Abstract
Leaf senescence is a natural age-dependent process that is induced prematurely by various environmental stresses. Typical alterations during leaf senescence include breakdown of chlorophyll, a shift to catabolism of energy reserves, and induction of senescence-associated genes, all of which can occur during submergence, drought, and constant darkness. Here, we evaluated the influence of the submergence tolerance regulator, SUBMERGENCE1A (SUB1A), in the acclimation responses during leaf senescence caused by prolonged darkness in rice (Oryza sativa). SUB1A messenger RNA was highly induced by prolonged darkness in a near-isogenic line containing SUB1A. Genotypes with conditional and ectopic overexpression of SUB1A significantly delayed loss of leaf color and enhanced recovery from dark stress. Physiological analysis revealed that SUB1A postpones dark-induced senescence through the maintenance of chlorophyll and carbohydrate reserves in photosynthetic tissue. This delay allowed leaves of SUB1A genotypes to recover photosynthetic activity more quickly upon reexposure to light. SUB1A also restricted the transcript accumulation of representative senescence-associated genes. Jasmonate and salicylic acid are positive regulators of leaf senescence, but ectopic overexpression of SUB1A dampened responsiveness to both hormones in the context of senescence. We found that ethylene accelerated senescence stimulated by darkness and jasmonate, although SUB1A significantly restrained dark-induced ethylene accumulation. Overall, SUB1A genotypes displayed altered responses to prolonged darkness by limiting ethylene production and responsiveness to jasmonate and salicylic acid, thereby dampening the breakdown of chlorophyll, carbohydrates, and the accumulation of senescence-associated messenger RNAs. A delay of leaf senescence conferred by SUB1A can contribute to the enhancement of tolerance to submergence, drought, and oxidative stress.
Leaf senescence is a natural developmental process that occurs near the end of leaf development. Senescence of leaves is also triggered prematurely by various environmental stresses, such as constant darkness, submergence, drought, salinity, and extreme temperature. Leaf senescence is a genetically programmed process that is actively regulated at various levels. Major physiological alterations include an increase in the breakdown of chloroplasts and a switch of carbon assimilation to catabolism of energy resources such as proteins, lipids, and nucleic acids. Recent molecular studies identified hundreds of Senescence-Associated Gene (SAG)-encoded mRNAs, which are differentially expressed during leaf senescence in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa; Lee et al., 2001; Gepstein et al., 2003; Lin and Wu, 2004; Liu et al., 2008). Consistent with physiological changes, genes involved in photosynthesis and other anabolic processes are down-regulated in senescing leaves, whereas genes responsible for macromolecule catabolism and transport are up-regulated. Genome-wide comparison of gene transcript data revealed that natural and induced senescence regulates overlapping but different sets of genes in Arabidopsis rosette leaves (Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006). Interestingly, the transcript profile of suspension culture cells exposed to Suc deficiency is more similar to that of senescing leaves during constant darkness than natural age-dependent senescence, suggesting that carbohydrate starvation can trigger alterations in transcript accumulation under the two conditions.
A number of genetic and biochemical studies revealed that various phytohormones are involved in the onset and progression of leaf senescence. Ethylene is a positive regulator of leaf senescence as well as flower senescence and fruit ripening. Transcriptome analysis revealed that genes associated with ethylene biosynthesis and signaling, including ACC SYNTHASE, ACC OXIDASE, ETHYLENE INSENSITIVE3 (EIN3), and ETHYLENE RESPONSIVE FACTOR1a (ERF1a), are up-regulated in Arabidopsis leaves during age-dependent and dark-induced senescence (van der Graaff et al., 2006). Application of ethylene decreases chlorophyll content and photochemical efficiency in Arabidopsis leaves (Woo et al., 2001; Jing et al., 2002). In addition, leaf senescence was delayed in the ethylene-insensitive mutants erecta, ethylene receptor1, and ein3, whereas the ethylene-hypersensitive mutants hyperrecombination protein1 and enhanced disease resistance1 exhibited an early-senescence phenotype (Zacarias and Reid, 1990; Grbić and Bleecker, 1995; Wawrzynska et al., 2008; Pan et al., 2012).
