Abstract
Mitochondria are essential organelles whose replication, development, and physiology are dependent upon coordinated gene interactions with both the mitochondrial and the nuclear genomes. The evolution of coadapted (CA) nuclear–mitochondrial gene combinations would be facilitated if such nuclear genes were located on the X-chromosome instead of on the autosomes because of the increased probability of cotransmission. Here, we test the prediction of the CA hypothesis by investigating the chromosomal distribution of nuclear genes that interact with mitochondria. Using the online genome database BIOMART, we compared the density of genes that have a mitochondrion cellular component annotation across chromosomes in 16 vertebrates. We find a strong and highly significant genomic pattern against the CA hypothesis: nuclear genes interacting with the mitochondrion are significantly underrepresented on the X-chromosome in mammals but not in birds. We interpret our findings in terms of sexual conflict as a mechanism that may generate the observed pattern. Our finding extends single-gene theory for the evolution of sexually antagonistic genes to nuclear–mitochondrial gene combinations.
Keywords: genomic conflict, gene transfer, sex chromosome, coadapted gene complex
Introduction
Mitochondria are essential organelles whose replication, development, and physiology depend upon coordinated interactions between gene combinations from both the mitochondrial and the nuclear genomes. When genes from both genomes interact, their differing modes of inheritance, bi-parental versus maternal, can result in genomic conflict (Werren 2011). There is direct evidence of the genomic conflict generated by epistatic interactions between the mitochondrial genes and nuclear genes from studies of mitochondria involvement in sperm development (Wang 2004; Rajender et al. 2010; Paoli et al. 2011). Furthermore, introgression of foreign mitochondria into Drosophila populations has revealed sex-biased epistatic effects with X-linked nuclear genes (Rand et al. 2001, 2006; Montooth et al. 2010; Aw et al. 2011). When organismal fitness depends on the epistatic interactions between the nuclear and mitochondrial genomes (Rand et al. 2001, 2006; Dowling et al. 2007), the cotransmission of nuclear–mitochondrial gene combinations facilitates epistatic selection (Wade and Goodnight 2006; Brandvain and Wade 2009). Cotransmission is important to epistatic selection because it maintains gene combinations across generations. By increasing the heritability of gene combinations, cotransmission allows epistatic selection to act more effectively on trans-genomic interactions.
The question remains, as to how these genome interactions have shaped the geography of the genome. Theory predicts that the nuclear genome should evolve in a way to maintain nuclear–mitochondrial gene combinations (Wade and Goodnight 2006; Brandvain and Wade 2009). One means of increasing the heritability of nuclear–mitochondrial gene combinations involves changing the physical location of nuclear genes within the nuclear genome as opposed to changing location between the nuclear and mitochondrial genomes. The probability of cotransmission of X-linked genes and mitochondria is significantly higher than that of mitochondria and autosomal genes (0.67 versus 0.50 in a randomly mating population) (Rand et al. 2001). Therefore, the evolution of coadapted (CA) nuclear–mitochondrial gene combinations would be facilitated if nuclear genes interacting with the mitochondria were located on the X-chromosome instead on the autosomes because of the increased probability of cotransmission (Rand et al. 2001).
The CA nuclear–mitochondrial gene hypothesis posits that selection for beneficial nuclear–mitochondrial gene combinations and against poor ones is facilitated by the location of nuclear genes located on the X chromosome relative to the autosomes. A clear prediction of the CA hypothesis is that nuclear genes which interact with the mitochondrion (N-mt genes), should be overrepresented on the X relative to autosomes. Here, we tested the prediction of the CA by investigating the chromosomal distribution of nuclear genes that interact with the mitochondria.
