Abstract
Epigenetic information is frequently erased near the start of each new generation (1). In some cases, however, epigenetic information can be transmitted from parent to progeny (epigenetic inheritance) (2). A particularly striking example of epigenetic inheritance is dsRNA-mediated gene silencing (RNAi) in C. elegans, which can be inherited for more than five generations (3–8). To understand this process we conducted a genetic screen for animals defective for transmitting RNAi silencing signals to future generations. This screen identified the gene heritable RNAi defective (hrde)-1. hrde-1 encodes an Argonaute (Ago) that associates with small interfering (si)RNAs in germ cells of the progeny of animals exposed to dsRNA. In nuclei of these germ cells, HRDE-1 engages the Nrde nuclear RNAi pathway to direct H3K9me3 at RNAi targeted genomic loci and promote RNAi inheritance. Under normal growth conditions, HRDE-1 associates with endogenously expressed siRNAs, which direct nuclear gene silencing in germ cells. In hrde-1 or nuclear RNAi deficient animals, germline silencing is lost over generational time. Concurrently, these animals exhibit steadily worsening defects in gamete formation and function that ultimately lead to sterility. These results establish that the Ago HRDE-1 directs gene-silencing events in germ cell nuclei, which drive multi-generational RNAi inheritance and promote immortality of the germ cell lineage. We propose that C. elegans uses the RNAi inheritance machinery to transmit epigenetic information, accrued by past generations, into future generations to regulate important biological processes.
We conducted a genetic screen to identify factors required for multi-generational RNAi inheritance. We mutagenized animals carrying a germline gfp reporter gene and screened for mutant animals that retained the ability to silence gfp when exposed directly to gfp dsRNA, but failed to silence gfp in subsequent generations. Among fourteen mutant alleles fulfilling these criteria, four of these alleles defined a gene we term here heritable RNAi defective (hrde)-1 (Fig. S2). hrde-1 mutant animals silenced GFP when exposed to gfp dsRNA, but failed to transmit this silencing to subsequent generations (Fig. 1a, and Fig. S3). Similarly, hrde-1 mutants silenced the germline expressed oma-1 gene when treated directly with oma-1 dsRNA, but were defective for silencing inheritance (Fig. S4). We conclude that hrde-1 promotes multi-generational RNAi silencing in the germline.
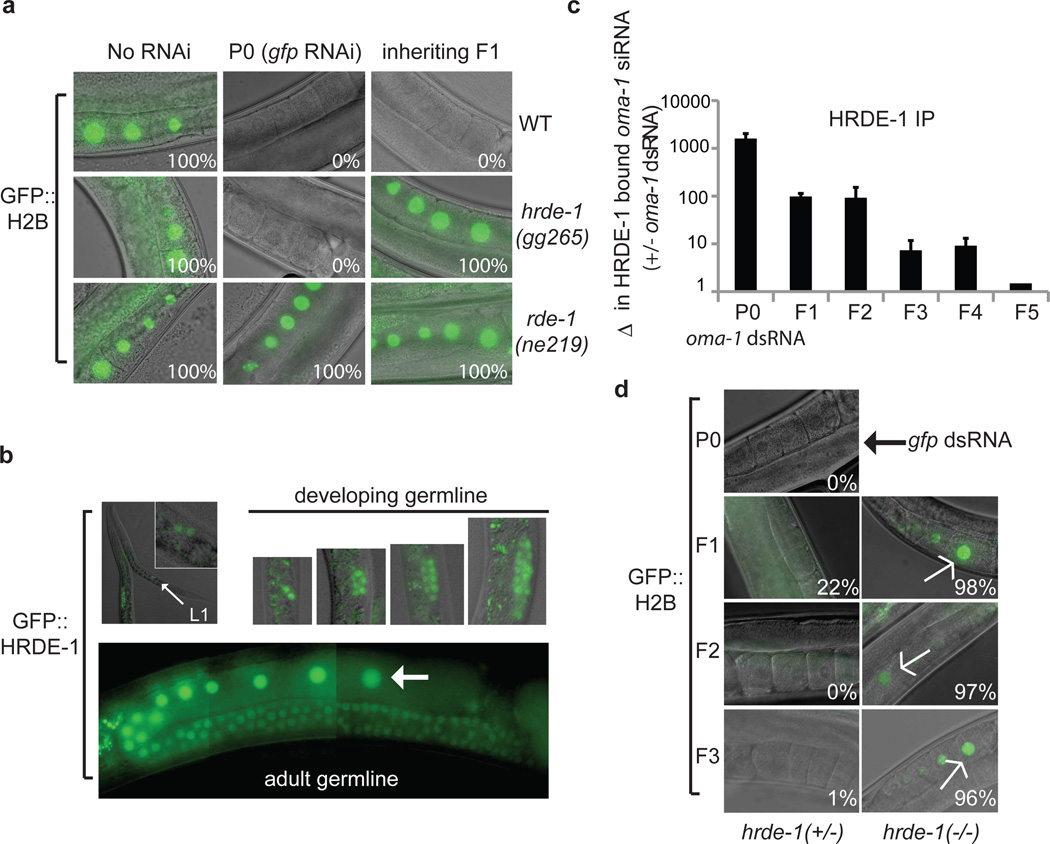
Figure 1. hrde-1 encodes a nuclear Ago that acts in inheriting generations to promote multi-generational germline RNAi inheritance.
