The expression and purification of Epstein–Barr virus BHRF1 as well as its co-crystallization with a peptidomimetic are described.
Keywords: BHRF1, apoptosis, peptidomimetic, intrinsic pathway
Abstract
BHRF1 is a pro-survival Bcl-2 homologue encoded by Epstein–Barr virus (EBV) that plays a key role in preventing premature host cell death during viral infection and may contribute to the development of malignancies associated with chronic EBV infections. The anti-apoptotic action of BHRF1 is based on its ability to sequester pro-apoptotic Bcl-2 family proteins, in particular Bim and Bak. These interactions have been previously studied in three dimensions by determining crystal structures of BHRF1 in complex with both Bim and Bak BH3 domains. Screening of a library of peptidomimetic compounds based on the benzoylurea scaffold that mimics critical Bim BH3 domain side chains against BHRF1 led to the identification of an inhibitor of BHRF1 that displays micromolar affinity. Single crystals were obtained from the co-crystallization of recombinant BHRF1 protein with this peptidomimetic compound. The crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 66.8, b = 91.1, c = 151.9 Å. Diffraction data were collected to 2.11 Å resolution on the MX2 beamline at the Australian Synchrotron.
1. Introduction
Viruses must overcome host apoptotic defences to ensure their own survival. Despite the complexity of mammalian cell-death processes, viruses have evolved numerous successful mechanisms for subverting the host-cell apoptotic machinery, which include the expression of homologues of the mammalian B-cell leukaemia/lymphoma 2 (Bcl-2) family of proteins (Galluzzi et al., 2008 ▶).
Bcl-2 family proteins have either pro-apoptotic or anti-apoptotic activity and regulate the mitochondrial (or intrinsic) pathway of apoptosis (Adams & Cory, 1998 ▶) by controlling the mitochondrial outer membrane permeabilization (MOMP) (Green & Kroemer, 2004 ▶). Members of the Bcl-2 family can be recognized by the presence of one or more of the Bcl-2 homology (BH) domains. Anti-apoptotic proteins (Bcl-2, Bcl-xL, Mcl-1, Bcl-w, Bcl-b and A1) contain four Bcl-2 homology (BH) domains and a transmembrane domain (Reed et al., 1996 ▶), although certain members of the family from poxviruses do not contain readily identifiable BH domains (Douglas et al., 2007 ▶; Kvansakul et al., 2007 ▶, 2008 ▶). Pro-apoptotic proteins can be subdivided into multi-domain proteins (Bax, Bak and Bok) that contain four BH domains, and BH3-only proteins (Bad, Bim, Puma, Bid, Noxa, Hrk and Bmf) that only possess the BH3 domain. BH3-only proteins selectively respond to proximal death and survival signals, requiring Bax or Bak to induce death (Cheng et al., 2001 ▶; Danial & Korsmeyer, 2004 ▶; Strasser, 2005 ▶). In higher organisms the pro-survival members maintain cell viability until the BH3-only proteins inactivate them, leading to the activation of Bax and Bak (Letai et al., 2002 ▶; Kuwana et al., 2005 ▶; Chen et al., 2005 ▶; Willis et al., 2005 ▶). Bax and Bak are only activated when the balance between pro-survival and pro-apoptotic Bcl-2 members is tilted towards apoptosis, resulting in MOMP and the activation of proteolytic caspases that mediate cellular demolition (Kuwana & Newmeyer, 2003 ▶; Breckenridge & Xue, 2004 ▶; Marsden et al., 2004 ▶; Green & Kroemer, 2004 ▶).
Epstein–Barr virus (EBV) is a member of the γ-herpesvirus family which infects the epithelium of the oropharynx and resting B cells. Acute infection manifests as infectious mononucleosis or glandular fever, whereas chronic EBV-associated transformation is associated with Burkitt lymphoma, Hodgkin’s disease and nasopharyngeal carcinoma (Crawford, 2001 ▶). EBV encodes BHRF1 (21.9 kDa, 191 amino acids), which is homologous to Bcl-2 in sequence, structure and function. It has been shown that BHRF1 is a potent inhibitor of apoptosis, and confers chemoresistance in mouse lymphoma models similar to mammalian Bcl-2, acting by sequestering pro-apoptotic Bcl-2 family members (Kvansakul et al., 2010 ▶).
