Abstract
Signaling of BMP ligands is antagonized by a number of extracellular proteins, including noggin, follistatin and members of the DAN family. Structural studies on the DAN family member sclerostin (a weak BMP antagonist) have previously revealed that the protein is monomeric and consists of an 8-membered cystine knot motif with a fold similar to TGF-β ligands. In contrast to sclerostin, certain DAN family antagonists, including Protein related to DAN and Cerberus (PRDC), have an unpaired cysteine that is thought to function in covalent dimer assembly (analogous to TGF-β ligands). Through a combination of biophysical and biochemical studies, we determined that PRDC forms biologically active dimers that potently inhibit BMP ligands. Furthermore, we showed that PRDC dimers, surprisingly, are not covalently linked, as mutation of the unpaired cysteine does not inhibit dimer formation or biological activity. We further demonstrated that the noncovalent PRDC dimers are highly stable under both denaturing and reducing conditions. This study was extended to the founding family member DAN, which also forms noncovalent dimers that are highly stable. These results demonstrate that certain DAN family members can form both monomers and noncovalent dimers, implying that biological activity of DAN family members might be linked to their oligomeric state.
Keywords: Transforming Growth Factor Beta (TGF-β), Bone Morphogenetic Protein (BMP), BMP antagonist, DAN family, Protein Related to Dan and Cerberus (PRDC), oligomerization
Introduction
The transforming growth factor-β (TGF-β) family consists of over thirty-three ligands that can be subdivided into three major classes, of which the bone morphogenetic protein (BMP) class is the largest. TGF-β ligands are involved in numerous cellular processes, such as cell proliferation and differentiation 1. Secreted ligands transduce cellular signals through the assembly of type I and II serine/threonine kinase receptors, which leads to the activation of Smad transcription factors 2.
TGF-β ligands are inhibited by extracellular antagonists, such as chordin, noggin, follistatin and the DAN (differential screening selected gene abberative in neuroblastoma) family of BMP antagonists 3; 4; 5; 6. Regulation of BMP ligands by extracellular antagonists is a critical mechanism for numerous developmental programs, including germ layer specification and temporospatial gradients important for the establishment of the dorsal-ventral axis and organ and skeletal formation 7; 8; 9; 10; 11; 12; 13. In addition to their role in development, BMP antagonists also have roles in the adult. Recent studies have indicated that increased levels of the BMP antagonist gremlin are associated with pulmonary fibrosis and diabetic nephropathy, possibly through countering of BMP7 signaling, which has been shown to attenuate renal fibrosis 14; 15; 16; 17. Furthermore, the role of BMP antagonists in cancer progression is mixed and context-dependent, as expected from the disparate nature of TGF-β ligands. For example, certain human tumors have been shown to express elevated levels of BMP antagonists, thereby blocking the anti-proliferative functions of BMP ligands 18; 19. On the other hand, elevated levels of BMP signaling (possibly through misregulation of extracellular antagonist expression) can support certain tumors 20; 21. Thus, attempts have been made to utilize extracellular BMP antagonists as tumor suppressors 22.
DAN family antagonists are small, single domain proteins (typically < 20 kDa) characterized by a core ‘DAN’ domain which contains a cystine knot motif 23. The knot structure is observed in a number of proteins including TGFβ ligands and consists of a conserved eight-residue ring formed by a pair of disulfide bonds that link two anti-parallel β-strands followed by an additional disulfide bond that travels through the ring 23. Members of the DAN family include DAN, gremlin, protein related to DAN and cerberus (PRDC), cerberus, sclerostin (SOST) and uterine sensitization-associated gene 1 protein (USAG1), among others. Most are glycoproteins 10; 24; 25 that antagonize BMP ligands with various affinities, and certain members can also antagonize Wnt ligands 4; 10; 24; 26. Deletion and replacement studies with the DAN domain indicate that this domain is essential for binding to BMP ligands 24; 27; 28. DAN family antagonists share structural characteristics with TGF-β family ligands 23. Recently, NMR structures of SOST revealed that the DAN domain consists of two long pairs of anti-parallel β-strands, typically referred to as ‘fingers’, joined by a coil segment with highly flexible N- and C-termini 29; 30. The structures confirmed that the disulfides formed a cystine knot arrangement, similarly to other growth factor proteins such as TGF-β,PDGF and NGF 31; 32; 33; 34; 35. Several growth factors that contain a cystine knot motif also have an unpaired cysteine that forms an intermolecular disulfide bond and links two monomers to form stable homo- or heterodimers. For TGF-β family ligands, this bond is necessary for maximal signaling activity, as mutation of the unpaired cysteine disrupts dimerization and significantly impairs ligand activity 36; 37. Interestingly, certain TGF-β family ligands, such as GDF9 and BMP15, lack an extra cysteine but still form functional homodimers 38. Recently, stability of the BMP15 dimer has been enhanced by introducing an unpaired cysteine in a location similar to other TGF-β family members, thereby forming a disulfide-bonded dimer 39. PRDC and most DAN family antagonists also contain an unpaired cysteine in a similar position to TGF-β family ligands, implying that DAN antagonists also form disulfide-linked dimers 10; 25. Unlike PRDC, the DAN family members SOST, USAG1 and DAN have an even number of cysteines. Interestingly, SOST, which contains 8 cysteine residues, has been shown to be monomeric, whereas DAN, which has 10 cysteine residues, has been suggested to form both covalent and noncovalent homodimers 10; 40.
Although DAN family antagonists have been thought to form disulfide-linked dimers, experimental evidence to support this has been lacking. Therefore, to investigate this we characterized both wild-type PRDC (PRDCWT) and PRDC where the unpaired cysteine was mutated to serine (PRDCC120S). In this study, we employ a variety of biophysical techniques to characterize the self-association, stability and biological activity of PRDCWT and PRDCC120S. We demonstrate that PRDC forms stable active dimers, which form independently of the unpaired cysteine and are also resistant to disulfide bond reduction. This study was extended to the antagonist DAN, which indicated that other DAN family members are non disulfide-linked dimers as well. Overall, the studies reported here provide insights into the domain structure and function of PRDC and other members of the DAN family.
Results
Structural comparison of DAN and TGF-β ligands
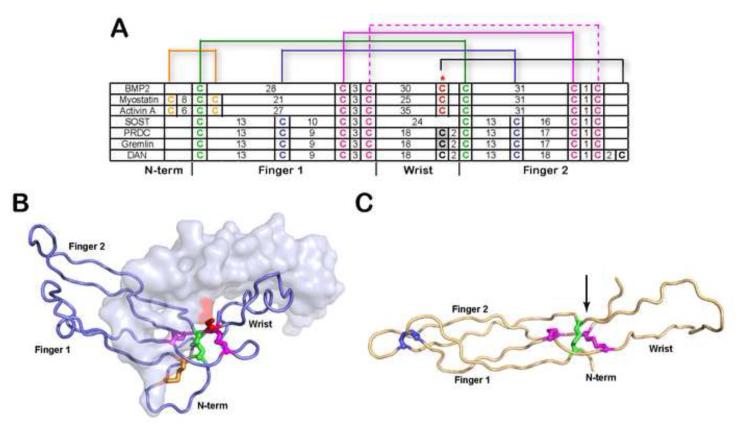
Based on the structural similarity to TGF-β ligands (distribution of cysteine residues, a cystine knot motif and the presence of an odd number of cysteines), PRDC has been suggested to exist as a disulfide-linked dimer 25. Alignment of the cysteine pattern of TGF-β ligands and DAN antagonists shows that the position of the unpaired cysteine is in a similar location to the cysteine that forms the intermolecular disulfide bond in TGF-β ligands (Fig. 1A). This is even more apparent when comparing the structures of TGF-β ligands, such as myostatin (Fig. 1B), and the DAN domain of the antagonist SOST (Fig. 1C). The structure of the DAN domain of SOST represents an adequate model for the DAN domain of PRDC since they share 26% sequence identity (residues 52-142 of PRDC compared to 79-169 of SOST). However, their overall sequence identity is only 18%, with significant differences occurring in both the sequence and length of the termini. Even though SOST does not contain an unpaired cysteine, the location of the putative unpaired cysteine (C120) for PRDC can be modeled onto the structure of SOST and is positioned after the wrist region, comparable to TGF-β ligands (Fig. 1C). This suggests that, if PRDC were to form an intermolecular disulfide bond similar to TGF-β ligands, the cysteine at position 120 would be the likely candidate.
Figure 1. Comparison of cysteine organization of DAN family antagonists to TGF-β ligands.
A) Disulfide linkage comparison of representative TGF-β ligands (BMP2, myostatin and activin A) and DAN family of antagonists. The number of amino acids spanning consecutive cysteines is indicated. The red asterisk indicates the cysteine responsible for dimerization in TGF-β ligands. For PRDC and gremlin, the putative unpaired cysteine implicated in dimerization is highlighted gray. Brackets represent disulfide-linkages. B) X-ray structure of myostatin (PDB:3HH2) depicting one monomer in surface and one in ribbon and C) NMR structure of SOST (PDB:2K8P). Disulfide bonds in (A-C) are colored consistently. The cystine knot is shown where two disulfide bonds on opposing β-strands (purple) form a ring structure with the polypetide chain linked through the center of the ring by a single disulfide bond (green). The arrow in (C) indicates the position of the unpaired cysteine in PRDC through alignment with SOST.