(a) pie-1::gfp::h2b expressing animals were exposed to gfp dsRNA. F1 progeny were grown in the absence of dsRNA, and gfp expression in oocytes was visualized with fluorescence microscopy. WT, wild-type. % fluorescent animals is indicated, n>100. rde-1 is required for RNAi (17). (b) GFP::HRDE-1 was visualized by fluorescent microscopy. Small arrow, L1 animal; inset, magnification showing GFP::HRDE-1 in primordial germ cells. Large arrow, oocyte nucleus. (c) 3xFLAG::HRDE-1 was immunoprecipitated with a-FLAG antibody and HRDE-1 co-precipitating RNA was isolated and oma-1 siRNAs were quantified with oma-1 TaqMan probe set. Data is expressed as a ratio (+/− oma-1 RNAi) (n=3, +/− s.d.). Note: RNA dependent RNA Polymerases likely maintain HRDE-1 siRNA populations during RNAi inheritance (see supplemental discussion) (d) pie-1::gfp::h2b fluorescence was scored in hrde-1(+/−)/(+/+) or hrde-1(−/−) inheriting progeny. The hrde-1(−) chromosome was marked with unc-93 (unc-93 is ~1.3 cM from hrde-1), and hrde-1 genotypes were inferred by Unc phenotypes. % fluorescent animals is indicated.
We mapped hrde-1 to a genomic region containing the gene c16c10.3. c16c10.3 is predicted to encode an Ago not known to contribute to gene-silencing (9,10). c16c10.3 encodes a predicted bipartite nuclear localization signal (NLS), a PAZ, and a PIWI domain (Fig. S5). We found that hrde-1 is c16c10.3 (Fig. S5). c16c10.3 was referred to once previously in the literature as worm Ago (wago)-9 (10). Henceforth, we refer to this Ago as hrde-1/wago-9. hrde-1 is a member of a worm-specific clade of Agos (wagos) (9). HRDE-1 appeared to be relatively unique amongst the WAGOs in its contribution to germline RNAi inheritance (Fig. S6). We constructed a fusion gene between gfp and a full-length genomic copy of hrde-1 (gfp::hrde-1). gfp::hrde-1 rescued RNAi inheritance in hrde-1(−) animals, indicating that GFP::HRDE-1 is functional (Fig. S5). GFP::HRDE-1 was expressed in nuclei of male and female germ cells (Fig. 1b, and Fig. S7). These data indicate that hrde-1 encodes a germline Ago that localizes to the nucleus.
HRDE-1 could conceivably promote multi-generational RNAi inheritance by acting in animals exposed directly to dsRNA (RNAi generation) or by acting in the progeny of these animals (inheriting generation). In C. elegans, dsRNA exposure induces the expression of siRNAs in inheriting generations ((7,8) and Fig S8). HRDE-1 co-precipitated with siRNAs for multiple generations after RNAi, consistent with the idea that HRDE-1 acts in inheriting generations to promote RNAi inheritance (Fig. 1c). Note: maintenance of HRDE-1 siRNA populations across generations is likely mediated by RdRPs (see supplemental discussion). The following genetic analyses confirmed that HRDE-1 acts in inheriting generations. Animals that were hrde-1(+/−) in the RNAi generation, but were hrde-1(−/−) in the F1 inheriting generation, failed to inherit RNAi silencing (Fig. 1d, and Table S1). Similarly, HRDE-1 activity was required in the F2 generation for F1 to F2 RNAi inheritance, and in the F3 generation for F2 to F3 RNAi inheritance (Fig. 1d). Conversely, animals that lacked HRDE-1 in the RNAi generation, but expressed HRDE-1 in the inheriting generation, were able to inherit RNAi silencing (Table S1). Thus, HRDE-1 acts in inheriting progeny to facilitate the memory of RNAi silencing events that occurred in previous generations. Altogether, these data establish that C. elegans possess machinery dedicated to propagating epigenetic information across generational boundaries.
