Abstract
Enzymatic loss in alkaptonuria (AKU), an autosomal recessive disorder, is caused by mutations in the homogentisate 1,2 dioxygenase (HGD) gene, which decrease or completely inactivate the function of the HGD protein to metabolize homogentisic acid (HGA). AKU shows a very low prevalence (1:100,000–250,000) in most ethnic groups, but there are countries with much higher incidence, such as Slovakia and the Dominican Republic. In this work, we report 11 novel HGD mutations identified during analysis of 36 AKU patients and 41 family members from 27 families originating from 9 different countries, mainly from Slovakia and France. In Slovak patients, we identified two additional mutations, thus a total number of HGD mutations identified in this small country is 12. In order to record AKU-causing mutations and variants of the HGD gene, we have created a HGD mutation database that is open for future submissions and is available online (http://hgddatabase.cvtisr.sk/). It is founded on the Leiden Open (source) Variation Database (LOVD) system and includes data from the original AKU database (http://www.alkaptonuria.cib.csic.es) and also all so far reported variants and AKU patients. Where available, HGD-haplotypes associated with the mutations are also presented. Currently, this database contains 148 unique variants, of which 115 are reported pathogenic mutations. It provides a valuable tool for information exchange in AKU research and care fields and certainly presents a useful data source for genotype–phenotype correlations and also for future clinical trials.
Electronic supplementary material The online version of this article (doi:10.1007/8904_2011) contains supplementary material, which is available to authorized users.
Introduction
Alkaptonuria (AKU) [MIM 203500] is an autosomal recessive disorder caused by the deficiency of homogentisate 1,2 dioxygenase [E.C.1.13.11.5] (HGD) activity (La Du et al. 1958). The enzymatic defect in AKU is caused by homozygous or compound heterozygous mutations within the HGD gene (Fernández-Cañón et al. 1996), which maps to the human chromosome 3q21–q23 (Pollak et al. 1993). This disease has a very low prevalence (1:100,000–250,000) in most ethnic groups, but it presents a remarkable allelic heterogeneity – about 96 different HGD mutations and 33 polymorphisms have already been reported (summarized in a review article Zatkova (2011) JIMD).
The HGD gene is a single-copy gene that spans 54,363 bp of genomic sequence split into 14 exons and coding for the HGD protein composed of 445 amino acids (Fernández-Cañón et al. 1996; Granadino et al. 1997).
By reexamination of the mutations and polymorphisms reported in HGD by 1999, Beltrán-Valero de Bernabé et al. showed that the “CCC” sequence motif and its inverted complement, “GGG” are preferentially mutated (Beltrán-Valero de Bernabé et al. 1999). Subsequently, nucleotide c.342+1G was also described as a mutational hotspot in HGD (Zatkova et al. 2000a). Therefore, this nucleotide and “CCC” triplets, together with CpGs, are considered to be mutational hotspots in the HGD gene.
The establishment of the crystal structure of the human HGD enzyme provided a framework for understanding the pathogenic effect of the AKU mutations. The active form of the HGD is organized as a hexameric protein, dimer of trimers: two disk-like trimers stacked base-to-base about twofold axes to form hexamers (Titus et al. 2000). Many noncovalent bonds between amino acid residues (hydrogen, salt, and hydrophobic bonds) are required to maintain the spatial structure of the monomer, of the trimer and finally of the hexamer. Thus, intersubunit interactions are important for the activity of the HGD enzyme and the complex structure of the functional HGD protein can be easily disrupted by mutations.
So far, 626 AKU patients have been reported in about 40 countries worldwide (Ranganath et al. 2011). Interestingly, countries such as Slovakia and the Dominican Republic exhibit an increased incidence in this disorder of up to 1:19,000 (Milch 1960; Srsen and Varga 1978). This increase is especially noticeable in Slovakia where 208 patients have been registered, including 110 children (Srsen et al. 2002). Ten different HGD mutations have currently been reported in Slovakia (Gehrig et al. 1997; Muller et al. 1999; Zatkova et al. 2000b; Zatkova et al. 2003; Zatkova et al. 2000c), and therefore it is difficult to explain the increased incidence of AKU in this relatively small country by a classical founder effect.
In this study, we report 11 novel HGD gene mutations identified in patients from different countries and discuss the genetic aspects of AKU in Slovakia. We inform also on a new HGD mutation database (http://hgddatabase.cvtisr.sk/) that we decided to create because of our involvement in the routine genetic screening of AKU patients sent to our laboratory from different countries and also because we saw that centralization of information regarding all reported patients with this rare error of metabolism would be useful. Additionally, the original AKU database (http://www.alkaptonuria.cib.csic.es/) located in Madrid has not been updated since 2001, and there was a need to adapt and correct the nomenclature of all known mutations according to the recommendations of the HGVS.
Material and Methods
Patients
Herein, 36 AKU patients and 41 family members from 27 families were analyzed. Of these, 18 patients were from 13 Slovak families, and the remaining 18 cases from 15 families from different countries were sent to our laboratory for mutation analysis (Supplementary Table 1).
Supplementary Table 1.
List of all patients and family members analyzed in the present study . AKU family code is indicated which enables the reader to identify members of identical family and each AKU chromosome in the HGD mutation database and in Supplementary Table 2. Old names are indicated in brackets for the mutations that have been reclassified according to the HGVS nomenclature recommendations. Relationships between the members of one family are indicated – always referring to the patient=P
| AKU family code | Status (P=patient) | Sex | Age (years) | HDG allele 1 | HDG allele 2 | Country | Notes |
|---|---|---|---|---|---|---|---|
| AKU_DB_113 | P | F | 39 | G123A | G123A | Algeria | |
| AKU_DB_113 | P | M | 28 | G123A | G123A | Algeria | |
| AKU_DB_117 | P | M | 62 | G123A | G123A | Algeria | |
| AKU_DB_118 | P | M | 78 | G123A | G123A | Algeria | |
| AKU_DB_108 | P^ | F | 57 | S287X | S287X | Algeria | Father and mother are cousins |
| AKU_DB_108 | Husband of ^ | M | 56 | L44F | wt | Algeria | |
| AKU_DB_108 | P (son of ^) | M | 23 | L44F | S287X | Algeria | |
| AKU_DB_97 | P | F | 38 | D153fs (G152fs) | A218fs | Algeria/France | |
| AKU_DB_97 | Mother | F | 58 | wt | A218fs | Algeria | |
| AKU_DB_97 | Son1 | F | 5 | wt | ? | Algeria/France | |
| AKU_DB_97 | Son2 | F | 7 | wt | ? | Algeria/France | |
| AKU_DB_96 | P | M | 52 | IVS7+2T>C | M368V | France | Father and mother are not consanguineous but come from the same region in north of France, he has affected sister |
| AKU_DB_111 | P | M | 61 | G360A | G360A | France | Parents are not consanguineous but come from the same region in south–west of France |
| AKU_DB_109 | P | F | 54 | G270R | M368V | France/Armenia | Father from Armenia, mother from France |
| AKU_DB_109 | Mother | F | 91 | wt | M368V | France | |
| AKU_DB_110 | P | M | 46 | G161R | V157fs | France/Serbia | |
| AKU_DB_110 | Father | M | 79 | G161R | wt | Serbia | |
| AKU_DB_110 | Son1 | M | 20 | G161R | wt | France/Serbia | |
| AKU_DB_110 | Son2 | M | 7 | wt | V157fs | France/Serbia | |
| AKU_DB_110 | Son3 | M | 3 | G161R | wt | France/Serbia | |
| AKU_DB_110 | Son4 | M | 16 | G161R | wt | France/Serbia | |
| AKU_DB_110 | Daughter | F | 12 | G161R | wt | France/Serbia | |
