Abstract
Paracoccidioidomycosis (PCM) cannot always be diagnosed by conventional means such as direct examination of histopathology or clinical samples, and serological methods, used as an alternative, still have many cases of cross-reactivity. In this scenario, molecular techniques seem to arise as a rapid approach, specific and direct that could be used in the diagnosis of this mycosis. In this study we analyzed 76 serum samples from patients in southern Bahia suspected of having paracoccidioidomycosis using a conventional PCR with primers for the ITS1 ribosomal DNA of P. brasiliensis. Of these 76 patients, 5 were positive for PCM by double immunodiffusion and/or direct examination and histopathology. To test specificity of PCR, we used human DNA and three isolates of P. lutzii (1578, 01 and ED01). Additionally, we analyzed by serial dilutions of DNA the limit of detection of the assay. The test of PCR proved specific, as only a 144 bp fragment of the three isolates of P. lutzii and no human DNA was amplified. Detection limit was 1.1 pg/µL of DNA. Despite the high detection limit and specificity of PCR none of the 76 serum samples were found positive by PCR, but a biopsy specimen obtained from one of the patients with PCM was positive. These results, albeit limited, show that PCR is not effective in detecting DNA of P. brasiliensis or P. lutzii in serum, but could perhaps be used with other types of clinical samples, especially in those instances in which conventional methods fail.
Author Summary
Paracoccidioidomycosis (PCM) has been included in such diseases neglected since this impact on public health have not been measured. Except in the South and Southeast of Brazil, there are no government programs for this mycosis. After 100 years of discovery of the disease there is the need to deploy an effective and continuous program for the prevention, diagnosis and treatment. PCM requires a prolonged treatment, generally greater than 1 year. Moreover, the patients with PCM are associated with other co morbidities such as alcoholism, smoking and malnutrition.
To aggravate this scenario the serological diagnosis of PCM in many cases results in cross reactivity with other mycoses. Thus, this study aimed to develop a PCR which can be used in association with the serological techniques to improve the diagnostic these individuals. We work with plasma samples from patients in northeastern Brazil, in an area where the disease has not been reported. None of the 76 plasma samples were positive by PCR, but a biopsy specimen obtained from a patient was positive. These results reinforce that PCR have the limitation when serum or plasma is used. However, PCR can be an important diagnostic tool when conventional diagnostic methods are not successful.
Introduction
Paracoccidioidomycosis (PCM) is a deep mycosis caused by the thermo-dimorphic fungus Paracoccidioides brasiliensis, endemic in some countries of Latin America, mainly in Brazil [1]. In this disease, the fungus can remain confined in the lungs, the primary focus of infection, or spread to other organs and tissues, resulting in different clinical manifestations. The form acute and sub acute occurs in young people of both sexes, mainly affecting the reticuloendothelial system. The chronic form of PCM predominates in adult males and is characterized by presence of the fungus restricted in the lungs and/or disseminated to the mucosa, skin and lymph nodes [2].
The conventional diagnosis of PCM is based on viewing and/or isolation of the fungus in clinical specimens. However, the specimen may not always be viewed and microbiological culture is time-consuming and mostly negative [3]. Serological techniques have been employed, but there are still many cases of cross-reactivity with other fungal species [4]. In addition, false-negative results can be obtained in cases where the patient has some type of immunodeficiency [5]. Nevertheless, the scene of the PCM have been changed since the discovery of a new species, P. lutzii, which has very distinct behaviors of P. brasiliensis [6].
Therefore, a molecular approach appears to be an excellent alternative in the diagnosis of PCM. However, so far this technique has not been used routinely in the diagnosis of PCM. To this end specific primers for conserved regions have been developed, i.e., for detection of 18S, 5.8S and 28S rDNA and their regions ITS1 and ITS2, of the gp43 gene and an antigenic protein of 27 kDa [7], [8], [9], [10]. Although the gp43 gene is one of genes used in the classification of cryptic species [11] and some studies show a relationship between polymorphisms of these gene and pathogenicity [9], we believe that perhaps not all isolates of P. brasiliensis or P. lutzii have the gp43 gene.
