Twenty-one human DHHC proteins formed acyl intermediates. Seventeen of the proteins exhibited protein acyltransferase activities. DHHC1, 10, 14, and 16 are novel protein acyltransferases. DHHC proteins are classified into three classes based on substrate specificities.
Abstract
Palmitoylation plays important roles in the regulation of protein localization, stability, and activity. The protein acyltransferases (PATs) have a common DHHC Cys-rich domain. Twenty-three DHHC proteins have been identified in humans. However, it is unclear whether all of these DHHC proteins function as PATs. In addition, their substrate specificities remain largely unknown. Here we develop a useful method to examine substrate specificities of PATs using a yeast expression system with six distinct model substrates. We identify 17 human DHHC proteins as PATs. Moreover, we classify 11 human and 5 yeast DHHC proteins into three classes (I, II, and III), based on the cellular localization of their respective substrates (class I, soluble proteins; class II, integral membrane proteins; class III, lipidated proteins). Our results may provide an important clue for understanding the function of individual DHHC proteins.
INTRODUCTION
Palmitoylation or, more specifically S-acylation, is a posttranslational protein modification that involves the attachment of palmitic acid to Cys residues through a thioester linkage. Protein acyltransferases (PATs), also known as palmitoyltransferases, catalyze this reaction by transferring the palmitoyl group from palmitoyl-CoA to the thiol group of Cys residues. Among the lipid modifications, palmitoylation is the most frequently observed for a wide range of proteins, such as G protein–coupled receptors, ion channels, small GTPases, and Src-family protein kinases (Dunphy and Linder, 1998; Resh, 1999).
Palmitoylation plays diverse physiological functions, especially in nervous and immune systems, through regulating protein activity, localization, and stability (Dunphy and Linder, 1998; Nadolski and Linder, 2007; Greaves and Chamberlain, 2011). Proteomic analyses on yeast, rat cortical embryonic neurons, human platelets, Jurkat T-cells, and prostate cancer DU145 cells have identified several hundred palmitoylated proteins (Roth et al., 2006; Kang et al., 2008; Martin and Cravatt, 2009; Yang et al., 2010; Dowal et al., 2012). Although there are no exact motifs for palmitoylation, in contrast to other lipid modifications, palmitoylated Cys residues are often found clustered at the following six distinct regions of proteins (Resh, 1999; Roth et al., 2006): 1) N-terminal regions of soluble proteins (N substrates), 2) C-terminal regions of soluble proteins (C substrates), 3) internal regions of soluble proteins (I substrates), 4) regions adjacent to other lipid modifications, including N-terminal myristoylation and C-terminal prenylation (L substrates), 5) cytosolic regions adjacent to membrane-spanning domains of single-transmembrane proteins (1TM substrates), and 6) cytosolic regions adjacent to membrane-spanning domains of multiple-transmembrane proteins (MTM substrates).
In 2002, two independent groups identified Akr1 and Erf2 as PATs for the casein kinase Yck2 and the small GTPase Ras2, respectively, using yeast genetics (Lobo et al., 2002; Roth et al., 2002). Both proteins are integral membrane proteins containing a common DHHC Cys-rich domain (DHHC, Asp-His-His-Cys), the signature motif of the PAT active site. The Cys residue within the DHHC domain forms a stable acyl intermediate and transfers the acyl chain to the Cys residue of a target protein (Roth et al., 2002; Mitchell et al., 2010). Many proteins containing the DHHC Cys-rich domain (DHHC proteins) have been identified in eukaryotes, including 7 in yeast (Akr1, Akr2, Erf2, Swf1, Pfa3, Pfa4, and Pfa5), 24 in mice (DHHC1–9, 10 [Zdhhc11; Z11], 11 [Zdhhc23; Z23], 12, 13 [Zdhhc24; Z24], 14–21, 22 [Zdhhc13; Z13], 23 [Zdhhc25; Z25], and 24 [Zdhhc22; Z22]), and 23 in humans (DHHC1–22 and 24; Supplemental Table S1; Fukata et al., 2004; Mitchell et al., 2006; Ohno et al., 2006; Nadolski and Linder, 2007; Greaves and Chamberlain, 2011). Among these DHHC proteins, 6 in yeast (Akr1, Erf2, Swf1, Pfa3, Pfa4, and Pfa5) and 16 in mammals (DHHC2–9, 12, 15, 17–21, and 22 [Z13]) have already been proven to be PATs (Mitchell et al., 2006; Roth et al., 2006; Greaves and Chamberlain, 2011; Lakkaraju et al., 2012). However, it remains unclear whether the rest of the DHHC proteins (yeast Akr2 and mammalian DHHC1, 10 [Z11], 11 [Z23], 13 [Z24], 14, 16, 23 [Z25], and 24 [Z22]) act as PATs.
The human DHHC genes are involved in several disorders, including cancers and neural diseases, such as DHHC2 in colorectal, liver, and non–small cell lung cancers; DHHC8 in schizophrenia; DHHC9 and DHHC15 in mental retardation; DHHC11 (Z23) in bladder cancer; and DHHC17 in Huntington's disease (Greaves and Chamberlain, 2011). In these patients, palmitoylation of certain proteins is suspected to be impaired, yet such proteins are mostly unidentified.
For the yeast DHHC proteins, comprehensive palmitoylation assays combined with mutant analyses have demonstrated distinct substrate specificities, with some overlap—for example, Akr1 for N and C substrates, Erf2 for L substrates, Swf1 for 1TM substrates, and Pfa4 for MTM substrates (Roth et al., 2006). On the other hand, an overall picture of substrate specificity for mammalian DHHC proteins remains poorly understood, even though the number of PATs responsible for some of the mammalian palmitoylated proteins being identified is increasing. This slow progress can be attributed to several technical difficulties. First, any single cell expresses many DHHC proteins and therefore displays high PAT activities. We previously reported that human testis and liver express >19 and 18 DHHC mRNAs, respectively (Ohno et al., 2006). Even a single cell line such as HeLa expresses 20 DHHC mRNAs (Lakkaraju et al., 2012). In general, PAT activities are determined based on the increased amount of palmitoylation products after the overexpression of certain mammalian DHHC proteins (Greaves and Chamberlain, 2011). However, the results are often ambiguous due to the preexisting palmitoylated proteins produced by endogenous PATs. Second, DHHC proteins show significant substrate redundancy (Roth et al., 2006; Hou et al., 2009). In yeast, depletion of six of seven gene products, with two different combinations, still did not eliminate palmitoylation of some substrate proteins (especially L substrates; Roth et al., 2006). Therefore it is likely that more multiple deletions are required to eliminate palmitoylation in mammals, which contain many more DHHC proteins than yeast. However, such multiple-gene knockouts in mammals are unrealistic. Third, in vitro palmitoylation assays are also difficult to perform due to the low activity of DHHC proteins in their solubilized state, the low accessibility between DHHC proteins and substrates (especially membrane protein substrates) in different micelles, and the difficulty in the preparation of substrate proteins without palmitoylation. To address these problems, we expressed human DHHC proteins in PAT-deficient yeast cells, in which the endogenous PAT activities were lowest, and analyzed their activities and substrate specificities. Almost all mammalian DHHC proteins were expressed in yeast and autopalmitoylated as in mammalian cells, suggesting that they were properly folded into their active structures in the yeast environment. The substrate specificity of an enzyme is determined not only by its affinity for a substrate, but also by localization of the enzyme and substrate. Therefore one could argue that differences in protein trafficking between yeast and mammals would interfere with the validity of our study. However, in our preliminary experiments, two well-studied mammalian proteins— postsynaptic scaffold protein PSD-95 and soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) protein SNAP25b—were palmitoylated by subsets of DHHC proteins in a manner essentially identical to that observed in the mammalian system. These results suggest the applicability of the yeast expression system in investigating the substrate specificities of DHHC proteins. Using six distinct classes of model substrates, including PSD-95 and SNAP25b, we identified 17 mammalian DHHC proteins possessing PAT activities, 4 of which (DHHC1, 10 [Z11], 14, and 16) are newly identified PATs. In addition, we categorized the mammalian and yeast DHHC proteins into three classes (I, II, and III).
