Abstract
The Plasmodium falciparum ortholog of the human cytokine, macrophage migratory inhibitory factor (PfMIF), is produced by the parasite during malaria infection and modulates the host’s immune response. As for other MIF orthologs, PfMIF has tautomerase activity, whose inhibition may influence the cytokine activity. To identify small-molecule inhibitors of the tautomerase activity of PfMIF, virtual screening has been performed by docking 2.1 million compounds into the enzymatic site. Assaying of 17 compounds identified four as active. Substructure search for the most potent of these compounds, a 4-phenoxypyridine analogue, identified four additional compounds that were purchased and also shown to be active. Thirty-one additional analogues were then designed, synthesized, and assayed. Three were found to be potent PfMIF tautomerase inhibitors with Ki values of ca. 40 nM; they are also highly selective with Ki values of >100 μM for human MIF.
INTRODUCTION
The parasite Plasmodium falciparum (P. falciparum) was responsible for the majority of the 287 million human cases of malaria in 2011 and the ca. 655,000 related deaths.1 Although the number of deaths appears to be decreasing (from ca. 781,000 for the year 2010), the incidence of infection is increasing with a projected 295 million total cases for 2012.1 Fortunately, the widespread use of bed nets, better diagnostics, and wider availability of effective antibiotics have greatly reduced mortality and the spread of the disease. To this day, artemisinin and artemisinin-analogues are the most effective drugs against malaria. Unfortunately, artemisinin mono-therapies have resulted in an alarmingly widespread emergence of artemisinin-resistant P. falciparum in Cambodia and surrounding regions. Thus, novel therapies are needed to fight against drug-resistant P. falciparum.
Upon infection, the parasite modulates the host immune defense through the release of the cytokine P. falciparum macrophage migration inhibitory factor (PfMIF).2 PfMIF is a homologue of the human immunoregulatory cytokine MIF3-5 (huMIF) and like huMIF, it is a homotrimer.6 The malarial cytokine is present in all P. falciparum life cycle stages and is released by infected cells.7 MIF family members have three tautomerase active sites formed by adjacent monomers. No physiologic functions have been attributed to MIF tautomerase activity, and cytokine function is believed to arise from protein-protein interactions, as deduced from site-directed mutagenesis and genetic studies in vivo.8 PfMIF also has been shown to bind to the mammalian MIF receptor, CD74, with comparable nM affinity as human MIF.2,9 Parasites may induce an inflammatory response, using the host immune system as a means to regulate the population of competing parasites within hosts. The latest evidence indicates that the key role of MIF orthologs in parasites is to promote depletion of CD4 T-cells resulting in degradation of immunological memory and response.2
The structure of PfMIF has significant structural similarity to huMIF with an average rmsd of 2.2 Å for the Cα atoms.10 They are both homotrimers with 115-residue monomer subunits. Interestingly PfMIF only has 30% sequence homology with huMIF. The PfMIF crystal structure, PDB code 2WKF, has two different trimers, signified by alternative conformations of the catalytic residue Pro1, one having a closed tautomerase site (PfMIF-B) and one having an open tautomerase site (PfMIF-A). The tautomerase site in PfMIF is larger than in huMIF, in part due to the reorientation of Tyr96 in comparison to the corresponding Tyr95 of huMIF.10 The tautomerase site of PfMIF is also more negatively charged than huMIF owing to the presence of Glu98 in PfMIF in place of Asn97 in huMIF.
PfMIF binds to the ectodomain of transmembrane protein CD74, presumably in a similar manner as huMIF. Inhibitors of the huMIF-CD74 interaction have previously been identified through virtual screening.11 Since a co-crystal structure for MIF bound to CD74 is not available, our strategy was to seek MIF tautomerase inhibitors, certain of which might also serve as inhibitors of the MIF-CD74 protein-protein interaction.11 The interference could stem from conformational changes or rigidification of MIF or protrusion of the tautomerase inhibitors from the surface of MIF. Support for this latter possibility was attained in a recent structural study of small molecule inhibitors of Ancylostoma ceylanicum MIF.12 There is evidence that the interaction between huMIF and CD74 occurs in the vicinity of the tautomerase active site and that there is some correlation between MIF tautomerase inhibition and MIF-CD74 inhibition.13 The same strategy has been followed here by first seeking tautomerase inhibitors of PfMIF. Selective inhibitors of PfMIF that do not interact with huMIF are particularly desired. They can be used as research tools in the study of immune response and possibly be developed into anti-malaria drugs.
METHODS
Protein Setup and Preparation of Ligands
The trimer of PfMIF-A was generated from the asymmetric unit of the crystal structure (PDB code: 2WKF)10 through symmetry operations using the program UCSF Chimera.14 This structure lacks coordinates for the C-terminal residues 102-115. Of the two Cys2 conformations in the PfMIF-A trimer, each with occupancy 0.50, Cys2A was kept. Glycerol and water molecules were removed. Hydrogens and other missing atoms were added and the protein was energy-minimized using the OPLS-AA 2005 force field within the Protein Preparation Wizard module.15,16 Pro1 was unprotonated to represent the catalytically active state for the tautomerase activity, yielding protein structure A. The Maybridge 2010 library was downloaded in sdf-format and subsequently processed with the program LigPrep,17 which generated tautomers and protonation states at pH 7 and up to 32 configurational stereoisomers for each compound with chiral centers. The Maybridge library was filtered by removing compounds with undesirable features for screening hits, namely compounds having i) a MW less than 200 or greater than 600, ii) a molecular charge higher than +2 or lower than −1, iii) more than one violation of the Lipinski rules,18 or iv) more than two violations of the rule of three or more than six #STARS, two indices calculated using the program QikProp.19 The 2006 ZINC “druglike” set of molecules20 was also used in a previously downloaded format that included multiple protonation and tautomerization states.11
All structures were docked and scored using Glide 5.5 in standard precision (SP)21 and extra precision (XP) modes.22 The receptor grid was prepared with a 20-Å side length with the centroid located between residues Tyr96A, Tyr37C, Pro1C, Ile65C, Met39C, Phe50A and with a ligand diameter midpoint box having a 12-Å side length. Expanded sampling was enabled to bypass elimination of initial poses in the rough scoring stage, and only Z-conformations were allowed for secondary amides. All other Glide options were kept at default settings. Maestro23 was used to display the protein-ligand complexes. From the top scoring compounds, hydrazones, hydrazides, hydrazines, α,ß-unsaturated ketones, catechols, imines, thiohydrazones, and compounds having four or more fused rings were eliminated as undesirable for reactivity concerns or complexity. Compounds whose poses had features associated with high internal energy, such as twisted ester or carboxylic acid groups, overly short non-bonded contacts, or energetically unfavorable E/Z isomers,24 were also removed from consideration.
