Crystal structures of the LpxA protein from A. baumannii were solved in apo forms that were suitable for structure-based antibacterial drug discovery.
Keywords: bacterial proteins, drug targets
Abstract
Acinetobacter baumannii is a Gram-negative pathogenic bacterium which is resistant to most currently available antibiotics and that poses a significant health threat to hospital patients. LpxA is a key enzyme in the biosynthetic pathway of the lipopolysaccharides that are components of the bacterial outer membrane. It is a potential target for antibacterial agents that might be used to fight A. baumannii infections. This paper describes the structure determination of the apo form of LpxA in space groups P212121 and P63. These crystal forms contained three and one protein molecules in the asymmetric unit and diffracted to 1.8 and 1.4 Å resolution, respectively. A comparison of the conformations of the independent protein monomers within and between the two crystal asymmetric units revealed very little structural variation across this set of structures. In the P63 crystal form the enzymatic site is exposed and is available for the introduction of small molecules of the type used in fragment-based drug discovery and structure-based lead optimization.
1. Introduction
Acinetobacter baumannii is a Gram-negative pathogenic bacterium that is inherently resistant to most currently available antibiotics. Immune-compromised hospital patients face a particularly large risk of becoming infected by this pathogen (Dent et al., 2010 ▶). Numerous epidemiological studies have been performed on hospital subpopulations in order to quantify the impact of A. baumannii infection on the mortality rate of critically ill patients, and these studies generally show a significantly higher mortality rate for patients with A. baumannii acquisition compared with patients in whom the bacterium is absent (Falagas et al., 2006 ▶, 2008 ▶). In recent years, A. baumannii infections have also emerged as a significant threat to soldiers wounded and hospitalized in Iraq and Afghanistan (Camp & Tatum, 2010 ▶). The critical and unmet need to develop new antibacterial therapies that target A. baumannii infections (Bassetti et al., 2008 ▶) suggests that structure-based approaches to drug discovery might be usefully applied to suitable protein targets within this organism.
The enzymes involved in the biosynthetic pathway of the lipopolysaccharides that form a component of bacterial outer membranes are potentially attractive drug targets because these proteins are critical for bacterial survival. The major components of these lipopolysaccharides are lipid A (a glucosamide-based saccharolipid) and KDO (3-deoxy-d-manno-oct-2-ulosonic acid). The synthesis of KDO2–lipid A occurs in the cytoplasm, where the first three enzymes in the pathway, LpxA, LpxC and LpxD, are all soluble proteins (Raetz & Whitfield, 2002 ▶). LpxC is a validated target for antibacterial drug discovery, with several inhibitors developed to the preclinical stage, but LpxA and LpxD do not appear to have been widely investigated as possible targets. The Binding Database (http://www.bindingdb.org/bind/index.jsp) does not contain any entries for ligands in complex with LpxA, and most of the ligands in the crystal structures of LpxA in the Protein Data Bank (http://www.rcsb.org/pdb) are uridine-5′-diphosphate (UDP) derivatives. In addition, LpxA is an appealing drug target because it does not show strong homology to human proteins. If the A. baumannii LpxA sequence is compared with human protein sequences in the UniProtKB database (http://www.uniprot.org), the most homologous protein (FUZ) is only half the length of LpxA and has an identity score of only 28%.
Crystal structures of LpxA have been determined for the proteins from Escherichia coli (Williams & Raetz, 2007 ▶; Ulaganathan et al., 2007 ▶; Williams et al., 2006 ▶; Raetz & Roderick, 1995 ▶), Helicobacter pylori (Lee & Suh, 2003 ▶), Leptospira interrogans (Robins et al., 2009 ▶) and Campylobacter jejuni. Most of these structures were solved at relatively high resolution and many of them contain ligands (Table 1 ▶). The amino-acid sequences of LpxA from these species are strongly homologous to the sequence from A. baumannii, with similarity scores in the range 58–70%. However, the resulting structural differences are sufficiently large for the different species to differentiate the length of the specific fatty acid that is utilized in lipid A biosynthesis (Williams & Raetz, 2007 ▶).
Table 1. LpxA crystal structures in the Protein Data Bank.
