash and 20 ng of pRL-CMV in addition to 12.5-50 ng of each expression construct. To knockdown mDvl2, each well of a 24-well plate was transfected with 33.3 nM of the siRNA (Sigma-Aldrich, Gillingham, UK) GGAAGAGAUCUCCGAUGAC (Lee et al., 2008). The Stealth RNAi siRNA negative control med GC duplex (Invitrogen) was used as a control. Twenty-four hours later, the cells were transfected with the required plasmids as above for luciferase assays and lysed after a further 24 hours. For the β-gal reporter assay, cells cultured in a six-well plate were transfected with a total of 1 μg DNA containing 200 ng of p10xRBPJκ-lacZ, 50 ng of pRL-CMV and 250-500 ng of each expression construct. For all immunoprecipitation assays, 500 ng of RBPJκ and mDvl2 constructs were transfected in a total of 2 μg DNA per 60 mm dish. pcDNA3.1(+) was used to ensure that the total amount of DNA remained constant.
Ligand co-culture assays
Recombinant Jagged1-Fc protein in PBS (R&D Systems) was bound to tissue culture plastic for 4 hours. PBS was removed and the dish dried before addition of cells. Cells were cultured for 24 hours on ligand-coated plastic before lysis.
Luciferase assays
Cells were washed twice in PBS and lysed by incubation for 20 minutes in 100 μl passive lysis buffer (Promega, Madison, USA) at room temperature. Luciferase assays were performed using the Dual Luciferase Reporter assay system (Promega) and a MicroLumatPlus plate reader (Berthold Technologies, Harpenden, UK). Experiments were performed in triplicate and the relative luciferase units (RLU) for each data point was normalised to the mean obtained with ΔN-mN1, VP16-RBPJκ, XSu(H)-ANK or Su(H)-VP16 alone.
qRT-PCR
Total RNA was extracted using the NucleoSpin RNA II Kit (Macherey Nagel, Düren, Germany). Total RNA (1 μg) was reverse-transcribed with the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, USA). HES1 transcripts were amplified using the LightCycler Taqman DNA Master Kit and a LightCycler 480 PCR machine (Roche Applied Science). Expression was normalised to the housekeeping gene HPRT1. α-tubulin and esr1 transcripts were amplified using the Fast SYBR Green Master Mix and a StepOnePlus Real-Time PCR machine (Applied Biosystems, Invitrogen). Expression was normalised to the housekeeping gene rpl8. Primers used are listed in supplementary material Table S2.
Injection of embryos and in situ hybridisation
Xenopus laevis embryos were obtained, dejellied and raised as previously described (Chalmers et al., 2002). To obtain Xenopus tropicalis embryos, male and female frogs were primed with 10 and 15 units of pregnant mare serum gonadotrophin (PMSG), respectively, 12-24 hours prior to ovulation. Mating was subsequently induced with 50 units of human chorionic gonadotrophin (hCG) in males and 75 units in females. Capped mRNA for injection was synthesised using the mMessage mMachine SP6 kit (Ambion, Invitrogen). Embryos were injected at the two-cell stage with 100 pg lacZ mRNA as a lineage tracer and 10 pg XNICD, 1 ng Xdvl2 or 1 ng Ds1 mRNA. Embryos were fixed in MEMFA [0.1 M MOPS (pH 7.4), 2 mM EGTA, 1 mM MgSO4, 3.7% formaldehyde] at stage 18 (Nieuwkoop and Faber, 1967) and processed for X-Gal staining and in situ hybridisation as described previously (Chalmers et al., 2002). The α-tubulin probe was provided by Eamon Dubaissi (University of Manchester, UK). XtDvl2 expression was reduced by injecting 17 ng of the following morpholino (Gene Tools, Philomath, USA) at the one-cell stage (MODvl2) ATGACTTTAGTCTCCGCCATCCTGC. A morpholino recognizing human β-globin was used as a control (MOC) CCTCTTACCTCAGTTACAATTTATA. For qRT-PCR analysis, embryos were lysed at stage 12.
Statistical analysis
Normalised luciferase data from at least three independent experiments were combined and the mean fold change (±s.e.m.) in RLU was calculated. The data were analysed with two-tailed t-tests and one-way ANOVA and Tukey's post-hoc tests using Prism software (GraphPad, La Jolla, USA). For Xenopus experiments, embryos from independent experiments were used for the analysis (n≥4 for β-gal control, n≥10 for experimental conditions). Ciliated cell precursors were counted within a standard area over the centre of each side of the embryos. Data are presented as mean±s.e.m. Precursor count data were analysed using two-tailed paired t-tests to compare the uninjected and injected sides of each embryo. Two-way ANOVA with Bonferroni's post-hoc tests were used to compare injected sides from different treatments. Morpholino data were analysed by one-way ANOVA with Tukey's post-hoc tests (n≥14).
Fractionation
Active transcription factors were enriched and separated from inactive transcription factors and chromatin by separation of the soluble nuclear fraction from the extraction-resistant fraction, as described (Schreiber et al., 1989) with the following adaptations: after washing with PBS, cells were detached in trypsin (Lonza) and centrifuged at 1500 g for 15 seconds; buffers A and C contained 1× Protease inhibitor cocktail (PIC) (Merck Chemicals), phosphatase inhibitors 10 mM NaF and 1 mM NaVO4 and lacked DTT. For the total nuclear extract, the nuclear pellet was sonicated in sample buffer [50 mM Tris-Cl (pH 6.8), 100 mM DTT, 2% SDS, 0.1% bromophenol blue, 10% glycerol].
Western blotting
Total cell lysis and subsequent immunoblotting were performed as described (Stylianou et al., 2006). Primary antibodies used are listed in supplementary material Table S3.
Immunoprecipitation
Cells were washed in PBS and lysed in 500 μl immunoprecipitation lysis buffer (1% NP-40, 10% v/v glycerol, 1× PIC, 1× PBS). Lysate was clarified by centrifugation at 21,000 g for 10 minutes at 4°C. For the immunoprecipitation of GFP-fusion proteins, GFP-Trap_A beads (Chromotek, Martinsried, Germany) were used. For all other proteins, the immunoprecipitating antibody was bound to Dynabeads (Invitrogen). To immunoprecipitate proteins from the nuclear fraction, cells were lysed on ice for 30 minutes in 10 mM Tris-Cl (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, 0.5% NP40, 1× PIC. Nuclei were isolated by centrifugation at 1500 g for 10 minutes at 4°C and resuspended in 10 mM Tris-Cl (pH 7.5), 0.5 mM EDTA, 0.5% NP40, 1× PIC, 1 μg/μl DNaseI) (Sigma-Aldrich) and agitated for 10 minutes at 4°C. NaCl was added to final concentration of 150 mM and the sample incubated at 4°C for 30 minutes. The soluble fraction was then added to antibody-coated Dynabeads.
Immunofluorescence
Cells cultured on coverslips were fixed in 4% formaldehyde for 10 minutes at room temperature. Following washing, coverslips were incubated with primary antibody diluted 1:100 in blocking solution [3% goat serum (Biosera, Sussex, UK), 0.1% Triton-X100 in TBS] in a humidified chamber for 1 hour, washed and incubated with Alexa488-conjugated or Alexa594-conjugated secondary antibody (Invitrogen) (1:400) and Hoechst (Sigma-Aldrich) (1:10,000) in blocking solution. Coverslips were mounted with Prolong anti-fade medium (Invitrogen). Images were captured with LAS AF confocal software using a TCS SP5 inverted confocal microscope (Leica, Milton Keynes, UK).