PRDC forms non-disulfide linked dimers
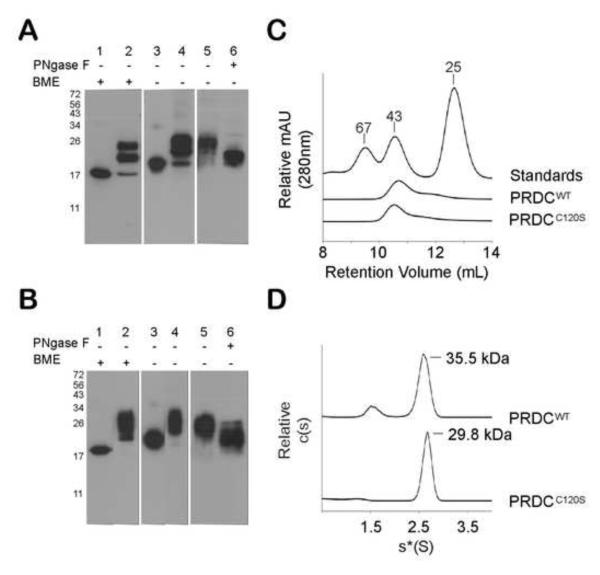
The characteristic disulfide-linked dimers of the TGF-β family are clearly identified through SDS-PAGE analysis and migrate at half the molecular weight under reducing compared to nonreducing conditions. Previous studies on PRDC and other DAN family members have not been as straightforward to interpret and often indicate the presence of both monomers and dimers under nonreducing conditions 10; 40. We previously demonstrated that bacterially derived PRDC refolded from inclusion bodies was biologically active 41. Under reducing conditions PRDCWT migrates as a single band at 17 kDa, which is consistent with the predicted MW of PRDC (Fig. 2A). However, under nonreducing conditions PRDCWT migrates at ~22 kDa. This indicates that either PRDC does not form disulfide-linked dimers or that disulfide-linked dimers migrate through the SDS gel at a smaller size than the expected MW of 34 kDa. To determine if this behavior was specific to bacterially derived PRDC, we transiently expressed PRDCWT with an N-terminal myc-his tag in HEK293F cells. PRDCWT derived from HEK293F cells showed three bands that migrated between the 17 and 26 kDa MW marker under reducing conditions. Similar to the bacterially derived PRDC, under nonreducing conditions the bands migrate at a slightly higher MW than the reducing conditions. Therefore, PRDC derived in HEK293F cells also does not exhibit a doubling in MW that would be expected of disulfide-bonded dimers (Fig. 2A). PRDC contains a single N-linked glycosylation consensus sequence and is glycosylated as treatment with PNGaseF lowers the MW of the protein (Fig. 2A). This is consistent with previous observations that PRDC is glycosylated and migrates slightly smaller than 26 kDa under both reducing and nonreducing conditions 25. Unless otherwise noted, bacterially produced PRDCWT was used for the remaining experiments.
Figure 2. PRDCWT and PRDCC120S are dimeric.
A) Western blot analysis of PRDCWT produced in E. coli (lanes 1 and 3) and in HEK293F (lanes 2, 4-6). If indicated, samples were treated with 5% 2-mercaptoethanol (BME) to reduce disulfide bonds prior to gel loading. PRDCWT produced in HEK293F cells was purified by His affinity resin (lane 5) and deglycosylated with PNGaseF (lane 6). B) Western blot analysis of PRDCC120S produced in E. coil and HEK293F similar to (A). A polyclonal anti-PRDC antibody was used for detection in (A) and (B). Size exclusion elution profile of PRDCWT, PRDCC120S and molecular weight standards. Purified proteins (100 μg) were applied to a Superdex 75 column. D) Sedimentation coefficient c(s) distribution profile of PRDCWT and PRDCC120S (1 mg/ml) determined by sedimentation velocity. After fitting for the frictional ratio (f/f0) the c(s) distribution was transformed into a c(M) distribution (not shown) to determine the molecular mass estimates (labeled).
The above data suggested that PRDC might not form a disulfide-linked dimer as expected but might be monomeric, similar to SOST. Therefore, we further characterized PRDCWT using size exclusion chromatography (SEC) to determine its oligomeric state. PRDCWT was applied to a Superdex 75 10/300 SEC column and the elution profile was compared to three MW standards (Fig. 2C). PRDCWT had a retention volume that eluted slightly smaller than the 43 kDa MW standard and near the MW of a dimer. Since molecular weight estimates from SEC can be skewed for non-spherical proteins, we pursued more definitive measurements of the molecular weight for PRDCWT using analytical ultracentrifugation sedimentation velocity. The c(s) distribution of the sedimentation profile (Fig. 2D) showed a species accounting for 82% of the observed molecules. Using the c(s) to fit the frictional ratio, the c(M) distribution was determined which resulted in a major peak with a predicted mass of 35.5 ± 5.7 kDa, which is compatible with the dimeric form of PRDCWT. Therefore, SEC and AUC data clearly indicate that PRDC is a dimeric protein.
To further rule out of the possibility that PRDC dimerization is mediated through a disulfide bond, we first mutated the putative free cysteine to serine (C120S). PRDCC120S was produced similarly to PRDCWT by refolding inclusion bodies in E. coli with the addition of a C-terminal 6x his tag, and the myc-tagged version was also expressed transiently in HEK293F cells. SDS-PAGE and Western blot analysis of both versions of PRDCC120S resulted in profiles similar to PRDCWT protein (Fig. 2B). This demonstrates that the increase in MW of PRDC under nonreducing conditions is not a result of disulfide bond formation through C120. Further analysis of PRDCC120S by SEC resulted in a peak that eluted in a similar retention volume to PRDCWT, indicating that PRDC dimers are still formed (Fig. 2C). Sedimentation velocity was also performed on PRDCC120S, which resulted in a sedimentation profile similar to PRDCWT. The velocity data indicated a single major sedimenting species with a calculated MW of 29.8 + 1.6 kDa (Fig. 2D). This data supports that PRDC forms dimers and that the putative free cysteine of PRDC is not involved in dimer formation.
PRDCWT and PRDCC120S bind BMPs with high affinity and specificity
In earlier coimmunoprecipitation studies, PRDC has been shown to inhibit BMP2 and BMP4 through direct interaction 25. To further characterize the binding of PRDCWT to BMP-type ligands and determine the effect that the PRDCC120S mutation has on binding, we analyzed BMP binding using surface plasmon resonance (SPR). PRDCWT and PRDCC120S protein were passed over a CM5 sensor chip coupled with BMP2, 4 or 7. Attempts to determine binding constants through kinetic analysis did not yield adequate fits, and therefore apparent KD values were estimated by analyzing the steady-state response at equilibrium (Table 1, Supplementary Fig. S1). Both PRDCWT and PRDCC120S have high affinity for BMP2 and BMP4, with apparent KD values less than 100 nM and a slightly lower affinity for BMP7. Similar to other TGF-β family antagonists, the PRDC-BMP complex dissociates slowly and requires short injections of GuHCl to fully clear PRDC from the BMP binding surface 42; 43. Furthermore, PRDC also exhibits strong specificity for BMP type ligands, as binding to a CM5 chip coupled with TGF-β1, activin A or myostatin was not detected (data not shown).
Table 1.
SPR binding analysis for the interactions of PRDCWT and PRDCC120S with BMP2, BMP4 and BMP7. KD (nM) values were determined by equilibrium analysis.
| Immobilized Ligand |
|||
|---|---|---|---|
| Analyte | BMP2 | BMP4 | BMP7 |
| PRDC WT | 54.1 ± 5.6 | 78.5 ± 5.8 | 107.5 ± 5.9 |
| PRDC C120S | 57.1 ± 7.8 | 70.7 ± 2.6 | 86.6 ± 3.7 |
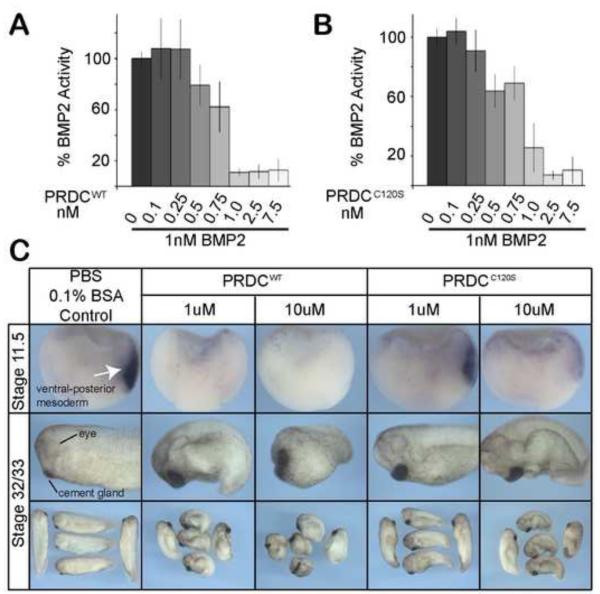
To determine if PRDCWT and PRDCC120S could also inhibit BMP signaling, we utilized a cell-based assay where C2C12 cells have been stably transfected with a luciferase-reporter gene under the control of a BMP responsive promoter 44. Cells were cultured with BMP2 ligand alone or ligand premixed with PRDCWT or PRDCC120S. PRDCWT effectively antagonized BMP2 at or near 1 nM (Fig. 3A). PRDCC120S also exhibited a similar dose response (Fig. 3B), indicating that the mutant did not alter BMP antagonism. To test the ability of PRDCWT and PRDCC120S to inhibit BMP signaling in vivo, we utilized a Xenopus embryological assay. In this assay inhibitors of endogenous BMP signaling can induce dorsalization and alter development by blocking the formation of BMP dependent ventral mesoderm tissue and inducing the formation of extra dorsal-anterior tissues, such as the head, resulting in a typical “dorsalized” embryo. Two concentrations (1 μM and 10 μM) of purified PRDCWT and PRDCC120S were injected into the blastocoel cavity of stage 9 blastula embryos. We assessed their ability to repress the BMP target gene sizzled at the late gastrula stage 45, and to induce a dorsalized phenotype at stage 32/33. Injection of both PRDCWT and PRDCC120S proteins resulted in a typical dorsalized phenotype with enlarged head and cement gland, and reduced tail (Fig. 3C). Furthermore, the expression of endogenous sizzled in the ventral-posterior mesoderm was completely inhibited by PRDCWT and dramatically reduced by PRDCC120S compared to the control injections (Fig. 3C). Overall, this data along with the SPR binding and luciferase reporter data supports that PRDCWT and PRDCC120S bind and inhibit BMP ligands with a similar activity. Therefore, the activity of PRDC is not dependent on the cysteine that was thought to be involved in dimer formation.