The crystal structures of BHRF1 in complex with Bim and Bak BH3 peptides have shown that BHRF1 interacts with these pro-apoptotic proteins in a manner similar to its mammalian counterparts such as pro-survival Bcl-xL (Kvansakul et al., 2010 ▶). Given the importance of BHRF1 in EBV-associated malignancies and in certain Burkitt lymphomas, the development of inhibitors of BHRF1 may be highly relevant for the development of novel treatment strategies for these diseases. The use of peptidomimetics has proven to be a useful chemical strategy as the basis for the development of such inhibitors (Kutzki et al., 2002 ▶; Letai et al., 2002 ▶; Yin & Hamilton, 2004 ▶; Lessene et al., 2008 ▶; Zheng et al., 2012 ▶; Bajwa et al., 2012 ▶). Screening of a library of peptidomimetics based on the benzoylurea scaffold against BHRF1 led to the identification of WEHI-271, a micromolar inhibitor of BHRF1 (Nhu et al., submitted). In this study, we report the overexpression, purification and co-crystallization of a C-terminally truncated form of BHRF1 in complex with WEHI-271, as well as its preliminary X-ray analysis.
2. Materials and methods
2.1. Protein expression and purification
BHRF1ΔC31 was cloned and transformed as previously described in Kvansakul et al. (2010 ▶). The C-terminally truncated form of BHRF1 was chosen to reduce protein flexibility. A small overnight culture (10 ml) grown at 310 K in 2×YT broth containing 100 µg ml−1 ampicillin was used to inoculate a 1 l culture. Cells were grown to an OD600 of 0.8. Expression of the recombinant protein was then induced by addition of IPTG to a final concentration of 1 mM. After 4 h induction at 310 K, the cells were harvested by centrifugation at 6000g for 20 min at 277 K. Subsequently, cells were homogenized using a FastPrep-24 instrument (MP Biomedicals) in lysis buffer (50 mM Tris–HCl buffer pH 8.5, 300 mM NaCl).
The soluble fraction was loaded onto a 1 ml HiTrap IMAC HP column (GE Healthcare) pre-charged with nickel and washed with 10 ml lysis buffer. The target protein was eluted with lysis buffer supplemented with 250 mM imidazole. Fractions containing BHRF1 were identified by SDS–PAGE analysis, pooled and concentrated to 1 ml for gel filtration.
Gel filtration was performed on a Superdex 200 10/300 GL column (GE Healthcare) connected to an ÄKTApurifier system (GE Healthcare) and pre-equilibrated with 25 mM HEPES buffer pH 7.5, 150 mM NaCl. Fractions containing the protein were pooled and concentrated to 5 mg ml−1 using a 10 kDa molecular-weight cutoff Amicon centrifugal concentrator (Millipore). The protein concentration was estimated from the absorption at 280 nm and the sample purity was estimated to be about 95% by SDS–PAGE.
2.2. Crystallization
Based on the known pro-survival/BH3-only protein structures, including the BHRF1–Bim structure (Kvansakul et al., 2010 ▶), a series of peptidomimetics based on a benzoylurea backbone were developed. Screening of this library revealed a number of hits with appreciable affinity for BHRF1 (Nhu et al., submitted). Amongst these small-molecule hits, WEHI-271 proved to be particularly interesting. A fivefold molar excess of peptidomimetic was added to the 5 mg ml−1 BHRF1 protein. Crystallization trials were carried out with the resultant complex at 293 K using the sitting-drop vapour-diffusion method by mixing 150 nl protein solution with an equal volume of mother liquor. The sparse-matrix protein crystallization screens used for initial screening at the CSIRO Collaborative Crystallization Centre (Melbourne, Australia) were Crystal Screen and Crystal Screen 2 (Hampton Research), Precipitant Synergy Screen (Emerald BioSciences), The PACT Suite and The Anions Suite (Qiagen).
The initial crystallization condition was established using the Precipitant Synergy Screen sparse-matrix protein crystallization screen (Emerald BioSciences) at the CSIRO Collaborative Crystallization Centre (Melbourne, Australia). The initial crystals, which were obtained in 0.05 M magnesium sulfate, 0.1 M imidazole pH 6.5, 2 M ammonium sulfate, 1%(v/v) glycerol, were clusters of very thin needles that appeared after 3 d (Fig. 1 ▶ a). Further optimization using a randomized approach in which a range of magnesium sulfate concentrations (0–0.1 M), ammonium sulfate concentrations (0.5–2 M), glycerol concentrations [0–5%(v/v)] and different pH values (6.2–7.4) were used did not yield a considerable improvement in crystal quality. Further optimization was carried out using the hanging-drop vapour-diffusion method by mixing 1 µl protein solution with an equal volume of mother liquor and equilibrating against 500 µl reservoir solution in a 24-well format at 293 K in conjunction with serial micro-seeding and streak-seeding techniques. This approach led to diffraction-quality single crystals after setting up 216 drops. The best crystals appeared after 1–3 d using streak-seeding in drops consisting of 0.1 M imidazole pH 6.2, 1 M ammonium sulfate, 2.5%(v/v) glycerol, 0.05 M magnesium sulfate (Fig. 1 ▶ b).