RESULTS
Dishevelled inhibits Notch signalling activity
To determine whether Wnt signalling inhibited Notch pathway activity in mammalian cells, we established a reporter gene assay using a SHEP cell line that stably carried an RBPJκ-dependent luciferase reporter. We activated endogenous Notch receptors with immobilised recombinant JAG1-Fc (supplementary material Fig. S1). To activate Wnt signalling, SHEP-RBPJκ-Luc cells were either transfected with a Wnt1 expression construct, or treated with conditioned medium from Wnt1-expressing cells. Compared with control cells, those expressing Wnt1 or treated with Wnt1-conditioned medium exhibited significantly reduced Notch signalling activity (Fig. 1A; supplementary material Fig. S1). Furthermore, the expression of the endogenous Notch target gene HES1 in JAG1-Fc-treated SHEP-RBPJκ-Luc cells was reduced when the cells expressed Wnt1 or were treated with Wnt1-conditioned medium (Fig. 1B; supplementary material Fig. S1). These data demonstrate that ligand-induced Wnt signalling can inhibit the endogenous Notch pathway in mammalian cells, extending previous observations from Drosophila (Axelrod et al., 1996; Rulifson et al., 1996; Brennan et al., 1999; Ramain et al., 2001; Strutt et al., 2002; Muñoz-Descalzo et al., 2010; Capilla et al., 2012).
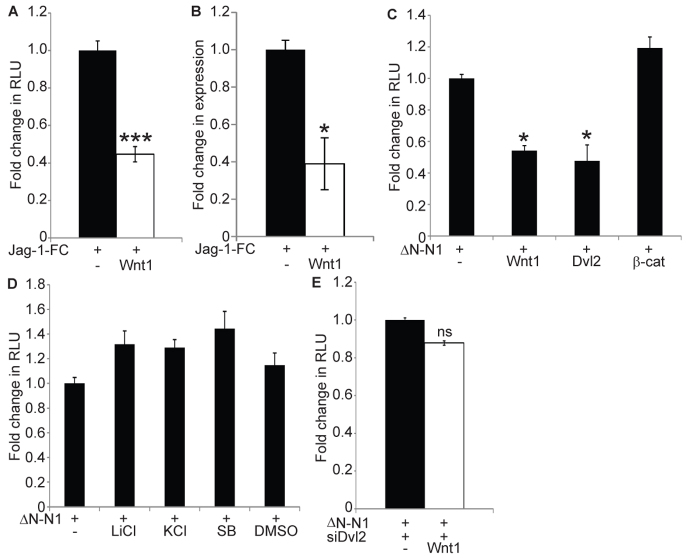
Fig. 1.
Wnt signalling inhibits Notch activity via Dishevelled. (A,B) Wnt inhibits endogenous Notch signalling. Control or Wnt1-expressing SHEP-RBPJκ-Luc cells were cultured on immobilised JAG1-Fc and Notch signalling was analysed by luciferase assays or qRT-PCR for the endogenous target Hes1. Data are presented as mean fold change (±s.e.m.) in relative luciferase units (RLU) (A) or HES1 expression normalised to HPRT1 (B), relative to control cells. Notch-induced luciferase and HES1 expression were significantly lower in Wnt1-expressing cells (two-tailed t-test, n=3). (C-E) Wnt signalling limits Notch-dependent transcriptional activity at the level of Dishevelled. CHO-K1 (C,D) and NIH3T3 (E) cells were transfected with the RBPJκ-reporter construct (RBPJκ-Luc) and a Renilla luciferase control construct (pRL-CMV). Notch signalling was activated by ΔN-mN1 expression. Data are presented as mean fold change (±s.e.m.) in RLU compared with ΔN-mN1 alone. (C) Wnt signalling was activated by expressing mWnt1, mDvl2 and S45F mβ-catenin (β-cat). Wnt1 and Dvl2 significantly inhibited Notch activity, whereas β-catenin did not (one-way ANOVA and Tukey's post-hoc tests, n=3). (D) Cells were treated with 20 mM LiCl or 10 μM SB216763 (SB) to inhibit GSK3β. Untreated cells (−) and control treatments of 20 mM KCl and DMSO are shown. None of the treatments significantly inhibited Notch activity (one-way ANOVA and Tukey's post-hoc tests, n=3). (E) Wnt1 was expressed in cells that had been previously transfected with an siRNA construct that recognises mDvl2. Dishevelled was required for Wnt-induced inhibition of Notch signalling (two-tailed t-test, n=3) (ns, P>0.05; *P<0.05; ***P<0.001).
To understand molecularly which point of the Wnt pathway inhibits Notch activity, we activated Wnt signalling at different levels by expressing Wnt1 or the downstream components mDvl2 and S45F β-catenin (a stabilised form of β-catenin) (Vécsey-Semjen et al., 2002). Notch signalling was initiated by expressing a constitutively active form of mouse Notch1 (ΔN-mN1) (Mizutani et al., 2001) and monitored by co-transfection with the RBPJκ-luciferase reporter construct. The expression of Wnt1 or Dvl2 significantly reduced ΔN-mN1 transcriptional activity, whereas S45F β-catenin did not (Fig. 1C). We confirmed the ability of Dvl2 to inhibit Notch signalling by repeating the JAG1-Fc-induced reporter assay with SHEP-RBPJκ-Luc cells (supplementary material Fig. S1). As Dishevelled activates β-catenin signalling by disrupting GSK3β activity (MacDonald et al., 2009); we examined whether blocking GSK3β function with LiCl or SB216763 inhibited Notch signalling. Neither the inhibitors nor the expression of a dominant-negative GSK3β (GSK3β K85R) (Fraser et al., 2002) attenuated Notch activity (Fig. 1D; supplementary material Fig. S2, S3). Last, to confirm that the Wnt-induced inhibition of Notch signalling is mediated by Dishevelled, we repeated the Notch reporter assay in cells transfected with an siRNA construct that targets mDvl2 (Lee et al., 2008). Wnt1 had no effect on Notch signalling in the absence of Dvl2 (Fig. 1E). Together, these data clearly place the point of crosstalk upstream of GSK3β at the level of Dishevelled and indicate that the crosstalk is independent of Wnt-induced transcription, as S45F β-catenin activates Wnt transcriptional activity to the same extent as mDvl2 (supplementary material Fig. S2).
Dishevelled regulates Notch signalling during cell fate decisions in vivo
Having established that Dishevelled inhibited Notch signalling in cultured cells, we then wanted to determine whether Dishevelled/Notch crosstalk affected cell fate decisions in vivo. The epidermis of the Xenopus embryo contains ciliated cells whose precursors are regularly spaced throughout the outer layer by a Notch-mediated lateral inhibition signal (Deblandre et al., 1999). The initial specification of these precursors is not altered by expressing β-catenin to activate downstream Wnt signalling (Ossipova et al., 2007).
We first examined whether Dishevelled was required for regulating Notch signalling during normal development. Embryos were injected at the one-cell stage with either a XDvl2 (MODvl2) morpholino to knock down XtDvl2 expression or a control (MOC) morpholino. Ciliated cell precursors were detected at stage 18 by the expression of α-tubulin using whole-mount in situ hybridisation (Deblandre et al., 1999), and the precursor number within a standard region over the central section of the embryo was quantified (Fig. 2D, see inset). MOC-injected embryos displayed no difference in the number or spacing of precursors, compared with uninjected controls (Fig. 2A,B,D) (P>0.05, n≥14). By contrast, MODvl2-injected embryos showed fewer precursors with increased spacing when compared with either the uninjected or MOC-injected embryos (Fig. 2A-D) (P<0.001, n≥19). We confirmed the reduction in α-tubulin expression by qRT-PCR and found that the Notch target gene esr1 gene was also increased (Fig. 2E). These results suggest that knock-down of XDvl2 increases endogenous Notch signalling and thereby reduces the number of ciliated cell precursors.
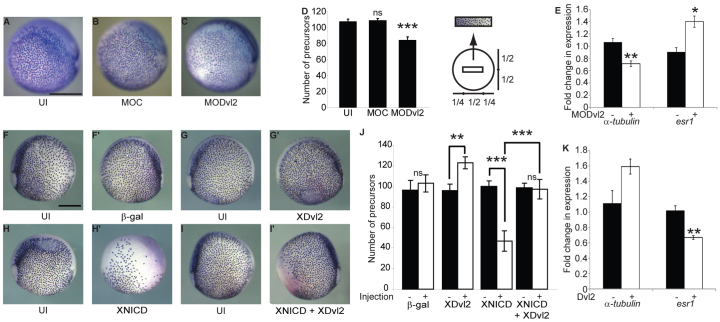
Fig. 2.