On the other hand, ribosomal DNA is present in all isolates and there are conserved regions within this structure, thus primers for these regions would be more appropriate in terms of sensitivity and specificity [12]. Thus, we aim to develop a conventional PCR using a pair of primers specific for the known ITS1 region of ribosomal DNA of P. brasiliensis [13], and know what is the viability of this PCR in serum samples. We also made an effort to determine whether this PCR assay could be used to detect DNA of P. lutzii.
Materials and Methods
Isolates of P. lutzii
We use three isolates of P. lutzii (1578, 01 and ED01) gently provided by Professor Carlos Taborda from the Laboratory of Pathogenic Dimorphic Fungi from the Institute of Biomedical Sciences II of Universidade de São Paulo (Sao Paulo, Brazil).
DNA extraction of isolates
DNA of the three isolates was extracted as described by Kennedy et al. [14], with some modifications. Briefly, 0.4 g yeast was macerated with liquid nitrogen and the resulting powder was transferred to a 2 mL microtube. Then 1.7 mL of extraction buffer (100 mM EDTA, 100 mM Tris, 1.5 M NaCl, 1% SDS, 2% CTAB) were added and the mixture was incubated for 20 min at 65°C (inverting the microtube every 5 min) before centrifugation (20 min at 4500× g, Centrifuge MiniSpin, Eppendorf-AG, Germany). The supernatant was collected and transferred to a new microtube. An equal volume of phenol-chloroform-isoamyl alcohol (25∶24∶1) was added, and after homogenization the tube's content was centrifuged at 4500× g for 10 min. The supernatant was collected and 0.7 volume of isopropanol (100%) and 0.1 volume of 3 M sodium acetate were added and the content mixed by gently inverting the microtube 10 times. After overnight storage at −20°C the sample was centrifuged for 10 min at 4500× g and the resulting pellet washed twice with 70% ethanol. The pellet was dried, resuspended in 200 µL MilliQ water and analyzed by electrophoresis in 2% agarose gel in TBE buffer (40 mM Tris base, 20 mM boric acid, 1 mM EDTA) at 100 V for 30 min and GelRed staining (Uniscience of Brazil, Brazil). Quantification was performed in GeneQuant pro (Amersham Bioscience, USA).
Primers
We used the primer pair described by Buitrago et al. [13] in a trial of Real-time PCR, but adapted for a classic PCR and using P. lutzii not P. brasiliensis. The direct primer (OliPbMB1) was 5′-ACCCTTGTCTATTCTACC-3′ and reverse primer (OliPbMB2) was 5′-TTACTGATTATGATAGGTCTC-3′, which generated a 144 bp fragment amplified from the region ITS1 of the rDNA of P. brasiliensis. Primers were synthesized by the Bioneer Corporation (CA, USA).
PCR
The reaction was performed in 0.2 mL sterile microtube containing 10 ng DNA, 75 mM tris-HCl (pH 8,8), 20 mM (NH4)2SO4, 1.5 mM MgCl2, 1.5 mM dNTP (Fermentas Inc., MA, USA), 0.4 pM of each primer, 1 U Taq polymerase (Fermentas Inc., MA, USA) and sterile milli-Q water, obtaining a final volume of 25 µL. The reactions were processed in the Veritas thermal cycler (Applied Biosystems, CA, USA) programmed as follows: 96°C for 5 min; 40 cycles of 55°C for 35 s, 72°C for 35 s and 96°C for 35 s; 72°C for 7 min. Positive control was DNA extracted from P. lutzii (isolate 01) and negative control was sterile water milli-Q. To visualize the reaction, 5 µL of the amplified product was applied in agarose gel 2% in TBE buffer, at 100 V for 1 h and stained with GelRed.
Detection limit and assay of specificity
Detection limit of the PCR assay was established by using serial dilutions of P. lutzii DNA (from 120 ng/µL to 0.5 pg/µL) and specificity was evaluated with DNA from humans and from the isolates of P. lutzii (1578,01 e ED01).