Methods
Data Collection
Using the online genome database BIOMART (http://www.biomart.org) via their MartView interface (Smedley et al. 2009), we collected genomic information on the following organisms using the available ENSEMBL GENES 63 datasets: Homo sapiens (GRCH37.p3) (International Human Genome Sequencing Consortium 2004), Pan troglodytes (CHIMP2.1) (Chimpanzee Sequencing and Analysis Consortium 2005), Gorilla gorilla (gorGor3) (Scally et al. 2012), Pongo pygmaeus (PPYG2) (Locke et al. 2011), Macaca mulatta (MMUL_1.0) (Rhesus Macaque Genome Sequencing Analysis Consortium et al. 2007), Callithrix jacchus (calJac3) (GenBank Assembly ID GCA_000004665.1), Mus musculus (NCBIM37) (Gregory et al. 2002), Rattus norvegicus (RGSC3.4) (Gibbs et al. 2004), Oryctolagus cuniculus (oryCun2.0) (Lindblad-Toh et al. 2011), Canis familiaris (CanFam_2.0) (Lindblad-Toh et al. 2005), Bos taurus (Btau_4.0) (Bovine Genome Sequencing Analysis Consortium et al. 2009), Sus scrofa (Sscrofa9) (GenBank Assembly ID GCA_000003025.4), Equus caballus (EquCab2) (Lindblad-Toh et al. 2011), Monodelphis domestica (monDom5) (Mikkelsen et al. 2007), Gallus gallus (WASHUC2) (Hillier et al. 2004), and Taeniopygia guttata (taeGut3.2.4) (Warren et al. 2010). We focused on mammalian datasets because they contain the greatest level of annotation, including chromosomal location. Also, we restricted our analysis to this single database so that we could make comparisons between genomes while minimizing data collection artifacts.
For each of the 16 genomes, we collected two gene sets: 1) complete list of all genes with annotated function; 2) including only those nuclear genes with mitochondrion annotation (N-mt genes) as specified by the Gene Ontology ID 0005739. From the database, we obtained for each gene: chromosomal location, ENSEMBLE gene name, and gene ID. Genes without a specific chromosomal location were excluded. We processed this output with scripts written in MATLAB (The Mathworks, Natick, MA) to calculate the counts of genes per chromosome in each genome for the two different datasets.
Statistical Analysis
We compared the density of N-mt genes on the X or Z chromosomes with that of the autosomes relative to the expectation based on overall gene density for 14 vertebrate genomes with male heterogamety and two vertebrate genomes with female heterogamety (the birds, G. gallus and T. guttata). We performed all of our statistical analysis in MATLAB. To determine if the set of N-mt genes was under- or overrepresented on each chromosome, we compared it with an expected count. For a particular genome, we used the following method to determine the expected number of N-mt genes per chromosome. We calculated the product of the total number of N-mt genes and the fraction of all annotated genes found on each chromosome. This expected gene count per chromosome assumes that the N-mt genes are distributed across the chromosomes in the same pattern as the total set of genes in the genome. The ratio of observed count to expected count will equal one when there is no under- or overrepresentation of N-mt genes on a particular chromosome. When the value is less (greater) than one, then there is an under representation (over representation) of N-mt genes on a chromosome.
We computed the comparisons as described above to limit any potential biases in how the data were collected. In order to create a bias in the density of this particular group of genes, some chromosomes would need to be over- or underannotated for N-mt genes in particular. We are unaware of any feature of the N-mt genes which make them more or less likely to be sequenced and subsequently recognized as genes relative to any other subset of genes. Our method is also robust to differences in annotation level between chromosomes within a genome. If for some reason there was a bias in the annotation level of autosomes compared with the sex chromosome, we would expect more genes to be annotated with the N-mt term as well. Our density of N-mt genes (rather than an absolute count) would not be affected.
We tested the significance of the underrepresentation on the sex chromosome by calculating a confidence interval of the mean over-/underrepresentation for the autosomes based on 10,000 replicate bootstrap samples and then determining if the sex chromosome representation fell below this confidence interval at a Šidák corrected alpha of 0.032, based on an individual alpha of 0.05 and 16 different comparisons (Šidák 1967).