Structure similarity is based on BLAST alignment using the RCSB PDB query interface over the given number of amino acids (Align). Only ligands larger than ten non-H atoms are listed. U21, uridine-5′-diphosphate-3-O-(R-3-hydroxydecanoyl)-N-acetyl-D-glucosamine; U20, uridine-5′-diphosphate-3-O-(R-3-hydroxymyristoyl)-N-acetyl-D-glucosamine; UD1, uridine-diphosphate-N-acetylglucosamine; SOG, 2-hydroxymethyl-6-octylsulfanyl-tetrahydropyran-3,4,5-triol; U22, uridine-5′-diphosphate-3-N-(R-3-hydroxylauroyl)-N-acetyl-D-glucosamine; S2N, S-[2-({N-[(2R)-2-hydroxy-4-{[(S)-hydroxy(methoxy)phosphoryl]oxy}-3,3-dimethylbutanoyl]-β-alanyl}amino)ethyl] (3R)-3-hydroxydodecanethioate.
| Type | Species | Identity (%) | Similarity (%) | Align | PDB entry | Resolution (Å) | Ligands |
|---|---|---|---|---|---|---|---|
| LpxA | E. coli | 52 | 70 | 260 | 2qiv | 1.85 | U21 |
| 2qia | 1.74 | U20 | |||||
| 2jf3 | 3.00 | UD1 | |||||
| 2jf2 | 1.80 | — | |||||
| 2aq9 | 1.80 | Pentadecapeptide | |||||
| 1lxa | 2.60 | — | |||||
| LpxA | H. pylori | 42 | 59 | 254 | 1j2z | 2.10 | SOG |
| LpxA | L. interrogans | 36 | 61 | 255 | 3i3x | 2.10 | U22 |
| 3i3a | 2.10 | S2N | |||||
| 3hsq | 2.10 | — | |||||
| LpxA | C. jejuni | 36 | 58 | 220 | 3r0s | 2.30 | — |
The biological unit for LpxA is the trimer; the available crystal structures contain either complete trimers in the asymmetric unit or single molecules from which trimers may be generated via crystallographic symmetry operations. The structure of LpxA from E. coli containing the product molecule UDP-3-O-(R-3-hydroxymyristoyl)-ClcNAc exemplifies the characteristic LpxA fold and identifies the location of the enzymatic binding site (Williams & Raetz, 2007 ▶; PDB entry 2qia). The typical topological aspects of LpxA structures are an N-terminal domain consisting of an extensive set of tightly wound β-strands with a triangular cross-section and a C-terminal domain containing a set of four α-helices. The enzymatic sites lie on the outside of the trimer between pairs of protein molecules. The fatty-acid portion of the ligand lies inside a hydrophobic groove between two N-terminal domains and parts of the remainder of the ligand are in contact with one of the C-terminal domains.
Structure-based drug-discovery methods have proved useful in the development of candidate molecules intended for use as antibacterial agents (Finn, 2012 ▶). This paper presents apo structures of LpxA from A. baumannii in crystal forms that may be used within structure-based drug-discovery and drug-development methodologies that include cocrystallography and fragment screening (Nienaber et al., 2000 ▶; Blundell et al., 2002 ▶).
2. Methods
2.1. Cloning, expression and purification
Wild-type LpxA from A. baumannii was cloned into a proprietary vector containing an N-terminal 6×His tag that is cleavable by TEV protease using primers 5′-TAT ATA GGT ACC AGC AAT CAC GAT TTA ATC C-3′ and 5′-TAT ATA CTC GAG TCA GCG CAC AAT TCC AC-3′. The restriction-enzyme sites were KpnI and XhoI. The sequence of the product was verified. The product was expressed in E. coli BL21 (DE3) cells and the cells were grown at 310 K to an OD600 of ∼0.6. The cells were subsequently induced with 1 mM IPTG at 298 K overnight, harvested and stored at 193 K until use.