Clinical samples
We analyzed serum samples from 76 patients (citizens from Itabuna and Ilhéus), suspected to have PCM and no conclusive diagnosis to another fungal disease/non-fungal. Five of these patients had been diagnosed with PCM in our laboratory by the method of double immunodiffusion alone (5) or in combination with direct examination of sputum and/or histopathology (3). Furthermore, the diagnosis was endorsed by a clinical experience of the physician responsible for the patient. These five patients had the chronic form of the disease and had not started treatment before blood collection. Only one of these had associated tuberculosis/paracoccidioidomycosis. The care and clinical monitoring of these patients was conducted in city of Itabuna at the Center for Health José Maria de Magalhaes or Santa Casa de Misericordia or at the Specialized Care Center III, located in Ilhéus.
Ethics statement
All individuals/patients were informed about the methodology and signed an informed consent according to ethical standards. The project was also approved by the Ethics in Research Committee of the Universidade Estadual de Santa Cruz (Text S1).
DNA extraction of clinical samples
DNA extraction from serum samples used QAamp DNA Mini Kit (Qiagen, Hilden, Germany). We used 200 µL of sample and smaller volumes were adjusted with phosphate buffer (PBS, pH 7.4). Purified DNA was eluted in 25 µL of elution buffer and analyzed by electrophoresis in 2% agarose gel in TBE buffer at 100 V for 30 min (stained with GelRed). Quantification of extracted DNA was performed in GeneQuant pro. We also extracted DNA from a biopsy sample of one the patients with confirmed PCM diagnosis. All DNA samples were then stored at −20°C until use in assay of PCR.
Double immunodiffusion
The test was performed in mesh citrate-agarose (1% agarose, 0.4% sodium citrate, 0.9% sodium chloride and 7.5% glycine). In the central well apply 10 µL of exoantigen of P. brasiliensis, 339 isolated, and the side wells 10 µL of serum from patients or healthy subjects. Slides were incubated in a humid chamber at room temperature for 48 hours, with readings every 24 hours. Subsequently, they were washed with sodium citrate to 5% and 0.85% physiological solution, dried at 60°C for 24 hours and stained with coomasie brilliant blue R-250 0.15% (Vetec, Rio de Janeiro, Brazil) in water-methanol-acetic acid (4∶4∶1) for 5 minutes. Positive samples were serially diluted (1/2 to 1/256) and re-subjected to the test ID for semiquantitative analysis. As a positive control we used the reaction hyperimmune serum of rabbit anti-exoantigens of P. brasiliensis, obtained from the Immunodiagnostic Laboratory of Mycoses of the Adolfo Lutz Institute (São Paulo) and kindly provided by Dr. Adriana Vicentini Pardini.
Results
Validation of PCR with DNA of isolates of P. brasiliensis and P. lutzii
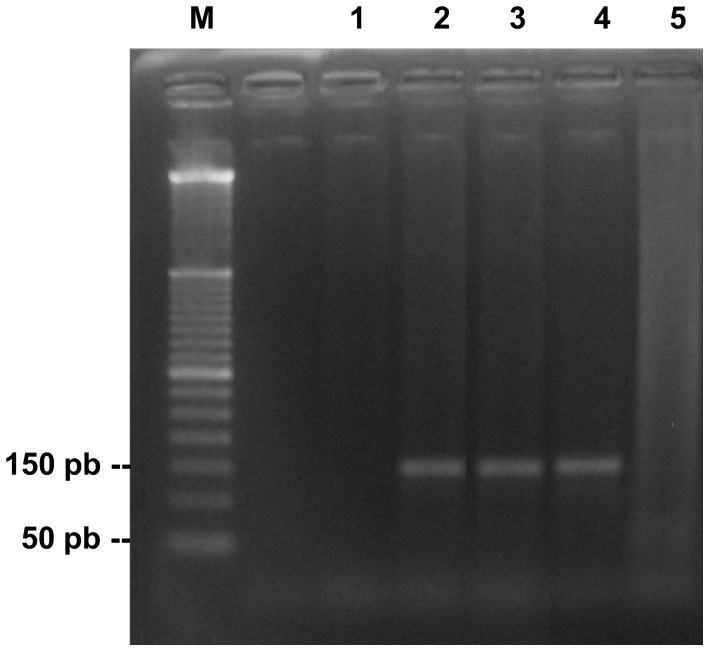
The functionality and specificity of PCR standardized by us can be proven by visualization of the 144 pb amplicon generated when using DNA from isolates of P. lutzii (Fig. 1). The reaction with P. brasiliensis (isolate 03) also generated the same amplicon (Fig. S1). Human DNA served as specificity control and was always negative.