RESULTS
Seventeen human DHHC proteins form acyl intermediates in mammalian cells
Twenty-four mammalian DHHC proteins are known, and 16 of them have been reported to exhibit PAT activities (Greaves and Chamberlain, 2011; Lakkaraju et al., 2012), whereas the PAT activities of the rest of the DHHC proteins (DHHC1, 10 [Z11], 11 [Z23], 13 [Z24], 14, 16, 23 [Z25], and 24 [Z22]) remain unknown. The DHHC proteins are known to form a stable acyl intermediate (autopalmitoylation) during the course of their catalytic process (Roth et al., 2002; Mitchell et al., 2010). To evaluate the PAT activity of each DHHC protein, we examined the formation of their acyl intermediates. Thus the HEK 293T cells expressing N-terminally His6-Myc–tagged human DHHC proteins were incubated with [3H]palmitic acid, and the resulting radioactive intermediates were detected by autoradiography. We observed the expressions of 19 DHHC proteins (except for DHHC1, 4, 16, and 24) by immunoblotting (Figure 1B), and 17 of them (DHHC2, 3, 5, 6–9, 10 [Z11], 12, 13 [Z24], 14, 15, 17, 18, 20, 21, and 22 [Z13]) were found to form the corresponding acyl intermediates (Figure 1A). Little or no activity was detected for DHHC11 (Z23) and DHHC19 (Figure 1A). The acyl intermediate levels varied among the DHHC proteins, with those of DHHC13 (Z24) and DHHC22 (Z13) being the lowest (Figure 1A). The PAT activities of DHHC10 (Z11), 13 (Z24), and 14 have not been reported until now.
FIGURE 1:
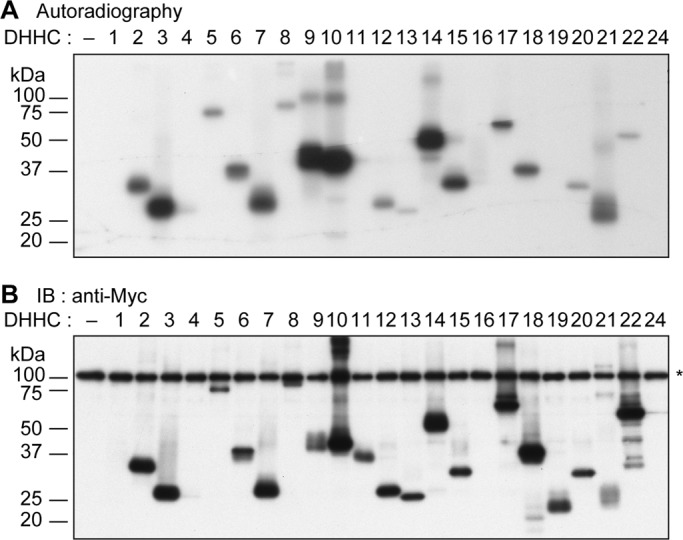
Seventeen human DHHC proteins form acyl intermediates in mammalian cells. (A, B) HEK 293T cells were transfected with the pCE-puro His6-Myc-1 (vector) or pCE-puro His6-Myc-DHHCx (His6-Myc-DHHCx, with x representing the DHHC number, x = 1–22 and 24) plasmid. Twenty-four hours after transfection, the culture medium was changed to serum-free DMEM, and cells were incubated for 1 h at 37°C. The cells were then labeled with 0.2 mCi of [3H]palmitic acid for 2 h at 37°C. Total cell lysates were prepared, and DHHC proteins were immunoprecipitated with anti-Myc antibodies. The precipitates were separated using SDS–PAGE and detected by autoradiography (A) or by immunoblotting with anti-Myc antibodies (B). Asterisk, nonspecific backgrounds.
Development of methods for measuring mammalian DHHC PAT activities using a yeast expression system
Detection of DHHC PAT activities in mammalian cells is often problematic due to the high activity of the endogenous DHHC PATs and the overlapping substrate specificity of DHHC PATs. Yeast is tractable to genetic manipulation and has only 7 DHHC genes, as opposed to 24 in mammals. Therefore the yeast PAT mutants, in which palmitoylation of the substrate protein of interest is kept to a minimum, should provide an ideal system for the detection of the ectopically expressed DHHC proteins. In the present study, human DHHC genes, as well as yeast DHHC genes for comparison, were N-terminally tagged with 3xFLAG and expressed in yeast cells. The immunoblot analysis demonstrated successful expression of the DHHC proteins in yeast, although the expression levels of DHHC1, 8, 22 (Z13), and 24 (Z22) were rather low (Supplemental Figure S1B). Because the N-terminally 3xFLAG-tagged DHHC18 was not expressed in yeast, the C-terminally 3xFLAG-tagged DHHC18 (18C) was used for further studies.
To investigate whether the human DHHC proteins form the acyl intermediates in yeast, we performed the acyl–biotinyl exchange (ABE) assay, in which the palmitate moiety of the palmitoylated proteins is exchanged to biotin for the detection of the palmitoylation site (Wan et al., 2007). Twenty human DHHC proteins (except for DHHC19, 22 [Z13], and 24 [Z22]) were found to form their corresponding acyl intermediates (Supplemental Figure S1A). The expression of DHHC1, 4, 16, and 24 (Z22) was detected in yeast but not in HEK 293T cells (Supplemental Figure S1B). Among them, DHHC1, 4, and 16 formed the acyl intermediates (Supplemental Figure S1A). The overall intermediate-forming activity of the human DHHC proteins in yeast was similar to that in mammalian cells. For example, the activities of DHHC11, 13 (Z24), 19, and 22 (Z13) were low or not detected in either HEK 293T cells or yeast cells (Figure 1A and Supplemental Figure S1A).
In theory, there remains a possibility that the observed palmitoylation of DHHC proteins may not represent the corresponding acyl intermediates (autopalmitoylation) but instead the products from palmitoylation of noncatalytic Cys residues by endogenous DHHC proteins (transpalmitoylation). Accordingly, we created the mutants of six DHHC proteins (DHHC3, 7, 10 [Z11], 14, 16, and 21) by replacing their catalytic Cys residues with Ala (i.e., DHHA mutants) and examined their palmitoylation. The ABE assay showed no or little formation of palmitoylated DHHA mutant proteins, in contrast to their corresponding wild-type DHHC proteins (Supplemental Figure S1C). This finding clearly excludes the possibility of transpalmitoylation, at least for the six DHHC proteins tested here.
The yeast DHHC genes were also cloned for comparison with mammalian genes and subjected to the same analysis. For the formation of the acyl intermediate, Akr1, Pfa3, and Pfa4 exhibited the highest activities, followed by Erf2, with Swf1 and Pfa5 exhibiting the lowest activities (Supplemental Figure S1A). The acyl intermediate of Akr2 was not detected.