Some compounds of interest were also docked using GOLD Suite v5.1.25 The active site was set to encompass all atoms within a 15-Å radius of the oxygen atom of Pro1. The starting geometries of the ligands were constructed using Maestro. Docking was performed using the GoldScore scoring function, and 2000 genetic-algorithm runs were executed for each ligand. All other settings were set to default values.
Docking Protocol Used for Compound Optimization
Analogues identified through substructure searches of virtual screening hits or subsequently designed analogues were docked using the following protocol. For each compound, a conformational analysis using torsional sampling was performed using the program MacroModel26 and the OPLS-AA 2005 force field. The implicit water Generalized-Born/Surface-Area model was applied. Extended torsion sampling options, 250 steps per rotatable bond, and 2500 maximum number of steps were used. Conformers were energy-minimized, duplicate conformers were removed, and up to 1000 unique conformers were allowed to be saved. The resulting conformers were rigidly docked with Glide as above.
Addition of Missing Residues (Structure B)
The Plasmodium berghei MIF (PbMIF) structure was also downloaded (PDB code: 2WKB10). It has all 115 residues resolved and a capping glycine. Chain C of PbMIF was aligned with one chain of the PfMIF PDB structure 2WKF, using UCSF Chimera. Residues 1-101 of the PbMIF monomer were deleted and the remaining residues 102-116 were added to the PfMIF chain. Residues 101 and 102 were manually connected. The new monomer was aligned with all three chains of the previously prepared PfMIF trimer and the old monomers from the prepared trimer were deleted, creating a new trimer where each chain has 116 residues. The modified structure was imported into Maestro. Hydrogen and missing atoms were added and the structure was energy-minimized, using the protein preparation wizard at default settings. A docking grid for structure B was set up in the same way as for structure A, using the same residues for the centroid and the same grid box size.
Synthetic Chemistry
The results of the virtual screening pointed to diphenyl ether and phenoxyazine derivatives as the most promising for optimization. The synthetic routes for the pursued series are summarized below; full details are in the Supplementary Information. The identities of all synthesized compounds were confirmed using 1H NMR, 13C NMR, and mass spectrometry. NMR spectra were recorded on Bruker Avance DRX-500 (500 MHz) or DRX-400 (400 MHz) instruments. Mass determination was performed using electrospray ionization on a Waters Micromass ZQ LC-MS. HRMS (ESI-TOF) analyses were performed on a Waters Xevo QTOF equipped with a Z-spray electrospray ionization source. The purity of all compounds was determined to be at least 95% by integration of the UV-trace from reverse phase HPLC, using a Waters 2487 dual λ absorbance detector with a Waters 1525 binary pump and a Phenomenex Luna 5μ C18(2) 250 × 4.6 mm column. Samples were run at 1 mL/min using gradient mixtures of 5-100% of water with 0.1% trifluoroacetic acid (TFA) (A) and 10:1 acetonitrile:water with 0.1% TFA (B) for 22 min followed by 3 min at 100% B.
Synthesis of 4-Phenoxy-Biphenyls and 2-Phenylpyridines
(4-bromophenoxy)(tert-butyl)dimethylsilane was coupled with substituted phenylboronic acids under Suzuki conditions, followed by desilylation with TBAF. The resulting substituted (1,1′-biphenyl)-4-ol was reacted with a second phenyl boronic acid using Cu(OAc)2 according to Evans’ method27 to provide the desired biaryl ether product (Scheme 1). The pyridine derivatives were synthesized through Suzuki coupling, followed by nucleophilic aromatic substitution (Scheme 2).
Scheme 1. Synthesis of 4-Phenoxy-1, 1′-biphenylsa.
aReagents: (a) Pd(PPh3)4, Na2CO3, toluene, H2O; (b) TBAF, THF; (c) (3-methoxyphenyl)boronic acid, Cu(OAc)2, TEA, DCM; (d) BBr3, DCM.
Scheme 2. Synthesis of 5-Phenoxy-2-phenylpyridinesa.
aReagents: (a) Pd(PPh3)4, Na2CO3, dioxane/H2O; (b) Cs2CO3, Cu2O, NMP.
Synthesis of Meta-Substituted Pyrimidines and Pyridines
Meta-substituted azines were also desired. Pyrimidine analogues were synthesized by refluxing benzamidine with ethyl 3-oxobutanoate, and sodium ethoxide was added to form the 2-phenylpyrimidine core. This was followed by chlorination with phosphoryl chloride and nucleophilic aromatic substitution with substituted phenols to provide the desired pyrimidine analogues (Scheme 3). Corresponding pyridines were prepared starting from 2-chloro-4-phenoxypyridines, followed by Suzuki cross coupling with substituted phenylboronic acids (Scheme 4).
Scheme 3. Synthesis of 4-Methyl-6-phenoxy-2-phenylpyrimidinesa.
aReagents: (a) Ethyl 3-oxobutanoate, NaOEt, EtOH; (b) POCl3; (c) Cs2CO3, DMF.
Scheme 4. Synthesis of 2-Phenyl-4-phenoxypyridinesa.
aReagents: (a) Cs2CO3, DMF/H2O, Pd(PPh3)4.
Replacement of the 2-phenyl group by morpholine was also considered. Nucleophilic aromatic substitution of 2,4-dichloropyridine with a substituted phenol yielded the 2-chloro-4-phenoxypyridines selectively for use in both Schemes 4 and 5. A second nucleophilic aromatic substitution at the 2-position, through reflux in neat morpholine, provided 2-(N-morpholinyl)-4-phenoxypyridines (Scheme 5).
Scheme 5. Synthesis of 2-(N-morpholinyl)-4-phenoxypyridinesa.
aReagents: (a) t-BuOK, DMA; (b) morpholine.