Purification of the target protein was performed in two- or three-column systems for the preparation of ‘uncleaved’ and ‘cleaved’ forms, respectively. The cell biomass was lysed by sonication in 50 mM Tris–HCl pH 7.8, 500 mM sodium chloride, 10% glycerol, 20 mM imidazole (buffer A) plus one Roche Complete Protease Inhibitor Tablet and 20 000 units of benzonase. The target protein was then extracted by binding to Ni2+-charged IMAC resin and eluted with 250 mM imidazole. The peak fractions were split into pools for the uncleaved protein (pool 1) and the cleaved protein (pool 2). Protein sample from pool 2 was cleaved with 3 mg TEV overnight in buffer A at 277 K; the cleaved protein was run over Ni2+-charged IMAC resin and the flowthrough was collected. Highly aggregated protein was removed from each pool by size-exclusion chromatography (S-200) in 20 mM potassium phosphate pH 8, 250 mM sodium chloride. At this size limit, the protein that assembles into trimers in solution was included in the fraction that was collected for concentration. The protein was then concentrated to 26.5 and 17.3 mg ml−1 for the uncleaved and cleaved forms, respectively. Compared with the native protein, the uncleaved form of the protein contains an additional 31 amino acids at the N-terminus and the cleaved form of the protein contains four additional amino acids at the N-terminus.
2.2. Crystallization and data collection
Crystals of uncleaved A. baumannii LpxA (LpxA-1) were obtained using the hanging-drop vapor-diffusion method by mixing 2 µl 15 mg ml−1 protein buffered in 20 mM potassium phosphate pH 8, 250 mM sodium chloride with 2 µl 0.2 M ammonium citrate tribasic pH 7.0, 20%(w/v) polyethylene glycol (PEG) 3350 at 293 K. Crystals appeared within 3–5 d. Prior to data collection, the crystals were transferred into a cryoprotectant solution consisting of 20%(v/v) glycerol in crystallization buffer and flash-cooled in liquid nitrogen. Diffraction data were collected on beamline 5.0.1 at the Advanced Light Source (ALS) using an ADSC Quantum 210 detector. Data were collected in 1° increments over 135 images.
Crystals of cleaved A. baumannii LpxA (LpxA-2) were obtained using the hanging-drop vapor-diffusion method by mixing 2 µl 25 mg ml−1 protein buffered in 20 mM potassium phosphate pH 8, 250 mM sodium chloride with 2 µl 1.8 M lithium sulfate, 0.1 M HEPES pH 7.5 at 293 K. Crystals with a side dimension of ∼0.4 mm that were morphologically distinct from the LpxA-1 form grew within 7 d. For data collection, the crystals were transferred into a cryoprotectant solution consisting of 20%(v/v) ethylene glycol in crystallization buffer and flash-cooled in liquid nitrogen. Diffraction data were collected on beamline 21ID-F at the Advanced Photon Source (APS) using a Rayonix MMX-225 detector. Data were collected in 0.5° increments over 360 images.
2.3. Structure solution and refinement
The data set from the LpxA-1 crystal form was integrated using the MOSFLM software (Leslie, 2006 ▶) and merged using the SCALA program (Evans, 2006 ▶) from the CCP4 program suite (Winn et al., 2011 ▶). The data set from the LpxA-2 crystal form was processed and reduced to merged intensities using the HKL-2000 software system (Otwinowski & Minor, 1997 ▶; Table 2 ▶).
Table 2. Crystal properties and refinement statistics for LpxA crystal structures from A. baumannii .
Values in parentheses are for the highest resolution shell. The percentage of amino acids in the allowed region of the Ramachandran plot is defined according to Kleywegt & Jones (1996 ▶) and was calculated using the PDB Validation Server (http://validate.rcsb.org/).
