Abstract
Niemann Pick type C (NPC) disease is an autosomal recessive disorder characterized by the lysosomal/late endosomal (LE) accumulation of unesterified cholesterol and other lipids due to a defect in the intracellular lipid trafficking. About 95% of patients present mutations in the NPC1 gene. Among the 290 mutations reported in the NPC1 gene, about 70% are missense. However, little information is available regarding the impact of missense mutations on NPC1 protein stability and function. In this study, we in vitro characterized the pathogenic effect of 7 NPC1 missense mutations. In all cases, the basal levels of mutant NPC1 expression were reduced with respect to wild type. Treatment of fibroblasts carrying NPC1 missense mutations in homo or hemizygosity, with the proteasome inhibitor MG132 or glycerol 10%, a chemical chaperone known to stabilize misfolded proteins, resulted in a significant increase of NPC1 protein levels in all cell lines, indicating that these mutants are subjected to proteasomal degradation due to protein misfolding The increment of NPC1 mutant protein induced by the proteasome inhibitor was associated with a localization of NPC1 protein within lysosomal/LE compartment. In cell lines carrying mutations p.N1156S, p.L1191F, p.V1165M, and p.I1061T, the increment of NPC1 mutant protein resulted in an improvement of the intracellular trafficking of cholesterol and GM1. These findings showed that it is possible to correct the NPC cellular phenotype by increasing the amount of endogenous NPC1 mutated protein, suggesting that at least some NPC1 mutations might be potentially rescued by small molecules-based chaperone therapy.
Introduction
Niemann Pick type C (NPC) disease (NPC1, MIM 257220; NPC2, MIM 607625) is an autosomal recessive neurovisceral disorder with an estimated incidence of 1:150,000 live birth and characterized by the accumulation of a broad spectrum of lipids including unesterified cholesterol, glycosphingolipids (GSLs), sphingosine and sphingomyelin within the lysosomes/late endosomes (LE) due to a defect in the intracellular lipid trafficking (Patterson et al. 2001; te Vruchte et al. 2004; Lloyd-Evans et al. 2008).
Clinically, NPC disease presents a highly variable phenotype ranging from fetal to adult age. Although it encompasses a continuous spectrum of phenotypes, it has classically been classified by the age at onset of neurological symptoms in a severe infantile form (onset before 2 years of age), a late infantile form (onset between 3 and 5 years of age), a juvenile form (onset between 5 and 16 years), and an adult form (onset at age > 16 years) (Patterson et al. 2001; Vanier and Millat 2003).
Two disease-causing genes, NPC1 (MIM#607623) and NPC2 (MIM#601015), have been identified (Vanier et al. 1996; Carstea et al. 1993; Naureckiene et al. 2000). NPC1 gene, located on chromosome 18q11-q12, encodes a large membrane glycoprotein of 1,278 aminoacids containing 13 transmembrane domains and located predominantly in late endosomes (Davies and Ioannou 2000). NPC2 gene is mapped to chromosome 14q24.3 and encodes a small soluble protein present in the lumen of the lysosomes (Naureckiene et al. 2000; Vanier and Millat 2004). Although it is known that both NPC1 and NPC2 bind unesterified cholesterol and both are involved in the egress of cholesterol and other lipids from the lysosomes, the precise mechanisms by which these proteins exert this function is not clear. Recent studies have begun to clarify the roles of NPC1 and NPC2 in the lysosomal export process. It has been demonstrated that a water soluble fragment of NPC1 binds cholesterol in an orientation opposite to NPC2 (Infante et al. 2008). Based on these results, it has been postulated that, after liberation from LDL, cholesterol is bound by NPC2 which carries it to the lysosomal membrane, where it transfers to the N-terminal domain of the membrane bound NPC1 (Kwon et al. 2009).
About 95% of human NPC disease is caused by mutations in the NPC1 gene, (Carstea et al. 1997), while the other 5% is due to mutations in the NPC2 gene.
To date, more than 290 mutations of NPC1 gene have been reported (http://npc.fzk.de/) (Runz et al. 2008), most of them are present in single families. Only three relative frequent mutations have been found within distinct patient groups: the p.G992W, almost exclusively seen in Acadian patients from Nova Scotia (Greer et al. 1998), the p.P1007A found in about 15% of mutated alleles in different European populations (Ribeiro et al. 2001; Fancello et al. 2009) and the p.I1061T that accounts for 15–20% of mutated alleles in Western Europe and USA (Millat et al. 1999; Sun et al. 2001; Park et al. 2003; Millat et al. 2005). However, a study performed in 44 Italian NPC patients showed that the p.I1061T mutation is much less common in Italy, representing only a 4.7% of the NPC1 alleles (Fancello et al. 2009).
Although the NPC1 mutational profile is exceedingly heterogeneous, nearly 70% of NPC1 alleles are due to point mutations distributed throughout the coding region of the NPC1 gene and resulting in codon replacements that may affect the correct folding of the protein (Runz et al. 2008).
In fact, it has been recently demonstrated that the NPC1-I1061T protein is recognized as misfolded by the endoplasmic reticulum (ER) quality control machinery and targeted for proteasomal degradation. Transient transfection of npc1-deficient CHO cells with GFP-tagged NPC1-I1061T led to lysosomal localization of the mutant protein and functional complementation of the NPC mutant phenotype. Therefore, it has been suggested that therapeutic strategies directed to enhance the amount of mutated protein, such as the use of chemical chaperones, may rescue the pathologic phenotype in patients carrying the p.I1061T mutation (Gelsthorpe et al. 2008).
In this study, we have characterized the effect of 7 NPC1 missense mutations on protein degradation and provided evidences indicating that by increasing the amount of endogenous mutated protein it is possible to correct the cellular phenotype. Our findings suggest that some NPC1 missense mutations, other than the p.I1061T might be potentially rescued by small molecules-based chaperone therapy.
Materials and Methods
Cell Culture and Treatments
Human fibroblasts were obtained from skin biopsies from seven patients affected with NPC disease and normal controls. All NPC patients presented with the classical biochemical phenotype characterized by massive lysosomal/LE accumulation of unesterified cholesterol in cultured fibroblasts. The diagnosis was confirmed by sequencing both NPC1 and NPC2 genes. All patients had mutations in the NPC1 gene. This study was approved by the ethical committee of the University Hospital “S. Maria della Misericordia,” and written consent was obtained from all subjects.
