Abstract
Regional differences in cardiomyocyte automaticity permit the sinoatrial node (SAN) to function as the leading cardiac pacemaker and the atrioventricular (AV) junction as a subsidiary pacemaker. The regulatory mechanisms controlling the distribution of automaticity within the heart are not understood. To understand regional variation in cardiac automaticity, we carried out an in vivo analysis of cis-regulatory elements that control expression of the hyperpolarization-activated cyclic-nucleotide gated ion channel 4 (Hcn4). Using transgenic mice, we found that spatial and temporal patterning of Hcn4 expression in the AV conduction system required cis-regulatory elements with multiple conserved fragments. One highly conserved region, which contained a myocyte enhancer factor 2C (Mef2C) binding site previously described in vitro, induced reporter expression specifically in the embryonic non-chamber myocardium and the postnatal AV bundle in a Mef2c-dependent manner in vivo. Inhibition of histone deacetylase (HDAC) activity in cultured transgenic embryos showed expansion of reporter activity to working myocardium. In adult animals, hypertrophy induced by transverse aortic constriction, which causes translocation of HDACs out of the nucleus, resulted in ectopic activation of the Hcn4 enhancer in working myocardium, recapitulating pathological electrical remodeling. These findings reveal mechanisms that control the distribution of automaticity among cardiomyocytes during development and in response to stress.
Keywords: Hcn4, cardiac automaticity, Mef2C, cis-regulatory element, atrioventricular bundle, electrical remodeling
Introduction
In the vertebrate heart, electrical properties of myocytes are distributed heterogeneously to optimize contraction and maintain electrical stability(Schram et al., 2002). Automaticity, or spontaneous cellular rhythmicity, is among the most fundamental of these properties. Although most cardiomyocytes can exhibit intrinsic rhythmicity under experimental conditions, a negative gradient of automaticity exists in vivo from sinoatrial (SA) to atrioventricular (AV) junctional to working myocytes. Under ordinary conditions, the rapid firing rate of SA nodal pacemaker cells and their efficient coupling to the atrial myocardium ensure that other potential cardiac pacemakers are silent.
Under abnormal conditions, extranodal areas of the heart exhibit automaticity. For example, when sinus arrest or heart block occurs, a subsidiary pacemaker at the atrioventricular junction serves as a ventricular pacemaker. Within the atria, regions of abnormal automaticity cause ectopic automatic tachycardias or trigger atrial fibrillation(Roberts-Thomson et al., 2006). The anatomic locations of ectopic atrial foci are non-random, with clustering in the posterior right and left atria, AV valvular annuli, pulmonary venous myocardium, coronary sinus myocardium, and near the AV junction(Kistler et al., 2006). In the ventricles, increased myocardial wall stress, myocardial hypertrophy, and heart failure lead to aberrant expression of genes associated with automaticity, creating a vulnerability to ventricular arrhythmias(Nattel et al., 2007; Stillitano et al., 2008). The gene regulatory networks that control normal and pathological variations in automaticity are not well understood.
At the cellular level, automaticity results from interactions among currents in the plasma membrane and sarcoplasmic reticulum(Lakatta et al., 2010). A key membrane component is an early diastolic, hyperpolarization-activated inward cation current (funny current; If) (DiFrancesco, 2010). If is carried by hyperpolarization-activated, cyclic nucleotide gated-ion channels (Hcn). Of the genes that encode these channels (Hcn1–4), Hcn4 is the most highly expressed in cardiac pacemaker tissue and regulates heart rhythm(Marionneau et al., 2005; Moosmang et al., 2001). Cardiac-specific deletion of murine Hcn4 causes embryonic lethality with fetal bradycardia(Stieber et al., 2003). Postnatal cardiac deletion of Hcn4 by different methods results in bradycardia of varying severity, from intermittent sinus pauses(Herrmann et al., 2007) to lethal bradycardia with AV block(Baruscotti et al., 2011). In humans, haploinsufficiency of HCN4 causes familial sinus bradycardia(Baruscotti et al., 2011; Milanesi et al., 2006). In addition to its role in sinoatrial automaticity, Hcn4 is expressed at the AV junction, where it regulates junctional pacemaker activity and AV conduction(Yamamoto et al., 2006). Hcn4 is also expressed in the inter-atrial groove and tricuspid annulus, and If is an important determinant of ectopic atrial rhythms that arise from these tissues(Shinohara et al., 2010;Yamamoto et al., 2006).
Understanding how regional expression of Hcn4 is controlled during development and in postnatal life would provide insight into the mechanisms of normal and abnormal automaticity. While previous studies have identified a few direct regulators of Hcn4 transcription using primarily cultured cells, including Sp1, NRSF, Mef2, and AP-1(Kuratomi et al., 2007; Kuratomi et al., 2009; Lin et al., 2009), it remains unclear how these widely expressed factors determine the highly patterned in vivo cardiac distribution of Hcn4.
The complexity of Hcn4 expression is one of many examples of fine patterning of gene expression in the heart. In several animal model systems, regionally and temporally restricted gene expression is controlled by conserved non-coding cis-regulatory elements (CREs) that activate or repress transcription when bound to specific combinations of activated transcription factors(Davidson, 2006). Functional analysis of CREs outside of their native genomic context has been fruitful for understanding regulatory hierarchies among cardiac transcription factors during development and for understanding how core cardiac structural and contractile proteins are transcriptionally regulated(Olson, 2006; Srivastava, 2006). However, it has been difficult to identify and validate CREs that control gene expression in subsets of functionally specialized cardiomyocytes as occur in the cardiac conduction system. In recent years, just one AV node CRE has been cloned and characterized (Munshi et al., 2009), a few enhancers with activity in the AV bundle and Purkinje Fibers have been described (Arnolds et al., 2012; Moskowitz et al., 2007; van den Boogaard et al., 2012), and no autonomous CREs with specific activity in the SA node have been identified, despite the functional importance of these structures, their unique patterns of gene expression, and numerous attempts(Stroud et al., 2007; Viswanathan et al., 2007). The difficulty in identifying CREs that recapitulate highly patterned gene expression may reflect a requirement for multiple interacting CREs.
To understand how regional variation of cardiac automaticity occurs in vivo, we investigated deeply conserved non-coding genomic regions around the Hcn4 locus, each containing previously identified transcription factor binding sites (TFBS). Each region was tested in vivo with a mouse transgenic enhancer-reporter assay. We found that a 5.7 kb region within the first intron of Hcn4 could direct reporter expression to areas of non-chamber myocardium in the developing heart, and to the AV junctional pacemaker in the postnatal heart. A deletion analysis revealed that two conserved domains from within this region were sufficient for enhancer activity. One of these domains contained a conserved Mef2 site, previously described in-vitro, that was essential for enhancer activity. The enhancer exhibited steep dosage-dependence on Mef2C levels during development, and displayed ectopic activity in ventricular chamber myocardium in response to HDAC inhibition and pathological stress. Taken together, our findings demonstrate that regional variation of Hcn4 expression is achieved, in part, through modular CREs whose output is dependent on the actions of signal-responsive transcription factors.
Methods
In-Situ Hybridization
A probe corresponding to amino acids 400–690 of mouse Hcn4 was cloned using PCR from mouse heart cDNA. RNA probe was generated by in-vitro transcription in the antisense direction, followed by labeling with the DIG RNA labeling kit (Roche). Embryos were fixed in 4% PFA, washed in PBS, permeabilized in PBS with 0.3% Triton (PBS.3), briefly treated with 20 µg/mL proteinase K, and post-fixed. Embryos were then incubated in prehybridization buffer (50% formamide, 4x SSC, 1x Denhardt’s solution, 0.5 µg/mL salmon sperm DNA, 0.25 µg/µL yeast tRNA, 50 µg/µl heparin) /50% PBS.3 for 3 h at 64°C. Labeled probe was added to pre-hyb buffer at 1 µg/mL and incubated with embryos at 64°C overnight. Embryos washed with post-hybridization buffer (50% formamide, 4x SSC, 1x Denhardt’s solution) at 64°C for 2 h, followed by PBS.3 at room temperature. Embryos were then incubated in 2% blocking reagent (20 mM maleic acid, 30 mM NaCl, and 2% BM blocking agent in PBS.3) at room temperature before application of anti-DIG antibody (1:2000). Embryos were developed in BM Purple for several hours before post-fixing and photography.
Mouse Transgenesis
Constructs were generated by PCR amplification of putative enhancer regions from a bacterial artificial chromosome containing the HCN4 genomic locus (BAC clone RP23-414K12, BACPAC, CHORI). Primers and genomic coordinates of putative enhancer regions are given in the Supplementary Table. Putative enhancer elements were cloned into the reporter vector pKS-hsp68LacZ (Kothary et al., 1989) by standard methods. DNA for injection was prepared from linearized enhancer-reporter constructs. The DNA was isolated on an agarose gel, purified with an ion exchange column (Elutip), ethanol precipitated, and taken up in 10 mM TRIS-HCL, 0.1 mM EDTA (pH 8.0) at a concentration of 2 ng/mL before pronuclear injection by the Gladstone Transgenic Core Facility. FVB/N mice were used for all transgenesis assays. Founders were maintained on the FVB/N background, except when crossed to Mef2C+/− animals, which were on a C57BL6 background. Mef2ctm1Eno (hereafter, Mef2C+/−) mice were generated by Eric Olson (UT Southwestern) and were kindly provided by Brian Black (UCSF) (Lin et al., 1997).
Histology
Embryos or hearts were collected in cold PBS, fixed for 1 hour in formalin, and stained at room temperature overnight with X-gal or bluo-gal staining solution (in mM): 5 potassium ferricyanide, 5 potassium ferrocyanide, 2 MgCl2, and 1 mg/ml X-gal or bluo-gal in PBS. Adult hearts were bisected before staining to facilitate access of the staining solution to the endocardial side of the heart. Embryos or hearts were then washed with PBS and post-fixed in formalin. Some embryos and hearts were dehydrated in methanol and cleared with a solution of 2:1 benzyl alcholol and benzyl benzoate before photography. Another group of embryos and hearts were embedded in paraffin, sectioned, and stained with either hematoxylineosin, Masson’s trichrome, or for acetylcholinesterase activity by standard methods. A third group of embryos and hearts was processed for immunohistochemistry after whole-mount bluogal staining.
Immunohistochemistry
Embryos or heart samples were fixed in 4% PFA, washed with PBS, and incubated in 30% sucrose before embedding in OCT for sectioning. A subset of hearts was already stained with bluo-gal before embedding in OCT. Sections were permeabilized with 0.3% Triton X-100, blocked with 10% normal goat serum, and incubated with primary antibody for Hcn4 (rabbit polyclonal, Alomone labs APC-052, 1:200), Connexin-43 (rabbit polyclonal, Sigma- Aldrich SAB4501173, 1:250), Connexin-40 (rabbit polyclonal, Alpha Diagnostics CX40-A, 1:200), or beta-Galactosidase (chicken polyclonal, AbCam ab9361, 1:200) overnight at 4°C. Slides were washed, incubated for one hour with the secondary antibody before mounting in Vectashield with DAPI (Vector Laboratories).
