Abstract
We present the Microfluidic Electrochemical Quantitative Loop-mediated isothermal AMPlification (MEQ-LAMP) platform for rapid, sensitive, and quantitative detection of pathogen genomic DNA at the point of care. DNA amplification is electrochemically monitored in real time within a monolithic microfluidic device, enabling the detection of as few as 16 copies of Salmonella genomic DNA via a single-step process in under an hour.
Keywords: Point-of-care, Microfluidics, Electrochemistry, Real-time, Loop-mediated isothermal amplification
Genetic detection of pathogens at the point of care (POC) has become increasingly important in applications ranging from molecular diagnostics[1, 2] and food safety testing[3, 4] to environmental monitoring[5, 6] and homeland security[7]. In such applications, an ideal detection method must be portable, rapid, and sensitive. It would also support real-time, quantitative analysis across a wide dynamic range, thus offer information (e.g., genome copy number) that conventional end-point detection methods fail to provide[8, 9]. Toward this end, microfluidic systems combined with electrochemical detection offer compelling advantages[10–13] because they enable facile integration of multiple assay steps (e.g., sample preparation, nucleic acid amplification and detection) into a monolithic and disposable device[14–18] and achieve their detection without the use of bulky optical components. However, despite significant progresses to date,[19–25] existing integrated electrochemical genetic detection systems have yet to match the performance of the (laboratory) gold standard method (i.e., optical, real-time PCR[8, 9]), ultimately preventing their wide-spread adoption toward pathogen detection at the point of care.
Motivated by this clear need, we have developed the Microfluidic Electrochemical Quantitative Loop-mediated isothermal AMPlification (MEQ-LAMP) system – an integrated microfluidic platform for the rapid, sensitive and quantitative detection of pathogenic DNA. Our system leverages loop-mediated isothermal amplification (LAMP)[26–28], a powerful alternative to PCR that offers advantages in terms of sensitivity, reaction speed and amplicon yield, and can be applied to non-denatured genomic DNA samples