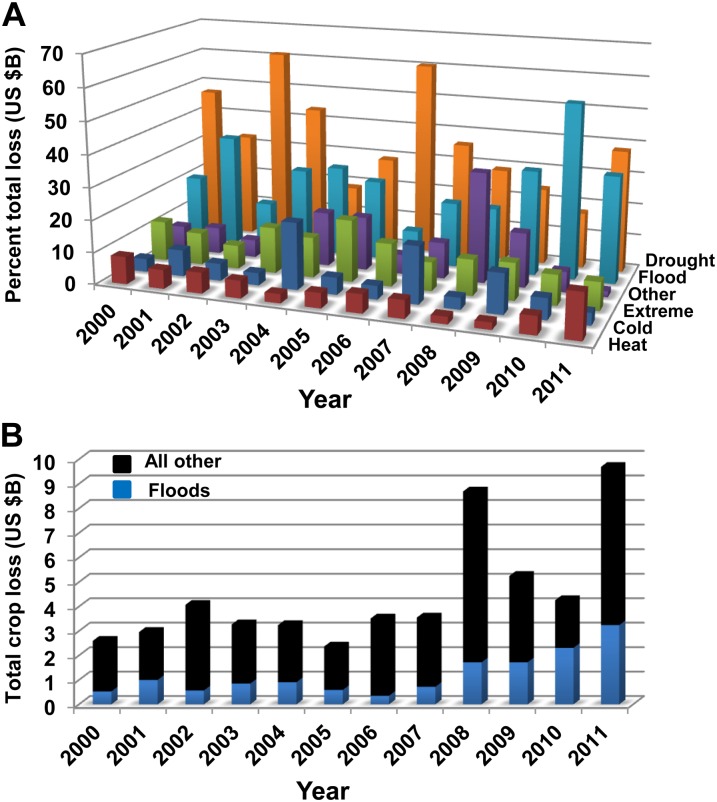
Extremes in water availability (droughts and floods) have increased in frequency and intensity over the past 50 years in farming regions throughout the world. In the United States, losses in crop production due to flooding were second to drought in many of the past 12 years, with these two abiotic stresses accounting for more than 70% of the reduction in harvests in 2011 (Fig. 1A). That year, insurance payouts due to floods exceeded $3 billion (Fig. 1B), with more than $1.6 billion in losses of maize (Zea mays) and soybean (Glycine max) in Midwestern fields. Over the same time period, flooding caused considerable crop loss in other parts of the world. For example, increased summer rainfall intensity in Europe caused numerous flooding and water stagnation events of economic consequence (Olesen et al., 2011), and floods in Australia impacted both production and prices of wheat (Triticum spp.). Exceptionally severe flooding in Pakistan inflicted $4.45 billion in damage to the wheat, cotton (Gossypium hirsutum), and rice (Oryza sativa) crops in 2010 (Arshad and Shafi, 2010). One of the most flooding-threatened crops is rice. At present, over 35% of the world’s rice acreage is flood prone, and much is in regions of Asia and Africa characterized as food insecure.
Figure 1.
Financial losses in crop production due to environmental stresses in the United States from 2000 to 2011. Yearly totals are in billions of U.S. dollars in loss of food, fiber, and fuel crops. Data are based on insurance indemnities paid to farmers. Values were determined from the U.S. Department of Agriculture Risk Management Agency Cause of Loss Historical Data. A, Percentage of total crop loss per environmental stress. Percentage losses are the quotient of the amount in billions of U.S. dollars of crop loss to each peril divided by the total crop loss for that given year. Drought, Water deficit stress; Flood, excess moisture, precipitation, or rain; Other, anthropogenic and biotic factors such as fire, insects, and plant disease; Extreme, cyclones, hail, hurricanes, and tropical depression events; Cold, cold weather, including frosts and freezes; Heat, excessive radiant heat and hot winds. B, Total crop losses and the proportion of total loss due to floods in billions of U.S. dollars.
A major international goal is to increase overall crop production to meet the anticipated needs of the world’s growing population. To achieve this goal, it is essential to develop and adopt germplasm that is better able to endure abiotic assaults. An example of this is the development of rice varieties that survive transient submergence and their noncommercial dissemination to farmers cultivating flood-prone paddies. In this case, robust submergence tolerance was transferred by marker-assisted breeding from a farmer’s landrace to modern varieties, through the initiatives of the International Rice Research Institute.
A detailed understanding of the developmental, morphological, physiological, and molecular mechanisms that underlie flooding tolerance should enable the translation of flooding survival strategies to additional crops to stabilize yields of food, feed, fiber, and fuel. The purpose of this Update is to provide a current overview of plant flooding survival strategies from the organismal to the molecular level, with a focus on crop improvement.
FLOODS AND FLOODING SURVIVAL MECHANISMS
Here, the term “flooding” is used to describe the inundation by water of all or part of a plant. We use “waterlogging” to describe flooding of the root system and “submergence” to describe the situation when most or all aerial tissue is under water. Floodwaters can be fresh, stagnant, or saline and affect a plant once or multiple times in a growing season. These inundation events can result from flash floods, seasonal rises in surface water at low elevations, or tidal surges. The sudden saturation of well-drained soil causes rhizosphere microbes to rapidly consume available oxygen, triggering changes in the soil ecosystem that can alter the fixation of nitrogen and the availability of other nutrients to the plant. Floods are often accompanied by a decrease in soil pH, which increases the solubility of toxic metals, including iron and manganese, as well as phosphorus and other elements (Setter et al., 2009; Michalcová et al., 2011). Importantly, the approximately 104-fold reduction in diffusion of gases in floodwaters limits the availability of oxygen and carbon dioxide for aerobic respiration and photosynthesis, respectively. If the water surrounding submerged aerial organs is turbid, light availability for photosynthesis is rapidly diminished. Therefore, flooded organs often must increase catabolic processes to meet ATP demands due to the inefficiency of anaerobic ATP production.
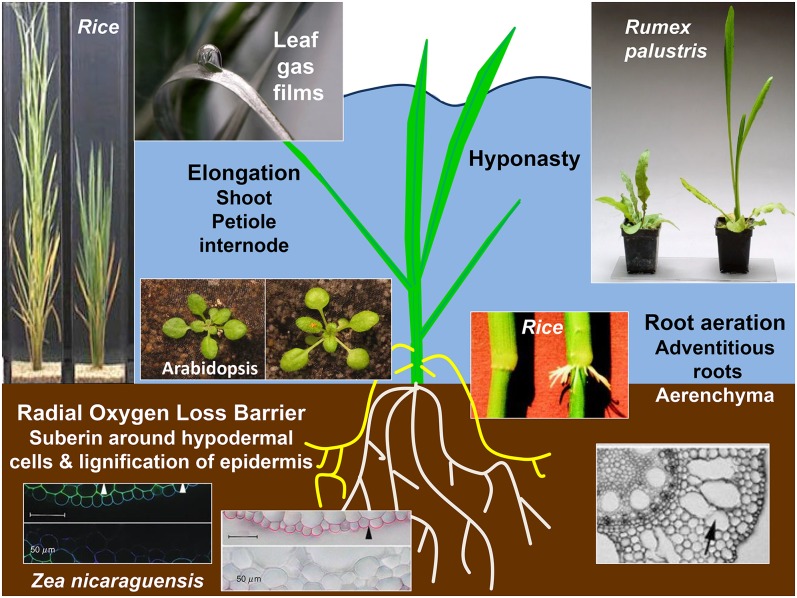
Over time, the hydrological signature of natural environments has fashioned plants to enable their habitation of environments with seasonal extremes in water availability (Fig. 2). For example, trees of the Amazon and other tropical floodplains endure several months of partial to complete submergence followed by months of moderate to severe water deficit (Parolin et al., 2010). Annual and perennial species native to riparian flood zones are adapted to flooding events of varying amplitude and duration. The species of these habitats have evolved mechanisms of “tolerance,” enabling the plant to withstand the stress, or “avoidance,” enabling the plant to circumvent the stress (Voesenek et al., 2006; Bailey-Serres and Voesenek, 2008; Colmer and Voesenek, 2009; Bailey-Serres et al., 2012). The two survival tactics that have emerged from physiological and molecular studies are (1) the low-oxygen quiescence strategy, characterized by a general restriction in cellular metabolism and growth, displayed by species that regularly endure deep floods of short duration, and (2) the low-oxygen escape strategy, characterized by rapid elongation of underwater stems or leaves to enable photosynthetic tissue to outgrow shallow floodwaters. Although antithetical, both strategies involve ethylene-mediated signal transduction. Plants that inhabit wetlands may also display adaptive features that facilitate the exchange of gases between aerial and flooded organs, including the ethylene-driven formation of constitutive or induced aerenchyma and adventitious roots as well as thinning of the leaf cuticle and reorientation of chloroplasts to optimize gas exchange. Additionally, leaves may have a sufficiently hydrophobic cuticle to maintain a surface layer of gas that sustains the rapid diffusion of oxygen and carbon dioxide to allow continued stomatal activity, benefiting underwater photosynthesis and respiration (Pedersen et al., 2009; Colmer et al., 2011; Winkel et al., 2011).
Figure 2.
Examples of survival traits displayed in flooded plants. Elongation growth response: deepwater (left) and lowland (right) rice cultivars after 7 d of partial submergence showing distinctions in internode elongation (Hattori et al., 2009); Arabidopsis (Columbia-0 ecotype) after 3 d of complete submergence in darkness showing petiole elongation (Mustroph et al., 2010); and 6-week-old R. palustris plants grown in air (left) or submerged for 14 d (right) showing petiole elongation (Colmer and Voesenek, 2009). Root aeration: adventitious root formation at the third internode in deepwater rice without (left) and with (right) ethephon treatment (Mergemann and Sauter, 2000); and increased lysigenous aerenchyma after 14 d of stagnant waterlogging (Abiko et al., 2012). Radial oxygen loss barriers formed after 14 d of stagnant waterlogging include suberin deposition in cell walls of hypodermal/exodermal layers and lignin deposition on outer epidermis. Z. nicaraguensis root images are 60 mm from the tip of an adventitious root. Leaf gas films cling to the surface of leaves of many semiaquatic species (Winkel et al., 2011).