Dishevelled regulates Notch-dependent cell fate decisions in vivo. (A-C) XDvl2 is required to regulate Notch signalling during development. Xenopus tropicalis embryos were injected at the one-cell stage with control morpholino (MOC) or one targeting Xdvl2 (MODvl2). Ciliated cell precursors were detected by α-tubulin expression (purple staining). (D) Precursors were counted within a box of standard area drawn over the centre of each embryo (see inset). Data are presented as mean number of precursors counted (±s.e.m.). Uninjected (UI) and MOC embryos were indistinguishable (A,B), whereas MODvl2 embryos exhibited a significant reduction in precursor number (C) (one-way ANOVA with Tukey's post-hoc tests). (E) qRT-PCR analysis of α-tubulin and esr1 expression in MOC and MODvl2 embryos. Expression was normalised to rpl8. Data are presented as mean fold change (±s.e.m.) in normalised expression values relative to uninjected embryos. esr1 expression was significantly increased in the MODvl2 embryos (two-tailed t-test, n=3). (F-I′) XDvl2 expression inhibits endogenous Notch signalling and rescues the NICD gain-of-function phenotype. Xenopus laevis embryos were injected in one blastomere of the two-cell embryo with mRNA encoding β-gal (F,F′), β-gal and XDvl2 (G,G′), β-gal and XNICD (H,H′), or β-gal, XNICD and XDvl2 (I,I′). (J) Ciliated cell precursors were quantified as above (see D). X-Gal staining was performed to distinguish the injected side (pale red). Images of the uninjected and injected sides of the same embryo are shown. XDvl2 promoted (G′) and XNICD inhibited (H′) ciliated cell precursor specification (two-tailed paired t-test). XDvl2 completely rescued the XNICD phenotype (I′) and there was no significant difference between the uninjected sides of any condition, and the β-gal or XNICD + XDvl2 expressing sides (two-way ANOVA and Bonferroni's post-hoc test). (K) qRT-PCR analysis of α-tubulin and esr1 expression in embryos expressing GFP or XDvl2 conducted as above (see E). esr1 expression was significantly decreased in XDvl2-expressing embryos (two-tailed t-test, n=3). Scale bars: 500 μm in A-C,F-I′. **P<0.01; ***P<0.001; ns, P>0.05.
We next wanted to determine whether increased XDvl2 expression results in the converse Notch loss-of-function phenotype. Embryos were injected in one blastomere of a two-cell stage embryo with Xdvl2 mRNA and a lacZ tracer, or tracer alone. The injected side was then compared with the uninjected control side for each embryo. Injection of the lacZ tracer had no effect on precursor specification (P>0.05, n=4) (Fig. 2F,F′,J). However, expression of XDvl2 significantly increased precursor number, indicating that XDvl2 inhibited endogenous Notch signalling (P=0.0024, n=10) (Fig. 2G,G′,J). qRT-PCR analysis also demonstrated a gain in α-tubulin expression and a reduction of the Notch target esr1 (Fig. 2K). To confirm the inhibition of Notch signalling by Dishevelled, we investigated whether XDvl2 could rescue a Notch gain-of-function phenotype. Notch signalling was activated by injection of XNICD mRNA. XNICD caused a severe reduction in the number of precursors (P<0.001, n=10) (Fig. 2H,H′,J; supplementary material Fig. S4) (Deblandre et al., 1999). Strikingly, co-injecting Xdvl2 mRNA with XNICD mRNA rescued this Notch gain-of-function phenotype, with most of the epidermis being indistinguishable from the uninjected control side (P>0.05, n=11) (Fig. 2I,I′,J; supplementary material Fig. S4). Together, these data show clearly that Dishevelled regulates Notch signalling in vivo during Xenopus development.
Dishevelled inhibits all four Notch paralogues
The effect of crosstalk between Notch and other signalling pathways can vary depending on the Notch paralogue analysed (Foltz et al., 2002; Dahlqvist et al., 2003; Espinosa et al., 2003; Sun et al., 2005; Beres et al., 2011). We therefore investigated whether Dishevelled could inhibit each one of the four Notch receptors. Notch signalling was activated by expressing constitutively active forms of the human Notch paralogues (ΔN-hN1-4). We found that Dvl2 inhibited the activity of each construct (Fig. 3A), demonstrating that Dishevelled/Notch crosstalk results in pan-Notch inhibition and most likely targets a step in the signal transduction mechanism that is common to all four receptors.
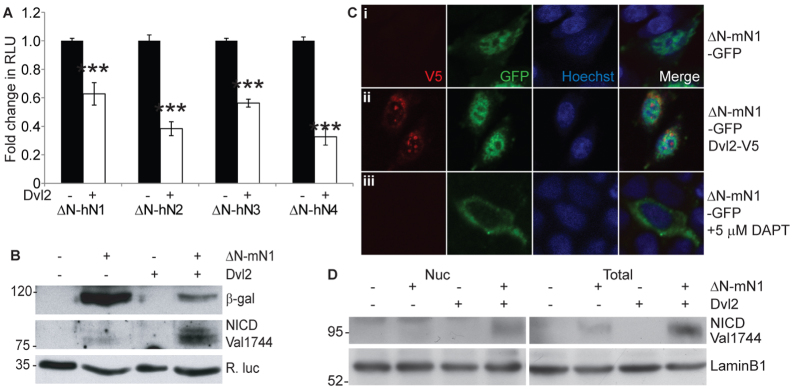
Fig. 3.
Dishevelled does not inhibit Notch cleavage or the nuclear translocation of NICD. (A) mDvl2 inhibits all four human Notch paralogues. CHO-K1 cells were transfected with RBPJκ-Luc and pRL-CMV. Notch signalling was initiated by expressing active forms of each human Notch paralogue (ΔN-hN1-4) in the presence or absence of mDvl2. Data are presented as mean fold change (±s.e.m.) in RLU compared with each Notch construct alone. Dvl2 inhibited each Notch paralogue (***P<0.001, one-way ANOVA and Tukey's post-hoc test, n=3). (B) mDvl2 does not inhibit Notch cleavage. Cells transfected with RBPJκ-lacZ and pRL-CMV were also transfected with vectors encoding ΔN-mN1 or mDvl2. Lysates were analysed by immunoblotting to examine β-gal reporter gene activity and ΔN-mN1 cleavage (NICD Val1744). R. luciferase is a loading control. (C,D) NICD released from ΔN-mN1 translocates to the nucleus even in the presence of mDvl2. (C) Cells expressing ΔN-mN1-GFP and mDvl2-V5, as indicated, were fixed and immunostained for GFP (green) and V5 (red) epitopes. DAPT treatment to inhibit γ-secretase function prevented nuclear translocation of NICD. (D) CHO-K1 cells expressing ΔN-mN1 and mDvl2, as indicated, were fractionated and nuclear accumulation of NICD was analysed by immunoblotting the nuclear fraction (Nuc) and total lysates (Total). LaminB1 is a loading control. Positions of molecular weight markers (in kDa) are indicated.
In Drosophila, it has been proposed that the region C-terminal to the Ankyrin repeats of Notch physically interacts with Dishevelled (Axelrod et al., 1996; Ramain et al., 2001; Strutt et al., 2002; Muñoz-Descalzo et al., 2010). To formally test whether the C terminus of mNotch1 is required for inhibition by Dishevelled, we generated deletion constructs: ΔN-mN1-ΔC238, -ΔC351 and -ΔC425. These mutants lack progressively more of the C terminus such that ΔN-mN1-ΔC425 lacks all the protein sequence C-terminal to the Ankyrin repeats (supplementary material Fig. S5). Using the RBPJκ-luciferase reporter assay, we found that none of these Notch deletions prevented the inhibition of signalling by Dishevelled (supplementary material Fig. S5). This result indicates that Dishevelled inhibited Notch independently of the C terminus, suggesting a distinct crosstalk mechanism than that previously suggested (Axelrod et al., 1996; Ramain et al., 2001; Strutt et al., 2002; Muñoz-Descalzo et al., 2010).