under isothermal reaction conditions[29]. As a further benefit, this amplification technique employs six different primers, conferring exquisite specificity and enabling MEQ-LAMP to readily distinguish pathogens of interest from non-target genomic DNA. Importantly, we have achieved real-time, quantitative electrochemical detection of LAMP amplification by monitoring the intercalation of DNA-binding methylene blue (MB) redox reporter molecules into newly formed amplicons with a set of integrated electrodes. This combination of functions represents a significant advancement in real-time genetic detection toward POC applications. As a demonstration of this platform’s effectiveness, we report the direct and quantitative detection of as few as 16 copies of genomic DNA of Salmonella enterica enterica Typhimurium – a pathogen that causes food poisoning – in under an hour.
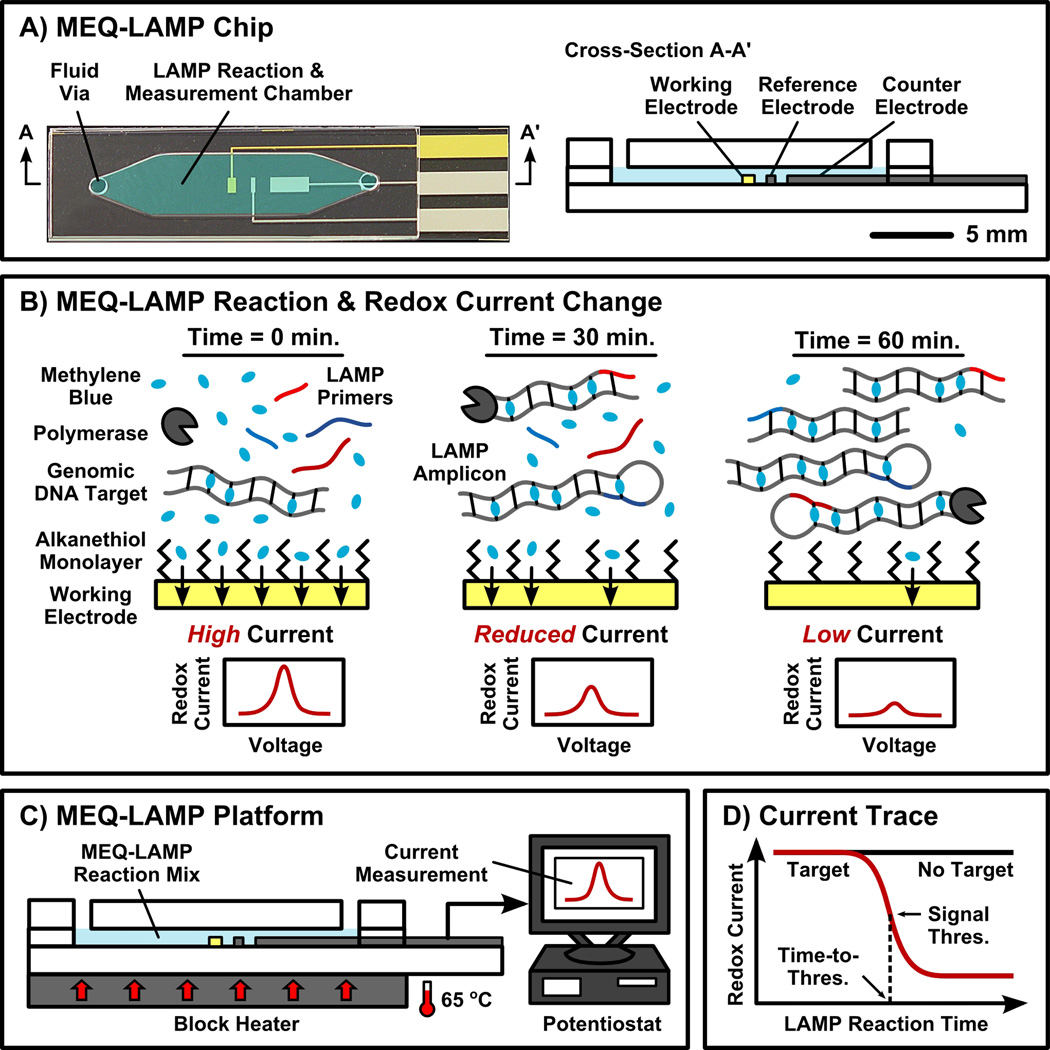
The MEQ-LAMP chip contains a single microfluidic chamber that serves as the LAMP reaction vessel as well as the electrochemical measurement cell (Figure 1A). The chip is constructed from glass and PDMS layers using standard microfabrication techniques (see Supporting Information for details). The microfluidic chamber (~20 µL) contains platinum counter and reference electrodes and a gold working electrode. Real-time monitoring of the LAMP reaction is achieved through in situ electrochemical interrogation and relies on two mechanisms: redox electron transfer between MB molecules and the working electrode, and the intercalation of MB within double-stranded LAMP reaction products (Figure 1B). Initially, the MB molecules within the LAMP reaction solution are free to encounter the gold working electrode and transfer electrons,[30, 31] producing a measurable current. During the LAMP reaction, the primers and the DNA strand-displacing polymerase enable the production of dumbbell-shaped intermediate structures and stem-loop and cauliflower-like amplicons in a continuous manner. As the reaction progresses, intercalation of the MB into double-stranded regions of LAMP amplicons reduces free MB concentration and thus decreases this redox current, providing a ready means of monitoring the reaction in real time.
Figure 1. Overview of MEQ-LAMP.
