B. cereus TubZ was cocrystallized with GDP and diffraction data were collected to 2.1 Å resolution.
Keywords: TubZ, Bacillus cereus
Abstract
TubZ is a structural homologue of tubulin and FtsZ GTPases, which are involved in the type III plasmid-partitioning system. TubZ assembles into polymers in a GTP-dependent manner and drives plasmid segregation as ‘cytomotive’ filaments. In this study, C-terminally truncated TubZ from Bacillus cereus was crystallized in the presence or absence of GDP by the hanging-drop vapour-diffusion method. The crystal of TubZ in complex with GDP belonged to the monoclinic space group P21, with unit-cell parameters a = 67.05, b = 84.49, c = 67.66 Å, β = 92.92°, and was non-isomorphous with GDP-bound TubZ previously crystallized in the presence of the slowly hydrolysable GTP analogue GTPγS. TubZ was also crystallized in the free form and the crystal belonged to space group P21, with unit-cell parameters a = 53.91, b = 65.54, c = 58.18 Å, β = 106.19°. Data were collected to 1.7 and 2.1 Å resolution for the free and GDP-bound forms, respectively.
1. Introduction
The bacterial cell-division protein FtsZ and its eukaryotic structural homologue tubulin are both cytoskeletal GTPases which assemble into polymers in a GTP-dependent manner (Löwe & Amos, 1998 ▶; Nogales et al., 1998 ▶; Erickson et al., 1996 ▶; Weisenberg et al., 1976 ▶). They possess a conserved GTPase domain at the N-terminus and a rather divergent C-terminal domain which plays a role as a GTPase activator when they form a polymer (Oliva et al., 2004 ▶). In the protofilament, each subunit makes longitudinal contacts whereby the GTP-binding site interacts with the T7 loop of the C-terminal domain of the neighbouring subunit, leading to GTP hydrolysis. Despite the similarity of the tertiary structure and the subunit–subunit contacts between FtsZ and tubulin, the filament architecture and functions are different. FtsZ, which forms a contractile ring in bacterial cytokinesis, assembles into protofilaments in vitro (Bi & Lutkenhaus, 1991 ▶; Mukherjee & Lutkenhaus, 1994 ▶). On the other hand, the tubulin heterodimer, which is a major component of the mitotic spindle, assembles into hollow cylindrical microtubules which consist of parallel protofilaments.
Recently, a new plasmid-partitioning system has been identified in the virulence plasmids pXO1 and pBtoxis from Bacillus anthracis (Ba) and B. thuringiensis (Bt), respectively, which utilizes one of the FtsZ/tubulin superfamily proteins, TubZ (Tinsley & Khan, 2006 ▶; Tang et al., 2006 ▶). The TubZ filament exhibits ‘treadmilling’, in which the filament grows at the plus end and simultaneously shrinks at the minus end (Larsen et al., 2007 ▶). A GTPase-deficient mutation of TubZ (a mutation of the conserved acidic residue in the T7 loop) eliminates treadmilling of the filaments and reduces the stability of pBtoxis, indicating that TubZ activity is required for plasmid maintenance in the cell. Furthermore, it has been suggested that TubZ is involved in the bacteriophage-partition system in Clostridium botulinum (Cb; Oliva et al., 2012 ▶). Since no motor proteins have been identified in bacteria to date, TubZ polymers are thought to work as a motor: the so-called ‘cytomotive’ filaments (Ni et al., 2010 ▶; Aylett et al., 2011 ▶).
Crystal structures of TubZ from Bt, Cb and B. cereus (Bc) have been determined (Ni et al., 2010 ▶; Aylett et al., 2010 ▶; Oliva et al., 2012 ▶; Hoshino & Hayashi, 2012 ▶). The tertiary structures resemble those of FtsZ and tubulin, with a slightly modified signature motif of the FtsZ/tubulin superfamily in the GTP-binding pocket [GGGTG(T/S)G for FtsZ, tubulin and Bc-TubZ, GGGVGTG for Bt-TubZ and AGGTGSG for Cb-TubZ]. Electron-microscopy studies showed that the filament of Bt-TubZ forms a double-stranded helix which is similar to the actin filament (Chen & Erickson, 2008 ▶; Aylett et al., 2010 ▶). Although Bt and Bc are genetically closely related, the sequences of TubZ coded by the virulence plasmids are divergent, showing ∼20% similarity to each other (Chen & Erickson, 2008 ▶; Hoshino & Hayashi, 2012 ▶). As described in our previous report, we have determined the crystal structures of Bc-TubZ in free, GDP-bound and GTPγS-bound forms at resolutions of 2.1, 1.9 and 3.3 Å, respectively (Hoshino & Hayashi, 2012 ▶). Since the crystals of the GDP-bound form were obtained in the presence of GTPγS, we proposed that the GDP-bound structure with protofilament-like intermolecular interactions reflects the post-GTP hydrolysis state of Bc-TubZ. In order to obtain more detailed information on Bc-TubZ in the GDP-bound form, crystallization trials were carried out with GDP. Here, we report the biochemical and crystallographic analysis of Bc-TubZ. C-terminally truncated Bc-TubZ (Bc-TubZΔ) was crystallized with or without a nucleotide. The attempt to crystallize Bc-TubZ in the presence of GDP resulted in a different crystal form from the previous GDP-bound Bc-TubZ crystal obtained with GTPγS (Hoshino & Hayashi, 2012 ▶). The crystal of GDP-bound Bc-TubZ diffracted to 2.1 Å resolution and that of the free form diffracted to 1.7 Å resolution.
