ETOC: NMI is a novel ARF-interacting protein identified in a yeast two-hybrid screen. NMI inhibits ULF-induced ubiquitin degradation of ARF protein. It mediates transcription-independent ARF regulation and is required for the stabilization and up-regulation of ARF in response to cellular stresses.
Abstract
The ARF tumor suppressor is a product of the INK4a/ARF locus, which is frequently mutated in human cancer. The expression of ARF is up-regulated in response to certain types of DNA damage, oncogene activation, and interferon stimuli. Through interaction with the p53 negative regulator MDM2, ARF controls a well-described p53/MDM2-dependent checkpoint. However, the mechanism of ARF induction is poorly understood. Using a yeast two-hybrid screen, we identify a novel ARF-interacting protein, N-Myc and STATs interactor (NMI). Previously, NMI was known to be a c-Myc–interacting protein. Here we demonstrate that through competitive binding to the ARF ubiquitin E3 ligase (ubiquitin ligase for ARF [ULF]), NMI protects ARF from ULF-mediated ubiquitin degradation. In response to cellular stresses, NMI is induced, and a fraction of NMI is translocated to the nucleus to stabilize ARF. Thus our work reveals a novel NMI-mediated, transcription-independent ARF induction pathway in response to cellular stresses.
INTRODUCTION
ARF (p14ARF in humans, p19ARF in mice), the product of an alternative open reading frame of the ARF/INK4a locus (Quelle et al., 1995), is a tumor-suppressor protein that is mutated or inactivated in a significant number of human tumors (Sherr, 2006). One of the best-defined functions of ARF is to control the p53/MDM2-dependent checkpoint in response to oncogenic signals. ARF is activated by hyperproliferative signals from oncogenes such as Myc (Zindy et al., 1998), E1A (de Stanchina et al., 1998), E2F1 (Bates et al., 1998), mutated Ras (Palmero et al., 1998), and v-Abl (Radfar et al., 1998). By antagonizing MDM2 (HDM2 in human) function, ARF triggers p53-dependent cell cycle arrest or apoptosis. MDM2 is a negative regulator of p53. ARF interacts directly with MDM2 and blocks MDM2-mediated ubiquitination, nuclear export, and subsequent degradation of p53 (Kamijo et al., 1998; Pomerantz et al., 1998; Stott et al., 1998; Zhang et al., 1998; Zhang and Xiong, 1999; Tao and Levine, 1999). It has also been proposed that ARF stabilizes nucleoplasmic p53 by binding and sequestering MDM2 in the nucleolus (Weber et al., 1999). However, other studies suggest that nucleolar relocalization of MDM2 is not required for p53 activation (Llanos et al., 2001; Korgaonkar et al., 2002). In addition, ARF can also stabilize and stimulate p53 activity by blocking the newly identified E3 ubiquitin ligase ARF-BP1/Mule-mediated ubiquitination and degradation of p53 (Chen et al., 2005).
Recent work has shown that ARF is implicated in more than just oncogene activation–induced p53 response. Several studies have implied a role for ARF in the DNA damage response. It has been demonstrated that ARF levels are increased by some forms of DNA damage (Khan et al., 2000; Eymin et al., 2006). ARF-null mice develop tumors more frequently than do wild-type mice after exposure to ionizing radiation (Kamijo et al., 1999). ARF−/− MEFs are partially defective in the DNA damage response and show reduced levels of p53 after ionizing radiation (Khan et al., 2000). Furthermore, the induction of ARF activates the ATM/ATR/CHK signaling pathways in response to genotoxic stresses and is required for the ability to induce p53 (Rocha et al., 2005; Eymin et al., 2006). ARF expression is also induced by interferons (IFNs) and after viral infection. These results reveal a novel role for ARF in viral infection surveillance (Sandoval et al., 2004; Garcia et al., 2006). These data demonstrate that ARF has a broader role in response to cellular stresses.
However, the mechanisms involved in ARF regulation in response to cellular stresses are not completely clear. Transcription is a regulatory component in the induction of ARF in some cases. As an example, ectopic expression of E2F1 in cancer cells could induce the transcription of ARF (Bates et al., 1998; Komori et al., 2005). Recently a specific E3 ligase for ARF called ubiquitin ligase for ARF (ULF) was identified. ARF is rapidly degraded by ULF in normal human cells. Oncogene activation such as that of c-Myc induces the stabilization of ARF by impairing the binding of ULF to ARF (Chen et al., 2010). Thus transcription-independent mechanisms are critically involved in ARF regulation during response to oncogenic stress. Here we report that the N-Myc and STATs interactor (NMI) is a novel ARF-interacting protein. By inhibiting the association of ULF and ARF, NMI promotes ARF stabilization in response to DNA damage, IFNs, and oncogenic stimuli. Our work suggests that ARF is subject to more dynamic controls than previously believed.
RESULTS
ARF interacts with NMI
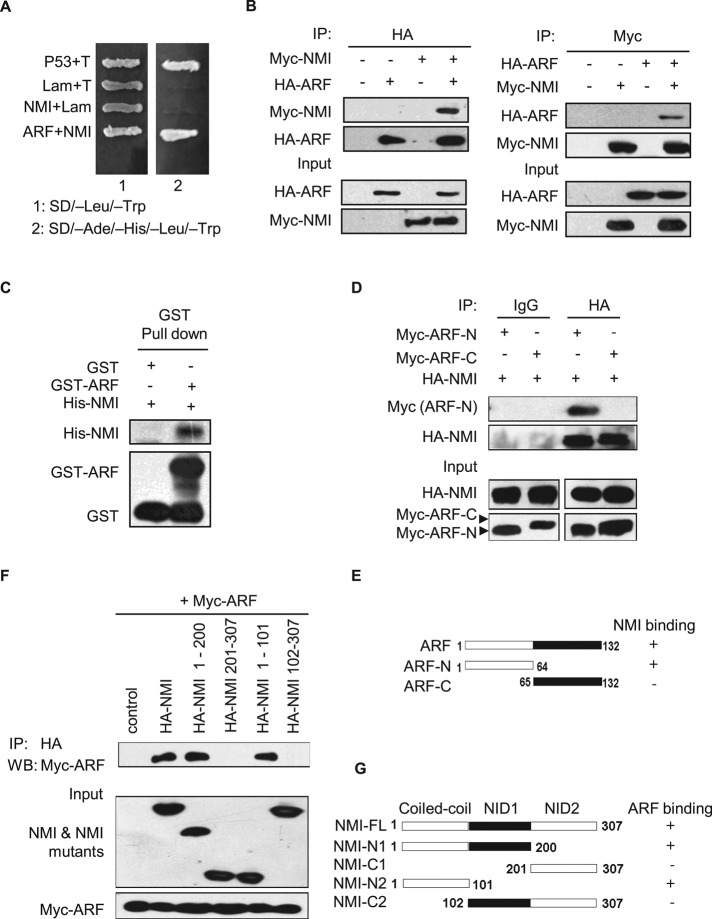
To better understand the regulation of ARF, we sought to identify novel ARF partners using a yeast two-hybrid screen. We screened a human bone marrow cDNA library with full-length human ARF as bait. From 2 × 106 yeast transformants screened, ∼100 putative positive clones were isolated. Among them, one clone encoding NMI, as confirmed by sequencing, was found to be a novel ARF-interacting protein (Figure 1A). Most of the other preys, such as COMMD1, p32, LZAP, p120E4F, and MIZ1, were previously reported to interact with ARF (Rizos et al., 2003; Wang et al., 2006; Huang et al., 2008; Itahana and Zhang, 2008; Herkert et al., 2010; Miao et al., 2010). Next the interaction between ARF and NMI in mammalian cells was verified by coimmunoprecipitation (CoIP) assay. As shown in Figure 1B, Myc-NMI can be observed in the hemagglutinin (HA)-ARF but not control immunoprecipitates. In a reciprocal coIP experiment, HA-ARF can be readily detected in the Myc-NMI immunoprecipitates. To investigate whether ARF directly interacts with NMI, we performed in vitro binding assays. In these binding assays we were able to consistently detect histidine (His)-NMI precipitated with glutathione S-transferase (GST)–ARF but not GST alone (Figure 1C). These data indicate that NMI and ARF can interact both in vivo and in vitro.