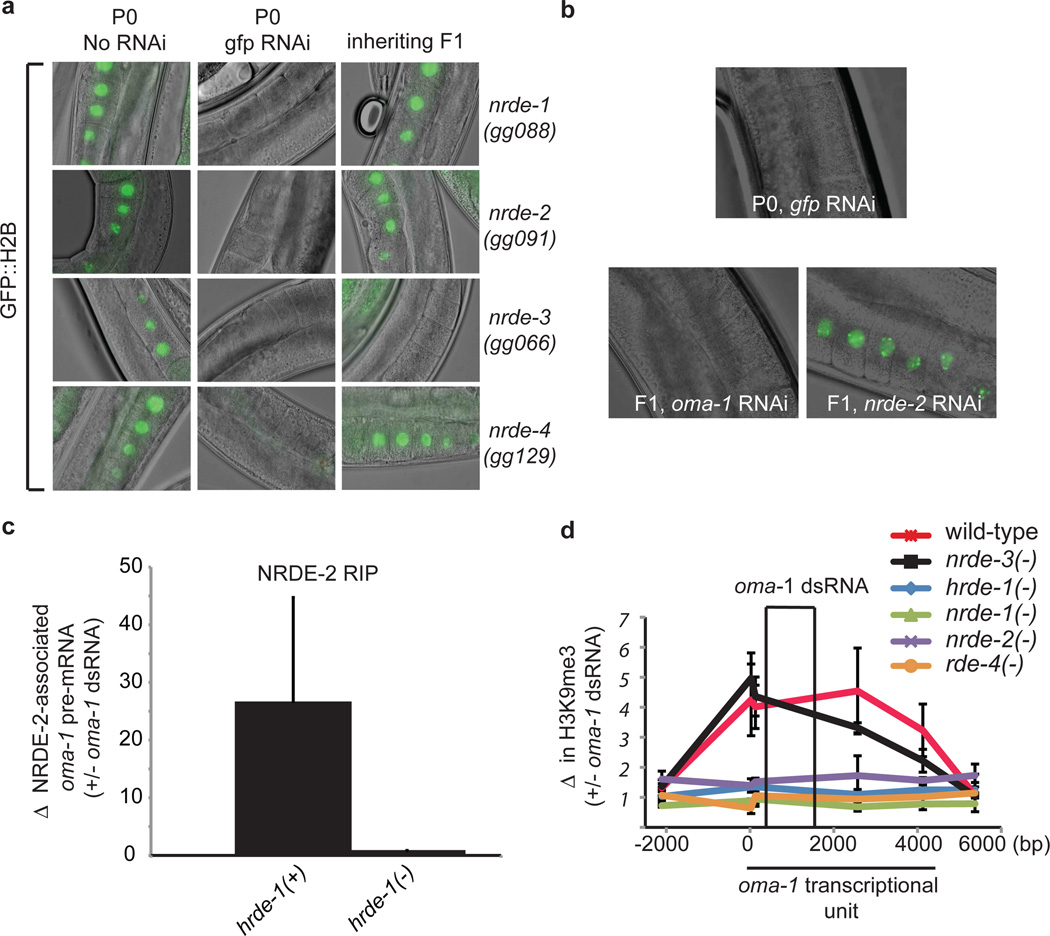
The nuclear RNAi factors NRDE-1/2/3/4 comprise a sub-branch of the C. elegans RNAi silencing machinery that is required for dsRNA-based silencing of nuclear-localized RNAs (11–13). According to our current model, siRNAs bound to the somatically expressed Ago NRDE-3 recognize and bind nascent RNA transcripts and recruit NRDE-1/2/4 (termed downstream Nrde factors) to genomic sites of RNAi in somatic cells. Together, the Nrde factors direct nuclear gene silencing events, which include the deposition of the repressive chromatin mark histone H3 lysine-9 me3 (H3K9me3), and the inhibition of RNA Polymerase II elongation (11–13). The Nrde factors contribute to heritable gene silencing events that are manifest in somatic cells (7). Five lines of evidence indicate that HRDE-1 engages the downstream Nrde factors to direct nuclear RNAi, and, consequently, RNAi inheritance in germ cells. First, the downstream Nrde factors were required for gfp or pos-1 germline RNAi inheritance (Fig. 2a, and Fig. S9). Second, like HRDE-1, the downstream Nrde factor NRDE-2 acted in inheriting generations to promote memory of RNAi in germ cells (Fig. 2b). Third, HRDE-1 was required for RNAi-mediated recruitment of NRDE-2 to a germline pre-mRNA, indicating that HRDE-1 acts as a specificity factor in germ cells to recruit a downstream Nrde factor to genomic sites of RNAi (Fig. 2c). Fourth, the ability of dsRNA to induce H3K9me3 was lost in mutant strains that eliminate hrde-1 or the downstream Nrde factors (Fig. 2d, Fig. S10, and see supplemental discussion). Fifth, consistent with the idea that hrde-1 and the downstream Nrde factors act together in the germline, hrde-1(−) animals share a germline mortality phenotype with nrde-1/2/4(−) animals (see below). These data indicate that NRDE-1/2/4 are required for multi-generational RNAi inheritance and support a model whereby HRDE-1 and NRDE-1/2/4 comprise a germline RNAi pathway that drives RNAi inheritance by inducing gene silencing in the nuclei of inheriting progeny. Henceforth, we refer to HRDE-1 and NRDE-1/2/4 as the germline RNAi inheritance machinery.