Dishevelled does not prevent the release or nuclear translocation of NICD
γ-Secretase is required for the cleavage of all four Notch paralogues to release the intracellular domain, which subsequently translocates to the nucleus to initiate transcription (Mizutani et al., 2001). Using a γ-secretase cleavage-specific Notch1 antibody (NICD Val1744), we investigated whether Dishevelled could inhibit N1ICD release from ΔN-mN1. A lacZ transcriptional reporter (RBPJκ-lacZ) was co-transfected to monitor Notch signalling. Surprisingly, we found that NICD Val1744 reactivity increased with Dvl2 expression, even as signalling was inhibited (Fig. 3B, compare lanes 2 and 4). This accumulation demonstrated that the cleavage of NICD was not inhibited by Dvl2.
This led to the possibility that Dvl2 might prevent NICD from translocating to the nucleus following release by γ-secretase cleavage. To address this hypothesis, cells were transfected with constructs encoding ΔN-mN1-GFP and Dvl2-V5 and subjected to immunofluorescence. N1ICD was detected in the nucleus of cells when ΔN-mN1-GFP was expressed alone or in combination with Dvl2 (Fig. 3C, i and ii), suggesting that Dvl2 did not inhibit nuclear translocation of NICD. Contrastingly, DAPT treatment to inhibit γ-secretase function prevented N1ICD nuclear translocation (Fig. 3C, part iii). Moreover, when ΔN-mN1- and Dvl2-expressing cells were fractionated, NICD Val1744 reactivity was increased in the nuclear-enriched fraction (Fig. 3D). Together, these data indicate that Dishevelled did not inhibit Notch signalling by preventing nuclear translocation of NICD.
The accumulation of NICD within the nucleus concomitantly with an inhibition of signalling can be explained by the lack of transcription-coupled degradation of NICD. When NICD forms an active transcriptional complex with RBPJκ and the co-activator MAML, it is phosphorylated by CDK8 and targeted for proteosomal degradation (Fryer et al., 2002; Fryer et al., 2004). This limits the duration of the Notch response by terminating the signal once transcription of target genes is initiated. We therefore reasoned that the accumulation of cleaved NICD observed following Dvl2 expression might represent the failure to form a functional NICD/RBPJκ/MAML transcriptional complex. This would explain the accumulation of NICD molecules that are unable to activate signalling.
Dishevelled inhibits Notch signalling at the level of RBPJκ downstream of NICD
As RBPJκ and MAML are required for NICD activity, we addressed whether Dishevelled affected either of these proteins. We first investigated whether Dvl2 inhibited MAML-mediated transcriptional activation or reduced the expression of MAML proteins, but found neither of these to be the case (supplementary material Fig. S6). Consequently, we examined whether Dvl2 could inhibit RBPJκ. Given CSL transcription factors act as repressors in the absence of Notch (Bray, 2006), we used an activated form of RBPJκ (VP16-RBPJκ) (Kato et al., 1997) to mimic the active NICD/RBPJκ/MAML transcriptional complex. We found that Dvl2 significantly inhibited VP16-RBPJκ activity, similar to its effect on ΔN-mN1 (Fig. 4A), suggesting that Dishevelled inhibited RBPJκ downstream of active Notch proteins. To establish whether Dvl2 inhibition of RBPJκ was independent of NICD, we used a VP16-RBPJκ molecule that carries a point mutation (K275M) to abolish the RBPJκ/NICD interaction (Fuchs et al., 2001). Dvl2 inhibited the transcriptional activity of both VP16-RBPJκ and VP16-RBPJκ-K275M (supplementary material Fig. S7), demonstrating that Dishevelled could inhibit RBPJκ independently of NICD.
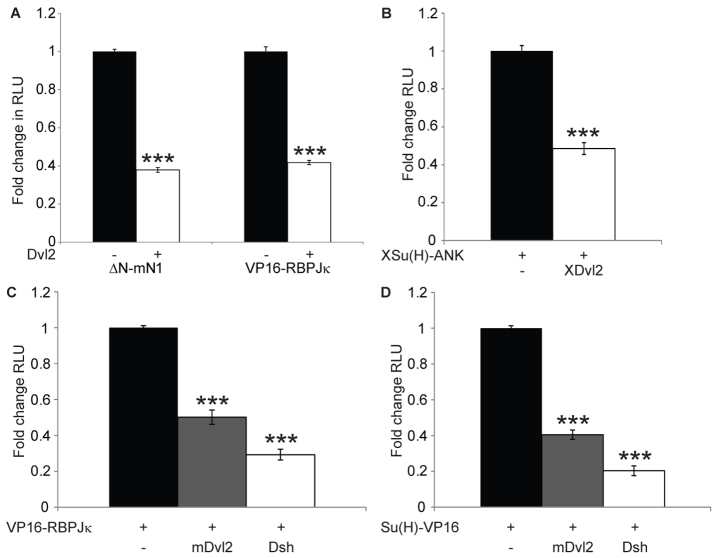
Fig. 4.
Dishevelled inhibits RBPJκ downstream of Notch. (A-D) CHO-K1 cells were transfected with RBPJκ-Luc (A,C,D) or NRE Su(H)-Luc (B) and pRL-CMV. Notch and Wnt signalling were activated by co-expression of ΔN-mN1 or an active form of RBPJκ, VP16-RBPJκ and mDvl2 (A), XSu(H)-ANK and XDvl2 (B), or VP16-RBPJκ, Su(H)-VP16, mDvl2 and Drosophila Dsh (C,D), as indicated. Data are presented as mean fold change (±s.e.m.) in RLU relative to each Notch pathway component alone. (A) mDvl2 inhibited both ΔN-mN1 and VP16-RBPJκ similarly. (B) XDvl2 inhibits XSu(H)-ANK. (C,D) Drosophila Dsh and mDvl2 inhibited VP16-RBPJκ (C) and Su(H)-VP16 (D). ***P<0.001, one-way ANOVA and Tukey's post-hoc test, n≥3.
To determine whether this Dvl-RBPJκ crosstalk was conserved across species, the effect of XDvl2 on the Xenopus CSL orthologue, and Drosophila Dsh on the Drosophila CSL orthologue was examined. Signalling was activated in luciferase reporter assays with VP16-RBPJκ, XSu(H)-ANK [a constitutively-active fusion of XSu(H) to the Notch Ankyrin repeats (Wettstein et al., 1997)] or Su(H)-VP16 [an activated form of the Drosophila CSL orthologue (Furriols and Bray, 2000)]. Expression of XDvl2 significantly inhibited XSu(H)-ANK activity (Fig. 4B), whereas Drosophila Dsh was able to inhibit VP16-RBPJκ to the same extent as mDvl2 (Fig. 4C). Moreover, we found that both Drosophila Dsh and mDvl2 inhibited Su(H)-VP16 activity (Fig. 4D). Together, these results suggest that the novel Dishevelled-CSL crosstalk we have identified is conserved between vertebrate and invertebrate homologues. Moreover, Dishevelled must regulate a key property of CSL proteins, as CSL transcriptional activity was inhibited irrespective of the method used to activate the CSL molecule [Notch ligand (Fig. 1A-B; Fig. 2G-G′;), active Notch proteins (Fig. 1C; Fig. 2H,I; Fig. 3A,B) or fusion to different exogenous activation sequences (Fig. 4)].