Unlike aquatic and wetland species, most crops are susceptible to flooding events of short duration, resulting in reductions in growth and yield (Table I). A notable exception is lowland rice, which is typically transplanted as a cluster of seedlings into paddies 5 to 15 cm or 10 to 50 cm deep that are maintained by irrigation or precipitation, respectively. Alternatively, pregerminated seeds are broadcast into shallow paddies. The ability to grow with a flooded root system is aided by the constitutive development of aerenchyma and physical barriers that limit oxygen loss by radial diffusion and the entry of soilborne toxins (Colmer and Voesenek, 2009). Some low-yielding rice traditionally cultivated by farmers is capable of surviving more extreme floods. For example, among the tens of thousands of landraces, some can escape a progressive seasonal flood by extensive underwater elongation of the culm internodes. These “deepwater” or “floating” rice varieties are not tolerant of complete submergence but maintain sufficient photosynthetic tissue in air to fuel growth and maturation. Conversely, some landraces are extremely submergence tolerant, with the capacity to survive drowning due to flash floods in turbid waters for more than 1 week (Bailey-Serres et al., 2010). There are also landraces that can be dry seeded directly into shallow paddies (less than 10 cm depth) that can become established despite limited oxygen availability (Angaji et al., 2010; Ismail et al., 2012). Although these condition and stage-specific flood survival strategies were excluded from modern cultivars, progress in the elucidation of their genetic determinants has already begun to enable their introduction into high-yielding varieties to produce even more “waterproof” rice (Septiningsih et al., 2009; Bailey-Serres et al., 2010).
Table I. Examples of flooding survival and response strategies of crop, wetland, and model species.
| Species | Condition | Survival (Duration or LT50) | Response Strategy Associated with Survival | Reference |
|---|---|---|---|---|
| Arabidopsis ecotypes (n = 8) | Submergence (day/night light regime) | LT50 > 15 d | Quiescence | Vashisht et al. (2011) |
| Arabidopsis ecotypes (n = 86) | Submergence (constant darkness) | LT50 = 4–12 d | Quiescence | Vashisht et al. (2011) |
| Maize | Complete submergence | 1–2 d | Unknown | E. Brinton and J. Bailey-Serres (unpublished data) |
| Maize | Stagnant waterlogging | <10 d | Aerenchyma, adventitious roots | Zaidi et al. (2004) |
| Lowland rice | Complete submergence | <7 d | Escape, shoot elongation | Fukao et al. (2006) |
| Deepwater/floating rice | Partial to shallow submergencea | <7 d | Escape | Catling (1992) |
| Rice Sub1 varieties | Complete submergence | >14 d | Quiescence | Fukao et al. (2006) |
| Oak (Quercus robur and Quercus petraea) | Waterlogging | >4–7 weeks | Hypertrophied lenticels, leaf hyponasty, slow oxygen consumption | Parelle et al. (2007) |
| Oenathe aquatic | Complete submergence | LT50 > 200 d | Quiescence | Mommer et al. (2007) |
| Rorippa sylvestris | Complete submergence | 100 d | Quiescence | Akman et al. (2012) |
| R. acetosa | Submergence (constant darkness) | LT50 = 19.5 ± 1.1 d | Quiescence | M. Akman (personal communication) |
| R. palustris | Submergencea (constant darkness) | LT50 = 20.4 ± 1.0 d | Escape, petiole elongation | M. Akman (personal communication) |
| Soybean | Waterlogging | 2–4 d | Secondary aerenchyma development, adventitious roots | Rhine et al. (2010) |
| Teosinte | Stagnant waterlogging | Life cycle | Constitutive aerenchyma, root radial oxygen barrier, adventitious roots | Abiko et al. (2012) |
| Winter wheat | Waterlogging | <25 d | Aerenchyma, adventitious roots | Setter and Water (2003) |
Cannot endure prolonged complete submergence. Submergence conditions under normal day/night cycle unless otherwise indicated.
ETHYLENE REGULATION OF ADAPTIVE GROWTH RESPONSES TO FLOODING
Ethylene is an essential upstream mediator of flooding responses (Bailey-Serres and Voesenek, 2008; Jackson, 2008). This gaseous hormone is constitutively synthesized in plant cells under normal growth conditions. Internal concentrations are maintained at low levels through rapid ventilation to the atmosphere. However, when an organ is surrounded by water, ethylene becomes trapped within cells, leading to signal transduction, which often includes further production of the hormone (Bailey-Serres and Voesenek, 2008). The characterization of two quantitative trait loci (QTLs) in rice demonstrated the importance of ethylene in flooding responses. These are the SUBMERGENCE1 (SUB1) locus isolated from a submergence-tolerant landrace (FR13A) of eastern India and the SNORKEL (SK) locus from a deepwater landrace (C9285) from Bangladesh (Xu et al., 2006; Hattori et al., 2009). Whereas SUB1 controls the tolerance of complete submergence by dampening underwater growth, SK controls the avoidance of submergence by promoting underwater elongation growth.
Quiescence Strategy of Submergence-Tolerant Rice
The SUB1 locus on chromosome 9 confers up to 69% of phenotypic variation in the tolerance of complete submergence of vegetative plants. Plants with the SUB1 region from FR13A are capable of surviving 2 weeks or longer of complete inundation. This multigenic locus includes two or three genes of the group VII subgroup of ethylene-response factor (ERF) transcription factors that were designated SUB1A, SUB1B, and SUB1C (Xu et al., 2006). It was determined that SUB1A is sufficient for submergence tolerance. Although SUB1B and SUB1C appear to be invariably present at the SUB1 locus, they are evidently not determinants of submergence tolerance by quiescence.
Among indica and aus accessions of rice with SUB1A, those with the SUB1A-1 allele are typically submergence tolerant, whereas those with the SUB1A-2 allele are typically submergence intolerant (Xu et al., 2006; Singh et al., 2010). These two alleles encode proteins that only differ at a single amino acid, Ser-186 in SUB1A-1 and Pro-186 in SUB1A-2, a putative mitogen-activated protein kinase (MPK) phosphorylation site in SUB1A-1 (Xu et al., 2006). To further assess the determination of submergence tolerance by SUB1A, Singh et al. (2010) evaluated SUB1A genotype and submergence tolerance in 76 rice accessions from a variety of geographic locations. Although tolerance was highly correlated with strong up-regulation of SUB1A-1 mRNA during submergence, there was an imperfect association between tolerance and SUB1A-1. For example, a moderately flooding-tolerant accession was identified that carries SUB1A-2. This accession displayed strong submergence induction of SUB1A-2 mRNA, in contrast to submergence-intolerant lines carrying SUB1A-2. These findings raised the hypothesis that it is the level of SUB1A expression, rather than allelic variation at the MPK phosphorylation site, that distinguishes tolerant and intolerant lines carrying SUB1A.