FIGURE 1:
ARF interacts with NMI. (A) ARF interacts with NMI in a yeast two-hybrid assay. AH109 yeast cells were cotransformed with the indicated plasmids and then plated on SD (minimal synthetic dropout medium)/–Leu/–Trp or SD/–Ade/–His/–Leu/–Trp selection media. Positive control pGBKT7-53 + pGADT7-T, negative control pGBKT7-Lam + pGADT7-T.(B) The 293T cells were cotransfected with the indicated plasmids, and cell lysates were immunoprecipitated with the indicated antibodies and immunoblotted with anti-HA or anti-Myc antibody. (C) In vitro binding assay. Purified His-NMI (2 μg) from E. coli BL21 was incubated with GST or GST-ARF bound to glutathione–Sepharose beads. Proteins retained on the beads were then blotted with the indicated antibodies. (D) Mapping of the ARF domains responsible for binding to NMI. The 293T cells were cotransfected with HA-NMI and the indicated truncated Myc-ARF mutants, and cell lysates were immunoprecipitated with anti-HA antibody and immunoblotted with anti-Myc antibody. (E) Schematic representation of ARF and ARF deletion mutants used in the experiment in D. +, interaction; –, no interaction. (F) Mapping of the NMI domains responsible for binding to ARF. The 293T cells were cotransfected with Myc-ARF and the truncated HA-NMI mutants as indicated, and cell lysates were immunoprecipitated with anti-HA antibody and then immunoblotted with anti-Myc antibody. (G) Schematic representation of NMI and NMI deletion mutants used in the experiment in F. +, interaction; –, no interaction.
To determine the region of ARF involved in the interaction with NMI, two deletion mutants of ARF were constructed (Figure 1E). As shown in Figure 1D, the N-terminal domain (amino acids 1–64) of ARF encoded by exon 1β was found to be sufficient for interaction with NMI, whereas the C-terminal domain (amino acids 65–132) showed no binding to NMI. The NMI protein consists of three major domains, with a coiled-coil domain in the N-terminal and two repeats of the NID domain in the C-terminal. To characterize the minimal region of NMI essential for ARF binding, we coexpressed various HA-tagged N-terminal and C-terminal truncation mutants of NMI with Myc-tagged ARF in 293T cells. Western blot analysis showed that the NMI N1 (1–200 amino acids [aa]) and NMI N2 (1–101 aa) mutants containing the coiled-coil domain retain the ability to coprecipitate Myc-tagged ARF, whereas the NMI C2 (102–307 aa) mutant containing both NID repeats and the NMI C1 (201–307 aa) mutant containing NID2 domain alone failed to complex with ARF (Figure 1F). These results indicate that amino acids 1–101 of NMI bearing the coiled-coil domain are necessary for its association with ARF (Figure 1G).
NMI regulates ARF turnover
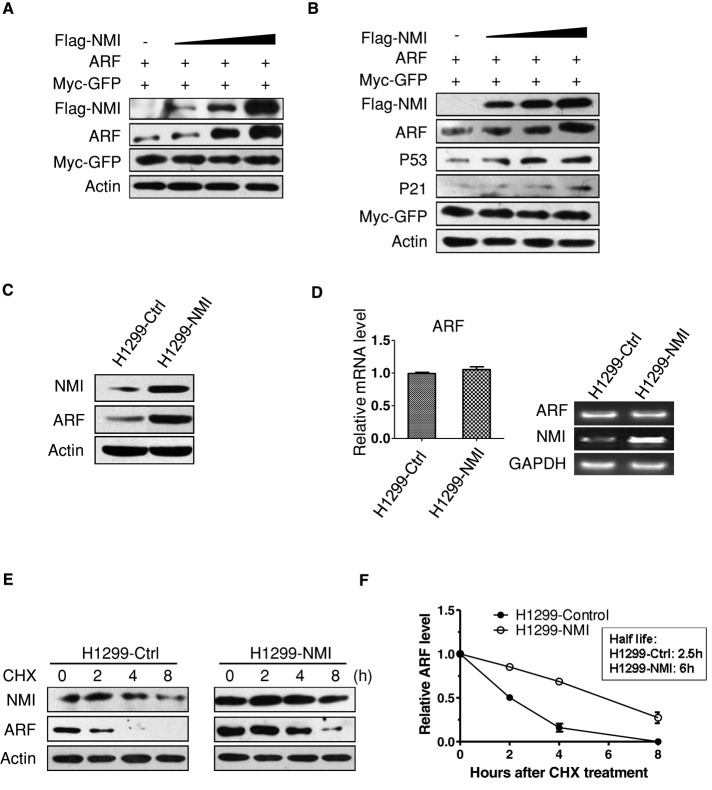
During our studies, we consistently observed that the level of ARF in cells transfected with NMI is higher than that in cells transfected with empty vector. Therefore we explored the possibility that NMI could regulate the abundance of ARF. To this end, we cotransfected 1 μg of ARF and increasing amounts of FLAG-NMI into 293T cells. As shown in Figure 2A, overexpression of NMI resulted in a dose-dependent increase in ARF levels. In p53-positive U2OS cells, we also found the accumulation of ARF with a concomitant increase in p53 and p21 levels (Figure 2B). Next we infected p53-null H1299 cells with retroviruses encoding NMI. Again, the endogenous ARF level was increased upon NMI expression (Figure 2C). This result indicates that NMI-mediated ARF induction is p53 independent. Quantitative real-time reverse transcription (RT)-PCR analysis revealed that ARF mRNA levels remain unchanged after NMI expression (Figure 2D), suggesting that the increased ARF levels observed in NMI-expressing cells might be due to enhanced ARF protein stability. Therefore we performed a protein half-life assay to examine the stability of ARF in H1299 cells expressing NMI. H1299-control and H1299-NMI cells were treated with cycloheximide (CHX) to block protein synthesis, and cell lysates were prepared at different times after CHX treatment. As shown in Figure 2, E and F, consistent with previous reports, the half-life of ARF is ∼3 h in H1299-control cells (Kuo et al., 2004; Peters et al., 2004; Pollice et al., 2004; di Tommaso et al., 2009). However, in H1299-NMI cells, the half-life of ARF was increased markedly, from ∼3 to 6 h. These results suggest that NMI regulates ARF stability.
FIGURE 2:
NMI regulates ARF turnover. (A, B) 293T (A) or U2OS (B) cells were cotransfected with 1 μg of ARF along with increasing amounts of FLAG-NMI. Cell lysates were prepared at 24 h posttransfection and analyzed for the indicated proteins. Myc-GFP expression was used as an internal control for transfection efficiency. Actin expression shows equal loading of samples. (C) Endogenous ARF levels in control and H1299-NMI cells analyzed by Western blotting with anti-ARF antibody. Actin was used as loading control. (D) Quantitative real-time RT-PCR analysis of ARF mRNA level. RNA was extracted from H1299-control and H1299-NMI cells. ARF mRNA was measured by quantitative RT-PCR, and levels relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA are shown. Results are the means ± SD of three independent experiments. (E) Half-life assay of endogenous ARF protein. H1299-control and H1299-NMI cells were treated with 30 μg/ml cycloheximide for the indicated durations, and protein levels were detected by Western blotting using anti-NMI, anti-ARF, and anti-actin antibodies. (F) Quantification of the experiment. The values shown are obtained from three independent experiments and are normalized to the actin control. For each experimental condition, the signal at the start of the experiment was set to 1.
