Abstract
Shoot branching in plants is regulated by many environmental cues and by specific hormones such as strigolactone (SL). We show that the GAT1_2.1 gene (At1g15040) is repressed over 50-fold by nitrogen stress, and is also involved in branching control. At1g15040 is predicted to encode a class I glutamine amidotransferase (GAT1), a superfamily for which Arabidopsis (Arabidopsis thaliana) has 30 potential members. Most members can be categorized into known biosynthetic pathways, for the amidation of known acceptor molecules (e.g. CTP synthesis). Some members, like GAT1_2.1, are of unknown function, likely involved in amidation of unknown acceptors. A gat1_2.1 mutant exhibits a significant increase in shoot branching, similar to mutants in SL biosynthesis. The results suggest that GAT1_2.1 is not involved in SL biosynthesis since exogenously applied GR24 (a synthetic SL) does not correct the mutant phenotype. The subfamily of GATs (GATase1_2), with At1g15040 as the founding member, appears to be present in all plants (including mosses), but not other organisms. This suggests a plant-specific function such as branching control. We discuss the possibility that the GAT1_2.1 enzyme may activate SLs (e.g. GR24) by amidation, or more likely could embody a new pathway for repression of branching.
Shoot branching plays an important role in establishing plant body plans during development and growth, also conferring the flexibility for plants to respond to environmental stresses. The control of bud growth/branching has been studied for many decades with much interest stemming from its value in agriculture. Indeed, many of our domesticated crops have been bred for modified branching to optimize yields. In early studies, auxin synthesized in the shoot apex was proposed to act indirectly to inhibit bud outgrowth, while cytokinin (CK) synthesized in the roots promoted bud outgrowth (Domagalska and Leyser, 2011). Studies on auxin inhibition suggested there should be another signal mediating bud growth control (Hayward et al., 2009; Stirnberg et al., 2010; Domagalska and Leyser, 2011). In the past decade, studies in Arabidopsis (Arabidopsis thaliana) and other plants have addressed this signal. Identification and characterization of mutants with increased branching in garden pea (Pisum sativum), Arabidopsis, rice (Oryza sativa), and Petunia hybrida demonstrated the existence of a long-distance signaling pathway that regulates shoot branching (Beveridge et al., 1996, 1997; Napoli, 1996; Stirnberg et al., 2002, 2007; Sorefan et al., 2003; Booker et al., 2004; Arite et al., 2007; Gomez-Roldan et al., 2008; Umehara et al., 2008, 2010; Lin et al., 2009; Liu et al., 2009, 2011; Zhang et al., 2010). Later, studies on pea (Gomez-Roldan et al., 2008) and rice (Umehara et al., 2008) demonstrated unequivocally that this hormone (or its precursor) is strigolactone (SL). Currently, it is proposed that SL acts downstream of auxin to regulate bud outgrowth (Brewer et al., 2009). It is also likely that SL and auxin have the capacity to modulate each other’s levels and distribution in a dynamic feedback loop required for the branching control (Ferguson and Beveridge, 2009; Hayward et al., 2009; Stirnberg et al., 2010). The interaction between SL and CK during bud outgrowth is less understood, although recent studies in pea indicate that SL and CK act antagonistically on bud growth (Dun et al., 2012).
Branching is also modulated in response to environmental conditions, including nutrient supply. Generally, nutrient deficiency in soil causes a reduction in shoot to root ratio, resulting in decreased shoot branching (Lafever, 1981). Under nitrogen or phosphate limitation, elevated levels of SL repress shoot branching in rice, tomato (Solanum lycopersicum), and Arabidopsis (Yoneyama et al., 2007; López-Ráez et al., 2008; Umehara et al., 2008, 2010; Kohlen et al., 2011), and possibly increase lateral root formation (Ruyter-Spira et al., 2011). This makes sense physiologically, diverting resources to roots from shoots to scavenge more nutrients. The basis for modulation of SL levels or nutrient-dependent branching control is not understood.
Here, we report a novel gene, GAT1_2.1 (At1g15040), predicted to encode a class I Gln amidotransferase (GAT1) in Arabidopsis, is highly repressed by long-term nitrogen stress (down 57-fold), and that mutation of this gene leads to an enhanced branching phenotype. Thus, this gene may present a link between the nitrogen stress response and branching control.
RESULTS
Mutation of a Nitrogen-Regulated GAT Gene Results in an Enhanced Branching Phenotype
We carried out an Agilent microarray to identify Arabidopsis genes that are controlled by long-term nitrogen stress (see “Materials and Methods”). Briefly, seedlings were grown for 15 d in Murashige and Skoog media with 30 mm NH4NO3 (+N) or with 1 mm NH4NO3 (−N). Based on gene expression profiling, 230 genes were activated or repressed greater than 8-fold (Supplemental Data S1). We selected 17 genes (mainly highly controlled regulatory genes or Gln-based enzymes) for which T-DNA insertion lines were available from The Arabidopsis Information Resource (TAIR; Supplemental Table S1). We were interested in whether any of these nitrogen-regulated genes are involved in specific traits displayed under nitrogen replete or limited growth. Among the T-DNA lines, the homozygous gat mutant was found to have an enhanced shoot branching phenotype (Fig. 1A). Figure 1I (top left) shows that GAT mRNA is not present in the gat mutant. The gat mutant line (SALK_031983C) carries a T-DNA insertion in the first exon of two exons in At1g15040. At1g15040 is annotated as a class I Gln amidotransferase-like superfamily gene and encodes a protein referred to as a GAT1_2 domain (Marchler-Bauer et al., 2011). Hence, we designate it GAT1_2.1 and will refer to this as GAT (or gat) in this article. At1g15040 is one of 30 putative genes encoding GAT1 in the Arabidopsis genome (see below).