Molecular Cloning and Production of Recombinant PfMIF
To the recombinant production of PfMIF, a full length nucleotide sequence optimized for codon usage by Escherichia coli was synthesized. The oligonucleotide was subcloned into the pCRT7 expression vector (Invitrogen) and transformed into BL21 (DE3) E. coli. A 2 liter production culture was induced by isopropyl-D-thiogalactoside to a final concentration of 1 mM at 37°C. The purification protocol was adapted from Kamir et al.28 Briefly, the induced bacterial culture was harvested and lysed in 20 mM Tris, pH 8.0 and 20 mM NaCl buffer using a French press. The recombinant PfMIF protein was purified by anion exchange chromatography using a Q-Sepharose resin (Amersham Bioscience) with a linear pH gradient to 30 mM Bis-Tris (pH 6.8) and 20 mM NaCl buffer, followed by a linear salt gradient from 20 mM Tris, pH 8.0 and 20 mM NaCl to 30 mM Tris pH 8.0, 1 M NaCl buffer. The remaining impurities were removed by size-exclusion chromatography with a Superdex 75 column (Amersham Bioscience), and the final amount of protein obtained was 3 mg. The production and purification of huMIF was previously described.29
Assay for PfMIF Tautomerase Activity
The following protocol was adapted from Taylor et al.30 The enol form of the substrate 4-hydroxyphenylpyruvate (4-HPP) was obtained by dissolution of 4-HPP in absolute EtOH (50 nM) and stored at −20°C until used. For the enzymatic inhibition assays, the reaction mixture was composed of PfMIF (75 nM of trimer) in 25 mM potassium phosphate buffer at pH 7.4, mixed with increasing concentrations of inhibitors (1.5, 5, and 10 μM) and incubated for 10 min. The optimal concentration of PfMIF for compound assessment was determined initially by measuring tautomerase activity at different concentrations of trimeric PfMIF (range: 50-100 nM). The negative control was PfMIF incubated with DMSO vehicle, which in all reactions was 140 mM (1%) and that did not itself influence tautomerase activity. The reaction was started by the addition of 4-HPP at different concentrations, and the formation of the keto form of 4-HPP was monitored by the decrease of absorbance at 306 nm, using an Infinite M200 (TECAN, Durham, NC) plate reader for 90 seconds. Calculation of initial velocities and nonlinear regression for the inhibition constant, Ki, was repeated three times and combined for analysis using the program Prism 5 (GRAPHPAD, La Jolla, CA), which provided the reported averages and standard deviations. For inhibition of huMIF, the identical protocol was used as previously described.11
The tautomerase inhibition can be gauged by monitoring either the enolization or ketonization process. For PfMIF, Michaelis-Menten analyses using the recombinant protein revealed a significantly higher Km value and lower kcat/Km for enolization than ketonization (Table 1). Thus, for PfMIF the sensitivity of the assay for discrimination of putative tautomerase inhibitors benefited from measuring the ketonization of the enol of 4-HPP.
Table 1. Kinetics Parameters for 4-HPP Tautomerization by PfMIF and huMIFa.
| PfMIF | huMIF | |||||
|---|---|---|---|---|---|---|
| Reaction | K m | k cat | kcat/Km | K m | k cat | kcat/Km |
| enolization | 674.6±46.9 | 17.98±0.7 | 2.7×104 | 1139±216.3 | 208.3±19.3 | 1.8×105 |
| ketonization | 186.7±50.6 | 32.90±5.3 | 1.8×105 | 192.4±47 | 86.62±11.7 | 4.5×105 |
Km in μM; kcat in s−1; kcat/Km in M−1s−1.
RESULTS AND DISCUSSION
Virtual Screening
The ZINC “drug-like” library (ca. 2.1 million compounds) and the Maybridge library (ca. 60,000 compounds) were docked into structure A using Glide SP. The 40,000 top-ranked compounds from ZINC and 4000 top-ranked ones from Maybridge from the SP docking were redocked using Glide XP. Sorted according to docking score, the top 1000 ZINC SP, 300 Maybridge XP, and 200 Maybridge SP complexes were displayed and visually inspected. These choices reflect a desire to seek balance between the scoring functions and compound libraries, while remaining cognizant of reasonable limits on human capacity to sit at a graphics station and view structures. All molecules containing unwanted structural features were removed such as those with readily hydrolizable and/or highly electrophilic functional groups. Final candidate structures were selected based on the docking scores, occurrence of favorable interactions with the receptor, prediction of physical properties from QikProp,19 and a preference for structural diversity. The predictions from QikProp that are especially noted are for aqueous solubility (S), log Po/w, Caco-2 cell permeability (PCaco), and number of primary metabolites. Compounds are avoided that have S less than 10−5 M, log Po/w greater than 4, PCaco less than 25 nm/s, and more than 4 primary metabolites.
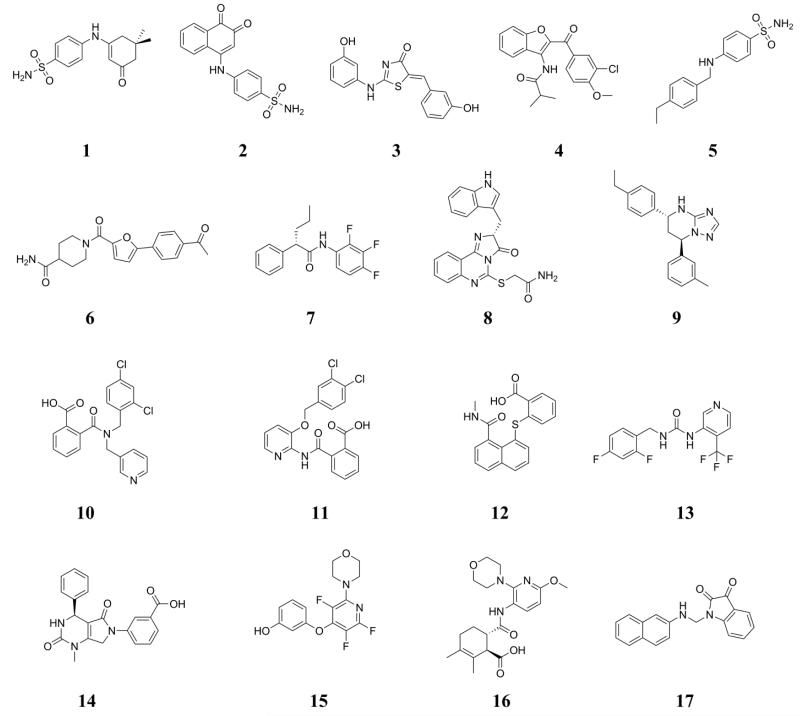
The structures of the 17 compounds which were ultimately purchased are shown in Figure 1. The identities of the compounds were verified by NMR and mass spectrometry, and the purity was found to be at least 95% by HPLC. The compounds were assayed for their ability to inhibit ketonization of the enol form of 4-HPP. The assay results and docking scores are summarized in Table 2. Four compounds (2, 3, 8, and 15) showed activity with Ki values of 99, 21, 25, and 9 μM, respectively. Illustrations of the docked structures for the active compounds are provided in Figure 2.
Figure 1.
Virtual screening led to the purchase and assaying of 1 – 17.