Human fibroblasts were cultured in RPMI 1640 medium supplemented with 10% (v/v) fetal calf serum (FCS), and 50 mg/ml penicillin/streptomycin (Gibco, Paisley, UK).
Fibroblasts were treated with vehicle or 15 μM of the protease inhibitor MG132 (Sigma, St Louis, MO, USA) for 24–48 h, with 10–1,000 nM of MG132 for 10 days or with glycerol 10% for 24 h.
Mutational Analysis
Genomic DNA from NPC patients was extracted from cultured skin fibroblasts and amplification of NPC1 gene was performed as previously described (Tarugi et al. 2002). After purification, the polymerase chain reaction (PCR) fragments were sequenced in an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Warrington, UK). In all cases the presence of the mutation has been confirmed in both parents who resulted to be heterozygous carriers.
Extraction of RNA, cDNA Synthesis and PCR Analysis
Total RNA was isolated from cultured fibroblasts using Trizol reagent (EuroClone Gibco, Paisley, UK) according to the manufacturer’s instructions. RNA was quantified spectrophotometrically at 260 nm in a Beckman Coulter DU®730 spectrophotometer (Fullertone, CA, USA). The RNA purity was evaluated by measuring the ratio A260/A280, considering RNA with appropriate purity those showing values between 1.8 and 2.0; its integrity was evaluated by gel electrophoresis. Total RNA (1 μg) was reverse transcribed using iScriptTM cDNA Synthesis kit BioRad according to manufacturer’s instructions. PCR analysis was performed as previously described (Di Leo et al. 2004) using appropriate sets of primers, which allow the amplification of ten overlapping fragments covering the whole NPC1 messenger RNA (mRNA) (Tarugi et al. 2002).
Real-Time Quantitative PCR
Real-Time quantitative PCR was performed in i-Cycler IQ; 18S and GAPDH were used as housekeeping genes. All primer pairs were synthesized by Sigma Genosys Ltd. (London Road, UK) and were designed using the software Beacon Designer 7.91 (PREMIER Biosoft International, Palo Alto, CA, USA). Primer sequences and references are specified in Table 1. PCR amplification was carried out in 25 μL reaction volume containing 25 ng of cDNA, 1x iQ SYBR Green Supermix [100 mM KCl; 40 mM Tris–HCl, pH 8.4; 0.4 mM each dNTP; 50 U/mL iTaq DNA polymerase; 6 mM MgCl2; SYBR Green I; 20 nM fluorescein; and stabilizers] and gene-specific sense and anti-sense primers. Standard curves using a “calibrator” cDNA (chosen among the cDNA samples) were prepared for each target and reference gene, and the efficiency was calculated. In order to verify the specificity of the amplification, a melt-curve analysis was performed, immediately after the amplification protocol. Nonspecific products of PCR were not found in any case. The relative quantification was made using the Pfaffl modification of the ΔΔCt equation, taking into account the efficiencies of individual genes. The results were normalized to 18S and GAPDH, the initial amount of the template of each sample (treated) was determined as relative expression versus its reference sample (untreated control) which in each case was considered the 1x sample. At least three different determinations for each gene were performed.
Table 1.
Set of primers used in real-time quantitative PCR
Western Blot Analysis
Twenty micrograms of protein extracts were resolved on 8% SDS PAGE gels and transferred to nitrocellulose membranes (Schleicher and Schuell, Keene, NH, USA). After overnight blocking with 5% nonfat dry milk in PBS-Tween 0.1% (PBS-T), the membranes were probed with anti-NPC1 polyclonal antibody (Novus Biologicals, Littleton, USA) overnight at 4°C. Anti-rabbit HPR conjugated antibody was used as a secondary antibody. Immunoreactive bands were detected by enhanced chemiluminescence ECL (Amersham). The signals were normalized to those obtained for actin using a polyclonal anti-actin antibody (Sigma, St Louis, MO, USA).
Filipin Staining
Filipin staining was performed using the method described by Blanchette–Mackie et al. (Blanchette-Mackie et al. 1988). Briefly, the cells were rinsed with PBS and fixed with 3% paraformaldehyde. After washing them with PBS, the cells were incubated with 1.5 mg of glycine/ml PBS for 10 min, stained with filipin (0.05 mg/ml, in PBS 10% FCS) for 2 h and examined using a Zeiss fluorescence microscope.
Immunolocalization of NPC1 Protein
Immunolocalization of NPC1 was performed in human NPC and normal fibroblasts.
Cells were grown as described above on glass cover slips. After 72 h, the cells were fixed with 4% paraformaldheyde (Sigma, St. Louis, MO, USA) and permeabilized with 0.2% Triton X-100. The cells were then blocked with 2% BSA (Sigma, St. Louis, MO, USA) and immunolabeling was carried out using an anti-NPC1 polyclonal antibody (Novus Biologicals, Littleton, USA). The secondary antibody was a FITC-conjugated anti-rabbit IgG (DAKO, Glostrup, Denmark). For colocalization studies, a monoclonal anti-LAMP-1 (Santa Cruz Biotechnology, Santa Cruz, California, USA) was used as a primary antibody, and an alexa fluor-conjugated anti-mouse (Invitrogen, Carlsbad, CA, USA) as a secondary antibody. The glass cover slips were mounted and analyzed using a Zeiss fluorescence microscope.
GM1 Staining
Cells were grown on glass cover slips and then fixed with 3% paraformaldheyde (Sigma, St. Louis, MO, USA) for 20 min, washed with PBS and permeabilized with 0.1% Triton X-100 in 2% BSA (Sigma, St. Louis, MO, USA) for 5 min. After washing the cells with PBS, they were incubated with 3 μg/ml cholera toxin B subunity FITC conjugated (Sigma, St. Louis, MO, USA) in 0,2% BSA for 30 min at room temperature. For colocalization studies, cells were washed with PBS and stained with filipin working solution (0,05 mg/ml, in PBS 10% FCS) for 2 h at room temperature (Blanchette-Mackie et al. 1988). The glass cover slips were mounted and analyzed using a Zeiss fluorescence microscope.
Results
The genotypes of the 7 NPC patients included in this study are summarized in Table 2. Genotypes of patients 1–6 had been already established and published in a previous study (Fancello et al. 2009). Patients 2–6 presented missense mutations in homozygosity (confirmed by the presence of the mutation in both parents), while patient 1 presented a missense mutation associated with a nonsense mutation in codon 670. The truncated protein that would be translated from this allele was not detected by western blot. Therefore, we considered that all the cell lines derived from patients 1–6 expressed only one NPC1-mutant variant.
