The SIN signaling pathway promotes cytokinesis and other late mitotic events. The terminal SIN kinase, Sid2, phosphorylates the kinesin-14 protein Klp2 to remove it from microtubules, which is important for efficient anaphase spindle elongation and telophase nuclear positioning.
Abstract
In Schizosaccharomyces pombe, a late mitotic kinase pathway called the septation initiation network (SIN) triggers cytokinesis. Here we show that the SIN is also involved in regulating anaphase spindle elongation and telophase nuclear positioning via inhibition of Klp2, a minus end–directed kinesin-14. Klp2 is known to localize to microtubules (MTs) and have roles in interphase nuclear positioning, mitotic chromosome alignment, and nuclear migration during karyogamy (nuclear fusion during mating). We observe SIN-dependent disappearance of Klp2 from MTs in anaphase, and we find that this is mediated by direct phosphorylation of Klp2 by the SIN kinase Sid2, which abrogates loading of Klp2 onto MTs by inhibiting its interaction with Mal3 (EB1 homologue). Disruption of Klp2 MT localization is required for efficient anaphase spindle elongation. Furthermore, when cytokinesis is delayed, SIN inhibition of Klp2 acts in concert with microtubules emanating from the equatorial microtubule-organizing center to position the nuclei away from the cell division site. These results reveal novel functions of the SIN in regulating the MT cytoskeleton and suggest that the SIN may have broader functions in regulating cellular organization in late mitosis than previously realized.
INTRODUCTION
Proper positioning and migration of nuclei in metazoans is important for cell migration, brain development, fertilization, and muscle function (Burke and Roux, 2009). The fission yeast Schizosaccharomyces pombe is a good model with which to study nuclear positioning because it has three defined cell cycle/developmental stages (interphase, karyogamy, and telophase) in which nuclear positioning must be regulated in distinct ways. Of these stages, interphase nuclear positioning is the best studied. In interphase, antiparallel microtubule (MT) bundles with minus ends anchored on the nuclear envelop grow toward opposite cell tips and exert pushing forces that keep the nucleus at the cell middle (Tran et al., 2001). Organization of these MT arrays depends on the minus end–directed kinesin-14 protein Klp2 and the antiparallel MT bundling protein Ase1 (Carazo-Salas and Nurse, 2006, 2007; Braun et al., 2009). During karyogamy, when two cells of different mating types fuse to undergo meiosis, the two haploid nuclei are pulled together and fuse. Nuclear congression is dependent on MTs and the motor proteins Klp2 and dynein (Troxell et al., 2001), which are believed to slide MTs emanating from each spindle pole body to pull the nuclei together. Unique mechanisms appear to regulate nuclear positioning in telophase, since the two nuclei are neither pulled together by Klp2 (as in karyogamy) nor pushed toward the cell middle by MT-mediated forces on cell ends (as in interphase). Instead, the nuclei maintain their position in the middle of the future daughter cells and away from the cell division apparatus. Although exclusion of nuclei from the cell middle is presumably important to keep them from being cleaved by the ingressing division septum, the mechanisms regulating telophase nuclear positioning are not known.
Some insight into telophase nuclear positioning can be gleaned from the phenotype of cytokinesis mutants. When cytokinesis is blocked or delayed due to defects in the contractile ring or septum synthesis, nuclei are positioned away from the cell middle in a manner dependent on the septation initiation network (SIN; Liu et al., 2000; Trautmann et al., 2001; Guertin et al., 2002; Mishra et al., 2004), a kinase signaling cascade that regulates cytokinesis and other late mitotic events (McCollum and Gould, 2001; Krapp and Simanis, 2008; Roberts-Galbraith and Gould, 2008). The SIN is turned on in anaphase, triggering activation of its terminal kinase, Sid2 (Sparks et al., 1999). Constitutive activation of the SIN leads to ectopic septation, resulting in cells with multiple septa that divide the cytoplasm in discrete chambers. Cells that lack SIN signaling fail cytokinesis, and the two daughter nuclei inappropriately cluster in the cell middle (aka kissing nuclei) in a process that requires the minus end–directed kinesin Klp2 (Okazaki and Niwa, 2008). The SIN is also required for formation of microtubule-organizing centers at the cell division site in late anaphase/telophase that nucleate the postanaphase array (PAA) of microtubules (Hagan and Hyams, 1988; Heitz et al., 2001; Pardo and Nurse, 2003). PAA microtubules have been proposed to be involved in keeping nuclei away from the cell center during cytokinesis (Hagan and Yanagida, 1997). Here we provide evidence that the SIN promotes telophase nuclear positioning by both negative regulation of Klp2 MT localization and promotion of PAA assembly. SIN-mediated phosphorylation of Klp2 prohibits its interaction with Mal3, an EB1 homologue that normally loads Klp2 on MT plus ends, preventing Klp2 from interfering with anaphase spindle elongation and telophase nuclear positioning.
RESULTS
SIN activity determines Klp2 localization
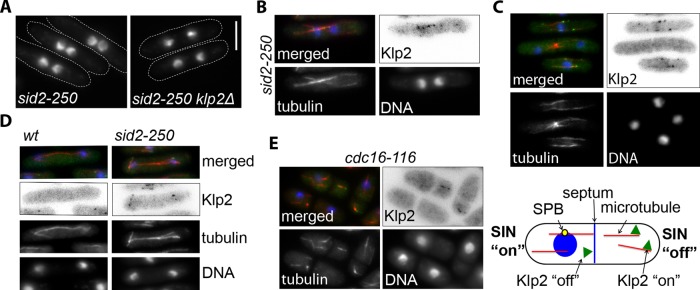
Wild-type S. pombe cells maintain daughter nuclei away from the division machinery, and if cytokinesis is delayed, the nuclei are positioned in the middle of the future daughter cells (Liu et al., 2000). To better understand how the SIN regulates telophase nuclear positioning, we tested the hypothesis that the SIN may act by inhibiting the motor Klp2 to prevent it from causing nuclear clustering. Consistent with a previous study (Okazaki and Niwa, 2008), we observed that deletion of klp2 prevents nuclei from clustering after cytokinesis failure in SIN-mutant cells (sid2-250; Figure 1A and Figure 3A later in the paper). Because Klp2 has been implicated in minus end–directed antiparallel MT sliding during interphase, we visualized Klp2 localization in sid2-250 cells after cytokinesis failure but before the two nuclei were completely clustered. Klp2-green fluorescent protein (GFP) was often seen as puncta on MT bundles that bridge the two sister nuclei (Figure 1B), suggesting that Klp2 promotes nuclear clustering in sid2-250 mutant cells through a mechanism similar to Kar3 in budding yeast karyogamy (Endow et al., 1994; i.e., by sliding antiparallel MTs emanating from each nucleus to pull the nuclei together). This observation suggests that the SIN might impede telophase nuclear clustering by inhibiting Klp2. To address this hypothesis, we tested whether Klp2-GFP localization was affected by the SIN. In wild-type cells, Klp2-GFP localized to the tips (presumably the plus ends) of cytoplasmic MTs during interphase, consistent with a previous report (Figure 1C and Supplemental Movie S1; Janson et al., 2007). However, Klp2 was not observed on MTs during anaphase and telophase, when the SIN is active. Consistent with these results, Klp2 remained on anaphase spindles of cells lacking a functional SIN (sid2-250 cells; Figure 1D). We also tested the effect of ectopic SIN activation on the localization of Klp2-GFP. Klp2-GFP was expressed in cells with a temperature-sensitive mutation in a negative regulator of the SIN, cdc16-116. When cdc16-116 cells are grown at restrictive temperature, the SIN is constitutively activated, causing ectopic septation and formation of compartments in the same cell that may or may not have a nucleus and spindle pole body (SPB; Minet et al., 1979). Because SIN signaling is centered at the SPB, which associates with the nucleus, compartments with a nucleus and therefore a SPB have active SIN signaling and those without them do not (Garcia-Cortes and McCollum, 2009; Figure 1E, scheme). Consistent with our earlier results, Klp2-GFP did not localize to MTs in cell compartments that contained a nucleus (active SIN, interphase cells) but decorated MTs of anucleate compartments with no SIN activity (Figure 1E). Taken together, these results show that the SIN inhibits localization of Klp2 to MTs.