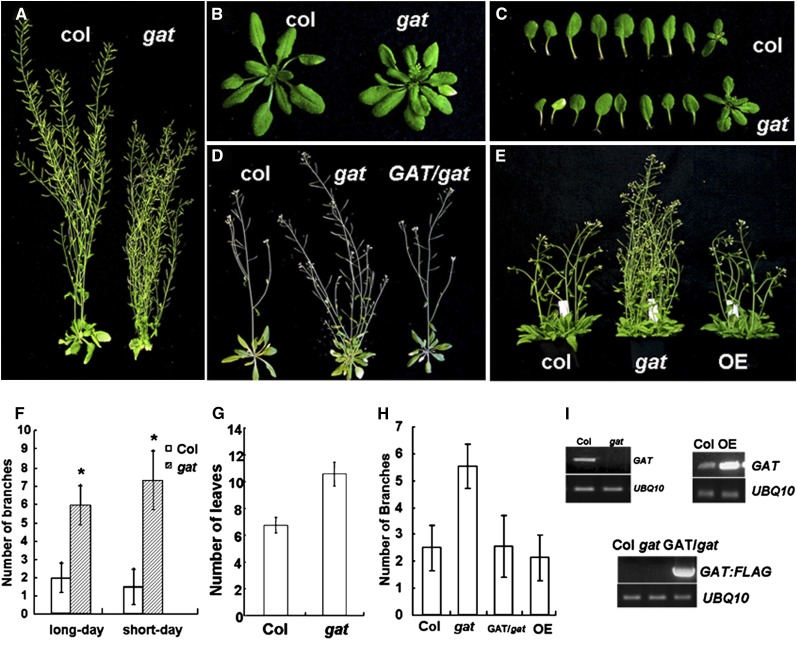
Figure 1.
The Arabidopsis gat (At1g15040) mutant phenotype. A, Phenotype of adult (8-week-old) gat mutant showing increased rosette branching. B, Phenotype of young seedlings (3 weeks old). C, Leaves and associated axillary shoots dissected from the shoot axis and displayed in the order of emergence, oldest leaf to the left. D, GAT complementary line with 35S-GAT:FLAG in gat background (GAT/gat; on right; 6 weeks old). E, Seedlings of the indicated lines (four seedlings per pot), including the GAT OE with 35S-GAT:FLAG (OE; on right; 4 weeks old). F, Number of rosette branches in the wild type and gat. G, Number of leaves in the wild type and gat. H, Number of rosette branches in the wild type, gat, complementary line (GAT/gat), and OE. I, RT-PCR results confirm gat mutant, complementary line (GAT/gat), and OE.
Under both long-day and short-day growth conditions, the gat mutant shows a significant increase in shoot branching compared with wild-type plants (Fig. 1, A and F). Due to early flowering in the gat mutant, young plants (4–6 weeks old) appear taller than the wild type (Fig. 1, D and E), while adult gat plants (older than 7–8 weeks) are shorter (Fig. 1A). The gat plants exhibit an increase in rosette leaves (Fig. 1, B and G) and these are smaller at the same developmental stage as the wild type (Fig. 1C). We conclude that GAT appears to control both axillary meristem initiation and axillary growth rate after meristem initiation.
The enhanced shoot branching of the gat mutant is due to a loss of function, since the coding region of GAT (FLAG tagged) complements the gat phenotype (Fig. 1, D and H). For complementation studies, six GAT:FLAG transgenic lines were randomly chosen for reverse transcription (RT)-PCR to test for transgene expression (see Supplemental Fig. S1). Three lines with relatively high expression of 35S:GAT:FLAG showed complementation in gat mutants, while the poorly expressed lines did not (data not shown). Figure 1I (bottom section) shows GAT:FLAG mRNA for the complemented line shown in Figure 1, D, E, and H. The shoot branch numbers of a GAT (FLAG tagged) overexpressor (OE) shows no significant differences from the wild type (Fig. 1, E and H).
gat Mutant Phenotype Is Similar But Not Identical to Known max Mutants
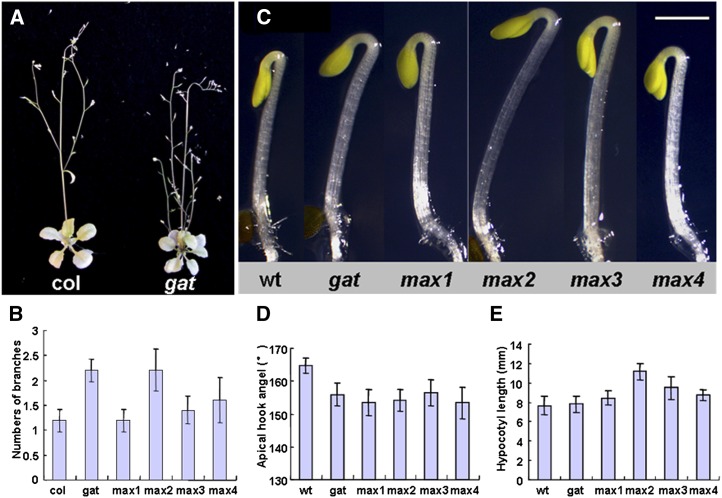
The phenotype described above for gat is similar to known branching mutants of Arabidopsis called max mutants (Stirnberg et al., 2002; Sorefan et al., 2003; Booker et al., 2004; Lazar and Goodman, 2006). To confirm known traits for max, and explore new ones, we analyzed selected properties in gat and max mutants, discovering that some traits are different (boldface in Table I). Table I summarizes traits of gat and max mutants, overall suggesting that gat and max exhibit similar traits. As described above, under nitrogen-replete conditions, gat exhibits increased shoot branching (see Fig. 1 and later). While under low nitrogen, gat mutant, and max2 as well, exhibit enhanced branching (Fig. 2, A and B), but not max1, max3, and max4 (Fig. 2B). To determine if gat also impacts hypocotyl development as shown for max2 (Shen et al., 2007; Nelson et al., 2011), we examined apical hook angle and hypocotyl length in gat and max mutants. Dark-grown Arabidopsis seedlings exhibit closed cotyledons and form an apical hook to protect the shoot meristem from damage (Guzmán and Ecker, 1990). Auxin response is known to be necessary in these processes (Strader et al., 2011). Three-day-old dark-grown gat seedlings were found to have a slightly decreased apical hook angle, similar to all four max mutants (Fig. 2, C and D). The hypocotyl length of gat, max1, max3, and max4 mutants is the same as the wild type, but max2 shows an increase in hypocotyl length (Fig. 2E). Supplemental Figure S5 shows that the primary root length in gat and max mutants under high nitrogen conditions were the same as the wild type. Under low nitrogen conditions, primary root length, as well as lateral root and root hair numbers, were the same for the gat mutant as the wild type (data not shown).
Table I. A summary of the gat mutant phenotype along with max mutants (compared with the wild type).