Table 2.
Genotypes of NPC patients included in the study
| Patient | Allele 1 | Allele 2 | Reference |
|---|---|---|---|
| NPC-1 | p.T1205K (c.3614C>A) | p.R607X (c.1819C>T) | Fancello et al. (2009) (patient NP10) |
| NPC-2 | p.V1023G (c.3068T>G) | p.V1023G (c.3068T>G) | Fancello et al. (2009) (patient NP4) |
| NPC-3 | p.Y1019C (c.3056A>G) | p.Y1019C (c.3056A>G) | Fancello et al. (2009) (patient NP29) |
| NPC-4 | p.N1156S (c.3467A>G) | p.N1156S (c.3467A>G) | Fancello et al. (2009) (patient NP14) |
| NPC-5 | p.L1191F (c.3571C>T) | p.L1191F (c.3571C>T) | Fancello et al. (2009) (patient NP2) |
| NPC-6 | p.I1061T (c.3182T>C) |
p.I1061T (c.3182T>C) | Fancello et al. (2009) (patient NP25) |
| NPC-7 | p.V1165M (c.3493G>A) | r.0 (c.2795+1G>C) | This study |
The lysosomal accumulation of unesterified cholesterol was demonstrated by filipin staining. All cells presented a classical biochemical phenotype characterized by massive lysosomal accumulation of unesterified cholesterol
Sequencing analysis of the entire coding region and the intronic flanking sequencing of NPC1 gene in patient 7 showed the presence of the known mutation p.V1165M (c.3493G>A) in one allele. The second allele presented a new intronic mutation c.2795+1G>C located at the 5′ donor splice site of intron 18. RT-PCR analysis of the NPC1 mRNA extracted from patient’s fibroblasts followed by sequencing of the PCR product showed that only the allele carrying the p.V1165M mutation was expressed even after 40 cycles of amplification. Based on this result, we assumed that patient 7 presents on one allele the p.V1165M mutation and on the other a splicing mutation that probably generates an unstable mRNA, which would be rapidly degraded, leading to the lack of the corresponding transcript. Therefore, we considered that the cell line derived from patient 7 also expressed only one NPC1-mutant variant.
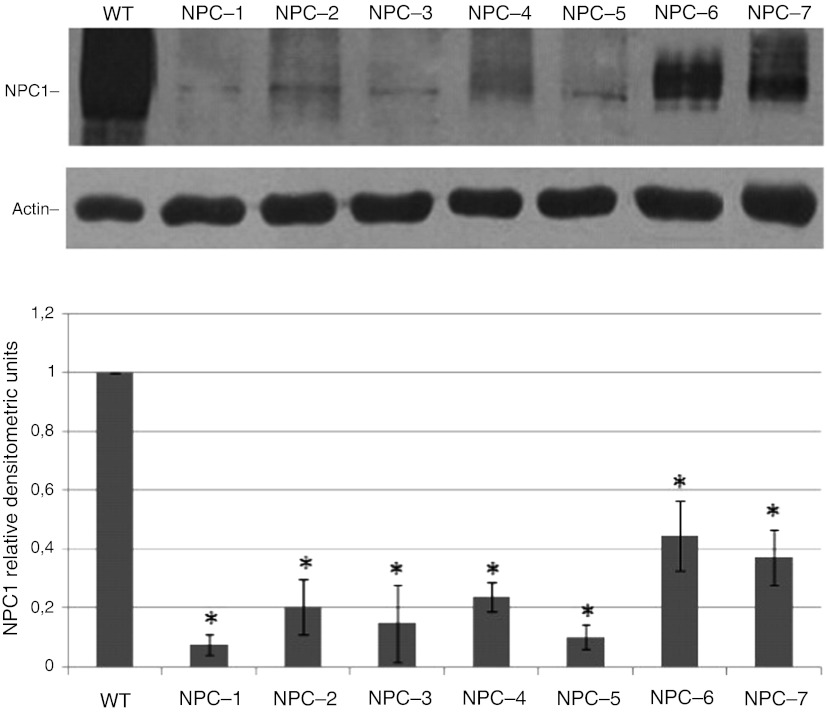
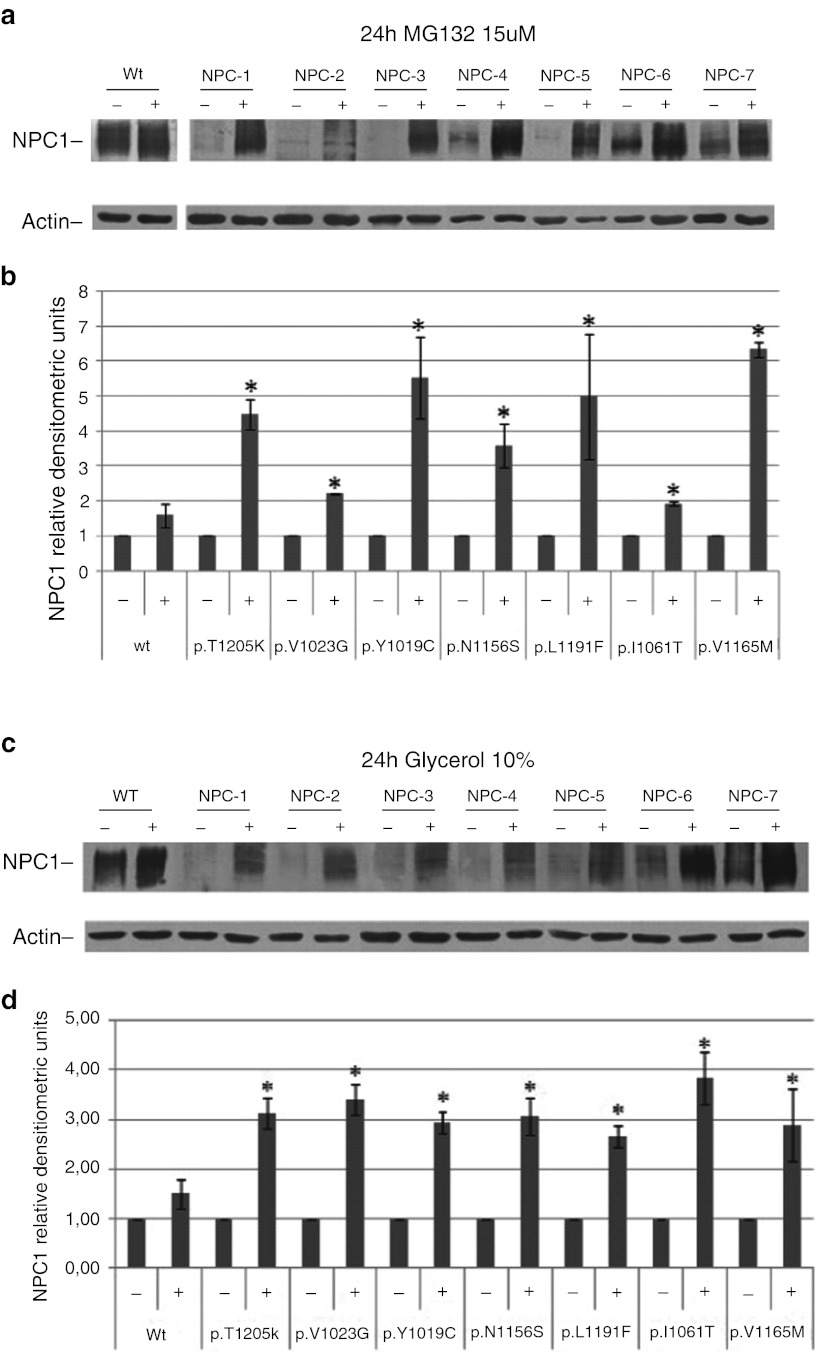
To analyze the impact of the missense mutations on protein abundance, we first analyzed the basal levels of NPC1 protein expression in fibroblasts cell lines by western blot. As shown in Fig. 1, the levels of mutant NPC1 expression were reduced in NPC fibroblasts with respect to normal controls. It has been well established, using other disease models, including lysosomal diseases, that some mutated misfolded proteins are retained in the ER, from where they are retro-translocated back to the cytosol to be eliminated by the ubiquitin–proteosomal pathway. This process is known as ER-associated degradation (ERAD) (Kopito 1997). In order to determine whether the reduction of NPC1 mutant protein is due, at least in part, to an increase in the rate of proteasomal degradation, normal and NPC fibroblast cell lines were treated for 24 h- with 15 μΜ of the proteasome inhibitor MG132 or vehicle and NPC1 protein abundance was determined by western blot (Fig. 2a). MG132 treatment resulted in a 2- to 6-fold increase of NPC1 mutant protein levels, suggesting that all these mutant variants are subjected to proteasomal degradation (Fig. 2b). An increase of wild-type NPC1 protein abundance was also observed after treatment with MG132. However, this effect was not statistically significant. In order to verify whether