Figure 3. PRDCWT and PRDCC120S are potent BMP inhibitors.
A-B) Inhibition of BMP2 by PRDCWT (A) and PRDCC120S (B) was analyzed in a luciferase promoter assay. A BMP-responsive BRE-luc stable cell line was treated with 1 nM of BMP2 alone or combined with PRDC proteins titrated from 0.1 nM to 7.5 nM. Luciferase activity was normalized (100%) to cells treated with BMP2 ligand alone. Errors bars represent the standard deviation of 4 replicates. C) Activities of PRDCWT and PRDCC120S were measured by their ability to inhibit BMP signaling in vivo. Control buffer (PBS, 0.1% BSA), PRDCWT or PRDCC120S were injected at low (1 μM) and high (10 μM) concentrations into the blastocoel cavity at blastula stage 9 of Xenopus embryos, which were assayed by in situ hybridization for expression of the BMP target gene sizzled at stage 11.5 or cultured to stage 32/33 to assess the whole embryo phenotype.
PRDC dimerization and activity are not dependent on disulfide bonds
Since our results show that PRDC does not dimerize through the unpaired cysteine (C120), we wanted to rule out the possibility that covalent dimerization occurs through a different cysteine. Therefore, we analyzed non-reduced PRDCWT by mass spectrometry (Supplementary Fig. S2). The mass profile of PRDCWT as measured by MALDI-TOF MS is consistent with a monomeric form detected primarily as the M+H form at 17,413 Da (Supplementary Fig. S2B) under oxidizing conditions and at 17,110 (which is the expected MW) when reduced with 10 mM DTT. This mass shift upon reduction is consistent with PRDCWT modified by a single glutathione. Furthermore, this is consistent with PRDCWT and PRDCC120S lacking a reactive cysteines as measured by the Ellman’s test (data not shown). Other forms labeled as M+2H, 2M+H, etc are consistent with the typical ionization and detection by MALDI-TOF as demonstrated with two well-characterized protein standards of apomyoglobin and carbonic anhydrase (Supplementary Fig. S2A and S2E). To rule out any likelihood that disulfide bonds could have been hydrolyzed during the MALDI process, activin A, a known disulfide-bonded dimer, was also examined (Supplementary Fig. S2C). The activin A spectrum was consistent with a disulfide-linked dimer at about 26 kDa (monomer MW = 13 kDa). As a final confirmation of the non-covalent status of PRDCWT, electrospray ionization (LC-ESI-MS) produced a charge-state distribution ranging from +12 to +19 protons with a corresponding transformed protein mass center at 17,406 Da, which is consistent with the monomeric mass of PRDCWT with four disulfide bonds and a single glutathionine addition (Supplementary Fig. S2F). Importantly, there is no continuous charge-state distribution that could account for a covalent dimer of the protein, thus ruling out the possibility of a substantial amount of a disulfide-linked dimer. Similar LC-ESI-MS results were also obtained after reduction of PRDCWT with DTT (data not shown).
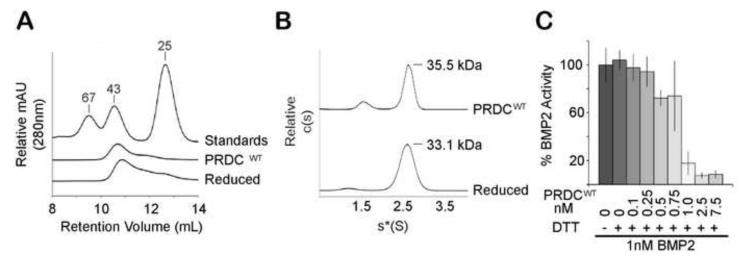
After determining that PRDC was not a covalently linked dimer, we next wanted to determine the significance of the intramolecular disulfide bonds on the dimerization mechanism and activity of PRDC. Since the DAN domain has been implicated for BMP inhibition, we anticipated that complete reduction of the disulfide bonds would disrupt the cystine knot motif and inactivate PRDC and possibly disrupt dimerization. Therefore, we assessed the self-association properties of PRDC under conditions where all disulfide bonds have been reduced. We first determined the concentration of DTT needed to completely reduce PRDCWT (Supplementary Fig. S3). Surprisingly, analysis of reduced and nonreduced PRDCWT by SEC showed that both had similar elution profiles with a peak corresponding to dimeric PRDC (Fig. 4A). In addition, we also performed sedimentation velocity experiments on reduced PRDCWT, which revealed one main species at a predicted MW of 33.1 ± 3.3 kDa (Fig. 4B). Therefore, both SEC and AUC velocity experiments indicate that fully reduced PRDCWT maintains a dimer structure. Unexpectedly, this data demonstrates that PRDC dimers still persist under conditions that reduce the cystine knot motif. Furthermore, since all cysteine residues are reduced, this data also supports that an intermolecular disulfide bond is not involved in dimer formation and is consistent with the mass spectrometry analysis. Following this observation, we also wanted to determine if fully reduced PRDCWT could still inhibit BMP2 in the C2C12 luciferase reporter assay. A stock solution of concentrated PRDCWT was fully reduced in 1 mM DTT and diluted upon mixing with BMP2 ligand. Since BMP2 consists of disulfide bonds necessary for robust signaling, we first confirmed BMP2 retained significant activity in the presence of the diluted DTT (Fig 4C). Interestingly, fully reduced PRDCWT still effectively inhibited BMP2 signaling at a similar concentration to that of nonreduced PRDC. To rule out the possibility that 1 mM DTT resulted in incomplete reduction, we performed a similar array of experiments after treating PRDCWT with 10 mM DTT and confirmed that PRDC retains BMP inhibition and remains dimeric when completely reduced (Supplementary Fig. S4). This data indicates that both dimerization and activity are not significantly altered when PRDC is fully reduced.
Figure 4. Reduced PRDCWT remains dimeric and biologically active.
A) PRDCWT and PRDCWT reduced with 1 mM DTT elute with a similar MW profile during SEC analysis on a Superdex 75 column. B. Sedimentation coefficient distribution profile of PRDCWT at 1 mg/ml reduced with 2-mercaptoethanol was determined by sedimentation velocity. C) Activity of reduced PRDCWT was measured in the luciferase reporter assay using the BRE-luc stable cell line by titrating PRDCWT from 0.1 nM to 7.5 nM against 1 nM BMP2. Prior to mixing with BMP2, PRDCWT was reduced with 1 mM DTT. Errors bars represent the standard deviation of 4 replicates.