Seventeen human DHHC genes suppress temperature-sensitive growth of Δakr1 cells
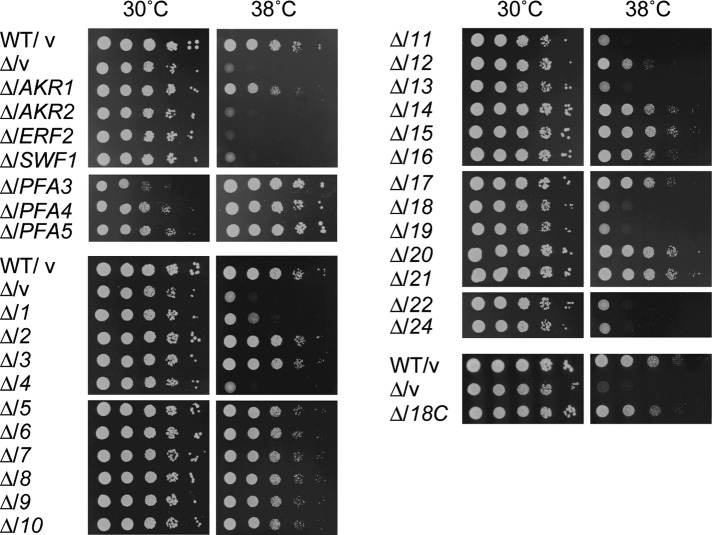
The Δakr1 cells exhibit temperature-sensitive growth (Pryciak and Hartwell, 1996). To evaluate the PAT activity of each DHHC protein, especially toward the substrates of Akr1, we first examined the complementation activity toward the temperature-sensitive growth of the Δakr1 cells. When each DHHC gene–encoding plasmid was introduced into the Δakr1 cells, the yeast Akr1, Pfa3, Pfa4, and Pfa5 and the human DHHC2, 3, 5–9, 10 (Z11), 14–18, 20, and 21 almost completely complemented the growth defect of the Δakr1 cells at 38°C (Figure 2, A and B). DHHC1 and DHHC12 exhibited partial complementation activities (Figure 2A). These results indicated that at least 4 of 7 yeast DHHC genes and 17 of 23 human DHHC genes function as PATs. Thus the complementation analysis has confirmed the newly identified PAT activities of DHHC1, 10 (Z11), 14, and 16.
FIGURE 2:
Seventeen human DHHC proteins restore the temperature-sensitive growth defects of the Δakr1 cells. (A, B) SEY6210 (wild type; WT) cells bearing the pAKNF314 (vector) or pLECF314 (vector) plasmid and KHY1045 (Δakr1) cells harboring the pAKNF314, pAKNF-AKR1 (3xFLAG-AKR1), pAKNF-AKR2 (3xFLAG-AKR2), pAKNF-SWF1 (3xFLAG-SWF1), pAKNF-ERF2 (3xFLAG-ERF2), pAKNF-PFA3 (3xFLAG-PFA3), pAKNF-PFA4 (3xFLAG-PFA4), pAKNF-PFA5 (3xFLAG-PFA5), pAKNF-DHHCx (3xFLAG-DHHCx, with x representing the DHHC number, x = 1–22 and 24), pLECF314, or pLECF-DHHC18 (DHHC18-3xFLAG) plasmid were grown to stationary phase. The cells were serially diluted at 1:10 and spotted onto plates of SC medium lacking tryptophan for 2 d at either 30 or 38°C. Δ, Δakr1; v, vector; number, human DHHC gene.
The yeast expression system is useful for determining substrate specificities of human DHHC proteins
To evaluate the effectiveness of the yeast expression system for determining the substrate specificity of the human DHHC proteins, two known substrate proteins—PSD-95 and SNAP25b—were subjected to the palmitoylation assay. The previous analysis using mammalian cultured cells has revealed that DHHC2, 3, 7, 8, 15, and 17 are PATs for PSD-95 (Fukata et al., 2004; Huang et al., 2004; Mukai et al., 2008). In the present assay, the palmitoylation of PSD-95, expressed in the wild-type yeast cells, was examined by the ABE assay. In agreement with the previous results, the expression of DHHC2, 3, 7, 8, 15, or 17 resulted in the palmitoylation of PSD-95 (Figure 3A). Among them, DHHC7 and DHHC8 exhibited the highest activities. In addition, DHHC5, 6, and 14 exhibited PAT activities toward PSD-95. We speculate that these DHHC proteins were negative in the previous analysis probably due to their low activities. Among the yeast DHHC proteins, Akr1, Pfa4, and Pfa5 were active toward PSD-95 (Figure 3A).
FIGURE 3:
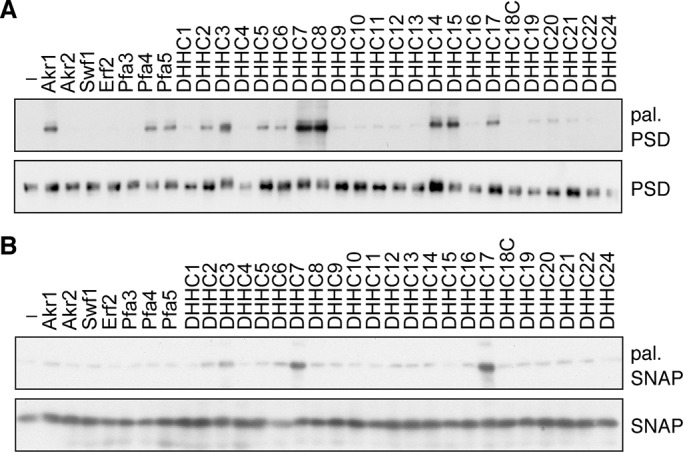
The yeast expression system is useful for determining PATs for PSD-95 and SNAP25b. (A, B) SEY6210 (wild type) cells harboring the pYU-PSD95 (PSD-95-9xMyc; A) or pAC-SNAP25b (9xMyc-SNAP25b; B) plasmid were transfected with the pAKNF314 (vector), pAKNF-AKR1 (3xFLAG-AKR1), pAKNF-AKR2 (3xFLAG-AKR2), pAKNF-SWF1 (3xFLAG-SWF1), pAKNF-ERF2 (3xFLAG-ERF2), pAKNF-PFA3 (3xFLAG-PFA3), pAKNF-PFA4 (3xFLAG-PFA4), pAKNF-PFA5 (3xFLAG-PFA5), pAKNF-DHHCx (3xFLAG-DHHCx, with x representing the DHHC number, x = 1–17, 19–22, and 24), or pLECF-DHHC18 (DHHC18-3xFLAG) plasmid. Total cell lysates were prepared and treated sequentially with NEM, hydroxylamine, and biotin-HPDP. (A) The biotinylated samples were subjected to immunoprecipitation with anti-Myc antibodies and detected either by streptavidin-HRP (top) or by immunoblotting with anti-Myc antibody (bottom). (B) The biotinylated samples were precipitated with immobilized avidin beads and detected by immunoblotting with anti-Myc antibody (top). The DHHC proteins in the total lysates were detected using immunoblotting with anti-Myc antibody (bottom). Each experiment was repeated three times, and representative results are shown. pal., palmitoylated; PSD, PSD-95; SNAP, SNAP25b.
DHHC3, 7, and 17 have previously been reported to be the PATs for SNAP25/23 in mammalian cultured cells (Greaves et al., 2010). As expected, these DHHC proteins also exhibited the PAT activities toward SNAP25b in our yeast assay system (Figure 3B). The other DHHC proteins exhibited little or no activity toward SNAP25b. The results obtained by using the yeast cells are quite compatible with those using the mammalian cells, suggesting the usefulness of our yeast assay system.