FIGURE 1:
Klp2 is required for nuclear clustering in SIN mutants, and its localization to MTs is modulated by the SIN. (A) klp2Δ sid2-250 and sid2-250 cells were grown to mid–log phase at 25°C and then shifted to 36°C for 2 h to inactivate Sid2. (Note that measurements of the distance between nuclei for both cell lines are shown in Figure 3A.) Cell outlines are marked with dashed lines. (B–E) Merged images show DAPI (blue), mCherry-Atb2 (α-tubulin; red), and Klp2-GFP (green). (B) sid2-250 mCherry-atb2 (tubulin) cells expressing Klp2-GFP were grown at 25°C and visualized after 90 min at 36°C. (C) Klp2-GFP was expressed in mCherry-atb2 cells. The selected field shows Klp2 localization during interphase (bottom and top cells) and during telophase (center). (D) Wild-type and sid2-250 cells expressing Klp2-GFP and mCherry-Atb2 were visualized during anaphase after 90 min at 36°C. (E) Klp2-GFP was visualized in cdc16-116 mCherry-atb2 cells after 2 h at 36°C. The scheme on the right represents the two different types of cell chambers generated in cdc16-116 cells after ectopic septation. In all cases, Klp2-GFP was expressed from the endogenous promoter. Cells were fixed with cold methanol. Images showing Klp2-GFP are shown inverted for clarity. DNA was stained with DAPI. Bar, 5 μm. All images are at the same magnification.
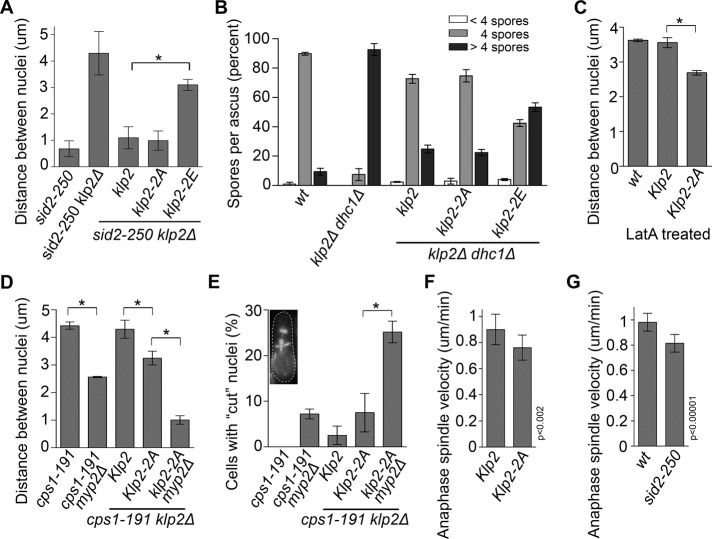
FIGURE 3:
Functional analysis of Klp2 phosphosite mutants. (A) Cells of the indicated genotypes were grown at 25°C and then shifted to 36°C for 2 h to measure the distance between nuclei. (B) Klp2-FLAG, Klp2-2A-FLAG, or Klp2-2E-FLAG was expressed in klp2Δ dhc1Δ (dynein heavy chain) cells of opposite mating types. Cells were then mated to the opposite mating type of the same genotype (including the wild-type and the parental cell line klp2Δ dhc1Δ) and allowed to enter meiosis. Asci were fixed with cold methanol, and DNA was stained with DAPI. The number of spores per ascus was estimated by visualizing the number of condensed DNA puncta. (C) Klp2-FLAG and Klp2-2A-FLAG were expressed in klp2Δ cells, and the distance between nuclei was measured after 30 min of incubation with 1.5 μM latrunculin A (Enzo) at 25°C. (D) Cells of the indicated genotypes were grown at 25°C and then shifted to 36°C for 3 h to measure the distance between nuclei. Only cells that were completely lacking primary septum, as determined by Calcofluor white staining, were scored. (E) Cells of the indicated genotypes were grown at 25°C and then shifted to 36°C for 3 h. Cells were fixed and stained with DAPI and calcofluor to visualize DNA and septa. Of cells displaying septa, the percentage with septa bisecting (inset) or directly over DAPI-stained nuclei were scored as having the “cut” phenotype. (F) The rates of anaphase spindle elongation of klp2Δ cells expressing the SPB marker cdc11-GFP and either Klp2-FLAG or Klp2-2A-FLAG and grown at 30°C were determined using time-lapse microscopy. (G) The rates of anaphase spindle elongation of wild-type or sid2-250 cells expressing the SPB marker sid4-GFP and grown at 36°C were determined using time-lapse microscopy. *p < 0.00001.
Klp2 is a substrate of the Sid2 kinase
To determine how the SIN regulates Klp2, we tested whether the SIN effector kinase Sid2 phosphorylates Klp2. In vitro kinase assays were performed by incubating purified recombinant 6His-Klp2 with Sid2-13Myc kinase purified from fission yeast in the presence of [γ-P32]ATP. Klp2 was specifically phosphorylated by Sid2 (Figure 2A), showing that the SIN might inhibit Klp2 through direct phosphorylation by Sid2. Phosphorylated recombinant Klp2 (glutathione S-transferase [GST]–Klp2) was also analyzed by mass spectrometry to identify sites of phosphorylation. Two sites (S113 and S123) matching the Sid2 phosphorylation consensus sequence (RXXS, where S is phosphorylated; Figure 2B and Supplemental Tables S2 and S3) were identified (Chen et al., 2008; Feoktistova et al., 2012). Although there are multiple RXXS motifs within Klp2 primary sequence, the two sites we identified as phosphorylated by Sid2 are close to each other and within the tail domain (Figure 2B and Supplemental Tables S2 and S3). To test whether these same sites were phosphorylated in vivo, we purified Klp2-TAP from cells with inactive SIN (sid2-250) and cells with constitutively active SIN (cdc16-116). Consistent with the in vitro phosphorylation results, the same two phosphoserines were identified in Klp2 purified from cdc16-116 cells but not in Klp2 purified from cells with inactive SIN (Supplemental Table S3). These phosphorylation sites were then mutated to either alanine or glutamic acid to generate nonphosphorylatable (Klp2-2A) and potentially phosphomimetic (Klp2-2E) versions of Klp2. The ability of Sid2 to phosphorylate recombinant Klp2-2A was tested using in vitro kinase assays. Klp2-2A phosphorylation was reduced to only 3% of the level measured for wild-type Klp2 (Klp2, 88.4, and Klp2-2A, 2.3, arbitrary units after background subtraction), indicating that these are the main Sid2 phosphorylation sites on Klp2 (Figure 2C).