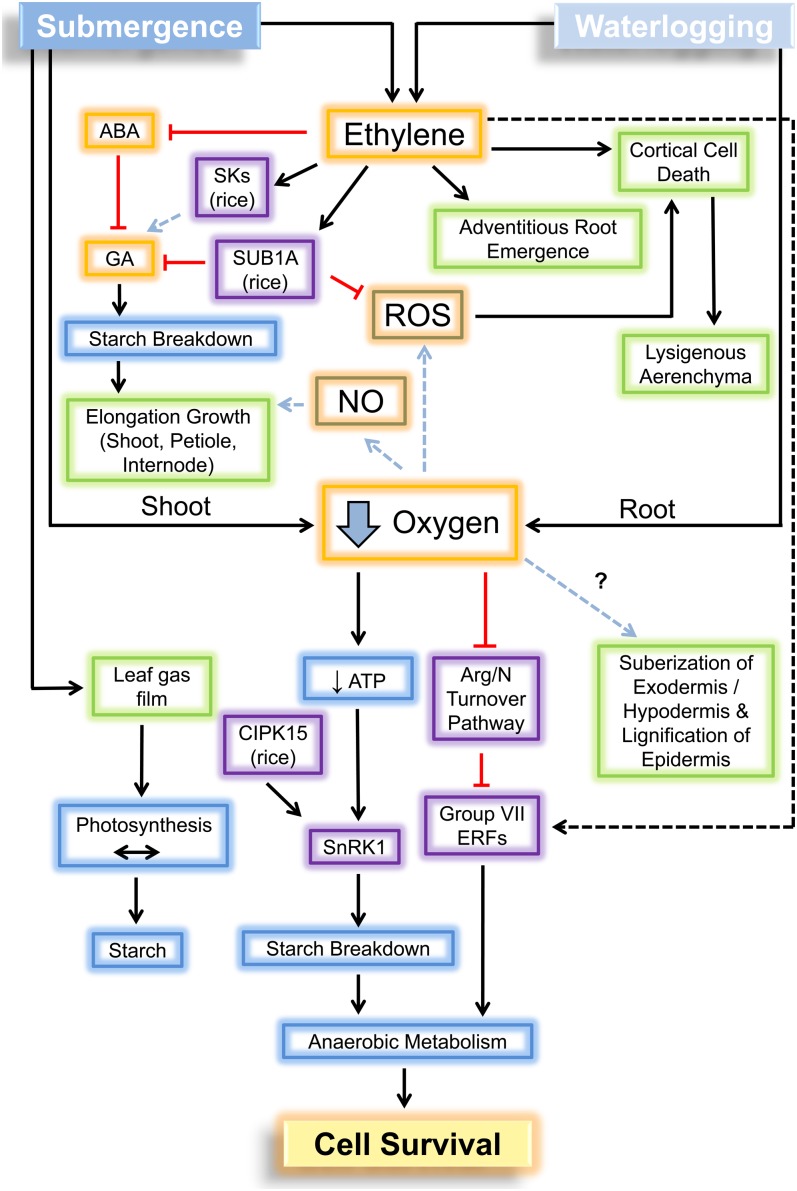
A series of mechanistic studies has shown that SUB1A-1 functions in a hierarchical hormonal pathway that regulates elongation growth during submergence (Fig. 3). Ethylene entrapped during submergence stimulates increased production of abscisic acid (ABA) 8′-hydroxylase, catalyzing the conversion of ABA to inactive dihydrophaseic acid (Saika et al., 2007). SUB1A-1 has no effect on this process, as ABA declines similarly in shoots of near isogenic lines that differ in the presence versus absence of SUB1A-1 [M202 and M202(Sub1); Fukao and Bailey-Serres, 2008]. The drop in ABA is followed by increased responsiveness to GA, due to a reduction in two GRAS domain transcription factors, the DELLA domain-containing protein SLENDER RICE1 (SLR1) and the DELLA domain-lacking protein SLR-LIKE1 (SLRL1; Fukao and Bailey-Serres, 2008; Sun, 2011). Remarkably, SLR1 and SLRL1 play key roles in submergence tolerance mediated by SUB1A. The first clue of this came from the observation that SUB1A-1 transgenics are semidwarf and display GA insensitivity throughout development (Fukao and Bailey-Serres, 2008). During submergence, SLRL1 transcript and protein accumulation were higher in SUB1A-1-containing genotypes [e.g. M202(Sub1) and Ubi:SUB1A-1 transgenics]. It was also found that treatment of seedlings with the ethylene precursor 1-aminocyclopropane-1-carboxylic acid inhibited GA-mediated elongation in M202(Sub1) and promoted elongation in M202 shoots. Based on these and other findings, it was proposed that the low-oxygen quiescence strategy is facilitated by ethylene-driven expression of SUB1A-1, which acts to limit GA-mediated starch breakdown, anaerobic metabolism, elongation growth, and leaf senescence (Fukao et al., 2006, 2012; Fukao and Bailey-Serres, 2008; Barding et al., 2012), all of which fuel elongation growth and submergence escape.
Figure 3.
Network of the regulation of submergence and waterlogging growth, adaptation, and metabolic responses by ethylene, low oxygen, ROS, and NO. Elongation growth is controlled by GA-mediated pathways, which act downstream of ethylene and ABA. In rice group VII ERFs, SK1 and SK2 positively regulate internode elongation, presumably through the stimulation of GA-mediated starch consumption and elongation growth. Conversely, the rice group VII ERF SUB1A inhibits shoot elongation by maintaining levels of the transcription factors SLR1 and SLRL1 to counterbalance GA responsiveness. In some species, underwater elongation involves hyponastic growth or the upward curvature of the leaf petiole or blade. The development of adventitious roots and aerenchyma is regulated by ethylene and ROS. The presence of a thin gas film on underwater leaves helps maintain photosynthesis and, therefore, starch production. Oxygen deficiency dampens ATP energy levels. In germinating seeds of rice, CIPK15 promotes SnRK1 activity to enhance carbohydrate catabolism. Low-oxygen stress enhances the consumption of soluble carbohydrates in anaerobic reactions that generate ATP to maintain cellular homeostasis. In Arabidopsis, the up-regulation of genes encoding enzymes required for anaerobic metabolism requires the stabilization of group VII ERFs. This occurs conditionally, as oxygen levels decline, by inhibition of the Arg/N branch of the N-end rule pathway of targeted proteolysis. All group VII ERFs of the Columbia-0 ecotype possess an N terminus that confers instability under aerated conditions and stability under oxygen deficiency. Orange, hormones/signaling molecules; green, physiological changes; purple, proteins/pathways; blue, metabolic components.
Recently, it has been shown that SUB1A also aids recovery from submergence. This is evident from the finding that the tolerant genotype M202(Sub1) displayed greater up-regulation of mRNAs encoding antioxidant enzymes during submergence (Jung et al., 2010; Mustroph et al., 2010) and less oxidative damage upon desubmergence than M202 (Fukao et al., 2011). Intriguingly, increased ABA responsiveness was recorded when SUB1A was submergence induced or constitutively expressed and correlated with reduced postsubmergence leaf dehydration as well as better reestablishment through tiller growth after severe water deficit (Fukao et al., 2011). It appears that the protection of meristems during submergence and drought enhance the ability of SUB1A-1 genotypes to recover after a stress event (Fukao et al., 2006, 2011; T. Fukao and J. Bailey-Serres, unpublished data). These findings also suggest that submergence and drought tolerance might be effectively pyramided into one genotype.
Underwater Escape by Deepwater Rice
Deepwater/floating varieties of rice have the capacity to elongate their submerged stem internodes by 25 cm per day, at pace with a slow-rising flood in a seasonal wetland (Kende et al., 1998). These plants can reach heights of 8 m but are typically low yielding due to the high investment of energy reserves in underwater biomass. In some regions of Asia or Africa, deepwater rice cultivation is extensive and can be effectively coupled with fish and oyster production. Thus, genetic identification of key loci controlling this flooding survival strategy can aid the breeding of more productive deepwater cultivars for farming wetlands. Toward this goal, QTL mapping of phenotypes associated with rapid underwater elongation growth identified three loci located on chromosomes 1, 3, and 12 (Hattori et al., 2009, 2011). A QTL on chromosome 12 conferring 30% of the phenotypic variation in underwater elongation was identified as a multigenic locus that encodes group VII ERFs. Remarkably, these genes are closely related to the SUB1s. They were aptly dubbed SK1 and SK2 for their role in maintaining the uppermost leaves and reproductive panicles above the air-water interface, a strategy that facilitates gas exchange between submerged and nonsubmerged tissues. The SKs also fit within the hormonal hierarchy that regulates underwater elongation (Fig. 3). SK1 and SK2 mRNAs are up-regulated by ethylene through binding of the transcription factor ETHYLENE-INSENSITIVE3 (EIN3)-like1b, the rice ortholog of Arabidopsis (Arabidopsis thaliana) EIN3 (Hattori et al., 2009). It is thought that the SKs alone or in combination with the chromosome 1 and 3 QTLs are responsible for the elevated GA and enhanced internode meristem cell division activity that underline the successful deepwater escape strategy.
The SKs are absent or nonfunctional in modern rice cultivars. However, the presence of one or both in the wild progenitors of domesticated rice (Oryza nivara and Oryza rufipogon) and floating rice of the Amazon (Oryza glumaepatula) indicates that this locus was excluded through human selection.
Quiescence and Escape in Other Species
Natural variation in the ethylene-mediated escape and quiescence strategies is also found in eudicots, as illustrated by studies with the genus Rumex and other species that inhabit wetlands (Table I). Underwater escape by Rumex palustris and quiescence of Rumex acetosa are conferred by distinctions in the elongation of leaf petioles involving the same ethylene, ABA, and GA hierarchy established for rice (Benschop et al., 2005; Bailey-Serres and Voesenek, 2008). For example, ABA insensitivity corresponded to greater underwater petiole elongation in R. palustris ecotypes (Chen et al., 2010). In this species, elongation growth toward the water surface is complemented by upward hyponastic curvature of the petiole. This process is triggered by ethylene entrapment in underwater leaves and requires ABA catabolism to derepress GA signaling (Cox et al., 2004) and converges with the shade-avoidance pathway triggered by a low ratio of red to far-red light at a downstream step involving GA regulation of cell expansion (Pierik et al., 2011).
Recent studies with Arabidopsis found that both petiole elongation and leaf hyponastic growth occur in rosette leaves in response to submergence in partial or complete darkness (Lee et al., 2011), very likely via an ethylene-dependent process. Natural variation in submergence survival in complete darkness (median lethal time [LT50]) was surveyed in 86 accessions of Arabidopsis (Table I; Vashisht et al., 2011). Although a modest inverse correlation between petiole elongation and LT50 was recorded, it was found that Arabidopsis could endure extremely prolonged (more than 40 d) periods of submergence, presumably via quiescence. The differences in survival of the accessions should be ample for genetic dissection of responsible loci.
MORPHOLOGICAL AND ANATOMICAL ADAPTATIONS THAT INCREASE FLOODING SURVIVAL
A shallow root system, a thickened root epidermis, aerenchymatous roots, and adventitious roots facilitate aeration in waterlogged soils and under partial submergence (Fig. 2). Rhizomes, found in numerous wetland and aquatic species, also facilitate aeration and provide starch reserves during prolonged periods of flooding. Of these anatomical adaptations, the development of aerenchyma and adventitious roots is controlled by ethylene.