Figure 1.
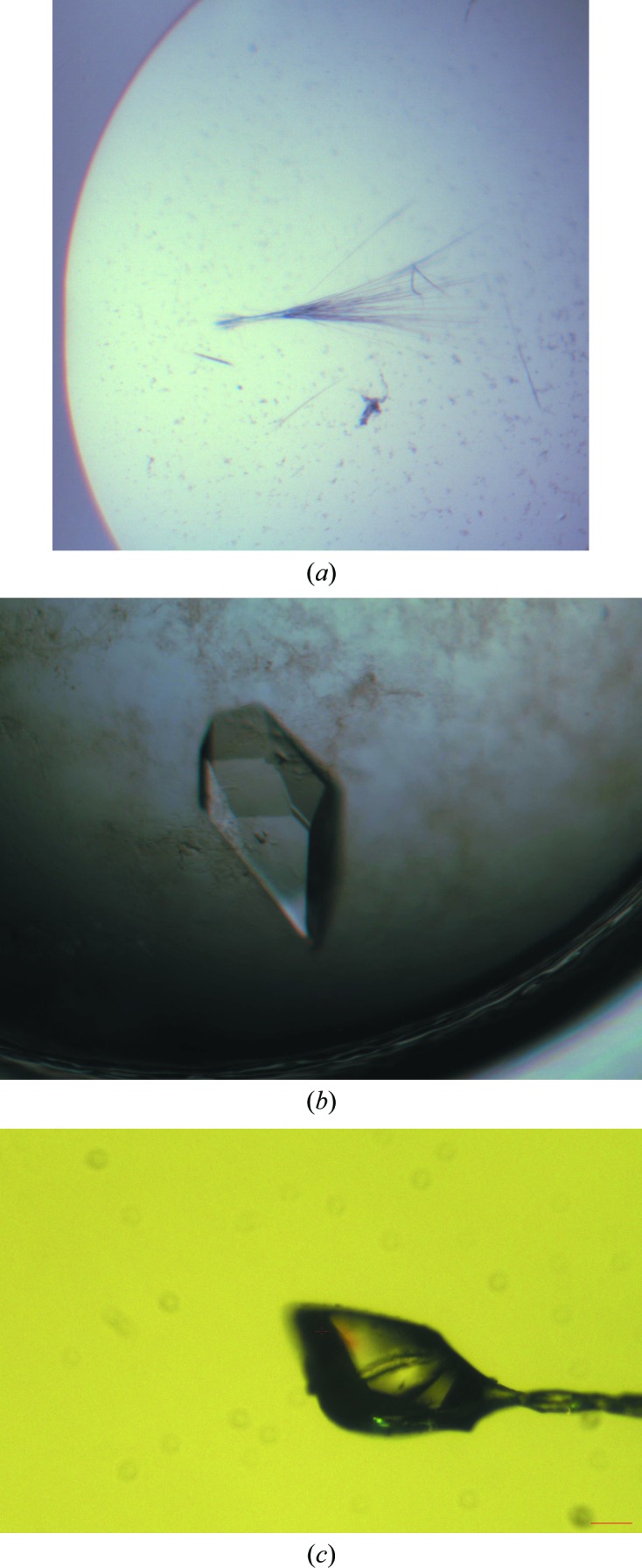
Crystals of BHRF1–peptidomimetic complex. (a) Crystals grown in the initial hit: 0.05 M magnesium sulfate, 0.1 M imidazole pH 6.5, 2 M ammonium sulfate, 1%(v/v) glycerol. (b) Crystals grown in 0.1 M imidazole pH 6.2, 1 M ammonium sulfate, 2.5%(v/v) glycerol, 0.05 M magnesium sulfate after streak-seeding in the well. (c) BHRF1–peptidomimetic complex crystal mounted on a 200 µm loop.