Methyl jasmonate (MeJA) and its precursor jasmonate (JA) also play important roles in the regulation of natural and stress-induced leaf senescence. For example, the level of endogenous JA significantly increases during natural and dark-induced senescence in Arabidopsis rosette leaves (He et al., 2002; Seltmann et al., 2010). Exogenous application of MeJA leads to the rapid reduction in chlorophyll content and induction of several SAGs in Arabidopsis and rice (Schenk et al., 2000; He et al., 2001; Woo et al., 2001; Kong et al., 2006). Moreover, leaf senescence is significantly delayed in JA-defective and -insensitive Arabidopsis mutants such as coronatine insensitive1, decanamide resistant root1, jagged and wavy-D, mitogen-activated protein kinase6, and mapk kinase9, emphasizing the significance of the JA pathway in the regulation of leaf senescence (He et al., 2002; Schommer et al., 2008; Zhou et al., 2009; Morquecho-Contreras et al., 2010). However, age-dependent and dark-induced senescence was not influenced by reduced accumulation of JA in RNA interference-knockdown lines of lipoxygenase2 (Seltmann et al., 2010), suggesting that leaf senescence is also modulated through a JA-independent pathway.
Salicylic acid (SA) is a positive regulator of leaf senescence. The concentration of endogenous SA is significantly greater in senescing leaves than in mature green leaves in Arabidopsis, and the levels of several SAG transcripts are reduced in SA-defective and -insensitive mutants during leaf senescence (Morris et al., 2000). A global-scale gene expression analysis confirmed the down-regulation of a subset of SAGs in SA-defective transgenic Naphthalene hydroxylase G (Buchanan-Wollaston et al., 2005). Low-light- and salinity-induced senescence is accelerated in the Arabidopsis senescence-associated ubiquitin ligase1 (saul1) mutant that overaccumulates SA and SAG transcripts under the stress (Vogelmann et al., 2012). An early-senescence phenotype in the mutant was rescued in the SA-deficient phytoalexin deficient4 background, indicating that SA is necessary for saul1-dependent premature senescence. Overall, these data indicate that multiple hormones are involved in the regulation of leaf senescence at molecular and physiological levels. However, the interplay of these hormones in the onset and progression of leaf senescence remains unknown.
Leaf senescence is a major visible symptom in plants exposed to prolonged submergence. For example, the loss of leaf color is clearly observed in Arabidopsis and rice plants after 5 to 7 d of complete submergence (Fukao et al., 2006; Lee et al., 2011; Vashisht et al., 2011). A rapid reduction in carbohydrate reserves also occurs in leaves of rice, Rumex palustris, and Rorippa spp. (Fukao et al., 2006; Chen et al., 2010; Akman et al., 2012; Barding et al., 2012). In rice, these physiological alterations are restrained by the submergence tolerance regulator SUBMERGENCE1A (SUB1A), an ERF domain-containing transcription factor. Only limited rice cultivars possess the SUB1A gene, and more specifically the SUB1A-1 allele, which is highly induced during submergence (Fukao et al., 2006; Xu et al., 2006). Genotypes containing SUB1A-1 (henceforth referred to as SUB1A) can endure complete submergence for prolonged periods with considerably less severe leaf senescence and recommence the development of new leaves and tillers after desubmergence. Functional characterization of SUB1A revealed that its submergence-induced expression restricts further ethylene production and dampens gibberellic acid responsiveness, causing shoot tissue to dampen carbohydrate consumption, chlorophyll breakdown, amino acid accumulation, and elongation growth (Fukao et al., 2006; Fukao and Bailey-Serres, 2008; Barding et al., 2012). This quiescence response to submergence aids the maintenance of carbohydrate reserves and the capability for photosynthesis. We hypothesized that avoidance of carbohydrate starvation may be involved in the less severe leaf senescence manifested during submergence in varieties bred to have SUB1A. In addition to submergence tolerance, SUB1A also enhances recovery from dehydration stress through the activation of reactive oxygen species detoxification and the induction of stress-specific transcription factors and downstream genes (Fukao et al., 2011).
Prolonged darkness has been used as a procedure to initiate synchronous senescence in leaves because it effectively induces the expression of SAGs, the breakdown of chlorophyll, and the catabolism of energy reserves (Lee et al., 2001; Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Seltmann et al., 2010). To dissect SUB1A-dependent regulatory mechanisms underlying stress-induced senescence in leaves, we evaluated the contribution of SUB1A to physiological and molecular adaptations to prolonged darkness. The role of SUB1A in the responsiveness and interaction of the senescence-regulating hormones ethylene, JA, and SA was also analyzed. The results demonstrate that SUB1A coordinates chlorophyll degradation, photosynthetic activity, and carbohydrate consumption via hormonal regulation, resulting in a significant postponement of leaf senescence during prolonged darkness.