Aerenchyma tissue is composed of low-resistance gas conduits in roots and stems that enable diffusion and exchange of oxygen and carbon dioxide from near the root apex to the uppermost submerged region of the root and into the stem (Jackson and Armstrong, 1999). Many wetland and aquatic species form primary aerenchymatous tissue by cell separation (schizogeny), differential expansion (expansigeny), or programmed cell death (lysigeny; Seago et al., 2005). Lysigenous aerenchyma can be formed constitutively in the root cortex, as observed in lowland rice, or can be induced by flooding, as seen in barley (Hordeum vulgare), maize, and wheat. Maize forms cortical lysigenous aerenchyma within 24 h of waterlogging through a programmed cell death process that involves ethylene, Ca2+, and reactive oxygen species (ROS; Fig. 3; Drew et al., 2000; Steffens et al., 2011). By laser-capture microdissection of root cell layers, Rajhi et al. (2011) profiled the transcriptomes of the cortex and stele of waterlogged or 1-methylcyclopropene-treated seedling roots of maize. This identified genes expressed preferentially in the cortex in a waterlogging- or ethylene-dependent manner that are associated with transcription, Ca2+ signaling, ROS regulation, and cell expansion. In rice, lysigenous cortical aerenchyma form constitutively in roots, even in well-drained soils, and in older internodes at the base of the culm (Steffens et al., 2011). Although aerenchyma formation was not directly linked to submergence tolerance in rice (Parlanti et al., 2011), it is important for waterlogging tolerance and may augment underwater photosynthesis and oxygen diffusion to the root system. In fact, some species with constitutive aerenchyma further improve aeration during waterlogging, as evidenced by the expansion of constitutively formed aerenchyma toward the tip of adventitious roots during flooding in a waterlogging-tolerant teosinte (Zea nicaraguensis; Abiko et al., 2012). Two QTLs responsible for 32% of phenotypic variation in this trait were mapped in backcross populations generated from a Z. nicaraguensis × maize cross (Mano et al., 2012). These loci might be used to improve waterlogging tolerance in maize.
In some species, secondary aerenchyma forms through a cell division process. For example, in soybean, aerenchyma arises through cell division of the phellogen to form a spongy parenchymaous cell layer between the cortex and epidermis (Thomas et al., 2005; Bailey-Serres and Voesenek, 2008). This arises after several days of waterlogging and enhances the aeration of roots and nodules necessary for growth and nitrogen fixation, respectively (Shimamura et al., 2010). The single cell layer cortex of Arabidopsis does not form aerenchyma, but waterlogging may promote the formation of lacunae in secondary xylem of the hypocotyl of mature rosettes (Mühlenbock et al., 2007), which could facilitate gas exchange.
Adventitious roots are those that emerge from stem tissue under conditions of partial to complete submergence (Fig. 2). These can replace compromised roots and provide efficient aerenchymatous connections between aerial shoot tissues and submerged organs. Adventitious roots can form via de novo meristem initiation or the emergence of preexisting root primordia. In the case of adventitious root emergence at lower stem internodes of flooded rice, the process involves signal transduction within the growing root and the overlying epidermal cells (Steffens and Sauter, 2009, 2010; Steffens et al., 2012). It was shown that in the adventitious root primordium, ethylene- and ROS-dependent signaling orchestrated the promotion of growth by signaling via mechanical force to the overlying epidermal cells. The force exerted on the tightly attached epidermal cells directly above the primordia triggered localized cell death through a process involving ethylene signaling and ROS production. This cell-to-cell mechanosignaling enabled emergence of the adventitious root. Remarkably, comparison of the transcriptomes of epidermal cells located directly above the primordium with those nearby indicated that there was spatial priming of programmed cell death prior to its elicitation (Steffens and Sauter, 2009). The mRNAs enriched above the subtending primordium were associated with ethylene biosynthesis, whereas the depleted transcripts included one encoding a metallothionein that negatively regulates cell death. Taken together, these studies on aerenchyma and adventitious roots confirm that ethylene regulates cell type-specific developmental processes of the cortex and epidermis that contribute to root aeration and flooding survival.
ALTERATIONS IN GENE EXPRESSION AND METABOLISM IN RESPONSE TO LOW OXYGEN AND FLOODING
Gene Transcript Regulation
Many studies have examined low-oxygen and flooding stress at the transcript and metabolite levels. Analyses of transcriptomes (total cellular mRNA) have been reported for Arabidopsis, cotton, poplar (Populus × canescens), rice, and soybean under hypoxia/anoxia (0%–8% oxygen) and flooding conditions (Branco-Price et al., 2005, 2008; Liu et al., 2005; Loreti et al., 2005; Lasanthi-Kudahettige et al., 2007; Kreuzwieser et al., 2009; Mustroph et al., 2009; Narsai et al., 2009; van Dongen et al., 2009; Christianson et al., 2010; Jung et al., 2010; Mustroph and Bailey-Serres, 2010; Lee et al., 2011; Nanjo et al., 2011). The strong down-regulation of protein synthesis and highly selective mRNA translation during low-oxygen stress has been investigated by comparative profiling of the total cellular mRNA content with the mRNAs engaged in translation (translatomes) during hypoxia (Branco-Price et al., 2005, 2008). By capturing and profiling ribosome-associated mRNAs from 21 cell types and developmental zones of seedlings, a core set of 49 hypoxia-responsive genes was recognized along with cell-specific distinctions in low-oxygen response (Mustroph et al., 2009). Thirty-four of the core response transcripts were also highly induced in shoots and roots of submerged Arabidopsis (Lee et al., 2011).
The low-oxygen/submergence-induced genes encode proteins associated with anaerobic metabolism, ethylene biosynthesis and response, and gene transcription. Notably, approximately 50% of these Arabidopsis genes encode proteins with no known biological function but that are likely to be involved in the low-oxygen survival. Orthologs of about one-half of these HYPOXIA-RESPONSIVE UNKNOWN PROTEIN (HUP) genes are low-oxygen regulated in other species, and the overexpression of several HUPs altered tolerance to hypoxia and/or submergence in Arabidopsis (Mustroph et al., 2010; Lee et al., 2011). Notably, the less dramatic up-regulation of hypoxia-responsive gene mRNAs in rosettes of submerged plants correlated with higher oxygen content in shoot tissues and the surrounding floodwaters as compared with that of the roots and soil (Lee et al., 2011).
An interesting question is whether waterlogging promotes changes in transcripts in nonflooded aerial organs. In waterlogged cotton, many core hypoxia-responsive gene mRNAs were up-regulated in both roots and shoots, whereas in waterlogged poplar, there was minimal effect on the shoot transcriptome (Kreuzwieser et al., 2009; Christianson et al., 2010). In waterlogged Arabidopsis, systemic up-regulation of genes in the shoot was associated with ABA biosynthesis and response (Hsu et al., 2011). In summary, adjustments of gene expression in response to low-oxygen regimes are influenced by oxygen level and/or energy homeostasis, cell type, and communication between stressed and unstressed organs.
Primary Metabolism
Evaluation of metabolic gymnastics in response to oxygen deprivation and flooding has been accomplished often in concert with the evaluation of transcriptomes and translatomes (Branco-Price et al., 2008; Kreuzwieser et al., 2009; van Dongen et al., 2009; Narsai et al., 2011; Barding et al., 2012). In a comparative study of rice and wheat, changes in metabolites were considered along with alterations in the proteome (Shingaki-Wells et al., 2011). These meta-analyses demonstrate common themes of low-oxygen response in monocots and eudicots. The primary response includes the up-regulation of genes and metabolites associated with enhanced glycolytic and fermentative pathways as well as the accumulation of Ala, γ-aminobutyric acid, and succinate (Mustroph et al., 2010; Narsai et al., 2011). Although anaerobic metabolism typically decreases ATP yield per mole of Glc from 34 to 36 to as few as 2, there is strong evidence that some plants enhance metabolism in a manner that boosts the net yield of ATP generated under anaerobiosis.
The conversion of soluble carbohydrates and starch to energy during oxygen deficiency and flooding varies at the cell type, organ, genotype, and species levels. The catabolism of leaf starch is promoted in submerged rice, but to a lesser extent in SUB1A genotypes (Fukao et al., 2006). In seeds germinated under low oxygen, starch catabolism is modulated in a Suc-dependent manner by calcineurin B-like-interacting protein kinase15 (CIPK15), which positively regulates the energy homeostasis sensor Suc nonfermenting1 (Snf1)-related protein kinase1 (SnRK1) to promote α-amylase production and ultimately starch breakdown (Lee et al., 2009). To enhance net anaerobic ATP production, the catabolism of Suc by invertase is limited and that by Suc synthase is enhanced (Bailey-Serres and Voesenek, 2008; Bailey-Serres et al., 2012). Further energy economization is accomplished in some plants by the elevation of enzymes that use inorganic pyrophosphate instead of ATP (Huang et al., 2008). For example, flooded rice induces mRNAs encoding pyruvate orthophosphate dikinase as well as a vacuolar inorganic pyrophosphate-dependent proton pump. The increased flux of carbon through glycolysis enables substrate-level ATP production (i.e. pyruvate kinase/pyruvate orthophosphate dikinase), which can only be maintained through the regeneration of NAD+ via pyruvate fermentation to lactate, ethanol, or Ala. These three products have different metabolic ramifications. Lactate production is disadvantageous because it rapidly leads to cytosolic acidosis unless actively effluxed out of the cell. Ethanol production is also disadvantageous because it allows carbon to be lost by diffusion. However, transport of ethanol from roots to shoots, where it escapes to the atmosphere, is a waterlogging tolerance mechanism in oak (Quercus spp.; Ferner et al., 2012). By contrast, increases in Ala aminotransferase, which catalyzes a transaminase reaction that converts pyruvate and Glu to Ala and 2-oxoglutarate, may conserve carbon and facilitate ATP production by the tricarboxylic acid cycle enzyme succinate-CoA ligase (Rocha et al., 2010; Sweetlove et al., 2010; Bailey-Serres et al., 2012).
