An estimated 11% of the human genome consists of Alu elements, which are short interspersed elements (SINEs) that derive from the gene encoding 7SL RNA of the signal recognition particle.1,2 Alu elements are ~300 base pairs and ~1.3 million copies exist per human genome.1,2 As examples, they can be transcribed by RNA polymerase III to generate Alu RNAs or embedded in protein-encoding genes and transcribed by RNA polymerase II as part of pre-mRNAs.3 Alu elements were long thought of as “junk DNA” at best and as “genomic parasites” at worst. However, recent studies have demonstrated that Alu elements can influence gene expression in unexpected and useful ways. As examples, Alu RNAs can repress the RNA polymerase II-mediated transcription of housekeeping genes in response to heat shock; Alu elements implanted into pre-mRNAs can constitute sites of alternative splicing; and inverted Alu elements implanted into pre-mRNAs can form a duplex that is targeted for A-to-I editing, resulting in nuclear retention of the spliced mRNA product.2,3
Remarkably, > 90% of the human genome is thought to be transcribed. While there are surprisingly few (~20,000) protein-encoding genes, there are surprisingly many (estimates range from 3,000 to 23,000) genes that produce long non-coding RNAs (lncRNAs).4,5 Based on their different complexities in different cell types and changes in their expression levels during development, at least some lncRNAs are not merely transcriptional “noise” but, instead, have been shown to modulate gene expression by inducing or repressing gene transcription, regulating the alternative splicing of pre-mRNA, generating endo-siRNA, regulating mRNA translation or affecting protein activity or localization.4,5
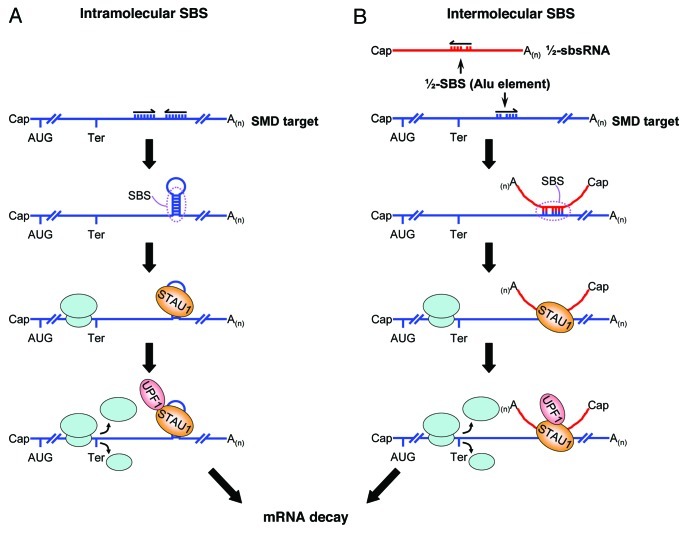
We recently uncovered a new role for Alu elements and lncRNAs: the transactivation of Staufen1 (STAU1)-mediated mRNA decay (SMD).6 STAU1 is a double-stranded (ds) RNA-binding protein. If an mRNA 3′-untranslated region (3′UTR) contains a STAU1-binding site (SBS) sufficiently downstream of its termination codon, then SMD will occur when translation terminates at that codon, because SBS-bound STAU1 can interact with the nonsense-mediated mRNA decay factor UPF1.7,8 Prior to Gong and Maquat (2011), the best-characterized human SBS derived from our studies of ADP-ribosylation factor 1 (ARF1) mRNA. The ARF1 SBS consists of a perfect 19-bp stem, the size of which is critical for STAU1 binding, and a 100-nt apex, which also contributes to STAU1 binding.8 Once we were unable to find a ≥ 19-bp stem in the 3′UTRs of other SMD targets, we noted that ~13% of the 3′UTRs in HeLa-cell mRNAs that are upregulated upon STAU1 downregulation contain an Alu element, whereas only ~4% of all HeLa-cell 3′UTRs contain one or more Alu elements.6 Another computational search, this time for transcripts that, at least in silico, could anneal to 3′UTR Alu elements, identified 378 Alu element-containing lncRNAs. Of the 378, those lncRNAs analyzed proved to be polyadenylated and almost exclusively cytoplasmic. The results of cellular RNP analyses indicated that individual lncRNAs indeed base pair with and trigger the translation-dependent and UPF1-dependent decay of a subset of SMD targets. Moreover, individual SMD targets were found to base pair with and be downregulated by a subset of Alu element-containing lncRNAs. Thus, SBSs can be created not only by intramolecular base pairing within a 3′UTR but also by intermolecular base pairing between ½-SBSs: one ½-SBS consists of the Alu element in an mRNA 3′UTR, and the other ½-SBS consists of a partially complementary Alu element in a lncRNA that we call a ½-sbsRNA (Fig. 1).