PRDC Dimers are highly stable
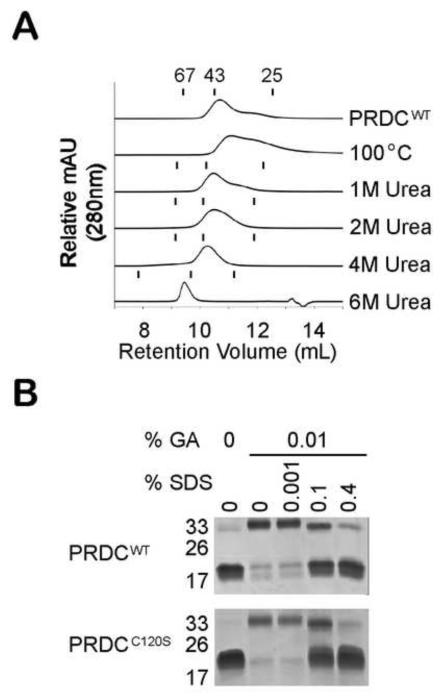
Since it was unexpected that PRDC remains dimeric in the presence of reducing agents, we also wanted to determine how stable the PRDC dimers are under different denaturing conditions. Since all SEC experiments were performed under high salt conditions (800 mM), ionic interactions do not appear to play a significant role in dimerization. We first incubated PRDC at 100°C for 5 minutes to thermally denature the protein. Surprisingly, PRDCWT eluted as a peak that corresponds to the dimer MW, although the peak appeared broad, possibly indicating the presence of both dimers and monomers (Fig. 5A). We further analyzed the stability of the PRDC dimer under chemical denaturation using urea. PRDCWT was incubated with 1 M, 2 M, 4 M and 6 M urea and analyzed by SEC with a column equilibrated in the corresponding molarity of urea. MW size standards were analyzed under different urea concentrations and compared to PRDC proteins (Fig. 5A). Up to 6M urea PRDCWT eluted at or around the 43 kDa MW marker, indicating that PRDC maintained a dimeric structure. In 6M urea, PRDC elutes at a larger than expected MW, indicating an increase in the hydrodynamic radius and possible denaturing. Similar results were observed for PRDCC120S under thermal and chemical denaturing conditions (data not shown). We next wanted to test if we could dissociate the dimer using detergents. Since detergents can significantly alter the protein behavior on SEC and hinder MW analysis, we performed a cross-linking experiment to measure the presence of PRDC dimer. Here we mixed PRDC with increasing amounts of sodium dodecyl sulfate (SDS) and cross-linked with glutaraldehyde prior to SDS-PAGE analysis. Without glutaraldehyde, PRDC migrates as a monomer under normal denaturing conditions (Fig. 5B). Dimeric PRDC was clearly visible when the cross-linking reaction was performed in a buffer without detergent. We then titrated increasing concentrations of SDS into the cross-linking reaction for both PRDCWT and PRDCC120S. At low concentrations of SDS, even up to 0.1% SDS, significant PRDC dimer was still observed. Not until we incubated PRDC with high concentrations of SDS (0.4%) did PRDC migrate as a monomer, further indicating that PRDC dimers are quite stable.
Figure 5. PRDCWT is a highly stable dimer.
A) Comparison of the elution profiles from SEC analysis of 100 μg of PRDCWT protein subjected to various denaturing conditions. For thermal analysis, PRDCWT was heated to 100°C and then analyzed at 25°C. For chemical denaturant analysis, PRDCWT was mixed with urea to a final concentration of 1 M, 2 M, 4 M or 6 M prior to loading onto a Superdex 75 column equilibrated under the same urea concentrations. The retention volume of the three MW standards (67, 43 and 25 kDa) under different urea concentrations are represented by tick marks above the corresponding elution profiles. B) SDS-PAGE analysis of PRDCWT and PRDCC120S cross-linked with 0.01% glutaraldehyde (GA) in the presence of increasing concentrations of SDS.
DAN also forms stable non-disulfide bonded dimers
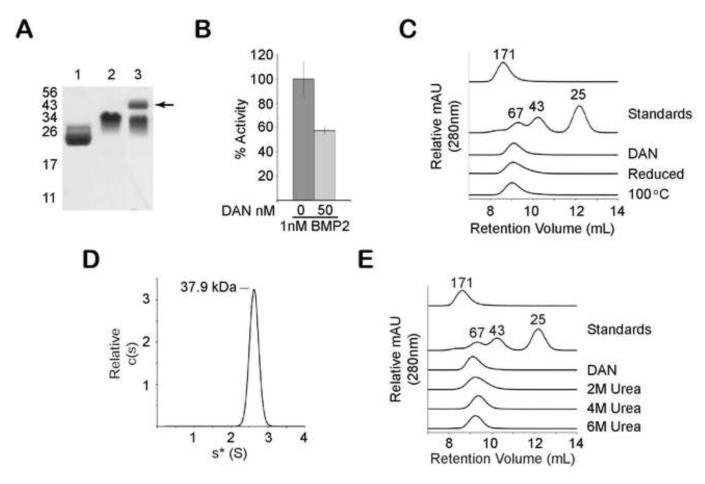
To determine if other DAN family members besides PRDC formed highly stable noncovalent dimers, we investigated the oligomeric state of the protein DAN - the founding family member. Unlike PRDC, DAN has an additional cysteine residue, which is thought to disulfide bond with the cysteine homologous to C120 of PRDC (Fig 1A). Previous studies have suggested that DAN forms a disulfide-linked dimer 10. More recently, cross-linking experiments have suggested that DAN forms a non-disulfide linked dimer 40. Since cross-linking experiments can sometimes be misleading, we wanted to resolve this discrepancy by analyzing the oligomeric state of DAN through more analytical techniques (e.g. SEC and AUC) Therefore, we generated a stable CHO cell line expressing the DAN protein linked to a cleavable C-terminal myc His tag. DAN was purified to homogeneity from conditioned medium and the epitope tag was removed by proteolysis (Fig 6A). Although not as potent as PRDC, purified DAN was shown to inhibit BMP signaling similarly to previously published results (Fig 6B) 40. SDS-PAGE analysis shows that DAN migrates at the expected monomer MW of 18 kDa under reducing conditions, but, similarly to PRDC, DAN migrates at a larger MW under nonreducing conditions (Fig. 6A). Preliminary cross-linking experiments followed by SDS-PAGE analysis of DAN revealed the presence of both monomers and dimers (Fig 6A). We then analyzed the purified DAN protein by SEC, which showed a single peak that corresponded to a size greater than the 67 kDa standard but smaller than the void volume (Fig. 6C). These results suggest that DAN is either a higher order oligomer or that DAN adopts a shape that significantly alters the elution profile and limits the accuracy of MW calculations. Therefore, sedimentation velocity analysis was used to more precisely determine the MW of DAN, which revealed a single major sedimenting species with a calculated MW of 37.9 + 3.0 kDa MW (Fig. 6D). This correlated closely with the MW of two molecules of DAN, indicating that DAN also forms dimers. We then tested whether DAN dimers also exhibited high stability. Similar to PRDC, DAN maintained its dimeric structure under high urea concentrations as measured by SEC (Fig 6E). Furthermore, DAN dimers were resistant to reducing conditions and thermal denaturation at 100°C (Fig. 6C). These results indicate that, similarly to PRDC, DAN forms a highly stable noncovalent dimer and are consistent with previous cross-linking results 40.
Figure 6. The protein DAN is also a highly stable dimer.
A) SDS-PAGE analysis of purified DAN from CHO-DG44 cells (lane 1-reduced, lane 2-nonreduced) and DAN cross-linked in 0.05% glutaraldehyde (lane 3). Arrow indicates the appearance of a band at the expected mass of a DAN dimer (~43 kDa) B) Inhibition of BMP2 activity using purified DAN in the BRE-luc stable cell reporter assay. C) SEC analysis comparing the elution profile of DAN to reduced and thermally denatured DAN. MW standards are indicated, including aldolase, which has a MW of 171 kDa and elutes in the void volume of the Superdex S75 column. D) Sedimentation coefficient distribution profile of DAN (1 mg/ml) was determined by sedimentation velocity. After fitting for the frictional ratio (f/f0) the c(s) distribution was transformed into a c(M) distribution (not shown) to determine the molecular mass estimates (labeled). E) SEC elution profile of DAN on a Superdex S75 column equilibrated at different urea concentrations (2 M, 4 M or 6 M). Profiles demonstrate that DAN remains dimeric under denaturing conditions.
Discussion
BMP ligands are regulated by a number of extracellular binding proteins. The DAN family of BMP antagonists represents the largest group of BMP inhibitors that all contain a similar structural core or ‘DAN domain’ that is defined by a cystine knot motif and a similar spacing of cysteine residues (Fig. 1). Interestingly, DAN family members exhibit variation in the total number of cysteine residues. Certain family members contain an unpaired cysteine that has been proposed to form an intermolecular disulfide bond, similar to the covalently linked TGF-β ligand dimers. In this study, we wanted to provide experimental evidence that DAN family members that contain an unpaired cysteine are indeed disulfide-bonded dimers. Our results show that PRDC does in fact form dimers, but, contrary to the proposed mechanism, dimerization is not dependent on disulfide-bond formation.
Even though the dimerization mechanism does not depend on covalent attachment, PRDC and DAN dimers are highly stable. This appears to be significantly different from TGF-β ligands and other disulfide linked dimers (e.g. PDGF) that form monomers when the intermolecular cysteine is disrupted 36; 37. We also determined that PRDC retains significant activity under strong reducing conditions, which indicates that the cystine knot motif can be reduced and PRDC still retains activity and maintains a dimeric structure. The basis for this dimerization remains unknown. Since the cystine knot motif is near one end of the molecule it is possible that the structural integrity of the DAN domain, specifically the anti-parallel β-strands, is maintained under reducing conditions. This is consistent with the data that indicates BMP binding activity is located within the anti-parallel β-strands of the DAN domain. Alternatively or in combination, regions outside the DAN domain could be important for dimerization and therefore, reduction of the DAN domain would not alter dimerization.