Dishevelled binds RBPJκ and reduces RBPJκ activity
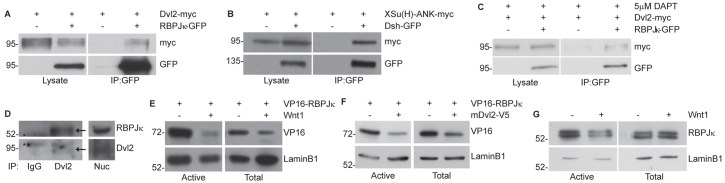
As Dishevelled did not appear to prevent the nuclear localisation of RBPJκ (supplementary material Fig. S7), we investigated whether mDvl2 could physically bind RBPJκ. Using GFP-tagged RBPJκ in a co-immunoprecipitation assay, we detected Dvl2-myc specifically in the immunoprecipitates from RBPJκ-GFP-expressing cells (Fig. 5A). This interaction was independent of the epitope tags used, as VP16-RBPJκ also bound to mDvl2-V5 in a reciprocal co-immunoprecipitation (supplementary material Fig. S7). Furthermore, a physical interaction between XDvl2-GFP and XSu(H)-ANK-myc was also detected (Fig. 5B). To confirm that the interaction is independent of NICD, we repeated the immunoprecipitations in the presence of DAPT to inhibit γ-secretase function and therefore NICD translocation to the nucleus (Fig. 3C, part iii; Fig. 5C), and with the VP16-RBPJκ-K275M mutant that cannot interact with NICD (supplementary material Fig. S7). The interaction between Dvl2 and RBPJκ was still observed in both conditions. Together, these data support a model whereby Dishevelled binds and inhibits CSL proteins independently of NICD, which limits the transcriptional activity of the NICD/CSL/MAML complex.
Fig. 5.
Dishevelled binds RBPJκ within the nucleus and reduces RBPJκ activity. (A-C) Dishevelled binds CSL proteins even in the absence of NICD. CHO-K1 cells expressing mDvl2-myc and RBPJκ-GFP (A,C) or XSu(H)-ANK-myc and XDvl2-GFP (B) were subjected to immunoprecipitation using GFP-Trap beads. Immunoprecipitation samples were analysed by immunoblotting for myc and GFP, alongside total lysates. (C) Cells were treated with 5 μM DAPT to prevent the generation of NICD by cleavage of the endogenous Notch protein (see Fig. 3C). (D) Dishevelled binds RBPJκ within the nucleus. The soluble nuclear fraction was isolated from CHO-K1 cells and subjected to immunoprecipitation with Dvl2 antibody or control IgG. Immunoprecipitation samples and nuclear input (Nuc) were analysed by immunoblotting with RBPJκ and Dvl2 antibodies (indicated by arrows; asterisk indicates a non-specific band). (E-G) Wnt signalling reduces the amount of RBPJκ found within the active transcription factor pool. Cells expressing Wnt1, mDvl2 and VP16-RBPJκ, as indicated, were fractionated to enrich for active transcription factors and subjected to immunoblotting with VP16 and RBPJκ antibodies. LaminB1 is a loading control. Wnt1 (E) and mDvl2 (F) reduce the amount of VP16-RBPJκ within the fraction containing active transcription factors. Wnt1 reduces the level of endogenous RBPJκ within the same fraction (G). Positions of molecular weight markers (in kDa) are shown.
To confirm the binding of Dvl2 and RBPJκ, we investigated the interaction between endogenous proteins within the nucleus. Using an antibody recognising Dvl2, we could co-immunoprecipitate RBPJκ from a soluble, nucleosol-enriched fraction (Fig. 5D). As the nucleosol-enriched fraction contains active transcription factors (Schreiber et al., 1989), we next examined whether Dishevelled reduced the level of RBPJκ within the active pool. We found that Wnt1 or Dvl2 expression reduced VP16-RBPJκ levels within this nuclear fraction (Fig. 5E,F). More importantly, Wnt1 expression also decreased endogenous RBPJκ within the same fraction (Fig. 5G). Together these results suggest that Dishevelled inhibits Notch signalling by reducing the amount of RBPJκ within the nuclear fraction that contains active transcription factors.
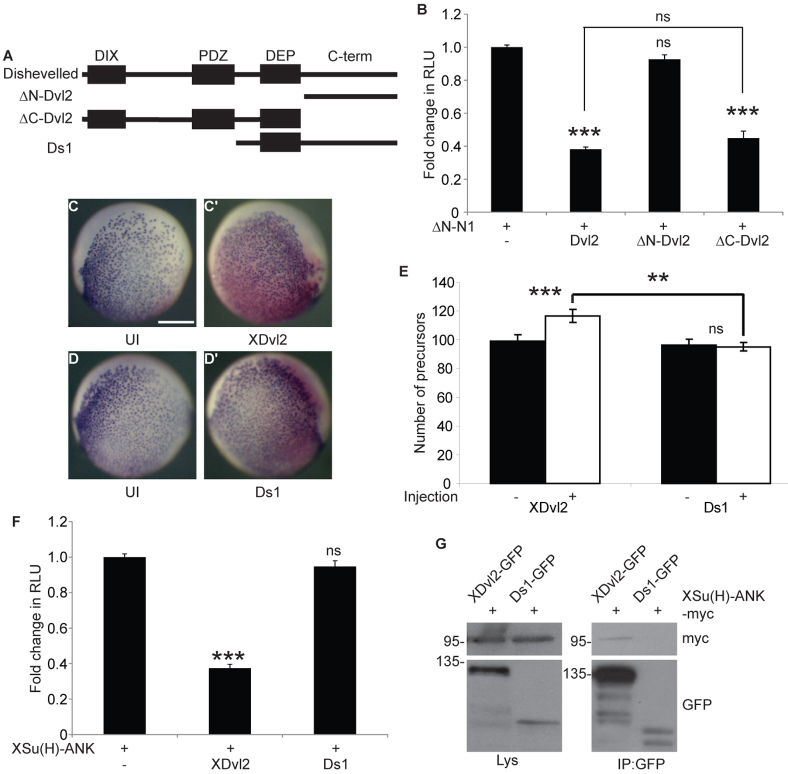
The DIX and PDZ domains of Dishevelled are required for RBPJκ inhibition
To further dissect the mechanism by which Dvl inhibits RBPJκ, we aimed to define the region of Dvl necessary for this activity. Dishevelled proteins contain three characterised domains (Fig. 6A); the DIX (Dishevelled, Axin), PDZ (PSD95, Discs, large ZO-1) and DEP (Dishevelled, EGL-10, Pleckstrin) domains, as well as short regions of homology within the C terminus (Wallingford and Habas, 2005). To determine which area of the Dvl2 molecule was required, two deletion constructs were generated: ΔN-mDvl2 and ΔC-mDvl2 (Fig. 6A). ΔN-mDvl2 contains the regions of homology within the C-terminal half and lacks the domains within the N terminus. ΔC-mDvl2 lacks the C-terminal region but contains the DIX, PDZ and DEP domains. Using ΔN-mN1 to activate Notch signalling in a luciferase reporter assay, we determined that ΔN-mDvl2 had no effect on Notch activity, whereas ΔC-Dvl2 inhibited signalling similarly to full-length Dvl2 (Fig. 6B). This suggests that the conserved sequence within the C-terminal half of Dishevelled is not required to inhibit Notch signalling.
Fig. 6.
Dishevelled DIX and PDZ domains are required for inhibition of RBPJκ. (A) Schematic of the structure of Dishevelled and the deletion constructs used. (B) The C terminus of mDvl2 is not required for inhibition of RBPJκ activity. CHO-K1 cells were transfected with RBPJκ-Luc and pRL-CMV, and vectors encoding ΔN-mN1 and the mDvl2 constructs illustrated. Data are presented as mean fold change (±s.e.m.) in RLU relative to ΔN-mN1 alone. Both ΔC-mDvl2 and mDvl2, but not ΔN-mDvl2, inhibit Notch signalling (one-way ANOVA and Tukey's post-hoc test, n≥3). (C-E) XDvl2 but not Ds1 inhibits endogenous Notch signalling in vivo. Xenopus laevis embryos were injected in one blastomere of a two-cell embryo with mRNA encoding β-gal and XDvl2 (C,C′) or β-gal and Ds1 (D,D′). Ciliated cell precursors were detected by α-tubulin expression and the injected side was determined by X-Gal staining. Images of the uninjected (UI) and injected sides of the same embryo are shown. (E) Ciliated cell precursors were quantified as in Fig. 2D. Data are presented as the mean number of precursors counted (±s.e.m.). XDvl2 significantly increased ciliated cell precursor specification but Ds1 did not (two-tailed paired t-test). XDvl2 also increased precursor specification compared with Ds1 (two-way ANOVA and Bonferroni's post-hoc test). (F) Ds1 does not inhibit XSu(H) activity. CHO-K1 cells were transfected with NRE Su(H)-Luc and pRL-CMV, and vectors encoding XSu(H)-ANK, XDvl2 or Ds1. Data are presented as mean fold change (±s.e.m.) in RLU relative to XSu(H)-ANK alone (one-way ANOVA and Tukey's post-hoc test, n=3). (G) Ds1 shows greatly reduced XSu(H) binding. CHO-K1 cells expressing XSu(H)-ANK-myc and XDvl2-GFP or Ds1-GFP were subjected to immunoprecipitation using GFP-Trap beads. Immunoprecipitation samples were analysed by immunoblotting for myc and GFP, alongside total lysates. Positions of molecular weight markers (in kDa) are shown. Scale bar: 500 μm. **P<0.01; ***P<0.001; ns, P>0.05.