| AKU_DB_110 | Daughter | F | 9 | G161R | wt | France/Serbia | |
| AKU_DB_112 | P | F | 56 | L116P | L116P | India | Parents are not consanguineous |
| AKU_DB_99 | P | M | A122V | A122V | India/Canada | ||
| AKU_DB_107 | P | F | 52 | W60X | W60X | Italy | Parents are not consanguineous, she has affected brother |
| AKU_DB_81 | P | M | 57 | G161R | G161R | Poland | |
| AKU_DB_100 | P | M | Q33R | G152A | South Korea | ||
| AKU_DB_100 | Mother | F | wt | G152A | South Korea | ||
| AKU_DB_100 | Father | M | Q33R | wt | South Korea | ||
| AKU_DB_98 | P | F | 14 | N219S | N219S | Turkey | Parents are not consanguineous, she has affected sister |
| AKU_DB_98 | P | F | 10 | N219S | N219S | Turkey | Parents are not consanguineous, she has affected sister, also has FMF |
| AKU_DB_98 | Father | M | 45 | wt | N219S | Turkey | |
| AKU_DB_98 | Mother | F | 40 | N219S | wt | Turkey | |
| AKU_DB_98 | Brother | M | 3 | N219S | wt | Turkey | Symptom free |
| AKU_DB_98 | Sister | F | 5 | N219S | wt | Turkey | Symptom free |
| AKU_DB_101 | P | F | 10 | G161R | D153fs (G152fs) | Slovakia | |
| AKU_DB_101 | Father | M | 45 | G161R | wt | Slovakia | |
| AKU_DB_101 | Mother | F | 37 | wt | D153fs (G152fs) | Slovakia | |
| AKU_DB_101 | Brother | M | 13 | G161R | wt | Slovakia | |
| AKU_ZAT_1 | P | M | 24 | G161R | G161R | Slovakia | |
| AKU_DB_102 | P | M | 24 | G161R | V300G | Slovakia | |
| AKU_DB_102 | P | M | 9 | G161R | V300G | Slovakia | |
| AKU_DB_102 | Brother | M | 11 | wt | wt | Slovakia | |
| AKU_DB_102 | Father | M | 52 | G161R | wt | Slovakia | |
| AKU_DB_102 | Mother | F | 46 | wt | V300G | Slovakia | |
| AKU_DB_103 | P | F | 8 | H371fs (P370fs) | G125fs | Slovakia | |
| AKU_DB_103 | Grandfather | M | 58 | H371fs (P370fs) | wt | Slovakia | |
| AKU_DB_103 | Mother | F | 37 | H371fs (P370fs) | wt | Slovakia | |
| AKU_DB_103 | Sister | F | 15 | H371fs (P370fs) | wt | Slovakia | |
| AKU_DB_104 | P | F | 53 | G161R | G161R | Slovakia | |
| AKU_DB_104 | P* | M | 51 | G161R | G161R | Slovakia | |
| AKU_DB_104 | Wife of* | F | 46 | wt | G161R | Slovakia | |
| AKU_DB_104 | P (son of*)# | M | 10 | G161R | G161R | Slovakia | |
| AKU_DB_104 | Cousin of# | M | 36 | wt | G161R | Slovakia | |
| AKU_DB_104 | Wife of cousin | F | 36 | wt | G161R | Slovakia | |
| AKU_DB_104 | P (son of cousin) | M | 9 | G161R | G161R | Slovakia | |
| AKU_DB_95 | P | F | 6 | P230S | M368V | Slovakia | |
| AKU_DB_95 | P | F | 7 | P230S | M368V | Slovakia | |
| AKU_DB_105 | P | F | 31 | G161R | G161R | Slovakia | |
| AKU_DB_106 | P | F | 32 | G161R | G161R | Slovakia | She has affected brother |
| AKU_DB_93 | P | F | 27 | E178G | G161R | Slovakia | |
| AKU_DB_94 | P | F | 52 | G161R | M368V | Slovakia | |
| AKU_DB_94 | Daughter | F | 32 | wt | M368V | Slovakia | |
| AKU_DB_94 | Son | M | 27 | G161R | wt | Slovakia | |
| AKU_DB_115 | P | M | 3m | G161R | G270R | Slovakia | |
| AKU_DB_115 | Father | M | 32 | wt | G270R | Slovakia | |
| AKU_DB_115 | Mother | F | 27 | G161R | wt | Slovakia | |
| AKU_DB_127 | P | M | 11 | G161R | G161R | Slovakia | |
| AKU_DB_127 | Father | M | 39 | G161R | wt | Slovakia | |
| AKU_DB_127 | Mother | F | 40 | wt | G161R | Slovakia | |
| AKU_DB_127 | Maternal grandfather | M | 67 | G161R | wt | Slovakia | |
| AKU_DB_127 | Maternal grandmother | F | 66 | wt | G161R | Slovakia | |
| AKU_DB_127 | Paternal grandfather | M | 76 | G161R | wt | Slovakia | |
| AKU_DB_127 | Paternal grandmother | F | 70 | wt | G161R | Slovakia | |
| AKU_DB_127 | Step brother (mother side) | M | 19 | G161R | wt | Slovakia | |
| AKU_DB_128 | P | F | 52 | G161R | G161R | Slovakia | He has affected siblings |
Mutation Analysis
Diagnostic tests were employed for 10 known Slovak AKU mutations in the Slovak patients (Zatkova et al. 2003). Concurrently, in 18 foreign patients and in 3 Slovak cases where diagnostic screening revealed only one mutated allele, all individual HGD exons were sequenced using commercial sequencing kits and ABI PRISM® 3100-Avant Genetic Analyzer (primer sequences are available upon request). Mutations are described according to the Human Genome Variation Society (HGVS) nomenclature additions (den Dunnen and Antonarakis 2000). The cDNA change position is based on coding DNA Reference Sequence NM_000187.3 with the first base of the Met-codon counted as position +1.
Haplotype Analysis
In order to construct HGD-haplotypes where possible, seven single nucleotide polymorphisms were analyzed by sequencing (IVS3-112C/T (c.176-112C/T), H80Q (c.240T/A), IVS4+31A/G (c.282+31A/G), IVS5+25T/C (c.342+25T/C), IVS6+46C/A (c.434+46C/A), IVS11+18A/G (c.879+18A/G)). Additionally, three dinucleotide repeats (HGO-3 /D3S4556, HGO-1 /D3S4496, HGO-2 /D3S4497) were ascertained using PCR with fluorescently labeled primers and subsequent fragment analysis on the ABI PRISM® 3100-Avant Genetic Analyzer.
Variant Verification
For novel missense mutations, the conservation of the affected amino acid position between Homo sapiens and Mus musculus, Rattus norvegicus, Danio rerio, Drosophila melanogaster, Arabinopsis thaliana and Aspergillus nidulans was checked, using ClustalW2.
Segregation of mutations was followed in the families wherever possible.
PolyPhen-2 (Polymorphism Phenotyping v2, http://genetics.bwh.harvard.edu/pph2/) (Adzhubei et al. 2010) and SNAP (Screening for nonacceptable polymorphisms, http://cubic.bioc.columbia.edu/services/SNAP/) (Bromberg and Rost 2007) programs were used to predict the possible effect of amino acid substitutions on the structure and function of the human HGD protein (NP_000178.2). PolyPhen-2 is a tool which uses straightforward physical and comparative considerations, and for a mutation it calculates the Naïve Bayes posterior probability that this mutation is damaging and reports estimates of false positive (the chance that the mutation is classified as damaging when it is in fact nondamaging) and true positive (the chance that the mutation is classified as damaging when it is indeed damaging) rates. The mutation is also appraised qualitatively, as benign, possibly damaging, or probably damaging based on the model’s false positive rate. We used HumVar-trained PolyPhen-2, which enables distinguishment of mutations with drastic effects from all the remaining human variation, including the abundant mildly deleterious alleles.
SNAP is a neural network-based method. Its reliability index (RI) ranges between 0 and 9, with higher reliability indexes strongly correlating with a higher accuracy of prediction. The expected accuracy at a given reliability index is the number of correctly predicted neutral or nonneutral samples in the SNAP testing set. This measure of accuracy establishes the likelihood that a given prediction is correct.
The effect of the splicing mutation was predicted using Splice Site Prediction by Neural Network (http://www.fruitfly.org/seq_tools/splice.html).
Database Construction
Data for the novel HGD-mutation database was summarized based on literature review. It includes all HGD variants and AKU patients reported so far, incorporating also the data from original AKU database with the agreement of Prof. Santiago Rodríguez de Córdoba. The new database is founded on the Leiden Open (Source) Variation Database (LOVD) system (Fokkema et al. 2005), and it is located in Bratislava. Submitted data is automatically forwarded to the curator and each variant receives a unique identifier as recommended (Claustres et al. 2002). Provided that there are no publication restrictions, all new variants will be entered in the database.
Results
Mutation Analysis
In all tested cases, two AKU-causing mutations were identified; from which 11 were novel (Table 1). The HGD mutations found in all AKU patients and their family members are summarized in Supplementary Table 1.
Table 1.