NMI stabilizes ARF by blocking the ULF–ARF interaction
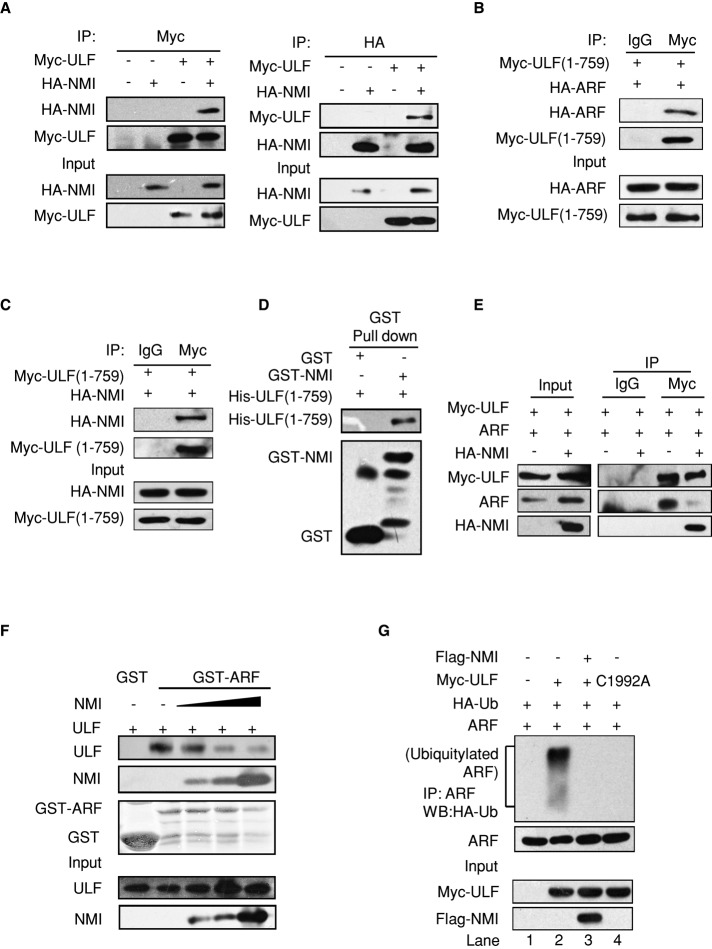
Recently ULF was identified as the specific E3 ubiquitin ligase for ARF (Chen et al., 2010). To investigate NMI-mediated regulation of ARF stability, we first examined whether NMI interacts with ULF. The 293T cells were cotransfected with expression vectors encoding HA-tagged NMI and Myc-tagged ULF proteins, and CoIP experiments were performed. As shown in Figure 3A, HA-NMI can be clearly observed in the Myc-ULF immunoprecipitates but not in the control immunoprecipitates. In a reciprocal coIP experiment, Myc-ULF can be readily detected in the HA-NMI immunoprecipitates. These data demonstrate that NMI interacts with ULF. A previous study reported that the N-terminal residues 1–759 of ULF are responsible for binding to ARF (Figure 3B; Chen et al., 2010). To investigate whether NMI binds to the same N-terminal region of ULF, we constructed the ULF (1–759) mutant. The 293T cells were transfected with HA-tagged NMI and Myc-tagged ULF (1–759), and the CoIP experiment was performed. As Figure 3C illustrates, HA-NMI was clearly detected in the Myc-ULF (1–759) immunoprecipitates, revealing that NMI binds to ULF (1–759). In subsequent in vitro binding assays with recombinant proteins, we demonstrated that NMI interacts directly with ULF (1–759) (Figure 3D). These results indicate that ULF interacts with NMI through its N-terminal residues 1–759. The observation that ULF interacts with NMI and ARF through the same N-terminal residues 1–759 of ULF suggested the possibility of a competition between NMI and ARF for binding to ULF. The prediction was that NMI would reduce the interaction between ULF and ARF. To analyze the effect of NMI on the ULF–ARF complex, we transiently transfected 293T cells with ULF and ARF together with NMI or empty vector, and performed a CoIP assay. As shown in Figure 3E, the expression of NMI caused a significant reduction in the amount of ULF-associated ARF. To further investigate the effect of NMI on the interaction between ULF and ARF, we performed a GST pull-down assay. 293T cells were transfected with constant amounts of ULF together with increasing amounts of NMI, and cell lysates were incubated with GST-ARF fusion protein in an in vitro binding assay. As shown in Figure 3F, with the increase of NMI, less ULF was coprecipitated with the recombinant ARF protein, indicating that NMI inhibits the ULF–ARF interaction.
FIGURE 3:
NMI inhibits the ULF–ARF interaction. (A) NMI binds to ULF. The 293T cells were transfected with the indicated plasmids, and cell lysates were immunoprecipitated with anti-Myc or anti-HA antibody and immunoblotted for the indicated proteins. (B) ARF interacts with ULF (1–759). The 293T cells were cotransfected with HA-ARF and the Myc-ULF (1–759) truncated mutant. Cell lysates were immunoprecipitated with anti-Myc antibody. Immunocomplexes were analyzed by Western blotting with anti-HA antibody. (C) NMI interacts with ULF (1–759). The 293T cells were cotransfected with HA-NMI and the Myc-ULF (1–759) truncated mutant. Cell lysates were immunoprecipitated with anti-Myc antibody and followed by Western blotting with anti-HA antibody. (D) Recombinant NMI binds to His-ULF (1–759) in vitro. A 2-μg amount of purified His-ULF (1–759) from E. coli BL21 were prepared and incubated with GST or GST-NMI immobilized on glutathione–Sepharose beads. Proteins retained on the beads were then blotted with the indicated antibodies. (E) NMI reduces the interaction between ULF and ARF. The 293T cells were transfected with the indicated combinations of expression vectors. Cell lysates were subject to immunoprecipitation with control immunoglobulin G or anti-Myc antibody. The immunoprecipitates were then blotted with the indicated antibodies. (F) The 293T cells were transfected with Myc-ULF and increasing amounts of Myc-NMI. Cells were lysed, and lysates were incubated with GST or GST-ARF Sepharose. Proteins retained on the beads were blotted with anti-Myc antibody. The levels of the GST-ARF fusion protein stained by Coomassie blue are shown. (G) NMI inhibits ULF-mediated ubiquitination of ARF. The 293T cells were transfected with the indicated combinations of expression vectors. At 24 h posttransfection, cells were treated with MG132 (5 μM) for 3 h and then lysed. Cell lysates were immunoprecipitated with anti-ARF antibody and immunoblotted with anti-HA or anti-ARF antibody.
These results raise the possibility that NMI might interfere with ULF-mediated ARF ubiquitination. To address this, we carried out an in vivo ubiquitination assay. Consistent with a previous report (Chen et al., 2010), ectopic expression of ULF induced the ubiquitination of ARF (Figure 3G, lane 2). As a control, expression of catalytically inactive ULF-C1992A did not induce ARF ubiquitination (Figure 3G, lane 4). Of interest, coexpression of NMI prevented the ubiquitination of ARF by ULF (Figure 3G, lane 3). Collectively, our data demonstrate that NMI promotes ARF stabilization by abrogating ULF-mediated ubiquitination by inhibiting the ULF–ARF interaction.
Oncogene activation, DNA damage, and IFNα stimulation induce NMI and ARF
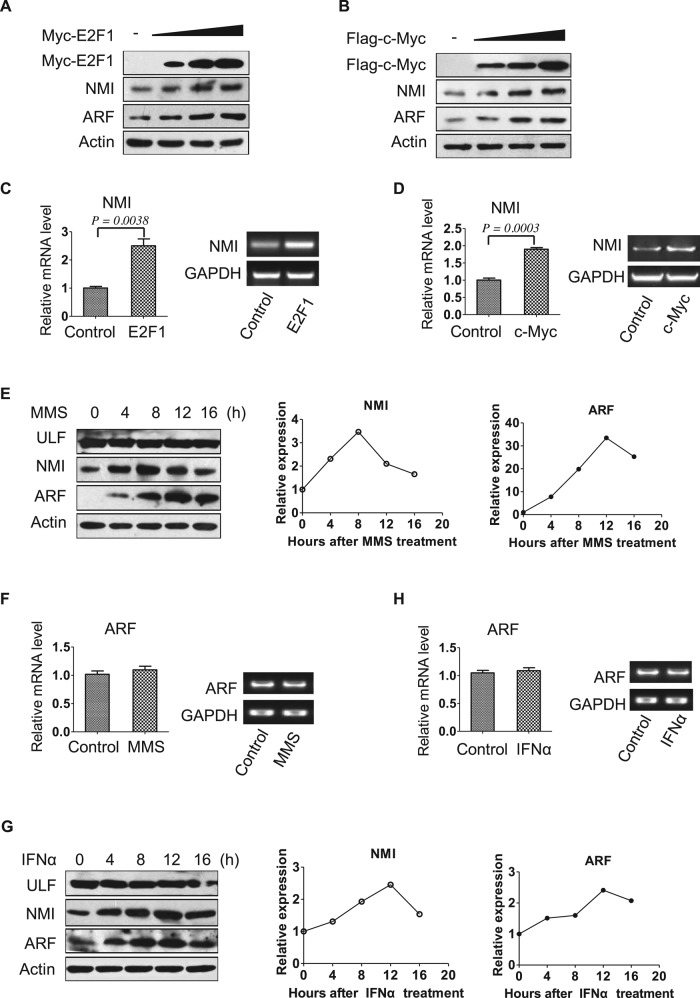
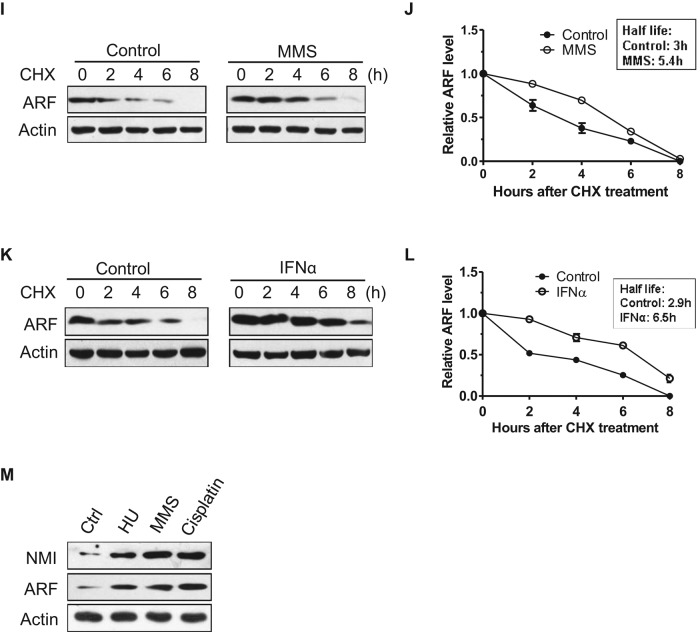