Another factor in the flooding survival equation is the regulation of energy use. In rice that is capable of rapid underwater elongation, energy is expended in cell division and growth in stem intercalary meristems (Kende et al., 1998). By contrast, energy-conserving measures are often invoked in response to severe oxygen deprivation by the down-regulation of ATP-demanding biosynthetic reactions such as ribosome biogenesis and cell wall biosynthesis. A programmatic repression of translation during oxygen deprivation in rice is calculated to conserve significant amounts of ATP (Edwards et al., 2012). Energy conservation was also reported in Arabidopsis, where hypoxia constrains translational initiation to a subset of cellular mRNAs (Branco-Price et al., 2005, 2008). The transcripts that were efficiently translated included the hypoxia-responsive core gene set. On the other hand, over 65% of total cellular mRNAs were stable but translationally repressed during the stress due to a sequestration mechanism that is rapidly reversed upon reoxygenation. In summary, adjustments manifested in response to low oxygen and flooding include adaptations to maximize anaerobic ATP production from limited energy reserves. The manipulation of genes that regulate metabolic flux and energy use could enable the production of more flooding-tolerant germplasm.
LOW-OXYGEN SENSING AND SIGNALING
Is low-oxygen sensing “direct” (i.e. determined by oxygen concentration) or “indirect” (i.e. as a consequence of a decline or increase in oxygen concentration)? This question has proven to be a longstanding challenge to plant biologists due to the inability to noninvasively monitor cellular oxygen concentration in concert with other cellular processes such as gene transcription and metabolic flux. Nonetheless, recent progress in this area indicates that plants are capable of both indirect and direct sensing of changes in oxygen availability.
Indirect Sensing
Indirect low-oxygen sensing is thought to involve the perception of dynamics in levels of adenylates, carbohydrates, and pyruvate as well as localized cellular changes in pH, Ca2+, ROS, and nitric oxide (NO; Bailey-Serres and Chang, 2005; Rhoads and Subbaiah, 2007; Bailey-Serres and Voesenek 2008; Blokhina and Fagerstedt, 2010). The decline in adenylates or carbohydrates is likely to trigger SnRK1-regulated carbon management (Baena-González, 2010). In Arabidopsis, the energy-sensing SnRK1s are KIN10 and KIN11, which manage carbon utilization under hypoxia and/or carbohydrate starvation (Baena-González et al., 2007). KIN10 positively regulates the S group of bZIP transcription factors, genes associated with carbohydrate and amino acid catabolism, nighttime starch breakdown, and leaf senescence (Baena-González et al., 2007; Cho et al., 2012). Included among the KIN10/11-regulated genes is EXORDIUM-LIKE1, a HUP that is necessary for carbon management under oxygen deprivation (Schröder et al., 2011). Consistently, up-regulation of the SnRK1 pathway via CIPK15 in germinating rice seeds enhanced starch breakdown and seedling coleoptile growth (Lee et al., 2009). The current view is that SnRK1 signaling is negatively regulated by Glc-6-P and/or trehalose-6-P and antagonizes the nutrient- and energy-sensing pathway regulated by Target of Rapamycin (TOR) kinase (Baena-González, 2010). A reasonable hypothesis is that low-energy sensing via the SnRK1 pathway promotes sufficient carbohydrate catabolism for the survival of low oxygen while inhibiting TOR signaling and thereby growth.
Indirect sensing mediated by changes in cytosolic Ca2+, ROS, and NO may be the consequence of the inhibition of mitochondrial electron transport (Bailey-Serres and Chang, 2005; Rhoads and Subbaiah, 2007). Recent studies with Arabidopsis showed that both severe oxygen deprivation and reoxygenation promote mitochondrial generation of ROS at complex III, which transiently activates MPK3, MPK4, and MPK6 (Chang et al., 2012). As noted for KIN10/11 signaling, MPK6 activation was not involved in the induction of hypoxia-induced mRNAs. Instead, its activation limited the number of hypoxia-reduced transcripts. This led to the suggestion that MPK6 may participate in maintaining poorly translated mRNAs during the stress so that they may be translated upon reoxygenation. Mitochondrial emission of NO also occurs under severe hypoxia (less than 1% oxygen) and was linked to signal transduction and metabolism (Hebelstrup et al., 2012; Hill, 2012). Hebelstrup et al. (2012) showed that NO contributes to ethylene-dependent leaf hyponasty. The levels of NO and the degree of hyponasty were reduced by the overexpression of class 1 nonsymbiotic hemoglobin genes, including HEMOGLOBIN1 (HB1/GLB1), a core hypoxia-responsive gene of Arabidopsis (Mustroph et al., 2009).
Direct Oxygen Sensing Regulates the Turnover of Transcription Factors
A direct oxygen-sensing mechanism is well defined in animals. The basic helix-loop-helix domain protein Hypoxia-Inducible Factor1α (HIF1α) is a key transcriptional regulator of hypoxia responses (Semenza, 2012). This protein is constitutively synthesized but targeted for proteasomal degradation when hydroxylated by oxygen-dependent prolyl hydroxylases (PHDs) at two Pro residues. This modification allows interaction with the von Hippel-Lindau tumor suppressor protein, enables an E3 ubiquitin ligase complex to bind HIF1α, and initiates proteasome-mediated turnover. The HIF1α PHDs have a relatively high Km (230–250 µm) for oxygen. Therefore, well before oxygen levels are fully depleted, HIF1α is stabilized and binds to its partner HIF1β to activate the transcription of genes that enable the survival of hypoxia. HIF1α orthologs are conserved within the animal kingdom (Loenarz et al., 2011) but absent from plants (Mustroph et al., 2010). To date, there is no evidence that plant PHDs function in oxygen sensing.
A direct oxygen-sensing mechanism that regulates the stability of the five group VII ERF transcription factors of Arabidopsis was recently reported in two independent studies (Gibbs et al., 2011; Licausi et al., 2011; for review, see Sasidharan and Mustroph, 2011). These Arabidopsis ERFs are in the same clade as the rice SUB1 and SK proteins and include HYPOXIA-RESPONSIVE ERF1 (HRE1), HRE2, RELATED TO AP2 2.2 (RAP2.2), RAP2.12, and RAP2.3/ETHYLENE-RESPONSIVE ELEMENT BINDING PROTEIN. The regulation of each of these genes is distinct in seedlings, with HRE1 and HRE2 strongly up-regulated by hypoxia, HRE1, RAP2.2, and RAP2.3 up-regulated by ethylene, and RAP2.12, RAP2.2, and RAP2.3 expressed constitutively (Mustroph et al., 2009; Hinz et al., 2010; Licausi et al., 2010; Hess et al., 2011; Yang et al., 2011). Evaluation of single and combinatorial mutants or RNA interference lines for several of these genes indicated that they are important for low-oxygen survival but overlap in function. Although RAP2.12 overexpression enhances low-oxygen survival, neither RAP2.12, RAP2.2, HRE1, nor HRE2 overexpression enhances the accumulation of hypoxia-responsive mRNAs under nonstress conditions (Papdi et al., 2008; Hinz et al., 2010; Licausi et al., 2010). This paradox is explained by the instability of the Arabidopsis group VII ERF proteins under oxygen-replete conditions.
The amino-end (N-end) rule pathway of targeted proteolysis determines the turnover of polypeptides with specific exposed N-terminal residues (Varshavsky, 2011). This hierarchical mechanism of proteasome-mediated turnover is well studied in mammalian and yeast (Saccharomyces cerevisiae) systems, whereas its function in plant development was established but no targets were known (Graciet et al., 2009, 2010; Graciet and Wellmer, 2010). The so-called arginylation-N (Arg/N) branch of the pathway governs the turnover of proteins with specific N-terminal residues, one of which is a Cys. Based on studies in animals, both NO and oxygen are required for the oxidation of an N-terminal Cys to either Cys-sulfinic (CysO2H) or Cys-sulfonic (CysO3H) acid. This modified N terminus is recognized by an arginyl-tRNA protein transferase that generates an Arg-Cysox protein or peptide (an N-recognin) that is targeted by an E3 ubiquitin ligase, ubiquitinated, and destroyed by the 26S proteasome (Varshavsky, 2011). In Arabidopsis and tobacco (Nicotiana tabacum), proteins with a Cys immediately following the first Met can be cleaved by a Met-amino peptide to generate an N-terminal Cys that targets the protein for turnover (Graciet and Wellmer, 2010). Arabidopsis has two arginyl transferases, ATE1 and ATE2, and at least one E3 ligase (PROTEOLYSIS6 [PRT6]) that recognizes Arg/N pathway N-recognins (Graciet et al., 2009). Since oxidation of the N-terminal Cys is a noncatalytic event, this subbranch of the Arg/N turnover pathway can function as a sensor of oxygen and NO (Varshavsky, 2011), as demonstrated in its regulation of three GTPase-activating proteins in embryos of mice (Hu et al., 2005; Lee et al., 2005).
It was recognized that prt6 and ate1 ate2 mutants of Arabidopsis constitutively express many core hypoxia-responsive genes under normal growth conditions (Gibbs et al., 2011). This led to the discovery that all five group VII ERFs are Arg/N targets. Independently, Licausi et al. (2011) deduced the importance of a conserved N-terminal motif characteristic of group VII ERFs, which includes a Cys at the second position (NH2-MCGGAI/L; Nakano et al., 2006), and its relationship to Arg/N pathway regulation. The importance of the second Cys was confirmed in several ways. First, mutation of the second Cys to an Ala stabilized all five Arabidopsis group VII proteins in an in vitro assay from rabbit reticulocytes that contain N-end rule activity (Gibbs et al., 2011). Second, deletion of the N terminus of RAP2.12 or mutation of Cys-2 to Ala-2 in RAP.12 and HRE2 was sufficient for protein stabilization under normal growth conditions (Gibbs et al., 2011; Licausi et al., 2011). Licausi et al. (2011) also found that RAP2.12 associates with Acyl-CoA-binding protein1 and -2 at the plasma membrane (Li and Chye, 2004) using yeast two-hybrid and bimolecular fluorescence complementation analyses. Based on imaging RAP2.12-GFP in plant cells, they were able to determine that a reservoir of this ERF accumulates at the plasma membrane under nonstress conditions that is relocated to the nucleus under low-oxygen stress and disappears upon reoxygenation.