Optical Projection Tomography
Samples to be imaged were embedded in agarose blocks and mounted, dehydrated with methanol, cleared with 2:1 benzyl alcolol:benzyl benzoate, and imaged with an optical projection tomography scanner according to the manufacturers instructions (Bioptonics, MRC Technology, Edinburgh, UK). Image processing and 3D reconstruction was performed with NR Recon (Bioptonics). Videos were created with Amira (Visage Imaging).
Luciferase Assays
Human embryonic kidney cells (HEK-293T) were transfected with lipofectamine reagent (Invitrogen) with 50 ng of transcription factor, 100 ng of luciferase reporter vector, and 10 ng of Renilla per well for each well of a 70% confluent 12-well tissue culture plate. Lipofectamine was removed after 4 h, and for experiments involving MC1568, either MC1568 (final concentration 4 µM) or vehicle (DMSO) was added after removal of transfection agent. The cells were lysed 24 h after transfection, and luciferase activity was measured using Dual-Luciferase Reporter Assay Kit (Promega).
Electromobility Shift Assays
Fluorescently labeled oligonucleotides were obtained from Integrated DNA Technologies, and diluted to 1 mg/mL. Oligos were incubated at 90°C for 10 min, 65°C for 10 min, 37°C for 10 min, and 22°C for 10 min. Mef2C protein was made using the TNT T7 Quick Coupled Translation/Transcription Kit (Promega). Samples were prepared using an EMSA Accessory Kit (Novagen), then run on a 10% acrylamide (20:1), non-denaturing 0.5x TBE gel which contained 0.1% APS and 0.04% TEMED.
Mouse Organ culture
Embryos were harvested at E11.5 and were dissected in a 1:1 mixture of F12/Ham’s media at 4 degrees. Hearts were removed from embryos with attachments to the torso intact and placed in pre-warmed DMEM/1%FBS at 37°C. Trichostatin-A (10 µM in DMSO) or DMSO alone were added 2 hours later, and embryos were incubated for 48 hours prior to fixation and staining.
Quantitative PCR
Hearts were dissected in nuclease-free PBS and homogenized in Tri-Reagent (Invitrogen) with a bead homogenizer (Bullet Blender, Next Advance). Total RNA was prepared from the aqueous fraction by isopropanol-ethanol precipitations. Quantiative PCR was performed on an Applied Biosystems 7900HT machine with the Taqman system (Applied Biosystems).
Thoracic Aortic Constriction
TAC was performed as described with minor modifications(Tarnavski et al., 2004). Adult R2R3LacZ mice were anesthetized with isoflurane, intubated endotracheally and mechanically ventilated. TAC was performed through a 0.5-cm incision over the left thorax via a mediastinal approach. A silk suture was tied around a 27-guage needle positioned adjacently to the aorta. Successful TAC was confirmed by Doppler measurement of pressure gradient 10 days after surgery. A VisualSonics small animal echo machine was used to obtain short-axis, M-Mode and Doppler images on sham-operated and post-TAC R2R3LacZ adult mice. Pressure gradients were calculated according the modified Bernoulli’s equation as 4v2, where v is blood velocity as determined by continuous wave Doppler. Parameters were averaged for each genotype, and comparisons were made with Student’s T-tests on two independent samples assuming a two-tailed distribution for each parameter.
Results
Expression Pattern of Hcn4
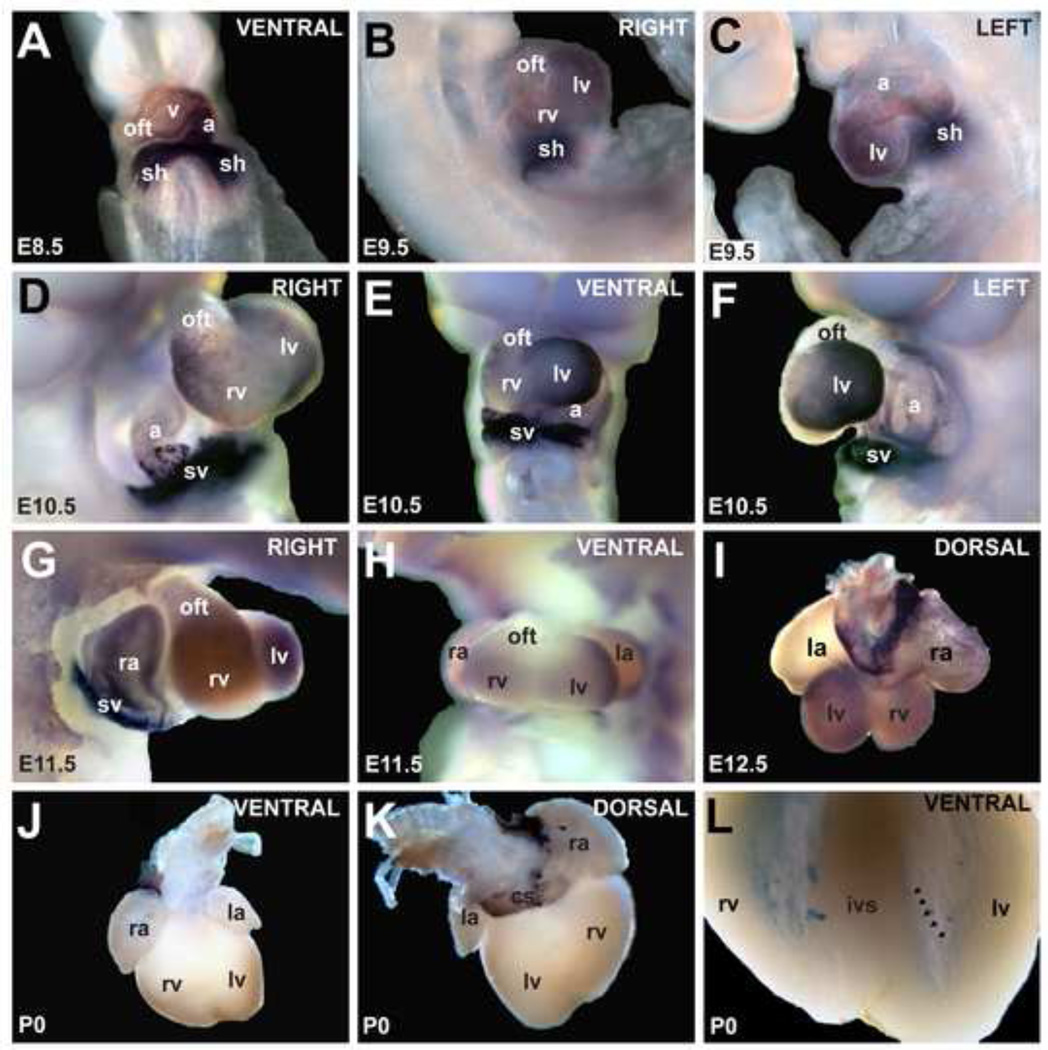
The expression patterns of Hcn4 during cardiac development and in the postnatal heart have been described, with several previous studies including areas outside the SAN and AVN (Aanhaanen et al., 2010; Garcia-Frigola et al., 2003; Herrmann et al., 2011; Yamamoto et al., 2006; Yanni et al., 2009). We used whole-mount in situ hybridization and immunohistochemistry at several developmental stages to characterize Hcn4 expression throughout the heart. Hcn4 was highly expressed in the sinus horns at embryonic day (E) 8.5 with a negative gradient extending to the arterial pole of the heart (Fig. 1A). More complex patterning developed between E9 and E11, with high expression in the sinus horns and left ventricle, lower expression in the inner curvature of the AV canal and posterior atria, and trace expression in the anterior atrium, right ventricle and outflow tract (Fig. 1B–F). By mid-development (E11–E13) Hcn4 was highly expressed in the right sinoatrial region and sinus venosus contribution to the atria. There was lower level expression in the ventricles, concentrated at the crest of the interventricular septum (Fig. 1G–I, Fig. S1A). Hcn4 expression in several of the areas outside the sinus node was maintained in postnatal animals, including the sinus venosus derivatives (coronary sinus and posterior atria), the atrioventricular conduction axis, the tricuspid annulus, and at lower intensity within the His-Purkinje system (Fig. 1J–L, Fig. S1B). Although cardiomyocytes in these areas have distinct developmental origins, they are often grouped together as non-chamber myocardium to distinguish them from the working myocardial cells, whose primary role is force generation. We found that, within the non-chamber myocardium, Hcn4 expression was highly patterned, with the following negative gradient: SA node > posterior atrium/atrial septum > AV node and nodal extensions > AV bundle > infra-nodal ventricular conduction system. Within the working myocyte compartment, Hcn4 expression waxes in early and mid-development before waning in late development. This expression pattern correlates well with established differences in automaticity among different subsets of cardiomyocytes, and suggests a complex cis-regulatory architecture for patterning of automaticity in the heart.
Figure 1. Expression pattern of Hcn4 mRNA by whole-mount in-situ hybridization.
(A) At E8.5, Hcn4 was expressed throughout the heart tube in a caudal-to-cranial gradient. By E9.5 (B,C), expression was less in the atrial chamber, right ventricle (rv), and outflow tract (oft) than in other areas. At E10.5 and E11.5 (D–H), Hcn4 was strongly expressed in the sinus horns (sh) 27 and their interface with the atria, and at lower levels in the atria and ventricles. At E12.5 (I), Hcn4 was expressed highly in the sinoatrial node, the sinus venosus contribution to the atria including the coronary sinus (cs), and at lower levels in the ventricles and right atrium. In the neonatal heart (J,K), Hcn4 was expressed in the SA node, the sinus venosus derivatives, and the AV ring tissue. In (L) the ventricles are shown after clearing. A low-intensity meshwork of staining is visible, consistent with the His-Purkinje system (arrowheads delineate an example of a Purkinje fiber strand in the left ventricle). Abbreviations: ra, right atrium; la, left atrium; v, ventricle; rv, right ventricle; lv, left ventricle; sh, sinus horns; sv, sinus venosus oft, outflow tract; cs, coronary sinus.