Results
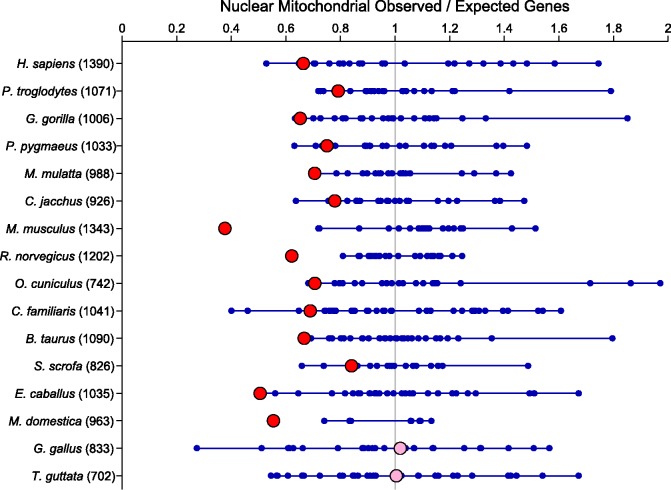
Using the online genome database BIOMART (http://www.biomart.org) via their MartView interface (Smedley et al. 2009), we examined the distribution of genes that have a mitochondrion cellular component annotation (Gene Ontology ID 0005739) across chromosomes in 16 vertebrates. We found a strong and significant underrepresentation of nuclear genes interacting with the mitochondrion on the sex chromosomes in all of the mammals (fig. 1) but in neither of the two birds. In mammals, although the sex chromosome did not always have the lowest density of N-mt genes relative to expectation, it was always in the lower end of the distribution of chromosomes. In both bird species, the sex chromosomes (Z) had the expected density of N-mt genes based on overall gene density.
Fig. 1.—
Underrepresentation of nuclear–mitochondrial genes on the sex chromosomes of mammals. Expected counts of genes are calculated based on the distribution of all annotated genes in the genome. The fraction of all genes present on a particular chromosome is then multiplied by the total number of N-mt genes in a particular genome to give the expected count. Ratios close to one show no over- or underrepresentation on a chromosome. Each chromosomal value for each genome is represented by a blue (autosomes) or red and pink (sex chromosome [X/Z]) circle. Lines connect the maximum and minimum autosomal ratios. Values in parenthesis after the genome name indicate the number of identified N-mt genes in the genome. Sex chromosomes that are significantly different from the autosomes are highlighted in red and nonsignificant values are pink (H. sapiens, P < 0.0001; P. troglodytes, P < 0.0001; G. gorilla, P < 0.0001; P. pygmaeus, P < 0.0001; M. mulatta, P < 0.0001; C. jacchus, P < 0.0001; M. musculus, P < 0.0001; R. norvegicus, P < 0.0001; O. cuniculus, P < 0.0001; C. familiaris, P < 0.0001; B. taurus, P < 0.0001; S. scrofa, P = 0.0001; E. caballus, P < 0.0001; M. domestica, P < 0.0001; G. gallus, P = 0.7621; and T. guttata, P = 0.5281).
Discussion
We have discovered a striking pattern in the distribution of genes among chromosomes: nuclear genes which interact with the mitochondrion are significantly underrepresented on the X-chromosome in mammals, but not in birds (fig. 1). These data are inconsistent with the predictions of the CA nuclear–mitochondrial gene hypothesis which proposes that due to the increased heritability of gene combinations, the N-mt genes should be overrepresented on the X-chromosome. We discuss other hypotheses that may help to explain the observed pattern of gene distributions.
We propose that the observed pattern may be explained by sexual conflict (SC). SC occurs when genes beneficial to the fitness of one sex are deleterious to the fitness of the other. The evolution of maternally transmitted organelles can lead to SC when mitochondrial mutations that interact epistatically with nuclear genes have favorable effects in females but are deleterious in males (Rice 1984; Rice et al. 2006; Vicoso and Charlesworth 2006). With epistatic selection, the effects of nuclear genes interacting with mitochondria manifest only on certain mitochondrial backgrounds and not on others (fig. 2A). Reciprocally, the effects of mitochondrial genes occur only in specific nuclear backgrounds. Importantly, changing the mitochondrial background alters the effects of the interacting nuclear genes without changing the nuclear gene sequence. Epistatic fitness interactions between nuclear genes and the mitochondrion can result in sexually antagonistic effects for nuclear genes.