FIGURE 2:
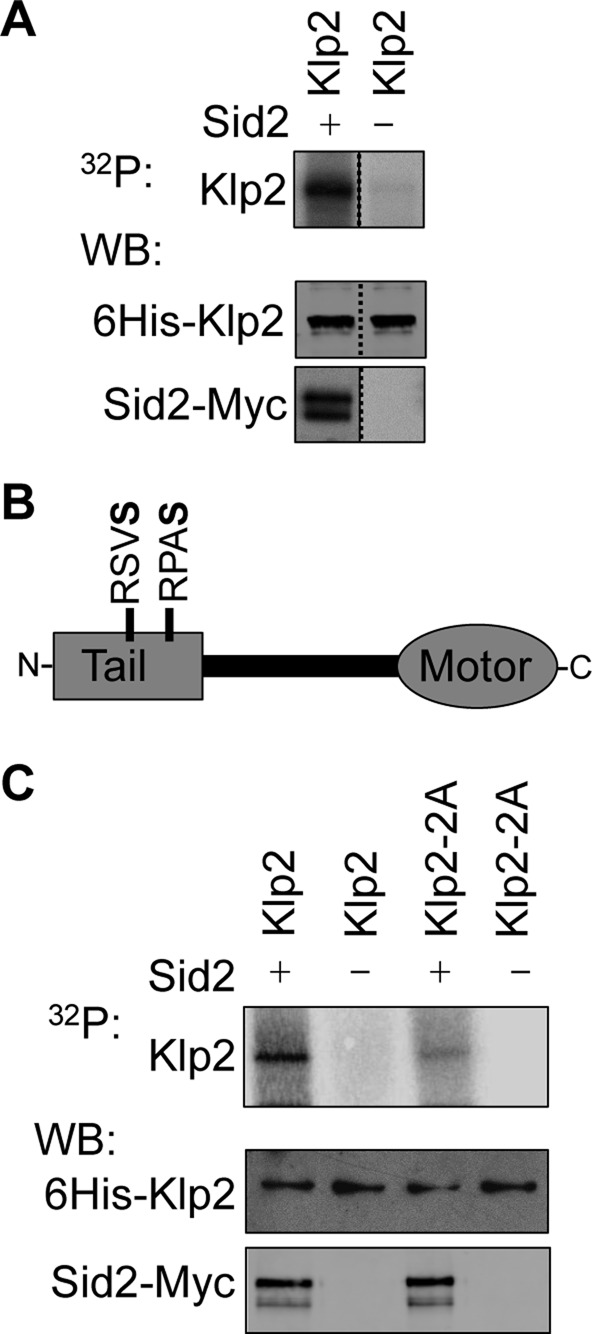
Klp2 is a direct Sid2 kinase substrate. (A) In vitro Sid2 kinase assays were performed using 6His-Klp2 purified from bacteria. 6His-Klp2 was incubated with Sid2 beads (+) or control beads (–). Half of the kinase reaction was used to detect phosphorylation by autoradiography (32P) and half was used in Western blots (WB) to determine the levels of 6His-Klp2 and Sid2. The dashed line indicates the deletion of irrelevant lanes of the same gel. (B) Schematic representation of Klp2 with the two identified “RXXS” phosphosites within the tail domain identified from in vitro and in vivo analysis by mass spectrometry. Note that for in vivo analysis, RXXS sites were identified on Klp2-3HA-TAP purifications from cdc16-116 (SIN “on”) and sid2-250 (SIN “off”) cells by LC-MS/MS. All RXXS sites were only detected in the SIN “on” purification (see Supplemental Table S3). Phosphoserines are in bold. (C) Sid2 kinase assays were performed using recombinant 6His-Klp2 and 6His-Klp2-2A as described in A.
Klp2 function is inhibited by phosphorylation
To understand the significance of Klp2 phosphorylation by Sid2 in vivo, we examined whether Sid2 phosphorylation regulates Klp2 function. Because loss of SIN activity leads to nuclear clustering after cytokinesis failure, we tested whether mimicking Sid2 phosphorylation on Klp2 would restore normal nuclear positioning in cells with disrupted SIN activity. Phosphomimetic Klp2-2E-FLAG, along with Klp2-FLAG and Klp2-2A-FLAG, was integrated in klp2Δ sid2-250 cells and expressed from the nmt1 promoter under repressed conditions (Supplemental Figure S1A). (Note that leaky expression from the nmt1 promoter under “repressed” conditions resulted in similar but slightly higher Klp2 levels than the endogenous promoter; Supplemental Figure S1B.) The distance between sister nuclei after cytokinesis failure was then measured and compared among the three cell lines (Figure 3A). Cells expressing Klp2-FLAG and Klp2-2A-FLAG clustered their nuclei at the cell middle in a similar manner to cells expressing endogenous Klp2, showing that the klp2-2A mutation did not interfere with Klp2 function. However, the distance between nuclei in cells expressing Klp2-2E-FLAG was significantly larger (p < 0.001) than the one measured for cells rescued with Klp2-FLAG. Furthermore, although the distance between nuclei in Klp2-2E-FLAG cells was somewhat less than for klp2Δ cells, the difference did not quite reach the level of statistical significance (p = 0.05), suggesting that Sid2 phosphorylation inhibits Klp2.
As another way to assess the impact of SIN phosphorylation on Klp2, we tested whether the phosphorylation-site mutants were functional for the karyogamy stage of meiosis. Yeast cells can undergo meiosis after fusing with another cell of the opposite mating type. The two haploid nuclei then migrate toward each other, fuse, and form a diploid nucleus that divides meiotically, producing four spores. In contrast to telophase, where nuclei must be kept apart, during karyogamy Klp2 functions together with dynein to pull nuclei together to mediate fusion. Klp2-2E-FLAG, Klp2-FLAG (wild type), or Klp2-2A-FLAG was expressed in klp2Δ dhc1Δ (dynein heavy chain) cells. These cells typically fail karyogamy and enter meiosis with two haploid nuclei that each undergoes meiotic divisions to form aneuploid supernumerary spores (Troxell et al., 2001). Expression of Klp2-FLAG or Klp2-2A-FLAG in klp2Δ dhc1Δ cells reduced the percentage of cells with supernumerary spores per ascus from >90%, to just greater than 20% (Figure 3B). However, expression of Klp2-2E-FLAG only decreased the percentage of asci with supernumerary spores to ∼55%. These data indicate that the function of phosphomimetic Klp2-2E is impaired compared with wild-type Klp2, or Klp2-2A, consistent with our earlier results. The decrease of asci with supernumerary spores from klp2Δ dhc1Δ cells to cells rescued with Klp2-2E-FLAG was statistically significant (p < 0.0001), suggesting that the phosphomimetic mutant retained some residual function.
The SIN acts through Klp2 and PAA microtubules to keep nuclei away from the cell division site during telophase delays
In contrast to karyogamy, SIN phosphorylation of Klp2 during telophase is likely involved in keeping nuclei apart. To test this notion, we assayed whether expression of Klp2-2A-FLAG influenced the position of nuclei during telophase. Because we did not observe an obvious effect of Klp2-2A-FLAG on telophase nuclear positioning in normally cycling cells (see Discussion), we looked at cells where a “telophase-like” state was extended by either inhibiting actomyosin ring constriction and septum deposition (using a cps1-191 mutant; Liu et al., 2000) or by slowing down actomyosin ring constriction using a low dose of the actin-depolymerizing drug latrunculin A (Mishra et al., 2004). In both latrunculin A–treated wild-type cells and cps1-191 mutant cells, the distance between nuclei in Klp2-2A-FLAG–expressing cells was significantly reduced (p < 0.00001) compared with those expressing wild-type Klp2-FLAG (Figure 3, C and D), confirming our hypothesis that SIN inhibition of Klp2 contributes to maintaining proper nuclear positioning when cytokinesis is delayed. Although Klp2-2A-FLAG caused the nuclei in cps1-191 cells to move closer to each other, the nuclei still did not cluster like they do when the SIN is inactivated, suggesting that the SIN may have additional targets involved in keeping the nuclei apart. The PAA MTs, which are nucleated near the actomyosin ring in late anaphase, have been proposed to help position telophase nuclei (Hagan and Yanagida, 1997). Because the SIN is required for formation of the PAA (Heitz et al., 2001), we investigated the involvement of PAAs in nuclear positioning. Cells lacking the unconventional myosin Myp2 fail to assemble PAAs but have no apparent defects in other types of MTs (Samejima et al., 2010). Deletion of myp2 in cps1-191–arrested cells caused a decrease in internuclei distance similar to that caused by klp2-2A (Figure 3D), confirming that PAA microtubules have a role in nuclear positioning. To determine whether klp2-2A would have an additive effect with myp2∆, we expressed Klp2-2A-FLAG in cps1-191 klp2Δ myp2Δ cells and measured the distance between nuclei after incubation at restrictive temperature. Strikingly, the distance between nuclei in these cells was similar to that observed in SIN-mutant cells (Figure 3, D, compare to A), suggesting that the nuclear clustering in SIN-mutant cells could be explained by loss of two SIN functions: Sid2 phosphorylation of Klp2 and SIN-dependent assembly of PAAs. We observed that some cps1-191 cells form partial or complete septa even at the restrictive temperature. Of interest, because of the nuclear positioning defects in cps1-191 klp2Δ myp2Δ cells, we often observed nuclei being “cut” by the ingressing septum (Figure 3E). “Cut” phenotypes were not observed in these cells at permissive temperature for cps1-191. Thus the SIN-dependent nuclear positioning system seems to be important for cells suffering delays in cytokinesis but dispensable under optimal growth conditions.