Figure 1. PCR assay revealing the target fragment of 144 pb in the DNA of P. lutzii.
M, molecular marker of 50 bp; 1, negative control; 2, 3 and 4, DNA of P. lutzii isolates 01, ED01 and 1578, respectively; 5, human DNA.
Detection limits
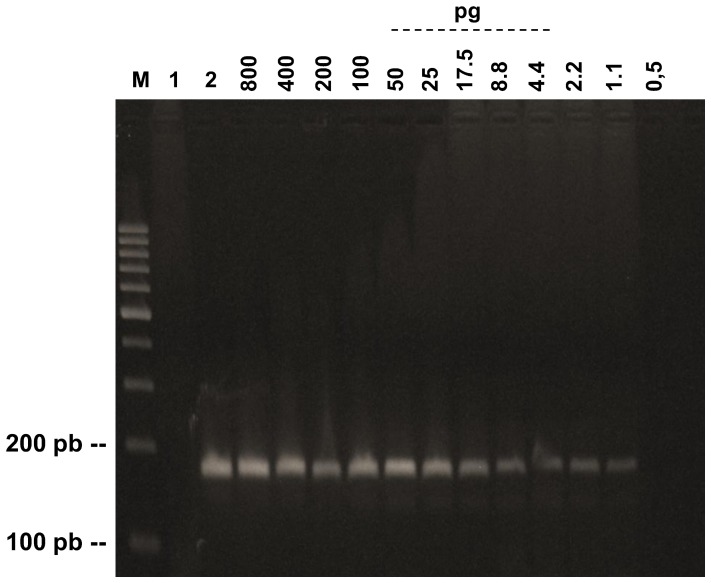
Our standardized PCR test was able to detect until 1.1 pg of DNA of P. lutzii (Fig. 2). The range of detection of the test was from 1.1 pg/µL to 60 ng/µL of DNA.
Figure 2. Detection limit of PCR assay with different amounts of DNA of P. lutzii.
M, 100 bp DNA ladder; 1, negative control; 2, positive control.
Application of PCR in serum samples
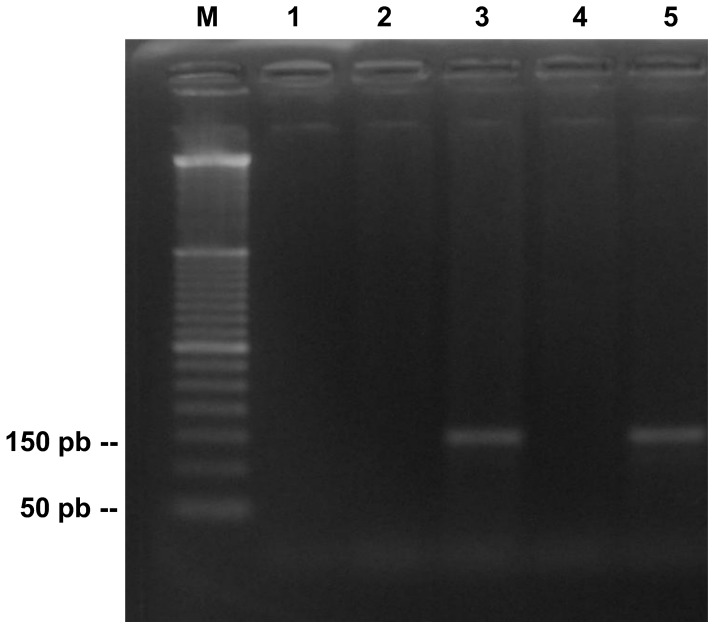
We analyzed seventy-six serum samples of patients with suspected PCM. Five of these patients has been diagnosed with PCM by double immunodifusion (Fig. S2), and three of them were confirmed by direct examination of sputum or histopathology (Fig. S3). In the PCR, none of the seventy-six serum specimens were positive. However, the single biopsy specimen that was tested was positive, revealing the expected 144 pb fragment (Fig. 3).