It has been demonstrated that ARF is induced by cellular stresses (Khan et al., 2000; Sandoval et al., 2004; Eymin et al., 2006; Garcia et al., 2006). We sought to determine whether NMI is involved in ARF induction in response to cellular stresses. To this end, we first examined whether oncogenic stress induces NMI. As shown in Figure 4, A and B, the levels of endogenous NMI and exogenous ARF proteins were up-regulated after c-Myc or E2F1 expression in 293 cells. Quantitative real-time RT-PCR analysis revealed that c-Myc or E2F1 expression induced NMI transcription (Figure 4, C and D). As to ARF induction, previous studies found that E2F1 could directly activate ARF transcription (Bates et al., 1998; Komori et al., 2005), but the use of exogenous ARF in our study suggests a posttranscriptional mechanism. Next, to investigate whether DNA damage induces NMI, we treated H1299 cells with the DNA damage agent methyl methanesulfonate (MMS) and harvested cell extracts at different times. Of interest, Figure 4E shows that after induction of DNA damage by MMS treatment, the levels of endogenous NMI increased, and, in agreement with a previous report (Eymin et al., 2006), this was accompanied by an increase in endogenous ARF levels. In contrast, DNA damage did not alter the levels of ULF (Figure 4E). Similar to what we found for oncogene activation and DNA damage, IFNα also induced endogenous NMI and ARF (Figure 4G), which is in accordance with previous studies (Zhou et al., 2000; Sandoval et al., 2004; Garcia et al., 2006). Quantitative real-time RT-PCR analysis revealed that ARF mRNA levels were unchanged after MMS or IFNα treatment (Figure 4, F and H). Examination of ARF stability demonstrated that the half-lives of ARF upon MMS and IFNα treatments increase from 3 to 5.4 and 6.5 h, respectively (Figure 4, I–L), in line with the results obtained in H1299-NMI cells (Figure 2F). The half-life of NMI is also extended after MMS and IFNα treatments (Supplemental Figure 1, A and B). Previous study showed that oncogenic stress such as c-Myc abrogates ULF-mediated ARF ubiquitination and induces transcription-independent accumulation of ARF protein (Chen et al., 2010). Our data showed that similar to oncogenic stress, MMS and IFNα treatments also up-regulate ARF levels through transcription-independent mechanisms. We also tested whether other DNA-damaging agents induce NMI. As shown in Figure 4M, cisplatin and hydroxyurea (HU) treatments clearly increased endogenous NMI and ARF levels in H1299 cells. Taken together, these data reveal that NMI is induced by various kinds of cellular stresses, and the induction of NMI is associated with a significant increase in ARF levels.
FIGURE 4:
Oncogene activation, DNA damage, and IFNα stimulation induce NMI and ARF. (A, B) Oncogenes induce NMI and ARF. The 293 cells were transfected with ARF and increasing amounts of Myc-E2F1 (A) or FLAG-c-Myc (B), and lysates prepared from cells 36 h after transfection were probed with anti-ARF or anti-NMI antibodies. (C, D) The 293 cells were transfected with Myc-E2F1 (C) or FLAG-c-Myc (D), and 36 h later, cells were collected and NMI mRNA was measured by quantitative real-time RT-PCR. Levels relative to GAPDH mRNA are shown. Results are the means ± SD of three independent experiments. (E, G) H1299 cells were treated with 0.2 mM MMS (E) or 1000 U/ml IFNα (G) for the indicated times, and lysates were probed for the indicated proteins. Quantification of the NMI and ARF protein levels relative to β-actin from the experiments are shown on the right. (F, H) H1299 cells were treated with 0.2 mM MMS or 1000 U/ml IFNα for 12 h. ARF mRNA was measured by quantitative RT-PCR, and levels relative to GAPDH mRNA are shown. (I, K) H1299 cells were treated with 0.2 mM MMS (I) or 1000 U/ml IFNα (K) for 6 h before the addition of CHX (30 μg/ml). Cells were harvested at the indicated times after CHX addition, and cell lysates were blotted with the indicated antibodies. (J, L) Quantification of the ARF protein levels relative to β-actin from the experiments shown in I and K. (M) NMI and ARF levels after treatment with DNA-damaging agents. H1299 cells were treated for 12 h with the indicated cytotoxic agents (0.2 mM MMS, 0.5 mM HU, and 5 μM cisplatin), and whole-cell extracts were subjected to Western blotting using anti-ARF or anti-NMI antibody.
NMI is required for ARF induction after oncogene activation and MMS and IFNα treatments
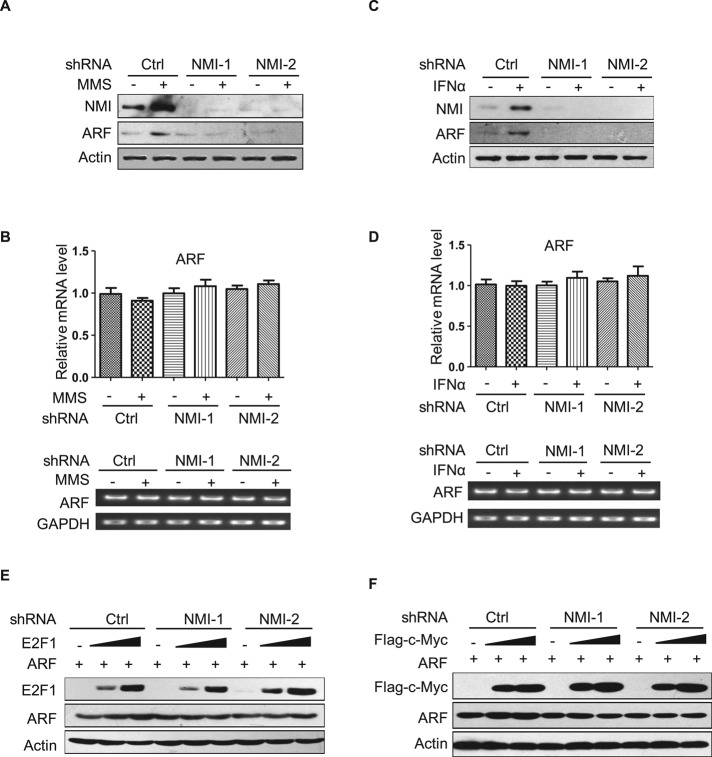
We next determined whether the inactivation of NMI has any effect on the induction of ARF after DNA damage or IFNα treatment. As shown in Figure 5, A and C, up-regulation of endogenous ARF expression was clearly observed in H1299 cells treated with MMS or IFNα. Under these conditions, neutralization of NMI with short hairpin RNA (shRNA) abolished the accumulation of endogenous ARF. To exclude off-target effects, we also treated H1299 cells with another NMI shRNA, NMI shRNA-2, which recognizes different region of the NMI mRNA; similar results were obtained (Figure 5, A and C). Quantitative RT-PCR analysis showed that loss of NMI did not affect the mRNA levels of ARF (Figure 5, B and D). As shown in Figure 5, E and F, NMI shRNA also prevented E2F1- and c-Myc–mediated up-regulation of exogenously expressed ARF in 293 cells.
FIGURE 5:
NMI is required for ARF induction after oncogene activation or MMS and IFNα treatments. (A, C) H1299 cells were infected with lentiviruses expressing either control or shRNA targeting NMI. After 48 h, cells were treated with 0.2 mM MMS (A) or 1000 U/ml IFNα (C) for 12 h. Cells were harvested, and lysates were prepared. The levels of endogenous NMI and ARF proteins were analyzed by Western blotting. (B, D) Expression of ARF mRNA by quantitative RT-PCR. H1299-control and H1299-NMI-shRNA cell lines were treated with 0.2 mM MMS (B) or 1000 U/ml IFNα (D) for 12 h, and mRNAs were extracted and subjected to quantitative RT-PCR. Quantification of ARF transcript levels is shown. (E, F) The 293 cells were infected with control viruses or those expressing NMI shRNA. At 48 h after infection, cells were transfected with ARF together with increasing amounts of Myc-E2F1 (E) or FLAG-c-Myc (F), and 24 h later, cells were harvested and lysates were probed with the indicated antibodies.
