ABSTRACT
WNT4 is required for normal ovarian follicle development and female fertility in mice, but how its signal is transduced remains unknown. Fzd1 encodes a WNT receptor whose expression is markedly induced in both mural granulosa cells and cumulus cells during the preovulatory period, in a manner similar to Wnt4. To study the physiological roles of FZD1 in ovarian physiology and to determine whether it serves as receptor for WNT4, Fzd1-null mice were created by gene targeting. Whereas rare Fzd1−/− females were sterile because of uterine fibrosis and ovarian tubulostromal hyperplasia, most were subfertile, producing ≈1 fewer pup per litter on average relative to controls. Unlike WNT4-deficient mice, ovaries from Fzd1−/− mice had normal weights, numbers of follicles, steroid hormone production, and WNT4 target gene expression levels. Microarray analyses of granulosa cells from periovulatory follicles revealed few genes whose expression was altered in Fzd1−/− mice. However, gene expression analyses of cumulus-oocyte complexes (COCs) revealed a blunted response of both oocyte (Zp3, Dppa3, Nlrp5, and Bmp15) and cumulus (Btc, Ptgs2, Sema3a, Ptx3, Il6, Nts, Alcam, and Cspg2) genes to the ovulatory signal, whereas the expression of these genes was not altered in WNT4-deficient COCs from Wnt4tm1.1Boer/tm1.1Boer;Tg (CYP19A1-cre)1Jri mice. Despite altered gene expression, cumulus expansion appeared normal in Fzd1−/− COCs both in vitro and in vivo. Together, these results indicate that Fzd1 is required for normal female fertility and may act in part to regulate oocyte maturation and cumulus cell function, but it is unlikely to function as the sole ovarian WNT4 receptor.
Keywords: cumulus cells, fertility, granulosa cells, mouse, ovary
Frizzled 1 (Fzd1) is induced in periovulatory granulosa and cumulus cells; a null mutation blunts the induction of genes associated with cumulus expansion and results in reduced female fertility.
INTRODUCTION
The WNTs are an expansive family of secreted glycoproteins known mostly for their roles in embryonic development and tumorigenesis [1, 2]. To transduce their signal, WNTs bind to a receptor complex consisting of a Frizzled (FZD) seven-transmembrane-spanning cell surface receptor and the lipoprotein-related receptor proteins LRP5 and LRP6, which act as coreceptors. This interaction can activate three distinct intracellular pathways known as the planar cell polarity (c-jun kinase), WNT/Ca2+, and WNT/CTNNB1 (or canonical) pathways [3–5]. The activation of the latter pathway results in the hypophosphorylation and stabilization of CTNNB1 (β-catenin), which accumulates in the cell and translocates to the nucleus, where it serves as a transcriptional coregulator of a number of target genes, the identity of which varies according to cell type and physiological context [6].
The roles of WNT signaling during the embryonic development of the female gonad are now well established. Indeed, WNT4 and the WNT signaling effectors CTNNB1 and RSPO1 are essential for female sex determination, and they act by controlling female somatic cell differentiation, germ cell commitment to meiosis, gonadal cell migration and sorting, and sex-specific vasculogenesis [7–17]. In addition to its functions during embryogenesis, a growing body of evidence now indicates that WNT signaling plays multiple important roles in the adult ovary as well. For instance, CTNNB1 has been shown to regulate CYP19 expression in a human granulosa cell line and in primary cultures of rat granulosa cells [18]. Further in vitro experiments using the Cre/LoxP system to knock down CTNNB1 expression in primary cultured granulosa cells confirmed its requirement for follicle-stimulating hormone (FSH)-induced estrogen synthesis [19]. Likewise, CTNNB1 has been shown to regulate StAR expression in luteal cells and can enhance luteinizing hormone (LH)-stimulated progesterone secretion [20]. A recent study also demonstrated that sustained overexpression of CTNNB1 in granulosa cells in vivo promotes preovulatory follicular development but represses LH-induced gene expression [21]. WNT2 stimulates proliferation in cultured mouse granulosa cells and acts via the CTNNB1 pathway [22]. Beyond its roles in gonadal development, WNT4 has also recently been shown to play important roles in follicle development. Its conditional inactivation in granulosa cells in the Wnt4tm1.1Boer/−;Amhr2tm3(cre)Bhr/+ model resulted in reduced fertility linked to inadequate antral follicle development [23]. Moreover, in vivo and in vitro experiments demonstrated that WNT4 regulates the expression of the steroidogenic enzymes StAR, CYP19, and CYP11A1, and Wnt4tm1.1Boer/−;Amhr2tm3(cre)Bhr/+ mice also had reduced circulating progesterone levels. Although StAR and CYP19 are CTNNB1 transcriptional targets as mentioned above, whether or not WNT4 signals via the canonical pathway in granulosa cells has not yet been determined [24].
Little is known of the functions of FZDs in the mammalian ovary, with the exception of FZD4. Female Fzd4-null mice are sterile, at least in part because of improper formation and function of their corpora lutea [25]. FZD4 does, however, appear to be dispensable for follicle development, rendering unclear which FZD(s) may serve as receptors for follicular WNTs, such as WNT2 and WNT4. One strong candidate is FZD1, because its expression in granulosa cells is markedly induced prior to ovulation, in a manner similar to WNT4 [23, 26]. We therefore hypothesized that FZD1 serves to transduce the WNT4 signal in preovulatory follicles, and that loss of FZD1 would result in ovarian defects similar to those observed in the Wnt4tm1.1Boer/−;Amhr2tm3(cre)Bhr/+ model. To test this hypothesis, we generated Fzd1 knockout mice and assessed their fertility, follicular development, cumulus expansion, ovarian gene expression, and WNT signaling pathway activity.
MATERIALS AND METHODS
Gene Targeting
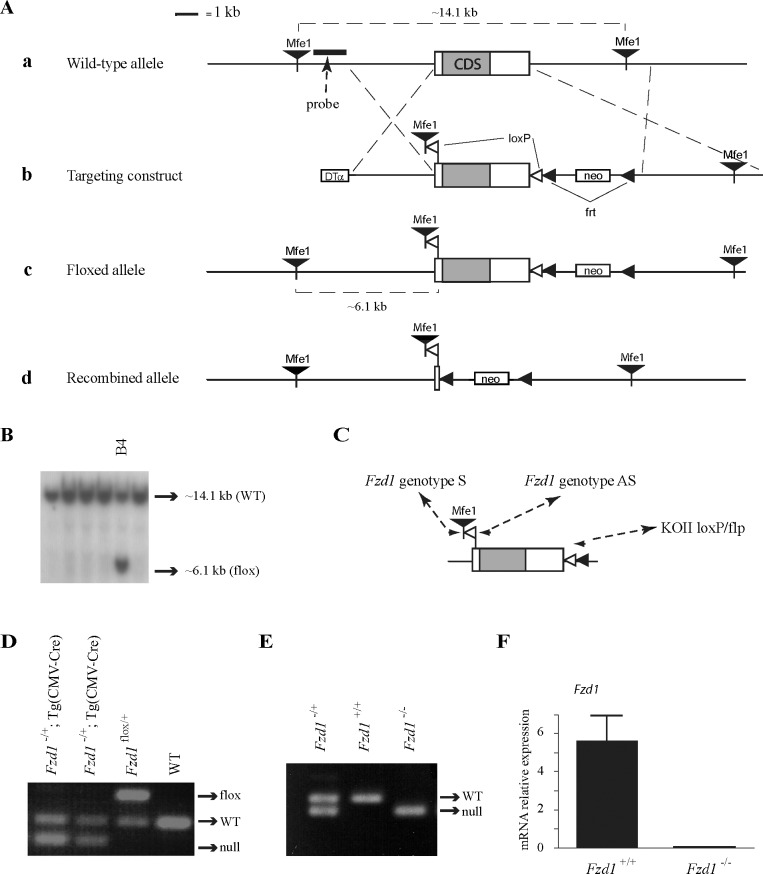
A floxed Fzd1 allele (Fig. 2Ac) was created using a standard gene targeting approach. Briefly, a targeting construct was built using the pKOII vector [27], into which was inserted a modified genomic fragment containing the single Fzd1 exon flanked by two loxP sites. An MfeI restriction site was also included for Southern blot screening of embryonic stem (ES) cell clones (Fig. 2Ab). All genomic sequences were cloned from R1 ES cell genomic DNA by PCR using the Expand Long Template PCR system (Roche Molecular Biochemicals, Laval, QC, Canada); LoxP and restriction sites were included in the primers used to amplify the genomic fragments. The targeting vector was linearized prior to electroporation into in R1 ES cells. Clonal selection was achieved by growing the cells in a medium containing 400 μg/ml Geneticin (Invitrogen, Burlington, ON, Canada) for 8–9 days. Selected colonies were cultured in triplicate, and duplicate sets were analyzed by Southern blotting to screen for proper homologous recombinants as previously described [28]. A total of 3 of 384 colonies screened showed a second band consistent with the expected size for the targeted allele (Fig. 2B). All three targeted cell lines were subsequently microinjected into blastocysts and transferred to pseudopregnant recipients according to standard protocols. Chimeric males derived from all cell lines sired pups heterozygous for the targeted allele.