Figure 1. Two mechanisms of SBS formation. An SBS that triggers SMD can be formed by (A) intramolecular base pairing within an mRNA 3′UTR or (B) intermolecular base-pairing between a ½-SBS within an mRNA 3′UTR and a partially complementary ½-SBS Alu element within a lncRNA (½-sbs RNA). Those ½-SBSs characterized to date derive from Alu elements. While the features of either intramolecular or intermolecular SBSs have yet to be determined, intermolecular SBSs, which have been found to vary in size from 86 to 264 base pairs (but may have sizes outside of this range), contain multiple bulges, each of which does not exceed two nucleotides. When translation terminates sufficiently upstream of an SBS, so the terminating ribosome does not displace SBS-bound STAU1, the interaction of SBS-bound STAU1 with UPF1 triggers SMD. While not shown, it is conceivable that more than one STAU1 molecule binds to a given SBS in a way that depends on SBS length.
The complicated post-transcriptional gene regulatory network that can now be ascribed to Alu element-containing lncRNAs is reminiscent of the microRNA network, not only in its sequence complexity, but also in its unpredictability. As examples, there appear to be differences in ½-sbsRNA binding-site accessibility among different mRNAs, and there is likely to be competition among distinct ½-sbsRNAs for binding to the same 3′UTR Alu element. These and other parameters undoubtedly contribute to defining distinct cell types, developmental stages and phases of the cell cycle.
While Alu elements are restricted to primates, the newly revealed orangutan genome sequence indicates a slower rate of genomic evolution in orangutans compared with humans as evidenced, in part, by surprisingly fewer Alu elements, orangutans have only ~250 recent insertions compared with ~5,000 in man and ~2,300 in chimpanzees, the latter of which is more closely related to man than the orangutan.9 As a consequence, it is likely that SMD that involves intermolecular Alu element-mediated base pairing contributes to post-transcriptional control in humans more than it does in our primate relatives.
SMD also occurs in non-primate mammals such as the mouse.10 In the mouse, SINEs called B elements exhibit some functional overlap with Alu elements.2,11 However, B1 elements, which, like Alu elements, derive from 7SL RNA, are present at only about 0.5 million copies per mouse genome, and there are ~9-fold fewer 3′UTR B1 elements in the mouse, as there are 3′UTR Alu elements in humans.12 It will be interesting to define B element-containing lncRNAs and determine if any can function as ½-sbsRNAs. Nevertheless, it is clear from what is currently known that Alu elements and the lncRNAs that contain them shape the human genome to be functionally distinct from that of non-primate mammals as well as other primates and, by so doing, contribute to human-specific phenotypes.
lncRNA-based gene silencing is emerging as an important regulator of mammalian cell characteristics. lncRNA-mediated silencing of protein-encoding genes at the transcriptional level can be achieved via pathways that modulate the epigenetic status of chromatin structure.13 Our results reveal a new mechanism of lncRNA-mediated gene silencing that is post-transcriptional. It is conceivable that our findings using Alu element-containing ½-sbsRNAs may extend to human lncRNAs that do not contain a repetitive element.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/15589
References
- 1.Cordaux R, et al. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger A, et al. Prog Mol Subcell Biol. 2011;51:119–46. doi: 10.1007/978-3-642-16502-3_6. [DOI] [PubMed] [Google Scholar]
- 3.Walters RD, et al. IUBMB Life. 2009;61:831–7. doi: 10.1002/iub.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilusz JE, et al. Genes Dev. 2009;23:1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lipovich L, et al. Biochim Biophys Acta. 2010;1799:597–615. doi: 10.1016/j.bbagrm.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Gong C, et al. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim YK, et al. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 8.Kim YK, et al. EMBO J. 2007;26:2670–81. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locke DP, et al. Nature. 2011;469:529–33. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong C, et al. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner SD, et al. Mol Cell Biol. 2010;30:91–7. doi: 10.1128/MCB.01115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sela N, et al. Genome Biol. 2007;8:R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercer TR, et al. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]