(A) The MEQ-LAMP reaction is performed within a single-chamber microfluidic chip, which functions as both the LAMP reaction well and the electrochemical measurement cell. (B) The MEQ-LAMP reaction solution contains an electrochemically-active DNA-binding compound methylene blue (MB). Prior to amplificaiton, MB is free in solution and thus generates a redox current via its rapid diffusion to the surface of the gold working electrode (left). As the LAMP reaction progresses, MB intercalates into the newly formed, double-stranded product (middle), decreasing the observed redox current (right). (C) In the complete MEQ-LAMP system, the microfluidic chip is connected to a potentiostat for current measurement as well as a block heater to maintain optimal LAMP conditions. (D) Real-time redox current measurements produce a current trace as a function of the reaction time. The initial amount of target DNA can be quantitatively determined by measuring the time required to reach the signal threshold, the point at which amplification occurs with peak efficiency, in a manner analogous to optical real-time PCR methods.
The MEQ-LAMP assay is well suited for POC applications because it is performed in a single-step: the genomic DNA and reaction mixture are loaded into the microfluidic chip, and the reaction immediately commences as the device is heated to a constant 65 °C (Figure 1C). Concurrently, the MB redox current is continuously measured and recorded (e.g., at 1-minute intervals) with a potentiostat to obtain the concentration of the LAMP product in real-time (Figure 1D). By defining a threshold signal and the corresponding time-to-threshold, it is then possible to determine the initial target quantity in a manner analogous to optical real-time PCR methods. It is important to note that the single-step convenience is credited to the fact that LAMP can be applied directly to genomic DNA without fragmentation or high-temperature denaturation,[29] which also eliminates the need for the intermediate supplementation of reagents (e.g., polymerase), making the process ideal for microfluidic integration.
We have employed S. Typhimurium as our model target and employed a set of six primers to target its InvA gene[32] (see Supporting Information for primer sequences). We first tested the specificity of our primers by performing MB-supplemented LAMP reactions in Eppendorf PCR tubes in a bench-top heat block with genomic DNA of Escherichia coli or Shigella flexneri – both enteric bacteria that are closely related to Salmonella – and compared the results with S. Typhimurium via gel electrophoresis. We observed strong bands of amplified DNA from the S. Typhimurium samples, but no product from the E. coli or S. flexneri samples (Figure S1), thus demonstrating the high specificity of our primers.
In order to test the fidelity of our on-chip MB-LAMP reaction, we performed standard (MB-free) LAMP and MB-LAMP reactions in PCR tubes while also performing an equivalent MB-LAMP reaction in our MEQ-LAMP chip. We used the same target DNA concentrations (approximately 2×104 genome copies per 25 µL reaction) in all three cases, and compared the results via gel electrophoresis. We observed similar distributions of bands and band intensities among the three reactions (Figure S2), demonstrating that effective and reliable LAMP reactions are performed in the presence of MB and in the microfluidic chip.
The MB redox current measured by the on-chip electrochemical module offered an effective indicator for the LAMP amplicon concentration. For example, at the beginning of a reaction starting with approximately 1.6×104 copies of S. Typhimurium genomic DNA, we measured a relatively large peak current due to the high concentration of free MB in the reaction mixture (Figure 2A; black, 0 min.). At the end of the reaction, however, the binding of MB to double-stranded LAMP amplicon significantly reduced the concentration of free MB in the sample solution and thereby diminished the peak current (Figure 2A; red, 60 min.).