2. Materials and methods
2.1. Identification of a crystallizable Bc-TubZ fragment
Wild-type Bc-TubZ and Bc-TubZΔ were purified as previously reported, with minor modifications (Hoshino & Hayashi, 2012 ▶). The detailed protocol is described in the Supplementary Material1. 10 µg Bc-TubZ was subjected to limited proteolysis. 0.05 µg trypsin (Sigma), chymotrypsin (Sigma) or thermolysin (Wako, Japan) was mixed with the protein in a buffer consisting of 10 mM Tris pH 8.0, 0.15 M NaCl, 1 mM DTT. After incubation for 30 min at room temperature, the reaction mixtures were analysed by 12% SDS–PAGE electrophoresis with Coomassie Blue staining. The protein digested with thermolysin was further analysed by N-terminal sequencing and matrix-assisted laser desorption/ionization–time-of-flight (MALDI–TOF) mass spectrometry.
2.2. GTPase activity measurement
2 µM Bc-TubZ was polymerized at 310 K in a buffer consisting of 50 mM HEPES pH 7.7, 0.1 M potassium acetate, 5 mM magnesium acetate, 1 mM EGTA, 0.5 mM GTP. The reaction mixture also included 0.4 mM phosphoenolpyruvate, 0.3 mM NADH, 20 U ml−1 pyruvate kinase and 20 U ml−1 lactate dehydrogenase in order to measure the GTP hydrolysis activity by monitoring the decrease in NADH absorption at 340 nm using a Jasco V-630BIO UV–Vis spectrophotometer (Chen & Erickson, 2008 ▶). The effect of the GTP analogue was examined by the addition of 50 µM of the slowly hydrolysable GTP analogue guanosine 5′-O-3-(thio)triphosphate (GTPγS), the nonhydrolysable GTP analogue 5′-guanylyl-imidodiphosphate (GMPPNP) or GDP to the reaction mixture.
2.3. Crystallization
Crystal Screen, Crystal Screen 2 (Hampton Research), The JCSG+ Suite (Qiagen) and laboratory solutions were used for crystallization trials of Bc-TubZΔ in the presence or absence of GDP. Optimization of the conditions was carried out using the hanging-drop vapour-diffusion method at 293 K. The best crystal of the free form was grown in 50 mM bis-tris pH 5.5, 0.2 M MgCl2, 25% PEG 4000. GDP-bound Bc-TubZΔ crystals were obtained using 0.1 M MES pH 5.5, 1 mM MgCl2, 20% PEG 400, 1 mM GDP.
2.4. Data collection and processing
All crystals were transferred into a cryoprotectant solution consisting of 30% glycerol in the crystallization buffer and flash-cooled in liquid nitrogen. Diffraction data were collected at 100 K on beamlines BL17A and NW12A at the Photon Factory (PF), Tsukuba, Japan. Diffraction images were indexed, integrated and scaled using the HKL-2000 suite (Otwinowski & Minor, 1997 ▶) (Table 1 ▶).
Table 1. Data-collection statistics.