Eleven novel HGD mutations identified in our cohort as well as eight novel mutations found recently in the United Kingdom. The mutational hot spot is indicated, as well as segregation in the family, conservation of amino acids and PolyPhen-2 and SNAP predictions for missense mutations (HGD protein sequence NP_000178.2). DNA numbering system is based on cDNA (NM_000187.3), with +1 corresponding to the A of the ATG
| Exon | Short name | Nucleotide change | Protein change | Hot-spot | Country of origin | AKU chromosome code | Segregation in family | Conserved amino acid | PolyPhen-2 predictions, score (HumVar) | SNAP predictions (reliability Index; expected accuracy) |
|---|---|---|---|---|---|---|---|---|---|---|
| 03 | Q33R | c.98A>G | p.(Gln33Arg) | South Korea | AKU_DB_100a | Yes (from father) | Yes (except Arabidopsis thaliana) | Possibly damaging with a score of 0.804 (sensitivity: 0.74; specificity: 0.82) | Neutral (1;60%) | |
| 03 | L44F | c.130C>T | p.(Leu44Phe) | Algeria | AKU_DB_108c | Yes (from father) | Yes (except Arabidopsis thaliana) | Possibly damaging with a score of 0.675 (sensitivity: 0.79; specificity: 0.78) | Neutral (3;78%) | |
| 04 | W60X | c.179G>A | p.(Trp60X) | Italy | AKU_DB_107a,b | |||||
| 06 | G115R | c.343G>C | p.(Gly115Arg) | “CCC” triplet | United Kingdom | AKU_DB_124a | Yes | Probably damaging with a score of 0.99 (sensitivity: 0.07; specificity: 0.99) | Non-neutral (5;87%) | |
| 06 | L116P | c.347T>C | p.(Leu116Pro) | France (Indian origin) | AKU_DB_112a,b | Yes | Probably damaging with a score of 0.995 (sensitivity: 0.33; specificity: 0.96) | Non-neutral (3;78%) | ||
| 06 | G123A | c.368G>C | p.(Gly123Ala) | Algeria | AKU_DB_113a,b | Two affected sibs | Yes | Probably damaging with a score of 0.965 (sensitivity: 0.58; specificity: 0.90) | neutral (0;53%) | |
| 07 | G152A | c.455G>C | p.(Gly152Ala) | “CCC” triplet | South Korea | AKU_DB_100b | Yes (from mother) | Yes | Probably damaging with a score of 0.980 (sensitivity: 0.52; specificity: 0.92) | Neutral (4;85%) |
| 08 | V157fs | c.469_470dupA | p.(Val157AspfsX22) | France | AKU_DB_110b | Yes (from mother) | ||||
| 08 | V157fs* | c.470-1_494del25 | p.(Val157GlufsX11) (predicted skipping of entire exon) | United Kingdom | AKU_DB_123b | |||||
| 08 | F169L | c.507T>G | p.(Phe169Leu) | United Kingdom | AKU_DB_122b | Yes (except Arabidopsis thaliana and Aspergillus nidulans) | Benign with a score of 0.103 (sensitivity: 0.92; specificity: 0.58) | Neutral (6;92%) | ||
| 08 | E178G | c.533A>G | p.(Glu178Gly) | Slovakia | AKU_DB_93b | Yes | Probably damaging with a score of 0.997 (sensitivity: 0.20; specificity: 0.98) | Non-neutral (2;70%) | ||
| 09 | R197G | c.589A>G | p.(Arg197Gly) | United Kingdom | AKU_DB_120a | Yes | Probably damaging with a score of 0.998 (sensitivity: 0.13; specificity: 0.99) | Non-neutral (3;78%) | ||
| 10 | N219S | c.656A>G | p.(Asn219Ser) | Turkey | AKU_DB_98a,b | Yes | Yes | Probably damaging with a score of 0.880 (sensitivity: 0.70; specificity: 0.84) | Non-neutral (2;70%) | |
| 11 | K276N | c.828G>C | p.(Lys276Asn) | United Kingdom | AKU_DB_121a | Yes | Probably damaging with a score of 0.995 (sensitivity: 0.33; specificity: 0.96) | Non-neutral (1;63%) | ||
| 11 | S287X | c.860C>A | p.(Ser287X) | Algeria | AKU_DB_108a,b | |||||
| 13 | G360A | c.1079G>C | p.(Gly360Ala) | “CCC” triplet | France | AKU_DB_111a,b | Yes (except Aspergillus nidulans) | Probably damaging with a score of 0.960 (sensitivity: 0.60; specificity: 0.90) | Neutral (2;69%) | |
| 13 | G361R | c.1081G>A | p.(Gly361Arg) | “CCC” triplet | United Kingdom | AKU_DB_121b, AKU_DB_126b | Yes | Probably damaging with a score of 0.998 (sensitivity: 0.13; specificity: 0.99) | Non-neutral (5;87%) | |
| 13 | D374H | c.1120G>C | p.(Asp374His) | United Kingdom | AKU_DB_125b | Yes | Probably damaging with a score of 0.987 (sensitivity: 0.46; specificity: 0.94) | Non-neutral (2;70%) | ||
| 14 | K431fs | c.1282_1292delGAG CCACTCAA | p.(Lys431HisfsX11) | United Kingdom | AKU_DB_124b |
^, #, * these are the symbols that refer to the patient from the specific family. the patients are labeled for example P^ and all his relatives are described as mother of patient ^, etc..
The conservation of amino acid positions affected by novel missense mutations is summarized in Table 1, and it can be also viewed in Supplementary Fig. 1. PolyPhen2 predicted that all but one novel missense mutations have a “possibly” or a “probably damaging effect” (Table 1). SNAP predicted Q33R, L44F, G123A, G152A, and G360A to be neutral. F169L found in the patient in United Kingdom was predicted benign by both programs.
Fig. 1.
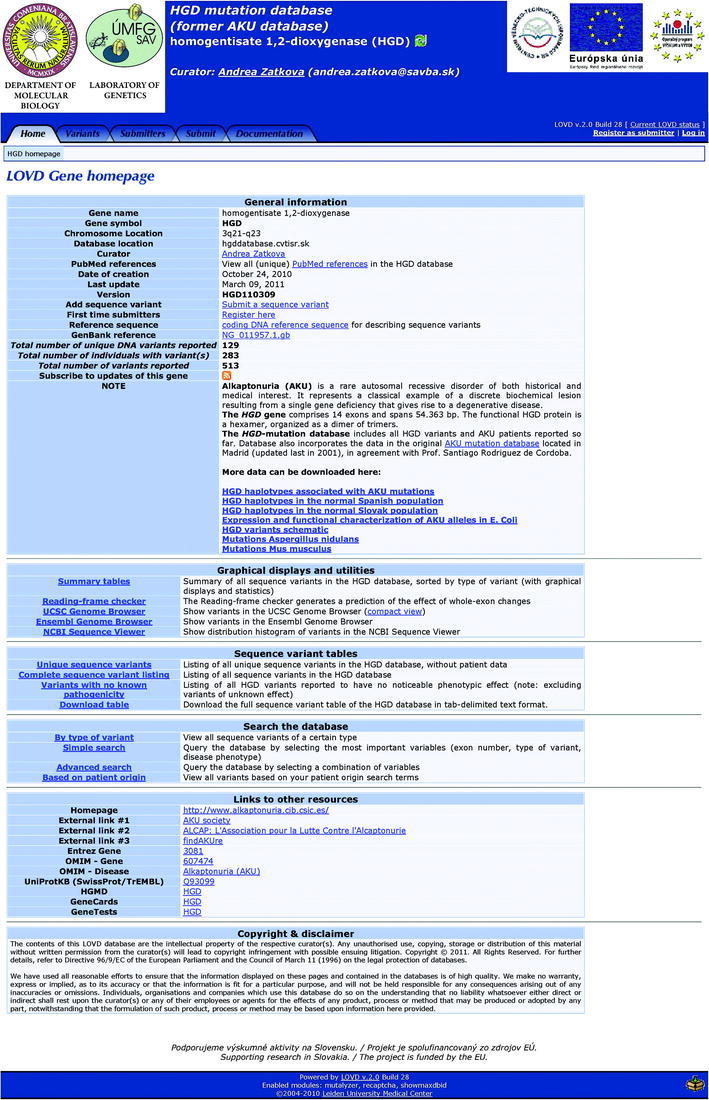
HGD mutation database home page divided into five sections with corresponding links that are described in the text
We also analyzed 136–140 control chromosomes from the Slovak population by small amplicon-based high-resolution melting (HRM) assays, for the presence of the novel mutations Q33R, L116P, G152A, E178G, and N219S (data not shown). None of the tested variants was identified among healthy individuals, which further indicates that these do not represent rare variants, but rather that they are pathogenic mutations.
Allelic associations (haplotypes) of all novel HGD mutations from this study and also all mutations currently recognized in Slovakia are summarized in Supplementary Table 2.
Supplementary Table 2.