FIG. 2.
Targeting of Fzd1. A) Illustration of the strategy used to generate Fzd1−/− mice. a) Placement of the genomic DNA probe used for Southern blotting. b) In the targeting construct, LoxP sites were inserted upstream and downstream of the single Fzd1 exon. c) Fzd1flox allele. d) Fzd1− (i.e., Cre-recombined) allele. B) Generation of ES cell lines heterozygous for the targeted Fzd1 allele. Presence of the targeted allele was detected as a 6.1-kb MfeI restriction fragment by Southern blotting. One representative positive clone (B4) is shown alongside five negative clones. C) PCR genotyping analysis strategy. Oligonucleotides (small black arrowheads) were designed to generate PCR products of 175 bp for the Fzd1+ allele, 299 bp for the Fzd1flox allele, and 136 bp for the Fzd1− allele. D) PCR genotype analysis of pups generated by the mating of Fzd1flox/+ with Tg (CMV-cre)1Cgn/J mice. E) PCR analysis of pups generated by backcross of Fzd1+/− mice. F) Quantitative RT-PCR analysis of Fzd1 expression in granulosa cells isolated from immature eCG-treated Fzd1+/+ and Fzd1−/− mice 12 h after the administration of an ovulatory dose of hCG, confirming the abolition of Fzd1 expression in the Fzd1-null mice. Data were normalized to the housekeeping gene Rpl19 and are expressed as means ± SEM (columns and error bars); n = 4–5 animals per genotype. DTα, diphtheria toxin α chain; neo, neomycin resistance cassette; WT, wild type.
Animal Model
To generate a null Fzd1 allele (Fig. 2Ad), Fzd1flox/+ mice were mated to the Tg (CMV-cre)1Cgn/J “cre deleter” strain (The Jackson Laboratory, Bar Harbor, ME), which expresses Cre in all tissues, including the germline. After PCR verification of the Fzd1− allele (Fig. 2D), the Cre transgene was eliminated from Fzd1−/+;Tg (CMV-cre)1Cgn/J mice in the following generation by mating to C57BL/6J wild-type mice (The Jackson Laboratory). Fzd1-null mice were then obtained by backcrossing Fzd1+/− mice. Genotyping analyses were performed by PCR on DNA obtained from tail biopsies, using the oligonucleotide primers 5′-CCTGGACCACTCTTGCTCTCCT-3′ (Fzd1 genotype S), 5′-GGGTGGTGGCTCCCACTGTGAT-3′ (Fzd1 genotype AS), and 5′-TAGGAACTTCAATTCCCCGCAAGA-3′ (KOII loxP/flp), using the following PCR conditions; 1 min at 95°C for one cycle, 45 sec at 95°C, 45 sec at 58°C, and 1 min at 72°C for 35 cycles, and 7 min at 72°C for one cycle (Fig. 2C). Primers were designed to generate PCR products of ∼175 bp for the wild-type Fzd1 allele, ∼299 bp for the floxed allele, and ∼136 bp for the knockout (i.e., Cre-recombined) allele (Fig. 2, C–E). All animal procedures were approved by the institutional animal care and use committee and conformed to the International Guiding Principles for Biomedical Research Involving Animals as promulgated by the Society for the Study of Reproduction.
Mating Trial
The mating trial was conducted using 8-wk-old Fzd1−/− (n = 7) and control littermate Fzd1−/+ (n = 9) female mice. Adult C57BL/6J males were placed in the cages for 12 mo, and the experiment was terminated 22 days after their removal to allow for the final litter. Males were changed after 6 mo to avoid a possible decline in fertility.
Follicle Counting
Left ovaries from 6-wk-old and 1-yr-old mice were collected, fixed in Bouin fixative for 24 h, and washed for 3 days with 70% ethanol before being paraffin embedded. Serial sections were prepared at a thickness of 6 μm and every seventh (42-day-old) or fifth (1–yr-old) section was counterstained with hematoxylin and eosin. Photomicrographs of all follicles containing a visible oocyte nucleus were taken for all of the sections. Blind classification of the follicles based on the Pedersen system was then performed as previously described [23].
Tissue Collection
Ovaries, granulosa cells, and cumulus-oocyte complexes (COCs) were obtained from 23- to 26-day-old mice 48 h after treatment with equine chorionic gonadotropin (eCG; 5 IU i.p.; Folligon; Intervet, Whitby, ON, Canada), and with or without the subsequent administration of an ovulatory dose of human chorionic gonadotropin (hCG; 5 IU i.p.; Chorulon; Intervet). Intact ovaries (isolated 24 h after hCG) were frozen at −80°C until the time of RNA extraction. Ovaries from mice obtained 0, 4, 6, 8, 10, or 12 h after hCG were placed in Hanks Balanced Salt Solution (HBSS) and punctured to release granulosa cells and COCs as previously described [29]. The COCs were then isolated from granulosa cells by pipette, pooled, centrifuged at 1000 × g and frozen at −80°C until RNA or protein extraction. For in vivo gene expression analysis of COCs at 12 and 20 h after hCG, oviducts were dissected, placed in HBSS, and flushed to release their contents. The COCs were collected by pipette, pooled, and frozen at −80°C until RNA or protein extraction. The COCs isolated at 12 h after hCG were collected by both methods (ovary puncture plus oviduct flushing) because not all follicles had ovulated at that time.
To evaluate Fzd1 expression in the uterus, uterine tissue was obtained from nonpregnant mice and at different times during gestation. C57BL/6J female mice were housed with males and checked in the morning for vaginal plugs. Females were killed 9, 11, or 14 days after mating, uteri were collected, and fetal tissues were flushed from the uterine horns. Uterine tissue was placed immediately in lysis buffer, and RNA was extracted as described below.
Real-Time RT-PCR and Microarray Analyses
RNA from whole ovaries, granulosa cells, COCs, and uteri was purified using the RNeasy mini kit (Qiagen, Mississauga, ON, Canada), except for RNA from granulosa cells used to determine Fzd1 expression (Fig. 2F), which was purified with the RNeasy micro kit. The RT-PCR analyses of granulosa cell (Fig. 2F only) and uterine gene expression were performed using the SuperScript III Platinum Two-Step RT-PCR kit with SYBR Green (Invitrogen) using the oligonucleotides listed in Table 1. Quantitative PCR analysis was done using the Rotor-Gene RG-3000 apparatus (Corbett Research, Mortlake, Australia), and gene expression was determined using Rotor-Gene 6.0 software (Corbett Research). Relative gene expression was calculated by dividing individual gene expression values by their corresponding Rpl19 housekeeping control gene expression values. All other RT-PCR analyses (i.e., Figures 1, 5, and 6) were done using the SuperScript ViloTM cDNA synthesis kit (Invitrogen) and Power SYBR Green PCR Master Mix (Applied Biosystems, Streetsville, ON, Canada), with the primers listed in Table 1. These analyses were conducted on an ABI Prism 7300 instrument (Applied Biosystems), and gene expression was normalized to the reporter gene Rpl19. The RT-PCR data obtained with this method were presented as fold increases in gene expression relative to a reference sample for each gene analyzed.
TABLE 1.
Oligonucleotides for RT-PCR analysis.

FIG. 1.
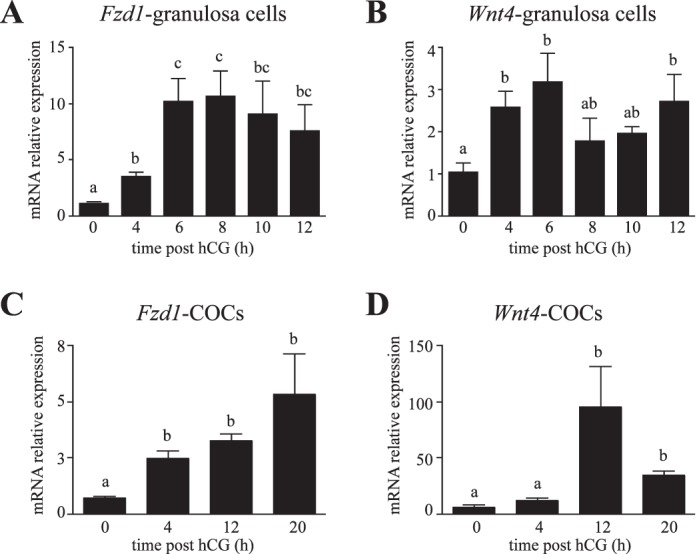
Similar patterns of Fzd1 and Wnt4 mRNA regulation by hCG in granulosa cells and COCs. Quantitative RT-PCR analysis of Fzd1 (A) and Wnt4 (B) mRNA levels in granulosa cells isolated from immature eCG-treated mice without or 4, 6, 8, 10, or 12 h after the administration of an ovulatory dose of hCG. Analyses were also conducted on isolated COCs (C and D) obtained from immature eCG-treated mice at different times after hCG administration. The COCs were collected by follicle puncture (0 and 4 h), by flushing the oviducts (20 h), or by using both methods (12 h). For all analyses, n = 3–5 samples per time point, with each sample consisting of a pool of cells collected from two to five animals. All data were normalized to the housekeeping gene Rpl19 and are expressed as means ± SEM (columns and error bars). Columns that are not labeled with common letters are significantly different (P < 0.05).