RESULTS
Prolonged Darkness Increases the Transcript Levels of the Three SUB1 Locus Genes
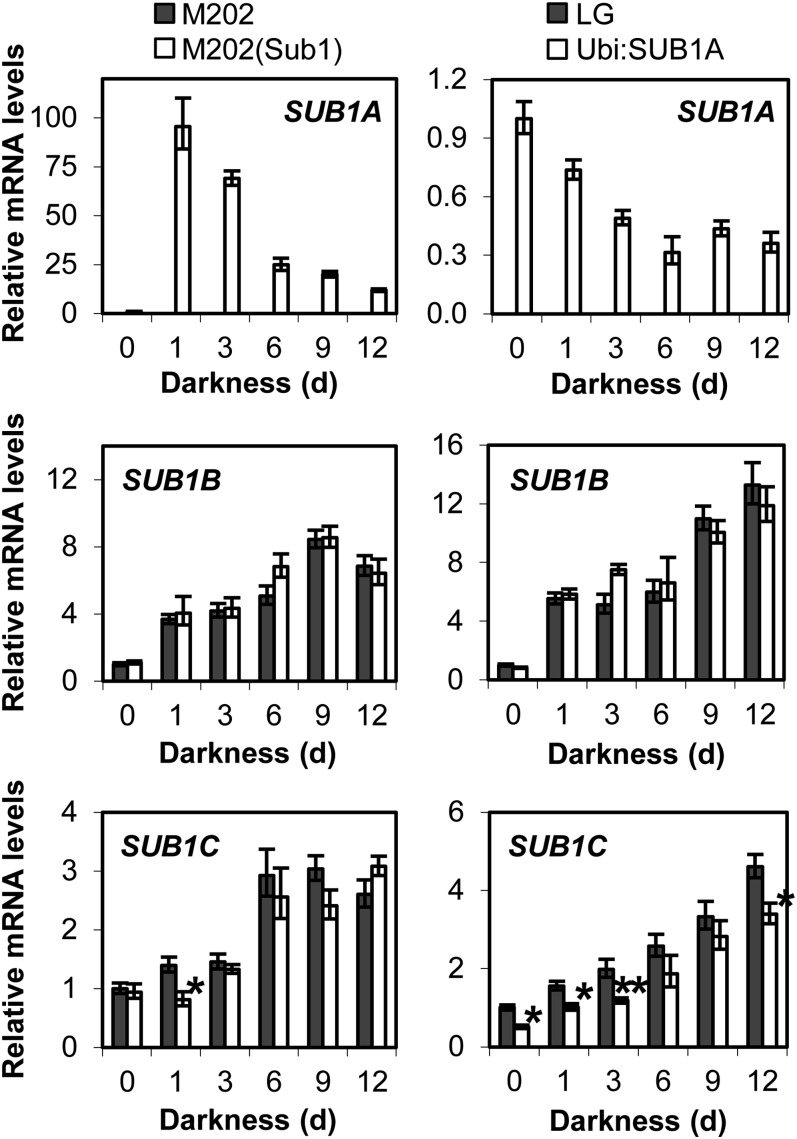
The multigenic locus SUB1 encodes three ERF domain-containing transcription factors, SUB1A, SUB1B, and SUB1C, all of which are submergence inducible (Fukao et al., 2006; Xu et al., 2006). To discern whether prolonged darkness affects the levels of SUB1 gene transcripts, relative levels of the three mRNAs were monitored in aerial tissue of plants exposed to complete darkness for up to 12 d in the near-isogenic M202 and M202(Sub1) lines, nontransgenic Liaogeng (LG), and the SUB1A overexpression line Ubi:SUB1A (Fig. 1). Our quantitative reverse transcription (qRT)-PCR analysis revealed that the abundance of SUB1A transcript was considerably elevated by 1 d of darkness, and the transcript level gradually decreased in a time-dependent manner in M202(Sub1). SUB1B and SUB1C mRNA levels increased in response to the stress in both M202 and M202(Sub1). The levels of these transcripts were elevated or maintained during the stress period, but the degree of induction was significantly lower for these genes than SUB1A. In the SUB1A overexpression line, SUB1A mRNA constitutively accumulated under nonstressed conditions, but exposure to prolonged darkness decreased the level of the transcript. Consistent with the observations in M202 and M202(Sub1), the abundance of SUB1B and SUB1C transcripts increased in response to prolonged darkness with similar trends in LG and Ubi:SUB1A. The level of SUB1C mRNA was significantly lower in Ubi:SUB1A at multiple time points during dark treatment, confirming previously reported SUB1A-dependent negative regulation of SUB1C mRNA accumulation (Xu et al., 2006; Fukao and Bailey-Serres, 2008; Fukao et al., 2011).