Values in parentheses are for the highest shell.
| Bc-TubZΔ (free form) | Bc-TubZΔ (GDP-bound form) | |
|---|---|---|
| Beamline | BL17A, PF | NW12A, PF |
| Space group | P21 | P21 |
| Unit-cell parameters (Å, °) | a = 53.19, b = 65.54, c = 58.18, β = 106.19 | a = 67.05, b = 84.49, c = 67.66, β = 92.92 |
| Resolution range (Å) | 50–1.7 | 50–2.1 |
| Wavelength (Å) | 0.9800 | 1.0000 |
| No. of unique reflections | 42723 | 44458 |
| Completeness (%) | 99.9 (99.6) | 99.9 (99.9) |
| Multiplicity | 3.6 (3.5) | 5.5 (4.9) |
| 〈I/σ(I)〉 | 22.4 (1.8) | 28.7 (4.2) |
| R merge † | 0.052 (0.628) | 0.054 (0.378) |
| Molecules in asymmetric unit | 1 | 2 |
R
merge = 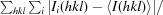
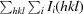 , where I
i(hkl) is the intensity measurement and 〈I(hkl)〉 is the mean intensity for multiply recorded reflections.
, where I
i(hkl) is the intensity measurement and 〈I(hkl)〉 is the mean intensity for multiply recorded reflections.
3. Results
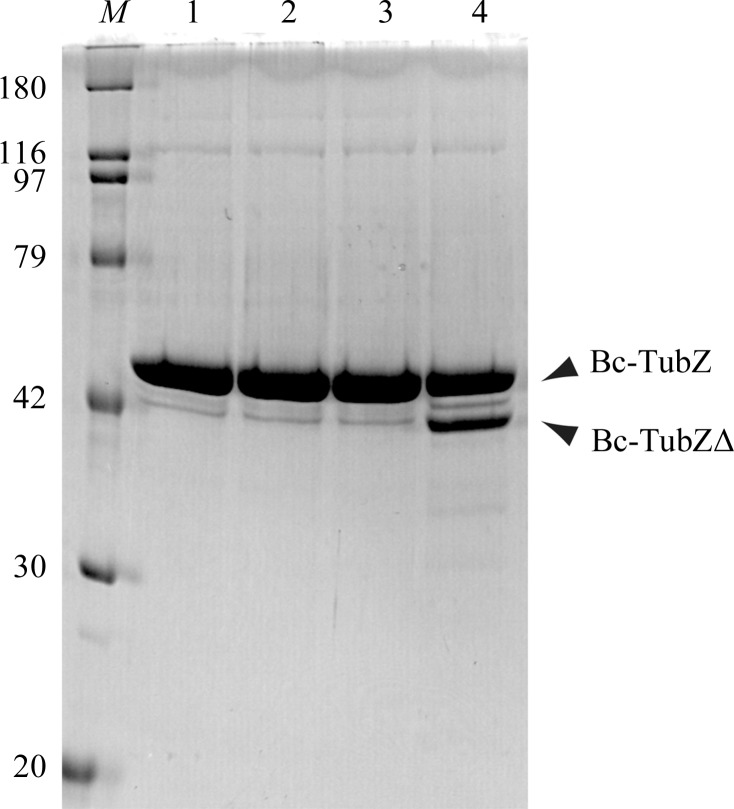
Since Bc-TubZ was found to be susceptible to proteolysis during the purification process, limited proteolysis of Bc-TubZ was carried out in order to define a stable construct for crystallization. Full-length Bc-TubZ was subjected to proteolysis with trypsin, chymotrypsin and thermolysin (Fig. 1 ▶). While the C-terminus of Bc-TubZ and that of Bt-TubZ possess a basic residue cluster and are thought to be involved in association with the centromere-binding protein TubR, Bc-TubZ is resistant to trypsin digestion, implying that the C-terminal tail is involved in the intramolecular interaction (Ni et al., 2010 ▶; Hoshino & Hayashi, 2012 ▶). Only thermolysin treatment yielded a shorter fragment. N-terminal sequencing of the fragment resulted in a sequence of ten amino acids (AGNFSEIESQ) which corresponds to residues 2–11 of Bc-TubZ. Mass-spectrometric analysis indicated a molecular mass of 43 383 Da, suggesting that the fragment encompasses residues 1–389 (Bc-TubZΔ). Bc-TubZΔ was expressed as a His-tagged form, purified to homogeneity and used for crystallization.
Figure 1.
Limited proteolysis of Bc-TubZ. Digested samples were resolved by 12% SDS–PAGE with Coomassie Blue staining. The major fragment is labelled with an arrow. The sizes of the molecular-mass marker (lane M) are indicated on the left in kDa. Lane 1, intact Bc-TubZ; lane 2, Bc-TubZ digested with trypsin; lane 3, Bc-TubZ digested with chymotrypsin; lane 4, Bc-TubZ digested with thermolysin.