Fig. 2.—
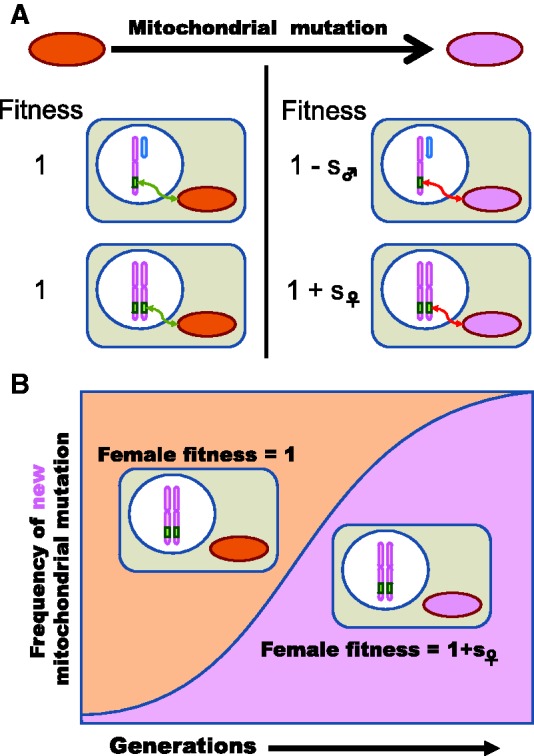
Mitochondrial mutations as drivers of SC. A schematic of the hypothesized process generating selection for the movement of N-mt genes off the X chromosome. (A) Some nuclear genes interact with the mitochondrion (green box). This interaction produces epistatic fitness effects (green arrow). A mitochondrial mutation occurs (purple circle) with epistatic sexually antagonistic fitness effects (red arrow) on the current nuclear genetic background. (B) Adaptive spread of the mitochondrial mutation leads to the eventual fixation of the female beneficial mutant mitochondria despite male deleterious effects.
With nuclear–mitochondrial epistasis, the force of genomic conflict (between the nucleus and the mitochondrion) is aligned with that of intralocus SC (between males and females). Because mitochondria have nearly exclusive maternal inheritance, selection can favor those mitochondrial genotypes that enhance female fitness, even if at the expense of male fitness (Partridge and Hurst 1998; Rand et al. 2006; Montooth et al. 2010; but see Wade and Brandvain 2009). That is, mitochondrial mutations with epistatic effects favorable to females will spread owing to maternal inheritance (fig. 2B). Because the mitochondrial mutations are not selected on the basis of their effect in males, even mutations which carry unfavorable effects in males will spread as long as those mutations are favorable in females. As a result, a nuclear gene interacting with the mitochondria will become more deleterious to males over time.
There is direct evidence of sexually antagonistic epistatic interactions between the mitochondrial and nuclear genes from studies of the involvement of mitochondria in sperm development (Wang 2004; Rajender et al. 2010; Paoli et al. 2011). Furthermore, introgression of foreign mitochondria into Drosophila populations has revealed sex-biased epistatic effects for X-linked nuclear genes (Rand et al. 2001, 2006; Montooth et al. 2010; Aw et al. 2011).
Although SC based on segregating nuclear genes depends upon the nature of allelic dominance (Rice 1984), selection on the mitochondrial mutations is independent of the dominance relationships among alleles of its nuclear partners, requiring only positive fitness effects on females in some nuclear backgrounds. Furthermore, the SC process we are proposing does not depend on the simultaneous polymorphism of both nuclear and mitochondrial genes or the cotransmission of nuclear–mitochondrial gene combinations, which was a key to the alternative CA hypothesis. SC due to the spread of mitochondrial mutations with epistatic effects with nuclear loci can occur whether the nuclear partner of the mitochondrion is autosomal or X-linked.
As a mitochondrial mutation causing the SC spreads within the population, there is increasing selective pressure to resolve the SC. Several mechanisms for the nuclear resolution of the SC have the potential to alter the genomic distribution of N-mt genes. An evolutionary means for resolving such SC (among many) is via gene movement off of the X and onto the autosomes. This is a process involving several steps, including gene duplication, fixation and subsequent gene loss, and requiring significant chromosomal bias at one or all of these steps to account for the observed pattern (Wu and Yujun Xu 2003). An alternative process is the creation of duplicate copies on the autosomes with sex-specific or sex-biased expression (Connallon and Clark 2011; Gallach and Betrán 2011). Both proposed mechanisms involve the fixation of nuclear gene duplications which ameliorate the male detrimental effects caused by the mitochondrial mutation.