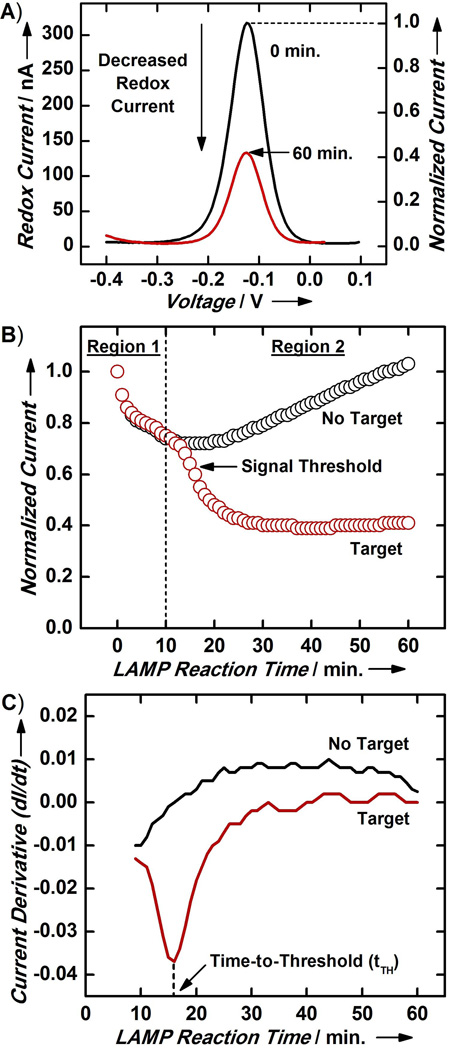
Figure 2. Electrochemical Monitoring of MEQ-LAMP.
(A) The on-chip electrochemical measurement module readily detects MB-LAMP amplification of S. Typhimurium target genomic DNA. All current measurements are normalized to the initial current measurement (i.e., 0 min.) to account for chip-to-chip variations. (B) Normalized real-time current traces for a negative control (no DNA) sample and sample containing S. Typhimurium genomic DNA exhibit clear differences. Only the latter displays a sharp current decrease in Region 2 of the current trace. The amplification kinetics in this second region shows a sigmoid behavior, similar to typical real-time PCR and real-time LAMP kinetics. We designate the point of highest reaction efficiency (steepest decrease in current) as the signal threshold. (C) By taking the derivative of the current trace, the signal threshold is revealed as the local minimum in the curve, facilitating reliable determination of the time-to-threshold (tTH, vertical dashed line), defined as the reaction time required to reach the signal threshold.
Time-course current traces of MEQ-LAMP reactions acquired from real-time monitoring provided valuable information about the sample that end-point detection cannot provide. For example, the current trace for the no-target, negative control reaction revealed that the trace exhibited two distinct regions: Region 1 (t < 10 min), where the current initially decreased, and Region 2 (t > 10 min), where the current subsequently rebounded and gradually increased (Figure 2B, black). The decrease in current observed in Region 1 presumably occurs because heating the device from room temperature (~22 °C) to 65 °C forces MB molecules initially entrapped within the alkanethiol passivation monolayer to dissociate into solution[33, 34]. This dissociation lowers the local MB concentration at the electrode surface, thus reducing the redox current. At later time-points, the elevated reaction temperature gradually breaks the gold-thiol bond between the alkanethiol passivation monolayer and the electrode surface.[35–37] Electron transfer is enhanced as the monolayer dissociates, causing the current rise observed in Region 2. We verified this model by performing control experiments in which we incubated MEQ-LAMP chips at either 22 °C or 65 °C and measured redox currents, before, during, and after these thermal treatments (Figures S3 and S4, and see Supporting Information for details).
Region 1 of the real-time current trace from the sample containing 1.6×104 copies of S. Typhimurium genomic DNA (Figure 2B, red) resembled the negative control, with a similar dip at t < 10 min arising due to dissociation of MB from the monolayer as described above. However, the two traces differed dramatically in region 2; as LAMP amplification products increasingly sequestered free MB, the current decreased in a sigmoid pattern that resembles the reaction kinetics and phases observed with typical real-time PCR[8, 9] and real-time LAMP[27, 28] reactions. However, since the amplification is being measured based on decreased signalling current, the orientation of our curve is inverted relative to a standard real-time PCR sigmoid (Figure 2B; red, Region 2). In the early phases of the reaction, LAMP amplicons are generated efficiently and accumulate rapidly, resulting in equally rapid sequestration of free MB and yielding a sharp drop in the current trace. As LAMP reagents get consumed, the reaction rate decreases, resulting in a more gradual decrease in the peak current until the current trace finally flattens when the reaction reaches its plateau and the concentration of free MB achieves equilibrium.