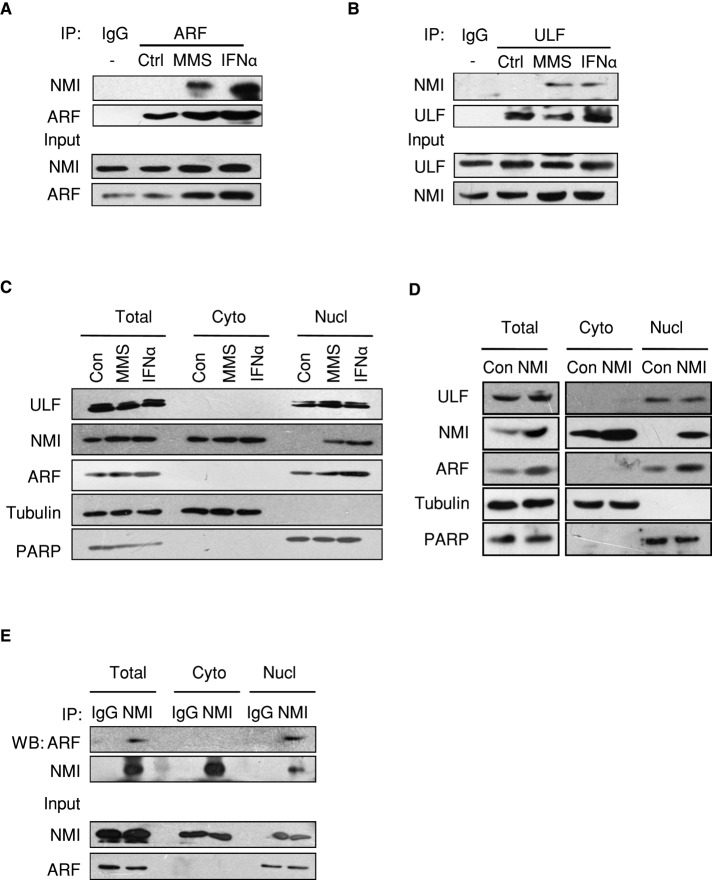
To further elucidate the role of NMI in ARF induction, we examined NMI–ARF and NMI–ULF complex formation in response to cellular stresses. H1299 cells were first treated with MMS or IFNα, and CoIP assays were then performed to assess changes in the NMI–ARF and NMI–ULF associations. As shown in Figure 6A, endogenous ARF was coprecipitated with endogenous NMI in cells treated with MMS or IFNα. We also found that endogenous NMI associated with ULF in MMS or IFNα-treated cells (Figure 6B). These results indicate that MMS and IFNα induce the endogenous NMI–ARF and NMI–ULF associations. Next we fractionated H1299-NMI cells and performed the CoIP assays with cell lysates from different cellular fractions. As shown in Figure 6E, the NMI–ARF association was detected in the nuclear fraction. Of interest, cellular stresses appear to induce NMI translocation from the cytoplasm to the nucleus. In unstressed cells, NMI is predominantly localized in the cytoplasm. On MMS or IFNα treatment, increased amounts of endogenous NMI were detected in the nucleus (Figure 6C). A portion of exogenously expressed NMI in the H1299-NMI cells also accumulated in the nucleus (Figure 6D). These results suggested that NMI translocates to the nucleus to interact with ARF and ULF in response to cellular stresses.
FIGURE 6:
NMI interacts with ARF and ULF in response to cellular stresses. (A) Endogenous NMI interacts with ARF after MMS and IFNα treatments. H1299 cells were treated with MMS or IFNα for 12 h. Cells were then harvested for a CoIP assay with control immunoglobulin G (IgG) or ARF antibody. (B) Endogenous NMI binds to ULF upon MMS and IFNα treatments. H1299 cells were treated as in A, and cell lysates were immunoprecipitated with control IgG or anti-ULF antibodies and immunoblotted with anti-NMI antibody. (C) Cellular stresses induce NMI nucleus translocation. H1299 cells were treated with MMS or IFNα for 12 h. Cells were then harvested and fractionated into nuclear and cytoplasmic fractions, and the fractions were immunoblotted with anti-NMI and anti-ARF antibodies. (D) Subcellular distribution of NMI and ARF in H1299-NMI cells. Nuclear or cytoplasmic fractions of H1299-control and H1299-NMI cells were analyzed by Western blotting with anti-NMI and anti-ARF antibodies. The cytoplasmic marker protein tubulin and the nuclear marker protein PAPR were used as controls. (E) NMI interacts with ARF in the nucleus. H1299-NMI cells were fractionated into nuclear and cytoplasmic fractions, and the fractions were immunoprecipitated with anti-NMI antibody and immunoblotted with anti-ARF or anti-NMI antibody.
We also treated H1299 cells with MMS and examined the nucleophosmin (NPM/B23)–ARF association, which is responsible for the stabilization of ARF in cancer cells. As shown in Supplemental Figure 2A, the amount of ARF-associated NPM decreased in MMS-treated cells, which is in accordance with previous work showing that DNA damage disrupts the NPM–ARF complex (Lee et al., 2005). Under these conditions, c-Myc rapidly decreased with elevation of NMI (Supplemental Figure 2B). This suggests that the enhanced stability of ARF is not due to NPM or c-Myc binding after MMS treatment.
Collectively, these findings provide evidence that NMI contributes to ARF accumulation after DNA damage, oncogene activation, and IFNα treatment.
NMI inhibits cell proliferation and induces G2/M arrest
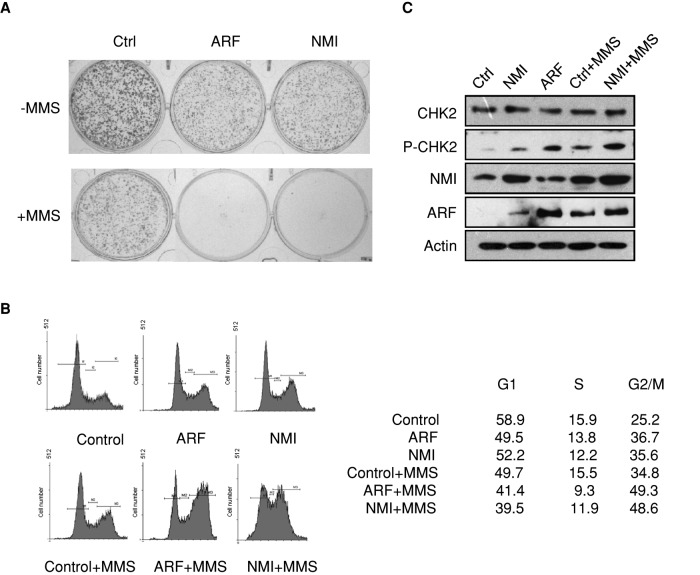
As a tumor suppressor, ARF induces p53-dependent and p53-independent cell growth arrest (Sherr, 2006). To further investigate NMI-mediated ARF regulation, we examined the effect of high levels of NMI on cell proliferation and survival. As shown in Figure 7A and Supplemental Figure 3, the rate of proliferation of H1299-NMI cells was reduced relative to that of normal H1299 cells, consistent with the presence of elevated levels of ARF in H1299-NMI cells (Figure 2C). Previously it was shown that ARF overexpression induced G2/M cell cycle arrest in H1299 cells (Chen et al., 2005; Eymin et al., 2006). Cell cycle analysis revealed that overexpression of NMI in H1299 cells increased the G2/M population in a manner reminiscent of ARF induction (Figure 7B). Recent work suggests that overexpression of ARF induces the activation of CHK2 and that activation of the ATM/CHK2 signaling pathway contributes to ARF-mediated G2/M arrest in p53-deficient cells (Eymin et al., 2006). In our study, we found that NMI up-regulates ARF levels. We then sought to determine whether NMI overexpression would lead to the activation of the ATM/CHK2 signaling pathway. As shown in Figure 7C, the overexpression of NMI did induce the activation of CHK2. Next we treated H1299-control, H1299-NMI, and H1299-ARF cells with MMS to induce DNA damage. MMS treatment induced ARF and activated the ATM/CHK2 signaling pathway, resulting in G2/M cell cycle arrest in H1299 cells (Figure 7, B and C). After MMS treatment, H1299-NMI and H1299-ARF cells exhibited enhanced G2/M cell cycle arrest compared with H1299-control cells (Figure 7B). Western blotting revealed that the enforced expression of NMI in H1299-NMI cells increased MMS-induced activation of the ATM/CHK2 signaling pathway. Consistent with the foregoing results, H1299-NMI and H1299-ARF cells did not recover in clonogenic assays after MMS treatment, in contrast to control cells (Figure 7A). Taken together, these data demonstrate that NMI inhibits cell proliferation and induces activation of the ATM/CHK2 signaling pathway.