Figure 3. Analysis of clinical samples (serum and biopsy) of PCM by PCR.
Lane M, 50 bp DNA ladder; lanes 1 and 2, samples of serum; lane 3, biopsy specimen; lane 4 e 5, negative control and positive, respectively.
Discussion
In those cases where the microscopic observation of P. brasiliensis is not successful and when the levels of antigen or antibody are very low, the use of molecular techniques such as PCR has been shown to be effective [15]. There is much antigenic variation among isolates of P. brasiliensis from the different parts of Brazil and Latin America, and as consequence differences in serologic reactivity have been observed, showing the need to work with antigens specific for isolates from a defined region [16], [17].
Considering these aspects and the fact that we use antigens of isolates from other regions in the double immunodiffusion test to detect cases of PCM in Southern Bahia and that we have not yet succeeded in isolating the fungus, the use of PCR in detection of PCM seemed more appropriate.
For these purpose, the primers pairs specific for the ITS1 region of rDNA of P. brasiliensis described by Buitrago et al. [13] seemed more suitable for application of PCR as a diagnostic tool. Therefore, this primes pairs allowed the detection of both DNA P. lutzii as P. brasiliensis, being generated the same 144 pb amplicon. However, because to this characteristic, it is not possible to distinguish which of the two species is the DNA amplified from single biopsy sample tested.
Our results reinforce the fact that PCR and PCR-based molecular techniques have a limitation when serum or plasma is used [18], [19]. This can be explained because of the fungus is rarely in the blood stream [20]. Another possibility is that in this body part the yeast and DNA of P. brasiliensis is readily phagocytosed by leukocytes, reducing the chances of finding these elements there [21].
Despite these issues, we conducted an experiment where we evaluated the possibility of directly using the serum/plasma, without extraction of DNA, in the PCR reaction (data not shown). In this case, serum/plasma could contain the yeast P. brasiliensis or P. lutzii and/or its DNA dispersed, or none at all of these elements. Considering the case of the yeast to be integrated, we mixed a plasma sample a predetermined amount of the yeast P. brasiliensis and submitted to the PCR reaction but the result was negative.
Interestingly, when conducting another test with a plasma sample received 10 ng of DNA of P. brasiliensis, the reaction was positive. This experiment leads us to two hypothesis: that in plasma not exist inhibitory elements of the PCR reaction or that these elements exist but are at low levels and are further diluted when mixed with other components of the PCR reaction mix. More tests are needed to confirm this finding to be applied in the PCR reactions in general.
In summary, we include some information about the limitations (Text S2) and experimental design of this study (Fig. S4). We believe that PCR can be used as a diagnostic tool in diagnosis of PCM, especially when conventional diagnostic methods are not successful, but much remains to be done to make this molecular test secure. Moreover, we know that quantitative molecular methods, such as real-time PCR, would be more appropriate to predict disease or infection. However, this is an expensive technique and would not be performed at some locations. Thus, more studies are needed that aim to refine and increase the discriminatory power of more feasible molecular techniques as conventional PCR or nested-PCR.
Supporting Information
The primer pair OliPbMB1/OliPbMB2 generate fragments of the same size both in P. brasiliensis as in P. lutzii . M, molecular marker of 100 bp; 1, negative control; 2 and 3, DNA of P. lutzii (isolate 01) and P. brasiliensis (isolate 03), respectively.
(TIF)
Double immunodiffusion test revealed antibody anti- P. brasiliensis in the sera of five patients(P1–5). The slides were stained with Coomasie Brilliant Blue R-250 0.15%. Ag: exoantigens of P. brasiliensis, isolate 339; C+: rabbit serum anti-exoantigens of P. brasiliensis.
(TIF)
Three patients positive by immunodiffusion were confirmed by direct examination or histopathology. The arrow show P. brasiliensis in multiple budding in direct examination (A) and as yeast single in histopatology (B). Direct examination was performed on sputum samples and using 20% KOH solution as a clarifier. Histopathology was performed only in one patient, from a liver biopsy and stained with hematoxylin and eosin. in this photo, 400× magnification.