Figure 1.
Relative levels of SUB1 gene transcripts in aerial tissue under prolonged darkness. Developmentally matched 14-d-old [M202, M202(Sub1), and LG] and 21-d-old (Ubi:SUB1A) plants were exposed to complete darkness for up to 12 d, and aerial tissue was analyzed by qRT-PCR using gene-specific primers. The relative level of each mRNA was calculated by comparison with the nonstressed control [M202(Sub1) or Ubi:SUB1A at day 0 for SUB1A; M202 or LG at day 0 for SUB1B and SUB1C]. The data represent means ± se from three biological replicates. Asterisks indicate significant differences between the two genotypes (*P < 0.05, **P < 0.01).
SUB1A Enhances Survival of Prolonged Darkness through the Maintenance of Chlorophyll and Carbohydrate Reserves in Aerial Tissue
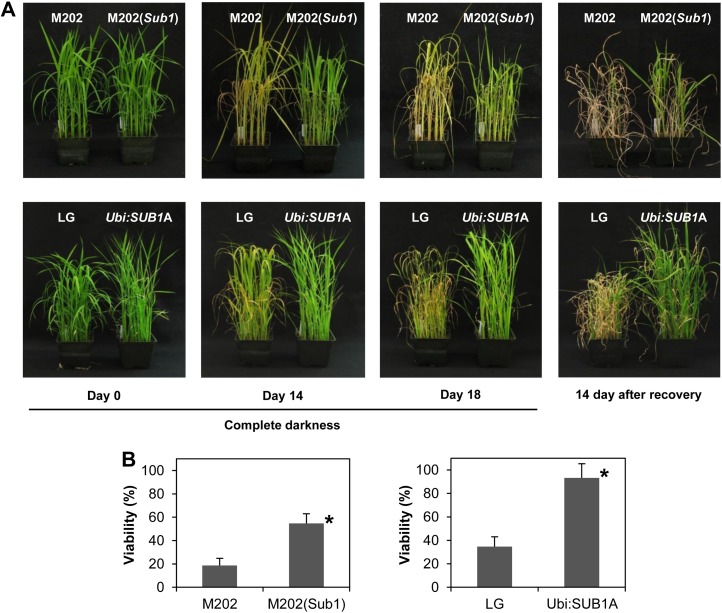
Prolonged darkness discontinues energy conversion by photosynthesis, resulting in an acceleration of carbohydrate catabolism and chlorophyll degradation. To evaluate the contribution of SUB1A to survival of dark stress, rice plants were transferred to complete darkness for 18 d and recovered under regular growth conditions for 14 d (Fig. 2A). The green leaf color of both M202 and LG plants turned to pale green or yellow after 14 d of dark treatment, whereas the color of M202(Sub1) and Ubi:SUB1A leaves was maintained. By day 18, most leaves of M202 and LG were yellowish and wilted, but the two SUB1A-containing genotypes sustained green leaves. During the recovery period, significantly more plants established new leaves from apical meristems in M202(Sub1) and Ubi:SUB1A as compared with M202 and LG (Fig. 2B). Notably, almost all Ubi:SUB1A plants (93.3%) recovered from 18 d of complete darkness. These data indicate that SUB1A delays leaf senescence promoted during prolonged darkness and significantly enhances survival of dark stress.
Figure 2.
SUB1A enhances tolerance to prolonged darkness. A, Photographs of M202, M202(Sub1), LG, and Ubi:SUB1A plants exposed to complete darkness for up to 18 d and recovered under regular growth conditions for 14 d. B, Viability of plants treated with 18 d of complete darkness. The survival of each genotype was evaluated 14 d after recovery. Plants were counted as viable if new leaves appeared during recovery. The data represent means ± sd from three biological replicates (n = 75). Asterisks indicate significant differences between the two genotypes (P < 0.01).