Allelic associations of all HGD mutations found so far in Slovakia (A) and of novel mutations identified in the present study (B). All haplotypes are compared to those described so far. Differences in the haplotypes are indicated by color. The AKU allele code serves for identification of each allele in the HGD mutation database (http://hgddatabase.cvtisr.sk/). An empty box indicates that polymorphism has not been analyzed. Two different alleles in one box show that it was not possible to establish it unambiguously. The position of the mutation within the haplotype is marked by a thick black line. The short name, with the original name in brackets, is indicated for the mutations where the numbering has changed due to nomenclature recommendations. The DNA numbering system is based on cDNA (NM_000187.3), with +1 corresponding to the A of the ATG
| Exon/intron | Mutation | Short name (original name) | Nucleotide change | IVS2+35 | IVS2-218 | IVS3-112 | ex4 (c.240) | IVS4+31 | HGO-3 | HGO-1 | IVS5+25 | IVS6+46 | IVS11+18 | HGO-2 | Origin | Allele code in AKU database | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (A) | |||||||||||||||||
| 01i | Ivs1-1G>A | INV1-1G>A | c.16-1G>A | A | A | T | T | A | 201 | 161 | T | C | A | 181 | Algeria | AKU_DB_43a,b | Zatkova et al. (2000a) |
| A | A | T | T | A | 201 | 161 | T | C | A | 181 | Poland | AKU_DB_22a | Beltrán-Valero de Bernabé et al. (1999) | ||||
| A | A | T | T | A | 201 | 161 | T | C | A | 181 | Slovakia | AKU_DB_53c | Zatkova et al. (2000a) | ||||
| A | A | T | T | A | 201 | 161 | T | C | A | 181 | Slovakia | AKU_DB_54a | Zatkova et al. (2000a) | ||||
| A | A | T/C | T | A | 201/189 | 161 | T | C | A | 181/187 | Slovakia | AKU_DB_71a | Zatkova et al. (2000a) | ||||
| 03 | p.Ser47Leu | S47L | c.140C>T | A | T | T | A | A | 197 | 161 | T | C | A | 197 | Slovakia | AKU_DB_57a | Zatkova et al. (2000a) |
| 03 | p.Ser59AlafsX52 | S59fs (R58fs) | c.175delA | A | A | T | T | A | 193 | 161 | T | C | A | 181 | India | AKU_DB_49a | Zatkova et al. (2000a) |
| A | A | T | T | A | 197 | 161 | T | C | A | 181 | Finland | AKU_DB_40a,b | Beltrán-Valero de Bernabé et al. (1999) | ||||
| A | A | T | T | A | 197 | 161 | T | C | A | 181 | Slovakia | AKU_DB_66a | Zatkova et al. (2000a) | ||||
| T | T | A | 197 | 161 | T | C | A | 181 | Turkey | AKU_DB_89a,b | Uyguner et al. (2003) | ||||||
| T | T | A | 197 | 161 | T | C | A | 181 | Turkey | AKU_DB_92a,b | Uyguner et al. (2003) | ||||||
| T | T | A | 197 | 161 | T | C | A | 181/189 | Turkey | AKU_DB_90a | Uyguner et al. (2003) | ||||||
| T/C | T | A | 197/195 | 161/163 | T/C | C/A | A | 181/187 | Turkey | AKU_DB_90b | Uyguner et al. (2003) | ||||||
| 05i | IVS5+1G>A | INV5+1G>A | c.342+1G>A | A | T | T | A | A | 193 | 161 | T | C | A | 187/191 | Slovakia | AKU_DB_41a | Zatkova et al. (2000a) |
| A | A | C | T | A | 189 | 161 | T | C | G | 183 | Slovakia | AKU_DB_51a,b | Zatkova et al. (2000a) | ||||
| 07 | p.Asp153GlyfsX26 | A153fs (G152fs) | c.457dupG | T | A | C | T | A | 191 | 161 | T | C | A | 187 | France | AKU_DB_39a,b | Zatkova et al. (2000a) |
| C | T | A | 191 | 161 | T | C/A | A | 187 | France | AKU_DB_97a | Present report | ||||||
| T | A | C | T | A | 191 | 161 | T | C | A | 187 | Italy | AKU_DB_28a,b | Porfirio et al. (2000) | ||||
| A | A | C | T | A | 189 | 161 | T | C | A | 187 | Slovakia | AKU_DB_50a | Zatkova et al. (2000a) | ||||
| A | A | C | T | A | 189 | 161 | T | C | A | 187 | Slovakia | AKU_DB_51c | Zatkova et al. (2000a) | ||||
| A | A | C | T | A | 189 | 161 | T | C | A | 187 | Slovakia | AKU_DB_55a,b | Zatkova et al. (2000a) | ||||
| A | A | C | T | A | 189 | 161 | T | C | A | 187 | Slovakia | AKU_DB_58a | Zatkova et al. (2000a) | ||||
| A | A | C | T | A | 189 | 161 | T | C | A | 187 | Slovakia | AKU_DB_60a,b | Zatkova et al. (2000a) | ||||
| A | A | C | T | A | 189 | 161 | T | C | A | 187 | Slovakia | AKU_DB_61a | Zatkova et al. (2000a) | ||||
| A | A | C | T | A | 189 | 161 | T | C | A | 187 | Slovakia | AKU_DB_70a,b | Zatkova et al. (2000a) | ||||
| A | A | C/T | T | A | 189/201 | 161 | T | C | A | 187/181 | Slovakia | AKU_DB_71b | Zatkova et al. (2000a) | ||||
| C | T/A | A | 193 | 161 | T | C | A | 187 | Slovakia | AKU_DB_101a | Present report | ||||||
| C/T | T | A/G | 199 | 161 | T | C | A | 187 | Slovakia | AKU_DB_103a | Present report | ||||||
| 08 | p.Gly161Arg | G161R | c.481G>A | T | A | A | 193/191 | 161/163 | T | C/A | A | 191/187 | Serbia | AKU_DB_110a | Present report | ||
| A | T | A | A | 193 | 161 | T | C | A | 191 | Poland | AKU_DB_81a,b | Present report | |||||
| A | T | T | A/T | A | 193 | 161 | T | C | A | 191/173 | USA | AKU_DB_45b | Zatkova et al. (2000a) | ||||
| T | A/T | A | 193 | 161 | T | C | A | 191 | Slovakia | AKU_DB_101b | Present report | ||||||
| A | T | T | A | A | 193 | 161 | T | C | A | 191 | Slovakia | AKU_DB_58b | Zatkova et al. (2000a) | ||||
| T | A | A | 193 | 161 | T | C | A | 191 | Slovakia | AKU_DB_104a,b,c,d | Present report | ||||||
| A | T | T | A | A | 193 | 161 | T | C | A | 191 | Slovakia | AKU_DB_62a,b | Zatkova et al. (2000a) | ||||
| T | A | A | 193 | 161 | T | C | A | 191 | Slovakia | AKU_DB_106a,b | Present report | ||||||
| A | T | T | A | A | 193 | 161 | T | C | A | 191/187 | Slovakia | AKU_DB_41b | Zatkova et al. (2000a) | ||||
| A | T | T | A | A | 193 | 161 | T | C | A | 191 | Slovakia | AKU_DB_63a,b | Zatkova et al. (2000a) | ||||
| T/C | A/T | A | 193/189 | 161/163 | T/C | C/A | A | 191 | Slovakia | AKU_DB_93a | Present report | ||||||
| T | A | A | 193 | 161 | T | C | A | 191 | Slovakia | AKU_DB_94a | Present report | ||||||
| T | A | A | T | C | A | Slovakia | AKU_DB_127a,b | Present report | |||||||||
| C | T | A | T | C | A | Slovakia | AKU_DB_115a | Present report | |||||||||
| C | T | A | 197 | 161 | T | C | A | 191 | Slovakia | AKU_DB_105a,b | present report | ||||||
| C | T | A | 197/189 | 161 | T | C | A | 191 | Slovakia | AKU_DB_102a | Present report | ||||||
| A | A | C | T | A | 197 | 161 | T | C | A | 191 | Slovakia | AKU_DB_53d | Zatkova et al. (2000a) | ||||
| A | A | C | T | A | 197 | 161 | T | C | A | 191 | Slovakia | AKU_DB_57b | Zatkova et al. (2000a) | ||||
| A | A | C | T | A | 197 | 161 | T | C | A | 191 | Slovakia | AKU_DB_59a | Zatkova et al. (2000a) | ||||
| A | A | C | T | A | 197 | 161 | T | C | A | 191 | Slovakia | AKU_DB_61b | Zatkova et al. (2000a) | ||||
| A | A | C | T | A | 197 | 161 | T | C | A | 191 | Slovakia | AKU_DB_67a | Zatkova et al. (2000a) | ||||
| C | T | A | T | C | A | Slovakia | AKU_DB_128a,b | Present report | |||||||||
| 08 | p.Glu178Gly | E178G | c.533A>G | T/C | T/A | A | 189/193 | 163/161 | T/C | C/A | A | 185/191 | Slovakia | AKU_DB_93b | Present report | ||
| 10 | p.Pro230Ser | P230S | c.688C>T | A | A | C | T | A | 189 | 163 | C | A | A | 185 | Slovakia | AKU_DB_64a,b | Zatkova et al., (2000a) |
| C | T | A | 189/199 | 163/161 | C | A/C | A | 185/183 | Slovakia | AKU_DB_95a | Present report | ||||||
| A | A | C | T | A | 189 | 163 | C | A | A | 185 | Spain | AKU_DB_1a,b | Beltrán-Valero de Bernabé et al. (1998) | ||||
| A | A | T | A | A | 191 | 163 | C | A | A | 185 | Spain | AKU_DB_2a | Beltrán-Valero de Bernabé et al. (1998) | ||||
| T | A | C | T | A | 191 | 163 | C | A | A | 185 | Turkey | AKU_DB_3a,b | Beltrán-Valero de Bernabé et al. (1998) | ||||
| 11 | p.Gly270Arg | G270R | c.808G>A | A | T | A | A | 191 | 163 | C | A | A | 189 | Rep Dom | AKU_DB_77a,b | Goicoechea de Jorge et al. (2002) | |
| T/C | T | A | 195/193 | 161 | T | C | A | 183/189 | France/Armenia | AKU_DB_109a | Present report | ||||||
| A | A | T | T | A | 195 | 161 | T | C | A | 181 | Slovakia | AKU_DB_52a | Zatkova et al. (2000a) | ||||
| A | A | T | T | A | 195 | 161 | T | C | A | 181 | Slovakia | AKU_DB_53a | Zatkova et al. (2000a) | ||||
| A | A | T | T | A | 195 | 161 | T | C | A | 181 | Slovakia | AKU_DB_59b | Zatkova et al. (2000a) | ||||