(TIF)
Flowchart showing the method of experimental analysis of patient samples submitted to the study.
(TIF)
Approval of the Ethics Committee in Research of Universidade Estadual de Santa Cruz.
(DOCX)
STARD Checklist.
(DOC)
Acknowledgments
The authors wish to thank Dr. Martin Brendel (Frankfurt University/Visitant Researcher State University of Santa Cruz) for their help in translation and preparation of this manuscript. We thank thank Dr. Carlos P. Taborda and Dra. Fernanda Dias (Laboratory of Pathogenic Dimorphic Fungi, Biomedical Sciences Institute II/University of São Paulo) for providing the isolates of P. brasiliensis and P. lutzii. We wanted also to thank MSc. Cintia Marques (Laboratory of Pharmacogenomics and Molecular Epidemiology/State University of Santa Cruz), for their assistance.
Funding Statement
The project was funded by the Coordination of Improvement of Higher Education Personnel (CAPES), http://www.capes.gov.br/. The Research Foundation of the State of Bahia (Fapesb), http://www.fapesb.ba.gov.br/, was responsible for providing scholarship to one of the researchers. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Blotta MHSL, Mamoni RL, Oliveira SJ, Nouér SA, Papaiordanou PM, et al. (1999) Endemic regions of paracoccidioidomycosis in Brazil: a clinical and epidemiologic study of 584 cases in the southeast region. Am J Trop Med Hyg 61: 390–394. [DOI] [PubMed] [Google Scholar]
- 2.Lacaz CS (1994) Historical evolution of the knowledge on paracoccidiodomycosis and its etiologic agent, Paracoccidioides brasiliensis. In: Franco M, Lacaz CS, Restrepo Moreno A, Del-Negro G, Paracoccidioidomycosis. Boca Raton, Florida.
- 3. Koishi AC, Vituri DF, Dionízio Filho PS, Sasaki AA, Felipe MS, et al. (2010) A semi-nested PCR assay for molecular detection of Paracoccidioides brasiliensis in tissue samples. Rev Soc Bras Med Trop 43: 728–730. [DOI] [PubMed] [Google Scholar]
- 4. Bialek R, Ibricevic A, Aepinus C, Najvar LK, Fothergill AW, et al. (2000) Detection of Paracoccidioides brasiliensis in tissues samples by a nested PCR assay. J Clin Microbiol 38: 2940–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marques SA, Robles AM, Tortorano AM, Tuculet MA, Negroni R, et al. (2000) Mycosis associated with AIDS in the third world. Med Mycol 38: 269–279. [PubMed] [Google Scholar]
- 6. Teixeira MM, Theodoro RC, de Carvalho MJ, Fernandes L, Paes HC, et al. (2009) Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Mol Phylogenet Evol 52 2: 273–283. [DOI] [PubMed] [Google Scholar]
- 7. Gomes GM, Cisalpino PS, Taborda CP, Camargo ZP (2000) PCR for diagnosis of paracoccidioidomycosis. J Clin Microbiol 38: 3478–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Imai T, Sano A, Mikami Y, Watanabe K, Aoki FH, et al. (2000) A new PCR primer for the identification of Paracoccidioides brasiliensis based on rRNA sequences coding the internal transcribed spacers (ITS) and 5.8S regions. Med Mycol 38: 323–326. [DOI] [PubMed] [Google Scholar]
- 9. Molinari-Madlum EE, Felipe MS, Soares CM (1999) Virulence of Paracoccidioides brasiliensis isolates can be correlated to groups defined by random amplified polymorphic DNA analysis. Med Mycol 37: 269–276. [PubMed] [Google Scholar]
- 10. Motoyama AB, Venancio EJ, Brandão GO, Petrofeza-Silva S, Pereira IS, et al. (2000) Molecular identification of Paracoccidioides brasiliensis by PCR amplification of ribosomal DNA. J Clin Microbiol 38: 3106–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matute DR, McEwen JG, Puccia R, Montes BA, San-Blas G, et al. (2006) Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Mol Biol Evol 23: 65–73. [DOI] [PubMed] [Google Scholar]