We applied a similar methodology as in real-time PCR and defined the signal threshold as the end of the early reaction phase of the MEQ-LAMP reaction – the point at which amplification was most efficient and the corresponding current decrease occurred most rapidly (Figure 2B, red). This threshold is reliably determined by identifying the local minimum in the corresponding current derivative trace (dI/dt; Figure 2C) for each MEQ-LAMP reaction. Indeed, the negative control reaction did not display any minima (Figure 2C, black), but the target-containing reaction showed a clear local minimum (Figure 2C, red). We subsequently defined the “time-to-threshold” (tTH) as the time required for a particular sample reaction to reach the signal threshold – analogous to the “threshold cycle” (CT) concept in real-time PCR – and determined that tTH = 16 min for this particular sample (Figure 2C, red).
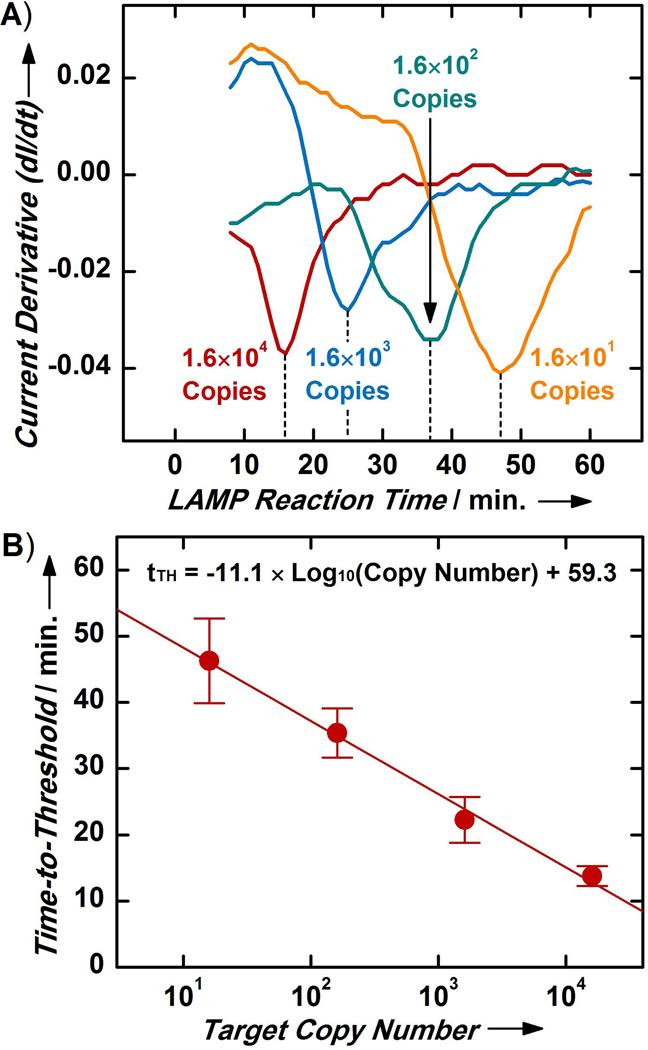
We verified the utility of tTH as a quantitative indicator of target DNA copy number by performing MEQ-LAMP reactions with different initial copy numbers of S. Typhimurium DNA (ten-fold serial dilutions from 1.6×104 to 1.6×101 copies). We observed temporally distinct local minima separated by approximately 10 minutes for each of the current derivative traces, with a distinct tTH for each initial target copy number (Figure 3A). Decreased quantities of the template DNA resulted in slower accumulation of LAMP amplicons, which were in turn reflected by longer periods of current rise (positive derivative) due to monolayer degradation and longer tTH. We also corroborated the MEQ-LAMP tTH measurements by performing bench-top MB-LAMP reactions in PCR tubes with the same initial target quantities, using their corresponding tTH as the reaction time (e.g., 1.6×104 copies for ~15 minutes). Gel electrophoresis analysis revealed similar distributions of amplicon bands and band intensities in all four cases, clearly supporting the tTH values measured in the MEQ-LAMP reactions (Figure S5).