Determination of substrate specificities of human DHHC proteins using model substrates
Palmitoylated proteins are mainly categorized into six groups (N, I, C, L, 1TM, and MTM substrates) based on the location of the palmitoylated Cys residue (Resh, 1999; Roth et al., 2006). It has been demonstrated that each yeast DHHC protein exhibits an activity toward a specific group of substrates (Roth et al., 2006), whereas the substrate specificities of the mammalian DHHC proteins are largely unknown. In the present study, we revealed the substrate specificity of the human DHHC proteins using six model substrates belonging to the respective groups. PSD-95 is a known N substrate, in which both Cys-3 and Cys-5 residues undergo palmitoylation (Topinka and Bredt, 1998). SNAP25b, comprising 206 amino acids, belongs to the I substrates, for which the palmitoylation takes place at Cys-85, Cys-88, Cys-90, and Cys-92 (Lane and Liu, 1997). For the last four groups, we used the yeast casein kinase Yck1 (C substrate; Roth et al., 2002), the yeast Gα protein Gpa2 myristrated at the N-terminus (L substrate; Harashima and Heitman, 2005), the yeast SNARE protein Sso1 (1TM substrate; Roth et al., 2006), and the yeast amino acid permease Agp1 (MTM substrate; Roth et al., 2006). We also prepared several yeast strains as hosts to minimize the palmitoylation of the substrates of interest. Among seven yeast DHHC genes, six genes (except the AKR1 gene) can be deleted (Δ6) without causing lethal effects. For each model substrate, we examined the palmitoylation activity in the Δ6, single, double, and triple DHHC gene-deletion mutants and selected suitable cells exhibiting the least palmitoylation activity.
Because the palmitoylation of the C substrate Yck1 was almost eliminated in the Δakr1 cells, as reported previously (Roth et al., 2002, 2006), the yeast and human DHHC genes were each introduced into the Δakr1 cells expressing N-terminally 9xMyc-tagged Yck1. The ABE assay revealed that Akr1 was the most active among the yeast DHHC proteins, and Pfa3, Pfa4, and Pfa5 exhibited weaker activities (Figure 4A). Among the human DHHC proteins, DHHC16 exhibited the highest activity, followed by DHHC7 and DHHC15 (Figure 4A). DHHC2, 3, 5, 6, 8, 9, 10 (Z11), 14, 17, 18, 20, and 21 exhibited weak or moderate activities.
FIGURE 4:
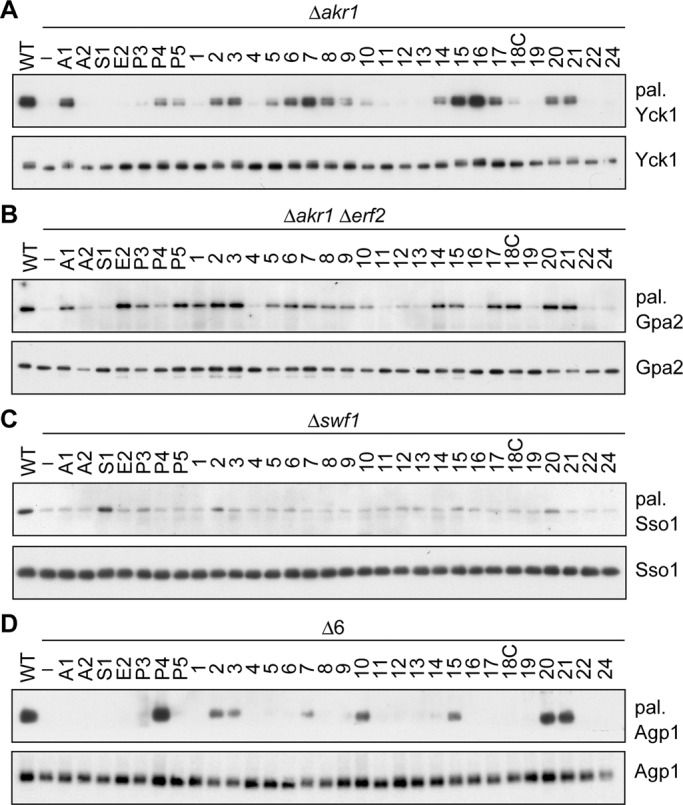
Determination of PATs for Yck1, Gpa2, Sso1, and Agp1. (A–D) The KAY39 (Δakr1 cells expressing N-terminally 9xMyc-tagged Yck1; A), KAY57 (Δakr1 Δerf2 cells expressing C-terminally 9xMyc-tagged Gpa2; B), KAY113 (Δswf1 cells expressing N-terminally 9xMyc-tagged Sso1; C), and IAY106 (Δ6 cells expressing N-terminally 9xMyc-tagged Agp1; D) cells were transfected with the pAKNF314 (vector), pAKNF-AKR1 (3xFLAG-AKR1), pAKNF-AKR2 (3xFLAG-AKR2), pAKNF-SWF1 (3xFLAG-SWF1), pAKNF-ERF2 (3xFLAG-ERF2), pAKNF-PFA3 (3xFLAG-PFA3), pAKNF-PFA4 (3xFLAG-PFA4), pAKNF-PFA5 (3xFLAG-PFA5), pAKNF-DHHCx (3xFLAG-DHHCx, with x representing the DHHC number; x = 1–17, 19–22, and 24), or pLECF-DHHC18 (DHHC18-3xFLAG) plasmid. Total cell lysates were prepared from the transfected cells, as well as the respective wild-type cells (A, KAY62 cells expressing 9xMyc-Yck1; B, KAY47 cells expressing Gpa2-9xMyc; C, KAY110 cells expressing 9xMyc-Sso1; D, KAY44 cells expressing 9xMyc-Agp1), and treated sequentially with NEM, hydroxylamine, and biotin-HPDP. The DHHC proteins were immunoprecipitated with anti-Myc antibodies, and the precipitates were detected by streptavidin-HRP (A, B, and D, top), Ultra Sensitive ABC Peroxidase Staining Kit (C, top), or immunoblotting with anti-Myc antibody (A–D, bottom). Each experiment was repeated three times, and representative results are shown. A1, Akr1; A2, Akr2; S1, Swf1; E2, Erf2; P3, Pfa3; P4, Pfa4; P5, Pfa5, number, human DHHC protein; pal., palmitoylated.
The Δakr1 Δerf2 double mutants were used for the palmitoylation analysis of the L substrate Gpa2 because palmitoylation of Gpa2 was almost eliminated in these mutants. We found that many DHHC proteins (4 yeast and 16 human DHHC proteins) were active toward Gpa2, suggesting a high substrate overlap among the DHHC proteins (Figure 4B). Such redundancy in the substrate specificity toward L substrates among PATs has already been reported (Roth et al., 2002; Hou et al., 2009; Greaves and Chamberlain, 2011). For the yeast DHHC proteins, Erf2 and Pfa5 were highly active, and Akr1 and Pfa3 exhibited moderate activities (Figure 4B). The human DHHC2, 3, 17, 18, 20, and 21 exhibited high activities. The activities of DHHC1, 5–9, 14, and 15 were moderate, and DHHC10 (Z11) and DHHC12 showed weak activities (Figure 4B).
To examine whether the enzyme–substrate specificity is conserved through evolution from yeast to mammals, we investigated mammalian Gαi3 protein, a homologue of Gpa2, as a substrate. Of the 16 mammalian DHHC proteins exhibiting activities toward Gpa2, 13 DHHC proteins (DHHC2, 3, 5–9, 10 [Z11], 14, 15, 17, 20, and 21) were also found to be active toward Gαi3 (Supplemental Figure S2). Therefore the enzyme–substrate specificity seems to be highly conserved between yeast and human in most, but not all, of the DHHC proteins.
The Δswf1 cells exhibited the least palmitoylation activity toward the 1TM substrate Sso1 among the yeast mutants tested and thus used for the PAT analysis. Unlike Gpa2, the palmitoylation of Sso1 was highly limited. Swf1 exhibited a high activity, as reported previously (Roth et al., 2006; Figure 4C). DHHC2, DHHC20, and Pfa3 showed weak activities (Figure 4C).
The Δ6 cells, in which all genes (except for the AKR1 gene) were deleted, were used for the palmitoylation assay of the MTM substrate Agp1. Yeast Pfa4 exhibited a high activity, as reported (Roth et al., 2006; Figure 4D). Among human DHHC proteins, DHHC20 and DHHC21 were most active, followed by DHHC2, 3, 7, 10 (Z11), and 15 (Figure 4D).