Boldface indicates unique trait of gat and/or max2.
| Trait | gat | max1 | max2 | max3 | max4 |
|---|---|---|---|---|---|
| Shoot branching | Enhanced (Fig. 1) | Enhanceda | Enhanceda | Enhancedb | Enhancedc |
| Adult plant height | Shorter (Fig. 1) | Shorterag | Shortera | Shorterbg | Shortercg |
| Leaf shape | Rounder, smaller (Fig. 1) | Rounder, smallerag | Rounder, smallerag | Rounder, smallerbg | Rounder, smallercg |
| Apical hook angle | Smaller (Fig. 2) | Smallerg | Smallerg | Smallerg | Smallerg |
| Primary root length | Wild-type level (Fig. 5) | Wild-type levelfg | Wild-type levelfg | Wild-type levelfg | Wild-type levelfg |
| Shoot branching/N− | Enhanced (Fig. 2) | Wild-type levelg | Enhancedg | Wild-type levelg | Wild-type levelg |
| GR24 correction | No (Fig. 6) | Yesg | Nodg | Yesdg | Yesdg |
| Hypocotyl length | Wild-type level (Fig. 2) | Wild-type leveleg | Longereg | Wild-type leveleg | Wild-type leveleg |
| Leaf no. | More (Supplemental Fig. S4) | Wild-type levelg | Wild-type levelg | Wild-type levelg | Wild-type levelg |
| Flowering time | Early (Fig. 1) | As wild typeg | As wild typeg | As wild typeg | As wild typeg |
Figure 2.
Other traits of gat and max mutants. A, Wild type and gat grown under low nitrogen condition. B, Mean rosette branch numbers (±se; n = 12–15) of the wild type, gat, max1, max2, max3, and max4. C, The gat mutant and four max mutants display decreased apical hook angle. Three-day-old seedlings grown in the dark, wild type, gat, max1, max2, max3, and max4. Bar = 2 mm. D, Mean apical hook angles (±se; n > 12) of wild type, gat, max1, max2, max3, and max4 mutants. E, Mean hypocotyl length (±se; n > 12) of wild type, gat, max1, max2, max3, and max4 mutants. [See online article for color version of this figure.]
GAT Belongs to the Class I Gln Amidotransferase Superfamily
A GAT enzyme has never been implicated in branching in any plant, so we analyzed class I Gln amidotransferase members in Arabidopsis. All GAT1 enzymes have a domain of approximately 250 residues that comprises the Gln amidotransferase domain (Kaplan et al., 1985; Zalkin, 1985; Mouilleron and Golinelli-Pimpaneau, 2007; see Figure 3, A and B, with members of GAT1_2 as examples). A completely conserved triad of residues (Cys-His-Glu) characteristic of the amidotransferase active site removes the side chain ammonia from Gln. This ammonia then acts as a nucleophile on a myriad of acceptors. The acceptor domain can be a part of the same GAT protein, or as another subunit (Mouilleron and Golinelli-Pimpaneau, 2007). Searching Arabidopsis annotation and various BLASTp analyses yielded 30 potential class I Gln amidotransferase proteins in Arabidopsis (Fig. 3C). Phylogenetic analysis based on the protein sequences of the GAT1 domain clustered Arabidopsis GAT1 members into six subgroups and six individual genes (Fig. 3C). Some of these likely encode known enzymatic activities (see “Discussion”). The At1g15040 (GAT1_2.1) clusters closely with two other similarly sized proteins (approximately 400 amino acids) of unknown function (Fig. 3, B and C). We discuss later this cluster’s unique presence in the plant kingdom, as well as the potential roles of other Arabidopsis GAT1 proteins. We now address the expression of GAT1_2.1 and the possible role(s) of this Gln amidotransferase in the Arabidopsis branching pathway.
Figure 3.
The GAT1 gene family in Arabidopsis. A, GAT1_2.1 encodes a class I Gln amidotransferase with a GATase1_2 domain and a possible acceptor domain. Triangles showing the conserved catalytic triad found in GAT family. B, Sequence alignment of GAT1_2.1 (At1g15040) with two close neighbors, At5g38200 and At1g66860. Framed residues show the conserved catalytic triad residues (Cys-His-Glu). C, Maximum likelihood tree showing relationships among the predicted Arabidopsis class I GATs. Framed members show the GAT1_2 subgroup. CPS, Carbamoyl phosphate synthase.
Expression of the GAT Gene
Real-time quantitative (q)PCR was carried out using both young seedlings (2 weeks old) and mature plants (7 weeks old) to analyze GAT expression patterns (Fig. 4A). For all real-time qPCR assays described above, UBQ10 was selected as internal control in different tissues and different stages based on a thorough comparison among 10 different reference genes (Tong et al., 2009) showing that UBQ10 is constitutive, in agreement with an earlier study that demonstrated highly stable expression of UBQ10 in Arabidopsis (Czechowski et al., 2005). Young seedlings grown on Murashige and Skoog media were separated into root and shoot, while different organs of mature plants (in soil) were analyzed. Both the Murashige and Skoog and soil conditioning were nitrogen replete. GAT is expressed in seedlings at significantly higher levels in roots than shoots. In mature plants, GAT is expressed at higher levels in flowers and young siliques, and shows relatively low expression in stems and leaves (Fig. 4A).
Figure 4.
Expression of GAT. A, Expression pattern of GAT. GAT mRNA quantification for each plant tissue type were determined by real-time qPCR, and UBQ10 gene was used as the internal control. B, Expression of GAT under nitrogen and phosphate stress conditions. Seven-day-old seedlings grown on Murashige and Skoog media were transferred to nitrogen stress media and grown for an additional 10 d. Expressions in root (left) and shoot (right).
Since gat was initially studied based on our results from nitrogen stress microarrays, GAT expression in response to both nitrogen and phosphate stress was analyzed (Fig. 4B). When 7-d-old seedlings grown on Murashige and Skoog media were treated under nitrogen stress for an additional 10 d, both roots and shoots show significantly down-regulated GAT under nitrogen stress (50-fold in shoots and 77-fold in roots, consistent with the 57-fold repression observed in the nitrogen microarray using whole seedlings; Supplemental Table S1). In contrast, phosphate limitation induced expression of GAT approximately 4-fold in roots and shoots (Fig. 4B). These results suggest a highly nitrogen-regulated control rather than general stress response (see “Discussion”).
Is GAT Involved in the Branching Hormone (SL) Pathway?