To substantiate this result, we expressed an N-terminal deletion construct of XDvl2, Ds1 [which lacks the DIX and PDZ domains (Fig. 6A) (Itoh et al., 2005)] in Xenopus embryos to determine its effects on endogenous Notch signalling in vivo during ciliated cell precursor specification. Expression of XDvl2 increased the number of precursors (P<0.001, n=14) (Fig. 6C,C′,E), whereas Ds1 had no effect on precursor specification (P>0.05, n=13) (Fig. 6D,D′,E). This result demonstrates that the N-terminal DIX and PDZ domains are required to inhibit XSu(H)-dependent Notch signalling in vivo. In keeping with this result, full-length XDvl2, but not Ds1, was able to inhibit XSu(H)-ANK activity in a luciferase reporter assay (Fig. 6F). Finally, we tested whether the inability of Ds1 to inhibit Notch/XSu(H) signalling was due to reduced binding to XSu(H). We expressed either XDvl2-GFP or Ds1-GFP with XSu(H)-ANK-myc and performed an immunoprecipitation with a GFP antibody. Compared with full-length XDvl2-GFP, we found that Ds1-GFP showed greatly reduced binding to XSu(H)-ANK-myc (Fig. 6G). Together, these data illustrate the N-terminal region of Dishevelled is required to bind and inhibit XSu(H) and thereby limit Notch signalling in vivo.
DISCUSSION
Here, we have shown that ligand-induced Wnt signalling not only activates downstream β-catenin signalling but also limits the Notch pathway through Dishevelled-mediated inhibition of RBPJκ. Moreover, we have elucidated the underlying mechanism of Notch inhibition showing that Dvl2 binds to CSL proteins to reduce their level within the active transcription factor pool. We also demonstrate that Dishevelled-CSL crosstalk is evolutionarily conserved. Thus, this study identifies a novel mode of inhibitory crosstalk between Wnt and Notch, and provides the first evidence of CSL transcription factors being a target for signalling crosstalk.
A recent analysis of how Notch interacts with other developmental signalling pathways shows that the points of crosstalk uncovered thus far predominantly affect individual Notch receptors or concern the regulation of shared target genes (Hurlbut et al., 2007). By contrast, crosstalk between Dishevelled and CSL transcription factors targets the key node of the Notch pathway found downstream of all four Notch receptors and ensures all Notch signalling is inhibited. This makes Dishevelled-CSL crosstalk a powerful means with which to limit Notch signalling and reveals a new paradigm of Notch regulation. Mechanistically, Dishevelled has been shown to act as a key molecular scaffold that regulates diverse processes such as the internalisation of Frizzled receptors (Yu et al., 2007), stabilisation of aPKC activity (Zhang et al., 2007), ubiquitylation of the polarity protein Prickle1 (Narimatsu et al., 2009) and the formation of transcriptional complexes (Gan et al., 2008). We speculate that Dishevelled may also be acting as a scaffold to regulate CSL activity.
The dual role of Dishevelled as an activator of downstream Wnt signalling and an inhibitor of Notch activity places it in a key position to regulate cell-fate decisions in which Wnt and Notch have opposing effects. This antagonism is often seen in the cell-fate decisions made by individual cells with Wnt signalling promoting the adoption of one cell fate and Notch another. For example, in both the skin and mammary gland, Wnt signalling promotes the maintenance of the stem cell fate whereas Notch signalling promotes lineage commitment and differentiation (Zhu and Watt, 1999; Lowell et al., 2000; Bouras et al., 2008; Zeng and Nusse, 2010). In this case, the inhibition of Notch signalling by Dishevelled would help to maintain the stem cell population. The Notch and Wnt pathways also have opposing effects at later steps within a cell lineage. For example, Notch and Wnt signalling influence terminal differentiation within the intestinal epithelium, with Notch activity biasing cells towards the absorptive fate and Wnt signalling favouring secretory cell differentiation (Fre et al., 2005; Stanger et al., 2005; van Es et al., 2005b; van Es et al., 2005a). In each of these cases, a clear distinction between a Notch response and Wnt response is vital for appropriate and robust cell-fate decisions to occur. Inhibitory crosstalk, such as inhibition of Notch/CSL by Dishevelled, reinforces these binary decisions.
Within a single lineage, the temporal transition from a state of Notch-ON/Wnt-OFF to Wnt-ON/Notch-OFF is also crucial for progression from precursor to terminally differentiated cell. A clear example of this requirement is during early embryonic myogenesis and in muscle repair in the adult (Brack et al., 2008; Rios et al., 2011). Here, an initial peak of Notch signalling is required to trigger the differentiation programme of progenitors and to expand the muscle progenitor pool prior to terminal differentiation. However, this Notch signal must be shut off and Wnt signalling must then be activated for terminal differentiation to occur appropriately. If this temporal switch does not occur and the Notch signal is maintained, then proper myogenesis fails to occur (Rios et al., 2011), even in the presence of active Wnt signalling (Brack et al., 2008). The dual role of Dishevelled in activating Wnt signalling while preventing concurrent Notch activity ensures a Wnt-ON/Notch-OFF output. This maximises the distinction between Wnt-ON and Notch-ON responses which is vital for ensuring regulated transitions between cell states within a lineage.
In addition to ensuring that different cell types are produced with the appropriate number and timing, Wnt/Notch interactions are also required for the correct positioning of Notch activity within a tissue. This occurs during the segmentation of the vertebrate hindbrain into rhombomeres (Cheng et al., 2004; Amoyel et al., 2005) and the specification of the dorsal (D) and ventral (V) compartments in the developing Drosophila wing (de Celis et al., 1996; Rulifson et al., 1996; Micchelli et al., 1997; Klein and Arias, 1998; Micchelli and Blair, 1999). In both cases, Notch signalling is activated in the cells that make up the boundary between compartments. These boundary cells then produce Wnt proteins that orchestrate the growth and patterning of the adjacent compartments. Within the developing Drosophila wing, Notch signalling is initially activated in a broad stripe at the D/V boundary, or wing margin. This induces the expression of the Wnt protein Wingless, which refines this stripe of Notch activity in two ways. Wingless signalling maintains Notch ligand expression in the cells that flank the margin so that they can signal back to the margin cells and maintain Notch activity (Micchelli et al., 1997). Wingless also inhibits Notch signalling in the cells outside of the margin through direct crosstalk that requires Dishevelled but not Armadillo (the Drosophila β-catenin homologue) (Rulifson et al., 1996; Brennan et al., 1999). Our data now suggest that Dishevelled-Su(H) crosstalk may contribute to this refinement of Notch activity.