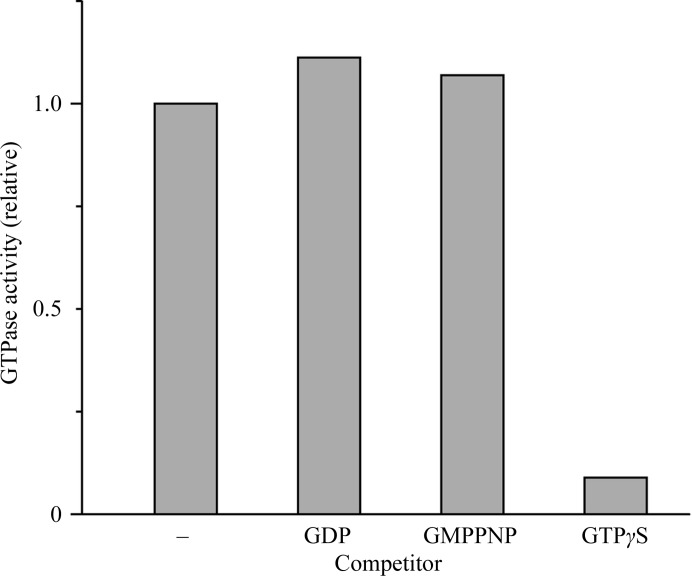
Next, we measured the relative GTPase activity of Bc-TubZ and analysed the effects of the GTP analogues GTPγS, GMPPNP and GDP, since FtsZ has different affinities for various GTP analogues (Scheffers et al., 2000 ▶; Fig. 2 ▶). The addition of 50 µM GTPγS drastically reduced GTP hydrolysis by Bc-TubZ, whereas GMPPNP and GDP had little effect on the Bc-TubZ activity, indicating that GTPγS is a substrate for Bc-TubZ, but not GDP or GMPPCP. Since the nucleotide in the filament of Ba-TubZ (98% identical to Bc-TubZ) has been shown to be GDP, we aimed to analyse the crystal structure of the inactive form of Bc-TubZ using GDP (Chen & Erickson, 2008 ▶).
Figure 2.
GTP hydrolysis of Bc-TubZ with or without GTP analogues. The competing nucleotides used were GDP, GMPPNP and GTPγS.
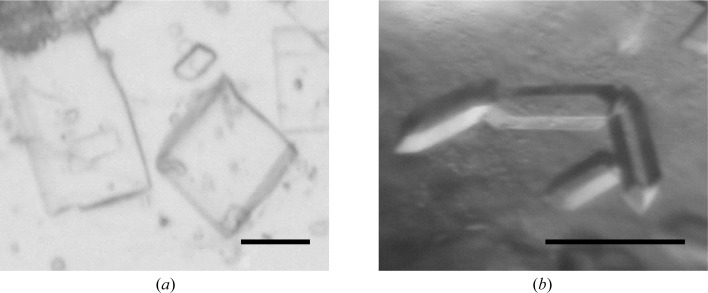
We previously determined the structure of Bc-TubZΔ in the free form at 2.1 Å resolution and electron density was not observed in the GTP-binding pocket (Hoshino & Hayashi, 2012 ▶). In this study, we obtained a crystal belonging to the monoclinic space group P21 (Fig. 3 ▶) with similar unit-cell parameters, a = 53.91, b = 65.54, c = 58.18 Å, β = 106.19°, that diffracted to 1.7 Å resolution. Electron-density maps were calculated by SIGMAA (Read, 1986 ▶) using the previously determined free-form structure of Bc-TubZΔ (PDB entry 4ei8, Hoshino & Hayashi, 2012 ▶). The crystal structure was solved; however, the GTP-binding pocket remained disordered, indicating that binding of the substrate is required for conformational stability of the catalytic site. The GDP–Bc-TubZΔ crystals appeared in an identical condition to that reported previously (pH 5.5, 1 mM magnesium ion and PEG 400 as a precipitant) except for the guanine nucleotide in the crystallization drop (Hoshino & Hayashi, 2012 ▶). The GDP–Bc-TubZΔ crystal, which was non-isomorphous to the previous GDP–Bc-TubZΔ crystal obtained in the presence of GTPγS, belonged to the monoclinic space group P21, with unit-cell parameters a = 67.05, b = 84.49, c = 67.66 Å, β = 92.92°. Diffraction data were collected to 2.1 Å resolution. The free-form structure of Bc-TubZΔ was used for phasing of the GDP–Bc-TubZΔ data, and the molecular-replacement trials gave a good solution. The crystal structure of GDP–Bc-TubZΔ was solved and two monomers were found in the asymmetric unit. The monomer structure of GDP–Bc-TubZΔ is identical to the previously determined structure of GDP–Bc-TubZΔ. Moreover, similar subunit–subunit interactions were observed between the two monomers in both GDP–Bc-TubZΔ structures: one subunit utilizes the nucleotide-binding region and the other utilizes the T7 loop for molecular contacts, as has also been observed in the protofilament-like crystal structure of FtsZ (Oliva et al., 2004 ▶). Notably, however, the residues involved in the subunit–subunit interactions differed in the two crystal forms. Given the facts that GTP hydrolysis is coupled to the polymerization activity of FtsZ-related proteins and that the previous GDP–Bc-TubZΔ crystal was obtained in the presence of GTPγS, the previous GDP–Bc-TubZΔ structure is likely to reflect the post-GTP hydrolysis state, whereas the intermolecular interactions in the present crystal structure seem to be caused by a crystal-packing effect (Mukherjee & Lutkenhaus, 1998 ▶; Hoshino & Hayashi, 2012 ▶). Information on the detailed structures will be published later.