The cystine knot motif appears frequently in growth factor families where the motif is thought to be important for proper folding. For instance, a missenese mutation of one of the cysteine residues in the cystine knot motif of the SOST gene results in protein that is retained in the endoplasmic reticulum and impairs activity. In most cases cystine knot motif proteins are dimeric with variations on how the monomers are joined to stablize dimers 46. For instance, in certain cases (e.g. VEGF, PDGF and TGF-β) monomers are covalently attached through a disulfide bond. Furthermore, certain families have a prodomain (e.g. NGF, PDGF and TGF-β) with chaperone-like properties required for the formation of the bioactive dimers. In the case of TGF-β, the prodomain also appears important for regulating homodimerzation, as each prodomain makes significant contact with the opposing mature monomer 47. This mechanism might be particularly important for ligands that appear to have an unstable dimer interface, which has been suggested from multiple structures of activin A and TGF-β3 5; 47; 48; 49; 50; 51. In contrast to TGF-β family ligands, we have shown that DAN family members assemble into dimers that are not disulfide-linked but are highly stable. Since DAN family members lack a prodomain, this stability may play an important role in formation of a proper bioactive dimer. One possibility is that the high dimer stability observed for DAN family proteins obviates the need for a prodomain. It is also possible that the stability of the dimer represents a low energy state that limits formation of heterodimers, which have not been reported for DAN family members. Similarly to DAN family members, the gonadotrophin hormones (e.g. LH and FSH) do not require a prodomain and also form noncovalently linked dimers. In this case, hormones form stable heterodimers where the common alpha subunit is ‘locked’ into place by the variable β-subunit 52; 53.
The mechanism of how DAN family members inhibit BMP ligands is poorly understood. With the exception of SOST, most DAN family members are strong to moderate inhibitors of BMP signaling 25; 26; 28; 41. Inhibitors have a relatively low sequence identity (Supplementary Fig. S5) making it difficult to pinpoint the residues that are important for strong BMP binding and/or dimerization. Interestingly, SOST is the only DAN family member identified to date that is monomeric 29; 30. Therefore, besides individual residues conferring BMP binding, it is possible that DAN family members need to be dimers in order to have an increased affinity for BMP ligands. This binding strategy would be consistent with other TGF-β family antagonists that neutralize dimeric TGF-β ligands. For instance, the antagonist follistatin, which is not dimeric, forms an inhibitory complex that requires two molecules of follistatin bound to one TGF-β family dimer 5; 54. On the other hand noggin, which is a disulfide-linked dimer, binds in a 1:1 complex with the dimeric ligand where each monomer of noggin binds to a monomer of ligand3. Therefore, a preformed dimer with two binding sites, as in PRDC and DAN, would have the advantage of increased affinity over the monomer through avidity effects. In fact, this principle is why the Fc-receptor fusion proteins have a much higher affinity than the individual domains (sometimes as much as 1000-fold) for the ligands 55; 56. If each monomer of PRDC and DAN could bind BMP, then dimerization would be a way to increase the overall affinity for BMP. Alternatively, dimerization might create a unique interface that specifically interacts with BMP. Due to the high stability of the dimer, monomers of PRDC and DAN cannot be isolated and therefore further characterization of the DAN:BMP interaction is needed to distinguish between these two possibilities.
For numerous signaling ligands and extracellular protein agonists and antagonists, free cysteines can play critical roles in defining protein activity and function. For example, in the Wnt signaling cascade acylation of Wnt ligands is critical for allowing these proteins to bind to their target frizzled (Fzd) receptors 57; 58. In addition, within the TGF-β signaling cascade, the latent TGF-β binding protein (LTBP) binds to latent signaling ligands through their associated propeptides through disulfide exchange, resulting in the formation of a covalent disulfide bond that ultimately promotes ligand signaling 59. Furthermore, the platelet integrin αIIbβ3 has been characterized to maintain free cysteine residues in vivo, in addition to numerous disulfide bonds, that are capable of undergoing regular disulfide exchange to allow for the interconversion between active and resting state integrin 60. Interestingly, these free cysteines in a number of cases can be modified to protect the protein from potential rearrangement, promote the active state stabilization, or be used for future degradation 61; 62. It is possible that glutathionylation, or another commonly occurring cysteine modification, may play a role in PRDC (and possibly gremlin and cerberus) activity and functioning in the greater scheme of TGF-β signaling 61; 62. From our data, it seems that this would be unlikely to occur for antagonism of BMP signaling, but may play a pertinent role in the cross-talk of the DAN family members with both the VEGF signaling and Wnt signaling cascades 63; 64; 65; 66. In contrast, the DAN protein has an even number of cysteines, which might inhibit this potential modification and distinguish it from the other DAN family members. Future studies will most certainly be aimed at addressing these questions and the potential in vivo role of cysteine modification for the DAN family proteins.
The data presented in this paper indicate that PRDC and DAN family members form stable noncovalent dimers. Interestingly, a number of unique dimerization mechanisms and structures exist for cystine knot growth factor families and play a critical role in defining their function. How DAN family members form noncovalent dimers should be the focus of future studies. It will also be interesting to compare the structures of dimeric DAN family members with the monomeric SOST protein, which could provide an explanation for high affinity BMP interaction.
Materials and Methods
Generation of DNA constructs
The gene sequence corresponding to amino acids 22 – 168 of mouse PRDC was amplified from pCS2-PRDC-6xMyc (a gift from Eek-hoon Jho, University of Seoul, Korea) by standard PCR. The insert was cloned into pET21a using NdeI and XhoI and designated pET21a-PRDC. The TGA stop codon was included in the 3′ primer, resulting in a protein product with no tags. Full length PRDC was amplified from pCS2-PRDC-6xMyc and cloned into pcDNA4, designated pcDNA4-mPRDC, using BamHI and XhoI. Mutation of Cys120 to Ser (C120S) was done using the QuikChange Mutagenesis (Stratagene) protocol with pET21a-mPRDC and pcDNA4-mPRDC. C120S in pET21a contained a 6xHis tag. The human Dan gene was purchased from Open Biosystems and cloned into pOptivec (Invitrogen) using XbaI and NotI. The 3′ primer included sequences that added a PreScission protease (PP) cleavage site (LEVLFQGP), myc tag and 6x histidine tag to the C-terminal end of the DAN protein, resulting in the construct pOptivec-DAN-PP-Myc-His.
Protein Expression and Purification
Recombinant PRDC was produced as previously described 41. Briefly, PRDCWT and PRDCC120S were expressed as inclusion bodies in E. coli. PRDC from the inclusion bodies was purified prior to refolding in 50 mM Tris, 150 mM NaCl, 5 mM GSH, 5 mM oxidized GSH, 0.5 mM cysteine, 5 mM EDTA, 0.5 M arginine, pH 8.5. Refolded PRDC was further purified with a C18 reverse phase HPLC column. Finally, pure PRDC was buffer exchanged with 20 mM HEPES, 150 mM NaCl, pH 7.5.
PRDCWT and PRDCC120S in pcDNA4 were transfected into HEK293F (Freestyle) cells (Invitrogen) using 293fectin. Media was collected after 6 days and purified through histidine affinity resin (His•Bind, Novagen) equilibrated with 50 mM NaH2PO4, 300 mM NaCl, pH 8 and eluted with 500 mM imidazole. Proteins produced in HEK293F were semi-quantified by western blot using a PRDC pAb (AF2069, R&D Systems) and known standards from bacterially derived PRDC. PRDC was deglycosylated in 0.1% SDS and heated to 100°C for 5 min prior to treatment with 0.75% NP-40 and overnight incubation with 5 μU of N-glycanase (Prozyme).
To generate the protein DAN, CHO-DG44 cells (obtained from Lawrence Chasin, Columbia University) were transfected with pOptivec-DAN-PP-Myc-His, and a high expressing clone was selected and amplified with increasing concentrations of methotrexate. Conditioned medium from the DAN expressing cells was applied to histidine affinity resin equilibrated with 50 mM NaH2PO4, 300 mM NaCl, pH 8 and eluted with the addition of 1 M imidazole. Protein was dialyzed in 50 mM NaH2PO4, 300 mM NaCl, pH 8 overnight at 4°C. The C-terminal tag was removed by digesting 1 mg of DAN with 60 μg of PreScission protease at 4°C for 16 h. DAN was further purified and separated from the tags by size exclusion chromatography on a Superdex S75 HR 10/300 column (GE Biosciences) in 20 mM HEPES, 250 mM NaCl, pH 7.5. DAN eluted in a single peak that was pooled, and the protein was quantified by the absorbance at 280 using an extinction coefficient of 0.826 M−1cm−1. The resulting full length DAN protein has the additional amino acids LEVLFQ added to the C-terminus.
Size Exclusion Chromatography (SEC)
PRDCWT, PRDCC120S and DAN were applied to a Superdex 75 HR 10/300 column at room temperature. The column was pre-equilibrated with 20 mM HEPES, 800 mM NaCl, pH 7.5 buffer. For analysis of samples under reducing conditions, 10 mM DTT and 1 mM tris(2-carboxyethyl)phosphine (TCEP) were added to the above buffer. For analysis of samples under denaturing conditions, different concentrations of urea ranging from 1 to 6 M were added to the above buffer. In all cases, 100 μg of protein was applied to the column and eluted at a flow rate of 0.5 ml/min. The molecular mass standards used for comparison were aldolase (171 kDa) bovine serum albumin (67 kDa), ovalbumin (43 kDa) and chymotrypsinogen (25 kDa).