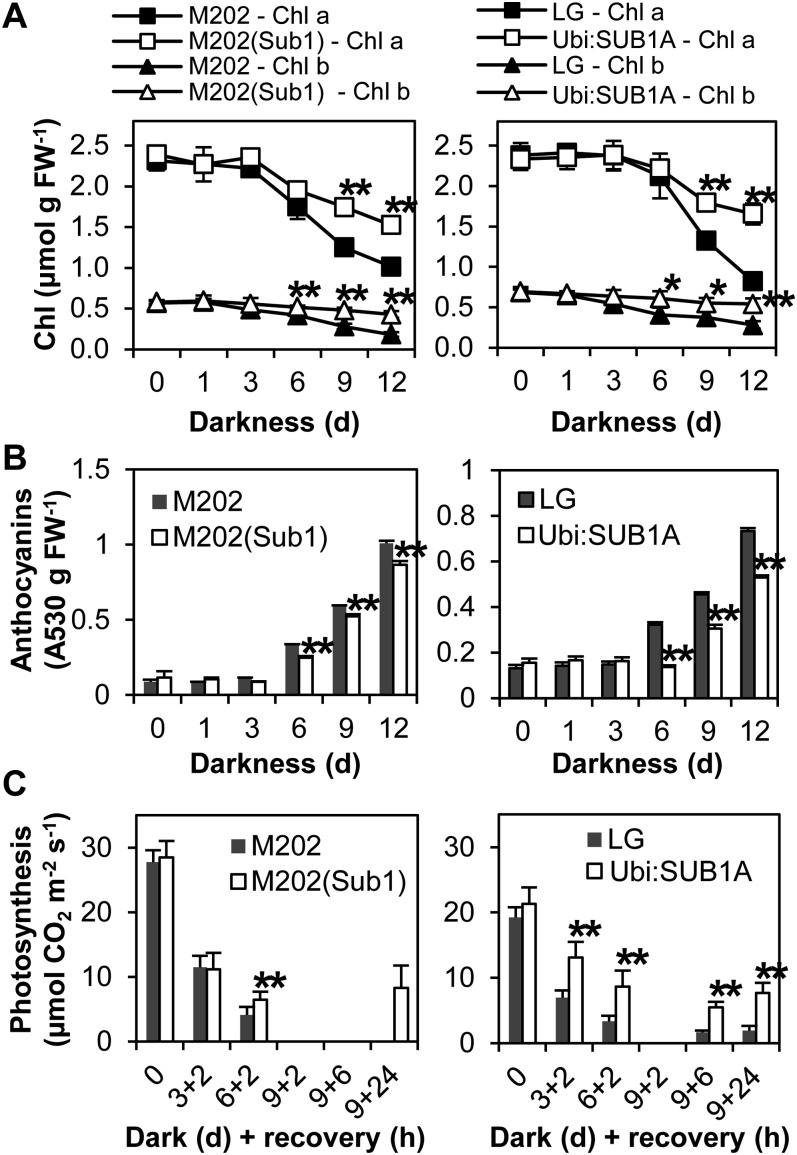
To quantify the alteration in chlorophyll content during dark treatment, the abundance of chlorophyll a and b was monitored in aerial tissue of plants exposed to complete darkness (Fig. 3A). Both were sustained for 3 d of darkness and then gradually decreased until day 12 in all four genotypes tested. However, more chlorophyll a and b were maintained in the genotypes with SUB1A during dark treatment, consistent with the visual inspection. Better maintenance of chlorophylls was also observed in SUB1A genotypes treated with prolonged submergence and oxidative stress (Fukao et al., 2006, 2011). Many environmental stresses trigger an increase in the level of anthocyanins in vegetative tissue, which is an indicator of cellular damage. We found that the abundance of anthocyanins increased 6 d after complete darkness in aerial tissue of M202 and M202(Sub1), with greater accumulation in the genotypes lacking SUB1A (Fig. 3B).
Figure 3.