| Crystal | LpxA-1 | LpxA-2 |
|---|---|---|
| PDB code | 4e6t | 4e6u |
| Space group | P212121 | P63 |
| Unit-cell parameters | ||
| a (Å) | 71.7 | 75.8 |
| b (Å) | 104.5 | 75.8 |
| c (Å) | 107.0 | 119.1 |
| Beamline | ALS 5.0.1 | APS 21ID-F |
| Resolution (Å) | 1.80 | 1.41 |
| Wavelength (Å) | 1.0 | 0.979 |
| R merge | 0.083 (0.372) | 0.064 (0.591) |
| Completeness (%) | 97.2 (86.4) | 99.8 (99.8) |
| Observed reflections | 360638 (35610) | 836805 (63291) |
| Unique reflections | 72800 (9257) | 73881 (7336) |
| 〈I/σ(I)〉 | 12.1 (3.0) | 36.1 (5.0) |
| Multiplicity | 5.0 (3.8) | 11.3 (11.2) |
| Resolution range (Å) | 47.61–1.80 | 36.09–1.41 |
| Total reflections for refinement (work + free) | 68976 | 70122 |
| R work | 0.178 | 0.175 |
| R free | 0.211 | 0.191 |
| Total protein atoms | 5890 | 1931 |
| Total ligand atoms | 26 | 46 |
| Total waters | 485 | 293 |
| Ligand PDB ID codes | FLC | SO4, EDO |
| Average B factor (Å2) | 15.6 | 20.8 |
| R.m.s. deviation from ideal | ||
| Bond lengths (Å) | 0.012 | 0.010 |
| Bond angles (°) | 1.268 | 1.291 |
| Ramachandran plot (% of residues in allowed regions) | 97.9 | 98.4 |
Molecular-replacement calculations on the LpxA-1 data set used a protein monomer from the E. coli LpxA structure (Williams & Raetz, 2007 ▶; PDB entry 2qia) as the search model and were performed using the Phaser program (McCoy et al., 2007 ▶). Molecular-replacement Z-scores and an absence of severe packing violations confirmed the space group as P212121, with three molecules in the asymmetric unit. Molecular-replacement calculations for the LpxA-2 data set used a monomer from the refined LpxA-1 structure as the search model and gave a very distinct solution in space group P63, with one molecule in the asymmetric unit.
The refinements of both structures were performed using the REFMAC5 software (Murshudov et al., 2011 ▶) run via the MIExpert graphical interface in the MIFit model-building program (http://code.google.com/p/mifit/; Table 2 ▶). Interactive model fitting was performed with the MIFit software. The initial model-rebuilding process of the LpxA-1 structure utilized the expansion of monomer models by noncrystallographic symmetry, but later in the refinement process the protein copies were independently refitted. Noncrystallographic symmetry restraints were not applied in the refinement. Rebuilding and refinement were guided by automated tests applied after each set of refinement cycles to locate specific amino acids that misfitted the density or contained cis-peptide bonds, ϕ–ψ angles corresponding to Ramachandran plot outliers, abnormal side-chain rotamer angles, strained covalent geometries or van der Waals violations. In the final LpxA-2 structure the close contacts that are flagged between some pairs of water molecules correspond to cases in which elongated water density features were modelled as split sites and the waters were assigned fractional occupancy values.
3. Results and discussion
3.1. Structure description
Electron-density maps for the LpxA-1 crystal were traced over amino acids 5–262 (copy A), 4–262 (copy B) and 5–262 (copy C) out of the 262 amino acids in the native protein sequence. The N-terminal tag was not cleaved in this protein preparation, and a six-amino-acid and a ten-amino-acid portion of the tag in protein copies A and B, respectively, was sufficiently well resolved to model into the electron-density map. Since the three copies of the protein are in different crystallographic environments, they make different inter-protein packing contacts and, evidently, these differences impact the position and degree of order in the structure of the tag sequence. Amino acids 5–262 were traced in the LpxA-2 crystal, which is almost identical to the range of the ordered region of all three proteins in the asymmetric unit of the LpxA-1 crystal. Thus, it appears likely that the majority of the LpxA A. baumannii sequence is intrinsically well ordered, with only the N-terminal 3–4 amino acids disordered.