Figure 2. HRDE-1 engages the Nrde nuclear RNAi pathway to direct multi-generational RNAi inheritance.
(a) pie-1::gfp::h2b fluorescence is shown. >98% of animals of each genotype exhibited phenotypes similar to that of image. (b) P0 animals expressing pie-1::gfp::h2b were exposed to gfp dsRNA. F1 progeny were exposed to nrde-2 or oma-1 dsRNA (n=3). (c) FLAG::NRDE-2 was precipitated with a-FLAG antibody and NRDE-2 co-precipitating oma-1 pre-mRNA was quantified with qRT-PCR using exon/intron primer sets. Data are expressed as ratio (+/−) oma-1 RNAi. (n=3, +/− s.e.m.). (d) The F1 progeny of oma-1 dsRNA-treated animals were subjected to H3K9me3 Chromatin Immunoprecipiation (ChIP). Co-precipitating oma-1 DNA was quantified with qRT-PCR. Data were normalized to co-precipitating eft-3 DNA and expressed as a ratio +/− oma-1 RNAi (1=no change). X-axis, 0 denotes predicted start codon of oma-1. (bp), base pair. rde-4 is required for RNAi (18) (n=3–4, +/− s.e.m.).
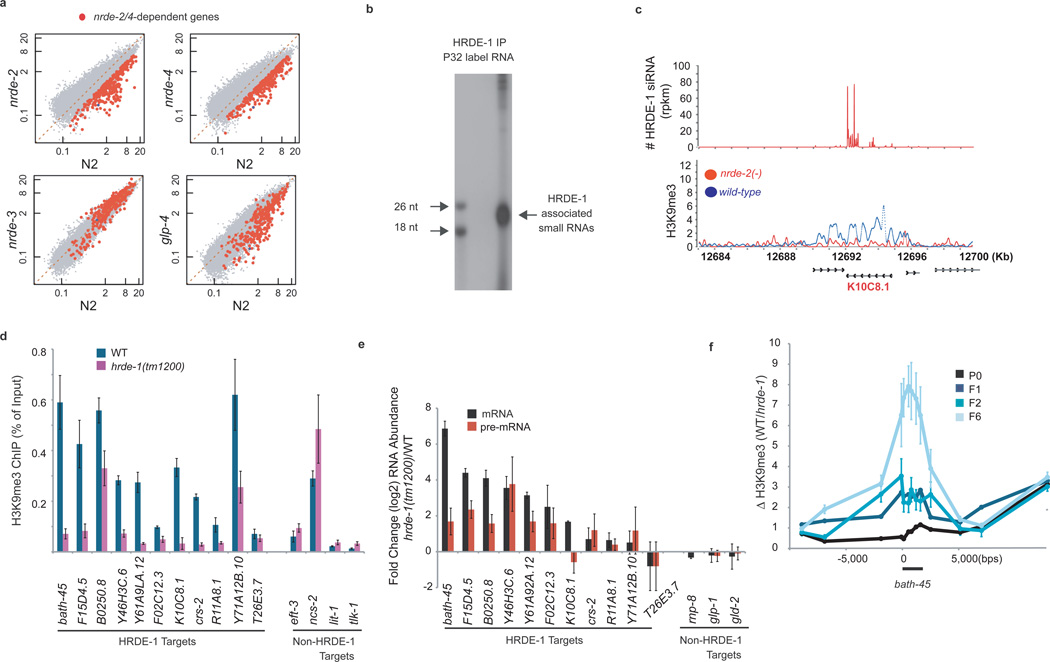
We asked if, under normal reproductive conditions, the germline RNAi inheritance machinery transmits endogenous RNAi silencing signals across generations. To test this idea, we first used H3K9me3 as a read-out for endogenous nuclear RNAi in germ cells. We isolated wild-type or nrde-2/3/4(−) animals, conducted H3K9me3 ChIP, and subjected H3K9me3 co-precipitating nucleosome core DNA to high-throughput sequencing (8). We identified 320 predicted genes that were depleted for H3K9me3 >2 fold in both nrde-2(−) and nrde-4(−) animals relative to wild-type (Fig. 3a, and Table S2). H3K9me3 ChIP, followed by directed qRT-PCR analysis, confirmed the nrde-2/4 dependence of H3K9me3 at four out of four of these loci (data not shown). Nrde-dependent H3K9me3 was present in germ cells; in glp-4(ts) mutants (14), which lack most germ cells, H3K9me3 was significantly reduced at most nrde-2/4-dependent sites (Fig. 3a, p-value 2×10−13). Together, these data show that nrde-1/2/4 contribute to H3K9me3 at multiple loci in germ cells. Henceforth, we refer to these loci as the endogenous Nrde “germline target genes”.