The SIN promotes spindle elongation through phosphorylation of Klp2
Because Klp2 mediates minus end–directed sliding of antiparallel MTs, active Klp2 in anaphase could interfere with spindle elongation, which is driven by plus end–directed motors (Fu et al., 2009). Therefore we tested whether Klp2-2A expression affected the rate of spindle elongation in anaphase. We used time-lapse microscopy to measure anaphase spindle elongation rates (using the SPB marker Cdc11-GFP) in Klp2-FLAG or Klp2-2A-FLAG cells. Anaphase spindles of cells expressing Klp2-2A-FLAG elongated 16% more slowly (p < 0.002, n = 12) than spindles in cells expressing wild-type Klp2-FLAG (n = 15; Figure 3F). Consistent with SIN-mediated inhibition of Klp2, sid2-250 (SIN off) mutant cells showed 18% slower spindle elongation (n = 23) compared with wild-type cells (n = 29; p < 0.00001; Figure 3G). These results suggest that SIN inhibition of Klp2 promotes anaphase spindle elongation and telophase nuclear positioning.
Sid2 phosphorylation inhibits localization of Klp2 to MTs
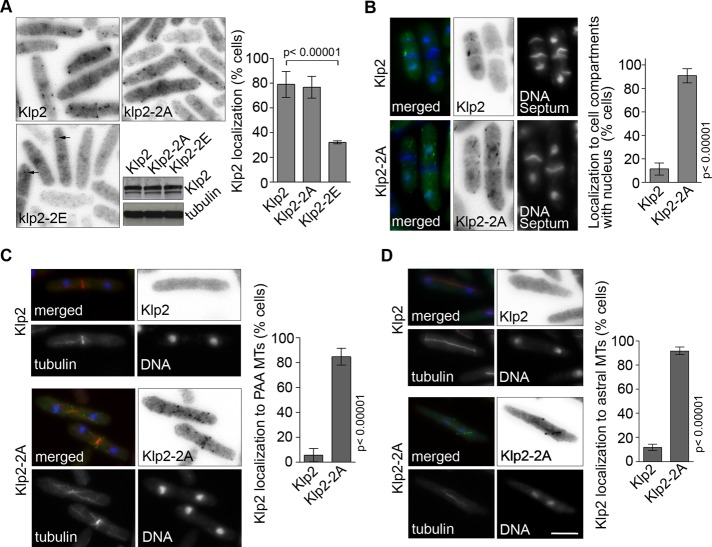
To determine whether Sid2 inhibits Klp2 function by regulating its localization to MTs, we integrated GFP versions of Klp2, Klp2-2A, and Klp2-2E expressed from the nmt1 promoter into klp2Δ cells and scored them for the presence of GFP puncta on MT tips. Similar to Klp2-GFP expressed from the endogenous promoter, Klp2-GFP and Klp2-2A-GFP expressed from the repressed nmt1 promoter were observed as puncta in ∼80% of the cells (Figure 4A, Supplemental Figure S1C, and Supplemental Movies S1–S3). However, Klp2-2E-GFP puncta were observed in only 30% of the cells and were fainter than those in Klp2-GFP and Klp2-2A-GFP–expressing cells, consistent with the loss of Klp2 from MTs when the SIN is active. We then examined how loss of SIN inhibition of Klp2 affects its localization in cdc16-116 cells (active SIN). Whereas wild-type Klp2-GFP does not localize to MTs in cell compartments with active SIN, we found that Klp2-2A-GFP localized to puncta in cell compartments with active SIN (Figure 4B). Localization of GFP puncta in compartments that contain a nucleus increased from 10% for klp2-GFP to 90% in klp2-2A-GFP cells. This dramatic increase in Klp2 localization within “SIN-on” compartments indicates that the SIN inhibits Klp2 MT localization primarily through phosphorylation of serine residues 113 and 123.
FIGURE 4:
Sid2 phosphorylation of Klp2 regulates its MT localization. (A) Klp2-GFP, Klp2-2A-GFP, and Klp2-2E-GFP were expressed in klp2Δ cells and visualized by microscopy. Klp2 localization was quantified as percentage of cells with fluorescent puncta (previously shown to localize to MTs). Arrows point at faint Klp2-GFP puncta. The relative levels of Klp2-GFP and the Klp2-2A and Klp2-2E are shown. (B) klp2Δ cdc16-116 cells expressing Klp2-GFP or Klp2-2A-GFP were grown at 25°C and shifted to 36°C for 2 h. Cells were scored for the presence of Klp2 or Klp2-2A puncta in cell compartments containing a nucleus (and presumably active SIN signaling). (C) Klp2-GFP and Klp2-2A-GFP were expressed in klp2Δ mCherry-atb2 cells. Cells were grown at 25°C and scored for the presence of Klp2 or Klp2-2A on PAAs of microtubules. (D) Klp2-GFP and Klp2-2A-GFP were expressed in klp2Δ mCherry-atb2 cells at 25°C. Anaphase cells were scored for the presence of Klp2 or Klp2-2A on anaphase MTs. (A–C) Values expressed in bar graphs represent an average and SD of three independent averages obtained from independent experiments. Cells were fixed with cold methanol. Images showing Klp2-GFP are shown inverted for clarity. DNA was stained with DAPI. Bar, 5 μm. All images are at the same magnification.
Klp2 is typically lost from MTs during late mitosis and cytokinesis when the SIN is active. Thus Klp2 is usually not observed on MTs characteristic of this stage of the cell cycle, such as the PAAs that originate at the cell middle, and anaphase spindles. Because mutation of the two identified phosphosites abrogated SIN control over Klp2 localization, we decided to observe whether Klp2-2A localized to those MT structures. Klp2-GFP and Klp2-2A-GFP were integrated in klp2Δ cells expressing a MT marker (mCherry-atb2) to better determine the precise localization of fluorescent puncta. Klp2-GFP puncta were detected on PAAs in only 6% of cells (n = 49), and in most cases were very faint (Figure 4C and Supplemental Movie S1). This was in marked contrast to the strong localization of Klp2-2A-GFP to PAAs in >85% of the cells (n = 58; also see Supplemental Movie S2). Of interest, unlike the case with wild-type Klp2, Klp2-2A was observed as puncta on astral microtubules associated with anaphase spindles (44 of 48 spindles; Figure 4D), similar to Klp2 localization in sid2-250 cells. Klp2-2A also faintly decorated intranuclear spindle MTs (see also Supplemental Movie S3). Instead of forming puncta, the Klp2-2A observed on intranuclear MTs was evenly distributed along the length of spindle and tended to fade quickly. The presence of Klp2-2A on spindles is consistent with our observation that klp2-2A cells have a decreased anaphase spindle elongation velocity.