MG132, in addition to its effect as a proteasome inhibitor, exerts also an effect of NPC1 mRNA expression, a quantitative real-time PCR in normal and NPC fibroblasts treated with vehicle or MG132 15 μM for 24 h was performed. A slight but not statistically significant increase of NPC1 mRNA expression was observed after treatment (fold of increase with respect to nontreated cells: 2.38 ± 1.49; p = 0.07). Taken together, these data demonstrate that the increase of NPC1 protein abundance after 24 h treatment with MG132 was mainly due to an inhibition of its proteasomal degradation.
Fig. 1.
NPC1 protein abundance in NPC fibroblasts. (a) Representative western blot analysis of NPC1 protein expression in NPC and normal fibroblast cell lines. (b) The intensity of the NPC1 signals was normalized against actin. The NPC1 protein content in NPC fibroblasts was expressed as a percentage of the NPC1 protein content found in fibroblasts from a normal control. Data are means ± SD of 3 independent experiments (*p < 0.05)
Fig. 2.
NPC1 protein abundance in normal and NPC fibroblasts after treatment with MG132 15 μM or glycerol 10% for 24 h. Representative western blot analysis of NPC1 protein expression in normal and NPC fibroblasts cell lines treated with MG132 for 24 h with 15 μM (panel a) or glycerol 10% (panel c). The intensity of the NPC1 signals were normalized against actin. The NPC1 protein content in NPC1 fibroblasts was expressed as a percentage of the NPC1 protein content found in vehicle treated cells (panels b and d). Data are means ± SD of 3 independent experiments (*p < 0.05). Treatment of NPC cells with MG132 15 μM or glycerol significant increase the content of NPC1 mutant protein
In addition, NPC cells were treated with glycerol, a chemical chaperon known to stabilize misfolded proteins, for 24 h and the NPC1 protein abundance was determined. As shown in Fig. 2(c and d), glycerol treatment resulted in a significant increase of NPC1 protein levels in all cell lines. These data provide further evidence indicating that these mutants are selected to ERAD due to protein misfolding.
We then wanted to address whether the increment of the intracellular levels of endogenous mutant protein leads to the clearance of accumulated free cholesterol.
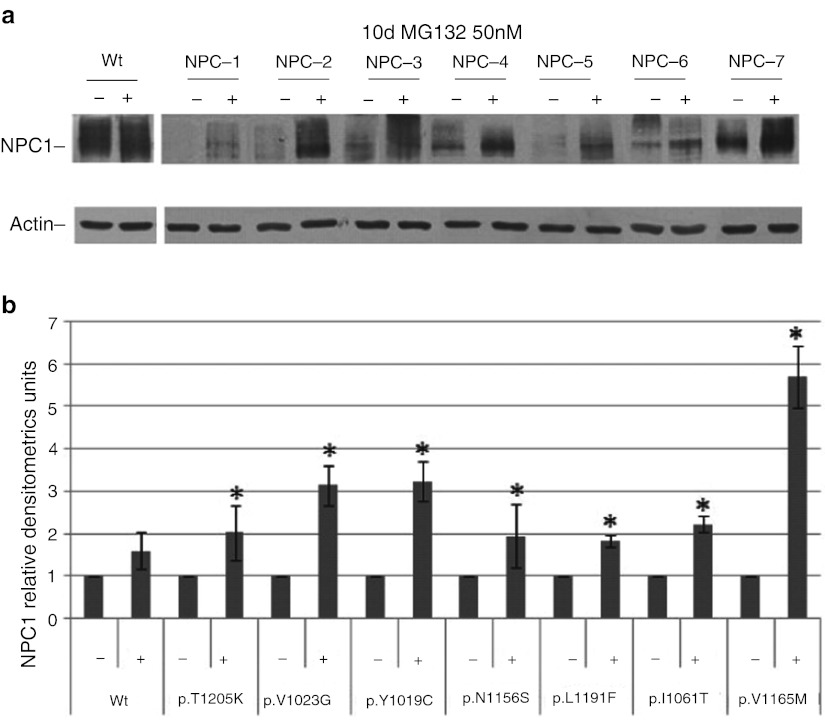
Treatment of NPC cell with 15 μΜ of MG132 for 24 h did not exert any effect on cholesterol accumulation (data not shown). However, experiments performed in our laboratory showed that NPC cells cultured in the absence of serum become negative to the filipin staining only after 3–5 days of starvation. Therefore, we hypothesized that the exposure of NPC cell to the proteasome inhibitor for 24 h may not be enough to cause a sensible reduction of the intracellular cholesterol accumulation. Since treatment of NPC fibroblasts with 15 μM of MG132 for more than 48 h resulted in a massive reduction of cell viability, NPC cells were treated with decreasing concentrations of MG132 (1,000, 500, 100, 50, and 10 nM) or vehicle for 10 days. Treatment of NPC cells with 500–1,000 nM of MG132 still resulted in a significant reduction of cell viability, while the minimum concentration of MG132 still able to cause an increase of NPC1 protein abundance was 50 nM (Fig. 3a, b).