Figure 3. Target Quantification with MEQ-LAMP.
(A) MEQ-LAMP reactions containing different initial copy numbers of S. Typhimurium genomic DNA (from 1.6×104 to 1.6×101 copies) display temporally distinct local minima, revealing a clear pattern wherein samples with higher initial target copy number yield shorter tTH. (B) The calibration curve between initial target copy number and tTH shows a log-linear relationship that spans four orders of magnitude (from 1.6×104 to 1.6×101 copies) with high sensitivity and reproducibility (R2 = 0.91). Values and error bars represent means and standard deviations from four separate measurements.
The strong correlation between the initial target copy number and the tTH of the MEQ-LAMP reaction enabled us to construct a calibration curve that can be used for the absolute quantification of targets of unknown concentration. Our calibration curve revealed a log-linear relationship between the initial target copy number and the tTH across the entire range we investigated (from 1.6×104 copies to 1.6×101 copies) (Figure 3B; n = 4; R2 = 0.91). This log-linear relationship suggests exponential reaction kinetics during the initial phase of the reaction, which is consistent with previous reports regarding the performance of optical real-time LAMP.[27, 28, 38–40] The MEQ-LAMP platform delivered highly reproducible tTH measurements and target quantification, evidenced by the small error bars from four separate measurements for each initial target copy number. Real-time monitoring also supports target quantification across at least a four order of magnitude range of initial copy numbers, a dynamic range that is beyond the scope of end-point detection methods. Finally, MEQ-LAMP achieved a detection limit of 16 copies of S. Typhimurium genomic DNA, corresponding to 4 fg/µL of DNA and approximately 0.8 copies/µL; this is approximately seven orders of magnitude more sensitive than previously reported electrochemical detection of LAMP products[25].
In summary, as a step toward rapid, sensitive and quantitative POC pathogen detection, we have demonstrated MEQ-LAMP, a microfluidic, electrochemical, real-time LAMP platform that can quantitatively detect very small quantities of pathogen genomic DNA with high specificity in a single step. Our MEQ-LAMP platform makes use of a single-chamber microfluidic device within which we can conduct the isothermal LAMP reaction. By supplementing the LAMP reaction mix with MB, an electrochemically-active, DNA-binding redox reporter, we can readily monitor the reaction in real-time by using integrated electrodes to measure decreases in current resulting from increasing binding of MB to LAMP reaction products. In parallel, we have devised reliable signal threshold and time-to-threshold metrics for target quantification, and demonstrate that our platform is capable of directly detecting as few as 16 copies of S. Typhimurium genomic DNA in under an hour.
By combining the advantages of real-time electrochemical read-out and LAMP within a microfluidic platform, our MEQ-LAMP method obviates the need for bulky and sophisticated optical detectors and temperature controls, while also ensuring robust microfluidic DNA amplification by eliminating the potential for high-temperature-induced reaction failures. Furthermore, our platform precludes manual handling steps and end-point analysis of reaction products and thus eliminates the need for additional fluid handling, reagent mixing and buffer switching. The MEQ-LAMP platform also offers considerable potential for further expansion. For example, we are currently exploring the integration of front-end sample processing modules to enable the direct detection of pathogens from complex, clinically relevant samples. Single-step, reverse-transcription LAMP for viral RNA detection could also be readily implemented in our platform. Finally, this microfluidic platform is readily compatible with multiplexing, as the chip could easily be modified to contain multiple chambers for the parallel detection of multiple targets. The current performance and the potential for expanded capability thus lead us to envision the MEQ-LAMP platform as a powerful tool for genetic detection at the point-of-care.