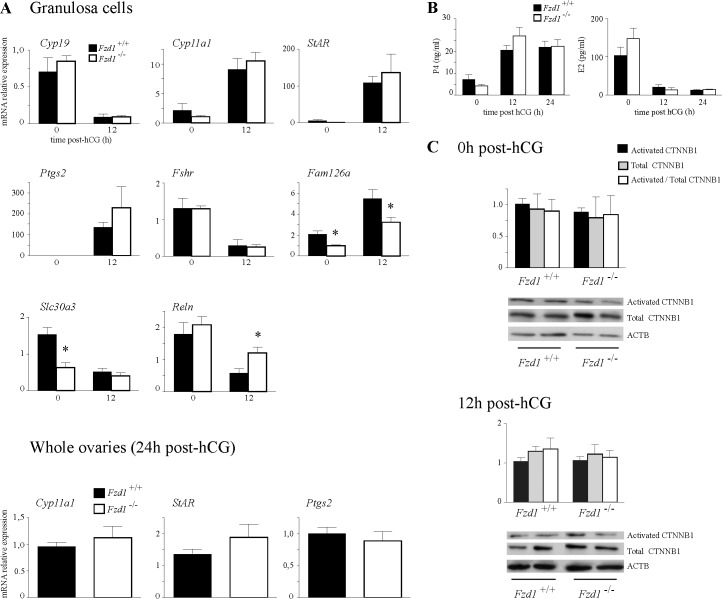
FIG. 5.
In vivo analysis of ovarian gene expression in Fzd1−/− mice. A) Quantitative RT-PCR analysis of known Wnt4-regulated genes and Fzd1-regulated genes identified by microarray analysis. Analyses were performed on RNA isolated from granulosa cells or whole ovaries from immature eCG-treated mice of the indicated genotypes, without or 12 h (granulosa cells) or 24 h (whole ovaries) after the administration of an ovulatory dose of hCG. For all analyses, n = 5–6 animals per genotype per time point. B) Serum P4 and E2 levels in mice of the indicated genotypes. Samples were taken from immature, eCG-treated mice either without or 12 or 24 h after the administration of an ovulatory dose of hCG; n = 11–27 animals per genotype per time point. C) Western blot analysis of activated and total CTNNB1 in protein extracts from granulosa cells collected from immature eCG-treated mice of the indicated genotypes without or 12 h after the administration of an ovulatory dose of hCG. ACTB (β-actin) was used as a loading control, and quantification of protein expression was done by densitometric analysis. The ratios of protein expression levels to ACTB and the ratios of activated to total CTNNB1 are illustrated; n = 3–5 animals per genotype per time point, and the experiment was repeated three times with similar results. Representative blots (n = 2 animals per genotype per time point) are shown below the graphs. All data are expressed as means ± SEM (columns and error bars). *P < 0.05.
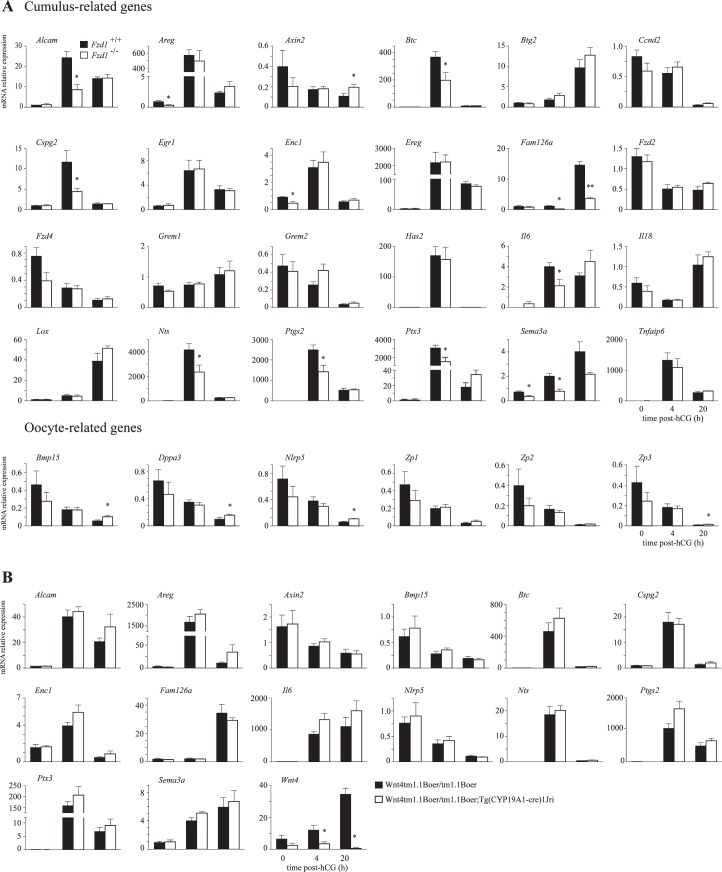
FIG. 6.
Regulation of COC gene expression by FZD1, but not by WNT4. A) Quantitative RT-PCR analysis of genes in COCs from Fzd1−/− mice and controls that are expressed in cumulus cells or oocytes whose expression is known to be regulated during COC expansion. B) COC genes whose expression was altered in Fzd1−/− mice were analyzed by quantitative RT-PCR in Wnt4tm1.1Boer/tm1.1Boer;Tg (CYP19A1-cre)1Jri and relevant control mice. For all RT-PCR analyses, n = 5 samples per genotype per time point, and each sample represents a pool of cells collected from two to five animals. All data were normalized to the housekeeping gene Rpl19 and are expressed as means ± SEM (columns and error bars). *P < 0.05; **P < 0.01.
Microarray analyses were done using triplicate RNA samples from granulosa cells collected from Fzd1−/− and Fzd1+/+ mice treated with eCG for 48 h and with hCG for 12 h, as described above. MouseRef-6 v.2.0 expression BeadChips technology (Illumina, San Diego, CA) was used, and all steps of RNA quality control, probe synthesis, hybridization, washing, and array scanning were done by the McGill University Génome Québec Innovation Center (Montréal, QC, Canada). Microarray data were analyzed using FlexArray 1.6.1 software (Génome Québec). Data from both genotypes were processed using the lumi package followed by a t-test. A P value threshold of 0.05 and 1.5-fold change cutoff values were used for identification of differentially expressed genes. All array data were deposited in the MIAME compliant database GEO, with accession number GSE38315.
Immunohistochemistry
Immunohistochemical analyses of StAR and TRP63 were done on formalin-fixed, paraffin-embedded, 7-μm ovary (StAR) or uterus (TRP63) sections using the Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA) as directed by the manufacturer. Sections were probed with a primary antibody against StAR (Santa Cruz Biotechnology Inc., Santa Cruz, CA) or against TRP63 (anti-p63; Santa Cruz Biotechnology), and staining was done using a 3,3′-diaminobenzidine peroxidase substrate kit (Vector Laboratories). Sections of ovaries were briefly counterstained with hematoxylin before mounting.
Immunoblotting
Frozen granulosa cells (corresponding to 0.5 ovary per sample) or COCs (25 COCs per sample), collected as described above, were lysed in SDS loading buffer (50 mM Tris-HCl, pH 6.8; 2% SDS; 10% glycerol; 1% β-mercaptoethanol; 12.5 mM ethylene diamine tetraacetic acid; and 0.02% bromophenol blue). Samples were resolved on 10% SDS-polyacrylamide gels and transferred to Hybond-P PVDF membrane (GE Amersham, Piscataway, NJ). The membrane was then sequentially probed with antibodies against active CTNNB1, total CTNNB1 (Cell Signaling Technology, Danvers, MA), and ACTB (Santa Cruz Biotechnology) diluted in 5% bovine serum albumin. After incubation with horseradish peroxidase-conjugated secondary anti-rabbit antibody (GE Amersham), the protein bands were visualized by chemiluminescence using the ECL Plus Western Blotting Detection Reagents (GE Amersham) and High Performance Chemiluminescence film (GE Amersham).