Table 2. Experimentally Determined Inhibition Constants (Ki) and Docking Scores for the Purchased Compounds 1-17 and the Initial Analogues of 15 (18-23).
| compd | Ki (μM)a) | SP Score | XP Score | GoldScore | Library |
|---|---|---|---|---|---|
| 1 | NA | −7.06 | −5.23 | 52.2 | ZINC |
| 2 | 99 | −7.05 | −6.55 | 57.4 | ZINC |
| 3 | 20.6 | −7.28 | −6.08 | 58.3 | ZINC |
| 4 | NA | −7.07 | −5.03 | 64.1 | ZINC |
| 5 | NA | −7.21 | −6.67 | 57.7 | ZINC |
| 6 | NA | −6.99 | −6.25 | 61.1 | ZINC |
| 7 | NA | −6.98 | −7.35 | 51.0 | ZINC |
| 8 | 25.4±2.9 | −6.97 | −6.11 | 77.5 | ZINC |
| 9 | NA | −7.61 | −7.83 | 62.3 | ZINC |
| 10 | NA | −6.69 | −8.28 | 63.9 | Maybridge |
| 11 | NA | −7.44 | −6.98 | 69.3 | Maybridge |
| 12 | NA | −6.24 | −6.98 | 58.7 | Maybridge |
| 13 | NA | −7.30 | −9.67 | 52.8 | ZINC |
| 14 | NA | −7.16 | −6.62 | 65.6 | ZINC |
| 15 | 8.6±7.2 | −7.16 | −6.38 | 52.7 | ZINC |
| 16 | NA | −6.39 | −8.09 | 55.7 | Maybridge |
| 17 | NA | −6.91 | −6.91 | 60.6 | Maybridge |
| 18 | 15.5±8.9 | −7.89 | −6.91 | 52.1 | − |
| 19 | 28.6±3.6 | −7.91 | −6.51 | 55.3 | − |
| 20 | 19.3±2.1 | −7.37 | −4.92 | 57.8 | − |
| 21 | 122.3±11.4 | −7.30 | −6.45 | 63.9 | − |
| 22 | 43.4±21 | −8.13 | −6.28 | 54.2 | − |
| 23 | 22.9±1.3 | −6.95 | −5.57 | 57.3 | _ |
NA indicates assayed but not active compounds.
Figure 2.
Docking poses for the four active compounds from the virtual screening.
The structures of the active compounds are diverse. The docked structure of 2 benefits from hydrogen bonds between Tyr96A and Glu98A and the sulfonamide group and an aryl-aryl interaction between Tyr37C and the naphthalenedione fragment. The ortho-quinone substructure provides some concern that the observed activity may involve covalent modification of the protein. Compound 3 has an exocyclic double bond in a rhodanine-related motif. This is a known frequent hitter in assays owing to Michael addition activity.31 Though a considerable list of undesirable substructures led to exclusions, our list of such features has further increased since the time when this docking study was carried out in 2010. We would also now exclude compounds such as 2 and 3 from consideration of screening, and we did not pursue analogues. Compound 8 is more complex with two polycyclic ring systems and a chiral center. It is predicted to form two hydrogen bonds via its terminal amido group with Arg93A and Ile95A and an aryl stacking interaction with Tyr37C (Figure 2). The identification of the interactions with Tyr37C, Arg93A, Ile95A, and Tyr96A for 2, 3, and 8 is a useful observation. If we were to pursue analogues of 8, we would first try to simplify the structure and consider, for example, 2,4-substituted quinolines, which retain the interactions with Tyr37C, Arg93A, and Ile95A.
The most promising hit that arose from the virtual screening is 15, a 4-phenoxypyridine analogue. It has no striking liabilities for an assay hit. The docking pose for 15 has a hydrogen bond to Glu98A via the terminal phenol, similar to the docking pose of 3. There are also possible hydrogen bonds with Tyr96A and Pro1C, and the central ring is in the hydrophobic region between Tyr37C and Tyr96A. The morpholinyl group in the pose appears to make little contact with the protein as it is oriented into the solvent. 15 was the most active compound at 8.6 μM and it was amenable for synthesis of analogues, so it was chosen for further development.
Initial Analogues of 15
To begin, substructure searches for commercially available analogues of 15 were performed using SciFinder Scholar. The 3-hydroxyphenoxy fragment was retained and alternatives were considered for the central ring. The closest analogues that were found and purchased are 18 - 21. 18 has just the 5-fluorine in 15 replaced by chlorine, while 19 is the piperidine replacing morpholine analogue of 15. 20 probes the possibility of a pyrimidine core with a 2-phenyl substituent and 21 incorporates a triazolopyridazine core. At this point, 22, the fluorine-less analogue of 15, was synthesized via Scheme 5 to gauge the importance of the fluorines for activity. There was also some concern that the 6-position in the fluorinated pyridines 15, 18, and 19 might be reactive towards nucleophilic aromatic substitution leading to potential false positives. In addition, the methyl ether analogue 23 was prepared to test the importance of the hydroxyl group and the putative hydrogen bond to Glu98A.
The six compounds were docked into structure A and due to the low rotational barriers around the aryl ether bonds many docked solutions were obtained. For the top scored poses of all six compounds, the 3-hydroxy/methoxy phenyl has similar orientation as for the docked pose of 15. The two most populated docked solutions for 18-20, 22, and 23 have the pyridine/pyrimidine flipped causing the ring on C2 to interact with Tyr37C as opposed to being sandwiched between Phe50A and Tyr96A. Figure 3 shows these alternative poses for 18. Less well scored poses would generally have the positions of the terminal rings interchanged. The top scoring poses for 20 and 21 also follow the motif favoring interaction of a terminal ring with Tyr37C (Figure 4).

Figure 3.
(a) Top pose of 18; the morpholine ring contacts Tyr37C. (b) The next most favorable pose of 18 with the morpholine ring placed between Phe50A and Tyr96A.
Figure 4.
(a) Top pose for 20. (b) Top pose for 21.
The six compounds, 18-23, were evaluated in the tautomerase assay (Table 2). It was gratifying that they all showed inhibitory activity confirming the viability of the core structure from 15. Considering the standard deviations of the measurements, 18, 19, 20, and 23 have similar activity as 15, which opens up a range of possibilities for the central and C2 rings. Exchange of the three fluorine atoms in 15 for hydrogen atoms (22) incurs a ca. five-fold penalty. Interestingly, the 3-methoxy substituent of 23 does not diminish the potency relative to 22, while the triazolopyridazine analogue 21 is significantly less active at 122±11 μM.
Table 2 also lists the SP and XP docking scores for 1 - 23. Overall, the SP scoring was superior with many of the best scoring compounds including 18 - 22 showing activity. The compounds with the best XP scores (9, 10, 13, 16) were inactive, while some with relatively poor XP scores such as 20 and 23 were active. GOLD was also applied to 1 - 23; the scoring in this case is such that the larger the number, the better the score. Qualitatively, the performance of Glide XP and GOLD were similar. The purchased compounds with the best GoldScores were generally inactive, e.g., 4, 6, 9, 10, 11, 14, and 17. The only exception was the active 8, which is the largest compound and had the best GoldScore. On the other hand, the key lead 15 had one of the worst GoldScores. Successful compound selection from virtual screening continues to require a significant human component in evaluating computed poses.