Analytical Ultracentrifugation (AUC)
Sedimentation velocity analysis was performed with a Beckman XL-I analytical ultracentrifuge (Beckman Coulter, Fullerton, CA). Protein samples were dialyzed to osmotic equilibrium against 20 mM HEPES at pH 7.5, 150 mM NaCl prior to loading. For analysis under reducing conditions, samples were dialyzed in the above buffer with 0.5 mM 2-mercaptoethanol. Three separate protein samples at a concentration of 0.25, 0.5 and 1 mg/ml, along with the reference buffer, were loaded into separate compartments. DAN protein was equilibrated in 50 mM NaH2PO4, 300 mM NaCl, pH 8 and sedimentation velocity experiments were performed at 1 mg/ml. The sedimentation velocity experiments were carried out at 20°C and 46,000 rpm (AN-60Ti rotor), and 250-300 scans were collected at 2 min intervals. Absorbance readings were measured at 230 nm, a wavelength with minimal buffer absorbance. The data were fitted to a continuous sedimentation coefficient (c(s)) distribution model with the program SEDFIT 67. Molecular mass estimates were determined after fitting the frictional ratio and are based on a continuous c(M) analysis in SEDFIT.
Cross-linking
Approximately 3 μg of protein samples were cross-linked with 0.01% glutaraldehyde for 20 min at room temperature. Native cross-linking reactions were performed in buffer alone (20 mM HEPES, 250 mM NaCl, pH 7.5) or buffer with the addition of SDS as indicated. The cross-linking reaction was neutralized with 1 M Tris pH 8 to a final concentration of 200 mM. Samples were normalized with the highest % SDS prior to PAGE analysis. All conditions were separated by SDS-15% PAGE under nonreducing conditions.
Luciferase-reporter assay
A C2C12 cell line stably transfected with the luciferase gene under control of a BMP-responsive promoter 44 (Kindly provided by Dr. Gareth Inman of the Beatson Institute for Cancer Research, UK) was used to measure the function of PRDC. Culture procedures have been previously described. Briefly, cells were plated at 1.6×104 cells/well in 96-well plates with DMEM/High glucose, 0.7 mg/ml G418, 10% FBS, penicillin and streptomycin and grown at 37°C in 5% CO2 for 18 hrs. The medium was replaced with DMEM/High glucose, 0.1% FBS, penicillin, streptomycin and incubated for 6 to 8 h. BMP2 was mixed with PRDC in DMEM/High glucose, 0.1% FBS, penicillin, and streptomycin, allowed to incubate at room temperature for 45 to 60 min and applied to the C2C12-BRE cells, which were then incubated overnight. To analyze PRDC under reducing conditions, the protein was incubated for 1 h with 1 mM DTT, then mixed with BMP2. Cells were lysed in 20 μl of Passive Lysis Buffer (Promega). Cell lysates were transferred to a 96-well isoplate (Perkin Elmer) then mixed with 40 μl of luciferase substrate (Promgea). Luminescence was measured using a BioTek Synergy H1 plate reader. Error bars indicate the standard deviation of fold activation from 4 individual wells.
Xenopus embryo BMP target gene assay
Embryo manipulations and microinjections were performed as previously described 68 and staged according to the normal table of development for Xenopus laevis 69. To assay PRDC activity in vivo the blastocoel cavities of stage 9 Xenopus embryos were injected with 1 μM or 10 μM of either PRDCWT or PRDCC120S. The total injection volume was adjusted to a constant 40 nl using PBS with 0.1% BSA. After injection, embryos were cultured at room temperature until stage 11.5, fixed overnight at 4°C in MEMFA and analyzed for expression of the BMP-target gene sizzled via whole-mount in-situ hybridization as previously described 68. The sizzled in-situ probe was prepared using T7 RNA polymerase with SalIlinearized pCMV-Sport6-sizzled plasmid template (IMAGE clone 4057152 obtained from Open Biosystems, cat.no. EXL1051-4538645).
Surface Plasmon Resonance (SPR)
SPR measurements were performed in a BIAcore 3000 system (GE Healthcare) and were analyzed using Scrubber 2 (Biologic Software). Recombinant BMP2, 4 & 7 were dissolved at 2.5 μg/ml in 10 mM sodium acetate (pH 5.0) and immobilized on a CM5 sensor chip, using random amine coupling, to a level of 2466, 2108 and 2050 response units, respectively. A mock flow cell was activated and blocked in the absence of protein. The mock cell was then used to subtract nonspecific binding and refractive index changes resulting from changes in bulk properties of the solution. Analyte binding and washes were performed at 25°C in 20 mM HEPES, pH 7.5, 500 mM NaCl, 3.4 mM EDTA, 0.005% (v/v) P20 surfactant. To assess binding affinity, equilibrium analyses were performed in which various concentrations of PRDC proteins were passed over the immobilized ligands. Each experimental cycle consisted of an initial 15-min analyte injection (association) into the respective flow cells at a flow rate of 5 μl/min, followed by a 10-min injection of buffer alone (dissociation). After each cycle, the surfaces of the chip were regenerated by a four short injections of buffer containing 2 M guanidine-HCl at a flow rate of 100 μl/min. Steady-state binding responses (RUmax) were plotted as a function of PRDC concentration (C) and then subjected to nonlinear regression analysis using the equation RUmax=Rmax/(1+ KD/C), where KD is equilibrium dissociation constant and Rmax is the maximal response at a saturating concentration of PRDC (GraphPad Prism software).
Mass Spectrometry
PDRC and Activin A samples were analyzed by both MALDI-TOF and LC-ESI-MS to determine if covalent dimers existed. For MALDI-TOF analysis, 200 ng of PRDC was mixed with 5 mg/ml sinapinic acid matrix and spotted onto a MALDI-TOF sample plate. Linear MALDI-TOF spectra were collected in positive ion mode on an AB Sciex 4800 TOF/TOF system with an average of 1000 spectra collected for each sample. For LC-ESI-MS, 120 ng of PDRC was injected onto a Vydac C4, 300Å, 5 μ, 15 cm x 300 μm capillary column in 10% acetonitrile containing 0.1% formic acid. The protein was separated and detected using an LCPackings Ultimate capillary HPLC coupled to the ThermoFisher LCQ Deca XP Max ion trap system. After a 3 min wash with 10% acetonitrile at 5 μl/min, the protein was eluted with a rapid gradient to 90% acetonitrile over 7 min. Mass spectra were collected at 4 kV in positive ion mode using Xcaliber ver 1.4, SR1 software. The resulting spectra are presented as the average charges-state distribution (25 scans) from the eluted protein and as a single mass profile by transforming the charge-state profile with the ProMass for Xcaliber ver 2.5, SR 1 algorithm (Novatia, LLC)
Supplementary Material
Highlights.
DAN family members are BMP inhibitors that have been suggested to form covalent dimers through an unpaired cysteine
PRDC and DAN function as a dimer.
The unpaired cysteine in PRDC is not responsible for dimerization.
PRDC and DAN dimers are maintained under reducing and denaturing conditions indicating that the dimers are highly stable.
DAN family members form stable, noncovalently linked dimers that antagonize BMP signaling.
Acknowledgements
This work was supported by research grants from the NIHGM R01 (GM084186) to T.B.T. Xenopus experiments were supported by NIDDK R01 (DK070858) to A.M.Z.