EB1 homologue Mal3 loads Klp2 on MTs
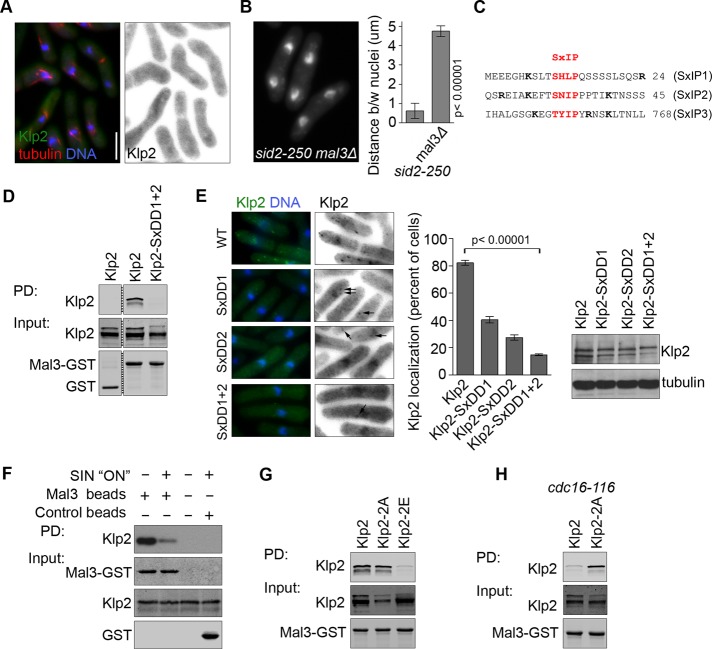
To determine how Sid2 phosphorylation inhibits localization of Klp2 to MTs, we investigated how Klp2 localizes to MT plus ends. EB1 (S. pombe Mal3), the archetypical MT plus end–tracking protein, has been implicated in loading multiple plus end factors onto MTs, including the kinesin Tea2 in fission yeast (Browning and Hackney, 2005). We tested whether Mal3 is required for localization of Klp2 to MTs. Klp2-GFP was expressed from the endogenous promoter in mal3Δ mCherry-atb2 (α-tubulin) cells. Strikingly, no Klp2 was observed on MTs, although its expression in this genetic background was slightly higher than in wild-type cells (Figures 5A and Supplemental Figure S1D). Like Klp2, Mal3 is required for nuclear clustering in the sid2-250 mutant (Figure 5B), and the average distance between nuclei after cytokinesis failure increased from 0.6 um (sid2-250) to 4.5 um in mal3Δ sid2-250 cells, echoing the phenotype we observed for Klp2-2E. Loss of Klp2 from MTs could explain the nuclear clustering defect in mal3∆ cells, although the reduced MT length in mal3∆ cells could also contribute to the phenotype (Beinhauer et al., 1997).
FIGURE 5:
Mal3 loading of Klp2 on MTs is regulated by Sid2. (A) Klp2-GFP was expressed from the endogenous promoter in mal3Δ mCherry-atb2 cells. (B) The distance between nuclei was quantified in sid2-250 and sid2-250 mal3Δ cells after 2 h at 36°C. (C) The protein sequence of Klp2 surrounding consensus EB1/Mal3 motifs (in red). Basic residues flanking the consensus SxIP are shown in bold. (D) Klp2 or Klp2-SxDD1+2 were expressed in klp2Δ cells and pulled down with recombinant purified Mal3-GST. Purified GST protein was used as control. Protein levels were tested by Western blotting. Irrelevant lanes were deleted from the panel (dashed line). (E) Klp2-GFP and the indicated SxIP mutants (also as –GFP fusions) were expressed in klp2Δ cells, and the quantifications reflect the percentage of cells with GFP puncta. Arrows point at faint Klp2-GFP puncta. Protein levels were tested by immunoblotting. (F) Recombinant purified Mal3-GST was used to pull down Klp2-GFP expressed in wild-type or cdc16-116 cells. Cells were grown at 25°C and shifted to 36°C for 2 h. Protein levels were tested by Western blotting. (G) Klp2, Klp2-2A, or Klp2-2E was expressed in klp2Δ cells and pulled down with recombinant purified Mal3-GST. Protein levels were tested by Western blotting. (H) Klp2-GFP or Klp2-2A-GFP was expressed in klp2Δ cdc16-116 cells and pulled down with recombinant purified Mal3-GST. Cells were grown at 25°C and shifted to 36°C for 2 h. In all cases, quantifications represent an average and SD of three independent experiments. Cells were fixed with cold methanol. Images showing Klp2-GFP are shown inverted for clarity. DNA was stained with DAPI. Bar, 5 μm. All images are at the same magnification.
Klp2 contains three putative EB1/Mal3-binding sites (SxIP; Figure 5C), two of which are located in the tail domain near the Sid2 phosphorylation sites. We tested whether Klp2 and Mal3 interact in vitro in a manner dependent on the two Mal3-binding motifs located in the tail domain (the third motif is located in a conserved region of the motor domain). The hydrophobic residues Ile–Pro within the SxIP motif provide hydrophobic interactions that are required for binding to EB1 (Honnappa et al., 2009); thus they were replaced by the hydrophilic residues Asp–Asp to generate mutants of the first and second SxIP motifs (denoted Klp2-SxDD1/2). Recombinant Mal3-GST was used as bait to pull down Klp2 or the SxDD tandem mutant (Klp2-SxDD1+2, i.e., the first [1] and second [2] SxIP motifs were both mutated to SxDD) from yeast lysates. Although wild-type Klp2 interacted with Mal3, Klp2-SxDD1+2 did not (Figure 5D). The importance of the Mal3-binding motifs within Klp2 was further tested by analyzing the localization of Klp2-SxDD mutants (Figure 5E). Localization of Klp2-SxDD1 and Klp2-SxDD2 to MT puncta was reduced by >50% when compared with wild-type Klp2, and loss of both Mal3-binding sites (Klp2-SxDD1+2) reduced the percentage of interphase cells with Klp2 puncta on MTs from 80% to <20%. This is consistent with findings that suggest that tandem EB1-binding sites act synergistically to increase EB1 binding efficiency (Honnappa et al., 2009).
We next tested whether SIN activity affected interaction of Klp2 with EB1/Mal3. Recombinant Mal3-GST was used to pull down Klp2-GFP from either wild-type cells or cells with activated SIN signaling (cdc16-116). Binding to Mal3 was strongly reduced in lysates from cells with constitutively activated SIN (cdc16-116 cells), supporting the idea that phosphorylated Klp2 cannot interact with Mal3 (Figure 5F). To assess the role of Klp2 phosphorylation more directly, we examined the interaction of Mal3-GST with phosphomimetic Klp2-2E-GFP, Klp2-2A-GFP, or wild-type Klp2-GFP (Figure 5G). This analysis revealed that binding of Klp2-2E-GFP to Mal3 was significantly reduced compared with Klp2-GFP or Klp2-2A-GFP. We then tested whether the interaction between Mal3 and nonphosphorylatable Klp2-2A-GFP was resistant to SIN activity. “SIN-on” cell lysates (cdc16-116) of cells expressing Klp2-GFP and Klp2-2A-GFP were incubated with Mal3-GST in pull-down experiments. Unlike wild-type Klp2-GFP, Klp2-2A-GFP was able to bind Mal3 even when the SIN was active (Figure 5H). Taken together, these results show that Sid2 phosphorylation of Klp2 prevents Klp2 binding to Mal3 and loading onto MTs and therefore Klp2 function in nuclear positioning.