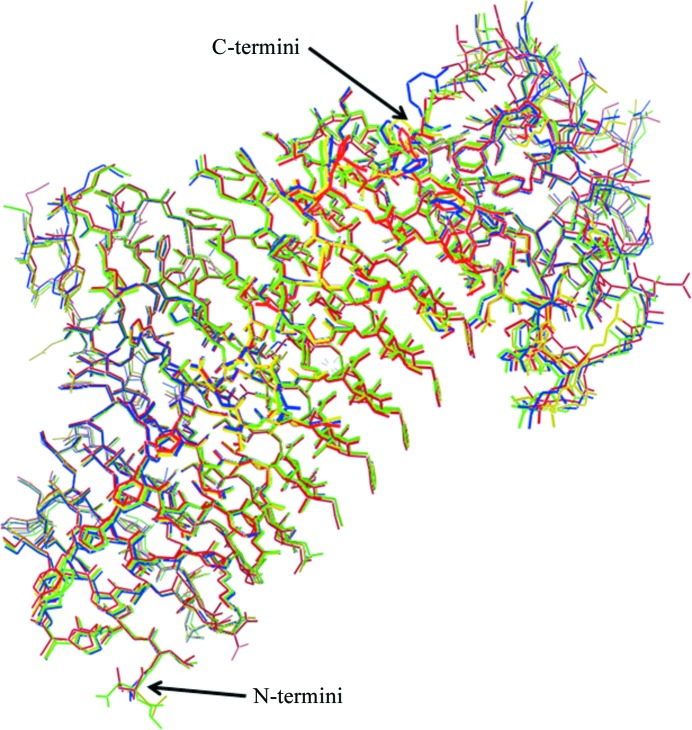
The structure of LpxA from A. baumannii follows the same general architecture as has been identified in the crystal structures of LpxA obtained from other bacterial species. Structure superpositions between the three proteins within the asymmetric unit of the LpxA-1 crystal and the unique molecule in the LpxA-2 crystal showed very little overall difference between backbone conformations and no regions exhibited large structural variability (Table 3 ▶, Fig. 1 ▶). Except for the tag regions in LpxA-1, the three monomers in the LpxA trimer either obey tight noncrystallographic symmetry (Fig. 2 ▶ a) or crystallographic symmetry (Fig. 2 ▶ d) and the different packing contacts outside the trimer do not induce significant conformational changes. For this reason, it seems likely that the LpxA trimer retains a symmetric character in solution.
Table 3. Comparison of equivalent Cα positions (Å) for the three independent monomers in LpxA-1 (copies A, B and C) and the unique monomer (A) in the LpxA-2 crystal asymmetric unit after structure superposition.
| LpxA-1 (A) | LpxA-1 (B) | LpxA-1 (C) | LpxA-2 (A) | |
|---|---|---|---|---|
| LpxA-1 (A) | — | 0.43 | 0.39 | 0.56 |
| LpxA-1 (B) | — | 0.33 | 0.64 | |
| LpxA-1 (C) | — | 0.54 |
Figure 1.
Structure superposition of the three protein monomers from the LpxA-2 crystal (A, yellow; B, green; C, blue) onto the protein monomer from the LpxA-1 crystal (red).
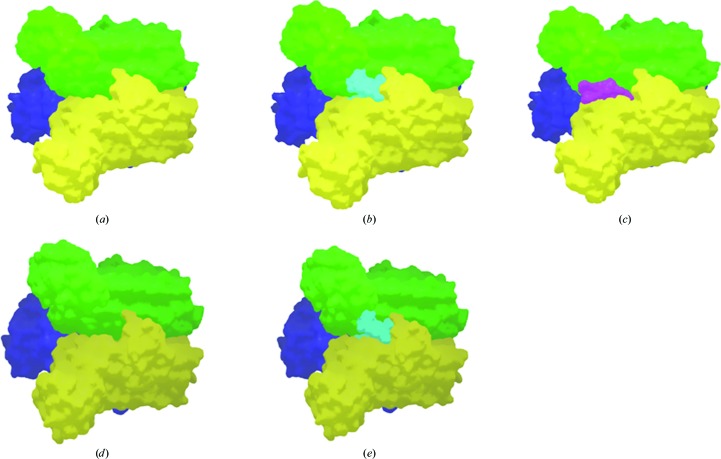
Figure 2.
(a) The LpxA trimer within the asymmetric unit of the LpxA-1 crystal form. The molecular surfaces of the three independent molecules are colored yellow, green and blue. (b) An LpxA-1 trimer in which the position of the uridine-5′-diphosphate-3-O-(R-3-hydroxymyristoyl)-N-acetyl-d-glucosamine molecule (pale blue) was assigned by superposition of the cocrystal structure from E. coli (Williams & Raetz, 2007 ▶; PDB entry 2qia). The location of this molecule serves to map the enzymatic site volume in the A. baumannii LpxA structure. (c) The LpxA trimer from the LpxA-1 crystal form with the largest visible portion of the of the uncleaved N-terminal tag from a neighboring trimer in the crystal (ten amino acids) displayed in purple. (d) An LpxA trimer for the LpxA-2 crystal form generated by application of crystallographic symmetry operations to the unique molecule in the asymmetric unit. The surfaces of the three individual proteins are colored yellow, green and blue. (e) An LpxA-2 trimer with the position of the uridine-5′-diphosphate-3-O-(R-3-hydroxymyristoyl)-N-acetyl-d-glucosamine molecule (pale blue) assigned by superposition of the cocrystal structure from E. coli (Williams & Raetz, 2007 ▶; PDB entry 2qia).