Figure 3.
Crystals of Bc-TubZΔ in the free form (a) and the GDP-bound form (b). The scale bars are 0.1 mm in length.
Supplementary Material
Supplementary material file. DOI: 10.1107/S1744309112045551/pu5385sup1.pdf
Acknowledgments
We thank the beamline staff at BL17A and NW12A of the PF for help with data collection. This work was supported by the Hayashi Memorial Foundation for Female Natural Scientists (to IH).
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: PU5385).
References
- Aylett, C. H., Löwe, J. & Amos, L. A. (2011). Int. Rev. Cell Mol. Biol. 292, 1–71. [DOI] [PubMed]
- Aylett, C. H., Wang, Q., Michie, K. A., Amos, L. A. & Löwe, J. (2010). Proc. Natl Acad. Sci. USA, 107, 19766–19771. [DOI] [PMC free article] [PubMed]
- Bi, E. F. & Lutkenhaus, J. (1991). Nature (London), 354, 161–164. [DOI] [PubMed]
- Chen, Y. & Erickson, H. P. (2008). J. Biol. Chem. 283, 8102–8109. [DOI] [PMC free article] [PubMed]
- Erickson, H. P., Taylor, D. W., Taylor, K. A. & Bramhill, D. (1996). Proc. Natl Acad. Sci. USA, 93, 519–523.
- Hoshino, S. & Hayashi, I. (2012). J. Biol. Chem. 287, 32103–32112. [DOI] [PMC free article] [PubMed]
- Larsen, R. A., Cusumano, C., Fujioka, A., Lim-Fong, G., Patterson, P. & Pogliano, J. (2007). Genes Dev. 21, 1340–1352. [DOI] [PMC free article] [PubMed]
- Löwe, J. & Amos, L. A. (1998). Nature (London), 391, 203–206. [DOI] [PubMed]
- Mukherjee, A. & Lutkenhaus, J. (1994). J. Bacteriol. 176, 2754–2758. [DOI] [PMC free article] [PubMed]
- Mukherjee, A. & Lutkenhaus, J. (1998). EMBO J. 17, 462–469. [DOI] [PMC free article] [PubMed]
- Ni, L., Xu, W., Kumaraswami, M. & Schumacher, M. A. (2010). Proc. Natl Acad. Sci. USA, 107, 11763–11768. [DOI] [PMC free article] [PubMed]
- Nogales, E., Downing, K. H., Amos, L. A. & Löwe, J. (1998). Nature Struct. Biol. 5, 451–458. [DOI] [PubMed]
- Oliva, M. A., Cordell, S. C. & Löwe, J. (2004). Nature Struct. Mol. Biol. 11, 1243–1250. [DOI] [PubMed]
- Oliva, M. A., Martin-Galiano, A. J., Sakaguchi, Y. & Andreu, J. M. (2012). Proc. Natl Acad. Sci. USA, 109, 7711–7716. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Read, R. J. (1986). Acta Cryst. A42, 140–149.
- Scheffers, D. J., den Blaauwen, T. & Driessen, A. J. (2000). Mol. Microbiol. 35, 1211–1219. [DOI] [PubMed]
- Tang, M., Bideshi, D. K., Park, H.-W. & Federici, B. A. (2006). Appl. Environ. Microbiol. 72, 6948–6954. [DOI] [PMC free article] [PubMed]
- Tinsley, E. & Khan, S. A. (2006). J. Bacteriol. 188, 2829–2835. [DOI] [PMC free article] [PubMed]
- Weisenberg, R. C., Deery, W. J. & Dickinson, P. J. (1976). Biochemistry, 15, 4248–4254. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material file. DOI: 10.1107/S1744309112045551/pu5385sup1.pdf