| A | A | T | T | A | 195 | 161 | T | C | A | 181 | Slovakia | AKU_DB_61c | Zatkova et al. (2000a) | ||||
| A | A | T | T | A | 195 | 161 | T | C | A | 181 | Slovakia | AKU_DB_66b | Zatkova et al. (2000a) | ||||
| A | A | T | T | A | 195 | 161 | T | C | A | 181 | Slovakia | AKU_DB_67b | Zatkova et al. (2000a) | ||||
| T | T | A | T | C | A | Slovakia | AKU_DB_115b | Present report | |||||||||
| T | A | C | T | A | 195 | 161 | T | C | A | 187 | Italy | AKU_DB_29a,b | Porfirio et al., (2000) | ||||
| T | T | A | 201 | 161 | T | C | A | 187 | Turkey | AKU_DB_91a,b | Uyguner et al. (2003) | ||||||
| A/T | T | T | C | A | United Kingdom | AKU_DB_125a | Present report | ||||||||||
| 12 | p.Val300Gly | V300G | c.899T>G | A | A | C | T | A | 189 | 163 | C | A | A | 187 | France | AKU_DB_5a | Beltrán-Valero de Bernabé et al. (1998) |
| A | A | C | T | A | 189 | 163 | C | A | A | 187 | Germany | AKU_DB_6a,b | Beltrán-Valero de Bernabé et al. (1998) | ||||
| A | C | T | A | 189 | 163 | C | A | A | 187 | Portugal | AKU_DB_84a,b | AKU database | |||||
| A | A | C | T | A | 189 | 163 | C | A | A | 187 | Spain | AKU_DB_2b | Beltrán-Valero de Bernabé et al. (1998) | ||||
| C | T | A | 189/197 | 163 | C | A | A | 187 | Slovakia | AKU_DB_102b | present report | ||||||
| T | A | C | T | A | 191 | 161 | T | A | A | 187 | Slovakia | AKU_DB_50b | Zatkova et al. (2000a) | ||||
| 13 | p.Met368Val | M368V | c.1102A>G | A | A | T | T | A | 195 | 161 | T | C | A | 183 | Finland | AKU_DB_36a,b | Beltrán-Valero de Bernabé et al. (1999) |
| A | A | T | T | A | 195 | 161 | T | C | A | 183 | Finland | AKU_DB_38a,b | Beltrán-Valero de Bernabé et al. (1999) | ||||
| A | A | T/C | T | A | 195 | 161 | T | C/A | A | 183/181 | France | AKU_DB_33c | AKU database | ||||
| T/C | T | A | 195/189 | 161/163 | T/C | C/A | A | 183/189 | France | AKU_DB_96b | present report | ||||||
| A | A | T | T | A | 195 | 161 | T | C | A | 183 | Germany | AKU_DB_7a,b | Beltrán-Valero de Bernabé et al. (1998) | ||||
| T | T | A | 195/193 | 161 | T | C | A | 183/189 | France/Armenia | AKU_DB_109b | Present report | ||||||
| A | A | T | T | A | 195 | 161 | T | C | A | 181 | Portugal | AKU_DB_47a,b | AKU database | ||||
| A | T | T | A | 195 | 161 | T | C | A | 181 | Portugal | AKU_DB_85a,b | AKU database | |||||
| T | T | A | 195 | 161 | T | C | A | 181 | Slovakia | AKU_DB_94b | Present report | ||||||
| T | A | T | T | A | 195 | 161 | T | C | A | 181 | France | AKU_DB_5b | Beltrán-Valero de Bernabé et al. (1998) | ||||
| A | A/T | T | T/A | A | 195 | 161 | T | C | A | 181 | The Netherlands | AKU_DB_25b | Beltrán-Valero de Bernabé et al. (1999) | ||||
| A/T | A | T/C | T | A | 195 | 161 | T | C/A | A | 181/179 | USA | AKU_DB_23a | Beltrán-Valero de Bernabé et al. (1999) | ||||
| A | A | T | T | A | 195 | 161 | T | C | A | 179 | Spain | AKU_DB_24a,b | Beltrán-Valero de Bernabé et al. (1999) | ||||
| A | A | T | T | A | 199 | 161 | T | C | A | 181 | Spain | AKU_DB_18a | Beltrán-Valero de Bernabé et al. (1999) | ||||
| A | T | T | A | 193 | 161 | T | A | A | 181/179 | France | AKU_DB_82a | AKU database | |||||
| C | T | A | 199/189 | 161/163 | T | C/A | A | 183/185 | Slovakia | AKU_DB_95b | Present report | ||||||
| 13 | p.His371ProfsX4 | H371fs (P370fs) | c.1111dupC | T/C | T | G/A | 199 | 161 | T | C | A | 179 | Slovakia | AKU_DB_103b | Present report | ||
| A | A | T | T | G | 199 | 161 | T | C | A | 179 | Slovakia | AKU_DB_52b | Zatkova et al. (2000a) | ||||
| A | A | T | T | G | 199 | 161 | T | C | A | 179 | Slovakia | AKU_DB_54b | Zatkova et al. (2000a) | ||||
| A | A | T | T | G | 199 | 161 | T | C | A | 179 | Slovakia | AKU_DB_69a,b | Zatkova et al. (2000a) | ||||
| A | A | C | T | A | 189 | 161 | T | C | A | 179 | Slovakia | AKU_DB_53b | Zatkova et al. (2000a) | ||||
| (B) | |||||||||||||||||
| 03 | p.Glu33Arg | Q33R | c.98G>A | C | T | A | 191 | 161 | T | C | A | 181 | Korea | AKU_DB_100a | Present report | ||
| 03 | p.Leu44Phe | L44F | c.130C>T | C | T | A | T | A | A | Algeria | AKU_DB_108c | Present report | |||||
| 04 | p.Trp60X | W60X | c.179G>A | T | A | A | 191 | 161 | T | C | A | 189 | Italy | AKU_DB_107a,b | Present report | ||
| 06 | p.Gly115Arg | G115R | c.343G>C | A | T/A | A | T | C | A/G | United Kingdom | AKU_DB_124a | Present report | |||||
| United Kingdom | AKU_DB_126a | Present report | |||||||||||||||
| 06 | p.Leu116Pro | L116P | c.347T>C | T | T | G | 191 | 161 | T | A | A | 183 | France (Indian origin) | AKU_DB_112a,b | Present report | ||
| 06 | p.Cys120Phe | C120F | c.359G>T | A | T | A | T/C | A | A | United Kingdom | AKU_DB_120a | Present report | |||||
| 06 | p.Ala122Val | A122V | c.365C>T | T | T | A | 197 | 161 | T | C | A | 191 | Canada (Indian origin) | AKU_DB_99a,b | Present report | ||
| A | A | T | T | A | 193 | 161 | T | C | A | 185 | India | AKU_DB_49b | AKU database | ||||
| 06 | p.Gly123Ala | G123A | c.368G>C | T | A | A | T | C | A | Algeria | AKU_DB_113a,b | Present report | |||||
| T | A | A | T | C | A | Algeria | AKU_DB_117a,b | Present report | |||||||||
| T | A | A | T | C | A | Algeria | AKU_DB_118a,b | Present report | |||||||||
| 07 | p.Gly152Ala | G152A | c.455G>C | C | T | A | 191 | 161 | T | A | A | 179 | Korea | AKU_DB_100b | Present report | ||
| 07i | IVS7+2T>C | INV7+2T>C | c.469+2T>C | T/C | T | A | 189/195 | 163/161 | C/T | C/A | A | 189/183 | France | AKU_DB_96a | Present report | ||
| A | T | A/G | C/T | C/A | A | United Kingdom | AKU_DB_122a | Present report | |||||||||
| 08 | p.Val157GlufsX11 | V157fs | c.470-1_494del25 | A/T | T | A | T | C | A | United Kingdom | AKU_DB_123b | Present report | |||||
| 08 | p.Val157AspfsX22 | V157fs | c.470-1_470insA | C | T/A | A | 191/193 | 163/161 | C | A/C | A | 187/191 | France | AKU_DB_110b | Present report | ||
| 08 | p.Pro158Leu | P158L | c.473C>T | C | T | A | T | A | A | Macedonia | AKU_DB_116a | Present report | |||||
| 08 | p.Phe169Leu | F169L | c.507T>G | A | T | A/G | T/C | C/A | A | United Kingdom | AKU_DB_122b | Present report | |||||
| 09 | p.Arg197Gly | R197G | c.589A>G | A | T | A | T/C | A | A | United Kingdom | AKU_DB_120b | Present report | |||||
| 10 | p.Ala218ProfsX11 | A218fs (G217fs) | c.652delG | A | T | T | A | 191 | 161 | C | A | A | 179 | Algeria | AKU_DB_83a | AKU database | |
| C | T | A | 191 | 161 | T | C/A | A | 181 | Algeria | AKU_DB_97b | Present report | ||||||
| 10 | p.Asn219Ser | N219S | c.656A>G | C | T | A | 191 | 161 | T | A | A | 187 | Turkey | AKU_DB_98a,b | Present report | ||
| 11 | p.Pro274Leu | P274L | c.821C>T | T | T | A | C | A | A | Macedonia | AKU_DB_116b | Present report | |||||
| 11 | p.Lys276Asn | K276N | c.828G>C | A | T | A | T | A | A | United Kingdom | AKU_DB_121a | Present report | |||||
| 11 | p.Ser287X | S287X | c.860C>A | T | A | A | 195 | 163 | C | A | A | 201 | Algeria | AKU_DB_108a,b | Present report | ||
| 13 | p.Gly360Ala | G360A | c.1079G>C | C | T | A | 193 | T | C | A | 197 | France | AKU_DB_111a,b | Present report | |||
| 13 | p.Gly361Arg | G361R | c.1081G>A | A | T | A | T | A | A | United Kingdom | AKU_DB_121b | Present report | |||||
| United Kingdom | AKU_DB_126b | Present report | |||||||||||||||
| 13 | p.Asp374His | D374H | c.1120G>C | A/T | T | T | C | A | United Kingdom | AKU_DB_125b | Present report | ||||||
| 14 | p.Lys431HisfsX11 | K431fs | c.1282_1292 delGAGCCACTCAA | A | T/A | A | T | C | A/G | United Kingdom | AKU_DB_121a | Present report | |||||
Described herein is the first AKU patient from South Korea who is a compound heterozygote for two novel missense mutations, Q33R (c.98A>G) and G152A (c.455G>C). Additionally, one novel haplotype associated with exon 10 mutation A218fs was described in an Algerian patient, and another associated with an A122V mutation was found in a patient from India (Supplementary Table 2). Accordingly, differences in haplotypes may indicate recurrent mutational events in these patients.