- 12. Lindsley MD, Hurst SF, Iqbal NJ, Morrison CJ (2001) Rapid identification of dimorphic and yeast-like fungal pathogens using specific DNA probes. J Clin Microbiol 39: 3505–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buitrago MJ, Merino P, Puente S, Gomez-Lopez A, Arribi A, et al. (2009) Utility of real-time PCR for the detection of Paracoccidioides brasiliensis DNA in the diagnosis of imported Paracoccidioidomycosis. Med Mycol 47: 879–882. [DOI] [PubMed] [Google Scholar]
- 14. Kennedy J, Codling CE, Jones BV, Dobson ADW, Marchesi JR (2008) Diversity of microbes associated with the marine sponge, Haliclona simulans, isolated from Irish waters and identification of polyketide synthase from the sponge metagenome. Environmental Microbiology 10: 1888–1902. [DOI] [PubMed] [Google Scholar]
- 15. Teles FRR, Martins ML (2011) Laboratorial diagnosis of paracoccidioidomycosis and new insights for the future of fungal diagnosis. Talanta 85: 2255–2264. [DOI] [PubMed] [Google Scholar]
- 16. Batista J Jr, Camargo ZP, Fernandes GF, Vicentini AP, Fontes CJF, et al. (2009) Is the geographical origin of a Paracoccidioides brasiliensis isolate important for antigen production for regional diagnosis of paracoccidioidomycosis? Mycoses 53: 176–180. [DOI] [PubMed] [Google Scholar]
- 17. Costa PF, Fernandes GF, Santos PO, Amaral CC, Camargo ZP (2009) Characteristics of environmental Paracoccidioides brasiliensis isolates. Mycopathologia 169: 37–46. [DOI] [PubMed] [Google Scholar]
- 18. Charbel CE, Levi JE, Martins JEC (2006) Evaluation of polymerase chain reaction for the detection of Paracoccidioides brasiliensis DNA on serum samples from patients with Paracoccidioidomycosis. Mem Inst Oswaldo Cruz 101: 219–221. [DOI] [PubMed] [Google Scholar]
- 19. Nakagawa Itano E, Uno J, Sano A, Yarita K, Kamei K, et al. (2002) Detection of the gp43 Gene and (1→3) -β-D-glucan of Paracoccidioides brasiliensis in the Blood of Experimentally Infected Mice. Nihon Ishinkin Gakkai Zasshi 43: 29–35. [DOI] [PubMed] [Google Scholar]
- 20. Singer-Vermes LM, Burger E, Calich VL, Modesto-Xavier LH, Sakamoto TN, et al. (1994) Pathogenicity and immunogenicity of Paracoccidioides brasiliensis isolates in the human disease and in an experimental murine model. Clin Exp Immunol 97: 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nishikaku AS, Burger E (2003) Evaluation of fungal burden in experimental paracoccidioidomycosis by using the fluorescent dye Blankophor. J Clin Microbiol 41: 3419–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The primer pair OliPbMB1/OliPbMB2 generate fragments of the same size both in P. brasiliensis as in P. lutzii . M, molecular marker of 100 bp; 1, negative control; 2 and 3, DNA of P. lutzii (isolate 01) and P. brasiliensis (isolate 03), respectively.
(TIF)
Double immunodiffusion test revealed antibody anti- P. brasiliensis in the sera of five patients(P1–5). The slides were stained with Coomasie Brilliant Blue R-250 0.15%. Ag: exoantigens of P. brasiliensis, isolate 339; C+: rabbit serum anti-exoantigens of P. brasiliensis.
(TIF)
Three patients positive by immunodiffusion were confirmed by direct examination or histopathology. The arrow show P. brasiliensis in multiple budding in direct examination (A) and as yeast single in histopatology (B). Direct examination was performed on sputum samples and using 20% KOH solution as a clarifier. Histopathology was performed only in one patient, from a liver biopsy and stained with hematoxylin and eosin. in this photo, 400× magnification.
(TIF)
Flowchart showing the method of experimental analysis of patient samples submitted to the study.
(TIF)
Approval of the Ethics Committee in Research of Universidade Estadual de Santa Cruz.
(DOCX)
STARD Checklist.
(DOC)