DISCUSSION
Since the identification of two different DHHC proteins, Akr1 and Erf2, as PATs in 2002 (Lobo et al., 2002; Roth et al., 2002), the PAT function of the DHHC proteins has been the focus of numerous investigations. The substrate specificity of the yeast DHHC proteins has already been identified by a comprehensive palmitoylation assay (Roth et al., 2006). However, the substrate specificity of the human DHHC proteins remains poorly understood. In addition, it is still not clear whether all the human DHHC proteins are PATs. The functional analysis of the mammalian DHHC proteins has been problematic due to the occurrence of numerous DHHC proteins in mammals and to some inherent technical problems. In the present study, we developed a yeast expression system for the PAT analysis and demonstrated its effectiveness for the investigation of the substrate specificity of each DHHC protein, as well as for the determination of the respective PATs for particular substrate proteins. In our assay system, the mammalian DHHC proteins expressed ectopically in yeast exhibited activities similar to those in the original mammalian cells with respect to both acyl intermediate–forming activity (Figure 1 and Supplemental Figure S1) and PAT activity (e.g., toward PSD-95 and SNAP25b; Figure 3, A and B).
In the present study, we detected the acyl intermediates of 21 human DHHC proteins (except for DHHC22 [Z13] and DHHC24 [Z22]), namely, 17 in mammalian cells and additional 3 in yeast cells (Figure 1 and Supplemental Figure S1). In addition, we identified the PAT activity of 17 human DHHC proteins, except for DHHC4, 11 (Z23), 13 (Z24), 19, 22 (Z13), and 24 (Z22), in the PAT assay using six model substrates, including PSD-95 (N substrate), SNAP25b (I substrate), Yck1 (C substrate), Gpa2 (L substrate), Sso1 (1TM substrate), and Agp1 (MTM substrate; Figures 3 and 4). On the basis of these results, we determined for the first time that the human DHHC1, 10 (Z11), 14, and 16 function as PATs. Their PAT activity was strongly substantiated by growth complementation analysis of Δakr1 cells (Figure 2). Among the yeast DHHC proteins, Akr2 was negative in all the assay systems examined. To our knowledge, the PAT activity of Akr2 has not been reported in the literature. It seems that Akr2 may negatively regulate the palmitoylation of the highly homologous Akr1 by interacting with the common substrates. Thus Akr1 is a PAT for the yeast long-chain base kinase Lcb4, whereas Akr2 binds to Lcb4 without palmitoylation and delivers it to the vacuole for degradation (Kihara et al., 2005).
Some considerations are needed for the human DHHC proteins that did not exhibit the PAT activity in our assay. First, it is possible that some of these DHHC proteins are indeed not PATs, but, like Akr2, they could possess some regulatory functions in relation to other DHHC proteins. Second, the negative assay results could be derived from their weak activities or their low expression levels. Third, cofactors or regulatory proteins essential for their PAT activities might be absent in yeast. Finally, these DHHC proteins might be PATs highly specific to the substrates other than the six model substrates. In particular, any of the last three possibilities could explain the negative results for DHHC4, 19, and 22 (Z13), since their PAT activities have been reported previously. DHHC19 is described as a highly active PAT toward phosphodiesterase 10A (Charych et al., 2010), and DHHC4 is reported to exhibit a weak activity toward the 1TM substrate BACE1 (Vetrivel et al., 2009). Mutations in the mouse Dhhc22 (Z13) gene cause alopecia, osteoporosis, and systemic amyloidosis in mice (Saleem et al., 2010). Recently the huntingtin protein has been suspected to be the substrate of DHHC22 (Z13) based on the observation that the palmitoylation of huntingtin was reduced in the mutant mice (Saleem et al., 2010).
The acyl intermediate levels varied among DHHC proteins. Various factors could affect the intermediate level, including the enzyme activity, the stability of the acyl intermediate, the amount of the intracellular substrate, and the cellular localization of the DHHC protein. It is noted, however, that DHHC4, 11 (Z23), 13 (Z24), 19, 22 (Z13), and 24 (Z22), which did not exhibit PAT activities, formed few or no intermediates (Figure 1 and Supplemental Figure S1). This finding seems to indicate that there is some correlation between PAT activity (Figures 3 and 4) and acyl intermediate level (Figure 1 and Supplemental Figure S1) but that the relationship is not strict.
Membrane localization is necessary for the activity of the yeast casein kinase Yck1 and Yck2, which are both substrates of Akr1. The temperature-sensitive phenotype of the Δakr1 cells is attributable to the dysfunction of Yck1 and Yck2 caused by their mislocalization to the cytosol (Roth et al., 2006). In the growth complementation assay, DHHC1 and DHHC12, whose PAT activities toward Yck1 were close to the detection limit (Figure 4A), partially complemented the temperature sensitivity of the Δakr1 cells (Figure 2). On the other hand, the PAT activities of Akr1, Pfa3, Pfa4, Pfa5, and DHHC2, 3, 5–9, 10 (Z11), 14–18, 20, and 21 toward Yck1 were detectable (Figure 4A), and these DHHC proteins almost fully complemented the growth of the Δakr1 cells at the restrictive temperature (Figure 2). Given the very weak activity of Pfa3, DHHC10 (Z11), and DHHC18, the palmitoylation level required for Yck1/Yck2 to complement the growth of the Δakr1 cells must be low, and therefore a small amount of palmitoylated Yck1/Yck2 may be sufficient to exert their function at high temperature.
Several yeast DHHC proteins have been shown to display a broad substrate specificity when overexpressed (Hou et al., 2009), which could raise a concern as to the validity of overexpression approaches to study substrate specificity of DHHC proteins. In this regard, it is worth noting that in our assay system, in which substrate proteins were also overexpressed under the strong TEF promoter in addition to DHHC proteins, Akr1, the true PAT for Yck1 (Roth et al., 2006), was indeed the most active of the yeast DHHC proteins (Figure 4A). Our system seems to provide the enzyme–substrate balance suitable for the specificity study.
Accordingly, in the present study, we also determined the substrate specificity of human and yeast DHHC proteins toward the six model substrates (Figures 3 and 4). Because the expression level of DHHC proteins varied, it is difficult to compare their PAT activities toward a certain substrate using the yeast expression system. It is also conceivable that in yeast some human DHHC proteins may not be able to display their full activities as a result of their ectopic expression. Differences in the intracellular localization of DHHC proteins and their substrates can also affect the apparent PAT activity. However, comparison of the activity of a certain DHHC protein toward different substrates is possible because the expression level of an individual DHHC protein was nearly identical, irrespective of the host yeast strains used. Furthermore, all the PAT activities were measured using the same conditions. Our results showed a wide range of substrate specificities among the DHHC proteins. For example, DHHC1, 16, and 18, Swf1, and Erf2 were highly specific toward only one model substrate, whereas DHHC2, 3, 7, and 15 were active toward several substrates (Figures 3 and 4). A number of substrates for DHHC2, 3, and 15 have also been reported in the literature (Greaves and Chamberlain, 2011). The highly specific DHHC proteins DHHC1 and 18 were active toward the L substrate Gpa2 (Figure 4B) but not toward Gαi3, a mammalian homologue of Gpa2 (Supplemental Figure S2), suggesting that these DHHC proteins recognize not only the lipidated regions, but also other structural features in the substrates.