As indicated, the enhanced branching phenotype of the gat mutant is reminiscent of Arabidopsis max mutants involved in SL synthesis (MAX1, MAX3, MAX4) and sensing (MAX2; Booker et al., 2005; Dun et al., 2009; Beveridge and Kyozuka, 2010; see Fig. 5A). It is formally possible that the gat defect is due to aberrant expression in gat of the known MAX genes or other genes known to be involved in synthesis of SL precursors, carotenoids. To test this, real-time qPCR was carried out using the gat mutant and wild-type plants to compare expression of carotenoid biosynthetic genes and MAX genes, including PSY, PDS, ZDS, βLCY, εLCY, βOHase, εOHase, CCD1, CCD4, CCD7 (MAX3), CCD8 (MAX4), MAX1, MAX2, and CCR2 (Cazzonelli et al., 2009). No differences greater than 2-fold were observed in the gat mutant (Supplemental Fig. S2).
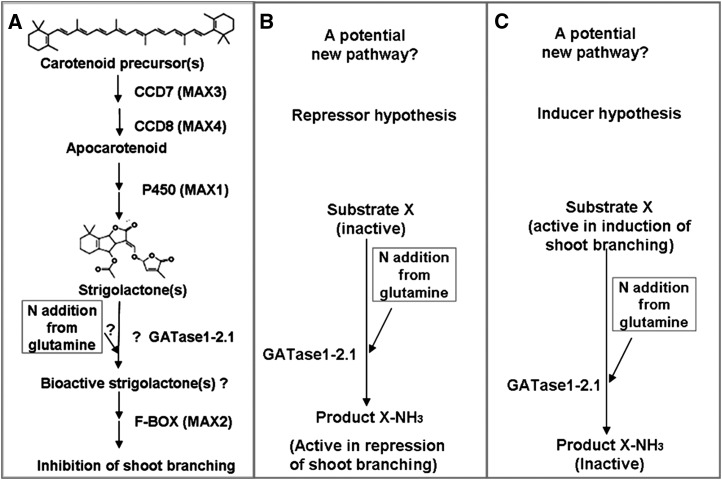
Figure 5.
The possible model of shoot branching inhibition with GAT1_2.1 (modified from Dun et al., 2009). A, SL synthesis from carotenoid precursors with a hypothetical GAT1_2.1 enzyme transferring a nitrogen group from Gln onto SL to activate its biological function. Diagram of the repressor model (B) and inducer model (C) described in the text.
An alternative possibility is that the known SL synthesis/regulation genes, MAX1 to MAX4, control expression of GAT. For example, MAX2 encodes an F-box regulator that is predicted to respond to the active SL, and mediate, by an unknown mechanism, the inhibition of shoot branching (see Fig. 5A). MAX2 might control GAT expression, which would then mediate branching inhibition. qPCR of GAT mRNA levels in max1 to max4 mutants also indicate no differences greater than 2-fold compared with the wild type (Supplemental Fig. S3).
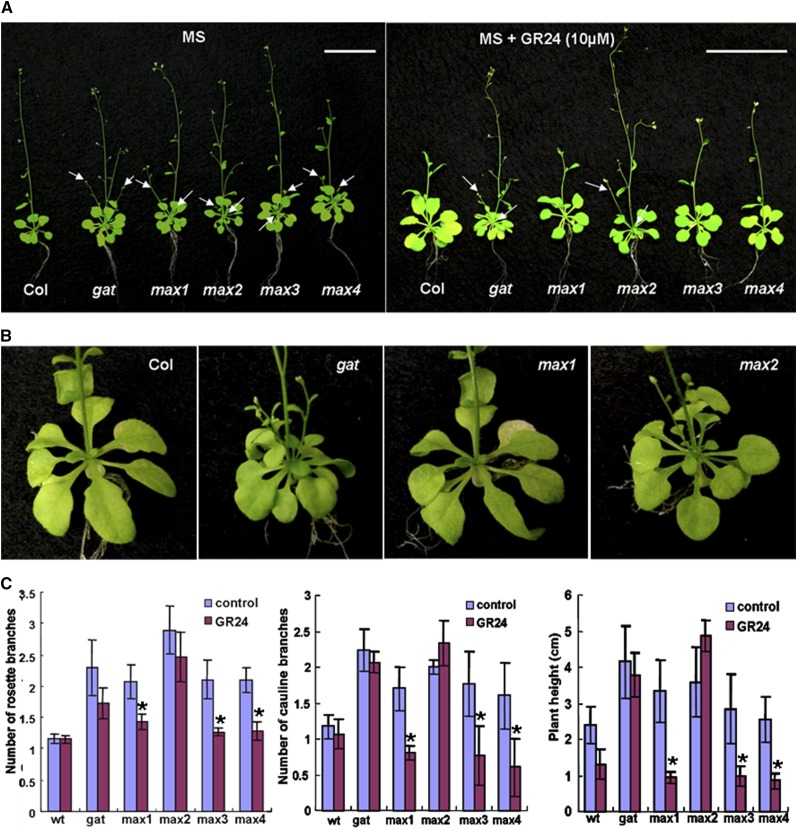
Recent studies using the SL analog, GR24, to complement max mutants have supported the assignment of MAX genes (proteins) in the pathway for SL synthesis and branching response shown in Figure 5A (Dun et al., 2009; Tsuchiya and McCourt, 2009; for review, see Umehara et al., 2008). That is, exogenous GR24 was shown to inhibit (complement) branching defects in Arabidopsis max1, max3, and max4 mutants but not in max2. Thus, max1, max3, and max4 are likely involved in biosynthesis of SL and max2 in response to SL (i.e. GR24). To determine whether GAT is involved in SL biosynthesis, GR24 was included in the growth media, and the gat mutant, the wild type, max1, max2, max3, and max4 mutants were tested for chemical correction of branching (Fig. 6). Consistent with published results (Umehara et al., 2008), exogenously applied GR24 corrects the branching phenotype of max1, max3, and max4 mutants, but not of max2 (Fig. 6). The gat phenotype (increased branching) is not complemented by GR24 (Fig. 6). In the “Discussion,” we speculate on the possible role of GAT in branching.
Figure 6.
GR24 does not rescue the gat phenotype. A, Wild type (Col), gat, max1, max2, max3, and max4 mutants grown on Murashige and Skoog control media (left) and Murashige and Skoog + 10 μm GR24 treatment media (right). Scale bar = 2 cm. B, Magnified photos of the axillary region to show rosette branches. C, Mean rosette branches (left), cauline branches (middle), and plant height (right). ±se; n = 45 on both control and treatment media.