3.2. Ligand-binding sites
The portions of the uncleaved tag sequences that were visualized for two of the three protein units in the LpxA-1 crystal asymmetric unit protrude into grooves formed between β-sheet domains of proteins in neighboring trimers in the crystal (Fig. 2 ▶ c). Structure superposition of LpxA from E. coli containing uridine-5′-diphosphate-3-O-(R-3-hydroxymyristoyl)-N-acetyl-d-glucosamine (Williams & Raetz, 2007 ▶; PDB entry 2qia) onto the monomers in the LpxA-1 crystal shows that this space is occupied by the alphatic tail region of the uridine-5′-diphosphate-3-O-(R-3-hydroxymyristoyl)-N-acetyl-d-glucosamine molecule, but that the sugar or phosphate group regions are not impacted. In addition, a citrate ion (a crystallization component) that was identified in two of the protein units in the LpxA-1 structure overlaps the space filled by uridine-5′-diphosphate-3-O-(R-3-hydroxymyristoyl)-N-acetyl-d-glucosamine in the structure from E. coli.
Superposition of the LpxA-2 crystal structure onto LpxA from E. coli (Williams & Raetz, 2007 ▶; PDB entry 2qia) also shows that the structure of the binding site is well conserved and that almost all of the space occupied by the bound uridine-5′-diphosphate-3-O-(R-3-hydroxymyristoyl)-N-acetyl-d-glucosamine in the E. coli structure is also accessible in this A. baumannii crystal form (Fig. 2 ▶ b). Part of the volume containing the aliphatic tail of the ligand molecule is observed to collect putative ethylene glycol molecules (the cryoprotectant) in the LpxA-2 crystal structure, further demonstrating the relatively hydrophobic character of this part of the ligand-binding site.
3.3. The role of LpxA crystals in structure-based drug discovery
The goal of this work was to develop a crystal form of LpxA from A. baumannii that would be suitable for ‘production’ cocrystallography for drug discovery, i.e. that would enable experiments that involve the soaking of many crystals in solutions of small-molecule compounds.
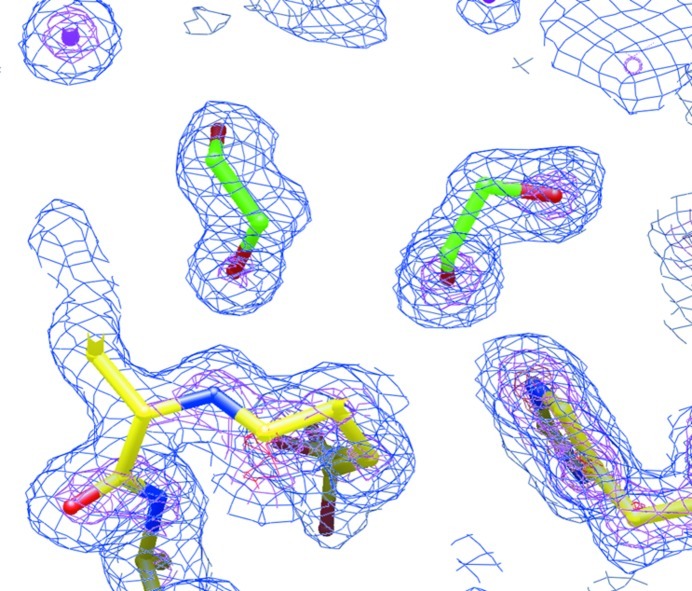
The two LpxA crystal forms that we have obtained are suitable for crystallographic small-fragment screening with screening-library compounds of molecular mass ∼160 Da (Hartshorn et al., 2005 ▶) in that they reproducibly diffract to high resolution and the enzymatic target sites are exposed. However, in the LpxA-1 crystal we visualized the occupation of parts of two of the three sites in the LpxA trimer by the uncleaved protein tags from neighboring proteins in the crystal, as well as a potential conflict with bound citrate ions. These observations suggest that the LpxA-2 crystal form would be a better vehicle for drug discovery with the enzymatic site as the target. The relatively high resolution of the LpxA-2 data set allowed us to identify putative cryoprotectant molecules in portions of the enzymatic binding site (Fig. 3 ▶) and these molecules might also potentially conflict with the binding of fragment-screening compounds. However, ongoing crystallographic work (not described here) has demonstrated the binding of compounds from fragment-screening libraries at this site provided that they are present at sufficiently high concentrations in the crystal soaking solutions.