COC Expansion Analysis
For in vitro COC expansion, nonexpanded COCs from immature mice treated for 48 h with eCG were plated (∼15 per well) in separate wells of a 24-well plate (Sigma-Aldrich, St. Louis, MO) in 50 μl of defined COC medium (minimum essential medium; 25 mM HEPES, 0.25 mM sodium pyruvate, 3 mM l-glutamine, 1 mg/ml bovine serum albumin, 100 U/ml penicillin, 100 μg/ml streptomycin, and 1% fetal bovine serum [Invitrogen]) with or without bovine FSH (100 ng/ml; National Hormone and Peptide Program, National Institute of Diabetes and Digestive and Kidney Diseases) or forskolin (10 μM; Cedarlane Laboratories, Burlington, ON, Canada). Expansion was assessed by microscopic examination 0, 8, 12, 16, and 20 h after the plating. The COC diameters were calculated as the means of four separate measurements per COC.
Steroid Hormone Measurement
Estradiol (E2) and progesterone (P4) levels were determined by Calbiotech (Spring Valley, CA) ELISA and radioimmunoassay, respectively. All assays were performed by the ligand assay and analysis core laboratory of the University of Virginia (Charlottesville, VA).
Statistical Analyses
Effects of genotype on total pups, total litters, litter sizes, ovary weights, and follicle numbers, as well as mRNA, protein, and steroid hormone levels, were analyzed by unpaired t-tests. Effects of hCG on Fzd1 and Wnt4 expression in both granulosa cells and COCs and Fzd1 expression in the uterus were analyzed by one-way ANOVA. Log transformation of data was performed before statistical analysis when unequal variances were detected using Bartlett test. P < 0.05 was considered statistically significant. Analyses were done using Prism 4.0a (GraphPad Software Inc., San Diego, CA) software.
RESULTS
Fzd1 Is Required for Normal Female Fertility
Our previous work identified the requirement for WNT4 for normal follicular development, ovarian steroidogenesis, and female fertility in mice [23]. To identify the receptor for WNT4 in ovarian granulosa cells, we first searched available databases and literature describing ovarian Fzd gene expression. Fzd1 is expressed in granulosa cells, and its expression was shown to be induced by hCG in the immature mouse model [26] in a manner similar to that of Wnt4 [23, 26]. Furthermore, microarray analyses identified Fzd1 as one of the genes that is most strongly induced in cumulus cells 8 h after hCG administration [30]. Our studies of Fzd1 expression in immature mouse ovarian granulosa cells demonstrated a significant induction of Fzd1 expression 4 h after hCG, with maximal levels attained by 6 h and maintained until ovulation (Fig. 1A). Wnt4 expression followed a similar pattern (Fig. 1B), confirming previous studies [26]. In COCs, Fzd1 expression was induced 4 h after hCG, and levels remained high after ovulation at 20 h after hCG (Fig. 1C), similar to results obtained in an earlier study [30]. As for Fzd1, we found that Wnt4 expression was induced at 4 h after hCG, but then further increased at 12 h and remained high in ovulated COCs (Fig. 1D). Together, these and previous studies indicate that Wnt4 and Fzd1 show similar expression patterns in granulosa and cumulus cells, and may therefore be functionally related.
To study the physiological functions of Fzd1, we performed conditional gene targeting. A targeting construct was devised to insert LoxP sites into the 5′ untranslated region and downstream of the coding region of the single exon (Fig. 2A). Three correctly targeted ES cell lines were obtained (Fig. 2B and data not shown) and used to generate chimeric mice, all of which gave germline transmission of the floxed allele. Mice bearing the floxed allele were mated to the Tg (CMV-cre)1Cgn/J “cre deleter” strain to obtain recombination of the floxed allele in the germline, and hence generate a knockout allele (Fig. 2, A and D–F). Fzd1-null mice were found to be viable, healthy, and otherwise indistinguishable from their wild-type littermates, and male mice were fertile (data not shown), confirming the results of a recent study [31].
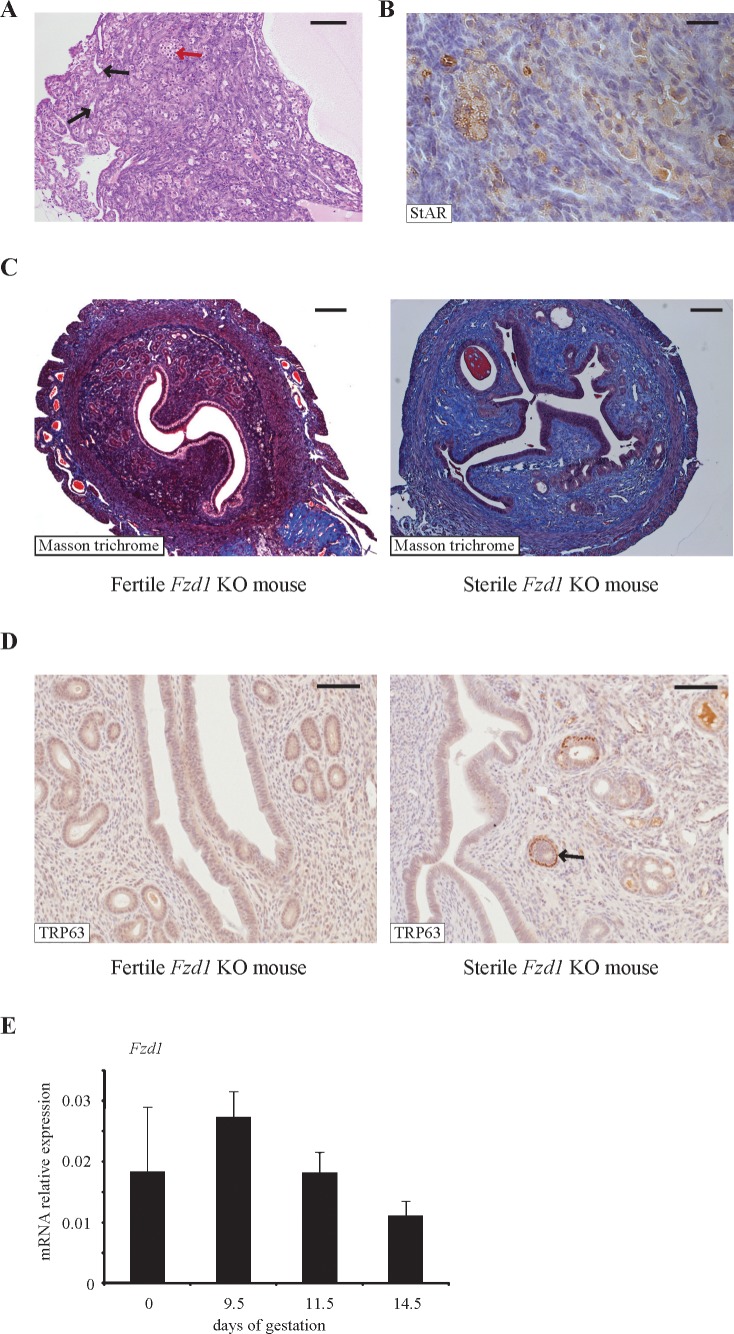
To assess the fertility of female Fzd1-null mice, 7-wk-old females were individually housed with fertile wild-type males for a period of 12 mo, and litter frequency and size were recorded. Whereas Fzd1−/− females produced a normal number of litters, their average litter size was ≈1 pup smaller (P < 0.05) than that of controls (Table 2), indicating that Fzd1 is required for normal female fertility. Interestingly, of the 17 initial mating pairs involving Fzd1−/− females that were formed to generate experimental animals, three (17.6%) failed to produce offspring. Histopathologic examination of the reproductive tissues of the sterile Fzd1−/− females revealed striking abnormalities. The ovaries of these animals were very small and were devoid of follicles (Fig. 3A). Numerous invaginations of the ovarian surface epithelium formed tubelike structures in the ovarian cortex (Fig. 3A), and multiple nests of StAR-positive cells resembling lutein cells were present throughout the ovary (Fig. 3B). This phenotype was diagnosed as tubulostromal hyperplasia, which is thought to be a consequence of premature follicle depletion in rodents [32]. The uteri of the sterile Fzd1−/− females were also grossly abnormal, having fewer glands and severe stromal fibrosis, as revealed by Masson trichrome staining (Fig. 3C). Some uterine glands (≈15%) in these mice featured an ectopic layer of cells immediately beneath the columnar epithelial layer that was positive for the basal cell marker TRP63 (Fig. 3D), an anomaly that also occurs in mice with conditional ablation of Wnt4 in the uterus [33]. Fzd1−/− females that did not have the sterility phenotype had ovaries and uteri that were indistinguishable from those of wild-type mice (Fig. 3 and data not shown). A uterine phenotype in Fzd1−/− mice was not expected, because Fzd1 expression in this tissue had not been previously characterized. We therefore examined uterine Fzd1 mRNA levels throughout gestation and found them to be expressed at high but unvarying levels in pregnant and nonpregnant mice (Fig. 3E).
TABLE 2.