DISCUSSION
In this study we showed that the SIN promotes spindle elongation and appropriate nuclear positioning through Sid2 phosphorylation of the kinesin Klp2. Sid2 phosphorylation keeps Klp2 off MTs by inhibiting its interaction with the EB1 homologue Mal3. Mal3 loads Klp2 on MT plus ends through binding to two SxIP motifs in a predicted unstructured region of the tail of Klp2. Sid2 phosphorylation of Klp2 at two sites proximal to the EB1/Mal3-binding motifs prevents this interaction. These results are reminiscent of several recent studies, which showed that phosphorylation near EB1/Mal3-binding motifs can disrupt association with EB1 by interfering with electrostatic interactions that contribute to binding (Honnappa et al., 2009; Kumar et al., 2009; Vacher et al., 2011). However, the inhibitory phosphorylation sites on Klp2 are not immediately flanking the EB1 binding sites as in the other examples, but instead are between 80 and 110 residues away. It is possible that the predicted unstructured nature of the Klp2 tail allows the phosphoserines to act at a greater distance to inhibit electrostatic interactions that contribute to binding. Alternatively, Klp2 phosphorylation could result in a structural transition, perhaps from a disordered to a more ordered state (Kissinger et al., 1999; Mendoza-Espinosa et al., 2009), which prevents Klp2 binding to Mal3 by making the SxIP binding motifs less accessible. The N-terminal sequence of myosin regulatory light chain is one example where phosphorylation results in a more stable domain. More detailed structural analysis will be required to determine the precise mechanism of phosphoinhibition of the Klp2–Mal3 interaction.
Our results showed that in cells delayed in telophase (cps1-191), loss of either SIN regulation of Klp2 (klp2-2A cells) or PAA microtubules (myp2∆ cells) caused a small displacement of nuclei toward the cell center. However, elimination of both SIN regulation of Klp2 and PAA microtubules (klp2-2A myp2∆ cells) caused a strong nuclear clustering defect similar to SIN mutants. Of interest, cps1-191 cells slowly form septa, and klp2-2A myp2∆ cps1-191 cells often showed mispositioned nuclei being “cut” by the ingressing septa. Surprisingly, klp2-2A myp2∆ cells did not display “cut” phenotypes or obvious mispositioning of nuclei toward the cell center in normally dividing cells, suggesting that SIN-dependent nuclear positioning is most important for preventing “cut” nuclei when cytokinesis has been delayed. It is possible that in normally dividing cells, where the septum ingresses quickly, the nuclei are kept away from the cell center by the interphase nuclear positioning system (Tran et al., 2001), which uses pushing forces from MTs associated with the nuclear envelope. In this scenario, ingression of the actomyosin ring and septum may provide a barrier for MTs associated with the nuclei to push against to keep the nuclei away from the division apparatus.
We presume that the SIN inhibits the MT localization of Klp2 to prevent its minus end–directed sliding activity from pulling telophase nuclei together. By this model, Klp2 would slide MTs emanating from opposite nuclei or the nucleus and PAA MTs to pull the nuclei toward the cell center. Our results also show that PAAs perform a Klp2-independent function in positioning the telophase nuclei. One possible function for PAA MTs could be that they interact with MTs associated with the nuclei via motor proteins to provide pushing forces to move the nuclei away from the cell center. One candidate motor could be the plus end–directed kinesin-6 Klp9, which transiently localizes at equatorial MTs in telophase before returning to the nucleoplasm (Fu et al., 2009).
Just as inappropriate minus end–directed MT sliding activity in telophase may pull the nuclei together, this same type of MT sliding activity would also be predicted to interfere with spindle elongation. Consistent with this model, Klp2-2A (in contrast to wild-type Klp2) was observed not just on astral MTs but also faintly on anaphase spindles. In addition, both SIN inhibition and the klp2-2A mutation slowed down spindle elongation in anaphase. Thus the SIN may inhibit Klp2 in anaphase to prevent its minus end–directed sliding activity from interfering with spindle elongation, although we cannot rule out other possibilities, such as subtle changes in MT organization or dynamics. It makes sense that the SIN might regulate anaphase spindle elongation, since the SIN becomes active in anaphase (Guertin and McCollum, 2001; Feoktistova et al., 2012). Furthermore, regulation of both spindle elongation and ingression of the actomyosin ring and septum by a single pathway may ensure proper coordination of these key events.
This work contributes to a broader understanding of kinesin-14 regulation and the dynamics involved in nuclear positioning. Klp2 plays a role in nuclear positioning during interphase and karyogamy but is inactivated during late mitosis to allow for rapid anaphase spindle elongation and telophase nuclear positioning. This study uncovered the mechanism governing EB1/Mal3-dependent loading of Klp2 onto MTs and SIN-mediated inhibition of the Klp2–Mal3 interaction. Furthermore, we also showed that phosphorylation at sites not immediately adjacent to the Mal3/EB1-binding motifs can regulate this interaction. This mode of regulation may be conserved in other kinesin-14 proteins because the Klp2 orthologue in Drosophila melanogaster, called ncd, localizes to MT plus ends via EB1 and is inhibited during anaphase B (Sharp et al., 2000; Goshima et al., 2005). It will be interesting to determine whether other members of the kinesin-14 family are regulated by phosphorylation like Klp2.
MATERIALS AND METHODS
Plasmid construction and cell culture
All fission yeast manipulation and culture were performed as previously described (Moreno et al., 1991). Yeast strains used in this study are listed in Supplemental Table S1. Except when noted, cells were grown in minimal media (EMM; MP Biomedicals) with appropriate supplements and 15 μM thiamine at 25°C. All cps1-191 mutant strains were grown in YE (yeast extract; MP Biomedicals) media. Where appropriate, temperature mutants were inactivated by shifting to 36°C. All experiments were carried out using integrated versions of Klp2 expressed from the endogenous promoter (klp2-pk-GFP) or from the nmt1 promoter (klp2-FLAG and klp2-GFP). For nmt1 constructs, klp2 was cloned in the GATEWAY entry vector (Invitrogen, Carlsbad, CA) and subcloned into pDUAL destination vectors (FFH1c or GFH1c; Matsuyama et al., 2004). Klp2-pDUAL constructs were then digested with NotI to excise the ARS1 sequence and the ura4+ marker and integrated at the leu-32 locus as previously described (Matsuyama et al., 2004). Point mutations were generated using the QuikChange II system (Agilent, Santa Clara, CA).
Microscopy
Cells were observed after fixation with −20°C methanol or as live cultures for time-lapse studies. Fixed cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) and calcofluor white (Sigma-Aldrich, St. Louis, MO) to visualize the nucleus and septum, respectively. Wide-field fluorescence images were acquired with a Nikon E600 microscope (Nikon, Melville, NY) equipped with an Orca-ER camera (Hamamatsu, Hamamatsu, Japan) and controller managed by IPlab Spectrum software (Scanalytics, Spectra Services, Ontario, NY). For live-cell imaging, cells were grown to an OD of 0.5 in minimal media, washed in the same media, and plated in lectin-coated, glass-bottom Petri dishes. Images were taken once per minute at room temperature using a Nikon TE 2000-E2 spinning-disk confocal microscope controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA). Image stacks were processed using ImageJ (Abramoff et al., 2004).
Statistical analysis
All phenotypes were quantified by triplicate in independent experiments, with at least 100 cells per replicate scored unless noted. Bar graphs show the mean and SD of the means of the three replicate experiments. The distance between nuclei was defined as the minimal distance between the DAPI-stained edges of the two nuclei. In experiments using the cps1-191 genetic background or latrunculin A to arrest cells in telophase, the distance between nuclei was measured only in cells completely lacking septa, as visualized by absence of calcofluor staining. For measuring the “cut” phenotype in cps1-191 cells, cells were scored as having the “cut” phenotype if the septa had either bisected the nuclear DNA or was on a path to bisect part of the nuclear DNA. For the quantification of Klp2-GFP (wild-type or mutant) puncta, all cells with at least a single focus of any intensity were scored as positive. Data quantifying phenotypes that were scored into categories (i.e., presence or absence of Klp2 on MTs) were statistically tested for significance with Fisher's exact test. Data sets with numerical values (i.e., distance between nuclei) were tested with Student's t test. All statistical tests were performed on pooled data sets containing all data points from the three replicate experiments. The p values were obtained from two-tailed analyses.