A 5.7-Kb Fragment of Hcn4 Intron1 Directs Reporter Expression to the Heart
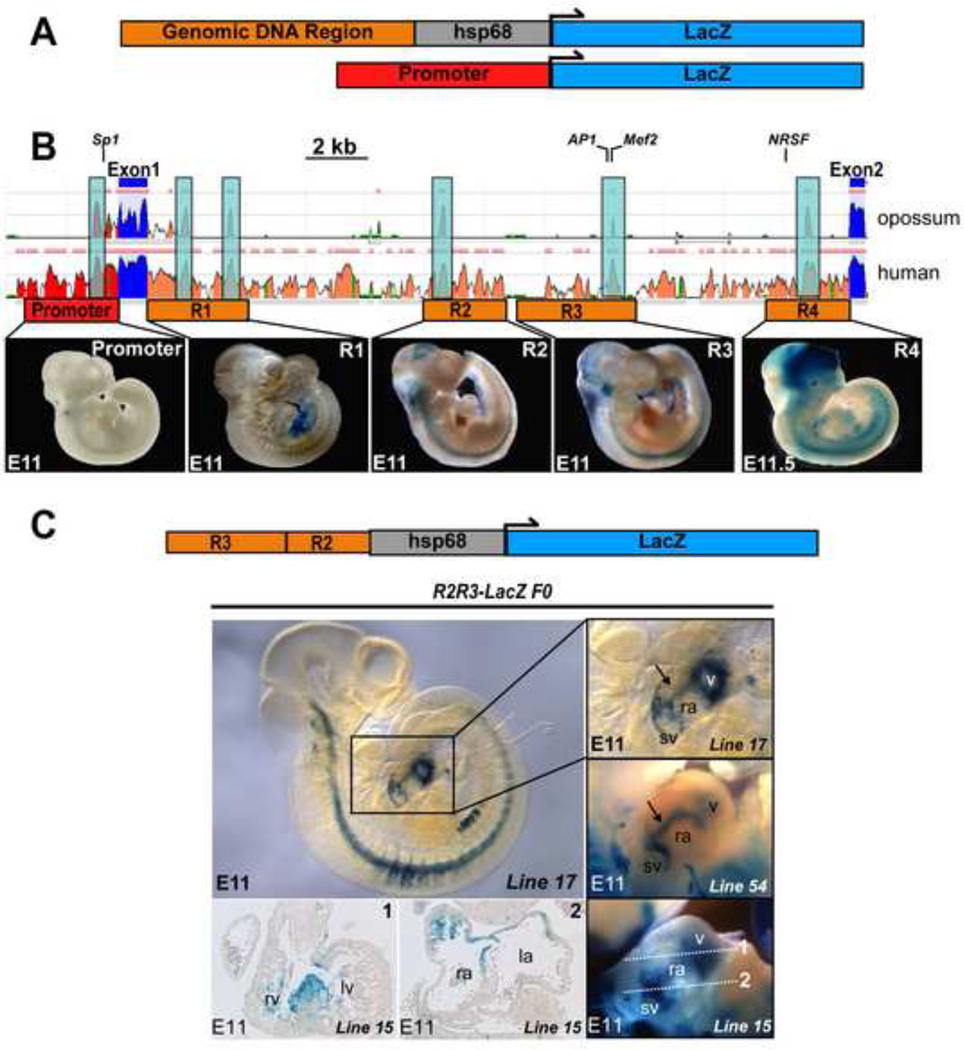
To identify conserved regions that might serve as CREs at the Hcn4 locus, we used the ECR Browser, an online multispecies sequence alignment tool (Fig. 2B)(Ovcharenko et al., 2004). The basal promoter and transcriptional start site of murine Hcn4 have been defined(Kuratomi et al., 2009), as have transcription factor binding sites (TFBS) located in intron 1 for Mef2, AP-1, and NRSF (Fig. 2B). Deeply conserved putative CREs were defined as segments of at least 100bp of non-coding genomic DNA with greater than 70% sequence homology among mouse, human, and opossum genomes (Fig. 2B). We defined five genomic regions of at least 2kb (Promoter region, R1–R4), each anchored by at least one deeply conserved segment. R1–R4 or the promoter region was each individually cloned from an Hcn4-containing BAC into either the basal promoter-containing hsp68LacZ reporter vector or directly upstream of LacZ (Supplementary Table). Enhancer-promoter-reporter constructs were linearized and injected into pronuclei of fertilized ova (Fig. 2A). The ova were implanted and founder embryos were harvested at E11.5 for staining with bluo-gal. At least 3 transgenic founders were examined for each construct. None of the constructs exhibited a consistent and robust pattern of reporter gene expression within the heart, indicating that none of these elements was sufficient to drive Hcn4 expression in the embryonic heart (Fig. 2B, Supplementary Table). To test whether larger genomic regions were required for enhancer activity, we investigated combinations of regions, beginning with R2 and R3 (a 5.7Kb contiguous piece of genomic DNA). In 12 of 13 R2R3-LacZ founder embryos, a consistent pattern of reporter activity at E11.5 was observed in discrete areas of the heart. While there was variability in overall intensity and in the extra-cardiac staining among founder embryos (reflecting different genomic integration sites), each of the founders exhibited reporter activity in a consistent pattern within the sinoatrial region, AV canal, and within the ventricles (Fig. 2C).
Figure 2. Identification and in vivo testing of Hcn4 regulatory elements.
(A) Genomic regions containing putative CREs were cloned upstream of hsp68LacZ or LacZ (basal promoter) to generate constructs for pronuclear injection. (B) Three species alignment at the Hcn4 locus from the ECR Browser, with mouse as the base genome against human and opossum. Peaks reflect degree of conservation (scale: 50–100%). Color code: red, upstream non-coding DNA; salmon, intronic DNA; blue, exons. Multiple conserved elements were selected for analysis (1–4) from Intron 1. Locations of previously validated transcription factor binding sites (TFBS) are indicated (NRSF, Mef2, AP-1, Sp1). Examples of founders from each construct injected are shown below the corresponding area on the Hcn4 genomic map. Although the presence of variable extra-cardiac reporter activity among different founders confirmed reporter insertion into permissive loci, none of the founders exhibited consistent reporter activity within the heart. (C) Left, an E11 embryo from an R2R3-LacZ transgenic line (line 17) stained with bluo-gal reporter activity within defined areas of the heart. Right, the cardiac region of line 17 in comparison with embryos from two additional transgenic founders (line 15 and line 22) shows similar cardiac reporter activity despite different genomic integration sites. By whole mount imaging, activity is concentrated at the junction of the sinus venosus and the right atrium, a linear area extending from the sinus venosus at the level of the interatrial groove to the ventricles (arrow), and within the ventricles. Sections from one founder embryo (line 15, bottom left) show reporter activity in the interventricular ring at the crest of the developing interventricular septum (section 1); and between the sinus venosus and atrium (section 2). Abbreviations: sv, sinus venosus; ra, right atrium; la, left atrium; v, ventricles; rv, right ventricle; lv, left ventricle.
Hcn4-R2R3 Enhancer Activity During Development
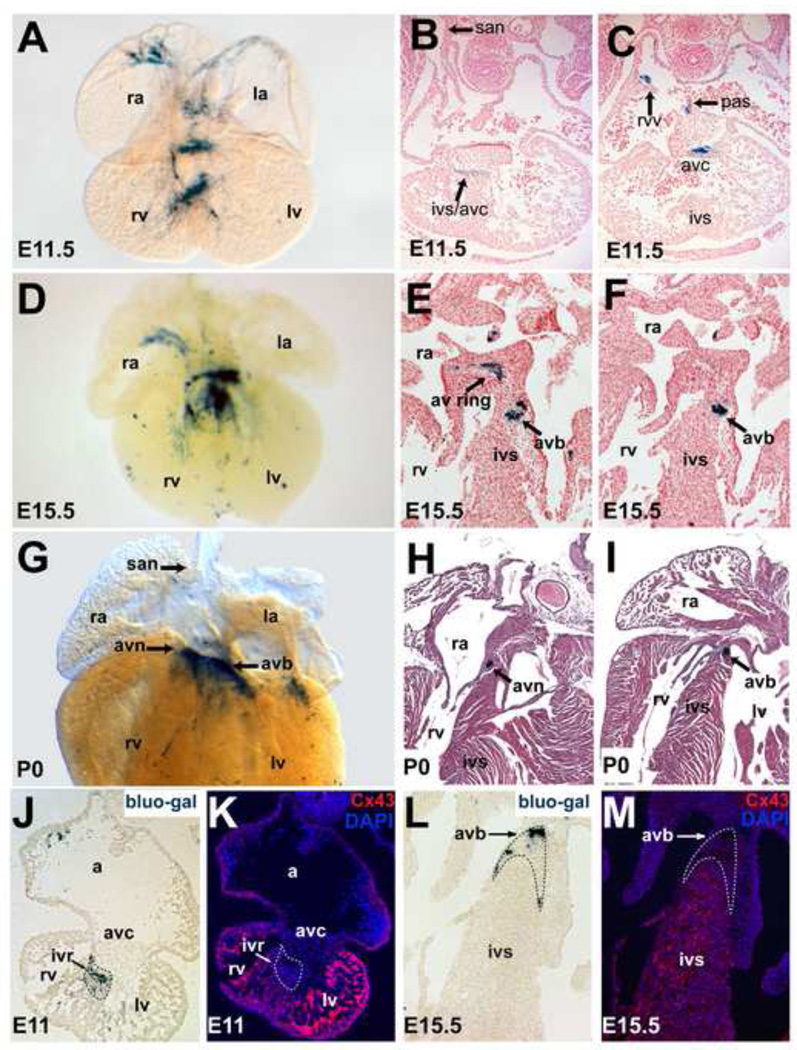
To characterize R2R3 activity in detail, we performed whole mount bluo-gal staining at several stages in two mouse lines derived from different R2R3-LacZ founder animals. Both lines exhibited the same spatiotemporal pattern of reporter activity in specific subdomains of the heart, but differed in the intensity of cardiac reporter activity and in extra-cardiac activity, reflecting distinct genomic integrations sites and likely different copy number. In both lines, reporter activity was visible in the inflow tract, caudal and posterior heart tube, and AV canal at E8.5 (Fig. S2). By E11.5, activity was detected only in non-chamber myocardium: the right venous valve, AV canal, posterior atrium, primary atrial septum, and crest of the developing interventricular septum (Fig. 3A–C; Video 1). By E15.5, there was less activity in atrial non-chamber regions, while high-level expression was preserved within the developing AV conduction system (Fig. 3D–F; Video 2). This pattern persisted in neonatal hearts, with loss of activity in atrial non-chamber myocardium and preserved expression in the AV conduction system (Fig. 3G–I). Hcn4 is expressed in all of these areas, indicating that R2R3 recapitulates the non-SA nodal, non-chamber myocardial component of the Hcn4 developmental expression pattern. Mutual exclusion of β-gal activity and expression of connexin-43 (Cx43), a marker of the chamber myocardium, confirmed restriction of enhancer activity to the non-chamber myocardium at E11.5 (Fig. 3J,K) and at E15.5 (Fig. 3L,M).
Figure 3. In vivo temporal reporter activity directed by the combined R2-R3 Hcn4 Enhancers.