We then classified 5 yeast and 11 mammalian DHHC proteins exhibiting substantial activities toward one of the six model substrates into three major classes (I, II, and III), based on the intracellular localization of their substrates (Figure 5). The class I DHHC proteins (Akr1 and DHHC3, 7, 8, and 14–17) exhibited high activities toward the soluble substrates (N, I, and C substrates), whereas the class II DHHC proteins (Swf1, Pfa4, and DHHC2, 10 [Z11], 20, and 21) were highly active toward the integral membrane substrates (1TM and MTM substrates). The class III DHHC proteins (Erf2 and Pfa5) palmitoylated only the lipidated substrate (L substrate). Many of the class I and II DHHC proteins also exhibited activities toward the L substrates. Such substrate redundancy toward L substrates was reported previously (Roth et al., 2006; Hou et al., 2009; Greaves and Chamberlain, 2011). For example, the palmitoylation of the yeast prenylated protein Ras2 could not be eliminated even by two different sextuple DHHC mutations (Roth et al., 2006). In addition, several yeast DHHC proteins, when overexpressed, appear to cause palmitoylation of the myristoylated protein Vac8 (Hou et al., 2009). Multiple DHHC proteins have also been identified as PATs for the mammalian myristoylated proteins eNOS, Fyn, and Lck (L substrates; Greaves and Chamberlain, 2011). The L substrates may display an intermediate property of soluble and membrane proteins due to a possible weak association with membranes through a single lipid modification (myristoylation or prenylation). We did not assign 1, 5, 6, 9, 12, and 18 and Pfa3 to any of the classes because they exhibited low activities in our assay system. We speculate that these DHHC proteins could be highly active PATs for some substrates other than those used in our assay. A recent study indicated that DHHC6 is highly specific to the endoplasmic reticulum chaperone calnexin (Lakkaraju et al., 2012).
FIGURE 5:
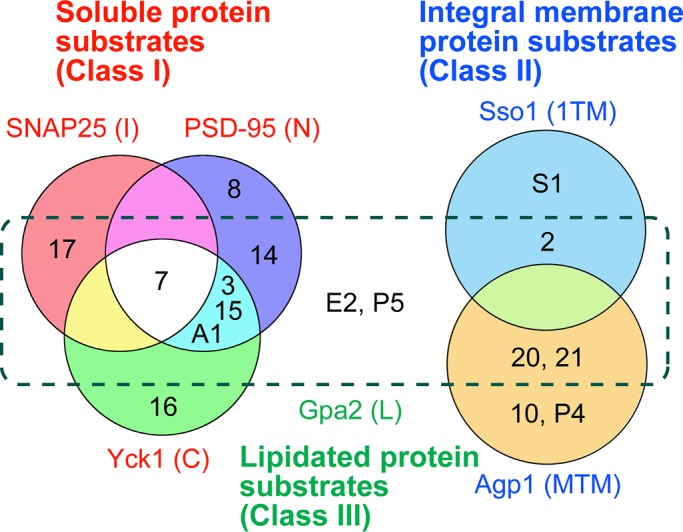
Classification of yeast and human DHHC proteins based on the substrate specificities. Each number corresponds to a human DHHC (e.g., 2 and 3 refer to DHHC2 and DHHC3, respectively). A1, Akr1; S1, Swf1; E2, Erf2; P4, Pfa4; P5, Pfa5. Class I DHHC proteins (Akr1, DHHC3, 7, 8, and 14–17) preferentially palmitoylate soluble proteins. Class II DHHC proteins (Swf1, Pfa4, and DHHC2, 10 [Z11], 20, and 21) exhibit high PAT activities toward integral membrane proteins. Class III DHHC proteins (Erf2 and Pfa5) are specifically active toward lipidated proteins.
The yeast expression system used in this study is a very promising system to identify or at least narrow down yet-unresolved PATs for hundreds of palmitoylated proteins, although the results must be confirmed in mammalian systems using overexpression and/or knockdown techniques. Several DHHC genes have been implicated in the pathology of human disease, including neural diseases and cancers (Greaves and Chamberlain, 2011). However, the disease-causing substrate proteins and mechanism are mostly unknown. The substrate specificities of DHHC PATs revealed in this study may help to identify such substrate proteins.
MATERIALS AND METHODS
Cell culture and transfection
Human embryonic kidney (HEK) 293T cells were grown in DMEM (Sigma-Aldrich, St. Louis, MO) containing 10% fetal calf serum and supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin using 0.3% collagen-coated dishes. Transfections were performed using Lipofectamine Plus Reagent (Invitrogen, Carlsbad, CA).
Yeast strains and media
The yeast Saccharomyces cerevisiae strains used in this study are listed in Supplemental Table S2. All yeast cells used were derived from SEY6210 or its isogenic SEY6211 cells. Cells were grown in either YPD medium (1% yeast extract, 2% bactopeptone, and 2% d-glucose) or synthetic complete medium (SC; 0.67% yeast nitrogen and 2% d-glucose) containing nutritional supplements.
Gene disruption was performed by homologous recombination. The Δakr2::KanMX4, Δswf1::KanMX4, Δpfa3::KanMX4, and Δpfa5::KanMX4 deletion mutations were created by replacing each entire gene with the drug-resistance marker KanMX4. The Δakr1::HIS3, Δerf2::LEU2, and Δpfa4::LEU2 cells were constructed by replacing the 1.2-kb MscI–AvrII region in the AKR1 gene, the 0.7-kb XbaI–NruI region in the ERF2 gene, and the 0.8-kb MscI–MscI region in the PFA4 gene, respectively, with the indicated markers. The RHY59 (Δ6) cells were constructed by repeating mating and spore formation using SEY6210 (MATα)– and SEY6211 (MATa)–derived DHHC gene-deletion mutants.
The Δakr1 cells exhibit severe growth defect, which was attributed to the dysfunction of the casein kinases Yck1 and Yck2 caused by their mislocalization from the plasma membrane to the cytosol (Roth et al., 2006). To eliminate the secondary effect of the slow growth, we constructed Δakr1 cells expressing Yck2-K-RasB, which is localized to the plasma membrane regardless of its palmitoylation status because of the membrane-localization sequence of human K-RasB (GKKKKKKSKTKCVIM) at the C-terminus of Yck2. The KHY1028 (SEY6210, Δyck2::YCK2-K-RASB URA3) cells, in which chromosomal YCK2 gene is replaced with YCK2-K-RASB, was constructed as follows. The YCK2-K-RASB fragment was amplified by PCR using yeast genomic DNA and primers YCK2-F1 and YCK2-R1 and cloned into the pGEM-T Easy vector (Promega, Madison, WI), producing the pAK981 plasmid. The pAK985 (YCK2-K-RASB + YCK2 3′-untranslated region [UTR]) plasmid was constructed by cloning the YCK2 3′-UTR, which had been amplified by PCR using yeast genomic DNA and primers YCK2-F2 and YCK2-R2, into the pAK981 plasmid. The URA3 marker was then inserted into the pAK985 plasmid, generating the pAK990 (YCK2-K-RASB + URA3 + YCK2 3′-UTR) plasmid. The SEY6210 cells were transfected with the YCK2-K-RASB + URA3 + YCK2 3′-UTR fragment from pAK990 and subjected to the selection on an SC-lacking-uracil plate. Among transformants, KHY1028 cells, in which chromosomal YCK2 was properly replaced with YCK2-K-RASB, were selected. The URA3 marker was then removed from the KHY1028 cells by transfection with the YCK2-K-RASB + YCK2 3′-UTR fragment from the pAK985 plasmid, followed by 5-fluoroorotic acid selection, generating KHY1031 (SEY6210, Δyck2::YCK2-K-RASB) cells. The KHY1046 (SEY6210, Δakr1::HIS3 Δyck2::YCK2-K-RASB) cells were constructed by introducing the Δakr1::HIS3 mutation into KHY1031 cells.