Theory and empirical evidence suggest that these duplicate copies accumulate on the autosomes (Connallon and Clark 2011; Gallach and Betrán 2011). The bias in the location of the duplicate copies creates the overrepresentation on the autosomes relative to the X chromosome. Female beneficial (male deleterious) epistatic effects with mitochondrial mutations are not favored equally at the X and autosomal locations because X-linked loci spend two-thirds of their time in females and only one-third in males (Rice 1984). Autosomal loci, on the other hand, spend half their time in males and such genes with male beneficial (female deleterious) epistatic effects would be expected to fix more easily on autosomes compared with the X chromosome. This autosomal-bias in selection efficiency may be particularly important for male traits involved in reproductive competition, where selection on male fertility can be several times stronger than opposing selection on females (Wade 1979; Shuster and Wade, 2003; Miller et al. 2006). Over time, the difference in selection efficiency would lead to a greater cumulative deterioration of X-linked relative to autosomal nuclear genes as a result of mitochondrial mutation pressure (fig. 2). Empirical support for this mechanism comes from Drosophila where Gallach et al. (2010) find that N-mt gene duplications accumulate additional copies on autosomes more readily than the X chromosome. Interestingly, these gene duplicates also show testis-specific expression that the authors suggest is evidence of SC resolution (Gallach et al. 2010).
Other hypotheses regarding the resolution of SC suggest that changes in the sex-specific expression of sexually antagonistic genes, possibly via cis-acting expression modifiers, resolve intralocus SC (Rice 1984; Rice and Chippindale 2001; Gibson et al. 2002; Connallon and Clark 2010; Stewart et al. 2010). Our data are based on the genomic distribution of N-mt genes and not on sex differences in gene expression. Thus, although our data in mammals are consistent with resolution of SC via gene movement or duplication, they do not exclude the possibility that SC antagonism may be resolved by alternative means. Our data suggest that mammalian N-mt genes may be useful for elucidating the mechanistic basis by which SC is resolved (Vibranovski et al. 2009; Gallach et al. 2011; Han and Hahn 2012).
We hypothesize that the pattern we observe in mammals differs from that observed in birds, because of one or a combination of differences affecting the proposed process based on mitochondrial mutation pressure and gene movement. First, there is reduced mitochondrial mutation pressure in birds (Ogburn et al. 1998; Hickey 2008). Second, there is a near absence of nuclear gene retro-transposition in birds relative to mammals (Haas et al. 2001; Warren et al. 2010) but see (Ellegren 2011). Third, if the fitness effect is caused by epistatic interactions with the mitochondrion, then strict maternal inheritance of the mitochondria results in reduced cotransmission of the mitochondria and Z chromosomes compared autosomes. Additionally, all of the cotransmission occurs from mother to sons at which point that mitochondrial-Z chromosome combination has a zero probability of being passed along to the next generation. The combination these factors suggests that our proposed process will be significantly weaker in birds, relative to mammals and that intralocus SC in birds may be resolved differently than it is in mammals.
We appreciate that our sample of genomes does not meet the criterion of phylogenetic independence. One could argue that we have a single contrast between two types of male heterogametic vertebrates (mammals and marsupials) and one type of female heterogametic vertebrate (birds). Although other vertebrate genomes are available, none meet the criteria of having sex chromosomes (e.g., Danio rerio and Fugu rubripes) and the insect gene annotations in this database are not coincident with their annotations in other databases (e.g., drosophilids GO annotation in BIOMART relative to Flybase). It is well recognized that genomes are highly fluid, with frequent translocations among chromosomes, even though a recognizable amount of synteny is present (Hillier et al. 2004; Ross et al. 2005). Thus, despite its taxonomic limitations, our data suggest that the signal of a lack of N-mt genes on the X chromosomes of mammals is present and has been maintained across a diversity of species for tens of millions of years.
Our results significantly extend the single-gene theory for the evolution of sexually antagonistic genes to involve epistatic interactions between nuclear and mitochondrial genes. It also synergistically combines SC theory with results from genomic conflict theory, wherein, mitochondrial genes with deleterious effects on males spread if they are favorable to females owing to uniparental, maternal transmission. The high rate of such mitochondrial mutations and their subsequent adaptive spread leads to the continual accumulation of male deleterious fitness effects on X-linked genes (Rand et al. 2001). Our theory does not preclude patterns of sex-limited gene expression for sex-linked genes with direct (instead of epistatic) fitness effects. Our empirical finding adds to our knowledge of the forces affecting gene clustering and gene expression on sex chromosomes. Our results are consistent with a synergistic interaction between the forces of SC and genomic conflict affecting the distribution of N-mt genes on the sex chromosomes relative to the autosomes in vertebrates.