Fig. 3.
NPC1 protein abundance in normal and NPC fibroblasts in the absence or presence of MG132 50 nM for 10 days. (a) Representative western blot analysis of NPC1 protein expression in normal and NPC fibroblasts cell lines treated with MG132 50 nM for 10 days. (b)The intensity of the NPC1 signals were normalized against actin. The NPC1 protein content in NPC1 fibroblasts was expressed as a percentage of the NPC1 protein content found in vehicle treated cells. Data are means ± SD of 3 independent experiments (*p < 0.05). Treatment of NPC cells with MG132 50 nM for 10 days significant increase the content of NPC1 mutant protein
It is worth of note that treatment of cells with MG132 50 nM for 10 days did not exert any effect on NPC1 mRNA expression, suggesting that the increase in protein abundance observed in cells treated with 50 nM for 10 days, is due to the inhibition of protein degradation.
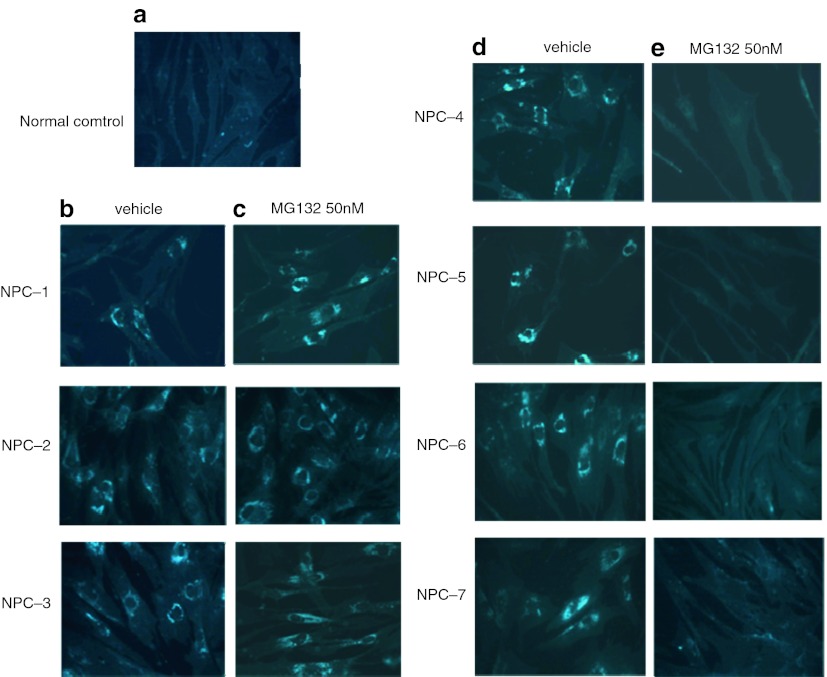
Under these experimental conditions MG132 treatment resulted in a significant reduction of cholesterol accumulation in fibroblasts cell lines carrying the p.N1156S, p.L1191F, p.V1165M, and p.I1061T NPC1 mutations. No effect on cell carrying the p.T1205K, p.V1023G and p. Y1019C was observed (Fig. 4). The same result was obtained even in cell treated with 100 nM of MG132 (data not shown).
Fig. 4.
Filipin staining of intracellular unesterified cholesterol. (a) Normal fibroblasts; (b and d) NPC fibroblasts treated for 10 days with vehicle showed a massive lysosomal accumulation of unesterified cholesterol; (c and e) NPC fibroblasts treated for 10 days with MG132 50 nM. Only in cells carrying the p.N1156S, p.L1191F, p.V1165M and p.I1061T NPC1 mutations (NPC-4, NPC-5, NPC-6, NPC-7) showed a reduction of intracellular cholesterol staining
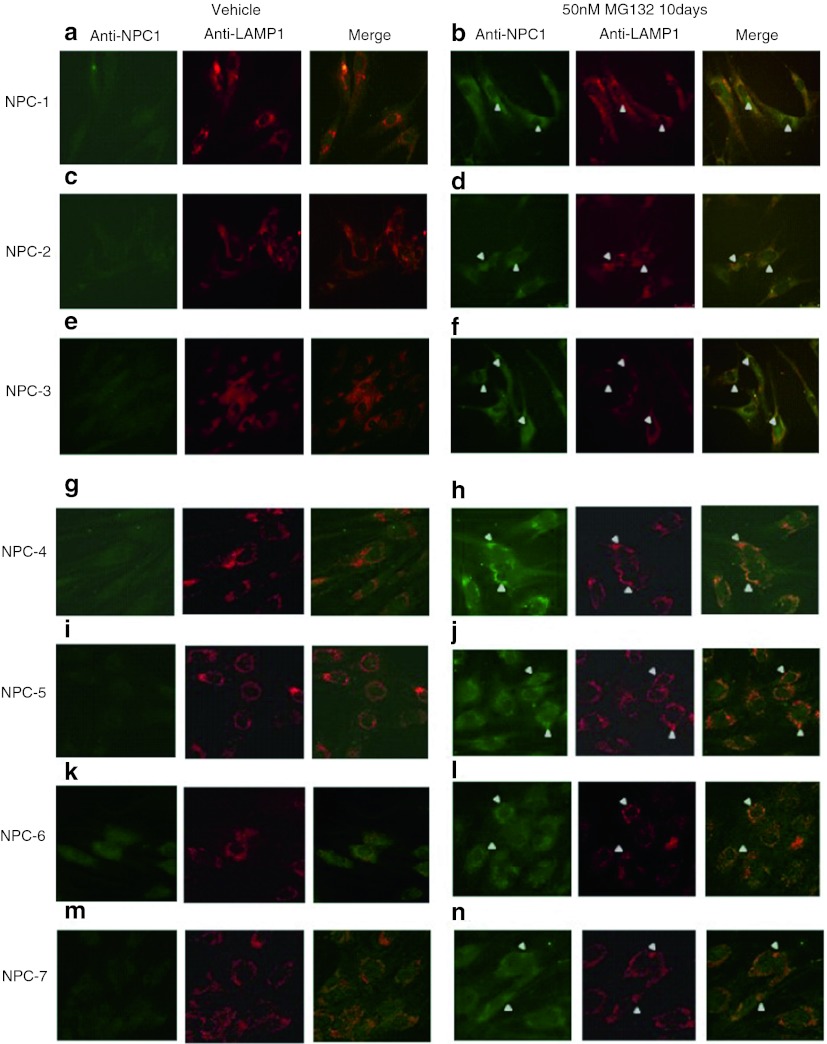
To investigate whether the reduction of cholesterol accumulation was associated with an increase of NPC1 protein within the lysosomal/LE compartment, the intracellular localization of the NPC1 mutant variants was assayed by immunofluorescensce. As shown in Fig. 5, under basal conditions the levels of mutant NPC1 protein were extremely low. Treatment of NPC cells for 10 days with low dose of MG132 resulted in a substantial increase of NPC1 mutant protein signal and a staining typical of lysosomal/LE localization, not only in fibroblasts carrying the p.N1156S, p.L1191F, p.V1165M and p.I1061T NPC1 mutations, but also in all 7 NPC studied cell lines. Furthermore, NPC1 colocalized with the lysosomal protein Lamp-1 providing further evidence that MG132 treatment improved the delivery of the NPC1 mutant to the lysosomal compartment. These data suggest that p.T1205K, p.V1023G, and p.Y1019C mutations may have an impact not only on NPC1 folding, trafficking, and degradation but also on the protein function itself.