In vitro kinase assay
Kinase assays were performed as previously described (Sparks et al., 1999). Expression of hexahistidine (6His)-Klp2 and 6His-Klp2-2A was induced with 1 mM isopropyl-β-d-thiogalactoside (IPTG) for 2 h at 25°C in BL21 DE3 Rosetta cells (EMD, San Diego, CA). Cells were lysed by sonication, and recombinant proteins were purified using nickel-nitriloacetic acid beads (Qiagen, Valencia, CA), following the manufacturer's directions. Then 6His-Klp2 and 6His-Klp2-2A were eluted by incubation with 500 mM imidazole (Sigma-Aldrich) for 30 min at 4°C. Sid2 kinase (Sid2-13Myc) was expressed from the endogenous promoter in cdc3-123 cdc16-116 cells grown to an OD of 0.4 at 25°C and then shifted to 36°C for 2 h to achieve maximum activity. Pellets of OD 50 were then collected and frozen in liquid nitrogen. Yeast cells were disrupted by bead beating (FastPrep), cleared by centrifugation, and immunoprecipitated with mouse anti-Myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA) in NP-40 buffer (1% NP-40, 150 mM NaCl, 2 mM EDTA, 6 mM Na2HPO4, and 4 mM NaH2PO4) supplemented with yeast protease inhibitor cocktail (Sigma-Aldrich), 1 mM phenylmethylsulfonyl fluoride, and phosphatase inhibitors (50 mM NaF and 100 μM NaVO4). Sid2-bound beads were then washed and suspended in kinase buffer (10 mM Tris-HCl, pH 7.4, 10 mM MgCl2) supplemented with yeast protease inhibitor cocktail (Sigma-Aldrich), and incubated with ∼0.5 μg of purified protein substrate, 2 μM ATP, and 5 μCi of [γ-32P]ATP for 30 min at 30°C. Kinase reactions were stopped by the addition of 20 μl of 2× SDS sample buffer, followed by boiling. Samples were subjected to SDS–PAGE, gels were dried, and the signal was quantified using a PhosphorImager (GE Healthcare, Piscataway, NJ).
Mal3-GST binding assays
Mal3-GST binding assays were carried out by incubating purified recombinant Mal3-GST with yeast lysates. Recombinant Mal3-GST expression was induced with 1 mM IPTG for 2 h at 25°C in BL21 DE3 Rosetta cells (EMD). Cells were lysed by sonication, and Mal3-GST was purified using glutathione beads (GE Healthcare), following the manufacturer's directions. Yeast lysates were obtained by bead beating (FastPrep) pellets of OD 20 with 0.5 ml of glass beads (Sigma-Aldrich) for 40 s. Beads were then extracted with 1 ml of NP-40 buffer and cleared at 21,000 × g for 15 min at 4°C. Yeast supernatants were incubated with Mal3-GST–bound beads for 1 h at 4°C. Beads were washed five times with 1 ml of NP-40 buffer and suspended in 2× SDS sample buffer, followed by boiling. Protein-binding levels were assessed by SDS–PAGE, followed by Western blotting.
Klp2 phosphosite identification
Phosphosite identification was performed using liquid chromatography–tandem mass spectrometry (LC-MS/MS) on Klp2 purified from yeast or recombinant GST-Klp2 protein after in vitro kinase reactions. Klp2-3HA-TAP was expressed from the endogenous promoter in wild-type cells, cells with active SIN (cdc3-123 cdc16-116), and cells with inactive SIN signaling (sid2-250). Klp2-3HA-TAP was purified from 2 l of 4× YE media as previously described (Gould et al., 2004). Klp2 was cloned in pDEST15 (Invitrogen) and transformed into BL21 Rosetta cells (EMD), and expression was induced for 2 h at 25°C with 1 mM IPTG. GST-Klp2 was purified with glutathione–Sepharose beads (GE Healthcare) following the manufacturer's directions, eluted with 20 mM glutathione, and subjected to kinase assays as described but using 25 μM ATP in the absence of [γ-32P]ATP. Purified Klp2 proteins were trichloroacetic acid (TCA) precipitated and analyzed by LC-MS/MS.
LC-MS/MS and MS data analysis
Klp2 TCA pellets were denatured with 8 M urea, reduced with Tris 2-carboxyethyl phosphine, alkylated with iodoacetamide, and digested overnight at 37°C with Trypsin Gold (Promega, Madison, WI) after diluting to 2 M urea with 50 mM Tris, pH 8.5. The resulting peptides were subjected to two-dimensional LC-MS/MS (MudPIT) on a Thermo LTQ as previously detailed (McDonald et al., 2002; Roberts-Galbraith et al., 2009). Thermo RAW files were converted to DTA files using Scansifter 2.0.13 (Ma et al., 2011), and spectra with <20 peaks were excluded from analysis. Protein identification was performed with the Sequest (TurboSequest, version 2.7, revision 12) algorithm (allowing two missed cleavages, carbamidomethylation, oxidation of methionine, and phosphorylation of serine, threonine, and tyrosine; peptide mass tolerance was 2.5 m/z) on a high-performance computing cluster (Advanced Computing Center for Research and Education, Vanderbilt University) using a concatenated FASTA database (10,352 total protein entries) containing forward and reverse protein sequences for all S. pombe proteins (PomBase, www.pombase.org/, May 2011) and common contaminants (e.g., immunoglobulin G and keratin). Peptide identifications were assembled into proteins and filtered in Scaffold 3, version 3.5 (Proteome Software, Portland, OR). Total sequence coverage for Klp2 ranged from 43 to 78% for all purifications (see Supplemental Table S2, showing all Klp2 peptides identified). Reported RXXS phosphosites (Figure 2) were manually interrogated to validate neutral loss (-98, phosphoric acid) and spectral quality (see Supplemental Table S3 for details) using Scaffold PTM, version 2.0 (Proteome Software).
Supplementary Material
Acknowledgments
We thank Richard McIntosh, Ursula Fleig, Andrew McAinsh, and Robert Cross for strains and plasmids and Takashi Toda for pointing out the presence of potential EB1 binding sites in Klp2. J.R.M. was supported by National Cancer Institute T32CA119925. This work was supported by the Howard Hughes Medical Institute, of which K.L.G. is an investigator, and National Institutes of Health Grant GM058406-14 to D.M.
Abbreviations used:
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- LC
liquid chromatography
- MS
mass spectrometry
- MT
microtubule
- PAA
postanaphase array
- SIN
septation initiation network
- WB
Western blot
- YE
yeast extract
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-07-0532) on October 19, 2012.
*Present address: Department of Biology, Boston College, Chestnut Hill, MA 02467.