2.3. Cryoprotection and data collection
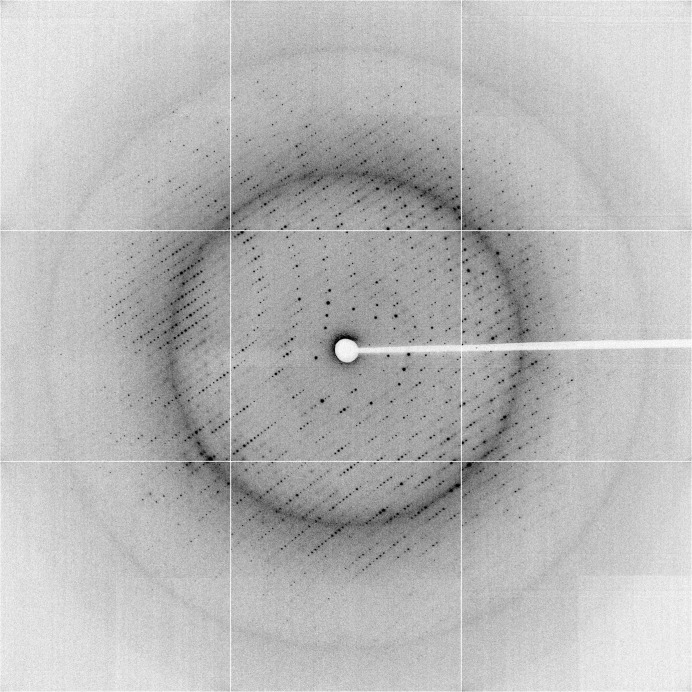
Single crystals were cryoprotected by adding 15%(w/v) glucose to the crystallization solution and flash-cooling at the Australian Synchrotron (beamline MX2). Diffraction data were collected at 100 K at a wavelength of 0.95370 Å (Figs. 1 ▶ c and 2 ▶) and processed with xia2 (Winter, 2010 ▶) using XDS (Kabsch, 1993 ▶); data-collection statistics are shown in Table 1 ▶. Examination of diffraction data and systematic absences indicated that the space group was P212121.
Figure 2.
X-ray diffraction image from a BHRF1–peptidomimetic complex crystal.
Table 1. Data-collection statistics for the BHRF1–peptidomimetic complex crystal.
Values in parentheses are for the highest resolution shell.
| Source | Australian Synchrotron MX2 |
| Detector | ADSC Quantum 315r |
| Space group | P212121 |
| Unit-cell parameters (Å, °) | a = 66.8, b = 91.1, c = 151.9, α = β = γ = 90 |
| Wavelength (Å) | 0.95370 |
| Resolution (Å) | 45.56–2.11 (2.16–2.11) |
| R merge † (%) | 10.3 (172) |
| R p.i.m. ‡ (%) | 2.0 (31.9) |
| Multiplicity | 28.9 (29.8) |
| Completeness (%) | 99.9 (99.9) |
| Mean I/σ(I) | 27.0 (4.8) |
| No. of unique reflections | 54148 (3952) |
R
merge = 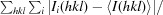
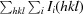 .
.
R
p.i.m. = 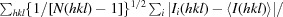
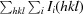 .
.
3. Results and discussion
BHRF1 is known to be a potent inhibitor of apoptosis that results in marked chemoresistance in mouse lymphoma models similar to mammalian Bcl-2, presumably by sequestering pro-apoptotic Bcl-2 family proteins (Kvansakul et al., 2010 ▶). In order to target this viral protein therapeutically, peptidomimetics have been developed based on the crystal structures of various pro-survival/BH3-only protein complexes. In this study, we expressed and purified recombinant BHRF1. We also established the co-crystallization conditions for the BHRF1–peptidomimetic complex in order to determine its structure and investigate its mode of binding. The determination of the binding mode will give important chemical bases for the development of BHRF1 inhibitors which may be highly relevant for the development of novel treatment strategies for EBV-associated malignancies.
Co-crystallization of the complex was carried out by the hanging-drop vapour-diffusion method at 293 K. After optimization and several rounds of micro-seeding and streak-seeding, the best crystals (Figs. 1 ▶ b and 1 ▶ c) were obtained with 0.1 M imidazole pH 6.2, 1 M ammonium sulfate, 2.5%(v/v) glycerol, 0.05 M magnesium sulfate after 1–3 d.
Native data were collected to 2.11 Å resolution (Fig. 2 ▶). The crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 66.8, b = 91.1, c = 151.9 Å, α = β = γ = 90°. The data-collection statistics are shown in Table 1 ▶. Phasing, model building and refinement are in progress.
Acknowledgments
We would like to thank the staff at the CSIRO Collaborative Crystallization Centre for the set-up of crystal screens and the staff of the MX team at the Australian Synchrotron for assistance with X-ray diffraction data collection. This work was funded by the NHMRC (fellowship to MK, project grant No. 637336).