Fig. 5.
Immunofluorescence staining of NPC1 protein in NPC fibroblast in the absence or presence of MG132. Under basal conditions negligible immunofluorescent labelling for NPC1 was observed (panels a, c, e, g, I, k, m). Treatment of NPC cells for 10 days with MG132 50 nM resulted in an increased immunofluorescence labelling for NPC1 and a colocalization with the lysosomal marker LAMP-1 (panels b, d, f, h, j, l, n)
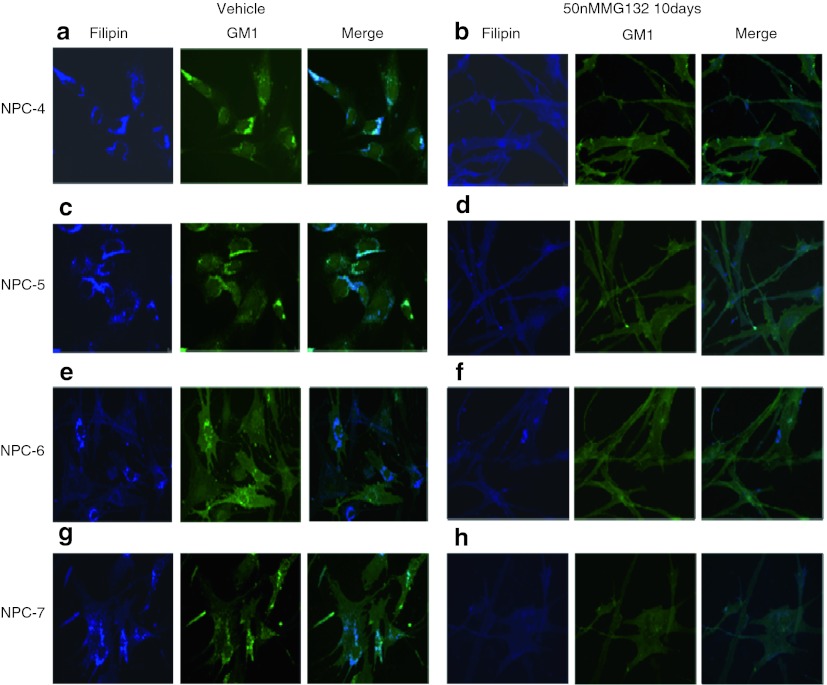
Since the storage profile in NPC cells include other lipids such as GSLs, we analyzed whether the increment of NPC1 protein results, not only in a clearance of unesterified cholesterol but also in the reduction of GM1 accumulation. Once again, treatment of NPC cell with 15 μΜ of MG132 for 24 h did not exert any effect on GM1 accumulation (data not shown). However, a significant reduction of GM1 accumulation was observed in fibroblasts cell lines carrying the p.N1156S, p.L1191F, p.V1165M, and p.I1061T NPC1 mutations after 10 days treatment with 50 nM of MG132 (Fig. 6).
Fig. 6.
Immunofluorescence staining of GM1 and filipin staining of intracellular unesterified cholesterol in “responsive” NPC fibroblasts in the absence and presence of MG132. Under basal conditions a massive accumulation of cholesterol and GM1 was observed (panels a, c, e, g). Treatment of NPC cells for 10 days with MG132 50 nM resulted in a concomitant reduction of both cholesterol and GM1 accumulation
Discussion
Loss of function diseases are often caused by the inability of a mutated protein to achieve a correct folding within the secretory pathway. In fact, it has been well documented that some mutant variants are recognized as misfolded by ER quality machinery, made up of molecular chaperones, which ensures that only properly folded proteins are secreted from the ER. Thus, these misfolded proteins are retained in the ER, from where they are retro translocated back to the cytosol to be eliminated by the ubiquitin–proteosomal pathway. This process is known as ER-associated degradation (ERAD) (Kopito 1997).
Protein misfolding has been implicated in many lysosomal storage disorders, such as Gaucher disease (Alfonso et al. 2005), Fabry disease (Fan et al. 1999), GM1 gangliosidosis (Tropak et al. 2004), and Pompe disease (Parenti et al. 2007).