Finally, the importance of proper regulation of the Notch and Wnt pathways is highlighted by aberrant Notch and Wnt signalling being linked to various human pathologies, including many cancers. Significantly, it has been shown that interactions between the pathways play a causative role in several of these conditions (Ayyanan et al., 2006; Rodilla et al., 2009). Thus, the results presented here will inform our understanding of how developmental mechanisms may be subverted in disease. For example, disease initiation or progression may be reliant on Wnt activation with the concomitant inhibition of Notch signalling mediated by crosstalk. Furthermore, treating a disease by limiting Wnt signalling at the level of the ligand may lead to detrimental activation of the Notch pathway, as Dishevelled-Notch/CSL crosstalk would also be lost; in this case, an inhibitor of the Wnt transcriptional response may be a better therapeutic tool (Chen et al., 2009; Gonsalves et al., 2011). Finally, the mimicking of inhibitory crosstalk also opens new avenues for therapeutic drug development by offering the promise of specificity in targeting signalling pathways.
Supplementary Material
Acknowledgements
We are grateful to Sìlvia Oliveira, Alfonso Martinez Arias, Mike Grant and Chris Thompson for constructive discussions. We thank the Papalopulu and Amaya groups, in particular Raphael Thuret, Eamon Dubaissi and Nitin Sabherwal, for help with Xenopus experiments; Ruth Lopez for molecular cloning; and all those who have gifted reagents.
Footnotes
Funding
This work was supported by grants from the Biotechnology and Biological Science Research Council [BB/J005983/1 to K.D.] and the Wellcome Trust [085073 to K.B. and G.M.C.]. Deposited in PMC for immediate release.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.081885/-/DC1
References
- Amoyel M., Cheng Y. C., Jiang Y. J., Wilkinson D. G. (2005). Wnt1 regulates neurogenesis and mediates lateral inhibition of boundary cell specification in the zebrafish hindbrain. Development 132, 775-785 [DOI] [PubMed] [Google Scholar]
- Axelrod J. D., Matsuno K., Artavanis-Tsakonas S., Perrimon N. (1996). Interaction between Wingless and Notch signaling pathways mediated by dishevelled. Science 271, 1826-1832 [DOI] [PubMed] [Google Scholar]
- Ayyanan A., Civenni G., Ciarloni L., Morel C., Mueller N., Lefort K., Mandinova A., Raffoul W., Fiche M., Dotto G. P., et al. (2006). Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc. Natl. Acad. Sci. USA 103, 3799-3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres B. J., George R., Lougher E. J., Barton M., Verrelli B. C., McGlade C. J., Rawls J. A., Wilson-Rawls J. (2011). Numb regulates Notch1, but not Notch3, during myogenesis. Mech. Dev. 128, 247-257 [DOI] [PubMed] [Google Scholar]
- Bouras T., Pal B., Vaillant F., Harburg G., Asselin-Labat M. L., Oakes S. R., Lindeman G. J., Visvader J. E. (2008). Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell 3, 429-441 [DOI] [PubMed] [Google Scholar]
- Brack A. S., Conboy I. M., Conboy M. J., Shen J., Rando T. A. (2008). A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell Stem Cell 2, 50-59 [DOI] [PubMed] [Google Scholar]
- Bray S. J. (2006). Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678-689 [DOI] [PubMed] [Google Scholar]
- Brennan K. R., Brown A. M. (2004). Wnt proteins in mammary development and cancer. J. Mammary Gland Biol. Neoplasia 9, 119-131 [DOI] [PubMed] [Google Scholar]
- Brennan K., Klein T., Wilder E., Arias A. M. (1999). Wingless modulates the effects of dominant negative notch molecules in the developing wing of Drosophila. Dev. Biol. 216, 210-229 [DOI] [PubMed] [Google Scholar]
- Brennan K., Gonzalez-Sancho J.M., Howe L.R., Brown A.M.C. (2004). Truncated mutants of the putative Wnt receptor LRP6/Arrow can stabilize beta-catenin independently of Frizzled proteins. Oncogene 23, 4873-4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capilla A., Johnson R., Daniels M., Benavente M., Bray S. J., Galindo M. I. (2012). Planar cell polarity controls directional Notch signaling in the Drosophila leg. Development 139, 2584-2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers A. D., Welchman D., Papalopulu N. (2002). Intrinsic differences between the superficial and deep layers of the Xenopus ectoderm control primary neuronal differentiation. Dev. Cell 2, 171-182 [DOI] [PubMed] [Google Scholar]
- Chen B., Dodge M. E., Tang W., Lu J., Ma Z., Fan C. W., Wei S., Hao W., Kilgore J., Williams N. S., et al. (2009). Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. C., Amoyel M., Qiu X., Jiang Y. J., Xu Q., Wilkinson D. G. (2004). Notch activation regulates the segregation and differentiation of rhombomere boundary cells in the zebrafish hindbrain. Dev. Cell 6, 539-550 [DOI] [PubMed] [Google Scholar]
- Dahlqvist C., Blokzijl A., Chapman G., Falk A., Dannaeus K., Ibâñez C. F., Lendahl U. (2003). Functional Notch signaling is required for BMP4-induced inhibition of myogenic differentiation. Development 130, 6089-6099 [DOI] [PubMed] [Google Scholar]
- de Celis J. F., Garcia-Bellido A., Bray S. J. (1996). Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development 122, 359-369 [DOI] [PubMed] [Google Scholar]
- Deblandre G. A., Wettstein D. A., Koyano-Nakagawa N., Kintner C. (1999). A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development 126, 4715-4728 [DOI] [PubMed] [Google Scholar]
- Espinosa L., Inglés-Esteve J., Aguilera C., Bigas A. (2003). Phosphorylation by glycogen synthase kinase-3 beta down-regulates Notch activity, a link for Notch and Wnt pathways. J. Biol. Chem. 278, 32227-32235 [DOI] [PubMed] [Google Scholar]
- Foltz D. R., Santiago M. C., Berechid B. E., Nye J. S. (2002). Glycogen synthase kinase-3beta modulates notch signaling and stability. Curr. Biol. 12, 1006-1011 [DOI] [PubMed] [Google Scholar]
- Fraser E., Young N., Dajani R., Franca-Koh J., Ryves J., Williams R. S., Yeo M., Webster M. T., Richardson C., Smalley M. J., et al. (2002). Identification of the Axin and Frat binding region of glycogen synthase kinase-3. J. Biol. Chem. 277, 2176-2185 [DOI] [PubMed] [Google Scholar]
- Fre S., Huyghe M., Mourikis P., Robine S., Louvard D., Artavanis-Tsakonas S. (2005). Notch signals control the fate of immature progenitor cells in the intestine. Nature 435, 964-968 [DOI] [PubMed] [Google Scholar]
- Fryer C. J., Lamar E., Turbachova I., Kintner C., Jones K. A. (2002). Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 16, 1397-1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer C. J., White J. B., Jones K. A. (2004). Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 16, 509-520 [DOI] [PubMed] [Google Scholar]
- Fuchs K. P., Bommer G., Dumont E., Christoph B., Vidal M., Kremmer E., Kempkes B. (2001). Mutational analysis of the J recombination signal sequence binding protein (RBP-J)/Epstein-Barr virus nuclear antigen 2 (EBNA2) and RBP-J/Notch interaction. Eur. J. Biochem. 268, 4639-4646 [DOI] [PubMed] [Google Scholar]
- Furriols M., Bray S. (2000). Dissecting the mechanisms of suppressor of hairless function. Dev. Biol. 227, 520-532 [DOI] [PubMed] [Google Scholar]
- Gan X. Q., Wang J. Y., Xi Y., Wu Z. L., Li Y. P., Li L. (2008). Nuclear Dvl, c-Jun, beta-catenin, and TCF form a complex leading to stabilization of beta-catenin-TCF interaction. J. Cell Biol. 180, 1087-1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalves F. C., Klein K., Carson B. B., Katz S., Ekas L. A., Evans S., Nagourney R., Cardozo T., Brown A. M., DasGupta R. (2011). An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc. Natl. Acad. Sci. USA 108, 5954-5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward P., Kalmar T., Arias A. M. (2008). Wnt/Notch signalling and information processing during development. Development 135, 411-424 [DOI] [PubMed] [Google Scholar]