A stable transgenic R2R3-LacZ mouse line was examined at E11.5 (A–C), at E15.5 (D–F) and at the neonatal stage (G–I). At E11.5, a whole mount bluo-gal stained embryo (A) and paraffin sections of bluo-gal stained embryos counterstained with nuclear fast red (B,C) showed reporter activity in the non-chamber embryonic myocardium including the AV canal, the interventricular ring at the crest of the developing interventricular septum, the posterior atrium/interatrial groove and right venous valve. At E15.5 (D–F), expression was confined to the atrial non-chamber myocardium and the developing AV conduction system. By late development, only faint expression was visible in atrial non-chamber myocardium while more robust expression was present within the AV conduction system (G–I). Of note, high-level activity was never noted in the SAN primordium or the perinatal SAN, indicating that R2R3 is not sufficient to direct expression to this tissue. Immunohistochemistry for Cx43, marking chamber myocardium, on bluo-gal stained cryosections (bottom row) showed mutual exclusion of Cx43 and β-gal activity in the ventricles at E11.5 (J,K) and at E15.5 (L,M). Abbreviations: san, sinoatrial node; avc, atrioventricular canal; rvv, right venous valve; pas, primary atrial septum; ra, right atrium; rv, right ventricle; ivs, interventricular septum; avn, atrioventricular node; avb, atrioventricular bundle; av ring, atrioventricular ring; a, common atrium; ivr, interventricular ring.
Hcn4-R2R3 Enhancer Activity in the Postnatal and Adult Heart
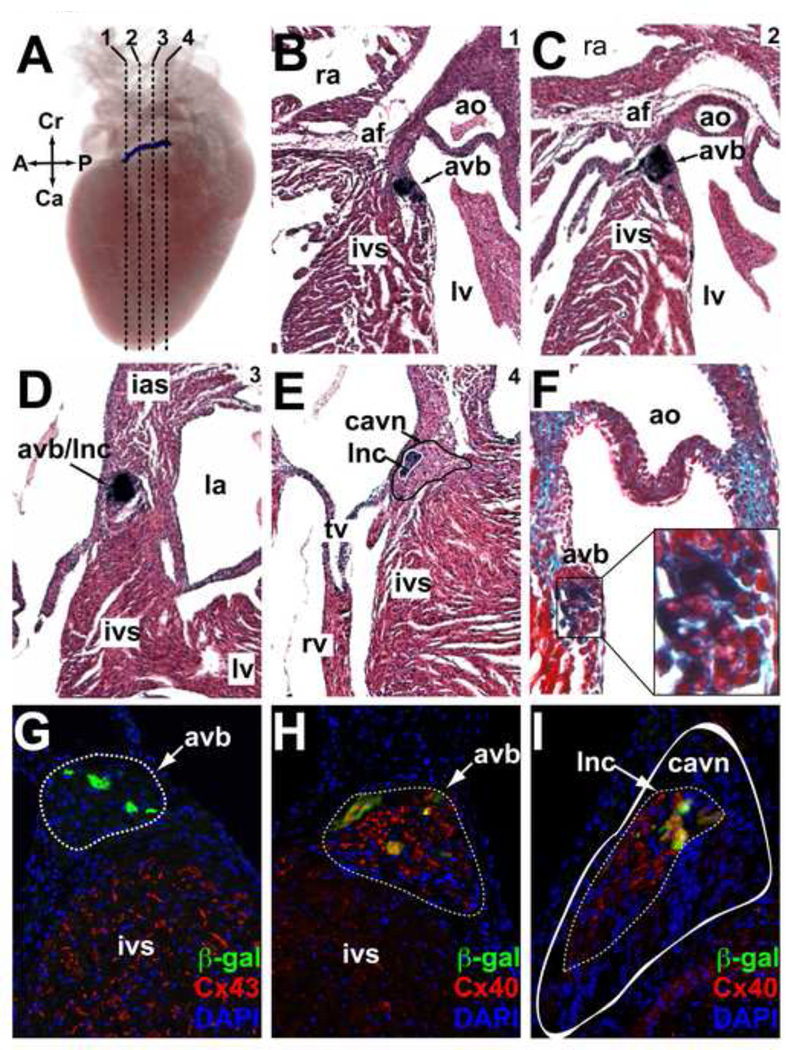
In the postnatal heart, R2R3 activity persisted in a continuous region extending from the interface of the compact AV node with the AV bundle (lower nodal cells) to the terminal portion of the AV bundle itself (Video 3). Expression was no longer present in the atrial non-chamber myocardial derivatives. As animals aged, the specificity of activity persisted with the AV bundle, but the intensity of activity gradually diminished into adulthood. Serial sections at the adult AV junction showed that expression was confined to the AV bundle/lower nodal cell (AVB/LNC) compartment, as evidenced by anatomic location of reporter activity and the presence of insulating fibrous tissue around β-gal positive cells (Fig 4A–F). The AVB/LNC is a single transcriptional domain that can be distinguished from the compact AV node by expression of connexin-40 (Cx40) and from working ventricular myocytes by the absence of connexin-43 (Cx43) (Aanhaanen et al., 2010). Immunohistochemistry for Cx43, Cx40, and β-gal at the AV junction of R2R3-LacZ adults, by which point enhancer activity had diminished in intensity, revealed persistent overlap between β-gal and Cx40 but mutual exclusion of β-gal and Cx43, confirming restriction of enhancer activity to the AVB/LNC into adulthood (Fig. 4G–I).
Figure 4. In vivo expression directed by the R2R3 Hcn4 enhancer in the post-natal AV bundle and its interface with the AV Node.
(A–E) A postnatal (P5) R2R3-LacZ heart was stained with bluo-gal, imaged with optical projection tomography in whole-mount, then sectioned and counterstained with hematoxylin-eosin. (A) An image from a 3D reconstruction of optical projection tomograms shows a discrete band of enhancer activity along the anterior-posterior axis of the AV junction. Dashed lines indicate the positions of the coronal sections shown in panels B-E proceeding from anterior-posterior (1–4). The band of reporter activity begins below the annulus fibrosis (af) anteriorly at the crest of the interventricular septum (ivs) just below the aortic valve (ao) (B). As it moves posteriorly (C,D) expression band proceeds superiorly and penetrates the af, eventually terminating within it. In section 4 (E), the solid black line demarcates the area of the compact atrioventricular node (cavn) + inferior nodal extension (ine) and the solid white line demarcates the atrioventricular bundle (avb) + lower nodal cells (lnc) where reporter activity was present. (F) Post-natal hearts were also sectioned and counterstained with trichrome to visualize fibrous tissue (green). A close-up view of the area of bluo-gal is shown to contain abundant insulating collagen fibers (light blue), consistent with its identity as the avb. (G,H) Immunohistochemical analysis of an R2R3-LacZ section from an adult heart at the level of the avb (outlined, dashed line) showed mutual exclusion of β-gal and Cx43, but co-expression of β-gal and Cx40 in the avb. (I) A more posterior section at the level of the cavn (outlined, solid white) showed β-gal expression within the Cx40+ lnc domain (outlined, dashed line) that comprises the interface between the avb and the avn. Abbreviations: A, anterior; P, posterior; Cr, cranial; Ca, caudal; lv, left ventricle; ra, right atrium; rv, right ventricle; tv, tricuspid valve; mv, mitral valve.
A Minimal Cis-Regulatory Element Contains Two Deeply Conserved Regions
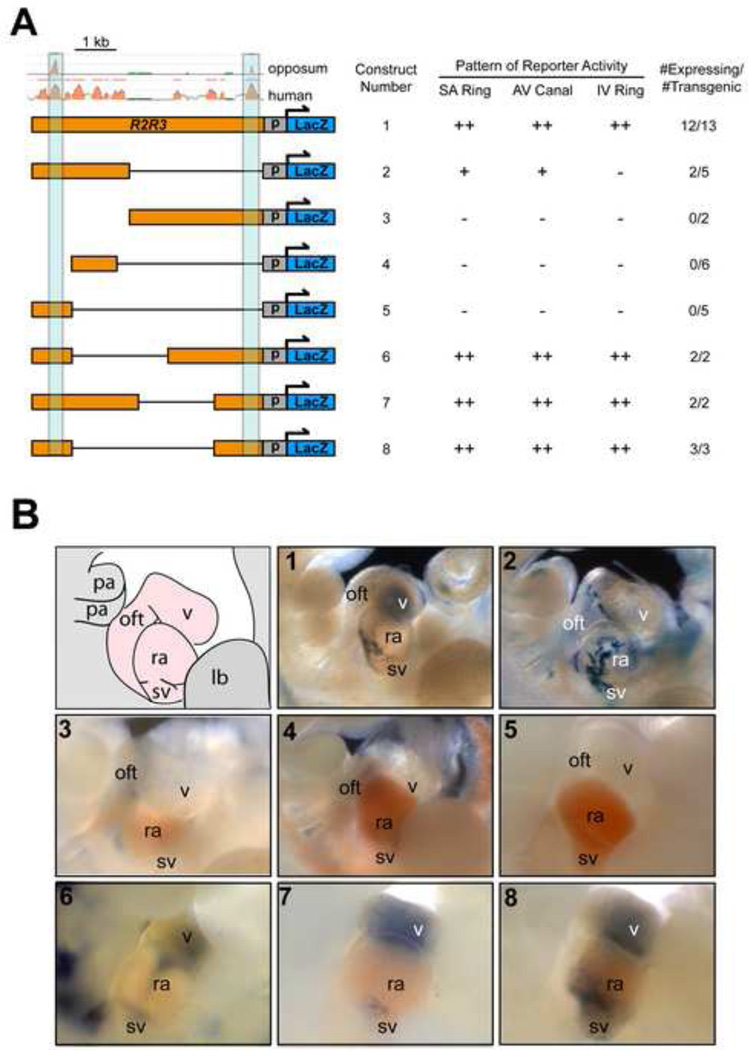
To define the minimal regulatory element capable of recapitulating the R2R3 pattern of enhancer activity, we performed a deletion analysis (Fig. 5A). While much of R2 and R3 could be deleted without affecting reporter activity, deletion of either of the most conserved segment of R2 or of R3 dramatically reduced reporter activity (Fig. 5B, constructs 2–5) as compared to the full-length construct (Fig. 5B, construct 1). The only deletion constructs that preserved enhancer activity specifically within the non-chamber myocardium of both atria and ventricles contained the two most highly conserved segments (Fig 5B, constructs 6–8), demonstrating that the presence of both conserved regions was necessary and sufficient for the full extent of reporter activity.
Figure 5. Identification of minimal enhancer regions within R2R3 that direct reporter activity in the Hcn4 expression domain.
(A) A deletion analysis of R2R3-LacZ revealed that only constructs bearing both deeply conserved sequences could direct robust reporter expression to the atrial and ventricular nonchamber myocardium at E11. (B) Representative 30 examples of E11 founder embryos from each construct is shown after bluo-gal staining and imaging of the heart in whole mount from the right side. A diagram is shown at the top left depicting the heart region. The pattern of full-length R2R3-LacZ is shown for comparison as construct 1. Of the deletion constructs tested that did not contain both conserved sequences (constructs 2–5), only construct 2 had some cardiac activity in atrial non-chamber myocardium in 2/5 embryos (D). In contrast, those with both conserved regions (constructs 6–8, bottom row) did preserve the activity of full length R2R3. The minimal enhancer capable of reconstituting the entire expression pattern of the full-length sequence, construct 8, consisted of the two conserved regulatory elements (light blue) without intervening sequence. Abbreviations: p, promoter; sv, sinus venosus; ra, right atrium; v, ventricle; oft, outflow tract; lb, limb bud; pa, pharyngeal arch; “++” denotes robust and consistent activity, “+” denotes inconsistent or lowlevel activity.