Mating trial.
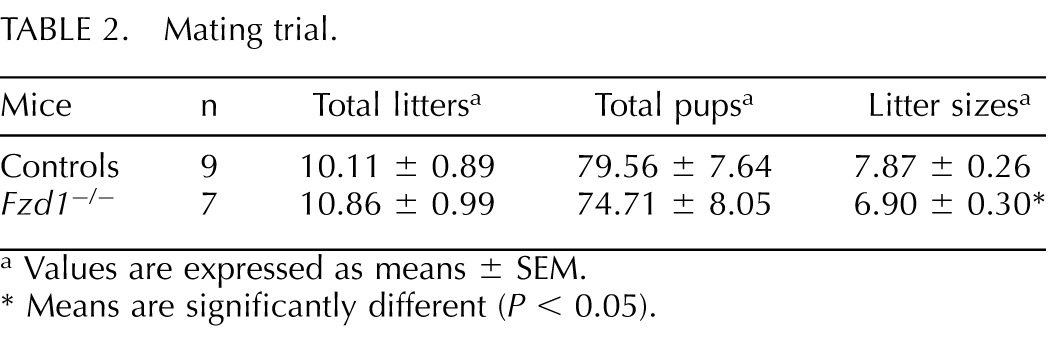
Values are expressed as means ± SEM.
Means are significantly different (P < 0.05).
FIG. 3.
A minority of Fzd−/− female mice are sterile and possess abnormalities in their reproductive tissues. A) Photomicrograph of the ovary of a sterile Fzd−/− mouse showing evidence of tubulostromal hyperplasia, characterized by the formation of tubelike structures in the stroma (black arrows) along with nests of lutein-like cells (red arrow). Bar = 100 μm. B) Immunohistochemical analysis revealed that the lutein-like cells express StAR. Bar = 25 μm. C) Masson trichrome staining revealed extensive stromal fibrosis (collagen fibers stain pale blue) and few glands in the uteri of sterile Fzd−/− mice compared with the uteri of fertile Fzd−/− mice. Bar = 200 μm. D) Many of the uterine glands in the sterile Fzd−/− mice had an ectopic layer of cells that stained positive for TRP63 (arrow), unlike those of fertile Fzd−/− females. Bar = 100 μm. E) Quantitative RT-PCR analysis of Fzd1 expression in uterine tissues of nonpregnant and Day 9.5, 11.5, or 14.5 pregnant mice (n = 5 per time point).
Follicle Development, Steroidogenesis, and Wnt4 Target Gene Expression Are Normal in Fzd1-null Mice
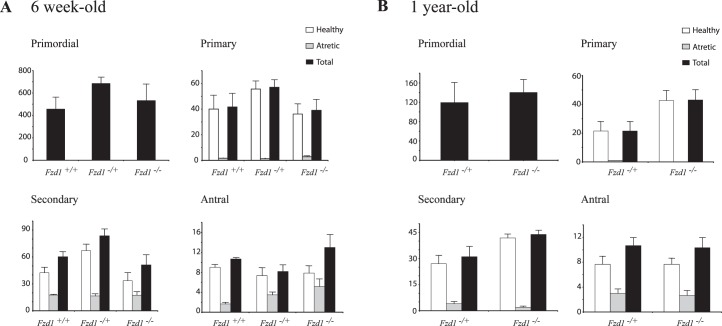
Mice with Wnt4-deficient granulosa cells are subfertile, which was attributed in part to their having reduced numbers of healthy antral follicles, with resultant decreased ovarian weights [23]. We therefore determined ovary weights and follicular reserves in Fzd1−/− mice. Fzd1−/− ovary weights at ages 6 and 28 wk were slightly lower than controls, although the difference was not statistically significant (Table 3). Likewise, no statistically significant difference was observed between 6-wk-old or 1-yr-old Fzd1-null and age-matched control animals regarding primordial, primary, secondary, or antral follicle abundance, or relative proportions of atretic follicles of any size or at either age (Fig. 4).
TABLE 3.
Ovary weights.
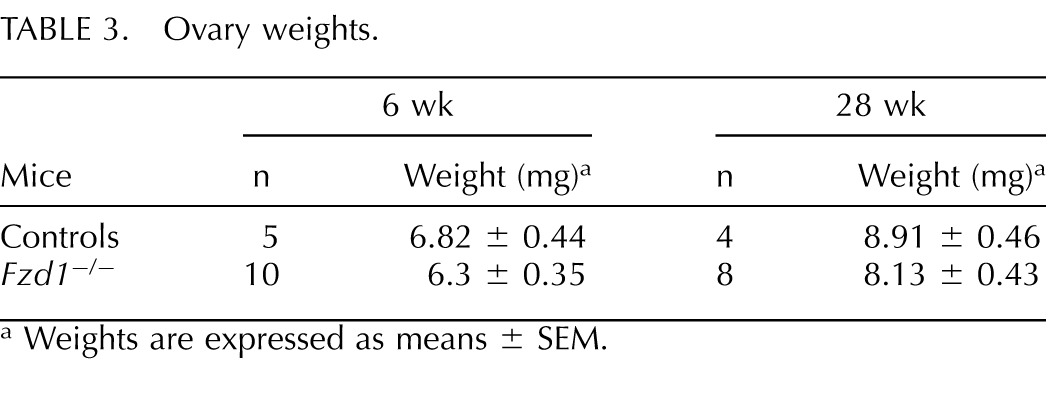
Weights are expressed as means ± SEM.
FIG. 4.
Quantitative analysis of ovarian follicles in peripubertal and 1-yr-old mice. The left ovaries of 6-wk-old (A) and 1-yr-old (B) mice of the indicated genotypes were serially sectioned, and all follicles from every fifth (1-yr-old) or seventh (6-wk-old) section were counted, categorized as primordial, primary, secondary, or antral, and scored as either healthy or atretic. Data represent raw follicle count numbers and were not adjusted to estimate the total ovarian follicle population; n = 5–6 ovaries per genotype. All data are expressed as means ± SEM (columns and error bars).
WNT4 was previously shown to regulate the expression of various granulosa cell genes, including Star, Cyp11a1, Cyp19, Ptgs2, and Fshr. WNT4 deficiency also resulted in reduced serum progesterone levels in eCG/hCG-treated immature Wnt4tm1.1Boer/−;Amhr2tm3(cre)Bhr/+ mice [23]. Analyses of gene expression in granulosa cells from eCG/hCG-treated immature Fzd1−/− mice revealed no differences in the expression of the aforementioned WNT4 target genes relative to controls (Fig. 5A). Expression of Cyp11a1, Cyp19, and Ptgs2 in whole ovaries from immature Fzd1−/− mice 24 h after hCG administration (i.e., after corpora lutea formation) was likewise unaffected (Fig. 5A). Analyses of circulating E2 and P4 levels in eCG-primed immature Fzd1−/− and control mice prior to and 12 or 24 h after an ovulatory dose of hCG revealed no differences between genotypes in levels of either hormone (Fig. 5B). Together, these data suggest that the decreased fertility observed in Fzd1−/− females was not due to a follicle development or steroidogenesis defect, and that FZD1 is unlikely to function as the sole WNT4 receptor in granulosa cells.