FIGURE 7:
Overexpression of NMI inhibits cell proliferation and induces G2/M cell cycle arrest. (A) Colony-formation assay was performed with H1299-control, H1299-ARF, and H1299-NMI cells treated with or without 0.2 mM MMS. In total, 500 cells were plated for each assay. Cells were stained with crystal violet after 10 d. (B) Fluorescence-activated cell sorting analysis. H1299-control, H1299-ARF, and H1299-NMI cells were treated with or without 0.2 mM MMS as indicated, and 12 h later, cells were harvested and stained with propidium iodide, and the cell cycle distribution was determined by flow cytometry. The percentage of cells in various phases of the cell cycle is shown. (C) Expression of NMI induces the activation of CHK2 kinase. H1299-control and H1299-NMI cells were treated with 0.2 mM MMS for 12 h or left untreated, and cell lysates were blotted with the indicated antibodies.
DISCUSSION
Until recently, ARF induction was believed to be mediated mainly at the transcriptional level. The promoter of the ARF gene contains an E2Fs binding site. E2F1 could directly activate ARF transcription in response to some oncogenic signals (Bates et al., 1998; Komori et al., 2005).
However, the mechanisms underlying ARF regulation now appear to be more complex than earlier conceptualized. An increasing number of studies are providing evidence that the expression of ARF is regulated at both the transcriptional and posttranslational levels. It has been demonstrated that although it lacks lysine, ARF protein can undergo N-terminal ubiquitination and is degraded by the proteasome (Kuo et al., 2004). Furthermore, several studies have shown that the stability of ARF is tightly regulated by NPM. In human tumor cells the majority of ARF protein resides within the nucleolus in a complex with NPM. The NPM–ARF interaction sequesters ARF in the nucleolus, thus preventing its nucleoplasmic degradation (Lindstrom et al., 2000; Itahana et al., 2003; Bertwistle et al., 2004; Kuo et al., 2004; Weber et al., 2004; Colombo et al., 2005; Chen et al., 2010). Recently the newly identified E3 ubiquitin ligase ULF was shown to interact with ARF and induce the lysine-independent ubiquitination and degradation of ARF. Of interest, NPM and c-Myc can promote ARF stabilization in cancer cells by abrogating ULF-mediated ARF ubiquitination. These findings suggest that ARF is also regulated through transcription-independent mechanisms during responses to oncogenic stress (Chen et al., 2010).
In this study, we provide new evidence that ARF regulation is under more dynamic control in response to cellular stresses. We show that 1) NMI is a novel ARF- and ULF-interacting protein; 2) NMI stabilizes ARF by blocking the ULF–ARF interaction and inhibiting ULF-mediated ARF ubiquitination and degradation; 3) MMS and IFNα treatments and oncogene activation such as c-Myc and E2F1 induce the expression of NMI and ARF; 4) neutralization of NMI abolishes the accumulation of ARF after MMS and IFNα treatments; 5) cellular stresses induce NMI nucleus translocation and NMI–ARF and NMI–ULF interactions; and 6) overexpression of NMI induces G2/M arrest in a manner reminiscent of ARF induction. These results reveal a novel mechanism of ARF regulation in response to certain types of DNA damage, oncogene activation, and IFNα stimulus.
Although NPM and c-Myc are involved in ARF stabilization by abrogating ULF-mediated ARF ubiquitination in response to oncogenic stress (Chen et al., 2010), the stabilization of ARF in response to genotoxic stress is more complicated. First, DNA damage triggers an immediate disruption of ARF's nucleolar interactions with NPM (Supplemental Figure 2A; Lee et al., 2005). Second, ARF redistributes transiently from the nucleolus to the nucleoplasm after DNA damage (Lindstrom et al., 2000; Lee et al., 2005; Huang et al., 2008). Third, c-Myc protein is degraded after DNA damage (Supplemental Figure 2B; Jiang et al., 2003; Britton et al., 2008). These studies raise a critical question: How is ARF stabilized in cells treated with DNA damage agents when ARF's nucleolar interactions with NPM are disrupted and c-Myc is degraded? We propose a model in which NMI stabilizes ARF in response to DNA damage to regulate cellular proliferation and apoptosis. In unstressed cancer cells, the stabilization of ARF is maintained by NPM by keeping ARF away from its nucleoplasmic E3 ubiquitin ligase, ULF. In addition, overexpressed c-Myc blocks the interaction between ULF and ARF and thereby promotes ARF stabilization (Chen et al., 2010). After DNA damage ARF's nucleolar interactions with NPM are disrupted, which enables ARF to redistribute to the nucleoplasm (Lee et al., 2005). In the nucleoplasm, NMI translocated from the cytoplasm might directly stabilize ARF by forming complexes with the free ARF released from the nucleolus. NMI can also indirectly stabilize ARF by interacting with ULF, which blocks the association of ULF and ARF (Figure 3E). Collectively, our work on NMI implicates a novel NPM- and c-Myc–independent pathway, the NMI/ULF/ARF pathway, for ARF stabilization in response to DNA damage and other cellular stresses.
NMI is an IFN-inducible protein that interacts with a variety of proteins, such as c-Myc, TIP60, some STAT family members, and BRCA1, which play important roles in tumorigenesis (Zhou et al., 2000; Bao and Zervos, 1996; Li et al., 2002; Zhang et al., 2007). Among them, TIP60, c-Myc, and BRCA1 are also ARF-binding proteins (Datta et al., 2004; Qi et al., 2004; Eymin et al., 2006; He et al., 2008). However, the function of NMI, particularly its potential role in tumorigenesis, has not been well characterized. One report showed that overexpression of NMI inhibits the Wnt/β-catenin signaling pathway by up-regulation of DKK1 and retards tumor cell growth (Fillmore et al., 2009). Other studies showed that NMI is expressed at high levels in certain myeloid leukemias, with c-Myc overexpression, and in some pancreatic ductal adenocarcinomas (Bao and Zervos, 1996; Lebrun et al., 1998; Chung et al., 2008). On the basis of our result that overexpression of c-Myc and E2F1 induces the expression of NMI and ARF (Figure 4, A and B), the high levels of NMI expression observed in some tumor cells might be the consequence of oncogene activation. We found that NMI is induced after DNA damage and oncogene activation and inhibits tumor cell proliferation by stabilizing ARF. These results suggest that NMI might play an important role in tumorigenesis. At present the underlying mechanism controlling NMI induction after DNA damage is not clear. Previous work showed that ultraviolet light induces the formation of Tip60–NMI complex in Jurkat cells, and the stability of NMI can be enhanced by its association with Tip60 (Zhang et al., 2007), which might contribute to the induction of NMI in response to DNA damage. Future studies are needed to test this hypothesis in a physiological or pathological setting.
In conclusion, in this study we found that NMI is up-regulated after DNA damage and oncogene activation and it acts to stabilize ARF by inhibiting the interaction between ULF and ARF. Our work reveals that NMI plays an important role in transcription-independent ARF regulation in response to cellular stresses.
MATERIALS AND METHODS
Yeast two-hybrid screen
PGBKT7-p14ARF was used to screen a human bone marrow MATCHmaker cDNA library cloned into pACT2 (Clontech, Mountain View, CA) according to the manufacturer's instructions. Briefly, the yeast strain AH109 was sequentially transformed with the pGBKT7-p14ARF vector and the library. An estimated 106 transformants were screened. Yeasts containing interacting proteins were identified by growth on selective media lacking leucine, tryptophan, and histidine and confirmed by β-galactosidase activity. Plasmids harboring interacting cDNAs were recovered from yeast by reintroduction into Escherichia coli, and the identities of putative interacting proteins were determined by sequencing and BLAST search of the National Center for Biotechnology Information database.