Hcn4-R2R3 Activity Requires a Conserved Mef2 Site
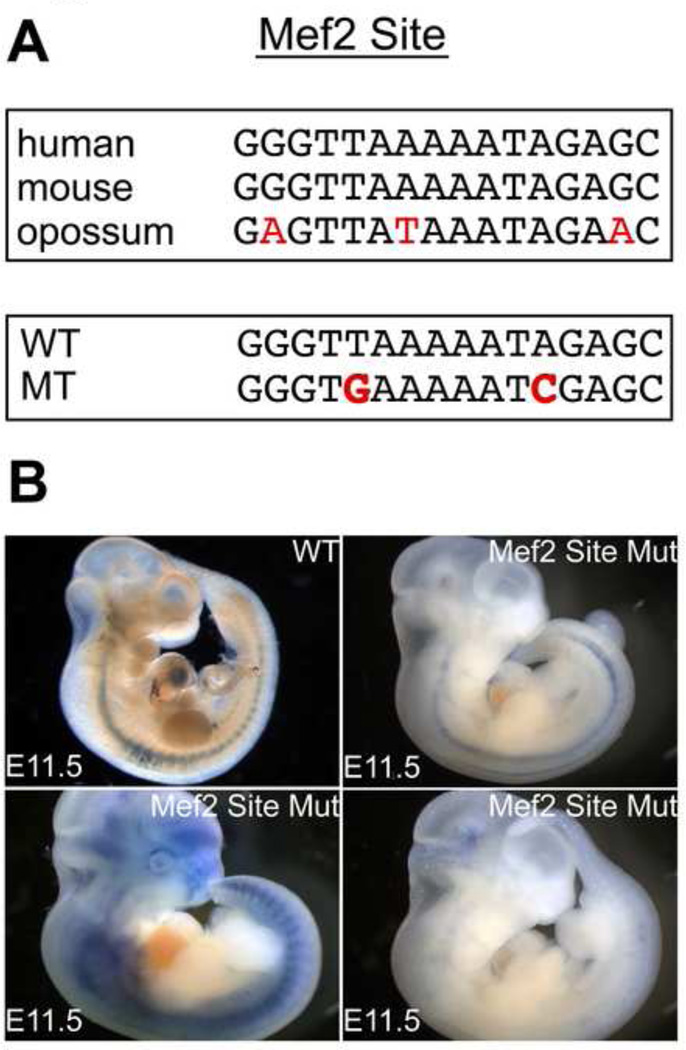
A conserved and functional Mef2 site in R3 has been well-characterized in vitro using binding assays and rat neonatal myocytes (Kuratomi et al., 2009). Moreover, mouse embryos expressing a dominant negative form of Mef2 have reduced levels of If and Hcn4. We therefore hypothesized that the previously characterized Mef2 site in R3 might be critical for activity of our enhancer. We first confirmed that the Mef2 site efficiently binds Mef2C transcription factor using an electromobility shift assay (EMSA) (Fig. S3A). To confirm the site is functional, we tested the ability of Mef2C conjugated to VP-16 to regulate R3-dependent luciferase activity. In HEK-293T cells, we observed robust activation of R3-luciferase by MEF2C-VP16 that was predominantly dependent on the conserved Mef2 site (Fig. S3B).
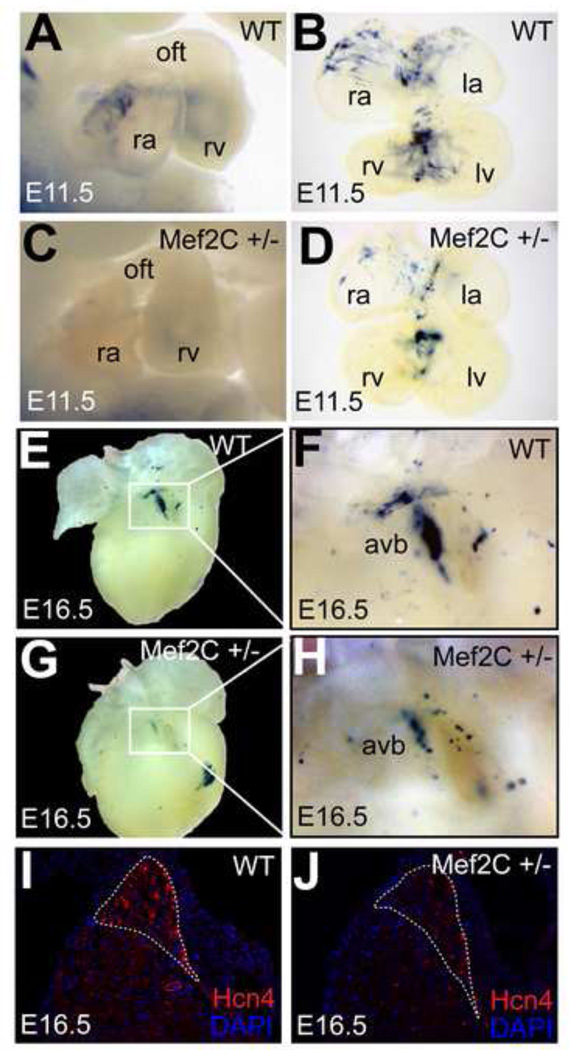
Since Mef2C is widely expressed throughout the embryonic heart, it is not clear how this site might regulate the dynamics of Hcn4 expression in vivo. Of the four Mef2 isoforms expressed in the heart, Mef2C predominates in the developing heart and declines relative to Mef2A and Mef2D after birth(Edmondson et al., 1994). We tested the importance of the Mef2 site in-vivo by mutating it in the context of full-length R2R3-LacZ. We found a loss of cardiac enhancer activity in four out of four transgenic founder embryos at E11.5 (Fig. 6A,B). The presence of extra-cardiac but not cardiac activity in these founders demonstrates insertion of the transgene into a permissive genomic locus and the importance of the Mef2 site for cardiac activity. To test whether R2R3 is regulated by Mef2C in vivo, we crossed R2R3-LacZ mice with Mef2C heterozygotes (Mef2C+/−). While the reporter expression pattern was preserved in the R2R3-LacZ; Mef2C+/− animals, the intensity of cardiac reporter activity was reduced in the nonchamber myocardium in all embryos examined (3 spearate litters) as compared to R2R3-LacZ littermates, confirming sensitivity of enhancer activity level to Mef2C dosage at E11.5 (Fig. 7A–D). This experiment was repeated in an R2R3-LacZ line with extensive extra-cardiac positional activity as well as activity in the nonchamber myocardium. Each of the Mef2C+/− embryos from this cross showed less cardiac reporter activity than WT, while the extra-cardiac activity was unchanged, confirming the specificity of the Mef2C dosage dependence (Fig S4). At E16, R2R3-LacZ ;Mef2C+/− hearts had less reporter activity in the AVB/LNC, indicating that Mef2C dosage sensitivity persisted late into development (Fig. 7E–H). Immunohistochemistry at the AV junction showed less Hcn4 expression in Mef2C+/− hearts at E16 (Fig. 7I,J), confirming the relevance of our enhancer findings to Hcn4 gene expression.
Figure 6. Mef2C regulates R2R3 Hcn4 enhancer activity via a conserved Mef2 Site.
(A) Top, a conserved binding site for Mef2 was present within R3 as shown by an alignment between homologous regions of mouse, human, and opposum genomic DNA. Mismatches, shown in red, were not predicted to prevent Mef2 binding. Bottom, the Mef2 site in R3 was mutated at sites required for binding (mutations in red). (B) Transient R2R3-LacZ transgenic embryos bearing the Mef2 binding site mutation lost cardiac reporter activity. A founder bearing the unmutated R2R3 is shown for comparison (top left). Non-specific extracardiac ectopic activity confirmed transgene insertion into a permissive locus in the three founder embryos shown that bear the mutated Mef2 site in the context of R2R3.
Figure 7. Mef2C regulates R2R3-LacZ and Hcn4 expression in vivo.
E11.5 embryos (A,C) and embryonic hearts (B,D) from Mef2C+/−;R2R3-LacZ compound heterozygous mice after whole-mount bluo-gal staining. Mef2C+/− embryos (C,D) had reduced cardiac staining. At E16.5, 31 the AV junction region of Mef2C+/− hearts (G) had reduced reporter activity compared to WT (E). (F,H) show higher magnification of the AV junction region. Immunohistochemistry for Hcn4 at the AV junction showed a moderate reduction in Hcn4 expression in Mef2C+/− embryos (J) compared to WT (I) at E16.5. The AV bundle is outlined (dashed line). Abreviations: avb, atrioventricular bundle; ra, right atrium; la, left atrium; rv, right ventricle; lv, left ventricle.
HDACs Activity Regulates the Spatial Extent of Enhancer Activity
Having established that Mef2C regulates the intensity of enhancer activity within the nonchamber myocardium, we explored two possible mechanisms that might explain the spatial restriction of enhancer activity to the nonchamber myocardium and AV bundle.
First, we sought to identify factors that might activate R2 in the nonchamber myocardium by searching for conserved TFBS within its most highly conserved segment. Using the MULAN multiple alignment tool and the RVISTA database of transcription factor position weight matrices, we identified several conserved putative TFBS for factors known to regulate development of the AV conduction system, including Gata, Nkx2.5, and Smad proteins. In addition, an E-box consensus site and a highly conserved consensus site for the downstream mediators of hedgehog signaling, Gli, were also identified. Each of these sites was mutated either singly or in tandem within R2R3. Although some alterations in the intensity of enhancer activity were observed with tandem mutation of Smad and Gli sites, and with mutation of the E-box site (Fig. S5), none of the mutants completely abrogated enhancer activity at E11.5 across multiple founders. This finding suggests redundancy in function of these sites or that these sites are not relevant to enhancer activity at E11.5.