Figure 3.
Final likelihood-weighted electron-density map of the LpxA-2 crystal structure showing two ethylene glycol molecules (green) in the enzymatic site. The map is contoured at 1× (blue), 3× (purple) and 5× (red) the r.m.s. density fluctuation of the map.
The close similarity of all of the independent protein structures obtained from the LpxA-1 and LpxA-2 crystal forms supports the view that the proteins in these crystals display biologically relevant conformations that are likely to be indicative of the structures found in vivo.
Supplementary Material
PDB reference: LpxA, 4e6t
PDB reference: 4e6u
Acknowledgments
The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Basic Energy Sciences of the US Department of Energy under Contract No. DE-AC02-05CH11231. We thank Rick Walter and Gina Ranieri (Shamrock Structures) for data collection at the Advanced Photon Source.
References
- Bassetti, M., Righi, E., Esposito, S., Petrosillo, N. & Nicolini, L. (2008). Future Microbiol. 3, 649–660. [DOI] [PubMed]
- Blundell, T. L., Jhoti, H. & Abell, C. (2002). Nature Rev. Drug Discov. 1, 45–54. [DOI] [PubMed]
- Camp, C. & Tatum, O. L. (2010). Lab. Med. 41, 649–657.
- Dent, L. L., Marshall, D. R., Pratap, S. & Hulette, R. B. (2010). BMC Infect. Dis. 10, 196. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Falagas, M. E., Bliziotis, I. A. & Siempos, I. I. (2006). Crit. Care, 10, R48. [DOI] [PMC free article] [PubMed]
- Falagas, M. E., Karveli, E. A., Siempos, I. I. & Vardakas, K. Z. (2008). Epidemiol. Infect. 136, 1009–1019. [DOI] [PMC free article] [PubMed]
- Finn, J. (2012). Methods Mol. Biol. 841, 291–319. [DOI] [PubMed]
- Hartshorn, M. J., Murray, C. W., Cleasby, A., Frederickson, M., Tickle, I. J. & Jhoti, H. (2005). J. Am. Chem. Soc. 48, 403–413. [DOI] [PubMed]
- Kleywegt, G. J. & Jones, T. A. (1996). Structure, 4, 1395–1400. [DOI] [PubMed]
- Lee, B. I. & Suh, S. W. (2003). Proteins, 53, 772–774. [DOI] [PubMed]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nienaber, V. L., Richardson, P. L., Klighofer, V., Bouska, J. J., Giranda, V. L. & Greer, J. (2000). Nature Biotechnol. 18, 1105–1108. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Raetz, C. R. & Roderick, S. L. (1995). Science, 270, 997–1000. [DOI] [PubMed]
- Raetz, C. R. & Whitfield, C. (2002). Annu. Rev. Biochem. 71, 635–700. [DOI] [PMC free article] [PubMed]
- Robins, L. I., Williams, A. H. & Raetz, C. R. (2009). Biochemistry, 48, 6191–6201. [DOI] [PMC free article] [PubMed]
- Ulaganathan, V., Buetow, L. & Hunter, W. N. (2007). J. Mol. Biol. 369, 305–312. [DOI] [PubMed]
- Williams, A. H., Immormino, R. M., Gewirth, D. T. & Raetz, C. R. (2006). Proc. Natl Acad. Sci. USA, 103, 10877–10882. [DOI] [PMC free article] [PubMed]
- Williams, A. H. & Raetz, C. R. (2007). Proc. Natl Acad. Sci. USA, 104, 13543–13550. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: LpxA, 4e6t
PDB reference: 4e6u