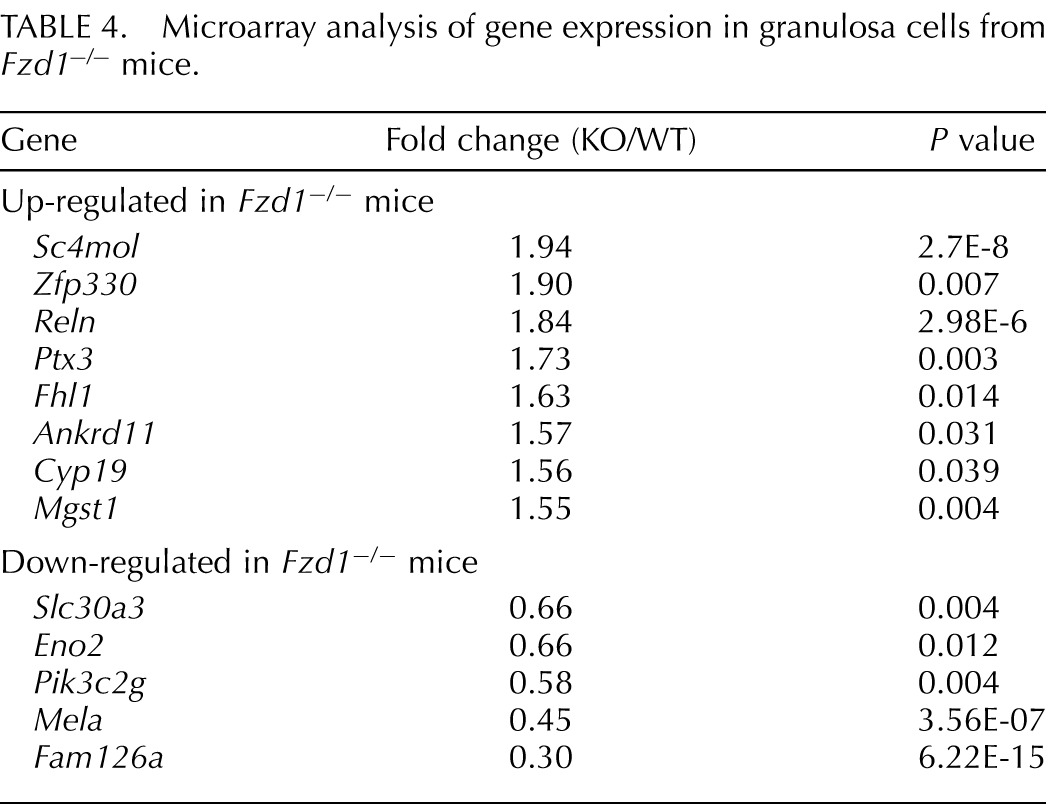
To identify genes whose expression was altered in Fzd1−/− ovaries, granulosa cells were isolated from immature Fzd1−/− and control mice 12 h after hCG, and mRNA from these cells was used for microarray analyses. Surprisingly, these analyses identified very few differentially expressed genes, with only 13 being at least 1.5-fold up-regulated or down-regulated in cells from Fzd1−/− mice (Table 4). Furthermore, real-time RT-PCR studies conducted using separate granulosa cell mRNA samples could only confirm the differential expression of 3 of the 13 genes identified by microarray: Fam126a and Slc30a3, whose expressions were down-regulated in Fzd1−/− mice, and Reln, whose expression was up-regulated (Fig. 5A and data not shown). Fam126a was previously characterized as a gene whose expression is down-regulated by CTNNB1 [34], suggesting that increased CTNNB1 signaling may occur in the granulosa cells of Fzd1−/− mice. To verify this, we compared total and “activated” (i.e., hypophosphorylated) CTNNB1 levels in Fzd1−/− and control granulosa cells isolated from eCG/hCG-treated immature mice. Results showed comparable levels of total and activated CTNNB1 protein in both models at all time points examined (Fig. 5C), suggesting that the level of WNT/CTNNB1 pathway signaling activity in granulosa cells was not altered by the loss of Fzd1, at least not at the level (or upstream) of CTNNB1.
TABLE 4.
Microarray analysis of gene expression in granulosa cells from Fzd1−/− mice.
Fzd1, but Not Wnt4, Is Required for Normal COC Gene Expression
Because Wnt4 and Fzd1 are similarly regulated in cumulus cells (Fig. 1), we next sought to determine whether FZD1 could serve as a WNT4 receptor specifically in that cell type, and whether cumulus expansion is altered by Fzd1 and/or Wnt4 loss. The COCs were harvested from eCG-primed immature Fzd1−/− and control ovaries prior to or 4 h after hCG administration, and they were collected from the oviducts 20 h after hCG. These COCs were then analyzed for the expression of a series of genes known to be regulated in oocytes or cumulus cells during cumulus expansion [30, 35]. Results revealed a number of genes involved in cumulus expansion to be underexpressed in Fzd1−/− COCs, including Areg, Btc, Ptgs2, Sema3a, Ptx3, Il6, Nts, Alcam, and Cspg2 (Fig. 6A), with the differences occurring mostly at 4 h after hCG. Expression of Fam126a was decreased and that of Axin2, a well-established CTNNB1 target gene, was increased in Fzd1−/− COCs, again suggesting increased CTNNB1 signaling activity. However, as for granulosa cells, we were not able to detect differences in total or activated CTNNB1 protein levels in COCs from Fzd1−/− and control animals at any of the time points examined (data not shown). We also analyzed the expression of a number of oocyte-specific genes whose expression is normally down-regulated by hCG during cumulus expansion (Hernandez-Gonzalez et al. [30] and Lapointe and Boerboom, unpublished observations). For a number of these genes (Zp3, Dppa3, Nlrp5, and Bmp15), hCG failed to down-regulate their expression to the same extent as the controls at 20 h after hCG (Fig. 6A). Together, these results indicate that the loss of Fzd1 delayed or blunted the gene expression changes associated with COC expansion in both the oocyte and the cumulus cells.
To verify whether WNT4 is similarly required for normal cumulus gene expression, we conditionally inactivated Wnt4 in granulosa and cumulus cells using strains bearing the Wnt4tm1.1Boer floxed allele [23] and the (CYP19A1-cre)1Jri transgene, in which Cre expression is driven by the human CYP19A1 promoter [36]. Wnt4 expression in COCs from Wnt4tm1.1Boer/tm1.1Boer;Tg (CYP19A1-cre)1Jri mice was efficiently knocked down, with reduction in mRNA levels attaining 98% at 20 h after hCG (Fig. 6B). The expression of the genes differentially expressed in Fzd1−/− COCs was then evaluated in Wnt4tm1.1Boer/tm1.1Boer;Tg (CYP19A1-cre)1Jri COCs. Results showed that the expression of none of the putative Fzd1 target genes was altered in Wnt4tm1.1Boer/tm1.1Boer;Tg (CYP19A1-cre)1Jri COCs relative to Wnt4tm1.1Boer/tm1.1Boer controls (Fig. 6B), indicating that WNT4 is not required for their normal expression. These results suggest WNT4 is unlikely to function as the sole ligand for FZD1 in cumulus cells.
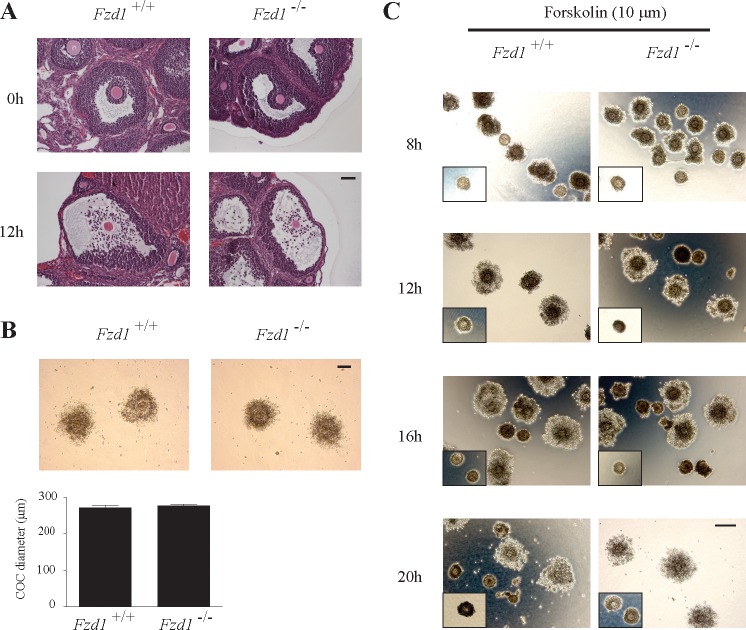
To study the functional consequences of altered COC gene expression in vivo, histological analyses of Fzd1−/− ovaries from immature mice were done throughout hCG-induced COC expansion and ovulation. Cumulus expansion occurred in Fzd1−/− follicles at the same apparent rate and to the same extent as in controls, without any obvious morphological differences (Fig. 7, A and B). Cumulus expansion was also studied in vitro on isolated COCs from eCG-treated Fzd1−/− and control mice. Upon treatment with either FSH or forskolin, Fzd1−/− COCs expanded in a manner indistinguishable from controls (Fig. 7C and data not shown).
FIG. 7.
Cumulus expansion is not altered in Fzd1−/− mice. A) Histological analysis of Fzd1−/− ovaries and controls collected from immature, eCG-treated mice either without or 12 h after the administration of an ovulatory dose of hCG. COCs from both genotypes were similarly expanded at 12 h after hCG. Bar = 50 μm. B) Photomicrographs and diameter measurements of COCs collected by flushing the oviducts of immature eCG-treated mice of the indicated genotypes 20 h after the administration of an ovulatory dose of hCG. Bar = 100 μm. Data are expressed as means ± SEM (columns and error bars); n = 15–19 COCs per genotype. C) An in vitro COC expansion assay did not reveal any differences in the rate or the extent of expansion between the two genotypes at 8, 12, 16, and 20 h after the addition of forskolin. Insets are representative scaled pictures of unexpanded COCs in an untreated control group. Bar = 100 μm.