Figure 3. The RNAi inheritance machinery transmits endogenous epigenetic information across generations.
(a) H3K9me3 in mutant (y-axis) vs. wild type (x-axis, N2). Levels of H3K9me3 at C. elegans genes are ratio of IP nucleosome/ input nucleosome counts. smg-1 (positive control) (8) (blue dot) and nrde-2/4-dependent genes (red) are highlighted. N2 and nrde-2 ChIP-seq data were published previously (accession number: GSE32631) (8). Other ChIP-seq data, and HRDE-1 siRNA data (accession number: GSE38041) (b) FLAG::HRDE-1 co-precipitating RNA was 32P-radiolabeled and analyzed by PAGE. (nt) nucleotide. (c) Example of HRDE-1 target gene. (d) qRT-PCR quantification of H3K9me3 ChIP (animals grown at 25°C) (+/− HRDE-1) at eleven genes targeted by HRDE-1 siRNAs (HRDE-1 targets). Four genes, which are not targeted by HRDE-1 siRNAs and exhibit Nrde-independent H3K9me3 in the germline (non-HRDE-1 targets), do not exhibit H3K9me3 loss, showing that H3K9me3 loss in hrde-1 mutants is not simply due to loss of germ cells. Data is expressed as % of input DNA recovered by ChIP (n=3, +/− s.d.). (e) Total RNA was isolated from +/− HRDE-1 animals (25°C) and qRT-PCR was used to quantify mRNA or pre-mRNA levels from eleven HRDE-1 targeted genes. Three genes not targeted by HRDE-1 siRNAs, but expressed in germ cells, are also shown. Data is normalized to nos-3 mRNA (germline only) and expressed as a ratio (hrde-1(−)/wild-type) (n=3, +/− s.d.). (f) dpy-17 or hrde-1; dpy-17 animals were out-crossed 5x, and Dpy adult animals (P0) and adult progeny (F1, F2, F6) were isolated (20°C) and H3K9me3 was quantified with qRT-PCR. Data are expressed as a ratio (wild-type/hrde-1(−)) (n=1 for P0-F1, and n=3 for F2 and F6, +/− s.e.m.). Note: in this panel increased H3K9me3 signal means loss of H3K9me3 in hrde-1(−) animals.