DISCUSSION
GAT Represses Shoot Branching and Is Repressed by Nitrogen Stress
The most striking trait of the gat mutant is the bushy appearance as a result of increased rosette branching (e.g. Fig. 1, A and E). The GAT open reading frame complements the gat mutant for enhanced branching. The GAT OE does not exhibit less branching than the wild type, suggesting that a maximum (optimum) amount of GAT product is already synthesized (Fig. 1, E and I). However, the wild type only exhibits approximately two rosette branches per plant, thus these growth conditions may not present the potential to observe a decrease upon overexpression.
High repression of GAT under nitrogen stress but not phosphate stress indicates a nitrogen-specific response. Unpublished microarray data from Genevestigator (Hruz et al., 2008) indicate a 4- to 8-fold reduction in GAT expression upon nitrogen limitation. These studies were carried out after 2 d (or less) of nitrogen stress whereas ours were carried out after 10 to 15 d, on 30 mm NH4NO3 compared with 1 mm NH4NO3. It is likely that GAT1_2.1 is a slow responder to nitrogen stress. Although the levels of internal Gln may not decrease dramatically in plants upon short-term nitrogen stress, plants in long-term stress (greater than 14 d) show significant reductions in Gln levels (Sugiharto and Sugiyama, 1992; Urbanczyk-Wochniak and Fernie, 2005). Thus, plants could possess two mechanisms to reduce the putative GAT1_2.1 product when nitrogen is severely limited, decreased transcription of GAT1_2.1 and enzymatic activity (via reduced substrate accordingly).
Under nitrogen stress conditions, only the gat and max2 mutants (not max1, max3, or max4) exhibit the enhanced branching (see Fig. 2B; Table I). This result argues against the amidation of SLs (i.e. activation) as GAT’s main role (Fig. 5A). Because GAT is a slow nitrogen responder, it is likely that when these increased buds emerge, the GAT enzyme and MAX2 protein are still operating (in the wild type). Again, whether GAT is directly linked to nitrogen-based control of branching, or is just coincidently repressed by long-term nitrogen stress awaits further investigation. We speculate that by the time long-term nitrogen limitation has advanced, a plant has paramount metabolic requirements before the initiation of budding, which it cannot satisfy at this stage. Thus, the metabolic precursors at this stage are in short supply (e.g. Urbanczyk-Wochniak and Fernie, 2005). We raise this issue again in the last paragraph of the “Discussion.” Other traits of gat are similar to the max mutants, particularly max2 (boldface in Table I). In fact, only three differences are noted: gat has more leaves and flowers earlier, and max2 has a longer hypocotyl. It is clear that GAT has an important and distinct role in branching (and traits associated with other branching mutants).
GAT Is Predicted to Encode a Class I Gln Amidotransferase Specific to the Plant Kingdom
As far as we are aware, no publications have described the distribution and families of GAT1 in plants. We uncovered a total of 30 class I GATs in Arabidopsis, and further phylogeny analysis placed this super family into six subgroups and six individual genes (Fig. 3C). Among these, GATase1_Anthranilate_Synthase and GATase1_1 subgroups have the shortest protein sequences (222–273 amino acids), only containing a GATase1 domain. While the function of the GAT1_1 group is unknown, it is likely that those in the anthranilate synthase family are involved in anthranilate synthesis (Li et al., 1974; Marchler-Bauer et al., 2011). GATase1_DJ-1 and GATase1_PfpI_1 subgroups have protein sequences varying from 392 to 472 amino acids, containing a N-terminal GATase1 domain (approximately 250 amino acids) and a C-terminal domain (approximately 200 amino acids) potentially for an unknown acceptor (for review, see Mouilleron and Golinelli-Pimpaneau, 2007). GATase_DJ-1 is found in humans, where it may be involved in multiple physiological processes including cancer, Parkinson’s disease, and male fertility (Honbou et al., 2003), but its acceptor is unknown. GATase_PfpI_1 may be involved in an ATP-independent intracellular proteolysis (Du et al., 2000), but again the acceptor and activity are unknown. GATase1_CTP_Synthase subgroup comprises the largest protein (556–600 amino acids), an N-terminal synthase domain and a C-terminal GATase1 domain (Fig. 3). This group is predicted to be a CTP synthase adding an amino group onto UTP to make CTP (Endrizzi et al., 2004).
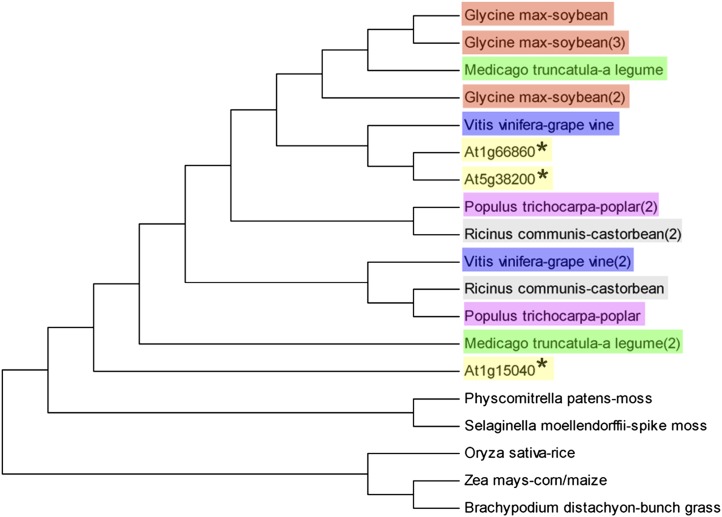
In the GATase1_2 subfamily, based on the sequence of the GAT domain, At1g15040 was clustered closely with At5g38200 and At1g66860 (Fig. 3C). The fact that the mutation in the first At1g15040 exon affects shoot branching indicates that the biological function of At1g15040 is not redundant to At5g38200 or At1g66860. We carried out a BLASTp search using the C-terminal sequence of At1g15040 (see Fig. 3A, sequence from 251–395 amino acids). It is likely that the C-terminal domain represents the acceptor binding site. The BLASTp analysis of this domain yielded significant orthologs only to open reading frames within the plant kingdom, including Arabidopsis, Medicago truncatula, grape (Vitis vinifera), corn (Zea mays), soybean (Glycine max), rice (Oryza sativa), castor bean (Ricinus communis), Populus trichocarpa, bunch grass (Brachypodium distachyon), spike moss (Selaginella moellendorffii), and moss (Physcomitrella patens; Fig. 7). This suggests that although At1g15040 possesses the conserved and ubiquitous GAT1 domain, it is involved in a highly conserved, plant-specific Gln amidotransferase reaction. We speculate below on what the acceptor may be.