REFERENCES
- Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Beinhauer JD, Hagan IM, Hegemann JH, Fleig U. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J Cell Biol. 1997;139:717–728. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M, Drummond DR, Cross RA, McAinsh AD. The kinesin-14 Klp2 organizes microtubules into parallel bundles by an ATP-dependent sorting mechanism. Nat Cell Biol. 2009;11:724–730. doi: 10.1038/ncb1878. [DOI] [PubMed] [Google Scholar]
- Browning H, Hackney DD. The EB1 homolog Mal3 stimulates the ATPase of the kinesin Tea2 by recruiting it to the microtubule. J Biol Chem. 2005;280:12299–12304. doi: 10.1074/jbc.M413620200. [DOI] [PubMed] [Google Scholar]
- Burke B, Roux KJ. Nuclei take a position: managing nuclear location. Dev Cell. 2009;17:587–597. doi: 10.1016/j.devcel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R, Nurse P. Sorting out interphase microtubules. Mol Syst Biol. 2007;3:95. doi: 10.1038/msb4100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas RE, Nurse P. Self-organization of interphase microtubule arrays in fission yeast. Nat Cell Biol. 2006;8:1102–1107. doi: 10.1038/ncb1479. [DOI] [PubMed] [Google Scholar]
- Chen CT, Feoktistova A, Chen JS, Shim YS, Clifford DM, Gould KL, McCollum D. The SIN kinase Sid2 regulates cytoplasmic retention of the S. pombe Cdc14-like phosphatase Clp1. Curr Biol. 2008;18:1594–1599. doi: 10.1016/j.cub.2008.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 1994;13:2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova A, Morrell-Falvey J, Chen JS, Singh NS, Balasubramanian MK, Gould KL. The fission yeast septation initiation network (SIN) kinase, Sid2, is required for SIN asymmetry and regulates the SIN scaffold, Cdc11. Mol Biol Cell. 2012;23:1636–1645. doi: 10.1091/mbc.E11-09-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Ward JJ, Loiodice I, Velve-Casquillas G, Nedelec FJ, Tran PT. Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev Cell. 2009;17:257–267. doi: 10.1016/j.devcel.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cortes JC, McCollum D. Proper timing of cytokinesis is regulated by Schizosaccharomyces pombe Etd1. J Cell Biol. 2009;186:739–753. doi: 10.1083/jcb.200902116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Nedelec F, Vale RD. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Ren L, Feoktistova AS, Jennings JL, Link AJ. Tandem affinity purification and identification of protein complex components. Methods. 2004;33:239–244. doi: 10.1016/j.ymeth.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Guertin DA, McCollum D. Interaction between the noncatalytic region of Sid1p kinase and Cdc14p is required for full catalytic activity and localization of Sid1p. J Biol Chem. 2001;276:28185–28189. doi: 10.1074/jbc.M103802200. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Trautmann S, McCollum D. Cytokinesis in eukaryotes. Microbiol Mol Biol Rev. 2002;66:155–178. doi: 10.1128/MMBR.66.2.155-178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Evidence for cell cycle-specific, spindle pole body-mediated, nuclear positioning in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1997;110(,):1851–1866. doi: 10.1242/jcs.110.16.1851. [DOI] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Heitz MJ, Petersen J, Valovin S, Hagan IM. MTOC formation during mitotic exit in fission yeast. J Cell Sci. 2001;114:4521–4532. doi: 10.1242/jcs.114.24.4521. [DOI] [PubMed] [Google Scholar]
- Honnappa S, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Janson ME, Loughlin R, Loiodice I, Fu C, Brunner D, Nedelec FJ, Tran PT. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 2007;128:357–368. doi: 10.1016/j.cell.2006.12.030. [DOI] [PubMed] [Google Scholar]
- Kissinger CR, Dunker AK, Shakhnovich E. Disorder in protein structure and function. Pacific Symp Biocomp. 1999:517–519. [Google Scholar]
- Krapp A, Simanis V. An overview of the fission yeast septation initiation network (SIN) Biochem Soc Trans. 2008;36:411–415. doi: 10.1042/BST0360411. [DOI] [PubMed] [Google Scholar]
- Kumar P, Lyle KS, Gierke S, Matov A, Danuser G, Wittmann T. GSK3beta phosphorylation modulates CLASP-microtubule association and lamella microtubule attachment. J Cell Biol. 2009;184:895–908. doi: 10.1083/jcb.200901042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang H, Balasubramanian MK. A checkpoint that monitors cytokinesis in Schizosaccharomyces pombe. J Cell Sci. 2000;113:1223–1230. doi: 10.1242/jcs.113.7.1223. [DOI] [PubMed] [Google Scholar]
- Ma Z-Q, et al. Supporting tool suite for production proteomics. Bioinformatics. 2011;27:3214–3215. doi: 10.1093/bioinformatics/btr544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Shirai A, Yashiroda Y, Kamata A, Horinouchi S, Yoshida M. pDUAL, a multipurpose, multicopy vector capable of chromosomal integration in fission yeast. Yeast. 2004;21:1289–1305. doi: 10.1002/yea.1181. [DOI] [PubMed] [Google Scholar]
- McCollum D, Gould KL. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001;11:89–95. doi: 10.1016/s0962-8924(00)01901-2. [DOI] [PubMed] [Google Scholar]
- McDonald W, Ohi R, Miyamoto D, Mitchison T, Yates JR. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int J Mass Spectrom. 2002;219:245–251. [Google Scholar]
- Mendoza-Espinosa P, Garcia-Gonzalez V, Moreno A, Castillo R, Mas-Oliva J. Disorder-to-order conformational transitions in protein structure and its relationship to disease. Mol Cell Biochem. 2009;330:105–120. doi: 10.1007/s11010-009-0105-6. [DOI] [PubMed] [Google Scholar]
- Minet M, Nurse P, Thuriaux P, Mitchison JM. Uncontrolled septation in a cell division cycle mutant of the fission yeast Schizosaccharomyces pombe. J Bacteriol. 1979;137:440–446. doi: 10.1128/jb.137.1.440-446.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M, Karagiannis J, Trautmann S, Wang H, McCollum D, Balasubramanian MK. The Clp1p/Flp1p phosphatase ensures completion of cytokinesis in response to minor perturbation of the cell division machinery in Schizosaccharomyces pombe. J Cell Sci. 2004;117:3897–3910. doi: 10.1242/jcs.01244. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Niwa O. Dikaryotic cell division of the fission yeast Schizosaccharomyces pombe. Biosci Biotechnol Biochem. 2008;72:1531–1538. doi: 10.1271/bbb.80035. [DOI] [PubMed] [Google Scholar]
- Pardo M, Nurse P. Equatorial retention of the contractile actin ring by microtubules during cytokinesis. Science. 2003;300:1569–1574. doi: 10.1126/science.1084671. [DOI] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Chen JS, Wang J, Gould KL. The SH3 domains of two PCH family members cooperate in assembly of the Schizosaccharomyces pombe contractile ring. J Cell Biol. 2009;184:113–127. doi: 10.1083/jcb.200806044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Gould KL. Stepping into the ring: the SIN takes on contractile ring assembly. Genes Dev. 2008;22:3082–3088. doi: 10.1101/gad.1748908. [DOI] [PubMed] [Google Scholar]
- Samejima I, Miller VJ, Rincon SA, Sawin KE (2010) Fission yeast Mto1 regulates diversity of cytoplasmic microtubule organizing centers. Curr Biol. 20:1959–1965. doi: 10.1016/j.cub.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks CA, Morphew M, McCollum D. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J Cell Biol. 1999;146:777–790. doi: 10.1083/jcb.146.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann S, Wolfe BA, Jorgensen P, Tyers M, Gould KL, McCollum D. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr Biol. 2001;11:931–940. doi: 10.1016/s0960-9822(01)00268-8. [DOI] [PubMed] [Google Scholar]
- Troxell CL, Sweezy MA, West RR, Reed KD, Carson BD, Pidoux AL, Cande WZ, McIntosh JR. pkl1(+)and klp2(+): two kinesins of the Kar3 subfamily in fission yeast perform different functions in both mitosis and meiosis. Mol Biol Cell. 2001;12:3476–3488. doi: 10.1091/mbc.12.11.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher H, Yang JW, Cerda O, Autillo-Touati A, Dargent B, Trimmer JS (2011) Cdk-mediated phosphorylation of the Kvbeta2 auxiliary subunit regulates Kv1 channel axonal targeting. J Cell Biol. 192:813–824. doi: 10.1083/jcb.201007113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.