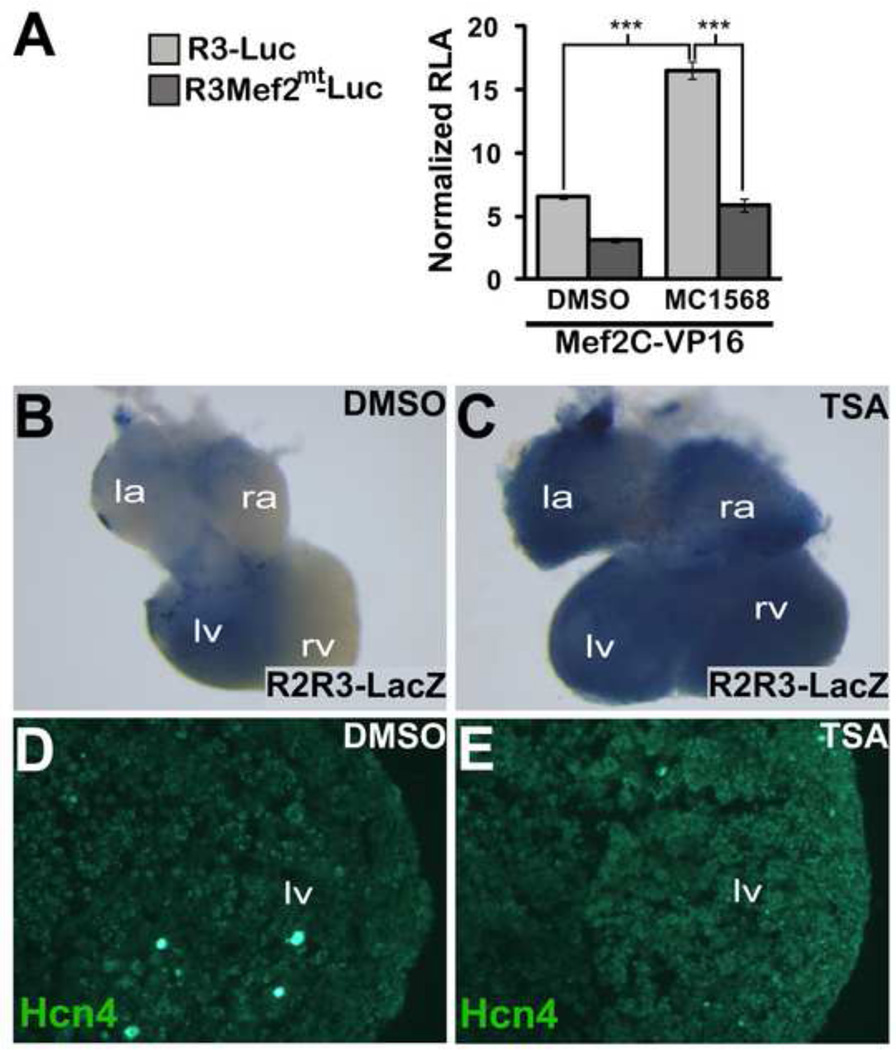
An alternative hypothesis is that domain restriction of R2R3 occurs in part by differential signaling to Mef2 within the non-chamber myocardium. Signal-responsiveness of Mef2-dependent transcriptional activity in cardiomyocytes depends on the activity of Class IIa HDAC proteins(Lu et al., 2000). We used the selective class II HDAC inhibitor MC1568 to test the Mef2 site in R3 for signal responsiveness in vitro using HEK cells. Transactivation of R3 by MEF2C-VP16 was augmented by MC1568, an effect that was abrogated by mutation of the Mef2 site (Fig. 8A). To determine whether differences in HDAC activity among chamber and non-chamber myocytes might contribute to the spatial restriction of R2R3 activity, we cultured R2R3-LacZ embryonic hearts and exposed them to HDAC inhibition. Because of limited solubility of MC1568, we used a less selective HDAC inhibitor with greater solubility, Trichostatin-A (TSA), for organ culture experiments. Embryos were cultured for 48 hours, and then were stained with bluo-gal to assess enhancer activity. We found a striking expansion of enhancer activity in cultured embryonic hearts exposed to TSA as compared to the hearts exposed to DMSO (Fig. 8B,C). To test whether Hcn4 expression was similarly affected, we performed immunohistochemistry for Hcn4 in these hearts, and found increased Hcn4 expression associated with HDAC inhibition (Fig. 8D,E). Taken together, these results demonstrate that HDAC activity is involved in restricting the spatial extent of enhancer activity and Hcn4 expression, and is therefore involved in the patterning of Hcn4 expression in the developing heart.
Figure 8. HDAC Activity Regulates Hcn4 R2R3.
(A) MEF2C-VP16 was co-transfected with WT R3-Luc or MT R3-Luc. The Class II HDAC inhibitor MC1568 or dimethyl sulfoxide (DMSO) vehicle was added after transfection. Relative luciferase activity (RLA) was normalized to the value obtained with co-transfection of WT R3-Luc and empty pcDNA1 in the absence of MC1568. Means were compared with two-tailed t-tests (*, p<0.05; ***, p<0.001) (B,C) Wholemount bluo-gal staining of E11.5 R2R3-LacZ embryonic hearts that were cultured for 48 hours in 10 µM Trichostatin-A (TSA) or DMSO. (D,E). Immunhistochemistry for Hcn4 in cryosections of the left ventricle of TSA or DMSO exposed cultured E11.5 embryonic hearts. R2R3-LacZ embryonic hearts exposed to TSA showed an expansion of enhancer activity and increased ventricular Hcn4 expression.
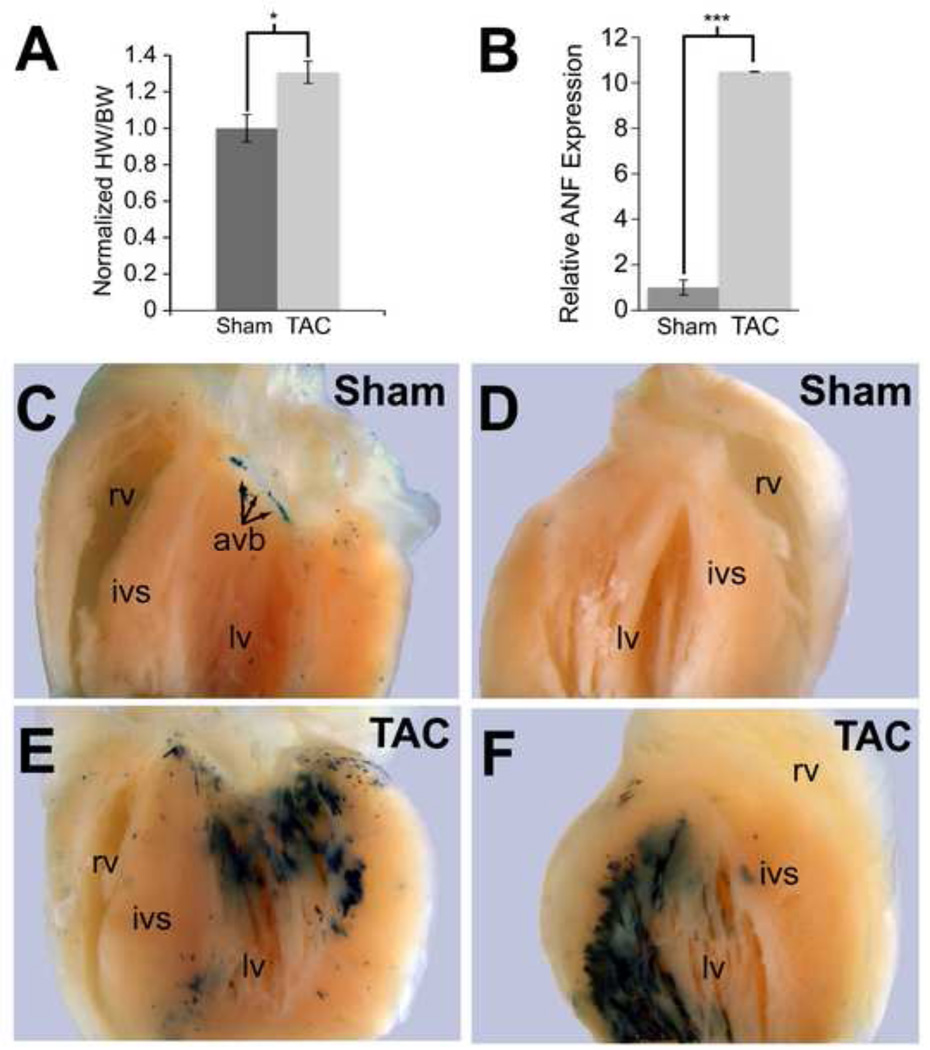
Hcn4-R2R3 Enhancer Is Activated Ectopically in a Model of Cardiac Hypertrophy
In human and mouse hearts, Mef2 activity increases with pathological remodeling, in part because class IIa HDACs are phosphorylated and exported from the nucleus in response to stress. In light of the HDAC responsiveness of R2R3 in-vitro and ex-vivo in organ culture, we hypothesized that the domain of R2R3 enhancer activity might expand outside of the AV bundle in response to stress. To test this hypothesis, six R2R3-LacZ mice were subjected either to transverse aortic constriction (TAC) or a sham operation. Development of cardiac hypertrophy in TAC animals was confirmed by echocardiography 10 days after the procedure (Figs. 9A and S6). Mice were sacrificed, and hearts stained with bluo-gal or processed for RNA preparation and quantitative PCR. Nppa expression increased markedly in TAC hearts, demonstrating activation of stress-responsive gene expression (Fig. 9B). Bluo-gal staining of TAC hearts showed dramatic broadening in areas that were subjected to increased wall stress but not in areas that were relatively protected from pressure overload (Fig. 9C–F). In contrast, enhancer activity in sham-operated animals was confined to the AVB/LNC. Thus, although R2R3 activity is tightly restricted to the AVB/LNC under normal conditions, stress conditions disrupt this fine patterning, providing an explanation for ectopic Hcn4 expression under pathological conditions.
Figure 9. Hcn4 R2R3-LacZ activity expands in response to ventricular pressure overload.
Hcn4 R2R3-LacZ transgenic animals subjected to transverse aortic constriction (TAC) had 30% greater heart weight to body weight (HW/BW) ratios compared to sham-operated animals (A) and had induction of ANF expression as assessed by quantitative PCR on whole ventricle RNA (B). Whole-mount bluo-gal staining showed no expansion of enhancer activity beyond the AVB/LNC in the sham-operated animals (C,D), whereas animals subjected to TAC showed an increase in enhancer activity throughout the subendocardial myocardium of the left ventricle (lv) where pressure load is the greatest (D,E). By comparison, the right ventricle (rv), which is not pressure-loaded in this experiment, did not show any increase in enhancer activity. Each value 32 is expressed as mean with error bars denoting standard error of the mean. Means were compared with two-tailed t-tests (*, p<0.05; ***, p<0.001). Abreviations: ivs, interventricular septum; avb, atrioventricular bundle.
Discussion
We investigated mechanisms that control the distribution of automaticity among cardiomyocytes during development and in response to stress using an in vivo CRE analysis at the Hcn4 locus. We found that a 5.7 kb segment of genomic DNA from Intron 1 of Hcn4 regulates regional expression of Hcn4 in-vivo. Detailed characterization of this enhancer showed expression restricted to embryonic non-chamber myocardium and the postnatal AV bundle. Enhancer activity required the presence of two deeply conserved fragments from within the full-length segment, one of which was sensitive to Mef2c dosage in vivo. Enhancer activity broadened in response to HDAC inhibition in cultured embryos and in response to a hypertrophic stimulus. Our data suggest that one mechanism by which regional restriction of Hcn4 expression to parts of the cardiac conduction system is achieved is through the HDAC-Mef2 system, and that stress signaling may disrupt this pathway, leading to pathological electrical remodeling in the diseased heart.