SUB1A restricts leaf senescence and maintains photosynthesis capability after prolonged darkness. A, Chlorophyll contents in aerial tissue during prolonged darkness. The levels of chlorophyll a and b were analyzed in aerial tissue of plants exposed to complete darkness. The data represent means ± sd (n = 3). FW, Fresh weight. B, Anthocyanin contents in aerial tissue during prolonged darkness. The level of anthocyanin was monitored in aerial tissue of plants exposed to complete darkness for 12 d. The data represent means ± sd (n = 3). C, Photosynthesis capability of plants treated with prolonged darkness. Photosynthetic assimilation of CO2 was measured in the uppermost leaves of plants exposed to complete darkness. After dark treatment, recovery under light conditions (50 µmol m−2 s−1) for more than 2 h was necessary to detect stable CO2 assimilation. The data represent means ± sd (n = 8). Asterisks indicate significant differences between the two genotypes (*P < 0.05, **P < 0.01).
To determine whether SUB1A affects photosynthetic activity after prolonged darkness, CO2 assimilation by photosynthesis was monitored in dark-treated plants (Fig. 3C). There was no significant difference in photosynthetic activity between M202 versus M202(Sub1) and LG versus Ubi:SUB1A plants under regular growth conditions. After more than 3 d of complete darkness, the rate of photosynthesis was below the detection limit in the four genotypes, and over 2 h of recovery in light was required to quantify CO2 assimilation. M202 plants could not recover after 9 d of darkness, whereas photosynthesis recommenced in M202(Sub1) after 24 h of recovery. A similar trend was observed in LG and Ubi:SUB1A, with quicker recovery after 9 d of darkness for the SUB1A overexpression line. These results indicate that SUB1A slows chlorophyll breakdown, thereby enabling a resumption of photosynthesis following a sublethal period of prolonged darkness.
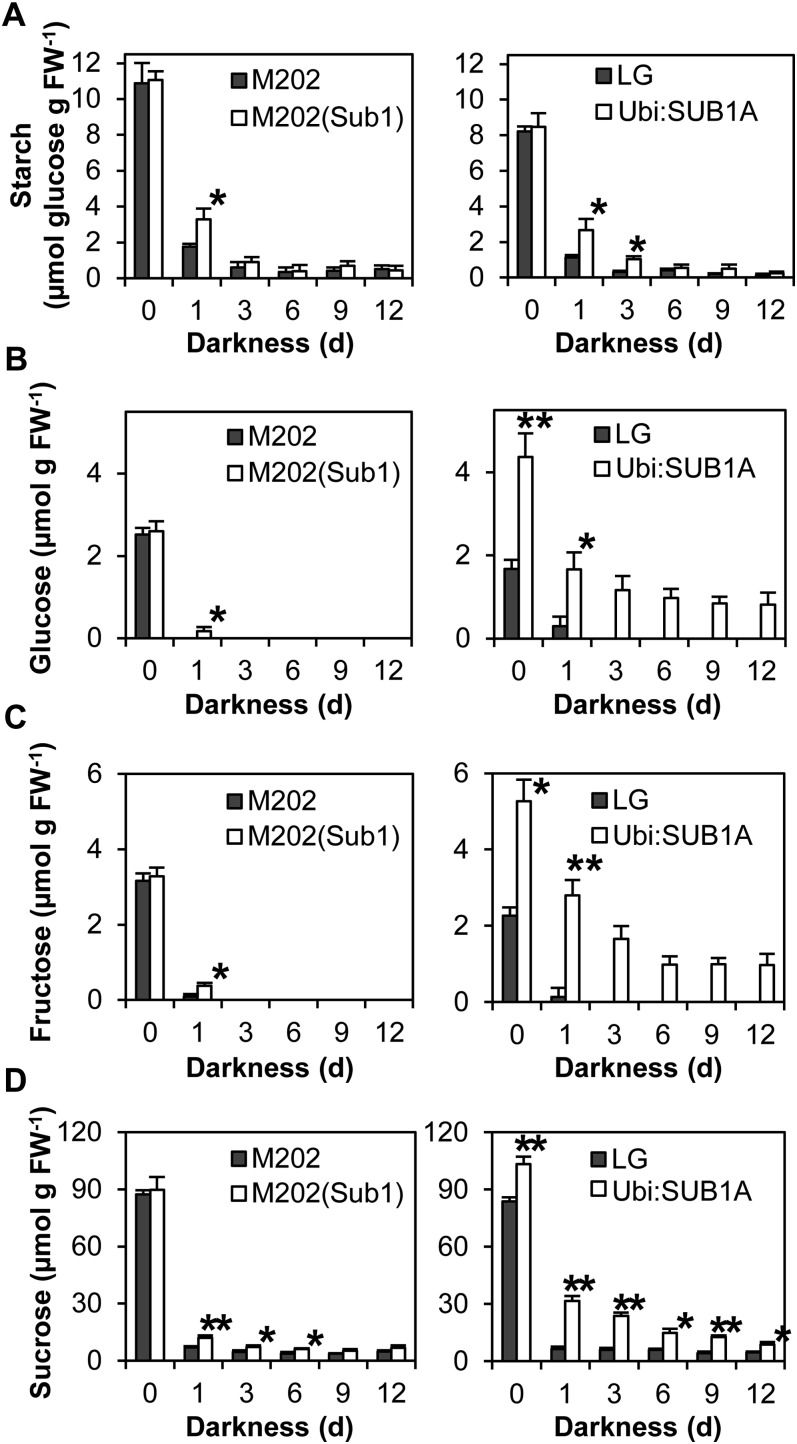