HRDE-1 co-precipitated with endogenous small RNAs (Fig. 3b). We sequenced these small RNAs and found that HRDE-1 bound 22G endogenous (endo) siRNAs, which were expressed in germ cells, and were antisense to ~1500 predicted coding genes (Table S2, and Fig. S11). HRDE-1 22G siRNAs also targeted pseudogenes and cryptic loci (Table S2). 22G siRNAs are synthesized by RNA dependent RNA Polymerases (RdRPs) acting on cellular RNAs templates (10,15), suggesting that the HRDE-1 22G siRNAs are synthesized via RdRP activity in germ cells (see supplemental discussion). Three lines of evidence link HRDE-1 to the regulation of gene expression at Nrde germline target genes. First, we observed a correlation between genomic sites homologous to the most abundant (top 200) HRDE-1 bound siRNAs and genomic sites depleted for H3K9me3 in nrde-2/4(−) animals (p-value 2×10−16) (Fig. 3c, Fig. S12, Table S2). Second, we conducted H3K9me3 ChIP on hrde-1(−) animals and quantified H3K9me3 at fourteen Nrde germline target genes. At thirteen of these loci, H3K9me3 was depleted in hrde-1(−) animals (Fig. 3d, and Fig. S13). Third, we observed increased pre-mRNA expression from many germline target genes in hrde-1(−) animals, indicating that the RNAi inheritance machinery silences germline target genes co-transcriptionally during the normal course of reproduction (Fig. 3e). These data indicate that HRDE-1 contributes to H3K9me3 in the germline and support a model whereby HRDE-1 uses 22G endo siRNAs as specificity factors to direct nuclear RNAi in germ cells.
We asked if HRDE-1-mediated nuclear RNAi at germline target genes was heritable. We out-crossed hrde-1(−) animals with wild-type animals, isolated hrde-1(−) progeny, and conducted H3K9me3 ChIP on these hrde-1(−) animals and their progeny. H3K9me3 at germline target genes was progressively lost over generations in hrde-1(−) animals (Fig. 3f, Fig. S14). Similar results were seen with nrde-1(−) animals (Fig. S14). Coincident with loss of H3K9me3, germline target gene over-expression became more pronounced in late generations hrde-1(−) animals (Fig. S15). These data show that the RNAi inheritance machinery transmits endogenous gene regulatory information across generational boundaries.
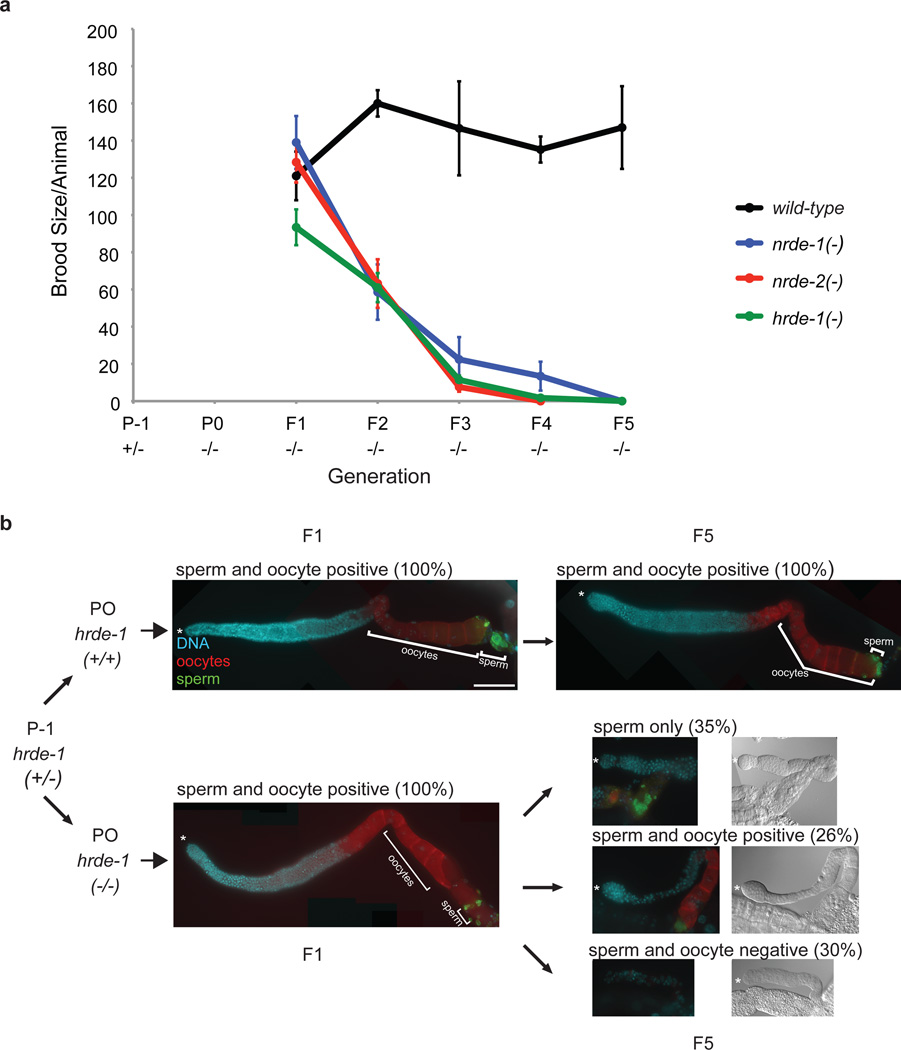
Why might an organism transmit gene regulatory information across generations? During the course of our studies, we noticed that our RNAi inheritance defective strains would periodically become sterile; stock plates would contain hundreds of adults, but no progeny. We hypothesized that the RNAi inheritance machinery might be required to maintain the integrity of the germ cell lineage. To test this idea, we out-crossed two independently isolated alleles each of hrde-1(−) and nrde-1/2/4(−) to wild-type and then monitored fertility across generations. After out-crossing hrde-1(−) and nrde-1/2/4(−) animals exhibited near wild-type fertility (early generations), but became sterile in subsequent generations (late generations) (Fig. 4a, and Fig. S16). These data show that RNAi inheritance defective animals exhibit a mortal germline (Mrt) phenotype (16). Animals lacking the somatic Ago NRDE-3 were not Mrt (Fig. S17). The Mrt phenotype of hrde-1(−) animals was temperature sensitive: hrde-1(−) animals were Mrt at 25°C, but not 20°C, indicating that growth at higher temperatures is required to reveal defects associated with loss of HRDE-1 (Fig. S18). Most late generation hrde-1(−) mutants (grown at 25°C) failed to produce mature oocytes or sperm, showing that one reason hrde-1(−) animals do not produce progeny is due to defects in gametogenesis (Fig. 4b, and Fig. S19). 