In this paper we demonstrated that, besides the p.I1061T NPC mutant protein, other 6 NPC1 mutant variants are unstable and subjected to an increased rate of proteasomal degradation. In fact, treatment of NPC fibroblasts carrying different missense mutations with the proteasome inhibitor MG132 resulted in a significant increment of mutant NPC1 protein levels and an improvement of the trafficking to the lysosomal/LE compartment. In NPC fibroblasts cell lines carrying mutations p.N1156S, p.L1191F, p.V1165M, and p.I1061T, the increment of NPC1 protein content led to a reduction of lysosomal/LE free cholesterol. Furthermore, we have demonstrated that by enhancing the NPC1 protein content, it is also possible to improve the trafficking of GSLs. Our data suggest that these mutations may render the NPC1 protein unstable and rapidly degraded but they would not completely abolish the NPC1 functionality. These finding are in line with those described by Gelsthorpe et al. (Gelsthorpe et al. 2008) showing that a twofold overexpression of GFP-tagged NPC1 I1061T in transient transfected npc1-deficient CHO cells was enough to partially restore the cholesterol trafficking. These authors postulated that a percentage of 2–5% of the nascent NPC1 I1061T mutant protein would be able to fold properly, escape the ER quality control and reach the lysosome/LE compartment. In cells overexpressing the mutant protein even if only a 2–5% of the protein achieves the correct folding, the quantity of correct folded mutant molecules would be enough to complement the mutant phenotype. A similar mechanism could be postulated to explain the effect of MG132 treatment on cholesterol and GSLs trafficking in NPC fibroblasts. Indeed, in this cellular model, the increase of NPC1 protein abundance was associated with a localization of NPC1 within the lysosomal/LE compartment. Under these conditions, the amount of NPC1 protein that reached the lysosomes/LE would be enough to reduce both cholesterol and GM1 accumulation in treated cells.
It has been shown that MG132 also enhances chaperone expression levels (Bush et al. 1997; Liao et al. 2006). Therefore, it is possible to hypothesize that the effect observed in NPC fibroblasts may be due to a direct inhibition of proteasomal degradation of NPC1 protein or due to an induction of endogenous chaperons which in turn stabilize NPC1 protein preventing its proteasomal degradation. The relative contribution of these mechanisms to the effect exerted by MG132 on NPC cells needs to be further investigated.
In 3 NPC1 cells lines (NPC-1, NPC-2, and NPC-3), 10 days treatment with MG132 50 nM resulted in an increase of protein content and trafficking to the lysosomal/LE compartment. However, this treatment did not result in a reduction of free cholesterol or GM1 accumulation. These data suggest that these mutations may have an impact not only on NPC1 folding, trafficking and degradation but also on the protein function itself.
In conclusion, our data provide evidence indicating that an increased degradation of mutant NPC1 protein, probably due to protein misfolding, would be implicated in the pathogenesis of NPC1 disease. Furthermore, it has been demonstrated that by enhancing the amount of endogenous mutant NPC1 protein, it would be possible to partially restore both cholesterol and GM1 trafficking in NPC cells.
These are promising results considering that up to date, two different approaches aimed to enhance protein stability and prevent mutant protein degradation, have been successfully assessed in other models of misfolding diseases, such as cystic fibrosis disease and Gaucher: the use of pharmacological chaperons (Alfonso et al. 2005) and the use of molecules that enhance the innate cellular protein homeostasis (Wang et al. 2006; Mu et al. 2008). Although molecules that may function as pharmacological chaperones for NPC1 protein need to be identified and the usefulness of these therapeutic approaches needs to be assessed in NPC cells, our data strongly suggest that, at least some NPC1 patients may benefit from these kind of therapeutic strategies.
Acknowledgments
The authors thank Silvia Cattarossi for the technical assistance.
Some samples were obtained from the “Cell Line and DNA Biobank from Patients Affected by Genetic Diseases” (G. Gaslini Institute) – Telethon Genetic Biobank Network (Project No. GTB07001A).
Synopsis
Proteasome inhibition results in a significant increase of NPC1 mutant protein levels and an improvement of cholesterol and GM1 trafficking in Niemann Pick C cells carrying different missense mutations.
Footnotes
Competing interests: None declared.
References
- Alfonso P, Pampin S, Estrada J, Rodriguez-Rey JC, Giraldo P, Sancho J, Pocovi M. Miglustat (NB-DNJ) works as a chaperone for mutated acid beta-glucosidase in cells transfected with several Gaucher disease mutations. Blood Cells Mol Dis. 2005;35(2):268–276. doi: 10.1016/j.bcmd.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Blanchette-Mackie EJ, Dwyer NK, Amende LM, et al. Type-C Niemann-Pick disease: low density lipoprotein uptake is associated with premature cholesterol accumulation in the Golgi complex and excessive cholesterol storage in lysosomes. Proc Natl Acad Sci USA. 1988;85(21):8022–8026. doi: 10.1073/pnas.85.21.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush KT, Goldberg AL, Nigam SK. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J Biol Chem. 1997;272(14):9086–9092. doi: 10.1074/jbc.272.14.9086. [DOI] [PubMed] [Google Scholar]
- Carstea ED, Polymeropoulos MH, Parker CC, et al. Linkage of Niemann-Pick disease type C to human chromosome 18. Proc Natl Acad Sci USA. 1993;90(5):2002–2004. doi: 10.1073/pnas.90.5.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstea ED, Morris JA, Coleman KG, et al. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- Davies JP, Ioannou YA. Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J Biol Chem. 2000;275(32):24367–24374. doi: 10.1074/jbc.M002184200. [DOI] [PubMed] [Google Scholar]
- Di Leo E, Panico F, Tarugi P, Battisti C, Federico A, Calandra S. A point mutation in the lariat branch point of intron 6 of NPC1 as the cause of abnormal pre-mRNA splicing in Niemann–Pick type C disease. Hum Mutat. 2004;24:44b. doi: 10.1002/humu.9287. [DOI] [PubMed] [Google Scholar]