A total number of 12 different AKU mutations were established in Slovakia in 104 AKU chromosomes from 50 families currently appearing in the literature and in this report, and this further underscores the allele heterogeneity of AKU in this country (Table 2). Individual allele frequencies observed in Slovakia were also compared with those observed elsewhere.
Table 2.
Summary of 12 HGD mutations identified in 50 Slovak families with Slovak origin (104 AKU chromosomes). Mutational hotspots are indicated. Allele frequencies in Slovakia are compared to those found in an additional 422 AKU chromosomes with mutations identified reported worldwide so far. References for all reported patients carrying specific mutation can be found in Supplementary Table 2. In the column “Where reported” the country of the first reported patient carrying relevant mutation is listed first. Numbers in brackets indicate the numbers of AKU chromosomes reported. Shaded in gray are mutations that most likely have their origin in Slovakia. The DNA numbering system is based on cDNA (NM_000187.3), with +1 corresponding to the A of the ATG. All 526 AKU chromosomes are reported in HGD mutation database (http://hgddatabase.cvtisr.sk/)
| HGD exon/intron | Short name (original description) | Nucleotide change (ATG=+1) | Protein change | Hot-spot | # of Slovak AKU chr. | % of 104 Slovak AKU chr. | # of AKU chr. in other countries | % of 422 AKU chr. from other countries | Where reported |
|---|---|---|---|---|---|---|---|---|---|
| 01i | IVS1-1G>A | c.16-1G>A | p.(Tyr6_Gln29del) | 5 | 4.8% | 7 | 1.7% | Poland, Algeria(2), Slovakia(5), Czech(2), USA(2) | |
| 03 | S47L | c.140C>T | p.(Ser47Leu) | CpG | 1 | 1.0% | 0 | 0.0% | Slovakia |
| 03 | S59fs (R58fs) | c.174_175delA | p.(Ser59AlafsX52) | 2 | 1.9% | 25 | 5.9% | Finland(2), India, Slovakia(2), Turkey(6), UAE(2), USA(11), La Reunion, UK(2) | |
| 05i | IVS5+1G>A | c.342+1G>A | p.(Leu95_Ser114del) | c.342+1 | 3 | 2.9% | 2 | 0.5% | Slovakia (3), Czech. rep, USA |
| 07 | D153fs (G152fs) | c.454_457insG | p.(Asp153GlyfsX26) | “CCC” triplet | 15 | 14.4% | 8 | 1.9% | Slovakia (15), Italy(2), USA(3), France(3) |
| 08 | G161R | c.481G>A | p.(Gly161Arg) | “CCC” triplet | 46 | 44.2% | 23 | 5.5% | Slovakia(44), Czech(4), Germany, USA(11), Slovakia/Hungary(2), Poland(2), France/Serbia, UK(4) |
| 08 | E178G | c.533A>G | p.(Glu178Gly) | 1 | 1.0% | 0 | 0% | Slovakia | |
| 10 | P230S | c.688C>T | p.(Pro230Ser) | “CCC” triplet | 5 | 4.8% | 9 | 2.1% | Spain(3), Turkey(2), Slovakia(5), USA(2), Canary Islands(2) |
| 11 | G270R | c.808G>A | p.(Gly270Arg) | “CCC” triplet, CpG | 8 | 7.7% | 10 | 2.5% | Italy(2), Slovakia(8), DomRep(2), Turkey(2), France/Armenia, USA(2), UK |
| 12 | V300G | c.899T>G | p.(Val300Gly) | 4 | 3.8% | 14 | 3.5% | France, Spain, Germany(2), Slovakia(4), Portugal(2), USA(4), La Reunion(3), UK | |
| 13 | M368V | c.1102A>G | p.(Met368Val) | 2 | 1.9% | 57 | 14.2% | Germany(9), France(4), France/Armenia, USA(28), The Netherlands, Finland(4), Portugal(4), Spain(3), Slovakia(2), Switzerland/Belgium, UK(2) | |
| 13 | H371fs (P370fs) | c.1111insC | p.(His371ProfsX4) | “CCC” triplet | 12 | 11.5% | 2 | 0.5% | Slovakia (12), USA(2) |
The HGD Mutation Database: Structure and Content
General Information
The database homepage (Fig. 1) contains the main section in which gene and database “General information” are summarized; e.g., the chromosomal and database location, the curator name and gene reference sequence that can be also downloaded. Using links in the NOTE, all users can access schematic drawings showing the location of the pathogenic variants and polymorphisms within the gene (“HGD variants schematic”), tables summarizing about 240 so far established and published HGD-haplotypes (“HGD haplotypes associated with AKU mutations”), as well as “HGD haplotypes in the normal Spanish and Slovak population”, “Expression and functional characterization of AKU alleles in E. Coli”, “Mutations Aspergillus nidulans” and “Mutations Mus musculus”. Part of this data has already been previously published and/or it was located in the original AKU database (Rodríguez et al. 2000; Zatkova et al. 2000a). In case of so far unpublished mutations, in the figure “HGD variants schematic” only the type of mutation is indicated. HGD haplotypes associated with AKU mutations were constructed based on the analysis of seven single nucleotide polymorphisms and three dinucleotide repeats as reported before (Beltrán-Valero de Bernabé et al. 1999; Zatkova et al. 2000a). Comparison of the haplotypes associated with the same mutations in the AKU patients from different countries enables studying the possible origin of each mutation as well as uncovering possible novel mutational events. General information section contains also links for registration and submission of the variants.
Graphic Displays and Utilities
“Graphic displays and utilities” include links to summary tables, UCSC, and Ensembl genome browsers and NCBI sequence viewer.
Sequence Variant Tables
The HGD mutation database currently in March 2011 contains 148 unique variants; of which 115 were reported to be pathogenic (107 are public at the present) occurring independently in 267 AKU families. It includes also 33 variants which were reported as nondisease-related polymorphisms either in the original AKU database or recently (Vilboux et al. 2009). The pathogenic mutations are distributed as follows: 77 missense, 7 nonsense (stop), 14 small deletions and insertions causing frameshift, 14 affecting splicing, 2 larger deletions, and 1 extension of the protein. For more statistics, see the review article Zatkova 2011 JIMD.
The “Sequence variant tables” section includes variant data set out in tables accessible via hyperlinks. In the “Unique sequence variant table,” the unique mutations are sorted by exon number without patient data being shown (Fig. 2); in the “Complete sequence variant table,” mutations associated with each patient are described in detail.
Fig. 2.
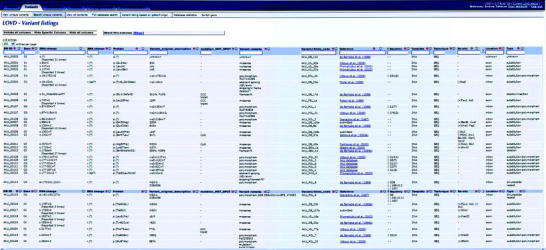
Unique variants view from HGD mutation database. All main columns described in the text are shown
Mutations are named according to the Human Genome Variation Society (HGVS) nomenclature additions (den Dunnen and Antonarakis 2000) and identified by a unique database ID. The cDNA change position is based on coding DNA Reference Sequence NCBI: NM_000187.3 with the first base of the Met-codon counted as position +1. All available data on each mutation is provided. In addition to standard database categories, an Allele code is included, which enables identification of all alleles from the same patients (i.e., patient with the Patient_ID AKU_AQR_11 has alleles AKU_AQR_11a and AKU_AQR_11b). The Database Identifier: AKU_00000 is used for all AKU patients alleles where no mutation was identified so far (unknown). Patients for whom HGD haplotypes associated with AKU mutations are reported can be recognized by their ID which begins with “AKU_DB_”.