The chromosomal YCK1, AGP1, and SSO1 genes were tagged with 9xMyc to express N-terminally 9xMyc-tagged proteins under the control of TEF promoter, generating KAY62 (pTEF-9xMYC-YCK1), KAY39 (Δakr1 pTEF-9xMYC-YCK1), KAY44 (pTEF-9xMYC-AGP1), IAY106 (Δ6 pTEF-9xMYC-AGP1), KAY110 (pTEF-9xMYC-SSO1), and KAY113 (Δswf1 pTEF-9xMYC-SSO1) cells, respectively. Similarly, the GPA2 gene was tagged with 9xMyc to express C-terminally 9xMyc-tagged proteins under the control of TEF promoter, creating KAY47 (pTEF-GPA2-9xMYC) and KAY57 (Δakr1 Δerf2 pTEF-GPA2-9xMYC) cells. Replacement of their own promoters with the TEF promoter and tagging of the chromosomal genes were performed as described previously (Janke et al., 2004) using primer sets (YCK1, YCK1-F and YCK1-R; GPA2, GPA2-F1, GPA2-R1, GPA2-F2, and GPA2-R2; AGP1, AGP1-F and AGP1-R; and SSO1, SSO1-F and SSO1-R; Supplemental Table S3).
Plasmids
The pCE-puro hexahistidine (His6)-Myc-1 plasmid is a mammalian expression vector designed to express an N-terminal, tandemly oriented (His6-Myc)-tagged protein. The pCE-puro His6-Myc-DHHCx plasmid (with x representing the DHHC number, x = 1–22) is a derivative of the pCE-puro His6-Myc-1 vector and encodes each human DHHC gene (Ohno et al., 2006). The human DHHC24 (Z22) cDNA was amplified by PCR using human brain cDNA (Clontech, TAKARA Bio, Shiga, Japan) and primers DHHC24-F and DHHC24-R (Supplemental Table S3) and cloned into the pCE-puro His6-Myc-1 vector, producing the pCE-puro His6-Myc-DHHC24 plasmid. The pAKNF314 (CEN, TRP1 marker) and pLECF314 (CEN, TRP1 marker) plasmids are yeast expression vectors designed to produce N-terminally and C-terminally 3xFLAG-tagged proteins, respectively, under the control of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) promoter. The pAKNF-DHHCx plasmid was constructed by cloning each DHHC gene from the pCE-puro His6-Myc-DHHCx plasmid into the pAKNF314 vector. The yeast AKR1, AKR2, SWF1, ERF2, PFA3, PFA4, and PFA5 genes were amplified by PCR using yeast genomes and primer sets (AKR1, AKR1-F and AKR1-R; AKR2, AKR2-F and AKR2-R; SWF1, SWF1-F and SWF1-R; ERF2, ERF2-F and ERF2-R; PFA3, PFA3-F and PFA3-R; PFA4, PFA4-F and PFA4-R; and PFA5, PFA5-F and PFA5-R; Supplemental Table S3) and cloned into the pAKNF314 vector, producing the pAKNF-AKR1, pAKNF-AKR2, pAKNF-SWF1, pAKNF-ERF2, pAKNF-PFA3, pAKNF-PFA4, and pAKNF-PFA5 plasmids, respectively. The human DHHC18 gene was amplified from the pCE-puro His6-Myc-DHHC18 plasmid using primers DHHC18C-F and DHHC18C-R and cloned into the pLECF314 vector, creating the pLECF-DHHC18 plasmid.
The pAC27 (2 μ, URA3 marker) and pYU248 (2 μ, URA3 marker) plasmids are yeast expression vectors designed to produce N-terminally and C-terminally 9xMyc-tagged protein, respectively, under the control of TEF promoter. The human PSD-95, SNAP25b, and Gαi3 genes were amplified by PCR using human brain (PSD-95 and SNAP25b) and kidney (Gαi3) cDNAs (Clontech, TAKARA Bio) and primers (PSD-95, PSD95-F and PSD95-R; SNAP25b, SNAP25b-F and SNAP25b-R; and Gαi3, Gαi3-F and Gαi3-R). The amplified PSD-95, SNAP25b, and Gαi3 fragments were cloned into the pYU248 (PSD-95 and Gαi3) and pAC27 (SNAP25b) vectors, generating the pYU-PSD95, pAC-SNAP25b, and pYU-Gαi3 plasmids, respectively.
Immunoblotting
Immunoblotting was performed as described previously (Kihara et al., 2003) using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, Waltham, MA). Anti-Myc PL14 antibodies (1 μg/ml; Medical and Biological Laboratories, Nagoya, Japan), anti-Myc 9E10 antibodies (1 μg/ml; Enzo Life Sciences, Farmingdale, NY), and anti-FLAG M2 antibodies (1 μg/ml; Stratagene, Agilent Technologies, La Jolla, CA) were used as primary antibodies. Peroxidase-conjugated anti-mouse immunoglobulin G F(ab′)2 fragments (diluted 1:7500 or 1:15,000; GE Healthcare Life Sciences, Piscataway, NJ) were used as secondary antibodies.
Detection of palmitoylation
[3H]Palmitic acid labeling was performed as described previously (Kihara et al., 2006). ABE assays were performed as described elsewhere (Roth et al., 2006; Wan et al., 2007). Yeast cells equivalent to 10 A600 were collected by centrifugation, washed with water, and suspended in 100 μl lysis buffer (LB; 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 5 mM EDTA) containing 10 mM NEM (Sigma-Aldrich), 2× protease inhibitor mixture (PI; Complete EDTA free; Roche Diagnostics, Indianapolis, IN), and 2 mM phenylmethylsulfonyl fluoride (PMSF). An equal volume of glass beads was added to the cell suspension and mixed vigorously by vortexing for 10 min at 4°C. After centrifugation, a 50-μl supernatant was recovered. An additional 75 μl of LB was added to the glass beads/cell debris fraction and mixed vigorously one more time. After centrifugation, a 74.5-μl supernatant was recovered and combined with the previous supernatant. The resulting total lysates were treated with 25.5 μl of 10% Triton X-100 for 1 h at 4°C with rotation. The solubilized proteins were then recovered as the supernatant after centrifugation (20,400 × g, 4°C, 3 min) and precipitated by the chloroform–methanol method (Wessel and Flugge, 1984). To block the free Cys residues, the protein precipitates were dissolved in 30 μl of 4% SDS buffer (4SB; 50 mM Tris-HCl, pH 7.4, 5 mM EDTA, and 4% SDS) containing 50 mM NEM, and the solution was incubated for 10 min at 37°C. After dilution with 90 μl of LB containing 0.2% Triton X-100, 1× PI, and 1 mM PMSF, the mixture was incubated overnight at 4°C with rotation. Proteins were precipitated by chloroform-methanol (×2) and suspended in 30 μl of 4SB buffer. The mixture was subsequently incubated with 90 μl of hydroxylamine/biotin buffer (H/B; 0.7 M hydroxylamine, pH 7.4, 0.2% Triton X-100, 1× PI, 1 mM PMSF, and 4 mM EZ-Link Biotin-HPDP; Thermo Fisher Scientific) for 1 h at room temperature with rotation. In this process, the palmitoyl thioester linkage was hydrolyzed by hydroxylamine and the exposed free Cys residue was cross-linked with biotin. After protein precipitation by chloroform–methanol (×2), the precipitates were dissolved in 30 μl of 2% SDS buffer (2SB; 50 mM Tris-HCl, pH 7.4, 5 mM EDTA, and 2% SDS), and the solution was subjected to immunoprecipitation with anti-Myc antibodies. Thus a 25-μl aliquot was diluted with 475 μl of LB containing 1× PI and 0.2% Triton X-100, and to this mixture was added 0.25 μl of anti-Myc MC045 antibodies (Nacalai Tesque, Kyoto, Japan) and 5 μl of protein G–Sepharose beads (GE Healthcare Life Sciences). After incubation overnight at 4°C with rotation, the beads were washed with 1 ml of LB containing 0.2% Triton X-100 (×2), and the bound proteins were eluted with 2× SDS sample buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, and a trace amount of bromophenol blue). The proteins were separated by SDS–PAGE, transferred to the Immobilon polyvinylidene difluoride membrane (Millipore, Billerica, MA), and subjected to either immunoblotting or biotin affinity blotting. For biotin affinity blotting, the membrane was blocked with 0.5% gelatin in TBST (20 mM Tris-HCl, pH 7.5, 137 mM NaCl, and 0.05% Tween 20) for 1 h at room temperature and incubated with streptavidin–horseradish peroxidase (HRP; Life Technologies, Carlsbad, CA) diluted in Can Get Signal solution 1 (TOYOBO, Osaka, Japan) for 1 h at room temperature. After washing with TBST three times, the proteins were detected by using Western blotting using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific). Ultra Sensitive ABC Peroxidase Staining Kit (Thermo Fisher Scientific) was also used to carry out the ABC staining method following the manufacturer's instructions. For the analysis of SNAP25b palmitoylation, because the immunoprecipitation efficiency toward anti-Myc antibodies was significantly low, the biotinylated 9xMyc-SNAP25b was precipitated using Immobilized Avidin (Thermo Fisher Scientific) before immunoblotting with anti-Myc antibodies.