Addition of Missing Residues at the C-terminus
Concern existed about possible influence of the missing residues 102-115 on the docking poses. Some of these residues are in proximity to Tyr96A and Glu98A. Fortunately, the PbMIF crystal structure has the terminal residues resolved and has 100% sequence identity with PfMIF for the last 25 residues, so it was used as a template to extend the PfMIF structure. The residues and the capping Gly116 were added and the new structure (structure B) was energy-minimized (Figure 5).
Figure 5.
The missing C-terminal residues (blue) have been added to the PfMIF structure (purple), which has been aligned with the unmodified crystal structure 2WKF for PfMIF (green). The addition had little effect on the positions of residues 1-101. 15 in orange is shown docked into the modified structure.
While the modified structure does not significantly differ from the crystal structure (0.44 Å rmsd between 746 atom pairs or 0.31 Å for 380 backbone atoms), there is some uncertainty about the position of Cys102. The backbone oxygen atom of Asp101 from the crystal structure overlaps the backbone nitrogen of Cys102 in the modified structure. Though there is a possibility that the nitrogen atom of Asp101 in the crystal structure has been misassigned as an oxygen, the issue causes small uncertainty in the positions of the added residues 102-115. With structure B, it emerges that there is additional room to extend analogues of 15 towards residues 103-108 by replacement of the 3-hydroxy group on the phenoxy ring with larger substituents.
4-Phenoxy-1,1′-biphenyls and 5-Phenoxy-2-phenylpyridines
The morpholine ring of 15 appeared to make little contact with the protein (Figure 2) and modeling suggested that hydrophobic interactions could be gained with Phe50A, Tyr57A, and Tyr96A by its replacement with a phenyl substituent para to the hydroxyphenoxy group. Thus, compounds containing either a 4-phenoxy-1,1′-biphenyl or a 5-phenoxy-2-phenylpyridine core were pursued (Table 3). In parallel, three 4-phenoxy-1,1′-biphenyls (24a-c) and two 5-phenoxy-2-phenylpyridines (24d and 24e) were synthesized that had hydroxyl groups in the 3- and/or 4-positions of the added phenyl ring. The idea was to establish additional hydrogen bonds with Arg93A and Ile95A, as for 3 and 8 in Figure 2. Four methoxylated intermediates (24f-i) from the syntheses of 24b-e had also been made in sufficient amounts to be included for biological evaluation.
Table 3. Experimentally Determined Inhibition Constants for Biphenyl and 2-Phenylpyridine Analogues.

| compd | X | R1 | R2 | R3 | Ki (μM) |
|---|---|---|---|---|---|
| 24a | CH | OH | OH | H | NA |
| 24b | CH | OH | H | OH | NA |
| 24c | CH | OH | OH | OH | NA |
| 24d | N | OH | OH | H | 6.9±1.5 |
| 24e | N | OH | H | OH | 67.6±19.4 |
| 24f | CH | OMe | H | OMe | NA |
| 24g | CH | OMe | OMe | OMe | NA |
| 24h | N | OMe | OMe | H | 57.2±3.4 |
| 24i | N | OMe | H | OMe | NA |
| 24j | N | CN | OH | H | 0.37±0.12 |
Among the five hydroxyl containing compounds, the compounds with a pyridine core, 24d and 24e, were active, while all compounds with the phenyl core (24a-c) were inactive. Having the meta hydroxyl group in 24d gave a tenfold boost in inhibitory activity compared to the para somer 24e.
We also noted that in protein structure B there might be a small polar cavity formed by the backbone oxygen atoms of Cys102C, Ser103C, and Asn106C. It was expected that there might be water molecules in this region, and that it might be beneficial to extend the inhibitor to interact with or displace some of the water.32 To this end, a compound with a 3-cyano substituent on the phenoxy ring, 24j, was designed and docked. The top pose has the cyano group oriented into the polar cavity, as desired (Figure 6). Thus, 24j was synthesized and found to be roughly 20 times more potent than the 3-hydroxyl analogue 24d (Table 3). The boost brought the project into the nanomolar realm and it also supported the structural models. The 3-pyridine core, however, did not offer a straightforward way to extend the compounds to benefit from hydrophobic contacts with Tyr37C, so attention turned to other series.
Figure 6.
Top pose for the 3-cyano containing 24j resulting from rigid Glide docking.
2-Morpholinyl-4-phenoxypyridines
Returning to the core of 15, the modeled active site cavity of structure B seemed large enough to allow additional substitution of the 3-methoxyphenyl group of 23. Modeling suggested that the 5-position would be most suitable for attachment of a second substituent. First a 5-methoxy substituent was explored with compound 25a in Table 4, though docking scores suggested that this might be too large of an addition. Indeed, 25a is five-fold less active than 23. However, the methyl and chloro derivatives, 25b and 25c, were prepared and both compounds gave a ca. four-fold boost in activity. Tolerance for extension of the 3-methoxy substituent to at least ethoxy seemed possible leading to preparation of 25d, which was found to be three-fold more active than 23.
Table 4. Experimentally Determined Inhibition Constants for Analogues of 23.

| compd | R1 | R2 | R3 | Ki (μM) |
|---|---|---|---|---|
| 23 | OMe | H | H | 22.9±11.2 |
| 25a | OMe | OMe | H | 122±48 |
| 25b | OMe | Me | H | 5.6±1.2 |
| 25c | OMe | Cl | H | 2.6±1.5 |
| 25d | OEt | H | H | 6.9±1.5 |
| 25e | OEt | H | Me | 7.2±1.1 |
| 25f | OMe | Me | Me | 1.2±1.9 |
| 25g | CN | H | Me | 11.3±1.5 |
| 25h | OMe | H | Me | 0.043±0.01 |
Next substitution at the 6-position on the pyridine ring was considered since fluorine or a methyl substituent was acceptable in 18 - 20. Thus, 6-methyl groups were added to 25d and 25b yielding 25e and 25f. Though the activity changed little in going to 25e, there was a four-fold improvement for 25f to 1 μM. In view of the increased substitution, docking yielded fewer alternative conformers for the complexes of 25e and 25f. It was conceivable that this might be detrimental from an entropic standpoint. Thus, compounds 25g and 25h were synthesized retaining the 6-methyl group, but with only a 3-cyano or 3-methoxy group in the phenoxy ring. The cyano idea came from 24j; however, docking of 25g indicated that the change in the core causes the cyano group to extend less well into the hydrophilic pocket near residues 102-106 (Figure 7a). Consistently, there is no activity gain for 25g over 25e. However, there is a large improvement with the methoxy analogue 25h, which is a 43-nM tautomerase inhibitor. It appears to embody comparatively optimal sizes and placement of the substituents. As a result of the introduction of the 6-methyl substituent, the morpholine ring prefers orientations towards Tyr37C according to the docking (Figure 7) as for 18 in Figure 3a. The docked structure for 25h (Figure 7b) also benefits from a hydrogen bond from Pro1C to the ether oxygen and improved contact of the phenoxy ring with Ile65C, which is shown to the left of Lys33C in Figure 7. It is noted that the enhanced activity of 25h relative to 23 is large for introduction of a methyl group.33 Such large effects normally require a conformational component and burial of the methyl group in a hydrophobic region. In the present case, the added methyl group is modeled to be in the hydrophobic region near Tyr96C and it appears to push the inhibitor up in the active site as illustrated yielding improved interactions for the phenoxy ring.