Figure 7.
At1g15040 (GAT1_2.1) is a plant-specific Gln amidotransferase. The C-terminal amino acid sequence of GAT1_2.1 (251–395 amino acids) was used for a BLASTp search among all organisms. Nineteen significantly similar sequences from 11 organisms are presented in this maximum likelihood tree, all members of the plant kingdom. Arabidopsis (in yellow) has three copies; soybean (in orange), M. truncatula (in green), grape vine (in blue), poplar (in pink), and castor bean (in gray) have two copies; and moss, spike moss, rice, corn, and bunch grass possess a single copy.
How Does GAT Inhibit Shoot Branching?
Given the plant-specific nature of the GAT1_2 family, and the branching phenotype, an obvious question is whether GAT is involved in biosynthesis of a plant hormone. SLs were originally isolated from plant root exudates as germination stimulants for root parasitic plants, and then became recognized as a new type of plant hormone that inhibits shoot branching (including a newly discovered SL-like plant hormone, carlactone; Chen et al., 2009; Xie et al., 2010; Alder et al., 2012). Since the gat mutant exhibits a similar phenotype as max mutants, and the GAT1_2.1 gene is expressed at higher levels in roots, one possibility is that, there could be a relationship between GAT and SLs. There is no difference in expression of MAX genes in gat. Moreover, unlike the Arabidopsis BRANCHED1 gene (Aguilar-Martínez et al., 2007), there is no reduction in expression of GAT in max mutants. The SL analog GR24 is not able to rescue the gat mutant phenotype, much like the max2 (Fig. 6) and branched1 mutant (Brewer et al., 2009). Our results suggest that GAT is not involved in SL biosynthesis, although we cannot completely rule out that GAT may act as an amidotransferase on SLs, which could be required for its biological function (see Fig. 5A). However, this hypothesis does not explain a few distinct traits for gat not observed in max mutants (e.g. more leaves, early flowering).
At present we do not believe GAT is involved directly in auxin function. We do not observe auxin-related phenotypes such as changes in root architecture. Additionally, exogenous auxin did not correct the gat mutant phenotype (data not shown). Previous studies have shown that there is a feedback up-regulation of MAX3, MAX4, and newly identified DWARF27 in max mutants (Hayward et al., 2009; Waters et al., 2012). Our qPCR results show that in the gat mutant, there is no significant up-regulation in MAX3 and MAX4 expression.
A preferred hypothesis for GAT function is its involvement in a unique pathway that controls branching, diagrammed in Figure 5, B and C. With this hypothesis, GAT functions by amidation of an unknown substrate that controls shoot branching. In Figure 5B, GAT substrate X is amidated to X-NH3, which behaves as a repressor, independent of SLs. In Figure 5C, GAT substrate X is an inducer of shoot branching, and amidation (to X-NH3) decreases the inducer concentration. Both models are consistent with the increased branching and other traits of a gat null mutant controlled by X (or X-NH3) might control. For the model in Figure 5C, the substrate X could be CK, but we feel this is unlikely for two reasons. The biosynthetic pathway for isoprenoid CKs is well understood, and does not involve a Gln amidotransferase (Hwang and Sakakibara, 2006). Second, phenotypes of CK mutants are also quite different than gat mutants. Models B and C are not mutually exclusive and both X and X-NH3 may have control functions.
It is possible that X and/or X-NH3 might act as an indicator and signal of the general nitrogen status of the plant, controlling the traits described here for the Arabidopsis gat mutant. Thus, bud initiation for example would only occur when nitrogen status is suitable. Establishing what the substrate is for the GAT1_2.1 enzyme, and whether GAT1_2.1 has the same role in other plants are important goals for the future.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All wild-type plants were Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0), all mutant seeds were obtained from the Arabidopsis Resource Center (Columbus, OH), gat as SALK_031983C, max1 as max1-1 (CS9564), max2 as max2-2 (CS9566), max3 as max3-9 (CS9567), and max4 as max4-1 (CS9568).
Soil-grown plants were grown in a Conviron growth chamber with 50% humidity and a light density of 125 mol m−2 s−1. Plants were grown under a 16-h-light/8-h-dark cycle as the long-day condition and an 8-h-light/16-h-dark cycle as the short-day condition. Plants are generally grown under the long-day condition except where noted. Plants were grown on sterile plates as previously described (Zhu et al., 2009).
Seeds were surface sterilized before placing on growth media. To examine hypocotyl development, plates were incubated under white light for 1 d and followed by 2 d in the dark. Seedlings were imaged, and apical hook angle was measured using ImageJ software.
Constructs and Plant Transformation
The coding region of GAT (At1g15040) was PCR amplified using KlenTaq LA DNA polymerase (Barnes, 1994) and primer set 5′-ACGGTACCATGGTTGTCGCCAATGAT-3′ (KpnI site was engineered in as underlined) and 5′-AACCCGGGATAGTTGAGAAAAAGGA-3′ (SmaI site as underlined), cloned into a 35S-promoter-driving plant binary vector pBAR-FAST (Ge et al., 2005) using KpnI/SmaI sites, and PCR-derived clones were confirmed by sequencing. Plasmids were then transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. Arabidopsis wild-type and mutant (gat) transformation was via A. tumefaciens-mediated transformation as previously described (Clough and Bent, 1998). Wild-type (Col-0) plants transformed with 35S:GAT:FLAG were designated OE line and gat mutant plants transformed were complementary lines labeled as GAT/gat in this article.
RT-PCR and Real-Time qPCR
Total RNA was extracted using Agilent plant RNA isolation mini kit (Agilent Technologies), 5 μg of RNA was DNaseI (Ambion)-treated and used to synthesize first-strand complementary DNA using oligo d(T) and Invitrogen SuperScript III reverse transcriptase following the manufacturer’s instruction (Life Technologies). The complementary DNA was used for RT-PCR and real-time qPCR. The relative expression level was quantified using Fermentas SYBR Green qPCR master mix (Thermo), and three technical replicates of each two to three biological replicates were performed by relative quantification using UBQ10 as a reference gene. An ABI7500 system was used to perform qPCR.