Supplementary Material
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (B) (23370057) from the Japan Society for the Promotion of Science. We are grateful to Y. Hiraga for the yeast strains used in this study. We also thank T. Toyokuni for scientific editing of the manuscript.
Glossary
Abbreviations used:
- ABE
acyl-biotinyl exchange
- HEK
human embryonic kidney
- His6
hexahistidine
- HRP
horseradish peroxidase
- LB
lysis buffer
- PAT
protein acyltransferase
- PI
protease inhibitor
- PMSF
phenylmethylsulfonyl fluoride
- SC
synthetic complete
- SNARE
soluble N-ethylmaleimide–sensitive factor attachment protein receptor
- UTR
untranslated region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-05-0336) on October 3, 2012.
*These authors contributed equally to the manuscript.
The authors declare no financial conflicts of interest.
REFERENCES
- Charych EI, Jiang LX, Lo F, Sullivan K, Brandon NJ. Interplay of palmitoylation and phosphorylation in the trafficking and localization of phosphodiesterase 10A: implications for the treatment of schizophrenia. J Neurosci. 2010;30:9027–9037. doi: 10.1523/JNEUROSCI.1635-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowal L, Yang W, Freeman MR, Steen H, Flaumenhaft R. Proteomic analysis of palmitoylated platelet proteins. Blood. 2012;118:e62–e73. doi: 10.1182/blood-2011-05-353078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy JT, Linder ME. Signalling functions of protein palmitoylation. Biochim Biophys Acta. 1998;1436:245–261. doi: 10.1016/s0005-2760(98)00130-1. [DOI] [PubMed] [Google Scholar]
- Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Greaves J, Chamberlain LH. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem Sci. 2011;36:245–253. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Greaves J, Gorleku OA, Salaun C, Chamberlain LH. Palmitoylation of the SNAP25 protein family: specificity and regulation by DHHC palmitoyl transferases. J Biol Chem. 2010;285:24629–24638. doi: 10.1074/jbc.M110.119289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima T, Heitman J. Gα subunit Gpa2 recruits kelch repeat subunits that inhibit receptor-G protein coupling during cAMP-induced dimorphic transitions in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:4557–4571. doi: 10.1091/mbc.E05-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, John Peter AT, Meiringer C, Subramanian K, Ungermann C. Analysis of DHHC acyltransferases implies overlapping substrate specificity and a two-step reaction mechanism. Traffic. 2009;10:1061–1073. doi: 10.1111/j.1600-0854.2009.00925.x. [DOI] [PubMed] [Google Scholar]
- Huang K, et al. Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron. 2004;44:977–986. doi: 10.1016/j.neuron.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Kang R, et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Anada Y, Igarashi Y. Mouse sphingosine kinase isoforms SPHK1a and SPHK1b differ in enzymatic traits including stability, localization, modification, and oligomerization. J Biol Chem. 2006;281:4532–4539. doi: 10.1074/jbc.M510308200. [DOI] [PubMed] [Google Scholar]
- Kihara A, Ikeda M, Kariya Y, Lee EY, Lee YM, Igarashi Y. Sphingosine-1-phosphate lyase is involved in the differentiation of F9 embryonal carcinoma cells to primitive endoderm. J Biol Chem. 2003;278:14578–14585. doi: 10.1074/jbc.M211416200. [DOI] [PubMed] [Google Scholar]
- Kihara A, Kurotsu F, Sano T, Iwaki S, Igarashi Y. Long-chain base kinase Lcb4 is anchored to the membrane through its palmitoylation by Akr1. Mol Cell Biol. 2005;25:9189–9197. doi: 10.1128/MCB.25.21.9189-9197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakkaraju AK, Abrami L, Lemmin T, Blaskovic S, Kunz B, Kihara A, Dal Peraro M, van der Goot FG. Palmitoylated calnexin is a key component of the ribosome-translocon complex. EMBO J. 2012;31:1823–1835. doi: 10.1038/emboj.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane SR, Liu Y. Characterization of the palmitoylation domain of SNAP-25. J Neurochem. 1997;69:1864–1869. doi: 10.1046/j.1471-4159.1997.69051864.x. [DOI] [PubMed] [Google Scholar]
- Lobo S, Greentree WK, Linder ME, Deschenes RJ. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J Biol Chem. 2002;277:41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Mitchell G, Ling Y, Budde C, Deschenes RJ. Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J Biol Chem. 2010;285:38104–38114. doi: 10.1074/jbc.M110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res. 2006;47:1118–1127. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- Mukai J, Dhilla A, Drew LJ, Stark KL, Cao L, MacDermott AB, Karayiorgou M, Gogos JA. Palmitoylation-dependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion. Nat Neurosci. 2008;11:1302–1310. doi: 10.1038/nn.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadolski MJ, Linder ME. Protein lipidation. FEBS J. 2007;274:5202–5010. doi: 10.1111/j.1742-4658.2007.06056.x. [DOI] [PubMed] [Google Scholar]
- Ohno Y, Kihara A, Sano T, Igarashi Y. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim Biophys Acta. 2006;1761:474–483. doi: 10.1016/j.bbalip.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Pryciak PM, Hartwell LH. AKR1 encodes a candidate effector of the Gβγ complex in the Saccharomyces cerevisiae pheromone response pathway and contributes to control of both cell shape and signal transduction. Mol Cell Biol. 1996;16:2614–2626. doi: 10.1128/mcb.16.6.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- Roth AF, Feng Y, Chen L, Davis NG. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J Cell Biol. 2002;159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, 3rd, Davis NG. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem AN, et al. Mice with alopecia, osteoporosis, and systemic amyloidosis due to mutation in Zdhhc13, a gene coding for palmitoyl acyltransferase. PLoS Genet. 2010;6:e1000985. doi: 10.1371/journal.pgen.1000985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topinka JR, Bredt DS. N-terminal palmitoylation of PSD-95 regulates association with cell membranes and interaction with K+ channel Kv1.4. Neuron. 1998;20:125–134. doi: 10.1016/s0896-6273(00)80440-7. [DOI] [PubMed] [Google Scholar]
- Vetrivel KS, et al. Alzheimer disease Aβ production in the absence of S-palmitoylation-dependent targeting of BACE1 to lipid rafts. J Biol Chem. 2009;284:3793–3803. doi: 10.1074/jbc.M808920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: purification and identification. Nat Protoc. 2007;2:1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR. Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol Cell Proteomics. 2010;9:54–70. doi: 10.1074/mcp.M800448-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.