- Hurlbut G. D., Kankel M. W., Lake R. J., Artavanis-Tsakonas S. (2007). Crossing paths with Notch in the hyper-network. Curr. Opin. Cell Biol. 19, 166-175 [DOI] [PubMed] [Google Scholar]
- Itoh K., Brott B. K., Bae G. U., Ratcliffe M. J., Sokol S. Y. (2005). Nuclear localization is required for Dishevelled function in Wnt/beta-catenin signaling. J. Biol. 4, 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Taniguchi Y., Kurooka H., Minoguchi S., Sakai T., Nomura-Okazaki S., Tamura K., Honjo T. (1997). Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development 124, 4133-4141 [DOI] [PubMed] [Google Scholar]
- Klein T., Arias A. M. (1998). Different spatial and temporal interactions between Notch, wingless, and vestigial specify proximal and distal pattern elements of the wing in Drosophila. Dev. Biol. 194, 196-212 [DOI] [PubMed] [Google Scholar]
- Lee Y. N., Gao Y., Wang H. Y. (2008). Differential mediation of the Wnt canonical pathway by mammalian Dishevelleds-1, -2, and -3. Cell. Signal. 20, 443-452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell S., Jones P., Le Roux I., Dunne J., Watt F. M. (2000). Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr. Biol. 10, 491-500 [DOI] [PubMed] [Google Scholar]
- MacDonald B. T., Tamai K., He X. (2009). Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli C. A., Blair S. S. (1999). Dorsoventral lineage restriction in wing imaginal discs requires Notch. Nature 401, 473-476 [DOI] [PubMed] [Google Scholar]
- Micchelli C. A., Rulifson E. J., Blair S. S. (1997). The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development 124, 1485-1495 [DOI] [PubMed] [Google Scholar]
- Mizutani T., Taniguchi Y., Aoki T., Hashimoto N., Honjo T. (2001). Conservation of the biochemical mechanisms of signal transduction among mammalian Notch family members. Proc. Natl. Acad. Sci. USA 98, 9026-9031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Descalzo S., Sanders P. G., Montagne C., Johnson R. I., Balayo T., Arias A. M. (2010). Wingless modulates the ligand independent traffic of Notch through Dishevelled. Fly (Austin) 4, 4 [DOI] [PubMed] [Google Scholar]
- Narimatsu M., Bose R., Pye M., Zhang L., Miller B., Ching P., Sakuma R., Luga V., Roncari L., Attisano L., et al. (2009). Regulation of planar cell polarity by Smurf ubiquitin ligases. Cell 137, 295-307 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P., Faber J. (1967). Normal Table of Xenopus Laevis (Daudin). Amsterdam: North-Holland Publishing Co. [Google Scholar]
- Ossipova O., Tabler J., Green J. B., Sokol S. Y. (2007). PAR1 specifies ciliated cells in vertebrate ectoderm downstream of aPKC. Development 134, 4297-4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires-daSilva A., Sommer R. J. (2003). The evolution of signalling pathways in animal development. Nat. Rev. Genet. 4, 39-49 [DOI] [PubMed] [Google Scholar]
- Raafat A., Goldhar A. S., Klauzinska M., Xu K., Amirjazil I., McCurdy D., Lashin K., Salomon D., Vonderhaar B. K., Egan S., et al. (2011). Expression of Notch receptors, ligands, and target genes during development of the mouse mammary gland. J. Cell. Physiol. 226, 1940-1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramain P., Khechumian K., Seugnet L., Arbogast N., Ackermann C., Heitzler P. (2001). Novel Notch alleles reveal a Deltex-dependent pathway repressing neural fate. Curr. Biol. 11, 1729-1738 [DOI] [PubMed] [Google Scholar]
- Reddy S., Andl T., Bagasra A., Lu M. M., Epstein D. J., Morrisey E. E., Millar S. E. (2001). Characterization of Wnt gene expression in developing and postnatal hair follicles and identification of Wnt5a as a target of Sonic hedgehog in hair follicle morphogenesis. Mech. Dev. 107, 69-82 [DOI] [PubMed] [Google Scholar]
- Rios A. C., Serralbo O., Salgado D., Marcelle C. (2011). Neural crest regulates myogenesis through the transient activation of NOTCH. Nature 473, 532-535 [DOI] [PubMed] [Google Scholar]
- Rodilla V., Villanueva A., Obrador-Hevia A., Robert-Moreno A., Fernández-Majada V., Grilli A., López-Bigas N., Bellora N., Albà M. M., Torres F., et al. (2009). Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc. Natl. Acad. Sci. USA 106, 6315-6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson E. J., Micchelli C. A., Axelrod J. D., Perrimon N., Blair S. S. (1996). wingless refines its own expression domain on the Drosophila wing margin. Nature 384, 72-74 [DOI] [PubMed] [Google Scholar]
- Schreiber E., Matthias P., Müller M. M., Schaffner W. (1989). Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17, 6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger B. Z., Datar R., Murtaugh L. C., Melton D. A. (2005). Direct regulation of intestinal fate by Notch. Proc. Natl. Acad. Sci. USA 102, 12443-12448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt D., Johnson R., Cooper K., Bray S. (2002). Asymmetric localization of frizzled and the determination of notch-dependent cell fate in the Drosophila eye. Curr. Biol. 12, 813-824 [DOI] [PubMed] [Google Scholar]
- Stylianou S., Clarke R. B., Brennan K. (2006). Aberrant activation of notch signaling in human breast cancer. Cancer Res. 66, 1517-1525 [DOI] [PubMed] [Google Scholar]
- Sun Y., Lowther W., Kato K., Bianco C., Kenney N., Strizzi L., Raafat D., Hirota M., Khan N. I., Bargo S., et al. (2005). Notch4 intracellular domain binding to Smad3 and inhibition of the TGF-beta signaling. Oncogene 24, 5365-5374 [DOI] [PubMed] [Google Scholar]
- Uyttendaele H., Soriano J. V., Montesano R., Kitajewski J. (1998). Notch4 and Wnt-1 proteins function to regulate branching morphogenesis of mammary epithelial cells in an opposing fashion. Dev. Biol. 196, 204-217 [DOI] [PubMed] [Google Scholar]
- van Es J. H., van Gijn M. E., Riccio O., van den Born M., Vooijs M., Begthel H., Cozijnsen M., Robine S., Winton D. J., Radtke F., et al. (2005a). Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959-963 [DOI] [PubMed] [Google Scholar]
- van Es J. H., Jay P., Gregorieff A., van Gijn M. E., Jonkheer S., Hatzis P., Thiele A., van den Born M., Begthel H., Brabletz T., et al. (2005b). Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat. Cell Biol. 7, 381-386 [DOI] [PubMed] [Google Scholar]
- Vécsey-Semjén B., Becker K. F., Sinski A., Blennow E., Vietor I., Zatloukal K., Beug H., Wagner E., Huber L. A. (2002). Novel colon cancer cell lines leading to better understanding of the diversity of respective primary cancers. Oncogene 21, 4646-4662 [DOI] [PubMed] [Google Scholar]
- Wallingford J. B., Habas R. (2005). The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development 132, 4421-4436 [DOI] [PubMed] [Google Scholar]
- Wettstein D. A., Turner D. L., Kintner C. (1997). The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development 124, 693-702 [DOI] [PubMed] [Google Scholar]
- Yu A., Rual J. F., Tamai K., Harada Y., Vidal M., He X., Kirchhausen T. (2007). Association of Dishevelled with the clathrin AP-2 adaptor is required for Frizzled endocytosis and planar cell polarity signaling. Dev. Cell 12, 129-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y. A., Nusse R. (2010). Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell 6, 568-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhu J., Yang G. Y., Wang Q. J., Qian L., Chen Y. M., Chen F., Tao Y., Hu H. S., Wang T., et al. (2007). Dishevelled promotes axon differentiation by regulating atypical protein kinase C. Nat. Cell Biol. 9, 743-754 [DOI] [PubMed] [Google Scholar]
- Zhu A. J., Watt F. M. (1999). beta-catenin signalling modulates proliferative potential of human epidermal keratinocytes independently of intercellular adhesion. Development 126, 2285-2298 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.