In the database was included a column indicating an involvement of the mutation hot-spots (“CCC”triplets, c.342+1G, CpG) that have been identified within HGD gene (Beltrán-Valero de Bernabé et al. 1999; Zatkova et al. 2000a). A creation or abolition of recognition site of some common restriction enzymes is included for easy identification of each mutation (Fig. 2).
In the “Variants with no known pathogenicity table,” all reported polymorphisms are shown. There are also the two variants ivs9-56G>A and ivs9-17G>A listed here. Although these were published as AKU causing mutations (Beltrán-Valero de Bernabé et al. 1998), Vilboux et al. (2009) considered that they most likely represent benign variants based on the negative predictions of their effect on splicing. Predictions of the potential effect of most of the reported missense and splicing mutations have recently been discussed and can be found in the supplementary material of (Vilboux et al. 2009) The full sequence variant table of the HGD database can be downloaded in tab-delimited text format.
Search the Database
The home page also provides a “Search the database” section for browsing data using simple search by type of variant and exon number, or search based on the patient’s origin. Through a more advanced search tool, the user can also mine data using sequence variation description, protein description or reference.
Links to the Other Resources
Furthermore, “Links to the other resources” include links to the three gene-related resources of MIM (http://www.ncbi.nlm.nih.gov/omim), HGMD (http://www.hgmd.cf.ac.uk) and Entrez (http://www.ncbi.nlm.nih.gov/Entrez). Connections to the websites of the AKU Society (http://www.alkaptonuria.info), French ALCAP (http://www.alcap.fr/), Italian AIMAKU (http://www.aimaku.it/) and findAKUre project (http://www.findakure.org) are also available. AKU Societies are support networks for AKU patients that provide them with the best information about the latest news, research and treatments of AKU, while the FindAKUre project is a joint collaborative research project of the AKU Society and the University of Liverpool.
Discussion
In this research, we identified 11 novel AKU causing mutations and we report on our new HGD gene mutation database. Recently, 8 novel mutations were identified in 21 AKU patients from the United Kingdom, which will be published soon (listed in Table 1), bringing the total number of known HGD mutations to 115 worldwide.
Since no functional studies were available, we used PolyPhen2 and SNAP analysis in order to validate the effect of novel missense variants. While PolyPhen2 predicted that all but one novel missense mutations have a “possibly” or a “probably damaging effect”, SNAP predicted Q33R, L44F, G123A, G152A, and G360A to be neutral. F169L found in one AKU patient was predicted benign by both programs. However, this and other novel mutations identified in United Kingdom will be reported separately.
In general, the performances of the prediction tools are estimated between 50% and 80% accurate (Bromberg and Rost 2007; Ng and Henikoff 2006). It is known that the HGD protein functions as a hexamer composed of two trimers (Titus et al. 2000). Although both PolyPhen-2 and SNAP programs use 3D protein structures, they have their limitation in considering the complexity of all inter-subunit interactions between the HGD monomers within the complex hexamer that can be easily affected by single residue change (Rodríguez et al. 2000). It is possible that amino acid substitutions, which would be benign if HGD functioned as a monomer, show deleterious effects due to disturbance to the higher organization of the functional hexamer. We presume that the same holds true for novel mutations predicted as neutral by SNAP. Evidence that all exons of the HGD gene with neighboring intronic sequences have been sequenced in the patients carrying the above-mentioned mutations, and no other pathogenic changes have been identified, also favors the pathogenic effect of these variants. Moreover, amino acids affected by substitutions are highly conserved among species, and mutations segregate in the families. Functional studies, which unfortunately are not currently available, would be required to confirm the functional consequences of these mutations.
Recently, also (Vilboux et al. 2009) assessed the potential effect of all missense variations on protein function; thus, their study and this report, together with the novel HGD mutation database, provide a valuable resource of complete information on the molecular analysis of AKU mutations, their origin and their possible effect on HGD function.
Slovak Aku Genetic Specificities
In Slovakia, a total number of 12 different AKU mutations have been established. This further underscores the allele heterogeneity of AKU in this country.
As already mentioned, the most frequently found were missense mutations, followed by splicing and/or frameshift mutations (Zatkova (2011) JIMD). Although distribution in Slovakia is similar, Slovak patients had more than twice the proportion of frameshift mutations. But this might just reflect the small mutation number and founder effect in this country.
In the previous study by (Zatkova et al. 2000a, b), and also herein, an allelic association was performed for 11 HGD intragenic polymorphisms in a total of 69 AKU chromosomes from 32 Slovak pedigrees. This was then compared to the HGD haplotypes of all AKU chromosomes carrying identical mutations characterized thus far in non-Slovak patients to study the possible origin of these mutations. Based on the analysis and comparison of haplotypes, two groups of HGD mutations were observed in Slovakia.
In the first group are the mutations such as P230S, V300G, S59fs (R58fs), M368V, and IVS1-1G>A which were shared by different populations. These mutations represent only 18/104, which accounts for 17.3% of the Slovak AKU chromosomes and thus provides a marginal contribution to the AKU gene pool in Slovakia. The most frequent European mutation M368V is present in one copy in only two unrelated Slovak families. Mutations of this group have most likely been introduced into Slovakia by the founder populations that spread throughout Europe (Zatkova et al. 2000a).
The second group consists of the remaining seven mutations established in 82.7% of Slovak patients. These include the most prevalent G161R, H371fs (P370fs), D153fs (G152fs), and G270R (Table 2), the splicing mutations IVS5+1G>A, and also the S47L and E178G mutations observed in only one patient and specific for Slovakia.
The Exon 8 mutation, G161R, is the most frequent, and it is found in 46 of 104 Slovak AKU chromosomes (44.2%). Four haplotypes, two of which are prevalent, have so far been shown to be associated with this mutation in Slovakia, exhibiting the differences in the 5′part (HGO-3 and distal polymorphisms), which can be explained by novel mutations events or by recombination (Supplementary Table 2). Patients with G161R reported in the USA, Poland, and France/Serbia share the same haplotype described in Slovakia. Mutation G270R in exon 11 was also found in Italy, Turkey, The Dominican Republic, and France/Armenia. The G270R-associated haplotypes in all these countries, except for France/Armenia, differ from Slovak ones in both the 5′and 3′ parts, indicating either recurrent mutation events in these countries or a high recombination rate. A similar situation is observed in the D153fs (G152fs) mutation, which is also seen in cases in Italy, France, and France/Algeria. The difference in haplotypes in these cases is, however, restricted to the 5′end and it can be explained by recombination (Supplementary Table 2).
The IVS5+1G>A mutation is present on two different haplotypes in Slovakia, indicating recurrent mutation (Zatkova et al. 2000a). This mutation was also found in one case in both the USA and The Czech Republic, but since no haplotypes have been described they cannot be compared.
In one out of five patients, the HGD haplotype associated with H371fs (P370fs) mutation differs from the remaining ones in the distal 5′end, but this can be explained by recombination (Supplementary Table 2). Although this mutation was also identified in two patients in the USA, it otherwise appears to be specific for Slovakia.
It is likely that mutations from this second group originated in Slovakia and spread into other countries with different migrations.
The distribution of the identified mutations within Slovak territory is also interesting. As previously reported, examination of the geographical origin of Slovak AKU mutations shows remarkable clustering in a small area in North-West Slovakia, with these mutations most likely originating in this area and spreading into other regions after the breakdown of genetic isolates in the 1950s (Zatkova et al. 2000a).
Sequence analysis shows that six of the seven prevalent Slovak AKU mutations are associated with hypermutated sequences in the HGD (“CCC” triplet, c.342+1, CpG; Table (1). In addition, as the haplotype analysis shows, one of the P230S, M368V and V300G alleles in Slovak patients may also represent a novel HGD mutational event (haplotypes show differences, Supplementary Table 2). Thus, 7 of the 12 (58.3%) AKU mutations which most likely originated in Slovakia are associated with hyper-mutated sequences in the HGD while worldwide it is 40/115 (34.8%) (HGD mutation database). Therefore, it is possible that an increased mutation rate in the HGD gene in a small geographical region is responsible for the high genetic heterogeneity in Slovak AKU (Zatkova et al. 2000b). However, it remains unclear which mechanism acted specifically on the HGD gene to increase its mutation rate, since similar targets are also present in other genes without evident elevated gene frequency in Slovakia (Srsen et al. 2002; Zatkova et al. 2000a).
It has been discussed that the Valachian colonization during the fourteenth to seventeenth centuries may also have played a role in the increased prevalence of AKU in Slovakia (Srsen et al. 2002; Zatkova et al. 2000a). Valachs were nomadic tribes who did not represent an ethnically defined group since they always mixed with the local populations. They came to Slovakia from The Balkans on the Carpathian Mountain curve through Romania and West Ukraine. No AKU cases from these countries have been reported so far, except for one recent young patient from Macedonia who, however, carries different mutations (P158L, P274L) (Gucev et al. 2011).