26% of late generation hrde-1(−) animals were able to produce sperm and oocytes (Fig. 4b). Most of these gametes, however, are unlikely to be functional as fecundity of hrde-1(−) animals in this late generation was only 1% that of wild-type animals (Fig. 4a). Finally, late generation hrde-1(−) animals exhibited a high incidence of male (Him) phenotype, suggesting that loss of RNAi inheritance may cause defects in chromosome pairing and/or segregation (Fig. S19). We conclude that the RNAi inheritance machinery is required to maintain the immortality of the germline and that, over generations, disabling the RNAi inheritance machinery causes progressive and diverse defects in germ cell formation and function.
Figure 4. The RNAi inheritance machinery promotes germline immortality.
(a) Animals of indicated genotypes were out-crossed to wild-type 2–4x and brood sizes scored across generations at 25°C. n=5, +/− s.e.m. Note: for unknown reasons, nrde-1 and nrde-4 mutants are Mrt at both 20°C and 25°C, but hrde-1 and nrde-2 mutants are only Mrt ~25°C (n=4). (b) hrde-1(−) animals were out-crossed 3x and (+/+) or (−/−) siblings were isolated. Gonads were isolated and stained with DAPI (DNA), and immuno-fluorescence was used to detect sperm (green) and oocytes (red) (see Methods) in F1 and F5 generations. Scale bar = 100µm.
Here we show that C. elegans possess dedicated regulatory machinery that promotes an epigenetic memory of RNAi-silencing events that occurred in distant ancestors (Fig. S1). The Ago HRDE-1 is at the heart of this process, binding heritable specificity determinants (siRNAs) to direct nuclear RNAi and promote RNAi inheritance in germ cells. Nuclear RNAi also promotes RNA inheritance in somatic cells (7), indicating that nuclear gene silencing events promote RNAi inheritance in both somatic and germ cells. Finally, we show that the germline RNAi inheritance machinery transmits endogenous epigenetic information across generational boundaries while promoting germline immortality (Fig. S1). Our data suggest a model in which endogenous heritable RNAs that engage HRDE-1 act as specificity factors to direct epigenomic maintenance and immortality of the germ cell lineage. Additional work is needed to determine how defects in epigenomic maintenance relate to germline mortality (see supplemental discussion). We note, however, that both processes depend upon the same complement of factors (hrde-1 and nrde-1/2/4), and in animals lacking these factors, defects in epigenome maintenance and defects in germ cell viability are coincident over generational time. Therefore, we propose that one biological function of the RNAi inheritance machinery is to transmit “germline immortality” small RNAs, selected during species evolution for their ability to promote fertility, across generational boundaries to promote fertility in future generations.