Expression vectors
Full-length ULF and E2F1 were amplified by PCR and subcloned into pCMV-Myc vector (Clontech). The pCMV-Myc-ULF (C1992A) mutant was generated with a QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA). FLAG-tagged c-Myc was PCR amplified and cloned into pCMV-Tag2 vector (Stratagene). PGBKT7-p14ARF was kindly provided by Hoi-Yeung Li. To construct the different ARF expression vectors, full-length ARF was amplified by PCR from the pGBKT7-p14ARF vector and cloned into pcDNA3.0, pCMV-Myc/HA, pBABE-puro, and pGEX4T vectors. For the NMI expression constructs, full-length NMI was amplified by PCR and subcloned into pCMV-Myc/HA, pCMV-Tag2, and pBABE-puro vectors. ARF and NMI deletion mutants were constructed by PCR and inserted into their respective expression vectors. All constructs derived from PCR products were verified by DNA sequencing.
Cell culture, transient transfection, and treatments
Human H1299, U2OS, HEK293, and 293T cells were cultured in DMEM (Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT) and 100 μg/ml penicillin and streptomycin. Transient transfections were carried out using a standard calcium phosphate method. MMS, HU, cisplatin, MG132, and cycloheximide were purchased from Sigma-Aldrich (St. Louis, MO) and added to subconfluent cells at the indicated doses. IFNα was from the Chinese Academy of Sciences, Biochemistry (Shanghai, China).
RNA interference
For RNA interference experiments we used a lentivirus-based vector, pLL3.7. Oligonucleotides targeting NMI (NMI shRNA-1, 5′-GGAGCATTCGCCAGATGAA-3′; NMI shRNA-2, 5′- GAGGACAGTGCTTCTGACA-3′) were cloned into the pLL3.7 vector. Recombinant lentiviral plasmids were cotransfected into 293T cells with the packaging plasmids VSV-G, RSV-REV, and pMDL, and after 48 h the viral supernatants were used to infect target cells in the presence of 6 μg/ml polybrene (Sigma-Aldrich).
Western blotting and immunoprecipitation
Total cell extracts were prepared in cell lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM NaF, 20 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg/ml leupeptin, 2 μg/ml aprotinin, 1% Triton X-100, and 1 mM EDTA) for immunoprecipitations. Immunocomplexes were resolved by SDS–PAGE, and Western blotting was performed with the following antibodies: anti-FLAG M2 and anti-actin monoclonal antibodies (Sigma-Aldrich); HA (Y-11), ULF/TRIP12 (E-14), NMI (N-16), Chk2 (H-300), and pChk2 (T68) polyclonal antibodies and anti-Myc (9E10) and p14ARF (DSC-240) monoclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA); anti-p14ARF (4C6/4) monoclonal antibody (Abcam, Cambridge, MA); anti-NPM monoclonal antibody (kindly provided by Q. F. Li, Xiamen University, Xiamen, China); and horseradish peroxidase–conjugated goat anti-mouse and anti-rabbit (Thermo Scientific, Waltham, MA).
Quantitative real-time RT-PCR
Total RNA was isolated from cells using RNA Simple Total RNA Kit (Tiangen Biotech, Beijing, China), and first-strand cDNA was synthesized from l μg of total RNA using the First-Strand cDNA Synthesis kit (Toyobo, Osaka, Japan) following the manufacturer's instructions. Prepared cDNA samples were amplified and analyzed by quantitative real-time RT-PCR with Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) using ABI 7500 Real Time PCR System (Applied Biosystems). Primer sequences were as follows: human ARF, 5′-GTGCGCAGGTTCTTGGTGACC-3′ (forward) and 5′-T CAGCCAGGTCCACGGGCAG-3′ (reverse); human GAPDH, 5′-GACATCAAGAAGGTGGTGAA-3′ (forward) and 5′-TGTCATACCAGGAAATGAGC-3′ (reverse).
Flow cytometry
Cells were washed twice in phosphate-buffered saline (PBS) and fixed in ice-cold 70% ethanol overnight. After two 1× PBS washes, the fixed cells were incubated at 37°C for 30 min with 100 μg/ml of RNase A and stained with propidium iodide (10 μg /ml in PBS) at 4°C overnight. DNA content was analyzed with a FACS Diva Flow Cytometer (Beckman Coulter, Brea, CA).
GST pull-down assay
GST-ARF and GST were expressed in E. coli and purified on glutathione–Sepharose (GE Healthcare, Piscataway, NJ). Lysates from 293T cells transiently transfected with pCMV-Myc-ULF and increasing amount of pCMV-Myc-NMI were prepared and incubated with GST or GST-ARF immobilized on glutathione–Sepharose beads. The beads were washed five times with binding buffer and 1× SDS loading buffer added and analyzed by Western blotting using anti-Myc (9E10) antibody.
In vivo ubiquitination assay
The 293T cells were cotransfected with expression vectors of ARF, NMI, HA-ubiquitin, ULF, or ULF (C1992A), as indicated. At 36 h after transfection, cells were treated with MG132 (5 μM) for 3 h and then lysed with RIPA buffer (0.2% SDS, 0.5% sodium deoxycolate, 0.5% Nonidet P-40, 10 mM NaF, 20 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM PMSF, 10 μg/ml leupeptin, and 2 μg/ml aprotinin). Ubiquitinated ARF was immunoprecipitated with anti-ARF antibody and then Western blot analyzed with anti-HA antibody (against HA-ubiquitin).
Cellular fractionation
Cells were washed with ice-cold PBS (pH 7.4) and resuspended in buffer A containing 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol, and 1 mM PMSF. Cells were incubated on ice for 10 min, and then 0.5% final concentration of NP-40 was added. Cell lysates were centrifuged at 15,000 × g for 15 min. The resulting supernatants were retained as the cytoplasmic fraction. The pellets were washed three times with buffer A and lysed in cell lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM NaF, 20 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM PMSF, 10 μg/ml leupeptin, 2 μg/ml aprotinin, 1% Triton X-100, and 1 mM EDTA). The lysates were then centrifuged at 3000 × g for 10 min, and the supernatants containing nuclear proteins were recovered.
Supplementary Material
Acknowledgments
We thank all the members of the S. Q. Zhang laboratory for their help and assistance. We also thank Hoi-Yeung Li for the pGBKT7-ARF construct, Q. Wu for the pCMV-HA-ubiquitin expression plasmid, and Q. F. Li for the anti-NPM antibody. This work was supported by grants from the National Natural Science Foundation of China (30971489), the Natural Science Foundation of Fujian Province of China (2008J0111), the 973 program (2007CB914602), the Science Planning Program of Fujian Province (2009J1010), the 111 Project (B06016), and the Program of Introducing Talents of Discipline to Universities (B12001).
Abbreviations used:
- ARF
alternative reading frame
- ATM
ataxia-telangiectasia-mutated kinase
- ATR
ATM and Rad3-related
- CHK
checkpoint kinase
- E2F
E2F transcription factor 1
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- HA
hemagglutinin
- HU
hydroxyurea
- IFN
interferon
- MDM2
murine double minute2
- MEF
mouse embryonic fibroblast
- MMS
methyl methanesulfonate
- Myc
myelocytomatosis oncogene
- NID
Nmi/IFP 35 domain
- NMI
N-Myc and STATs interactor
- NPM/B23
nucleophosmin
- RT-PCR
reverse transcription-PCR
- shRNA
short hairpin RNA
- STATs
signal transducers and activators of transcription
- ULF
ubiqutin ligase for ARF
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-04-0304) on October 3, 2012.
*These authors contributed equally to this work.
The authors declare no conflict of interest.