Supplementary Material
Footnotes
We are grateful for the financial support from the National Institutes of Health and the Institute of Collaborative Biotechnologies through the Army Research Office. We thank Prof. Ryan White for valuable discussions. We also thank the Plaxco Lab and the Turner Lab at UCSB for assistance in device preparation. Microfabrication was carried out in the Nanofabrication Facility at UCSB.
Supporting information for this article is available at http://www.angewandte.org.
Contributor Information
Kuangwen Hsieh, Department of Mechanical Engineering, University of California, Santa Barbara (USA).
Adriana S. Patterson, Department of Chemistry and Biochemistry and Biomolecular Science and Engineering Program, University of California, Santa Barbara (USA).
B. Scott Ferguson, Department of Mechanical Engineering, University of California, Santa Barbara (USA).
Kevin W. Plaxco, Department of Chemistry and Biochemistry and Biomolecular Science and Engineering Program, University of California, Santa Barbara (USA)
H. Tom Soh, Email: tsoh@engr.ucsb.edu, Materials Department and Department of Mechanical Engineering, University of California, Santa Barbara, Santa Barbara, CA 93106 (USA).
References
- 1.Yang S, Rothman RE. Lancet Infect. Dis. 2004;4:337. doi: 10.1016/S1473-3099(04)01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holland CA, Kiechle FL. Curr. Opin. Microbiol. 2005;8:504. doi: 10.1016/j.mib.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Palchetti I, Mascini M. Anal. Bioanal. Chem. 2008;391:455. doi: 10.1007/s00216-008-1876-4. [DOI] [PubMed] [Google Scholar]
- 4.Olsen JE. Food Res. Int. 2000;33:257. [Google Scholar]
- 5.Gardeniers JGE, van den Berg A. Anal. Bioanal. Chem. 2004;378:1700. doi: 10.1007/s00216-003-2435-7. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Rivas G, Cai X, Palecek E, Nielsen P, Shiraishi H, Dontha N, Luo D, Parrado C, Chicharro M, Farias PAM, Valera FS, Grant DH, Ozsoz M, Flair MN. Anal. Chim. Acta. 1997;347:1. [Google Scholar]
- 7.Wang J. Anal. Chim. Acta. 2004;507:3. [Google Scholar]
- 8.Higuchi R, Fockler C, Dollinger G, Watson R. Nat. Biotechnol. 1993;11:1026. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 9.Heid CA, Stevens J, Livak KJ, Williams PM. Genome Res. 1996;6:986. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 10.Wang J. Biosens. Bioelectron. 2006;21:1887. doi: 10.1016/j.bios.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 11.Drummond TG, Hill MG, Barton JK. Nat. Biotechnol. 2003;21:1192. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 12.Gooding JJ. Electroanalysis. 2002;14:1149. [Google Scholar]
- 13.Mir M, Homs A, Samitier J. Electrophoresis. 2009;30:3386. doi: 10.1002/elps.200900319. [DOI] [PubMed] [Google Scholar]
- 14.Lagally ET, Emrich CA, Mathies RA. Lab Chip. 2001;1:102. doi: 10.1039/b109031n. [DOI] [PubMed] [Google Scholar]
- 15.Liu YJ, Rauch CB, Stevens RL, Lenigk R, Yang JN, Rhine DB, Grodzinski P. Anal. Chem. 2002;74:3063. doi: 10.1021/ac020094q. [DOI] [PubMed] [Google Scholar]