Methods Summary
RNAi. RNAi experiments were conducted as described previously (11). The oma-1 and pos-1 constructs were taken from the Ahringer library.
RNA IP (RIP). RIPs were performed as described previously (11), with the exception that adult animals were used for all RIPs. Adult animals were frozen and dounced 10x prior to RIP. FLAG::NRDE-2 protein was immunoprecipitated with anti-FLAG M2 antibody (Sigma, A2220).
Chromatin IP (ChIP). ChIP experiments were performed as described previously (12), except that gravid adult animals were used. Worms were frozen prior to cross linking and were dounced 10x prior to sonicating. H3K9me3 antibody was from Upstate (07–523).
oma-1 siRNA TaqMan assay. TaqMan assay was performed as described previously (7). TaqMan probe set #1 was used in Fig. 2a (see Supplemental Methods).
Unless indicated otherwise, the following mutant alleles were used in this study: hrde-1(tm1200), nrde-1(gg088), nrde-2(gg091), nrde-3(gg066), nrde-4(gg129).
Supplementary Material
Acknowledgements
We thank Dr. Phil Anderson, Dr. Hopa Licious, and Dr. David Wassarman for thoughtful discussions. We thank Dr. Shawn Ahmed and member of the Ahmed lab for sharing unpublished data concerning the role of nrde-1 in germline immortality. This work was supported by grants from the Pew and Shaw scholar’s programs, and the National Institutes of Health GM88289 (S.K), GM37706 (A.F.), and (JK).
Footnotes
Full Methods and any associated references are available in the online version of this paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions: B.B. contributed to Fig. 1abc, 2bd, S3, S4, S5b, S6, S8, S10, S13, K.B. contributed to Fig. 2c, 3def, 4ab, S13, S14, S15, S16abc, S17, S18, S19c, S.G.G. and A.F. contributed to Fig. 3ac, Table S2, Figs. S11, S12, G.S. contributed to Fig. S2, S5A, S16d, A.K. and J.K contributed to Fig. 4b, S7, S19a, H.F. contributed to Fig. 4a, S16abc, S17, S18, S19c, S.K. to contributed to Fig. 1acd, 2a, 3b, Table S1, Figs. S2, S6b, S9, S10b, S19b. S.K, B.B, and K.B. wrote the manuscript.
Accession numbers: nrde-2 ChIP-seq data (published previously): GSE32631. Other ChIP-seq and HRDE-1 siRNA data: GSE38041
Competing financial interests: The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001 Aug 10;293(5532):1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 2.Jablonka E, Raz G. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q Rev Biol. 2009 Jun;84(2):131–176. doi: 10.1086/598822. [DOI] [PubMed] [Google Scholar]
- 3.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998 Feb 19;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 4.Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000 Mar 31;287(5462):2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- 5.Vastenhouw NL, Brunschwig K, Okihara KL, Muller F, Tijsterman M, Plasterk RH. Gene expression: long-term gene silencing by RNAi. Nature. 2006 Aug 24;442(7105):882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- 6.Alcazar RM, Lin R, Fire AZ. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics. 2008 Nov;180(3):1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. PNAS. 2011 doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet. 2012 Jan 8; doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006 Nov 17;127(4):747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 10.Gu W, Shirayama M, Conte D, Jr, Vasale J, Batista PJ, Claycomb JM, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009 Oct 23;36(2):231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, et al. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008 Jul 25;321(5888):537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010 Jun 24;465(7301):1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burkhart KB, Guang S, Buckley BA, Wong L, Bochner AF, Kennedy S. A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet. 2011 Aug;7(8):e1002249. doi: 10.1371/journal.pgen.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beanan MJ, Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 1992 Nov;116(3):755–766. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- 15.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007 Jan 12;315(5809):241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 16.Ahmed S, Hodgkin J. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature. 2000 Jan 13;403(6766):159–164. doi: 10.1038/35003120. [DOI] [PubMed] [Google Scholar]
- 17.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999 Oct 15;99(2):123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 18.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002 Jun 28;109(7):861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.