These findings have led to a homeostatic oxygen-sensing model for Arabidopsis (Bailey-Serres et al., 2012). The model proposes that Met-amino peptide activity exposes the N-terminal Cys, which is necessary for subsequent N-terminal arginylation by ATE1/ATE2 and then recognition by PRT6 or other E3 ligases. Missing data to solidify this model include the confirmation of Cys oxidation under normoxia or reoxygenation, knowledge of how RAP2.12 is released from the plasma membrane, and the identification of the direct gene targets of the individual ERFs. It seems reasonable to suggest that RAP2.12 and possibly other plasma membrane-bound group VII ERFs provide the first battalion for the activation of hypoxia-responsive genes, including HRE2, which may further promote the up-regulation of hypoxia-responsive transcripts. The up-regulation of two Arabidopsis group VII ERF genes by ethylene may provide a means for submerged shoots to prepare for oxygen deprivation upon submergence but prior to severe oxygen deprivation.
The modulation of ERF abundance by the Arg/N is likely to directly reflect the cellular oxygen content turnover pathway but might also be coordinated by dynamics in NO levels. It has been known for a considerable time that the NO scavenger HB1 contributes to the regulation of low-oxygen gene regulation and metabolism during seed development and transient hypoxia in several species (Hill, 2012). Therefore, low-oxygen-triggered NO production and HB1 scavenging may be involved in the regulation of group VII ERF turnover.
Rice SUB1A possesses most residues of the conserved N-terminal motif of group VII ERFs, raising the possibility that it is also Arg/N turnover pathway regulated. However, when assayed in a reticulocyte lysate system with N-end rule activity, SUB1A was stable, whereas a number of other plant group VII ERFs were unstable, including all five Arabidopsis family members (Gibbs et al., 2011; Bailey-Serres et al., 2012; E. Brinton and J. Bailey-Serres, unpublished data). This suggests that this key submergence survival regulator of rice may have evolved to evade Arg/N turnover pathway regulation (Gibbs et al., 2011; Bailey-Serres et al., 2012). To limit carbohydate consumption and growth during submergence when ethylene levels are high but oxygen levels are not depressed because of underwater photosynthesis, SUB1A may have acquired mutations that released it from oxygen-regulated turnover. SUB1C, which appears to function downstream of GA during submergence, completely lacks the N terminus typical of group VII ERFs. Perhaps other examples of selection in group VII ERFs to modulate flooding survival will be uncovered in wetland species by new genomic technologies.
IMPROVEMENT OF FLOODING SURVIVAL IN RICE AND OTHER CROPS
Over a period of 60 years, a flooding-resistant landrace (FR13A) of rice was collected, recognized as a genetic resource for submergence tolerance, used to identify a key stress tolerance gene, and crossed into advanced varieties (Bailey-Serres et al., 2010). The molecular identification of SUB1A greatly facilitated the establishment of an effective marker-assisted breeding strategy that utilizes single nucleotide polymorphisms in SUB1A and SUB1C along with markers elsewhere on the 12 rice chromosomes (Xu et al., 2006; Neeraja et al., 2007; Septiningsih et al., 2009; Iftekharuddaula et al., 2011). To date, the SUB1 locus of FR13A has been bred into 10 varieties favored by farmers in different locales of south and southeast Asia. The rapid adoption of Sub1 rice by farmers is attributed to its effectiveness, high similarity to the varieties it replaces, and involvement of farmers in the varietal selection (Singh et al., 2009; Manzanilla et al., 2011). However, additional loci that further improve submergence tolerance are present in rice germplasm and essential for developing robust flooding insurance (Septiningsih et al., 2012). It is anticipated that these and other survival traits, such as anaerobic germination, salinity tolerance, and drought tolerance, can be pyramided in cultivars to stabilize production in the rain-fed lowlands. For other crops, increased flooding tolerance might also be harnessed from within the species or wild relatives. For maize, loci from the teosinte Z. nicaraguensis may provide effective aerenchyma development, prolific adventitious rooting, and an effective radial oxygen loss barrier in roots without a yield penalty (Mano et al., 2012). Barley and wheat may benefit from genes from wetland species, such as the barley relative Hordeum marinum, which thrives in waterlogged and saline ecosystems. Encouragingly, wide hybridization of this species to the wheat Triticum aestivum resulted in amphiploid hybrids with improved root aeration in flooded conditions (Malik et al., 2011).
Although natural genetic variation may provide solutions for flooding stress for some crops, the engineering of survival strategies is also warranted, especially due to the urgent need to further grain production in food-insecure areas. The manipulation of group VII ERFs and components of the Arg/N branch of the N-end rule pathway provide promise (Gibbs et al., 2011; Licausi et al., 2011). Although overexpression of SUB1A in Arabidopsis did not confer submergence tolerance, it recapitulated phenotypes associated with ectopic expression of SUB1A in rice (Peña-Castro et al., 2011). Judicial selection of promoters with appropriate temporal and spatial regulation and consideration of posttranscriptional and posttranslational mechanisms of regulation are likely to be critical for successful engineering of flooding tolerance.
CONCLUSION
There is growing evidence of conserved strategies that enable flooding survival and involve signal transduction as a consequence of altered homoeostasis in ethylene, oxygen, and energy reserves. The ethylene-regulated processes interact with modules controlled by other hormones, including ABA, GA, and auxin, as well as ROS and NO, to control elongation growth and aeration. Evolution has tinkered with the key circuitry that regulates flooding tolerance to enable successful tolerance and avoidance strategies. It is anticipated that future studies that integrate genomic technologies with ecophysiological studies will prove instructive for the breeding and engineering of more waterproof crops.
Acknowledgments
We apologize to any of our colleagues whose work was not cited because of space constraints. Motoyuki Ashikari, Mikio Nakazono, Ole Pedersen, Margaret Sauter, and Rens Voesenek graciously contributed images.
Glossary
- QTL
quantitative trait locus
- ERF
ethylene-response factor
- ABA
abscisic acid
- LT50
median lethal time
- ROS
reactive oxygen species
- NO
nitric oxide
- PHDs
prolyl hydroxylases
- N-end
amino-end
References
- Abiko T, Kotula L, Shiono K, Malik AI, Colmer TD, Nakazono M. (2012) Enhanced formation of aerenchyma and induction of a barrier to radial oxygen loss in adventitious roots of Zea nicaraguensis contribute to its waterlogging tolerance as compared with maize (Zea mays ssp. mays). Plant Cell Environ 35: 1618–1630 [DOI] [PubMed] [Google Scholar]
- Akman M, Bhikharie AV, McLean EH, Boonman A, Visser EJ, Schranz ME, van Tienderen PH. (2012) Wait or escape? Contrasting submergence tolerance strategies of Rorippa amphibia, Rorippa sylvestris and their hybrid. Ann Bot (Lond) 109: 1263–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angaji SA, Septiningsih EM, Mackill DJ, Ismail AM. (2010) QTLs associated with tolerance of flooding during germination in rice (Oryza sativa L.). Euphytica 172: 159–168 [Google Scholar]
- Arshad RR, Shafi S. (2010) Pakistan Floods 2010: Preliminary Damage and Needs Assessment. Asian Development Bank and World Bank, Islamabad, Pakistan
- Baena-González E. (2010) Energy signaling in the regulation of gene expression during stress. Mol Plant 3: 300–313 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Chang R. (2005) Sensing and signalling in response to oxygen deprivation in plants and other organisms. Ann Bot (Lond) 96: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Fukao T, Gibbs DJ, Holdsworth MJ, Lee SC, Licausi F, Perata P, Voesenek LACJ, van Dongen JT. (2012) Making sense of low oxygen sensing. Trends Plant Sci 17: 129–138 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Fukao T, Ronald P, Ismail A, Heuer S, Mackill D. (2010) Submergence tolerant rice: SUB1’s journey from landrace to modern cultivar. Rice 3: 138–147 [Google Scholar]
- Bailey-Serres J, Voesenek LACJ. (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Barding GA, Jr, Fukao T, Béni S, Bailey-Serres J, Larive CK. (2012) Differential metabolic regulation governed by the rice SUB1A gene during submergence stress and identification of alanylglycine by 1H NMR spectroscopy. J Proteome Res 11: 320–330 [DOI] [PubMed] [Google Scholar]
- Benschop JJ, Jackson MB, Gühl K, Vreeburg RA, Croker SJ, Peeters AJ, Voesenek LACJ. (2005) Contrasting interactions between ethylene and abscisic acid in Rumex species differing in submergence tolerance. Plant J 44: 756–768 [DOI] [PubMed] [Google Scholar]
- Blokhina O, Fagerstedt KV. (2010) Oxidative metabolism, ROS and NO under oxygen deprivation. Plant Physiol Biochem 48: 359–373 [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J. (2008) Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56: 743–755 [DOI] [PubMed] [Google Scholar]
- Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J. (2005) Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann Bot (Lond) 96: 647–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catling D. (1992) Rice in Deepwater. MacMillan Press, London
- Chang R, Jang CJ, Branco-Price C, Nghiem P, Bailey-Serres J. (2012) Transient MPK6 activation in response to oxygen deprivation and reoxygenation is mediated by mitochondria and aids seedling survival in Arabidopsis. Plant Mol Biol 78: 109–122 [DOI] [PubMed] [Google Scholar]