- Fan JQ, Ishii S, Asano N, Suzuki Y. Accelerated transport and maturation of lysosomal alpha-galactosidase A in Fabry lymphoblasts by an enzyme inhibitor. Nat Med. 1999;5(1):112–115. doi: 10.1038/4801. [DOI] [PubMed] [Google Scholar]
- Fancello T, Dardis A, Rosano C, et al. Molecular analysis of NPC1 and NPC2 gene in 34 Niemann-Pick C Italian patients: identification and structural modeling of novel mutations. Neurogenetics. 2009;10(3):229–239. doi: 10.1007/s10048-009-0175-3. [DOI] [PubMed] [Google Scholar]
- Gelsthorpe ME, Baumann N, Millard E, Gale SE, Langmade SJ, Schaffer JE, Ory DS. Niemann-Pick type C1 I1061T mutant encodes a functional protein that is selected for endoplasmic reticulum-associated degradation due to protein misfolding. J Biol Chem. 2008;283(13):8229–8236. doi: 10.1074/jbc.M708735200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer WL, Riddell DC, Gillan TL, et al. The Nova Scotia (type D) form of Niemann-Pick disease is caused by a G3097– > T transversion in NPC1. Am J Hum Genet. 1998;63(1):52–54. doi: 10.1086/301931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante RE, Radhakrishnan A, Abi-Mosleh L, et al. Purified NPC1 protein: II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J Biol Chem. 2008;283:1064–1075. doi: 10.1074/jbc.M707944200. [DOI] [PubMed] [Google Scholar]
- Kopito RR. ER quality control: the cytoplasmic connection. Cell. 1997;88(4):427–430. doi: 10.1016/S0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Abi-Mosleh L, Wang ML, Deisenhofer J, Goldstein JL, Brown MS, Infante RE. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137(7):1213–1224. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Li X, Mancini M, Chan L. Proteasome inhibition induces differential heat shock protein response but not unfolded protein response in HepG2 cells. J Cell Biochem. 2006;99(4):1085–1095. doi: 10.1002/jcb.20996. [DOI] [PubMed] [Google Scholar]
- Lloyd-Evans E, Morgan AJ, He X, Smith DA, Elliot-Smith E, Sillence DJ, Churchill GC, Schuchman EH, Galione A, Platt FM. Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat Med. 2008;14(11):1247–1255. doi: 10.1038/nm.1876. [DOI] [PubMed] [Google Scholar]
- Millat G, Marcais C, Rafi MA, et al. Niemann-Pick C1 disease: the I1061T substitution is a frequent mutant allele in patients of Western European descent and correlates with a classic juvenile phenotype. Am J Hum Genet. 1999;65:1321–1329. doi: 10.1086/302626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millat G, Bailo N, Molinero S, Rodriguez C, Chikh K, Vanier MT. Niemann-Pick C disease: use of denaturing high performance liquid chromatography for the detection of NPC1 and NPC2 genetic variations and impact on management of patients and families. Mol Genet Metab. 2005;86:220–232. doi: 10.1016/j.ymgme.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mu TW, Ong DS, Wang YJ, Balch WE, Yates JR, 3rd, Segatori L, Kelly JW. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134(5):769–781. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naureckiene S, Sleat DE, Lackland H, et al. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 2000;290(5500):2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- Parenti G, Zuppaldi A, Pittis MG, et al. Pharmacological enhancement of mutated alpha-glucosidase activity in fibroblasts from patients with Pompe disease. Mol Ther. 2007;15(3):508–514. doi: 10.1038/sj.mt.6300074. [DOI] [PubMed] [Google Scholar]
- Park WD, O’Brien JE, Lundquist PA, et al. Identification of 58 novel mutations in Niemann-Pick disease type C: correlation with biochemical phenotype and importance of PTC1-like domains in NPC1. Hum Mutat. 2003;22:313–325. doi: 10.1002/humu.10255. [DOI] [PubMed] [Google Scholar]
- Patterson M, Vanier MT, Suzuki K, et al. Niemann Pick disease type C: a lipid trafficking disorder. In: Scriver CR, Beaudet AL, WS Sly, Valle D, et al., editors. The metabolic and molecular basis of inherited diseases. New York: Mc Graw-Hill; 2001. pp. 611–634. [Google Scholar]
- Ribeiro I, Marcao A, Amaral O, Sa Miranda MC, Vanier MT, Millat G. Niemann-Pick type C disease: NPC1 mutations associated with severe and mild cellular cholesterol trafficking alterations. Hum Genet. 2001;109(1):24–32. doi: 10.1007/s004390100531. [DOI] [PubMed] [Google Scholar]
- Runz H, Dolle D, Schlitter AM, Zschocke J. NPC-db, a Niemann-Pick type C disease gene variation database. Hum Mutat. 2008;29(3):345–350. doi: 10.1002/humu.20636. [DOI] [PubMed] [Google Scholar]
- Sun X, Marks DL, Park W, et al. Niemann-Pick C variant detection by altered shingolipid trafficking and correlation with mutations within specific domain of NPC1. Am J Hum Genet. 2001;68:1361–1372. doi: 10.1086/320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarugi P, Ballarini G, Bembi B, et al. Niemann–Pick type C disease. Mutations in NPC1 gene and evidence of abnormal expression of some mutant alleles in fibroblasts. J Lipid Res. 2002;43:1908–1919. doi: 10.1194/jlr.M200203-JLR200. [DOI] [PubMed] [Google Scholar]
- te Vruchte D, Lloyd-Evans E, Veldman RJ, Neville DC, Dwek RA, Platt FM, van Blitterswijk WJ, Sillence DJ. Accumulation of glycosphingolipids in Niemann-Pick C disease disrupts endosomal transport. J Biol Chem. 2004;279:26167–26175. doi: 10.1074/jbc.M311591200. [DOI] [PubMed] [Google Scholar]
- Tropak MB, Reid SP, Guiral M, Withers SG, Mahuran D. Pharmacological enhancement of beta-hexosaminidase activity in fibroblasts from adult Tay-Sachs and Sandhoff Patients. J Biol Chem. 2004;279(14):13478–13487. doi: 10.1074/jbc.M308523200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanier MT, Millat G. Niemann-Pick disease type C. Clin Genet. 2003;64(4):269–281. doi: 10.1034/j.1399-0004.2003.00147.x. [DOI] [PubMed] [Google Scholar]
- Vanier MT, Millat G. Structure and function of the NPC2 protein. Biochim Biophys Acta. 2004;1685(1–3):14–21. doi: 10.1016/j.bbalip.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Vanier MT, Duthel S, Rodriguez-Lafrasse C, Pentchev P, Carstea ED. Genetic heterogeneity in Niemann-Pick C disease: a study using somatic cell hybridization and linkage analysis. Am J Hum Genet. 1996;58(1):118–125. [PMC free article] [PubMed] [Google Scholar]
- Wang X, Venable J, LaPointe P, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127(4):803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]