Hcn4 Expression Is Dynamically Patterned during Development
We used regional expression level of Hcn4 as a marker for relative degree of automaticity. Hcn4 in situ hybridization data conformed to the well-established decreasing gradient of automaticity from sinoatrial to junctional to working myocytes. Recent studies on Hcn4 expression in avians and in humans support deep evolutionary conservation of the extranodal Hcn4 gradient(Sizarov et al., 2011; Vicente-Steijn et al., 2011). Although several studies demonstrate that If is involved in ectopic and junctional automaticity, the roles of Hcn channels in normal functioning of the AV junction and other areas have not been established. Cardiac-specific deletion of Hcn4 is embyronically lethal by E11.5, consistent with it playing a critical role in supporting embryonic cardiac electrical activity. In light of its wide area of expression during development and under stress, Hcn4 might play an important role in regulating electrical properties of working myocytes beyond its established role in supporting nodal automaticity.
Hcn4-R2R3 Activity Is Restricted to Specific Transcriptional Domains
Embryonic non-chamber myocardium includes precursors of the AV conduction system, the valvular annuli, and the interatrial septum. Myocytes in these areas have distinct gene expression profiles compared to embryonic chamber myocardium(Aanhaanen et al., 2010; Bakker et al., 2008b). The T-box transcription factor Tbx3 is specifically expressed in the non-chamber myocardium and regulates specification of the AV conduction system, largely by repressing the regulatory program of the working myocardium(Bakker et al., 2008a; Hoogaars et al., 2007). One paradigm for transcriptional activation in the AV conduction system is that widely expressed transcriptional activators such as Nkx2.5, Tbx5, and Gata4, acting in the presence of repressors of the chamber phenotype such as Tbx3, are sufficient to generate the phenotypic characteristics of the specialized conduction tissue(Bakker et al., 2011). In keeping with such a mechanism, although neither Mef2C expression nor Mef2 activity is confined to the non-chamber regions during development, we found that Mef2C-dependent R2R3 activity is restricted to non-chamber myocardium. R2R3 must, therefore, be under the control of factors or signaling pathways in addition to Mef2C. Mef2 activity may provide an activating signal by binding to R3, while other factors restrict the spatial extent of R2R3 activity, possibly by acting on R2.
The ventricular non-chamber myocardium, located at the crest of the developing interventricular septum, forms the AVB/LNC. R2R3 activity localizes to this tissue throughout development and in the adult heart. In explanted human heart preparations, the junctional rhythm originates at the interface of the AV node with the AVB, the area where we observe the highest intensity of enhancer activity (Fedorov et al., 2011). R2R3 and its upstream regulator Mef2 are therefore determinants of the unique gene expression profile of the junctional pacemaker.
Mef2C Regulates Hcn4 AV Junctional Gene Expression
Mef2C+/− animals have no reported phenotype, while Mef2C knockouts have early embryonic lethality with slow embryonic heart rate(Lin et al., 1997). Transgenic overexpression of a dominant negative Mef2 protein causes a reduction in Hcn4 levels in the embryonic heart (Kuratomi et al., 2009). The Mef2 site within R3 has been identified previously, but its specific in vivo role could not be defined because R3 did not have autonomous enhancer activity(Kuratomi et al., 2009). By combining R3 with other conserved segments, we showed that the Mef2 site has a role in regulating Hcn4 expression within specific domains during development and in the adult heart. Moreover, Mef2C can bind specifically to R3, and R2R3 activity was exquisitely sensitive to Mef2C dosage throughout development.
Mef2 binding sites are widely present on cardiac enhancers and regulate transcription of many cardiac genes. As suggested above, Mef2 might regulate spatially restricted R2R3 activity by interacting with other activators or repressors that are specific to the non-chamber myocardium and AVB/LNC. Alternatively, variations in basal signal strength between the AVB/LNC and working myocardium might be coupled to gene expression through HDAC-dependent Mef2 activity. If a basal signal is present in the AVB/LNC that causes nuclear export of class IIa HDACs, it might increase Mef2 activity enough to permit regionally restricted activity of R2R3.
Multiple Conserved Regions Are Required to Regulate Hcn4 Expression
None of the putative Hcn4 CREs we tested was sufficient to drive reporter expression when tested in isolation. These data comport with other studies of cardiac CREs, which defined a strikingly low rate of in vivo validation (2%) for conservation-based enhancer predictions(Pennacchio et al., 2006; Visel et al., 2008). This finding has been taken as evidence of the importance of weakly conserved CREs in the heart (Blow et al., 2010). An alternative explanation would be a requirement for combinations of conserved regions, which would not be detected using standard methods of in vivo enhancer analysis. Here, we showed that two spatially separated conserved genomic DNA segments, neither of which is sufficient to drive gene expression alone, promote domain-specific expression when combined, even with the intervening sequence removed. Whether this mechanism can be generalized to other instances of fine patterning in the heart remains to be determined.
Although Hcn4 is frequently used as a marker for specialized conduction tissue in studies of conduction system development(Aanhaanen et al., 2010), only a few studies explored transcriptional regulation of Hcn4 during development and in heart disease models (Kuratomi et al., 2007; Lin et al., 2009). Sp1, a ubiquitous transcription factor, is upregulated in cardiomyocytes in response to stress signaling and has many predicted binding sites in the core promoter region of Hcn4 that likely regulate Hcn4 expression (Lin et al., 2009). Although we tested the basal promoter region and found no evidence of autonomous in-vivo transcriptional activity at E11.5, TFBS in this region undoubtedly have important roles in regulating expression both during development and in response to stress. A binding site has also been defined for the neuron restricted silencing factor (NRSF) in intron 1 of Hcn4 (Kuratomi et al., 2007). This site is important for repressing Hcn4 transcription in perinatal ventricular myocardium, and relief of this repression is required for reactivation of Hcn4 expression in hypertrophic myocytes. This NRSF site was contained within R4, a region that did not exhibit autonomous transcriptional activity, consistent with its postulated role as a repressive regulatory element. We did not identify a conserved NRSF site in R2R3. However, the finding that R2R3 is not active in working ventricular myocardium under normal conditions but becomes active after TAC suggests that domain restriction of Hcn4 expression must be achieved through multiple transcriptional mechanisms. With the complexity of the Hcn4 locus, several overlapping and partially redundant mechanisms are likely required to confer robustness on the Hcn4 expression pattern, and by extension, the automaticity gradient.
The AVB/LNC Hcn4 enhancer is signal-responsive
In response to physiological signals and pathological stress, electrical properties of myocytes remodel through mechanisms that are under intensive investigation(Nattel et al., 2010). In animal models of heart disease and in human heart failure, Hcn4 becomes active in hypertrophied and failing ventricular myocytes and may promote arrhythmogenesis(Stillitano et al., 2008). The responsiveness of R2R3 to TAC led us to examine signaling pathways that regulate Mef2 activity in pathological ventricular remodeling. Class IIa HDACs act as transcriptional co-repressors by binding directly to Mef2 and silencing Mef2-dependent transcription(Haberland et al., 2009). Increased wall stress leads to nuclear export of class IIa HDACs by a calcium calmodulin kinase II–dependent pathway(Lu et al., 2000). In both our cell-culture system and in explanted hearts, we found that HDAC inhibition potentiates R2R3 activity, one possible explanation for the broadening of R2R3 activity after TAC. Although we did not directly test for an interaction between HDAC and Mef2 on the R2R3 enhancer and there are other possible mechanisms that might explain our findings, one unifying hypothesis is that activation of Hcn4 expression by Mef2 is regulated by HDAC activity. It will be interesting to determine if the increased activity of R2R3 in the setting of cardiac hypertrophy contributes to arrhythmogenicity by altering the domain of Hcn4 expression, and whether a specific interaction between class IIa HDACs and Mef2 directs this response.
Supplementary Material
3-dimensional reconstruction of an OPT scan of an E11.5 R2R3-LacZ heart. Visual light channel, which encodes the area of bluo-gal staining, is indicated in blue pseudocolor.The autofluorescence channel, in red pseudo-color, shows all cardiac surfaces. Bluo-gal positive areas include the sinoatrial ring, the primary atrial septum, the right AV ring, and the interventricular ring. These are the areas of non-chamber myocardium.
3-dimension reconstruction of an OPT scan of an E15.5 R2R3-LacZ heart. Bluo-gal staining (blue pseudocolor) is visible in the right venous valve, and more intensely in the AV ring and the crest of the interventricular septum. The latter two structures fuse during AV conduction system development to form the AV bundle and its attachment to the AVN.
3-dimensional reconstruction of an OPT scan of a postnatal day 5 R2R3-LacZ heart. Bluo-gal staining (blue pseudocolor) is specific to the AV bundle up to its attachment to the compact AVN. Atrial staining and staining in the valvular annuli are no longer present.
Highlights.
In-vivo transgenic enhancer assay reveals an enhancer for murine Hcn4 in intron 1.
Hcn4 enhancer has two short conserved domains that are required in combination.
Hcn4 enhancer is active in non-chamber myocardium and AV bundle but not SA node.
The Mef2C-HDAC system regulates Hcn4 enhancer and AV bundle Hcn4 expression.
The Hcn4 enhancer is ectopically activated in pathological cardiac hypertrophy.
Acknowledgments
We thank the Gladstone Transgenic Core Facility (S. Espineda, P. Swinton, J. Zhang) and Histology Core Facility (J.D. Fish, C. Miller) for pronuclear injection and histology, respectively; G. Galang for technical assistance; B. Black for providing the MEF2C-VP16 expression construct and Mef2C heterozygous mice. We also thank Brian Black, Benoit Bruneau, Nathalie Gaborit, Ian Harris, and Shan-Shan Zhang for helpful discussions. V.V. is supported by the NIH/NHLBI (K08 HL101989) and the GlaxoSmithKline Cardiovascular Research and Education 23 Foundation. D.S. is supported by grants from NIH/NHLBI, the California Institute of Regenerative Medicine, the Younger Family Foundation, the Eugene Roddenberry Foundation, and the L.K. Whittier Foundation. The J. David Gladstone Institutes received support from a National Center for Research Resources Grant RR18928-01.