Figure 7.
(a) Top pose of 25g from docking into structure B. The cyano-substituent does not extend as far into the polar cavity as the corresponding cyano-substituent of 24j. (b) Top pose of 25h from docking into structure B. The pyridine ring is farther up in the active site allowing improved contacts for the methoxyphenoxy group.
In this regard, the conformational flexibility provided by rotations about the aryl ether bonds is undoubtedly important in optimizing the non-bonded interactions. Though it is possible to rationalize the activity variations based on the docked structures, more confident analyses require experimental structure determinations.
6-Methyl-4-phenoxy-2-phenylpyrimidines and 4-Phenoxy-2-phenylpyridines
Encouraged by the activity of 20, replacement of the morpholine ring by a more easily substituted phenyl ring also warranted investigation. However, 20 with an activity of 19±2 μM is more than 400 times less potent than the best morpholine compound, 25h. Nevertheless, a goal was set to optimize 2-phenyl analogues to at least the level of 25h.
Mimicking the substitution pattern for 25h, the 2-phenyl pyrimidine analogue 26a was prepared (Table 5). It showed a solid gain over 20, but it is still far from the activity of 25h. The morpholinyl to 2-phenyl change may result in weaker hydrophobic interaction with Tyr37C. The 4-position on the phenoxy ring had not previously been explored and although structure B looked more crowded in that region, 26b and 26c were synthesized. Consistently, the 4-hydroxyl substituent of 26b was poorly tolerated (Ki = 93 μM), while the bulkier 4-methoxy substituent of 26c made the compound inactive. An analog of 20 with a 3-cyano substituent, 26d, was also considered. The top pose after conformational analysis and rigid docking suggested a complete change in binding mode with the phenoxy ring now aligned with Tyr37C and the 3-cyano substituent hydrogen-bonded to Lys33C (Figure 8). The docking procedure also generated other solutions where 26d had similar orientation as the top pose of 20 (Figure 4a), but none where the nitrile group extended as far as for 24j (Figure 6). This further illustrates that the protein cavity is large and symmetrical enough to accommodate some of the compounds in more than one binding mode. 26d was synthesized via Scheme 3 and turned out to improve the tautomerase inhibition by two or three-fold over 26a.
Table 5. Experimentally Determined Inhibition Constants for 2-Phenyl Pyridines and Pyrimidines.

| compd | X | R1 | R2 | R3 | R4 | R5 | Ki (μM) |
|---|---|---|---|---|---|---|---|
| 26a | N | OMe | H | H | Me | H | 5.5±2.3 |
| 26b | N | H | OH | H | Me | H | 93.3±18.2 |
| 26c | N | H | OMe | H | Me | H | NA |
| 26d | N | CN | H | H | Me | H | 1.6±2.6 |
| 26e | CH | OH | H | H | Me | H | 0.039±0.08 |
| 26f | CH | OMe | H | H | Me | H | 7.7±0.1 |
| 26g | CH | OH | H | H | H | OH | 3.0±2.5 |
| 26h | CH | OMe | H | H | H | OMe | NA |
| 26i | CH | OH | H | Me | H | OH | NA |
| 26j | CH | OMe | H | Me | H | OMe | 0.37±0.43 |
| 26k | CH | OMe | H | Me | Me | OMe | 0.038±0.09 |
Figure 8.
Top pose of 26d.
The N3 nitrogen in the pyrimidines in poses such as Figure 8 does not appear to be either solvent exposed or forming a hydrogen bond with the protein. Consequently, it was thought that greater activity would likely be obtained with pyridine derivatives. Compound 26f was prepared by analogy to 25h and 26a. 26f was also demethylated to give the 3-hydroxy analogue 26e. Though 26f has similar potency to 26a, a striking improvement to 39 nM was found for 26e. It was also deemed desirable to explore possible hydrogen bonding to the backbone carbonyl oxygen of Arg93A with this series, similarly to 3 (Figure 2) or 24j (Figure 6). Modeling suggested that a 4-hydroxy group in the 2-phenyl ring could reach Arg93A, but only if the 6-position in the pyridine ring were unsubstituted. The docked structure for 26g in Figure 9a shows the intention. Thus, 26g was prepared from 26h and both were assayed. 26g emerged with a Ki of 3 μM and 26h is inactive.
Figure 9.
(a) Top pose of 26g. (b) The nearly identical top pose of 26i.
The final effort explored analogues of 26g with a second substituent in the phenoxy ring as for 25f. Docking suggested that 26i with an added 5-methyl group would have a similar binding mode to 26g (Figure 9b). Compound 26j was obtained as an intermediate in the synthesis of 26i, and docking of 26j suggested that the compound would have a similar binding mode, though it would be pushed up in the binding site with the central pyridine ring closer to Pro1C (Figure 10a). While the top pose had the 4-methoxy substituted phenyl ring oriented towards Arg93A, lower scored poses had the phenyl ring aligned with Tyr37C. As it turned out, only 26j showed activity (Ki = 0.37 μM). It is clearly much more hydrophobic than 26i, which is expected to incur a significant desolvation penalty for the two hydroxyl groups. Continuing with 26j, the 6-methyl group was reintroduced into the pyridine ring to yield 26k. It was expected that this addition would enforce a binding mode where the 4-methoxyphenyl group would be aligned with Tyr37C (Figure 10b). This appeared to have the desired effect as 26k provided a ca. ten-fold increase in activity over 26j and has similar potency as 25h and 26e. The sensitivity of the computed binding mode to small structural changes is well illustrated in Figure 10. The uncertainties are magnified by the expected conformational flexibility of the tautomerase active site, which is known for huMIF.34 In the end, with moderate synthetic effort, the 9-μM docking hit 15 was progressed to these three ca. 40 nM tautomerase inhibitors.
Figure 10.
(a) Top pose of 26j. A hydrogen bond is predicted to occur between the biaryl ether oxygen and Pro1C. The 4-methoxyphenyl substituent remains aligned towards Arg93A. (b) Top pose of 26k. Addition of the 6-methyl group in the pyridine ring causes the 4-methoxyphenyl group to align with Tyr37C.