References
- Adams, J. M. & Cory, S. (1998). Science, 281, 1322–1326. [DOI] [PubMed]
- Bajwa, N., Liao, C. & Nikolovska-Coleska, Z. (2012). Expert Opin. Ther. Pat. 22, 37–55. [DOI] [PMC free article] [PubMed]
- Breckenridge, D. G. & Xue, D. (2004). Curr. Opin. Cell Biol. 16, 647–652. [DOI] [PubMed]
- Chen, L., Willis, S. N., Wei, A., Smith, B. J., Fletcher, J. I., Hinds, M. G., Colman, P. M., Day, C. L., Adams, J. M. & Huang, D. C. S. (2005). Mol. Cell, 17, 393–403. [DOI] [PubMed]
- Cheng, E. H.-Y. A, Wei, M. C., Weiler, S., Flavell, R. A., Mak, T. W., Lindsten, T. & Korsmeyer, S. J. (2001). Mol. Cell, 8, 705–711. [DOI] [PubMed]
- Crawford, D. H. (2001). Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 356, 461–473. [DOI] [PMC free article] [PubMed]
- Danial, N. N. & Korsmeyer, S. J. (2004). Cell, 116, 205–219. [DOI] [PubMed]
- Douglas, A. E., Corbett, K. D., Berger, J. M., McFadden, G. & Handel, T. M. (2007). Protein Sci. 16, 695–703. [DOI] [PMC free article] [PubMed]
- Galluzzi, L., Brenner, C., Morselli, E., Touat, Z. & Kroemer, G. (2008). PLoS Pathog. 4, e1000018. [DOI] [PMC free article] [PubMed]
- Green, D. R. & Kroemer, G. (2004). Science, 305, 626–629. [DOI] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst. 26, 795–800.
- Kutzki, O., Park, H. S., Ernst, J. T., Orner, B. P., Yin, H. & Hamilton, A. D. (2002). J. Am. Chem. Soc. 124, 11838–11839. [DOI] [PubMed]
- Kuwana, T., Bouchier-Hayes, L., Chipuk, J. E., Bonzon, C., Sullivan, B. A., Green, D. R. & Newmeyer, D. D. (2005). Mol. Cell, 17, 525–535. [DOI] [PubMed]
- Kuwana, T. & Newmeyer, D. D. (2003). Curr. Opin. Cell Biol. 15, 691–699. [DOI] [PubMed]
- Kvansakul, M., van Delft, M. F., Lee, E. F., Gulbis, J. M., Fairlie, W. D., Huang, D. C. S. & Colman, P. M. (2007). Mol. Cell, 25, 933–942. [DOI] [PubMed]
- Kvansakul, M., Wei, A. H., Fletcher, J. I., Willis, S. N., Chen, L., Roberts, A. W., Huang, D. C. S. & Colman, P. M. (2010). PLoS Pathog. 6, e1001236. [DOI] [PMC free article] [PubMed]
- Kvansakul, M., Yang, H., Fairlie, W. D., Czabotar, P. E., Fischer, S. F., Perugini, M. A., Huang, D. C. S. & Colman, P. M. (2008). Cell Death Differ. 15, 1564–1571. [DOI] [PubMed]
- Lessene, G., Czabotar, P. E. & Colman, P. M. (2008). Nature Rev. Drug Discov. 7, 989–1000. [DOI] [PubMed]
- Letai, A., Bassik, M. C., Walensky, L. D., Sorcinelli, M. D., Weiler, S. & Korsmeyer, S. J. (2002). Cancer Cell, 2, 183–192. [DOI] [PubMed]
- Marsden, V. S., Ekert, P. G., Van Delft, M., Vaux, D. L., Adams, J. M. & Strasser, A. (2004). J. Cell Biol. 165, 775–780. [DOI] [PMC free article] [PubMed]
- Reed, J. C., Zha, H., Aime-Sempe, C., Takayama, S. & Wang, H.-G. (1996). Adv. Exp. Med. Biol. 406, 99–112. [PubMed]
- Strasser, A. (2005). Nature Rev. Immunol. 5, 189–200. [DOI] [PubMed]
- Willis, S. N., Chen, L., Dewson, G., Wei, A., Naik, E., Fletcher, J. I., Adams, J. M. & Huang, D. C. (2005). Genes Dev. 19, 1294–1305. [DOI] [PMC free article] [PubMed]
- Winter, G. (2010). J. Appl. Cryst. 43, 186–190.
- Yin, H. & Hamilton, A. D. (2004). Bioorg. Med. Chem. Lett. 14, 1375–1379. [DOI] [PubMed]
- Zheng, C.-H., Yang, H., Zhang, M., Lu, S.-H., Shi, D., Wang, J., Chen, X.-H., Ren, X.-H., Liu, J., Lv, J.-G., Zhu, J. & Zhou, Y.-J. (2012). Bioorg. Med. Chem. Lett. 22, 39–44. [DOI] [PubMed]