For GAT expression profiling, soil-grown plant tissue was harvested from 7-week-old wild-type (Col-0) plants grown under long-day conditions. Once plants bolted, rosette stems (primary inflorescence minus flower and cauline leaves), cauline stems, rosette leaves, cauline leaves, rosette buds (with covering leaves and newly formed, unopened flowers), cauline buds, flowers, and young siliques (also newly developed, green, and tender) were collected. For sterile plate-grown seedlings, 2-week-old seedlings were carefully pulled from growth media and excised between root and shoot.
Stress Conditions
Wild-type seeds were germinated on one-half-strength Murashige and Skoog media for 7 d, and seedlings were then transferred onto stress media and grown for 10 d. For nitrogen stress, 1 mm nitrogen was used while the control media had 30 mm nitrogen as in one-half-strength Murashige and Skoog; while 10 μm H2PO4− was used as phosphate stress media.
Database Search and Phylogeny Analysis of Arabidopsis Class I Gln Amidotransferase
Initially, a search using the phrase glutamine amidotransferase was conducted at the TAIR Web site (www.arabidopsis.org). This step resulted in the retrieval of the predicted amino acid sequences of 23 class I GATs. In the second step, the protein sequences of these 23 GATs were each used as query sequence for Blastp searches at TAIR. Identification of additional predicted GATs was based on whether each has the conserved triad amino acids (Cys-His-Glu), and also verified by Blastp search on National Center for Biotechnology Information (NCBI) to confirm identified with GATs in other organisms. Seven additional predicted class I GATs were found in this step. In the third step, each of the newly retrieved Arabidopsis class I GATs was used as query sequences for another round of BLASTp searches on both TAIR and NCBI. However, no more class I GATs were found in this step, suggesting that the total number of class I Gln amidotransferase in Arabidopsis genome was likely to be 30.
Molecular Evolutionary Genetic Analysis (MEGA5) software (Tamura et al., 2011) was used for amino acid sequences alignment and phylogeny analysis. For the phylogeny trees shown in the “Results,” the amino acid sequences of GAT domain of the 30 Arabidopsis class I GAT members were first aligned using MUSCLE program included in MEGA5 software, and then the aligned file was phylogeny analyzed to make maximum likelihood tree.
Hormone Treatment
GR24 was bought from Chiralix (www.chiralix.com). For 10 μm GR24 treatment, GR24 was first dissolved in a small amount of dimethyl sulfoxide and then brought up to autoclaved one-half-strength Murashige and Skoog media; for control media, the same amount of dimethyl sulfoxide was added to serve as control. Seeds were germinated on a one-half-strength Murashige and Skoog media plate, and after 7 d seedlings were transferred to PhytaTray II (Sigma) filled with control media and GR24 treatment media, and grown for approximately 5 weeks until phenotype assay was carried out. Three biological replicates were included, for each biological replicate, a total of 15 plants grown in five PhytaTrays were counted for branch numbers.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: soybean (Glycine max): XP_003552718, XP_003555062, XP_003518630; Medicago truncatula: XP_003624379, XP_003592606; grape vine (Vitis vinifera): XP_002280944, XP_002279823; poplar (Populus trichocarpa): XP_002314840, XP_002301736; castor bean (Ricinus communis): XP_002527023, XP_002516767; moss (Physcomitrella patens): XP_001771273; spike moss (Selaginella moellendorffii): XP_002974123; rice (Oryza sativa): NP_001041973; corn/maize (Zea mays): NP_001140467; bunch grass (Brachypodium distachyon): XP_003568263.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. GAT complementation lines.
Supplemental Figure S2. Relative gene expression levels in gat mutant.
Supplemental Figure S3. Relative GAT expression level in the max mutants.
Supplemental Figure S4. Number of leaves of the gat and max mutants.
Supplemental Figure S5. Primary root length of the gat and max mutants.
Supplemental Table S1. T-DNA insertion lines obtained for seventeen selected genes
Supplemental Data S1. Microarray results (fold changes under nitrogen stress conditions).
Acknowledgments
We thank Dr. Lucia Strader for her critique on our manuscript and help with hypocotyl experiments. We acknowledge students in various semesters of Bio437 (Washington University, Department of Biology) for technical support, and Abha Khandelwaland Karen Fitzsimmons, who were involved in early studies on the Agilent microarray.
Glossary
- CK
cytokinin
- SL
strigolactone
- TAIR
The Arabidopsis Information Resource
- RT
reverse transcription
- OE
overexpressor
- q
quantitative
- Col-0
Columbia-0
- NCBI
National Center for Biotechnology Information
References
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S. (2012) The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351 [DOI] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51: 1019–1029 [DOI] [PubMed] [Google Scholar]
- Barnes WM. (1994) PCR amplification of up to 35 kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci USA 91: 2216–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Kyozuka J. (2010) New genes in the strigolactone-related shoot branching pathway. Curr Opin Plant Biol 13: 34–39 [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Murfet IC, Kerhoas L, Sotta B, Miginiac E, Rameau C. (1997) The shoot controls zeatin riboside export from pea roots: evidence from the branching mutant rms4. Plant J 11: 339–345 [Google Scholar]
- Beveridge CA, Ross JJ, Murfet IC. (1996) Branching in pea—action of genes Rms3 and Rms4. Plant Physiol 110: 859–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O. (2005) MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev Cell 8: 443–449 [DOI] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. (2009) Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol 150: 482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli CI, Cuttriss AJ, Cossetto SB, Pye W, Crisp P, Whelan J, Finnegan EJ, Turnbull C, Pogson BJ. (2009) Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 21: 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Zou JH, Zhang SY, Zaitlin D, Zhu LH. (2009) Strigolactones are a new-defined class of plant hormones which inhibit shoot branching and mediate the interaction of plant-AM fungi and plant-parasitic weeds. Sci China C Life Sci 52: 693–700 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O. (2011) Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol 12: 211–221 [DOI] [PubMed] [Google Scholar]