Specificity for PfMIF versus huMIF
The specificity of the three most potent compounds was determined by also measuring the tautomerase Ki values for the compounds with huMIF (Table 6). Excellent selectivity is apparent as the Ki values of the three compounds for huMIF are at least 2500 times greater than for PfMIF. The achieved selectivity is not surprising in view of the differences in the shapes of the huMIF and PfMIF tautomerase active sites, which is reflected in the differences in shape of huMIF inhibitors and the present ones. The huMIF site is smaller and more linear than the PfMIF site. The structural similarity of the three most active compounds found here is apparent in Scheme 6. The three compounds share the 6-methyl-4-phenoxypyridine core. They are distinct from the HPV substrate used in the tautomerase assays and from the three illustrated sub-μM huMIF inhibitors.4,35,36 Thus, it is should not be difficult to achieve selectivity, if the inhibitors are binding to the PfMIF and huMIF tautomerase active sites.
Table 6. Selectivity of the Most Potent PfMIF Tautomerase Inhibitors.
| compd |
PfMIF Ki (μM) |
huMIF Ki (μM) |
|---|---|---|
| 25h | 0.043±0.01 | NA |
| 26e | 0.039±0.08 | 101±55 |
| 26k | 0.038±0.09 | 270±170 |
Scheme 6. The Most Active PfMIF Tautomerase Inhibitors, the HPV Substrate, and Examples of Potent huMIF Tautomerase Inhibitors (27 - 29).
In what appears to be the only prior report of PfMIF tautomerase inhibitors,37 epoxyazadiradione and three other closely related liminoids from neem seed yielded IC50 values of 10, 46, 90, and 23 μM. However, they also gave very similar IC50s for inhibition of the tautomerase activity of huMIF, 6, 35, 85, and 18 μM, respectively. The relatively weak and non-selective activity could arise from non-specific surface binding to both MIFs. The compounds also contain multiple structural alerts including enones, epoxides, and carboxylic esters.31
CONCLUSION
Virtual screening of two commercial compound libraries was performed to seek tautomerase inhibitors of PfMIF. Four inhibitors were identified of which 15 was the most potent compound with a low-μM Ki. Its further development began with the purchase and evaluation of four analogues that were also found to be active. Missing residues in the PfMIF crystal structure were modeled from a structure for PbMIF, and using the modified structure, four series of analogues were designed, synthesized, and assayed. Three of the 31 synthesized compounds, 25h, 26e, and 26k, are potent (Ki = ca. 40 nM) and selective PfMIF tautomerase inhibitors. Analysis of computed structures of complexes of the inhibitors with PfMIF provided qualitative rationales for most of the activity variations, though experimental structural clarification is desired. The reported inhibitors may facilitate such studies since their binding may help stabilize the positions of residues 102-115.
Most importantly, the reported PfMIF tautomerase inhibitors can be used to further study the function of PfMIF and its effects on malaria infection. Additional studies in our laboratories will also address modulation of PfMIF-CD74 binding with the tautomerase inhibitors and further lead optimization. Given recent evidence for PfMIF’s role in suppressing anti-malarial immune responses, this approach may lead to novel anti-parasitic agents.
Supplementary Material
ACKNOWLEDGEMENTS
Support of this research by NIH grants GM32136 and AI042310 is gratefully acknowledged. Dr. Nathan D. Schley kindly executed the small molecule crystallography. MKD also expresses gratitude to the Sixten Gemzéus Foundation and IF’s Foundation for Pharmaceutical Research for postdoctoral fellowships.
ABBREVIATIONS USED
- PfMIF
Plasmodium falciparum macrophage migration inhibitory factor
- P. falciparum
Plasmodium falciparum
- huMIF
human macrophage migration inhibitory factor
- CD74
cluster of differentiation 74 (the MIF receptor)
- PbMIF
Plasmodium berghei macrophage migratory inhibition factor
- OPLS-AA
optimized potentials for liquid simulations all-atom
- SP
standard precision
- XP
extra precision
- TEA
triethylamine
- DCM
dichloromethane
- 4-HPP
4-hydroxyphenylpyruvate
- NA
Not Active.
Footnotes
Notes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information. Synthetic details, and 1H-NMR, 13C-NMR, and HRMS spectral data for all synthesized compounds, and X-ray crystal structures for 25f and 25g. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).WHO . World Malaria Report: 2011. WHO Press; Geneva, Switzerland: 2011. [Google Scholar]
- (2).Sun T, Holowka T, Song Y, Zierow S, Leng L, Chen Y, Xiong H, Griffith J, Nouraie M, Thuma PE, Lolis E, Janse CJ, Gordeuk VR, Augustijn K, Bucala RA. Plasmodium-encoded cytokine suppresses T-cell immunity during malaria. Proc. Nat. Acad. Sci. USA. 2012;109:E2117–E2126. doi: 10.1073/pnas.1206573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Bucala R. MIF and the Genetic Basis of Macrophage Responsiveness. Curr. Immunol. Revs. 2006;2:217–223. [Google Scholar]
- (4).Orita M, Yamamoto S, Katayama N, Fujita S. Macrophage migration inhibitory factor and the discovery of tautomerase inhibitors. Curr. Pharm. Des. 2002;8:1297–1317. doi: 10.2174/1381612023394674. [DOI] [PubMed] [Google Scholar]
- (5).Sanchez E, Gomez LM, Lopez-Nevot MA, Gonzalez-Gay MA, Sabio JM, Ortego-Centeno N, de Ramon E, Anaya JM, Gonzalez-Escribano MF, Koeleman BP, Martin J. Evidence of association of macrophage migration inhibitory factor gene polymorphisms with systemic lupus erythematosus. Genes Immun. 2006;7:433–436. doi: 10.1038/sj.gene.6364310. [DOI] [PubMed] [Google Scholar]
- (6).Cordery DV, Kishore U, Kyes S, Shafi MJ, Watkins KR, Williams TN, Marsh K, Urban BC. Characterization of a Plasmodium falciparum macrophage-migration inhibitory factor homologue. J. Infect. Dis. 2007;195:905–912. doi: 10.1086/511309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Shao D, Han Z, Lin Y, Zhang L, Zhong X, Feng M, Guo Y, Wang H. Detection of Plasmodium falciparum derived macrophage migration inhibitory factor homologue in the sera of malaria patients. Acta Trop. 2008;106:9–15. doi: 10.1016/j.actatropica.2007.12.008. [DOI] [PubMed] [Google Scholar]