Abbreviations
- Bone Morphogenetic Protein
(BMP)
- Differential Screening Selected Gene Abberative in Neuroblastoma
(DAN)
- Protein Related to DAN and Cerberus
(PRDC)
- Growth and Differentiation Factor
(GDF)
- Sclerostin
(SOST)
- Uterine Sensitization-associated Gene 1 protein
(USAG-1)
- PreScission protease
(PP)
- Surface Plasmon Resonance
(SPR)
- Analytical Ultracentrifugation
(AUC)
- Size Exclusion Chromatography
(SEC)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession NumbersGenBank accession nos. BAA29038.1, BAA92265.1, AAH07858
References
- 1.Hogan BL. Bone morphogenetic proteins in development. Curr Opin Genet Dev. 1996;6:432–8. doi: 10.1016/s0959-437x(96)80064-5. [DOI] [PubMed] [Google Scholar]
- 2.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–78. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 3.Groppe J, Greenwald J, Wiater E, Rodriguez-Leon J, Economides AN, Kwiatkowski W, Affolter M, Vale WW, Belmonte JC, Choe S. Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature. 2002;420:636–42. doi: 10.1038/nature01245. [DOI] [PubMed] [Google Scholar]
- 4.Hsu DR, Economides AN, Wang X, Eimon PM, Harland RM. The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol Cell. 1998;1:673–83. doi: 10.1016/s1097-2765(00)80067-2. [DOI] [PubMed] [Google Scholar]
- 5.Thompson TB, Lerch TF, Cook RW, Woodruff TK, Jardetzky TS. The structure of the follistatin:activin complex reveals antagonism of both type I and type II receptor binding. Dev Cell. 2005;9:535–43. doi: 10.1016/j.devcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Stamler R, Keutmann HT, Sidis Y, Kattamuri C, Schneyer A, Thompson TB. The structure of FSTL3.activin A complex. Differential binding of N-terminal domains influences follistatin-type antagonist specificity. J Biol Chem. 2008;283:32831–8. doi: 10.1074/jbc.M801266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearce JJ, Penny G, Rossant J. A mouse cerberus/Dan-related gene family. Dev Biol. 1999;209:98–110. doi: 10.1006/dbio.1999.9240. [DOI] [PubMed] [Google Scholar]
- 8.Xie J, Fisher S. Twisted gastrulation enhances BMP signaling through chordin dependent and independent mechanisms. Development. 2005;132:383–91. doi: 10.1242/dev.01577. [DOI] [PubMed] [Google Scholar]
- 9.Muller, Knapik EW, Hatzopoulos AK. Expression of the protein related to Dan and Cerberus gene--prdc--During eye, pharyngeal arch, somite, and swim bladder development in zebrafish. Dev Dyn. 2006;235:2881–8. doi: 10.1002/dvdy.20925. [DOI] [PubMed] [Google Scholar]
- 10.Stanley E, Biben C, Kotecha S, Fabri L, Tajbakhsh S, Wang CC, Hatzistavrou T, Roberts B, Drinkwater C, Lah M, Buckingham M, Hilton D, Nash A, Mohun T, Harvey RP. DAN is a secreted glycoprotein related to Xenopus cerberus. Mech Dev. 1998;77:173–84. doi: 10.1016/s0925-4773(98)00139-7. [DOI] [PubMed] [Google Scholar]
- 11.Glinka A, Wu W, Onichtchouk D, Blumenstock C, Niehrs C. Head induction by simultaneous repression of Bmp and Wnt signalling in Xenopus. Nature. 1997;389:517–9. doi: 10.1038/39092. [DOI] [PubMed] [Google Scholar]
- 12.Bouwmeester T, Kim S, Sasai Y, Lu B, De Robertis EM. Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann’s organizer. Nature. 1996;382:595–601. doi: 10.1038/382595a0. [DOI] [PubMed] [Google Scholar]
- 13.Chi L, Saarela U, Railo A, Prunskaite-Hyyrylainen R, Skovorodkin I, Anthony S, Katsu K, Liu Y, Shan J, Salgueiro AM, Belo JA, Davies J, Yokouchi Y, Vainio SJ. A secreted BMP antagonist, Cer1, fine tunes the spatial organization of the ureteric bud tree during mouse kidney development. PLoS One. 2011;6:e27676. doi: 10.1371/journal.pone.0027676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koli K, Myllarniemi M, Vuorinen K, Salmenkivi K, Ryynanen MJ, Kinnula VL, Keski-Oja J. Bone morphogenetic protein-4 inhibitor gremlin is overexpressed in idiopathic pulmonary fibrosis. Am J Pathol. 2006;169:61–71. doi: 10.2353/ajpath.2006.051263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMahon R, Murphy M, Clarkson M, Taal M, Mackenzie HS, Godson C, Martin F, Brady HR. IHG-2, a mesangial cell gene induced by high glucose, is human gremlin. Regulation by extracellular glucose concentration, cyclic mechanical strain, and transforming growth factor-beta1. J Biol Chem. 2000;275:9901–4. doi: 10.1074/jbc.275.14.9901. [DOI] [PubMed] [Google Scholar]
- 16.Dolan V, Murphy M, Sadlier D, Lappin D, Doran P, Godson C, Martin F, O’Meara Y, Schmid H, Henger A, Kretzler M, Droguett A, Mezzano S, Brady HR. Expression of gremlin, a bone morphogenetic protein antagonist, in human diabetic nephropathy. Am J Kidney Dis. 2005;45:1034–9. doi: 10.1053/j.ajkd.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Walsh DW, Roxburgh SA, McGettigan P, Berthier CC, Higgins DG, Kretzler M, Cohen CD, Mezzano S, Brazil DP, Martin F. Co-regulation of Gremlin and Notch signalling in diabetic nephropathy. Biochim Biophys Acta. 2008;1782:10–21. doi: 10.1016/j.bbadis.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Namkoong H, Shin SM, Kim HK, Ha SA, Cho GW, Hur SY, Kim TE, Kim JW. The bone morphogenetic protein antagonist gremlin 1 is overexpressed in human cancers and interacts with YWHAH protein. BMC Cancer. 2006;6:74. doi: 10.1186/1471-2407-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sneddon JB, Zhen HH, Montgomery K, van de Rijn M, Tward AD, West R, Gladstone H, Chang HY, Morganroth GS, Oro AE, Brown PO. Bone morphogenetic protein antagonist gremlin 1 is widely expressed by cancer-associated stromal cells and can promote tumor cell proliferation. Proc Natl Acad Sci U S A. 2006;103:14842–7. doi: 10.1073/pnas.0606857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langenfeld EM, Bojnowski J, Perone J, Langenfeld J. Expression of bone morphogenetic proteins in human lung carcinomas. Ann Thorac Surg. 2005;80:1028–32. doi: 10.1016/j.athoracsur.2005.03.094. [DOI] [PubMed] [Google Scholar]
- 21.Rothhammer T, Poser I, Soncin F, Bataille F, Moser M, Bosserhoff AK. Bone morphogenic proteins are overexpressed in malignant melanoma and promote cell invasion and migration. Cancer Res. 2005;65:448–56. [PubMed] [Google Scholar]
- 22.Virk MS, Petrigliano FA, Liu NQ, Chatziioannou AF, Stout D, Kang CO, Dougall WC, Lieberman JR. Influence of simultaneous targeting of the bone morphogenetic protein pathway and RANK/RANKL axis in osteolytic prostate cancer lesion in bone. Bone. 2009;44:160–7. doi: 10.1016/j.bone.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avsian-Kretchmer O, Hsueh AJ. Comparative genomic analysis of the eight-membered ring cystine knot-containing bone morphogenetic protein antagonists. Mol Endocrinol. 2004;18:1–12. doi: 10.1210/me.2003-0227. [DOI] [PubMed] [Google Scholar]
- 24.Lintern KB, Guidato S, Rowe A, Saldanha JW, Itasaki N. Characterization of wise protein and its molecular mechanism to interact with both Wnt and BMP signals. J Biol Chem. 2009;284:23159–68. doi: 10.1074/jbc.M109.025478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudo S, Avsian-Kretchmer O, Wang LS, Hsueh AJ. Protein related to DAN and cerberus is a bone morphogenetic protein antagonist that participates in ovarian paracrine regulation. J Biol Chem. 2004;279:23134–41. doi: 10.1074/jbc.M402376200. [DOI] [PubMed] [Google Scholar]
- 26.Piccolo S, Agius E, Leyns L, Bhattacharyya S, Grunz H, Bouwmeester T, De Robertis EM. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature. 1999;397:707–10. doi: 10.1038/17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dionne MS, Skarnes WC, Harland RM. Mutation and analysis of Dan, the founding member of the Dan family of transforming growth factor beta antagonists. Mol Cell Biol. 2001;21:636–43. doi: 10.1128/MCB.21.2.636-643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J, Zhuang FF, Mullersman JE, Chen H, Robertson EJ, Warburton D, Liu YH, Shi W. BMP4 activation and secretion are negatively regulated by an intracellular gremlin-BMP4 interaction. J Biol Chem. 2006;281:29349–56. doi: 10.1074/jbc.M603833200. [DOI] [PubMed] [Google Scholar]
- 29.Weidauer SE, Schmieder P, Beerbaum M, Schmitz W, Oschkinat H, Mueller TD. NMR structure of the Wnt modulator protein Sclerostin. Biochem Biophys Res Commun. 2009;380:160–5. doi: 10.1016/j.bbrc.2009.01.062. [DOI] [PubMed] [Google Scholar]
- 30.Veverka V, Henry AJ, Slocombe PM, Ventom A, Mulloy B, Muskett FW, Muzylak M, Greenslade K, Moore A, Zhang L, Gong J, Qian X, Paszty C, Taylor RJ, Robinson MK, Carr MD. Characterization of the structural features and interactions of sclerostin: molecular insight into a key regulator of Wnt-mediated bone formation. J Biol Chem. 2009;284:10890–900. doi: 10.1074/jbc.M807994200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daopin S, Piez KA, Ogawa Y, Davies DR. Crystal structure of transforming growth factor-beta 2: an unusual fold for the superfamily. Science. 1992;257:369–73. doi: 10.1126/science.1631557. [DOI] [PubMed] [Google Scholar]