DISCUSSION
Whereas the physiological roles of WNTs in ovarian function are gradually being defined, little is known of the mechanisms whereby their signals are transduced. In this report, we show that the WNT receptor FZD1 is required for normal female fertility, and that it appears to act (at least in part) by regulating the expression of genes involved in cumulus expansion. Although a recent study found Fzd1-null mice to be completely normal and fertile [31], the manner in which female fertility was assessed was not described, and it may have failed to detect the relatively mild subfertility phenotype reported herein. The requirement of Fzd1 for normal female fertility therefore represents the first nonredundant physiological role for Fzd1 described thus far. Furthermore, our discovery that Fzd1 regulates the expression of cumulus and oocyte genes represents the first functional evidence that WNT signaling may be involved in the regulation of cumulus expansion. Although Wnt4 and Fzd1 share similar LH/hCG-regulated expression patterns in both granulosa and cumulus cells, we found essentially no phenotypic similarities between Fzd1- and Wnt4-deficient mice at the level of follicle development, or in ovarian steroidogenesis or gene expression. It would therefore seem very unlikely that WNT4 and FZD1 function in a nonredundant manner as a ligand-receptor pair in the adult ovary. Further experiments will therefore be required to elucidate the mechanisms of FZD1 signaling in cumulus cells, as well as to define the various components of the signaling pathway(s) that it mediates, including its ligand(s).
Although we found that the COC response to hCG was blunted at the level of gene expression in Fzd1-null mice, the manner and extent to which this may have contributed to the subfertility phenotype remain unclear. Indeed, cumulus expansion proceeded unabated both in vitro and in vivo in the Fzd1−/− model. However, altered COC gene expression could lead to subtle changes in the composition of the cumulus extracellular matrix and/or the maturation of the oocyte that would be difficult to detect, and that could lead to altered survival, transport in the oviduct, fertilization and/or postfertilization development. Further studies will be required to determine whether any of the aforementioned processes are compromised in Fzd1−/− mice. Another possibility is that additional functional defects occur outside the ovary. Indeed, we found Fzd1 to be highly expressed in the uterus throughout gestation, indicating that it may play a role in implantation or some aspect of fetal development. Although we were not able to detect any histological anomalies in the uteri of Fzd1−/− mice (other than in the rare subset that was sterile), this certainly does not exclude the possibility of more subtle defects.
The up-regulation of the canonical WNT signaling target gene Axin2 and the down-regulation of Fam126a in the COCs of Fzd1−/− mice strongly suggest that FZD1 normally acts to repress CTNNB1 signaling in cumulus cells. Whereas some reports have characterized FZD1 as a possible canonical pathway agonist [37, 38], repression of CTNNB1 signaling by FZD1 has also been reported [39], perhaps indicating that the nature of the signal transduced by FZD1 can vary according to factors such as the WNT ligand that it binds, cell type, and physiological context. In spite of altered Axin2 and Fam126a expression, we found that levels of active (hypophosphorylated) CTNNB1 were not changed in Fzd1−/− COCs, suggesting that ovarian FZD1 signaling does not involve the canonical pathway. However, the mechanism whereby FZD1 represses the canonical pathway may not involve the modulation of CTNNB1 phosphorylation. Indeed, noncanonical WNTs such as WNT5a do not alter CTNNB1 expression, but nonetheless repress CTNNB1-mediated transcriptional activity [40, 41]. FZD1 may therefore act downstream of CTNNB1 to modulate the expression of Axin2, Fam126a, and potentially the other target genes identified in this study. That FZD1 does this by activating a noncanonical signaling pathway (i.e., the WNT/Ca2+ or planar cell polarity pathway) is a distinct possibility but remains to be determined.
Our observation of severe ovarian and uterine defects in a small subset of Fzd1−/− mice was somewhat paradoxical, as it could be compatible with FZD1 functioning as a receptor for WNT4. Indeed, Wnt4-null mice are characterized by the depletion of nearly their entire oocyte reserve [8], but do not survive beyond 48 h after birth because of kidney defects, precluding any study of adult ovaries. Because the tubulostromal hyperplasia observed in the ovaries of sterile Fzd1−/− mice may be the consequence of premature follicle depletion [32], it may be related to the phenotype observed in Wnt4-null mice, suggesting that FZD1 may function as a WNT4 receptor in the prenatal/perinatal ovary. Likewise, the conditional inactivation of Wnt4 in the mouse uterus results in a decreased number of uterine glands and the ectopic appearance of TRP63-positive basal cells in a manner similar to what we observed in the sterile Fzd1−/− mice [33]. Fzd1 has also previously been shown to be expressed in the epithelium and mesenchyme of the Müllerian duct (the anlage that gives rise to the uterus and other structures), and was proposed to be the receptor for WNT4 in that tissue [42]. Together, these observations support the notion that FZD1 functions as a uterine WNT4 receptor, at least during a limited developmental period. However, the reason why the severe ovarian and uterine phenotypes occur so infrequently in Fzd1−/− mice remains a mystery. Because our Fzd1−/− mice were created on a mixed genetic background, we postulated that other FZD(s) may compensate for the loss of Fzd1 during development in most strains, but certain genetic backgrounds may be more susceptible to the loss of Fzd1. Our efforts to breed our Fzd1−/− mice into a balb/c or C57BL/6 genetic background failed to increase the frequency of the sterility phenotype or the ovarian/uterine anomalies, and in fact seemed to make them rarer still (data not shown). Presuming that functional redundancy occurs in both the ovary and uterus, the ultimate identification of the FZDs involved in transducing the WNT4 signal will likely require mice with the concomitant inactivation of multiple Fzd genes.
In summary, we have determined that Fzd1 expression is required for normal female fertility, probably in part because of its regulation of genes required for cumulus expansion. Fzd1−/− mice did not reproduce the ovarian phenotype of Wnt4-deficient mice, indicating that WNT4 and FZD1 are unlikely to function nonredundantly as a ligand-receptor pair in the adult ovary. Additional studies will be required to define the mechanisms and signaling pathways whereby FZD1 regulates COC gene expression, and whether FZD1 regulates reproductive processes other than cumulus expansion.
ACKNOWLEDGMENT
The authors thank Ms. Meggie Girard for assistance with mouse colony management. The R1 embryonic stem cell line was provided by Dr. Andras Nagy, Reka Nagy, Dr. Janet Rossant, and Dr. Wanda Abramow-Newerly (University of Toronto). We also thank Dr. A.F. Parlow and the National Hormone and Peptide Program, National Institute of Diabetes and Digestive and Kidney Diseases, for providing bovine FSH.
Footnotes
Supported by an operating grant from the Canadian Institutes of Health Research and the Canada Research Chair in Ovarian Molecular Biology and Functional Genomics. The University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core is supported by the National Institutes of Health/National Institute of Child Health and Human Development (SCCPRR) grant U54-HD28934.