- Chen X, Pierik R, Peeters AJ, Poorter H, Visser EJ, Huber H, de Kroon H, Voesenek LACJ. (2010) Endogenous abscisic acid as a key switch for natural variation in flooding-induced shoot elongation. Plant Physiol 154: 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Hong JW, Kim EC, Yoo SD. (2012) Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol 158: 1955–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JA, Llewellyn DJ, Dennis ES, Wilson IW. (2010) Global gene expression responses to waterlogging in roots and leaves of cotton (Gossypium hirsutum L.). Plant Cell Physiol 51: 21–37 [DOI] [PubMed] [Google Scholar]
- Colmer TD, Voesenek LACJ. (2009) Flooding tolerance: suites of plant traits in variable environments. Funct Plant Biol 36: 665–681 [DOI] [PubMed] [Google Scholar]
- Colmer TD, Winkel A, Pedersen O. (2011) A perspective on underwater photosynthesis in submerged terrestrial wetland plants. AoB Plants 2011: plr030
- Cox MCH, Benschop JJ, Vreeburg RAM, Wagemaker CAM, Moritz T, Peeters AJM, Voesenek LACJ. (2004) The roles of ethylene, auxin, abscisic acid, and gibberellin in the hyponastic growth of submerged Rumex palustris petioles. Plant Physiol 136: 2948–2960, discussion 3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC, He CJ, Morgan PW. (2000) Programmed cell death and aerenchyma formation in roots. Trends Plant Sci 5: 123–127 [DOI] [PubMed] [Google Scholar]
- Edwards JM, Roberts TH, Atwell BJ. (2012) Quantifying ATP turnover in anoxic coleoptiles of rice (Oryza sativa) demonstrates preferential allocation of energy to protein synthesis. J Exp Bot 63: 4389–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferner E, Rennenberg H, Kreuzwieser J. (2012) Effect of flooding on C metabolism of flood-tolerant (Quercus robur) and non-tolerant (Fagus sylvatica) tree species. Tree Physiol 32: 135–145 [DOI] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J. (2008) Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc Natl Acad Sci USA 105: 16814–16819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J. (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18: 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Yeung E, Bailey-Serres J. (2011) The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23: 412–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Yeung E, Bailey-Serres J. (2012) The submergence tolerance gene, SUB1A, delays leaf senescence under prolonged darkness through hormonal regulation in rice. Plant Physiol 160: 1795–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey-Serres J, et al. (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graciet E, Mesiti F, Wellmer F. (2010) Structure and evolutionary conservation of the plant N-end rule pathway. Plant J 61: 741–751 [DOI] [PubMed] [Google Scholar]
- Graciet E, Walter F, Maoiléidigh DO, Pollmann S, Meyerowitz EM, Varshavsky A, Wellmer F. (2009) The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc Natl Acad Sci USA 106: 13618–13623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graciet E, Wellmer F. (2010) The plant N-end rule pathway: structure and functions. Trends Plant Sci 15: 447–453 [DOI] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Ashikari M. (2011) Rice growth adapting to deepwater. Curr Opin Plant Biol 14: 100–105 [DOI] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hebelstrup KH, van Zanten M, Mandon J, Voesenek LACJ, Harren FJ, Cristescu SM, Møller IM, Mur LA. (2012) Haemoglobin modulates NO emission and hyponasty under hypoxia-related stress in Arabidopsis thaliana. J Exp Bot 63: 5581–5591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess N, Klode M, Anders M, Sauter M. (2011) The hypoxia responsive transcription factor genes ERF71/HRE2 and ERF73/HRE1 of Arabidopsis are differentially regulated by ethylene. Physiol Plant 143: 41–49 [DOI] [PubMed] [Google Scholar]
- Hill RD. (2012) Non-symbiotic haemoglobins: what’s happening beyond nitric oxide scavenging? AoB Plants 2012: pls004
- Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R. (2010) Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol 153: 757–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu FC, Chou MY, Peng HP, Chou SJ, Shih MC. (2011) Insights into hypoxic systemic responses based on analyses of transcriptional regulation in Arabidopsis. PLoS ONE 6: e28888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu RG, Sheng J, Qi X, Xu Z, Takahashi TT, Varshavsky A. (2005) The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437: 981–986 [DOI] [PubMed] [Google Scholar]
- Huang S, Colmer TD, Millar AH. (2008) Does anoxia tolerance involve altering the energy currency towards PPi? Trends Plant Sci 13: 221–227 [DOI] [PubMed] [Google Scholar]
- Iftekharuddaula KM, Newaz MA, Salam MA, Ahmed HU, Mahbub MAA, Septiningsih EM, Collard BCY, Sanchez DL, Pamplona AM, Mackill DJ. (2011) Rapid and high-precision marker assisted backcrossing to introgress the SUB1 QTL into BR11, the rainfed lowland rice mega variety of Bangladesh. Euphytica 178: 83–97 [Google Scholar]
- Ismail AM, Johnson DE, Ella ES, Vergara GV, Baltazar AM. (2012) Adaptation to flooding during emergence and seedling growth in rice and weeds, and implications for crop establishment. AoB Plants 2012: pls019
- Jackson MB. (2008) Ethylene-promoted elongation: an adaptation to submergence stress. Ann Bot (Lond) 101: 229–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MB, Armstrong W. (1999) Formation of aerenchyma and the processes of plant ventilation in relation to soil flooding and submergence. Plant Biol 1: 274–287 [Google Scholar]
- Jung KH, Seo YS, Walia H, Cao P, Fukao T, Canlas PE, Amonpant F, Bailey-Serres J, Ronald PC. (2010) The submergence tolerance regulator Sub1A mediates stress-responsive expression of AP2/ERF transcription factors. Plant Physiol 152: 1674–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H, van der Knaap E, Cho HT. (1998) Deepwater rice: a model plant to study stem elongation. Plant Physiol 118: 1105–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzwieser J, Hauberg J, Howell KA, Carroll A, Rennenberg H, Millar AH, Whelan J. (2009) Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol 149: 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasanthi-Kudahettige R, Magneschi L, Loreti E, Gonzali S, Licausi F, Novi G, Beretta O, Vitulli F, Alpi A, Perata P. (2007) Transcript profiling of the anoxic rice coleoptile. Plant Physiol 144: 218–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Chen PW, Lu CA, Chen S, Ho TH, Yu SM. (2009) Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci Signal 2: ra61. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Tasaki T, Moroi K, An JY, Kimura S, Davydov IV, Kwon YT. (2005) RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci USA 102: 15030–15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Mustroph A, Sasidharan R, Vashisht D, Pedersen O, Oosumi T, Voesenek LACJ, Bailey-Serres J. (2011) Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol 190: 457–471 [DOI] [PubMed] [Google Scholar]
- Li HY, Chye ML. (2004) Arabidopsis acyl-CoA-binding protein ACBP2 interacts with an ethylene-responsive element-binding protein, AtEBP, via its ankyrin repeats. Plant Mol Biol 54: 233–243 [DOI] [PubMed] [Google Scholar]
- Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LACJ, Perata P, van Dongen JT. (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479: 419–422 [DOI] [PubMed] [Google Scholar]
- Licausi F, van Dongen JT, Giuntoli B, Novi G, Santaniello A, Geigenberger P, Perata P. (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J 62: 302–315 [DOI] [PubMed] [Google Scholar]
- Liu F, Vantoai T, Moy LP, Bock G, Linford LD, Quackenbush J. (2005) Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol 137: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenarz C, Coleman ML, Boleininger A, Schierwater B, Holland PW, Ratcliffe PJ, Schofield CJ. (2011) The hypoxia-inducible transcription factor pathway regulates oxygen sensing in the simplest animal, Trichoplax adhaerens. EMBO Rep 12: 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P. (2005) A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol 137: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik AI, Islam AK, Colmer TD. (2011) Transfer of the barrier to radial oxygen loss in roots of Hordeum marinum to wheat (Triticum aestivum): evaluation of four H. marinum-wheat amphiploids. New Phytol 190: 499–508 [DOI] [PubMed] [Google Scholar]
- Mano Y, Omori F, Takeda K. (2012) Construction of intraspecific linkage maps, detection of a chromosome inversion, and mapping of QTL for constitutive root aerenchyma formation in the teosinte Zea nicaraguensis. Mol Breed 29: 137–146 [Google Scholar]
- Manzanilla DO, Paris TR, Vergara GV, Ismail AM, Pandey S, Labios RV, Tatlonghari GT, Acda RD, Chi TTN, Duoangsila K, et al. (2011) Submergence risks and farmers’ preferences: implications for breeding Sub1 rice in southeast Asia. Agric Syst 104: 335–347 [Google Scholar]
- Mergemann H, Sauter M. (2000) Ethylene induces epidermal cell death at the site of adventitious root emergence in rice. Plant Physiol 124: 609–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalcová D, Gilbert JC, Lawson CS, Gowing DJG, Marrs HM. (2011) The combined effect of waterlogging, extractable P and soil pH on α-diversity: a case study on mesotrophic grasslands in the UK. Plant Ecol 212: 879–888 [Google Scholar]
- Mommer L, Lenssen JPM, Huber H, Visser EJW, De Kroon H. (2007) Ecophysiological determinants of plant performance under flooding: a comparative study of seven plant families. J Ecol 94: 1117–1129 [Google Scholar]