Acknowledgments
This research was improved by discussion with the θ working group at Indiana University and by comments from Kristi Montooth, Mira Han, and anonymous reviewers. This work was supported by National Institute of General Medical Sciences at the National Institutes of Health (grant numbers R01GM084238, R01GM065414 to M.J.W.). We thank the Marmoset Genome Sequencing Consortium for making their data publicly available and the Baylor College of Medicine—Human Genome Sequencing Center for providing the Marmoset Genome Project data. We thank the Swine Genome Sequencing Consortium for making their data publicly available and University of Illinois—Institute for Genomic Biology for providing the Sus scrofa data.
Literature Cited
- Aw WC, Correa CC, Clancy DJ, Ballard JWO. Mitochondrial DNA variants in Drosophila melanogaster are expressed at the level of the organismal phenotype. Mitochondrion. 2011;11:756–763. doi: 10.1016/j.mito.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Bovine Genome Sequencing Analysis Consortium, Elsik CG, Tellam RL, Worley KC. 2009. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science 324:522–528. [DOI] [PMC free article] [PubMed]
- Brandvain Y, Wade MJ. The functional transfer of genes from the mitochondria to the nucleus: the effects of selection, mutation, population size and rate of self-fertilization. Genetics. 2009;182:1129–1139. doi: 10.1534/genetics.108.100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Connallon T, Clark AG. Sex linkage, sex-specific selection, and the role of recombination in the evolution of sexually dimorphic gene expression. Evolution. 2010;64:3417–3442. doi: 10.1111/j.1558-5646.2010.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Clark AG. The resolution of sexual antagonism by gene duplication. Genetics. 2011;187:919–937. doi: 10.1534/genetics.110.123729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling DK, Friberg U, Hailer F, Arnqvist G. Intergenomic epistasis for fitness: within-population interactions between cytoplasmic and nuclear genes in Drosophila melanogaster. Genetics. 2007;175:235–244. doi: 10.1534/genetics.105.052050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Emergence of male-biased genes on the chicken Z-chromosome: sex-chromosome contrasts between male and female heterogametic systems. Genome Res. 2011;21:2082–2086. doi: 10.1101/gr.119065.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach M, Betrán E. Intralocus sexual conflict resolved through gene duplication. Trends Ecol Evol. 2011;26:222–228. doi: 10.1016/j.tree.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach M, Chandrasekaran C, Betrán E. Analyses of nuclearly encoded mitochondrial genes suggest gene duplication as a mechanism for resolving intralocus sexually antagonistic conflict in Drosophila. Genome Biol Evol. 2010;2:835–850. doi: 10.1093/gbe/evq069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach M, Domingues S, Betrán E. Gene duplication and the genome distribution of sex-biased genes. Intl J Evol Biol. 2011;2011:989438. doi: 10.4061/2011/989438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Gibson JR, Chippindale AK, Rice WR. The X chromosome is a hot spot for sexually antagonistic fitness variation. Proc R Soc Lond Ser B Biol Sci. 2002;269:499–505. doi: 10.1098/rspb.2001.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SG, et al. A physical map of the mouse genome. Nature. 2002;418:743–750. doi: 10.1038/nature00957. [DOI] [PubMed] [Google Scholar]
- Haas NB, et al. Subfamilies of CR1 non-LTR retrotransposons have different 5' UTR sequences but are otherwise conserved. Gene. 2001;265:175–183. doi: 10.1016/s0378-1119(01)00344-4. [DOI] [PubMed] [Google Scholar]
- Han MV, Hahn MW. Inferring the history of interchromosomal gene transposition in Drosophila using n-dimensional parsimony. Genetics. 2012;190:813–825. doi: 10.1534/genetics.111.135947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey AJR. An alternate explanation for low mtDNA diversity in birds: an age-old solution? Heredity. 2008;100:443–443. doi: 10.1038/hdy.2008.6. [DOI] [PubMed] [Google Scholar]