- 16.Liu RH, Yang J, Lenigk R, Bonanno J, Grodzinski P. Anal. Chem. 2004;76:1824. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- 17.Ferguson BS, Buchsbaum SF, Swensen JS, Hsieh K, Lou X, Soh HT. Anal. Chem. 2009;81:6503. doi: 10.1021/ac900923e. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson BS, Buchsbaum SF, Wu TT, Hsieh K, Xiao Y, Sun R, Soh HT. J. Am. Chem. Soc. 2011;133:9129. doi: 10.1021/ja203981w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee TMH, Carles MC, Hsing IM. Lab Chip. 2003;3:100. doi: 10.1039/b300799e. [DOI] [PubMed] [Google Scholar]
- 20.Yeung SSW, Lee TMH, Hsing I-M. Am. Chem. Soc. 2006;128:13374. doi: 10.1021/ja065733j. [DOI] [PubMed] [Google Scholar]
- 21.Yeung SSW, Lee TMH, Hsing I-M. Anal. Chem. 2008;80:363. doi: 10.1021/ac071198+. [DOI] [PubMed] [Google Scholar]
- 22.Defever T, Druet M, Rochelet-Dequaire M, Joannes M, Grossiord C, Limoges B, Marchal D. J. Am. Chem. Soc. 2009;131:11433. doi: 10.1021/ja901368m. [DOI] [PubMed] [Google Scholar]
- 23.Fang TH, Ramalingam N, Xian-Dui D, Ngin TS, Xianting Z, Lai Kuan AT, Peng Huat EY, Hai-Qing G. Biosens. Bioelectron. 2009;24:2131. doi: 10.1016/j.bios.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Kivlehan F, Mavre F, Talini L, Limoges B, Marchal D. Analyst. 2011;136:3635. doi: 10.1039/c1an15289k. [DOI] [PubMed] [Google Scholar]
- 25.Nagatani N, Yamanaka K, Saito M, Koketsu R, Sasaki T, Ikuta K, Miyahara T, Tamiya E. Analyst. 2011;136:5143. doi: 10.1039/c1an15638a. [DOI] [PubMed] [Google Scholar]
- 26.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mori Y. J. Biochem. Biophys. Methods. 2004;59:145. doi: 10.1016/j.jbbm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Mori Y, Nagamine K, Tomita N, Notomi T. Biochem. Biophys. Res. Commun. 2001;289:150. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 29.Nagamine K, Watanabe K, Ohtsuka K, Hase T, Notomi T. Clin. Chem. 2001;47:1742. [PubMed] [Google Scholar]
- 30.Svetlicic V, Clavilier J, Zutic V, Chevalet J. J. Electroanal. Chem. 1991;312:205. [Google Scholar]
- 31.Ju HX, Zhou J, Cai CX, Chen HY. Electroanalysis. 1995;7:1165. [Google Scholar]
- 32.Harakudo Y, Yoshino M, Kojima T, Ikedo M. FEMS Microbiol. Lett. 2005;253:155. doi: 10.1016/j.femsle.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 33.Sagara T, Kawamura H, Nakashima N. Langmuir. 1996;12:4253. [Google Scholar]
- 34.Grumelli D, De Leo LPM, Bonazzola C, Zamlynny V, Calvo EJ, Salvarezza RC. Langmuir. 2010;26:8226. doi: 10.1021/la904594p. [DOI] [PubMed] [Google Scholar]
- 35.Day BS, Fiegland LR, Vint ES, Shen WQ, Morris JR, Norton ML. Langmuir. 2011;27:12434. doi: 10.1021/la202444s. [DOI] [PubMed] [Google Scholar]
- 36.Phares N, White RJ, Plaxeo KW. Anal. Chem. 2009;81:1095. doi: 10.1021/ac8021983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakata T, Maruyama S, Ueda A, Otsuka H, Miyahara Y. Langmuir. 2007;23:2269. doi: 10.1021/la0616193. [DOI] [PubMed] [Google Scholar]
- 38.Lee SY, Huang JG, Chuang TL, Sheu JC, Chuang YK, Holl M, Meldrum DR, Lee CN, Lin CW. Sens. Actuators B. 2008;133:493. doi: 10.1016/j.snb.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gandelman OA, Church VL, Moore CA, Kiddle G, Carne CA, Parmar S, Jalal H, Tisi LC, Murray JAH. PLoS One. 2010;5:e14155. doi: 10.1371/journal.pone.0014155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang XE, Liu YY, Kong JL, Jiang XY. Anal. Chem. 2010;82:3002. doi: 10.1021/ac1000652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.