- Mühlenbock P, Plaszczyca M, Plaszczyca M, Mellerowicz E, Karpinski S. (2007) Lysigenous aerenchyma formation in Arabidopsis is controlled by LESION SIMULATING DISEASE1. Plant Cell 19: 3819–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Bailey-Serres J. (2010) The Arabidopsis translatome cell-specific mRNA atlas: mining suberin and cutin lipid monomer biosynthesis genes as an example for data application. Plant Signal Behav 5: 320–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, Yaghoubi-Masihi A, Fukao T, Bailey-Serres J. (2010) Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 152: 1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo Y, Maruyama K, Yasue H, Yamaguchi-Shinozaki K, Shinozaki K, Komatsu S. (2011) Transcriptional responses to flooding stress in roots including hypocotyl of soybean seedlings. Plant Mol Biol 77: 129–144 [DOI] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Carroll A, Ivanova A, Millar AH, Whelan J. (2009) Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol 151: 306–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Rocha M, Geigenberger P, Whelan J, van Dongen JT. (2011) Comparative analysis between plant species of transcriptional and metabolic responses to hypoxia. New Phytol 190: 472–487 [DOI] [PubMed] [Google Scholar]
- Neeraja CN, Maghirang-Rodriguez R, Pamplona A, Heuer S, Collard BC, Septiningsih EM, Vergara G, Sanchez D, Xu K, Ismail AM, et al. (2007) A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor Appl Genet 115: 767–776 [DOI] [PubMed] [Google Scholar]
- Olesen JE, Trnka M, Kersebaum KC, Skjelvåg AO, Seguin B, Peltonen-Sainio P, Rossi F, Kozyra J, Micale F. (2011) Impacts and adaptation of European crop production systems to climate change. Eur J Agron 34: 96–112 [Google Scholar]
- Papdi C, Abrahám E, Joseph MP, Popescu C, Koncz C, Szabados L. (2008) Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol 147: 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelle J, Brendel O, Jolivet Y, Dreyer E. (2007) Intra- and interspecific diversity in the response to waterlogging of two co-occurring white oak species (Quercus robur and Q. petraea). Tree Physiol 27: 1027–1034 [DOI] [PubMed] [Google Scholar]
- Parlanti S, Kudahettige NP, Lombardi L, Mensuali-Sodi A, Alpi A, Perata P, Pucciariello C. (2011) Distinct mechanisms for aerenchyma formation in leaf sheaths of rice genotypes displaying a quiescence or escape strategy for flooding tolerance. Ann Bot (Lond) 107: 1335–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolin P, Lucas C, Piedade MT, Wittmann F. (2010) Drought responses of flood-tolerant trees in Amazonian floodplains. Ann Bot (Lond) 105: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen O, Rich SM, Colmer TD. (2009) Surviving floods: leaf gas films improve O2 and CO2 exchange, root aeration, and growth of completely submerged rice. Plant J 58: 147–156 [DOI] [PubMed] [Google Scholar]
- Peña-Castro JM, van Zanten M, Lee SC, Patel MR, Voesenek LACJ, Fukao T, Bailey-Serres J. (2011) Expression of rice SUB1A and SUB1C transcription factors in Arabidopsis uncovers flowering inhibition as a submergence tolerance mechanism. Plant J 67: 434–446 [DOI] [PubMed] [Google Scholar]
- Pierik R, De Wit M, Voesenek LACJ. (2011) Growth-mediated stress escape: convergence of signal transduction pathways activated upon exposure to two different environmental stresses. New Phytol 189: 122–134 [DOI] [PubMed] [Google Scholar]
- Rajhi I, Yamauchi T, Takahashi H, Nishiuchi S, Shiono K, Watanabe R, Mliki A, Nagamura Y, Tsutsumi N, Nishizawa NK, et al. (2011) Identification of genes expressed in maize root cortical cells during lysigenous aerenchyma formation using laser microdissection and microarray analyses. New Phytol 190: 351–368 [DOI] [PubMed] [Google Scholar]
- Rhine M, Stevens G, Shannon G, Wrather A, Sleper D. (2010) Yield and nutritional responses to waterlogging of soybean cultivars. Irrig Sci 28: 135–142 [Google Scholar]
- Rhoads DM, Subbaiah CC. (2007) Mitochondrial retrograde regulation in plants. Mitochondrion 7: 177–194 [DOI] [PubMed] [Google Scholar]
- Rocha M, Licausi F, Araújo WL, Nunes-Nesi A, Sodek L, Fernie AR, van Dongen JT. (2010) Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol 152: 1501–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saika H, Okamoto M, Miyoshi K, Kushiro T, Shinoda S, Jikumaru Y, Fujimoto M, Arikawa T, Takahashi H, Ando M, et al. (2007) Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol 48: 287–298 [DOI] [PubMed] [Google Scholar]
- Sasidharan R, Mustroph A. (2011) Plant oxygen sensing is mediated by the N-end rule pathway: a milestone in plant anaerobiosis. Plant Cell 23: 4173–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder F, Lisso J, Müssig C. (2011) EXORDIUM-LIKE1 promotes growth during low carbon availability in Arabidopsis. Plant Physiol 156: 1620–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seago JL, Jr, Marsh LC, Stevens KJ, Soukup A, Votrubová O, Enstone DE. (2005) A re-examination of the root cortex in wetland flowering plants with respect to aerenchyma. Ann Bot (Lond) 96: 565–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. (2012) Hypoxia-inducible factors in physiology and medicine. Cell 148: 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Septiningsih EM, Pamplona AM, Sanchez DL, Neeraja CN, Vergara GV, Heuer S, Ismail AM, Mackill DJ. (2009) Development of submergence-tolerant rice cultivars: the Sub1 locus and beyond. Ann Bot (Lond) 103: 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Septiningsih EM, Sanchez DL, Singh N, Sendon PM, Pamplona AM, Heuer S, Mackill DJ. (2012) Identifying novel QTLs for submergence tolerance in rice cultivars IR72 and Madabaru. Theor Appl Genet 124: 867–874 [DOI] [PubMed] [Google Scholar]
- Setter T, Water I. (2003) Review of prospects for germplasm improvement for waterlogging tolerance in wheat, barley and oats. Plant Soil 253: 1–34 [Google Scholar]
- Setter TL, Waters I, Sharma SK, Singh KN, Kulshreshtha N, Yaduvanshi NP, Ram PC, Singh BN, Rane J, McDonald G, et al. (2009) Review of wheat improvement for waterlogging tolerance in Australia and India: the importance of anaerobiosis and element toxicities associated with different soils. Ann Bot (Lond) 103: 221–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura S, Yamamoto R, Nakamura T, Shimada S, Komatsu S. (2010) Stem hypertrophic lenticels and secondary aerenchyma enable oxygen transport to roots of soybean in flooded soil. Ann Bot (Lond) 106: 277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingaki-Wells RN, Huang S, Taylor NL, Carroll AJ, Zhou W, Millar AH. (2011) Differential molecular responses of rice and wheat coleoptiles to anoxia reveal novel metabolic adaptations in amino acid metabolism for tissue tolerance. Plant Physiol 156: 1706–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Dang TT, Vergara GV, Pandey DM, Sanchez D, Neeraja CN, Septiningsih EM, Mendioro M, Tecson-Mendoza EM, Ismail AM, et al. (2010) Molecular marker survey and expression analyses of the rice submergence-tolerance gene SUB1A. Theor Appl Genet 121: 1441–1453 [DOI] [PubMed] [Google Scholar]
- Singh S, Mackill DJ, Ismal AM. (2009) Responses of SUB1 rice introgression lines to submergence in the field: yield and grain quality. Field Crops Res 113: 12–23 [Google Scholar]
- Steffens B, Geske T, Sauter M. (2011) Aerenchyma formation in the rice stem and its promotion by H2O2. New Phytol 190: 369–378 [DOI] [PubMed] [Google Scholar]
- Steffens B, Kovalev A, Gorb SN, Sauter M. (2012) Emerging roots alter epidermal cell fate through mechanical and reactive oxygen species signaling. Plant Cell 24: 3296–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B, Sauter M. (2009) Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. Plant Cell 21: 184–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B, Sauter M. (2010) G proteins as regulators in ethylene-mediated hypoxia signaling. Plant Signal Behav 5: 375–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP. (2011) The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol 21: R338–R345 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Beard KF, Nunes-Nesi A, Fernie AR, Ratcliffe RG. (2010) Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci 15: 462–470 [DOI] [PubMed] [Google Scholar]
- Thomas AL, Guerreiro SM, Sodek L. (2005) Aerenchyma formation and recovery from hypoxia of the flooded root system of nodulated soybean. Ann Bot (Lond) 96: 1191–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JT, Fröhlich A, Ramírez-Aguilar SJ, Schauer N, Fernie AR, Erban A, Kopka J, Clark J, Langer A, Geigenberger P. (2009) Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann Bot (Lond) 103: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. (2011) The N-end rule pathway and regulation by proteolysis. Protein Sci 20: 1298–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashisht D, Hesselink A, Pierik R, Ammerlaan JM, Bailey-Serres J, Visser EJ, Pedersen O, van Zanten M, Vreugdenhil D, Jamar DC, et al. (2011) Natural variation of submergence tolerance among Arabidopsis thaliana accessions. New Phytol 190: 299–310 [DOI] [PubMed] [Google Scholar]
- Voesenek LACJ, Colmer TD, Pierik R, Millenaar FF, Peeters AJ. (2006) How plants cope with complete submergence. New Phytol 170: 213–226 [DOI] [PubMed] [Google Scholar]
- Winkel A, Colmer TD, Pedersen O. (2011) Leaf gas films of Spartina anglica enhance rhizome and root oxygen during tidal submergence. Plant Cell Environ 34: 2083–2092 [DOI] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Yang CY, Hsu FC, Li JP, Wang NN, Shih MC. (2011) The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol 156: 202–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi PH, Rafique S, Rai PK, Singh NN, Srinivasan G. (2004) Tolerance to excess moisture in maize (Zea maize L): susceptible crop stages and identification of tolerant genotypes. Field Crops Res 90: 189–202 [Google Scholar]