Abbreviations
- SA
Sinoatrial
- AV
Atrioventricular
- AVB/LNC
AV bundle and lower nodal cells
- R
Genomic region
- CRE
Cis-regulatory element
- TFBS
Transcription factor binding site
- EMSA
Electromobility shift assay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aanhaanen WT, Mommersteeg MT, Norden J, Wakker V, de Gier-de Vries C, Anderson RH, Kispert A, Moorman AF, Christoffels VM. Developmental origin, growth, and three-dimensional architecture of the atrioventricular conduction axis of the mouse heart. Circ Res. 2010;107:728–736. doi: 10.1161/CIRCRESAHA.110.222992. [DOI] [PubMed] [Google Scholar]
- Arnolds DE, Liu F, Fahrenbach JP, Kim GH, Schillinger KJ, Smemo S, McNally EM, Nobrega MA, Patel VV, Moskowitz IP. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J Clin Invest. 2012;122:2509–2518. doi: 10.1172/JCI62617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker ML, Boukens BJ, Mommersteeg MT, Brons JF, Wakker V, Moorman AF, Christoffels VM. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ Res. 2008a;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- Bakker ML, Boukens BJ, Mommersteeg MTM, Brons JF, Wakker V, Moorman AFM, Christoffels VM. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circulation Research. 2008b;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- Bakker ML, Moorman AF, Christoffels VM. The atrioventricular node: origin, development, and genetic program. Trends Cardiovasc Med. 2011;20:164–171. doi: 10.1016/j.tcm.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Baruscotti M, Bucchi A, Viscomi C, Mandelli G, Consalez G, Gnecchi-Rusconi T, Montano N, Casali KR, Micheloni S, Barbuti A, DiFrancesco D. Deep bradycardia and heart block caused by inducible cardiac-specific knockout of the pacemaker channel gene Hcn4. Proc Natl Acad Sci U S A. 2011;108:1705–1710. doi: 10.1073/pnas.1010122108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Bristow J, Ren B, Black BL, Rubin EM, Visel A, Pennacchio LA. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Burlington, MA: Academic Press; 2006. [Google Scholar]
- DiFrancesco D. The role of the funny current in pacemaker activity. Circ Res. 2010;106:434–446. doi: 10.1161/CIRCRESAHA.109.208041. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Lyons GE, Martin JF, Olson EN. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–1263. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- Fedorov VV, Ambrosi CM, Kostecki G, Hucker WJ, Glukhov AV, Wuskell JP, Loew LM, Moazami N, Efimov IR. Anatomic Localization and Autonomic Modulation of AV Junctional Rhythm in Failing Human Hearts. Circ Arrhythm Electrophysiol. 2011 doi: 10.1161/CIRCEP.111.962258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Frigola C, Shi Y, Evans SM. Expression of the hyperpolarization-activated cyclic nucleotide-gated cation channel HCN4 during mouse heart development. Gene Expr Patterns. 2003;3:777–783. doi: 10.1016/s1567-133x(03)00125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann S, Layh B, Ludwig A. Novel insights into the distribution of cardiac HCN channels: an expression study in the mouse heart. J Mol Cell Cardiol. 2011;51:997–1006. doi: 10.1016/j.yjmcc.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Herrmann S, Stieber J, Stockl G, Hofmann F, Ludwig A. HCN4 provides a 'depolarization reserve' and is not required for heart rate acceleration in mice. EMBO J. 2007;26:4423–4432. doi: 10.1038/sj.emboj.7601868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, Ravesloot JH, Moorman AF, Verheijck EE, Christoffels VM. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21:1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler PM, Roberts-Thomson KC, Haqqani HM, Fynn SP, Singarayar S, Vohra JK, Morton JB, Sparks PB, Kalman JM. P-wave morphology in focal atrial tachycardia: development of an algorithm to predict the anatomic site of origin. Journal of the American College of Cardiology. 2006;48:1010–1017. doi: 10.1016/j.jacc.2006.03.058. [DOI] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- Kuratomi S, Kuratomi A, Kuwahara K, Ishii TM, Nakao K, Saito Y, Takano M. NRSF regulates the developmental and hypertrophic changes of HCN4 transcription in rat cardiac myocytes. Biochem Biophys Res Commun. 2007;353:67–73. doi: 10.1016/j.bbrc.2006.11.119. [DOI] [PubMed] [Google Scholar]
- Kuratomi S, Ohmori Y, Ito M, Shimazaki K, Muramatsu S, Mizukami H, Uosaki H, Yamashita JK, Arai Y, Kuwahara K, Takano M. The cardiac pacemaker-specific channel Hcn4 is a direct transcriptional target of MEF2. Cardiovasc Res. 2009;83:682–687. doi: 10.1093/cvr/cvp171. [DOI] [PubMed] [Google Scholar]
- Lakatta EG, Maltsev VA, Vinogradova TM. A coupled SYSTEM of intracellular Ca2+ clocks and surface membrane voltage clocks controls the timekeeping mechanism of the heart's pacemaker. Circ Res. 2010;106:659–673. doi: 10.1161/CIRCRESAHA.109.206078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Xiao J, Luo X, Chen G, Wang Z. Transcriptional control of pacemaker channel genes HCN2 and HCN4 by Sp1 and implications in re-expression of these genes in hypertrophied myocytes. Cell Physiol Biochem. 2009;23:317–326. doi: 10.1159/000218178. [DOI] [PubMed] [Google Scholar]
- Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, McKinsey TA, Nicol RL, Olson EN. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci U S A. 2000;97:4070–4075. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marionneau C, Couette B, Liu J, Li H, Mangoni ME, Nargeot J, Lei M, Escande D, Demolombe S. Specific pattern of ionic channel gene expression associated with pacemaker activity in the mouse heart. J Physiol. 2005;562:223–234. doi: 10.1113/jphysiol.2004.074047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanesi R, Baruscotti M, Gnecchi-Ruscone T, DiFrancesco D. Familial sinus bradycardia associated with a mutation in the cardiac pacemaker channel. N Engl J Med. 2006;354:151–157. doi: 10.1056/NEJMoa052475. [DOI] [PubMed] [Google Scholar]
- Moosmang S, Stieber J, Zong X, Biel M, Hofmann F, Ludwig A. Cellular expression and functional characterization of four hyperpolarization-activated pacemaker channels in cardiac and neuronal tissues. Eur J Biochem. 2001;268:1646–1652. doi: 10.1046/j.1432-1327.2001.02036.x. [DOI] [PubMed] [Google Scholar]
- Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, Seidman JG, Seidman CE. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- Munshi NV, McAnally J, Bezprozvannaya S, Berry JM, Richardson JA, Hill JA, Olson EN. Cx30.2 enhancer analysis identifies Gata4 as a novel regulator of atrioventricular delay. Development. 2009;136:2665–2674. doi: 10.1242/dev.038562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nattel S, Frelin Y, Gaborit N, Louault C, Demolombe S. Ion-channel mRNA-expression profiling: Insights into cardiac remodeling and arrhythmic substrates. J Mol Cell Cardiol. 2010;48:96–105. doi: 10.1016/j.yjmcc.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Nattel S, Maguy A, Le Bouter S, Yeh YH. Arrhythmogenic ion-channel remodeling in the heart: heart failure, myocardial infarction, and atrial fibrillation. Physiol Rev. 2007;87:425–456. doi: 10.1152/physrev.00014.2006. [DOI] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Roberts-Thomson KC, Kistler PM, Kalman JM. Focal atrial tachycardia I: clinical features, diagnosis, mechanisms, and anatomic location. Pacing Clin Electrophysiol. 2006;29:643–652. doi: 10.1111/j.1540-8159.2006.00413.x. [DOI] [PubMed] [Google Scholar]
- Schram G, Pourrier M, Melnyk P, Nattel S. Differential distribution of cardiac ion channel expression as a basis for regional specialization in electrical function. Circ Res. 2002;90:939–950. doi: 10.1161/01.res.0000018627.89528.6f. [DOI] [PubMed] [Google Scholar]
- Shinohara T, Joung B, Kim D, Maruyama M, Luk HN, Chen PS, Lin SF. Induction of atrial ectopic beats with calcium release inhibition: Local hierarchy of automaticity in the right atrium. Heart Rhythm. 2010;7:110–116. doi: 10.1016/j.hrthm.2009.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizarov A, Devalla HD, Anderson RH, Passier R, Christoffels VM, Moorman AF. Molecular Analysis of the Patterning of the Conduction Tissues in the Developing Human Heart. Circ Arrhythm Electrophysiol. 2011 doi: 10.1161/CIRCEP.111.963421. [DOI] [PubMed] [Google Scholar]
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Stieber J, Herrmann S, Feil S, Loster J, Feil R, Biel M, Hofmann F, Ludwig A. The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc Natl Acad Sci U S A. 2003;100:15235–15240. doi: 10.1073/pnas.2434235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillitano F, Lonardo G, Zicha S, Varro A, Cerbai E, Mugelli A, Nattel S. Molecular basis of funny current (If) in normal and failing human heart. J Mol Cell Cardiol. 2008;45:289–299. doi: 10.1016/j.yjmcc.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Stroud DM, Darrow BJ, Kim SD, Zhang J, Jongbloed MR, Rentschler S, Moskowitz IP, Seidman J, Fishman GI. Complex genomic rearrangement in CCS-LacZ transgenic mice. Genesis. 2007;45:76–82. doi: 10.1002/dvg.20267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnavski O, McMullen JR, Schinke M, Nie Q, Kong S, Izumo S. Mouse cardiac surgery: comprehensive techniques for the generation of mouse models of human diseases and their application for genomic studies. Physiol Genomics. 2004;16:349–360. doi: 10.1152/physiolgenomics.00041.2003. [DOI] [PubMed] [Google Scholar]
- van den Boogaard M, Wong LY, Tessadori F, Bakker ML, Dreizehnter LK, Wakker V, Bezzina CR, t Hoen PA, Bakkers J, Barnett P, Christoffels VM. Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J Clin Invest. 2012;122:2519–2530. doi: 10.1172/JCI62613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Steijn R, Passier R, Wisse LJ, Schalij MJ, Poelmann RE, Gittenberger-de Groot AC, Jongbloed MR. Funny current channel HCN4 delineates the developing cardiac conduction system in chicken heart. Heart Rhythm. 2011 doi: 10.1016/j.hrthm.2011.03.043. [DOI] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S, Burch JB, Fishman GI, Moskowitz IP, Benson DW. Characterization of sinoatrial node in four conduction system marker mice. J Mol Cell Cardiol. 2007;42:946–953. doi: 10.1016/j.yjmcc.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Dobrzynski H, Tellez J, Niwa R, Billeter R, Honjo H, Kodama I, Boyett MR. Extended atrial conduction system characterised by the expression of the HCN4 channel and connexin45. Cardiovasc Res. 2006;72:271–281. doi: 10.1016/j.cardiores.2006.07.026. [DOI] [PubMed] [Google Scholar]
- Yanni J, Boyett MR, Anderson RH, Dobrzynski H. The extent of the specialized atrioventricular ring tissues. Heart Rhythm. 2009;6:672–680. doi: 10.1016/j.hrthm.2009.01.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3-dimensional reconstruction of an OPT scan of an E11.5 R2R3-LacZ heart. Visual light channel, which encodes the area of bluo-gal staining, is indicated in blue pseudocolor.The autofluorescence channel, in red pseudo-color, shows all cardiac surfaces. Bluo-gal positive areas include the sinoatrial ring, the primary atrial septum, the right AV ring, and the interventricular ring. These are the areas of non-chamber myocardium.
3-dimension reconstruction of an OPT scan of an E15.5 R2R3-LacZ heart. Bluo-gal staining (blue pseudocolor) is visible in the right venous valve, and more intensely in the AV ring and the crest of the interventricular septum. The latter two structures fuse during AV conduction system development to form the AV bundle and its attachment to the AVN.
3-dimensional reconstruction of an OPT scan of a postnatal day 5 R2R3-LacZ heart. Bluo-gal staining (blue pseudocolor) is specific to the AV bundle up to its attachment to the compact AVN. Atrial staining and staining in the valvular annuli are no longer present.