REFERENCES
- Bao J, Zervos AS. Isolation and characterization of Nmi, a novel partner of Myc proteins. Oncogene. 1996;12:2171–2176. [PubMed] [Google Scholar]
- Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton S, Salles B, Calsou P. c-MYC protein is degraded in response to UV irradiation. Cell Cycle. 2008;7:63–70. doi: 10.4161/cc.7.1.5111. [DOI] [PubMed] [Google Scholar]
- Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Chen D, Shan J, Zhu WG, Qin J, Gu W. Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature. 2010;464:624–627. doi: 10.1038/nature08820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JC, Oh MJ, Choi SH, Bae CD. Proteomic analysis to identify biomarker proteins in pancreatic ductal adenocarcinoma. ANZ J Surg. 2008;78:245–251. doi: 10.1111/j.1445-2197.2008.04429.x. [DOI] [PubMed] [Google Scholar]
- Colombo E, Bonetti P, Lazzerini Denchi E, Martinelli P, Zamponi R, Marine JC, Helin K, Falini B, Pelicci PG. Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol Cell Biol. 2005;25:8874–8886. doi: 10.1128/MCB.25.20.8874-8886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Nag A, Pan W, Hay N, Gartel AL, Colamonici O, Mori Y, Raychaudhuri P. Myc-ARF (alternate reading frame) interaction inhibits the functions of Myc. J Biol Chem. 2004;279:36698–36707. doi: 10.1074/jbc.M312305200. [DOI] [PubMed] [Google Scholar]
- de Stanchina E, et al. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Tommaso A, Hagen J, Tompkins V, Muniz V, Dudakovic A, Kitzis A, Ladeveze V, Quelle DE. Residues in the alternative reading frame tumor suppressor that influence its stability and p53-independent activities. Exp Cell Res. 2009;315:1326–1335. doi: 10.1016/j.yexcr.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymin B, Claverie P, Salon C, Leduc C, Col E, Brambilla E, Khochbin S, Gazzeri S. p14ARF activates a Tip60-dependent and p53-independent ATM/ATR/CHK pathway in response to genotoxic stress. Mol Cell Biol. 2006;26:4339–4350. doi: 10.1128/MCB.02240-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillmore RA, Mitra A, Xi Y, Ju J, Scammell J, Shevde LA, Samant RS. Nmi (N-Myc interactor) inhibits Wnt/beta-catenin signaling and retards tumor growth. Int J Cancer. 2009;125:556–564. doi: 10.1002/ijc.24276. [DOI] [PubMed] [Google Scholar]
- Garcia MA, Collado M, Munoz-Fontela C, Matheu A, Marcos-Villar L, Arroyo J, Esteban M, Serrano M, Rivas C. Antiviral action of the tumor suppressor ARF. EMBO J. 2006;25:4284–4292. doi: 10.1038/sj.emboj.7601302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Fan C, Ning X, Feng X, Liu Y, Chen B, Tang D. Interaction of p14ARF with Brca1 in cancer cell lines and primary breast cancer. Cell Biol Int. 2008;32:1302–1309. doi: 10.1016/j.cellbi.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Herkert B, et al. The Arf tumor suppressor protein inhibits Miz1 to suppress cell adhesion and induce apoptosis. J Cell Biol. 2010;188:905–918. doi: 10.1083/jcb.200908103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wu M, Li HY. Tumor suppressor ARF promotes non-classic proteasome-independent polyubiquitination of COMMD1. J Biol Chem. 2008;283:11453–11460. doi: 10.1074/jbc.M708544200. [DOI] [PubMed] [Google Scholar]
- Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- Itahana K, Zhang Y. Mitochondrial p32 is a critical mediator of ARF-induced apoptosis. Cancer Cell. 2008;13:542–553. doi: 10.1016/j.ccr.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang MR, Li YC, Yang Y, Wu JR. c-Myc degradation induced by DNA damage results in apoptosis of CHO cells. Oncogene. 2003;22:3252–3259. doi: 10.1038/sj.onc.1206501. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Bodner S, van de Kamp E, Randle DH, Sherr CJ. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59:2217–2222. [PubMed] [Google Scholar]
- Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SH, Moritsugu J, Wahl GM. Differential requirement for p19ARF in the p53-dependent arrest induced by DNA damage, microtubule disruption, and ribonucleotide depletion. Proc Natl Acad Sci USA. 2000;97:3266–3271. doi: 10.1073/pnas.050560997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori H, Enomoto M, Nakamura M, Iwanaga R, Ohtani K. Distinct E2F-mediated transcriptional program regulates p14ARF gene expression. EMBO J. 2005;24:3724–3736. doi: 10.1038/sj.emboj.7600836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar C, Zhao L, Modestou M, Quelle DE. ARF function does not require p53 stabilization or Mdm2 relocalization. Mol Cell Biol. 2002;22:196–206. doi: 10.1128/MCB.22.1.196-206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo ML, den Besten W, Bertwistle D, Roussel MF, Sherr CJ. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 2004;18:1862–1874. doi: 10.1101/gad.1213904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun SJ, Shpall RL, Naumovski L. Interferon-induced upregulation and cytoplasmic localization of Myc-interacting protein Nmi. J Interferon Cytokine Res. 1998;18:767–771. doi: 10.1089/jir.1998.18.767. [DOI] [PubMed] [Google Scholar]
- Lee C, Smith BA, Bandyopadhyay K, Gjerset RA. DNA damage disrupts the p14ARF-B23(nucleophosmin) interaction and triggers a transient subnuclear redistribution of p14ARF. Cancer Res. 2005;65:9834–9842. doi: 10.1158/0008-5472.CAN-05-1759. [DOI] [PubMed] [Google Scholar]
- Li H, Lee TH, Avraham H. A novel tricomplex of BRCA1, Nmi, and c-Myc inhibits c-Myc-induced human telomerase reverse transcriptase gene (hTERT) promoter activity in breast cancer. J Biol Chem. 2002;277:20965–20973. doi: 10.1074/jbc.M112231200. [DOI] [PubMed] [Google Scholar]
- Lindstrom MS, Klangby U, Inoue R, Pisa P, Wiman KG, Asker CE. Immunolocalization of human p14(ARF) to the granular component of the interphase nucleolus. Exp Cell Res. 2000;256:400–410. doi: 10.1006/excr.2000.4854. [DOI] [PubMed] [Google Scholar]
- Llanos S, Clark PA, Rowe J, Peters G. Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat Cell Biol. 2001;3:445–452. doi: 10.1038/35074506. [DOI] [PubMed] [Google Scholar]
- Miao L, Song Z, Jin L, Zhu YM, Wen LP, Wu M. ARF antagonizes the ability of Miz-1 to inhibit p53-mediated transactivation. Oncogene. 2010;29:711–722. doi: 10.1038/onc.2009.372. [DOI] [PubMed] [Google Scholar]
- Palmero I, Pantoja C, Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- Peters G, Rodway H, Llanos S, Rowe J. Stability of nucleolar versus non-nucleolar forms of human p14(ARF) Oncogene. 2004;23:6186–6192. doi: 10.1038/sj.onc.1207854. [DOI] [PubMed] [Google Scholar]
- Pollice A, Nasti V, Ronca R, Vivo M, Lo Iacono M, Calogero R, Calabro V, La Mantia G. Functional and physical interaction of the human ARF tumor suppressor with Tat-binding protein-1. J Biol Chem. 2004;279:6345–6353. doi: 10.1074/jbc.M310957200. [DOI] [PubMed] [Google Scholar]
- Pomerantz J, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature. 2004;431:712–717. doi: 10.1038/nature02958. [DOI] [PubMed] [Google Scholar]
- Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Radfar A, Unnikrishnan I, Lee HW, DePinho RA, Rosenberg N. p19(Arf) induces p53-dependent apoptosis during Abelson virus-mediated pre-B cell transformation. Proc Natl Acad Sci USA. 1998;95:13194–13199. doi: 10.1073/pnas.95.22.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizos H, Diefenbach E, Badhwar P, Woodruff S, Becker TM, Rooney RJ, Kefford RF. Association of p14ARF with the p120E4F transcriptional repressor enhances cell cycle inhibition. J Biol Chem. 2003;278:4981–4989. doi: 10.1074/jbc.M210978200. [DOI] [PubMed] [Google Scholar]
- Rocha S, Garrett MD, Campbell KJ, Schumm K, Perkins ND. Regulation of NF-kappaB and p53 through activation of ATR and Chk1 by the ARF tumour suppressor. EMBO J. 2005;24:1157–1169. doi: 10.1038/sj.emboj.7600608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval R, Xue J, Pilkinton M, Salvi D, Kiyokawa H, Colamonici OR. Different requirements for the cytostatic and apoptotic effects of type I interferons. Induction of apoptosis requires ARF but not p53 in osteosarcoma cell lines. J Biol Chem. 2004;279:32275–32280. doi: 10.1074/jbc.M313830200. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- Stott FJ, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci USA. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, He X, Luo Y, Yarbrough WG. A novel ARF-binding protein (LZAP) alters ARF regulation of HDM2. Biochem J. 2006;393:489–501. doi: 10.1042/BJ20050960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Brady SN, Yu Y, Maggi LB. ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Mol Cell Biol. 2004;24:9327–9338. doi: 10.1128/MCB.24.21.9327-9338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Zhang K, Zheng G, Yang YC. Stability of Nmi protein is controlled by its association with Tip60. Mol Cell Biochem. 2007;303:1–8. doi: 10.1007/s11010-007-9449-y. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- Zhou X, Liao J, Meyerdierks A, Feng L, Naumovski L, Bottger EC, Omary MB. Interferon-alpha induces nmi-IFP35 heterodimeric complex formation that is affected by the phosphorylation of IFP35. J Biol Chem. 2000;275:21364–21371. doi: 10.1074/jbc.M003177200. [DOI] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.