REFERENCES
- Nusse R, Fuerer C, Ching W, Harnish K, Logan C, Zeng A, ten Berge D, Kalani Y. Wnt signaling and stem cell control. Cold Spring Harb Symp Quant Biol 2008; 73: 59 66 [DOI] [PubMed] [Google Scholar]
- Lustig B, Behrens J. The Wnt signaling pathway and its role in tumor development. J Cancer Res Clin Oncol 2003; 129 (4): 199 221 [DOI] [PubMed] [Google Scholar]
- Karner C, Wharton KA, Jr,, Carroll TJ. Planar cell polarity and vertebrate organogenesis. Semin Cell Dev Biol 2006; 17 (2): 194 203 [DOI] [PubMed] [Google Scholar]
- Kohn AD, Moon RT. Wnt and calcium signaling: beta-catenin-independent pathways. Cell Calcium 2005; 38 (3–4): 439 446 [DOI] [PubMed] [Google Scholar]
- Huang H, He X. Wnt/beta-catenin signaling: new (and old) players and new insights. Curr Opin Cell Biol 2008; 20 (2): 119 125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archbold HC, Yang YX, Chen L, Cadigan KM. How do they do Wnt they do?: regulation of transcription by the Wnt/beta-catenin pathway. Acta Physiol (Oxf) 2012; 204 (1): 74 109 [DOI] [PubMed] [Google Scholar]
- Jaaskelainen M, Prunskaite-Hyyrylainen R, Naillat F, Parviainen H, Anttonen M, Heikinheimo M, Liakka A, Ola R, Vainio S, Vaskivuo TE, Tapanainen JS. WNT4 is expressed in human fetal and adult ovaries and its signaling contributes to ovarian cell survival. Mol Cell Endocrinol 2010; 317 (1–2): 106 111 [DOI] [PubMed] [Google Scholar]
- Vainio S, Heikkilä M, Kispert A, Chin N, McMahon AP. Female development in mammals is regulated by Wnt-4 signalling. Nature 1999; 397 (6718): 405 409 [DOI] [PubMed] [Google Scholar]
- Jeays-Ward K, Hoyle C, Brennan J, Dandonneau M, Alldus G, Capel B, Swain A. Endothelial and steroidogenic cell migration are regulated by WNT4 in the developing mammalian gonad. Development 2003; 130 (16): 3663 3670 [DOI] [PubMed] [Google Scholar]
- Heikkilä M, Peltoketo H, Leppaluoto J, Ilves M, Vuolteenaho O, Vainio S. Wnt-4 deficiency alters mouse adrenal cortex function, reducing aldosterone production. Endocrinology 2002; 143 (11): 4358 4365 [DOI] [PubMed] [Google Scholar]
- Yao HH, Matzuk MM, Jorgez CJ, Menke DB, Page DC, Swain A, Capel B. Follistatin operates downstream of Wnt4 in mammalian ovary organogenesis. Dev Dyn 2004; 230 (2): 210 215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CF, Parker K, Yao HH. WNT4/beta-catenin pathway maintains female germ cell survival by inhibiting activin betaB in the mouse fetal ovary. PLoS One 2010; 5 (4): e10382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassot AA, Ranc F, Gregoire EP, Roepers-Gajadien HL, Taketo MM, Camerino G, de Rooij DG, Schedl A, Chaboissier MC. Activation of beta-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum Mol Genet 2008; 17 (9): 1264 1277 [DOI] [PubMed] [Google Scholar]
- Tomizuka K, Horikoshi K, Kitada R, Sugawara Y, Iba Y, Kojima A, Yoshitome A, Yamawaki K, Amagai M, Inoue A, Oshima T, Kakitani M. R-spondin1 plays an essential role in ovarian development through positively regulating Wnt-4 signaling. Hum Mol Genet 2008; 17 (9): 1278 1291 [DOI] [PubMed] [Google Scholar]
- Chassot AA, Gregoire EP, Lavery R, Taketo MM, de Rooij DG, Adams IR, Chaboissier MC. RSPO1/beta-catenin signaling pathway regulates oogonia differentiation and entry into meiosis in the mouse fetal ovary. PLoS One 2011; 6 (10): e25641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatouk DM, DiNapoli L, Alvers A, Parker KL, Taketo MM, Capel B. Stabilization of beta-catenin in XY gonads causes male-to-female sex-reversal. Hum Mol Genet 2008; 17 (19): 2949 2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CF, Bingham N, Parker K, Yao HH. Sex-specific roles of beta-catenin in mouse gonadal development. Hum Mol Genet 2009; 18 (3): 405 417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parakh TN, Hernandez JA, Grammer JC, Weck J, Hunzicker-Dunn M, Zeleznik AJ, Nilson JH. Follicle-stimulating hormone/cAMP regulation of aromatase gene expression requires beta-catenin. Proc Natl Acad Sci U S A 2006; 103 (33): 12435 12440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez Gifford JA, Hunzicker-Dunn ME, Nilson JH. Conditional deletion of beta-catenin mediated by Amhr2cre in mice causes female infertility. Biol Reprod 2009; 80 (6): 1282 1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy L, McDonald CA, Jiang C, Maroni D, Zeleznik AJ, Wyatt TA, Hou X, Davis JS. Convergence of 3′,5′-cyclic adenosine 5′-monophosphate/protein kinase A and glycogen synthase kinase-3beta/beta-catenin signaling in corpus luteum progesterone synthesis. Endocrinology 2009; 150 (11): 5036 5045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, O'Connor A, Shitanaka M, Shimada M, Liu Z, Richards JS. Beta-catenin (CTNNB1) promotes preovulatory follicular development but represses LH-mediated ovulation and luteinization. Mol Endocrinol 2010; 24 (8): 1529 1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HX, Li TY, Kidder GM. WNT2 regulates DNA synthesis in mouse granulosa cells through beta-catenin. Biol Reprod 2010; 82 (5): 865 875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer A, Lapointe E, Zheng X, Cowan RG, Li H, Quirk SM, DeMayo FJ, Richards JS, Boerboom D. WNT4 is required for normal ovarian follicle development and female fertility. FASEB J 2010; 24 (8): 3010 3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe E, Boerboom D. WNT signaling and the regulation of ovarian steroidogenesis. Front Biosci (Schol Ed) 2011; 3: 276 285 [DOI] [PubMed] [Google Scholar]
- Hsieh M, Boerboom D, Shimada M, Lo Y, Parlow AF, Luhmann UF, Berger W, Richards JS. Mice null for Frizzled4 (Fzd4-/-) are infertile and exhibit impaired corpora lutea formation and function. Biol Reprod 2005; 73 (6): 1135 1146 [DOI] [PubMed] [Google Scholar]
- Hsieh M, Johnson MA, Greenberg NM, Richards JS. Regulated expression of Wnts and Frizzleds at specific stages of follicular development in the rodent ovary. Endocrinology 2002; 143 (3): 898 908 [DOI] [PubMed] [Google Scholar]
- Bardeesy N, Sinha M, Hezel AF, Signoretti S, Hathaway NA, Sharpless NE, Loda M, Carrasco DR, DePinho RA. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature 2002; 419 (6903): 162 167 [DOI] [PubMed] [Google Scholar]
- Sambook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Zeleznik AJ, Midgley AR, Jr,, Reichert LE., Jr. Granulosa cell maturation in the rat: increased binding of human chorionic gonadotropin following treatment with follicle-stimulating hormone in vivo. Endocrinology 1974; 95 (3): 818 825 [DOI] [PubMed] [Google Scholar]
- Hernandez-Gonzalez I, Gonzalez-Robayna I, Shimada M, Shimada M, Wayne CM, Ochsner SA, White L, Richards JS. Gene expression profiles of cumulus cell oocyte complexes during ovulation reveal cumulus cells express neuronal and immune-related genes: does this expand their role in the ovulation process? Mol Endocrinol 2006; 20 (6): 1300 1321 [DOI] [PubMed] [Google Scholar]
- Yu H, Smallwood PM, Wang Y, Vidaltamayo R, Reed R, Nathans J. Frizzled 1 and frizzled 2 genes function in palate, ventricular septum and neural tube closure: general implications for tissue fusion processes. Development 2010; 137 (21): 3707 3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr U. International Classification of Rodent Tumors: The Mouse. Hannover, Germany: Springer; 2001. [Google Scholar]
- Franco HL, Dai D, Lee KY, Rubel CA, Roop D, Boerboom D, Jeong JW, Lydon JP, Bagchi IC, Bagchi MK, DeMayo FJ. WNT4 is a key regulator of normal postnatal uterine development and progesterone signaling during embryo implantation and decidualization in the mouse. FASEB J 2011; 25 (4): 1176 1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasoe T, Furukawa Y, Daigo Y, Nishiwaki T, Ishiguro H, Fujita M, Satoh S, Miwa N, Nagasawa Y, Miyoshi Y, Ogawa M, Nakamura Y. Isolation and characterization of a novel human gene, DRCTNNB1A, the expression of which is down-regulated by beta-catenin. Cancer Res 2000; 60 (13): 3354 3358 [PubMed] [Google Scholar]
- Assou S, Anahory T, Pantesco V, Le Carrour T, Pellestor F, Klein B, Reyftmann L, Dechaud H, De Vos J, Hamamah S. The human cumulus–oocyte complex gene-expression profile. Hum Reprod 2006; 21 (7): 1705 1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel PL, Leone G. Expression of Cre recombinase in early diploid trophoblast cells of the mouse placenta. Genesis 2007; 45 (3): 129 134 [DOI] [PubMed] [Google Scholar]
- Flahaut M, Meier R, Coulon A, Nardou KA, Niggli FK, Martinet D, Beckmann JS, Joseph JM, Mühlethaler-Mottet A, Gross N. The Wnt receptor FZD1 mediates chemoresistance in neuroblastoma through activation of the Wnt/beta-catenin pathway. Oncogene. 2009; 28 (23): 2245 2256 [DOI] [PubMed] [Google Scholar]
- Neumann J, Schaale K, Farhat K, Endermann T, Ulmer AJ, Ehlers S, Reiling N. Frizzled1 is a marker of inflammatory macrophages, and its ligand Wnt3a is involved in reprogramming Mycobacterium tuberculosis-infected macrophages. FASEB J 2010; 24 (11): 4599 4612 [DOI] [PubMed] [Google Scholar]
- Roman-Roman S, Shi DL, Stiot V, Haÿ E, Vayssière B, Garcia T, Baron R, Rawadi G. Murine Frizzled-1 behaves as an antagonist of the canonical Wnt/beta-catenin signaling. J Biol Chem 2004; 279 (7): 5725 5733 [DOI] [PubMed] [Google Scholar]
- Yuzugullu H, Benhaj K, Ozturk N, Senturk S, Celik E, Toylu A, Tasdemir N, Yilmaz M, Erdal E, Akcali KC, Atabey N, Ozturk M. Canonical Wnt signaling is antagonized by noncanonical Wnt5a in hepatocellular carcinoma cells. Mol Cancer 2009; 8: 90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol 2006; 4 (4): e115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher E. Hung-Chang Yao H. Essential roles of mesenchyme-derived beta-catenin in mouse Mullerian duct morphogenesis. Dev Biol 2007; 307 (2): 227 236 [DOI] [PMC free article] [PubMed] [Google Scholar]