- Du XL, Choi IG, Kim R, Wang WR, Jancarik J, Yokota H, Kim SH. (2000) Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-A resolution. Proc Natl Acad Sci USA 97: 14079–14084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun EA, Brewer PB, Beveridge CA. (2009) Strigolactones: discovery of the elusive shoot branching hormone. Trends Plant Sci 14: 364–372 [DOI] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA. (2012) Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol 158: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi JA, Kim HS, Anderson PM, Baldwin EP. (2004) Crystal structure of Escherichia coli cytidine triphosphate synthetase, a nucleotide-regulated glutamine amidotransferase/ATP-dependent amidoligase fusion protein and homologue of anticancer and antiparasitic drug targets. Biochemistry 43: 6447–6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BJ, Beveridge CA. (2009) Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiol 149: 1929–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XC, Dietrich C, Matsuno M, Li GJ, Berg H, Xia YJ. (2005) An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis. EMBO Rep 6: 282–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR. (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O. (2009) Interactions between auxin and strigolactone in shoot branching control. Plant Physiol 151: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honbou K, Suzuki NN, Horiuchi M, Niki T, Taira T, Ariga H, Inagaki F. (2003) The crystal structure of DJ-1, a protein related to male fertility and Parkinson’s disease. J Biol Chem 278: 31380–31384 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sakakibara H. (2006) Cytokinin biosynthesis and perception. Physiol Plant 126: 528–538 [Google Scholar]
- Kaplan JB, Merkel WK, Nichols BP. (1985) Evolution of glutamine amidotransferase genes: nucleotide sequences of the pabA genes from Salmonella typhimurium, Klebsiella aerogenes and Serratia marcescens. J Mol Biol 183: 327–340 [DOI] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Liu Q, Bours R, Domagalska MA, Beguerie S, Verstappen F, Leyser O, Bouwmeester H, Ruyter-Spira C. (2011) Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol 155: 974–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafever HN. (1981) Genetic-differences in plant-response to soil nutrient stress. J Plant Nutr 4: 89–109 [Google Scholar]
- Lazar G, Goodman HM. (2006) MAX1, a regulator of the flavonoid pathway, controls vegetative axillary bud outgrowth in Arabidopsis. Proc Natl Acad Sci USA 103: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SL, Hanlon J, Yanofsky C. (1974) Structural homology of the glutamine amidotransferase subunits of the anthranilate synthetases of Escherichia coli, Salmonella typhimurium and Serratia marcescens. Nature 248: 48–50 [DOI] [PubMed] [Google Scholar]
- Lin H, Wang RX, Qian Q, Yan MX, Meng XB, Fu ZM, Yan CY, Jiang B, Su Z, Li JY, et al. (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21: 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WZ, Wu C, Fu YP, Hu GC, Si HM, Zhu L, Luan WJ, He ZQ, Sun ZX. (2009) Identification and characterization of HTD2: a novel gene negatively regulating tiller bud outgrowth in rice. Planta 230: 649–658 [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang QS, Ding YF, Li GH, Xu JX, Wang SH. (2011) Effects of external ABA, GA(3) and NAA on the tiller bud outgrowth of rice is related to changes in endogenous hormones. Plant Growth Regul 65: 247–254 [Google Scholar]
- López-Ráez JA, Charnikhova T, Gómez-Roldán V, Matusova R, Kohlen W, De Vos R, Verstappen F, Puech-Pages V, Bécard G, Mulder P, et al. (2008) Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol 178: 863–874 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu SN, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. (2011) CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39: D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouilleron S, Golinelli-Pimpaneau B. (2007) Conformational changes in ammonia-channeling glutamine amidotransferases. Curr Opin Struct Biol 17: 653–664 [DOI] [PubMed] [Google Scholar]
- Napoli C. (1996) Highly branched phenotype of the petunia dad1-1 mutant is reversed by grafting. Plant Physiol 111: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM. (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 108: 8897–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, et al. (2011) Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol 155: 721–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Luong P, Huq E. (2007) The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol 145: 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogné K, Goussot M, Bainbridge K, Foo E, Chatfield S, Ward S, Beveridge C, Rameau C, et al. (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17: 1469–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Ottoline Leyser HM. (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J 50: 80–94 [DOI] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HMO. (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141 [DOI] [PubMed] [Google Scholar]
- Stirnberg P, Ward S, Leyser O. (2010) Auxin and strigolactones in shoot branching: intimately connected? Biochem Soc Trans 38: 717–722 [DOI] [PubMed] [Google Scholar]
- Strader LC, Wheeler DL, Christensen SE, Berens JC, Cohen JD, Rampey RA, Bartel B. (2011) Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23: 984–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiharto B, Sugiyama T. (1992) Effects of nitrate and ammonium on gene expression of phosphoenolpyruvate carboxylase and nitrogen metabolism in maize leaf tissue during recovery from nitrogen stress. Plant Physiol 98: 1403–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZG, Gao ZH, Wang F, Zhou J, Zhang Z. (2009) Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol Biol 10: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, McCourt P. (2009) Strigolactones: a new hormone with a past. Curr Opin Plant Biol 12: 556–561 [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Magome H, Takeda-Kamiya N, Yamaguchi S. (2010) Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol 51: 1118–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200 [DOI] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, Fernie AR. (2005) Metabolic profiling reveals altered nitrogen nutrient regimes have diverse effects on the metabolism of hydroponically-grown tomato (Solanum lycopersicum) plants. J Exp Bot 56: 309–321 [DOI] [PubMed] [Google Scholar]
- Waters MT, Brewer PB, Bussell JD, Smith SM, Beveridge CA. (2012) The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol 159: 1073–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XN, Yoneyama K, Yoneyama K. (2010) The strigolactone story. Annu Rev Phytopathol 48: 93–117 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Xie XN, Kusumoto D, Sekimoto H, Sugimoto Y, Takeuchi Y, Yoneyama K. (2007) Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 227: 125–132 [DOI] [PubMed] [Google Scholar]
- Zalkin H. (1985) Glutamine amidotransferases. Methods Enzymol 113: 263–264 [DOI] [PubMed] [Google Scholar]
- Zhang SY, Li G, Fang J, Chen WQ, Jiang HP, Zou JH, Liu X, Zhao XF, Li XB, Chu CC, et al. (2010) The interactions among DWARF10, auxin and cytokinin underlie lateral bud outgrowth in rice. J Integr Plant Biol 52: 626–638 [DOI] [PubMed] [Google Scholar]
- Zhu HF, Fitzsimmons K, Khandelwal A, Kranz RG. (2009) CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol Plant 2: 790–802 [DOI] [PubMed] [Google Scholar]