Under conditions in which carbohydrate production through photosynthesis is unavailable, proper management of carbohydrate reserves is key for survival. To evaluate the influence of SUB1A in carbohydrate consumption during prolonged darkness, the amount of starch, Glc, Fru, and Suc was monitored in aerial tissue (Fig. 4). Before stress treatment, the levels of starch, Glc, Fru, and Suc were similar in aerial tissue of M202 and M202(Sub1). The same trend was also observed in the starch content of LG and Ubi:SUB1A. By contrast, overexpression of SUB1A significantly increased the accumulation of Glc, Fru, and Suc under nonstress conditions. Complete darkness rapidly decreased the abundance of carbohydrates in all genotypes, but M202(Sub1) and Ubi:SUB1A plants maintained more starch, Glc, Fru, and Suc during the stress. This SUB1A-dependent maintenance of carbohydrate reserves was also observed in aerial tissue of plants exposed to prolonged submergence (Fukao et al., 2006; Barding et al., 2012).
Figure 4.
Carbohydrate contents in aerial tissue during prolonged darkness. The levels of starch (A), Glc (B), Fru (C), and Suc (D) were monitored in aerial tissue of plants treated with complete darkness for up to 12 d. The content of starch was quantified after digestion into Glc. The data represent means ± sd (n = 3). Asterisks indicate significant differences between the two genotypes (*P < 0.05, **P < 0.01). FW, Fresh weight.
Transcript Accumulation of SAGs Is Dampened by SUB1A
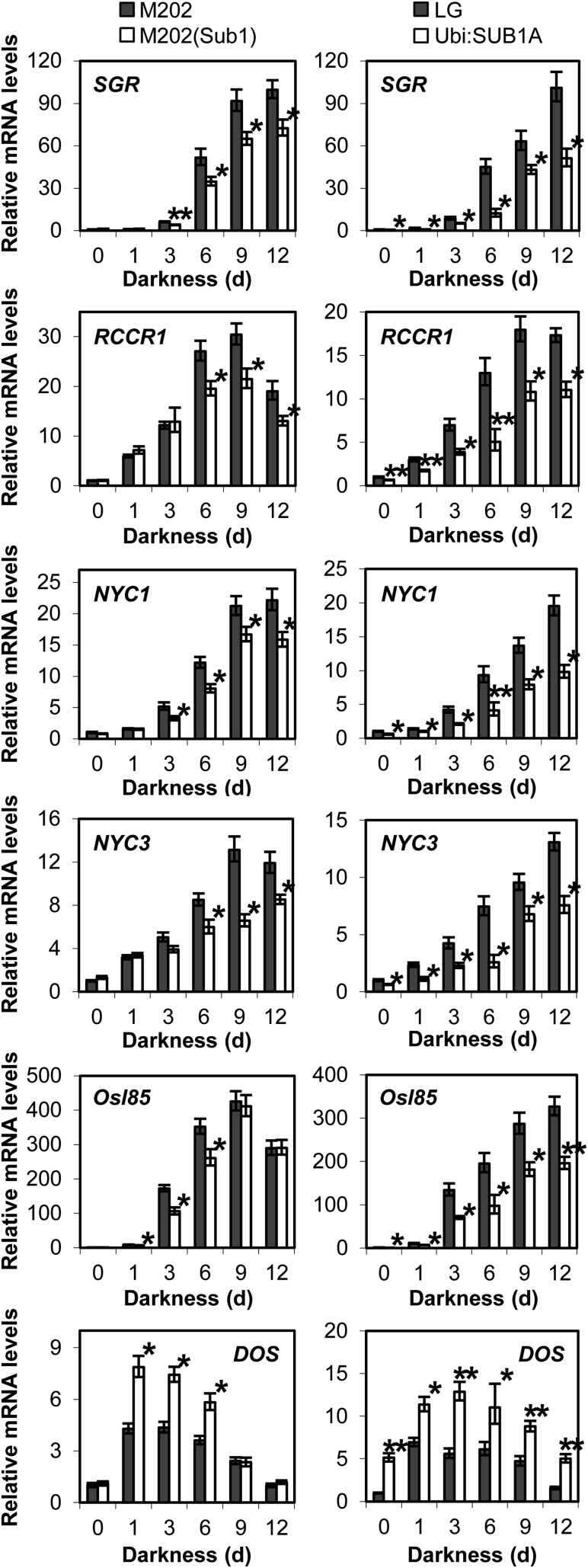
Leaf senescence occurs when plants are exposed to stressful conditions for prolonged periods. A subset of the genes involved in age-dependent senescence are induced by a variety of stresses such as prolonged darkness, carbohydrate deficiency, and submergence in rice and Arabidopsis (Lee et al., 2001; Buchanan-Wollaston et al., 2005; Rolland et al., 2006; Lim et al., 2007; Wang et al., 2007; Jung et al., 2010; Mustroph et al., 2010). To discern whether SUB1A affects the accumulation of genes associated with senescence, mRNA levels of representative SAGs were monitored in aerial tissue of plants exposed to complete darkness by qRT-PCR (Fig. 5). STAY-GREEN (SGR) encodes a novel chloroplast-located protein that is necessary for chlorophyll degradation in light-harvesting complex II in Arabidopsis and rice (Jiang et al., 2007; Park et al., 2007; Hörtensteiner, 2009). Red Chlorophyll Catabolite Reductase (RCCR) functions at the last step of chlorophyll degradation in chloroplasts (Pruzinská et al., 2007). Of the three RCCR genes in rice, RCCR1 