- 32.Radaev S, Zou Z, Huang T, Lafer EM, Hinck AP, Sun PD. Ternary complex of transforming growth factor-beta1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily. J Biol Chem. 285:14806–14. doi: 10.1074/jbc.M109.079921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald NQ, Lapatto R, Murray-Rust J, Gunning J, Wlodawer A, Blundell TL. New protein fold revealed by a 2.3-A resolution crystal structure of nerve growth factor. Nature. 1991;354:411–4. doi: 10.1038/354411a0. [DOI] [PubMed] [Google Scholar]
- 34.Oefner C, D’Arcy A, Winkler FK, Eggimann B, Hosang M. Crystal structure of human platelet-derived growth factor BB. EMBO J. 1992;11:3921–6. doi: 10.1002/j.1460-2075.1992.tb05485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlunegger MP, Grutter MG. An unusual feature revealed by the crystal structure at 2.2 A resolution of human transforming growth factor-beta 2. Nature. 1992;358:430–4. doi: 10.1038/358430a0. [DOI] [PubMed] [Google Scholar]
- 36.Husken-Hindi P, Tsuchida K, Park M, Corrigan AZ, Vaughan JM, Vale WW, Fischer WH. Monomeric activin A retains high receptor binding affinity but exhibits low biological activity. J Biol Chem. 1994;269:19380–4. [PubMed] [Google Scholar]
- 37.Brunner AM, Lioubin MN, Marquardt H, Malacko AR, Wang WC, Shapiro RA, Neubauer M, Cook J, Madisen L, Purchio AF. Site-directed mutagenesis of glycosylation sites in the transforming growth factor-beta 1 (TGF beta 1) and TGF beta 2 (414) precursors and of cysteine residues within mature TGF beta 1: effects on secretion and bioactivity. Mol Endocrinol. 1992;6:1691–700. doi: 10.1210/mend.6.10.1448117. [DOI] [PubMed] [Google Scholar]
- 38.Liao WX, Moore RK, Shimasaki S. Functional and molecular characterization of naturally occurring mutations in the oocyte-secreted factors bone morphogenetic protein-15 and growth and differentiation factor-9. J Biol Chem. 2004;279:17391–6. doi: 10.1074/jbc.M401050200. [DOI] [PubMed] [Google Scholar]
- 39.Pulkki MM, Mottershead DG, Pasternack AH, Muggalla P, Ludlow H, van Dinther M, Myllymaa S, Koli K, ten Dijke P, Laitinen M, Ritvos O. A covalently dimerized recombinant human bone morphogenetic protein-15 variant identifies bone morphogenetic protein receptor type 1B as a key cell surface receptor on ovarian granulosa cells. Endocrinology. 2012;153:1509–18. doi: 10.1210/en.2010-1390. [DOI] [PubMed] [Google Scholar]
- 40.Hung WT, Wu FJ, Wang CJ, Luo CW. DAN (NBL1) Specifically Antagonizes BMP2 and BMP4 and Modulates the Actions of GDF9, BMP2, and BMP4 in the Rat Ovary. Biol Reprod. 2012 doi: 10.1095/biolreprod.111.096172. [DOI] [PubMed] [Google Scholar]
- 41.Kattamuri C, Luedeke DM, Thompson TB. Expression and purification of recombinant protein related to DAN and cerberus (PRDC) Protein Expr Purif. 2012;82:389–95. doi: 10.1016/j.pep.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cash JN, Angerman EB, Kattamuri C, Nolan K, Zhao H, Sidis Y, Keutmann HT, Thompson TB. The structure of myostatin:follistatin-like 3: N-terminal domains of follistatin-type molecules exhibit alternate modes of binding. J Biol Chem. doi: 10.1074/jbc.M111.270801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kondas K, Szlama G, Trexler M, Patthy L. Both WFIKKN1 and WFIKKN2 have high affinity for growth and differentiation factors 8 and 11. J Biol Chem. 2008;283:23677–84. doi: 10.1074/jbc.M803025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herrera B, Inman GJ. A rapid and sensitive bioassay for the simultaneous measurement of multiple bone morphogenetic proteins. Identification and quantification of BMP4, BMP6 and BMP9 in bovine and human serum. BMC Cell Biol. 2009;10:20. doi: 10.1186/1471-2121-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marom K, Fainsod A, Steinbeisser H. Patterning of the mesoderm involves several threshold responses to BMP-4 and Xwnt-8. Mech Dev. 1999;87:33–44. doi: 10.1016/s0925-4773(99)00137-9. [DOI] [PubMed] [Google Scholar]
- 46.Iyer S, Acharya KR. Tying the knot: the cystine signature and molecular-recognition processes of the vascular endothelial growth factor family of angiogenic cytokines. FEBS J. 278:4304–22. doi: 10.1111/j.1742-4658.2011.08350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi M, Zhu J, Wang R, Chen X, Mi L, Walz T, Springer TA. Latent TGF-beta structure and activation. Nature. 474:343–9. doi: 10.1038/nature10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harrington AE, Morris-Triggs SA, Ruotolo BT, Robinson CV, Ohnuma S, Hyvonen M. Structural basis for the inhibition of activin signalling by follistatin. EMBO J. 2006;25:1035–45. doi: 10.1038/sj.emboj.7601000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenwald J, Vega ME, Allendorph GP, Fischer WH, Vale W, Choe S. A flexible activin explains the membrane-dependent cooperative assembly of TGF-beta family receptors. Mol Cell. 2004;15:485–9. doi: 10.1016/j.molcel.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 50.Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, Hinck AP. Crystal structure of the human TbetaR2 ectodomain--TGF-beta3 complex. Nat Struct Biol. 2002;9:203–8. doi: 10.1038/nsb766. [DOI] [PubMed] [Google Scholar]
- 51.Thompson TB, Woodruff TK, Jardetzky TS. Structures of an ActRIIB:activin A complex reveal a novel binding mode for TGF-beta ligand:receptor interactions. EMBO J. 2003;22:1555–66. doi: 10.1093/emboj/cdg156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pierce JG, Parsons TF. Glycoprotein hormones: structure and function. Annu Rev Biochem. 1981;50:465–95. doi: 10.1146/annurev.bi.50.070181.002341. [DOI] [PubMed] [Google Scholar]
- 53.Lapthorn AJ, Harris DC, Littlejohn A, Lustbader JW, Canfield RE, Machin KJ, Morgan FJ, Isaacs NW. Crystal structure of human chorionic gonadotropin. Nature. 1994;369:455–61. doi: 10.1038/369455a0. [DOI] [PubMed] [Google Scholar]
- 54.Cash JN, Rejon CA, McPherron AC, Bernard DJ, Thompson TB. The structure of myostatin:follistatin 288: insights into receptor utilization and heparin binding. EMBO J. 2009;28:2662–76. doi: 10.1038/emboj.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alt A, Miguel-Romero L, Donderis J, Aristorena M, Blanco FJ, Round A, Rubio V, Bernabeu C, Marina A. Structural and functional insights into endoglin ligand recognition and binding. PLoS One. 2012;7:e29948. doi: 10.1371/journal.pone.0029948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Komesli S, Vivien D, Dutartre P. Chimeric extracellular domain type II transforming growth factor (TGF)-beta receptor fused to the Fc region of human immunoglobulin as a TGF-beta antagonist. Eur J Biochem. 1998;254:505–13. doi: 10.1046/j.1432-1327.1998.2540505.x. [DOI] [PubMed] [Google Scholar]
- 57.Coudreuse D, Korswagen HC. The making of Wnt: new insights into Wnt maturation, sorting and secretion. Development. 2007;134:3–12. doi: 10.1242/dev.02699. [DOI] [PubMed] [Google Scholar]
- 58.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science. 2012;337:59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Y, Ali T, Todorovic V, O’Leary J M, Kristina Downing A, Rifkin DB. Amino acid requirements for formation of the TGF-beta-latent TGF-beta binding protein complexes. J Mol Biol. 2005;345:175–86. doi: 10.1016/j.jmb.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 60.Yan B, Smith JW. A redox site involved in integrin activation. J Biol Chem. 2000;275:39964–72. doi: 10.1074/jbc.M007041200. [DOI] [PubMed] [Google Scholar]
- 61.Jacob C, Battaglia E, Burkholz T, Peng D, Bagrel D, Montenarh M. Control of oxidative posttranslational cysteine modifications: from intricate chemistry to widespread biological and medical applications. Chem Res Toxicol. 2012;25:588–604. doi: 10.1021/tx200342b. [DOI] [PubMed] [Google Scholar]
- 62.Dalle-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 63.Mitola S, Ravelli C, Moroni E, Salvi V, Leali D, Ballmer-Hofer K, Zammataro L, Presta M. Gremlin is a novel agonist of the major proangiogenic receptor VEGFR2. Blood. 2010;116:3677–80. doi: 10.1182/blood-2010-06-291930. [DOI] [PubMed] [Google Scholar]
- 64.Chiodelli P, Mitola S, Ravelli C, Oreste P, Rusnati M, Presta M. Heparan sulfate proteoglycans mediate the angiogenic activity of the vascular endothelial growth factor receptor-2 agonist gremlin. Arterioscler Thromb Vasc Biol. 2011;31:e116–27. doi: 10.1161/ATVBAHA.111.235184. [DOI] [PubMed] [Google Scholar]
- 65.Li X, Zhang Y, Kang H, Liu W, Liu P, Zhang J, Harris SE, Wu D. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem. 2005;280:19883–7. doi: 10.1074/jbc.M413274200. [DOI] [PubMed] [Google Scholar]
- 66.Semenov M, Tamai K, He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J Biol Chem. 2005;280:26770–5. doi: 10.1074/jbc.M504308200. [DOI] [PubMed] [Google Scholar]
- 67.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sive HL, Grainger RM, Harland RM. Early Development of Xenopus laevis: A Laboratory Manual. 1 edit Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. [Google Scholar]
- 69.Nieuwkoop P. D. a. F., J. Normal Table of Xenopus laevis (Deaden): A Systematical and Chronological Survey of the Development from the Fertilized Egg till the End of Metamorphosis. Garland Publishing; New York: 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.