The increased number of mutations could also be the result of random accumulation of mutations in the region. The preservation of the most prevalent AKU variants in Slovakia may then be the result of a founder effect and genetic drift, due to the geographic isolation of villages in North-West Slovakia.
Perspectives
Since also some external factors, such as the use of minocycline for treatment of dermatologic or rheumatologic disorders may mimic AKU phenotype (Vilboux et al. 2009), identification of two HGD mutations represents the final confirmation of AKU diagnosis. Distribution on mutations and studying their haplotype background can contribute also to the understanding the genetics of the studied population. The presented HGD mutation database provides a valuable tool for information exchange in AKU research and care fields. It certainly presents a useful data source for genotype–phenotype correlations and also for future clinical trials.
Acknowledgment
We thank Dr. Klara Srsnova for help in summarizing information concerning Slovak AKU patients. We also thank Prof. Santiago Rodríguez de Córdoba for his initial help, and advice, and especially for making available original data from the AKU database. We highly appreciate Jacoppo Celli (Department of Human Genetics, Center for Human and Clinical Genetics, Leiden University Medical Center, Leiden, The Netherlands) and Jana Pigosova (Slovak Centre of Scientific and Technical Information (SCSTI), Bratislava, Slovakia) for their expertise in database construction and installation.
Details of the Contributions of Individual Authors
AZ performed majority of work, including the analysis of patients, haplotype constructions, database construction and writing the manuscript. TS, MN, and HP contributed to the mutation analysis, JR contributed to the CA-repeat analysis for haplotypes, RA, ID provided patients DNA. JLU performed mutation analysis in patients from United Kingdom. All authors approved the content of the final version of the manuscript.
Funding
This research was funded by IMPG SAS and FNS UK Bratislava, Slovakia and the project “Infrastructure for research and development- data center for research and development” with the financial support of European fund for regional development (project code: 26210120001, 26230120001).
Ethics Approval
No special ethic approval was needed. All patients signed informed consent for DNA analysis prior to a peripheral blood sample was taken from them.
Footnotes
Competing interests: None declared.
References
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrán-Valero de Bernabé D, Granadino B, Chiarelli I, et al. Mutation and polymorphism analysis of the human homogentisate 1, 2-dioxygenase gene in alkaptonuria patients. Am J Hum Genet. 1998;62:776–784. doi: 10.1086/301805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrán-Valero de Bernabé D, Jimenez FJ, Aquaron R, Rodríguez de Córdoba S. Analysis of alkaptonuria (aku) mutations and polymorphisms reveals that the ccc sequence motif is a mutational hot spot in the homogentisate 1,2 dioxygenase gene (hgo) Am J Hum Genet. 1999;64:1316–1322. doi: 10.1086/302376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, Rost B. Snap: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claustres M, Horaitis O, Vanevski M, Cotton RG. Time for a unified system of mutation description and reporting: a review of locus-specific mutation databases. Genome Res. 2002;12:680–688. doi: 10.1101/gr.217702. [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Fernández-Cañón JM, Granadino B, Beltrán-Valero de Bernabé D, et al. The molecular basis of alkaptonuria. Nat Genet. 1996;14:19–24. doi: 10.1038/ng0996-19. [DOI] [PubMed] [Google Scholar]
- Fokkema IF, den Dunnen JT, Taschner PE. Lovd: easy creation of a locus-specific sequence variation database using an “lsdb-in-a-box” approach. Hum Mutat. 2005;26:63–68. doi: 10.1002/humu.20201. [DOI] [PubMed] [Google Scholar]
- Gehrig A, Schmidt SR, Muller CR, Srsen S, Srsnova K, Kress W. Molecular defects in alkaptonuria. Cytogenet Cell Genet. 1997;76:14–16. doi: 10.1159/000134501. [DOI] [PubMed] [Google Scholar]
- Goicoechea De Jorge E, Lorda I, Gallardo ME, et al. Alkaptonuria in the dominican republic: Identification of the founder aku mutation and further evidence of mutation hot spots in the hgo gene. J Med Genet. 2002;39:E40. doi: 10.1136/jmg.39.7.e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granadino B, Beltrán-Valero de Bernabé D, Fernández-Cañón JM, Peñalva MA, Rodríguez de Córdoba S. The human homogentisate 1,2-dioxygenase (hgo) gene. Genomics. 1997;43:115–122. doi: 10.1006/geno.1997.4805. [DOI] [PubMed] [Google Scholar]
- Gucev Z, Slaveska N, Laban N et al (2011) Early-onset ocular ochronosis in a girl with alkaptonuria (aku) and a novel mutation in homogentisate 1,2-dioxygenase (hgd). Prilozi XXXII:263–268 [PubMed]
- La Du BN, Zannoni VG, Laster L, Seegmiller JE. The nature of the defect in tyrosine metabolism in alcaptonuria. J Biol Chem. 1958;230:251–260. [PubMed] [Google Scholar]
- Milch RA. Studies of alcaptonuria: inheritance of 47 cases in eight highly inter-related dominican kindreds. Am J Hum Genet. 1960;12:76–85. [PMC free article] [PubMed] [Google Scholar]
- Muller CR, Fregin A, Srsen S, et al. Allelic heterogeneity of alkaptonuria in central europe. Eur J Hum Genet. 1999;7:645–651. doi: 10.1038/sj.ejhg.5200343. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Pollak MR, Chou YH, Cerda JJ, et al. Homozygosity mapping of the gene for alkaptonuria to chromosome 3q2. Nat Genet. 1993;5:201–204. doi: 10.1038/ng1093-201. [DOI] [PubMed] [Google Scholar]
- Porfirio B, Chiarelli I, Graziano C, et al. Alkaptonuria in Italy: Polymorphic haplotype background, mutational profile, and description of four novel mutations in the homogentisate 1,2-dioxygenase gene. J Med Genet. 2000;37:309–312. doi: 10.1136/jmg.37.4.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganath L, Taylor AM, Shenkin A, et al. Identification of alkaptonuria in the general population: A united kingdom experience describing the challenges, possible solutions and persistent barriers. J Inherit Metab Dis. 2011;34(3):723–730. doi: 10.1007/s10545-011-9282-z. [DOI] [PubMed] [Google Scholar]
- Rodríguez JM, Timm DE, Titus GP, et al. Structural and functional analysis of mutations in alkaptonuria. Hum Mol Genet. 2000;9:2341–2350. doi: 10.1093/oxfordjournals.hmg.a018927. [DOI] [PubMed] [Google Scholar]
- Srsen S, Varga F. Screening for alkaptonuria in the newborn in slovakia. Lancet. 1978;2:576. doi: 10.1016/S0140-6736(78)92910-0. [DOI] [PubMed] [Google Scholar]
- Srsen S, Muller CR, Fregin A, Srsnova K. Alkaptonuria in slovakia: thirty-two years of research on phenotype and genotype. Mol Genet Metab. 2002;75:353–359. doi: 10.1016/S1096-7192(02)00002-1. [DOI] [PubMed] [Google Scholar]
- Titus GP, Mueller HA, Burgner J, Rodríguez de Córdoba S, Peñalva MA, Timm DE. Crystal structure of human homogentisate dioxygenase. Nat Struct Biol. 2000;7:542–546. doi: 10.1038/76756. [DOI] [PubMed] [Google Scholar]
- Uyguner O, Goicoechea de Jorge E, Cefle A, et al. Molecular analyses of the hgo gene mutations in turkish alkaptonuria patients suggest that the r58fs mutation originated from central asia and was spread throughout europe and anatolia by human migrations. J Inherit Metab Dis. 2003;26:17–23. doi: 10.1023/A:1024063126954. [DOI] [PubMed] [Google Scholar]
- Vilboux T, Kayser M, Introne W, et al. Mutation spectrum of homogentisic acid oxidase (hgd) in alkaptonuria. Hum Mutat. 2009;30:1611–1619. doi: 10.1002/humu.21120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatkova A, Beltrán-Valero de Bernabé D, Polakova H, et al. High frequency of alkaptonuria in slovakia: evidence for the appearance of multiple mutations in hgo involving different mutational hot spots. Am J Hum Genet. 2000;67:1333–1339. doi: 10.1016/s0002-9297(07)62964-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatkova A, Polakova H, Micutkova L, et al. Novel mutations in the homogentisate-1,2-dioxygenase gene identified in slovak patients with alkaptonuria. J Med Genet. 2000;37:539–542. doi: 10.1136/jmg.37.7.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatkova A, Chmelikova A, Polakova H, Ferakova E, Kadasi L. Rapid detection methods for five hgo gene mutations causing alkaptonuria. Clin Genet. 2003;63:145–149. doi: 10.1034/j.1399-0004.2003.00027.x. [DOI] [PubMed] [Google Scholar]
- Zatkova A (2011) An update on molecular genetics of Alkaptonuria (AKU). J Inherit Metab Dis. Jul 1. [Epub ahead of print] [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