- (8).Fingerle-Rowson G, Kaleswarapu DR, Schlander C, Kabgani N, Brocks T, Reinart N, Busch R, Schütz A, Lue H, Du X, Liu A, Xiong H, Chen Y, Nemajerova A, Hallek M, Bernhagen J, Leng L, Bucala R. A tautomerase-null macrophage migration-inhibitory factor (MIF) gene knock-in mouse model reveals that protein interactions and not enzymatic activity mediate MIF-dependent growth regulation. Mol. Cell Biol. 2009;29:1922–1932. doi: 10.1128/MCB.01907-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Augustijn KD, Kleemann R, Thompson J, Kooistra T, Crawford CE, Reece SE, Pain A, Siebum AH, Janse CJ, Waters AP. Functional characterization of the Plasmodium falciparum and P. berghei homologues of macrophage migration inhibitory factor. Infect. Immun. 2007;75:1116–1128. doi: 10.1128/IAI.00902-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Dobson SE, Augustijn KD, Brannigan JA, Schnick C, Janse CJ, Dodson EJ, Waters AP, Wilkinson AJ. The crystal structures of macrophage migration inhibitory factor from Plasmodium falciparum and Plasmodium berghei. Protein Sci. 2009;18:2578–2591. doi: 10.1002/pro.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Cournia Z, Leng L, Gandavadi S, Du X, Bucala R, Jorgensen WL. Discovery of human macrophage migration inhibitory factor (MIF)-CD74 antagonists via virtual screening. J. Med. Chem. 2009;52:416–424. doi: 10.1021/jm801100v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Cho Y, Vermeire JJ, Merkel JS, Leng L, Du X, Bucala R, Cappello M, Lolis E. Drug repositioning and pharmacophore identification in the discovery of hookworm MIF inhibitors. Chem. Biol. 2011;18:1089–1101. doi: 10.1016/j.chembiol.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Senter PD, Al-Abed Y, Metz CN, Benigni F, Mitchell RA, Chesney J, Han J, Gartner CG, Nelson SD, Todaro GJ, Bucala R. Inhibition of macrophage migration inhibitory factor (MIF) tautomerase and biological activities by acetaminophen metabolites. Proc. Natl. Acad. Sci. U.S.A. 2002;99:144–149. doi: 10.1073/pnas.011569399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- (15).Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- (16).Schrödinger Suite. 2009 Protein Preparation Wizard. Schrödinger, LLC; New York, NY: 2009. [Google Scholar]
- (17).LigPrep . version 2.5. Schrödinger, LLC; New York, NY: 2011. [Google Scholar]
- (18).Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- (19).Jorgensen WL. QikProp, version 3.0. Schrodinger, LLC; New York: 2006. [Google Scholar]
- (20).Irwin JJ, Shoichet BK. ZINC-a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1 Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- (22).Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision Glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- (23).Maestro, version 9.2. Schrödinger, LLC; New York, NY: 2011. [Google Scholar]
- (24).Terhorst JP, Jorgensen WL. E/Z energetics for molecular modeling and design. J. Chem. Theory Comput. 2010;6:2762–2769. doi: 10.1021/ct1004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25)).Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- (26).MacroModel, version 9.8. Schrödinger, LLC; New York, NY: 2010. [Google Scholar]
- (27).(a) Evans DA, Katz JL, West TR. Synthesis of diaryl ethers through the copper-promoted arylation of phenols with arylboronic acids. An expedient synthesis of thyroxine. Tet. Lett. 1998;39:2937–2940. [Google Scholar]; (b) Tzschucke CC, Murphy JM, Hartwig JF. Arenes to anilines and aryl ethers by sequential iridium-catalyzed borylation and copper-catalyzed coupling. Org. Lett. 2007;9:761–764. doi: 10.1021/ol062902w. [DOI] [PubMed] [Google Scholar]
- (28).Kamir D, Zierow S, Leng L, Cho Y, Diaz Y, Griffith J, McDonald C, Merk M, Mitchell RA, Trent J, Chen Y, Kwong Y-KA, Xiong H, Vermeire J, Cappello M, McMahon-Pratt D, Walker J, Bernhagen J, Lolis E, Bucala R. A Leishmania ortholog of macrophage migration inhibitory factor modulates host macrophage responses. J. Immunol. 2008;180:8250–8261. doi: 10.4049/jimmunol.180.12.8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Bernhagen J, Mitchell RA, Calandra T, Voelter W, Cerami A, Bucala R. Purification, bioactivity, and secondary structure analysis of mouse and human macrophage migration inhibitory factor (MIF) Biochemistry. 1994;33:14144–14155. doi: 10.1021/bi00251a025. [DOI] [PubMed] [Google Scholar]
- (30).Taylor AB, Johnson WH, Jr., Czerwinski RM, Li HS, Marvin L, Hackert ML, Whitman CP. Crystal structure of macrophage migration inhibitory factor complexed with (E)-2-fluoro-p-hydroxycinnamate at 1.8 Å resolution: Implications for enzymatic catalysis and inhibition. Biochemistry. 1999;38:7444–7452. doi: 10.1021/bi9904048. [DOI] [PubMed] [Google Scholar]
- (31).Baell JB, Holloway GA, A G. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- (32).Michel J, Tirado-Rives J, Jorgensen WL. Energetics of Displacing Water Molecules from Protein Binding Sites: Consequences for Ligand Optimization. J. Am. Chem. Soc. 2009;131:15403–15411. doi: 10.1021/ja906058w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Leung CS, Leung SSF, Tirado-Rives J, Jorgensen WL. Methyl effects on protein-ligand binding. J. Med. Chem. 2012;55:4489–4500. doi: 10.1021/jm3003697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Cho Y, Crichlow GV, Vermeire JJ, Leng L, Du X, Hodsdon ME, Bucala R, Cappello M, Gross M, Gaeta F, Johnson K, Lolis EJ. Allosteric inhibition of macrophage migration inhibitory factor revealed by ibudilast. Proc. Natl. Acad. Sci. USA. 2010;107:11313–11318. doi: 10.1073/pnas.1002716107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Hare AA, Leng L, Gandavadi S, Du X, Cournia Z, Bucala R, Jorgensen WL. Optimization of N-benzyl-benzoxazol-2-ones as inhibitors of macrophage migration inhibitory factor (MIF) Bioorg. Med. Chem. Lett. 2010;20:5811–5814. doi: 10.1016/j.bmcl.2010.07.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Jorgensen WL, Gandavadi S, Du X, Hare AA, Trofimov A, Leng L, Bucala R. Receptor agonists of macrophage migration inhibitory factor. Bioorg. Med. Chem. Lett. 2010;20:7033–7036. doi: 10.1016/j.bmcl.2010.09.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Alam A, Haldar S, Thulasiram HV, Kumar R, Goyal M, Iqbal M, Pal C, Dey S, Bindu S, Sarkar S, Pal U, Maiti NC, Bandyopadhyay U. Novel Anti-inflammatory Activity of Epoxyazadiradione against Macrophage Migration Inhibitory Factor. J. Biol. Chem. 2012;287:24844–24861. doi: 10.1074/jbc.M112.341321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.