- Hillier LW, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, et al. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Miller PM, Gavrilets S, Rice WR. Sexual conflict via maternal-effect genes in ZW species. Science. 2006;312:73. doi: 10.1126/science.1123727. [DOI] [PubMed] [Google Scholar]
- Montooth KL, Meiklejohn CD, Abt DN, Rand DM. Mitochondrial-nuclear epistasis affects fitness within species but does not contribute to fixed imcompatibilities between species of Drosophila. Evolution. 2010;64:3364–3379. doi: 10.1111/j.1558-5646.2010.01077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogburn CE, et al. Cultured renal epithelial cells from birds and mice: enhanced resistance of avian cells to oxidative stress and DNA damage. J Gerontol Ser A Biol Sci Med Sci. 1998;53:B287–B292. doi: 10.1093/gerona/53a.4.b287. [DOI] [PubMed] [Google Scholar]
- Paoli D, et al. Mitochondrial membrane potential profile and its correlation with increasing sperm motility. Fertil Steril. 2011;95:2315–2319. doi: 10.1016/j.fertnstert.2011.03.059. [DOI] [PubMed] [Google Scholar]
- Partridge L, Hurst LD. Sex and conflict. Science. 1998;281:2003–2008. doi: 10.1126/science.281.5385.2003. [DOI] [PubMed] [Google Scholar]
- Rajender S, Rahul P, Mahdi AA. Mitochondria, spermatogenesis and male infertility. Mitochondrion. 2010;10:419–428. doi: 10.1016/j.mito.2010.05.015. [DOI] [PubMed] [Google Scholar]
- Rand DM, Clark AG, Kann LM. Sexually antagonistic cytonuclear fitness interactions in Drosophila melanogaster. Genetics. 2001;159:173–187. doi: 10.1093/genetics/159.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Fry A, Sheldahl L. Nuclear-mitochondrial epistasis and Drosophila aging: introgression of Drosophila simulans mtDNA modifies longevity in D-melanogaster nuclear backgrounds. Genetics. 2006;172:329–341. doi: 10.1534/genetics.105.046698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhesus Macaque Genome Sequencing Analysis Consortium, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Rice WR. Sex-chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Rice WR, Chippindale AK. Intersexual ontogenetic conflict. J Evol Biol. 2001;14:685–693. [Google Scholar]
- Rice WR, et al. Assessing sexual conflict in the Drosophila melanogaster laboratory model system. Phil Trans R Soc B Biol Sci. 2006;361:287–299. doi: 10.1098/rstb.2005.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MT, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally A, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster SM, Wade MJ. Mating systems and strategies. Princeton, NJ: Princeton University Press; 2003. [Google Scholar]
- Šidák Z. Rectangular confidence region for the means of multivariate normal distributions. J Am Stat Assoc. 1967;62:626–633. [Google Scholar]
- Smedley D, et al. BioMart—biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AD, Pischedda A, Rice WR. Resolving intralocus sexual conflict: genetic mechanisms and time frame. J Heredity. 2010;101:S94–S99. doi: 10.1093/jhered/esq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Zhang Y, Long MY. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 2009;19:897–903. doi: 10.1101/gr.088609.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Wade MJ. Sexual selection and variance in reproductive success. Am Nat. 1979;114:742–747. doi: 10.1086/424531. [DOI] [PubMed] [Google Scholar]
- Wade MJ, Brandvain Y. Reversing mother’s curse: selection on male mitochondrial fitness effects. Evolution. 2009;63:1084–1089. doi: 10.1111/j.1558-5646.2009.00614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MJ, Goodnight CJ. Cyto-nuclear epistasis: two-locus random genetic drift in hermaphroditic and dioecious species. Evolution. 2006;60:643–659. [PubMed] [Google Scholar]
- Wang PJ. X chromosomes, retrogenes and their role in male reproduction. Trends Endocrinol Met. 2004;15:79–83. doi: 10.1016/j.tem.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Warren WC, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc Natl Acad Sci U S A. 2011;108:10863–10870. doi: 10.1073/pnas.1102343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-I, Yujun Xu E. Sexual antagonism and X inactivation—the SAXI hypothesis. Trends Genet. 2003;19:243–247. doi: 10.1016/s0168-9525(03)00058-1. [DOI] [PubMed] [Google Scholar]