is most highly induced by dark-induced and age-dependent senescence in leaves (Tang et al., 2011). NON-YELLOW COLORING1 (NYC1) and NYC3 were isolated through mutant screening of a stay-green phenotype in rice (Kusaba et al., 2007; Morita et al., 2009). NYC1 encodes chlorophyll b reductase, which catalyzes the conversion of chlorophyll b to chlorophyll a, whereas NYC3 is a chloroplast-located α/β-hydrolase family protein involved in chlorophyll degradation. Osl85 encodes isocitrate lyase, which is highly induced by prolonged darkness, natural senescence, sugar starvation, and submergence in leaves and suspension-cultured cells of rice (Lee et al., 2001; Wang et al., 2007; Mustroph et al., 2010). Our qRT-PCR studies revealed that the transcript abundance of these senescence-regulating genes was elevated in response to prolonged darkness in all four genotypes (Fig. 5). However, accumulation of these transcripts was significantly repressed in those encoding SUB1A. Notably, the overexpression of SUB1A constitutively restricted the accumulation of SAG mRNAs.
Figure 5.
Relative mRNA levels of genes associated with leaf senescence. Transcripts of representative genes were quantified in aerial tissue of plants exposed to complete darkness by qRT-PCR. The relative level of each mRNA was calculated by comparison with nonstressed M202 or LG. The data represent means ± se from three biological replicates. Asterisks indicate significant differences between the two genotypes (*P < 0.05, **P < 0.01).
DELAY OF THE ONSET OF SENESCENCE (DOS) has been identified as a negative regulator of leaf senescence in rice (Kong et al., 2006). DOS encodes a nucleus-localized CCCH-type zinc finger protein, and its mRNA level declines during age-dependent leaf senescence. In contrast with SAGs, overexpression of DOS delays chlorophyll degradation during prolonged darkness and age-dependent senescence. We found that the abundance of DOS mRNA was elevated in response to complete darkness, with higher accumulation in genotypes containing SUB1A. These data demonstrate that SUB1A enhances up-regulation of the key senescence-regulating gene, concomitant with delaying senescence during prolonged darkness.
SUB1A Enhances Responsiveness to the Senescence Regulatory Hormones JA and SA
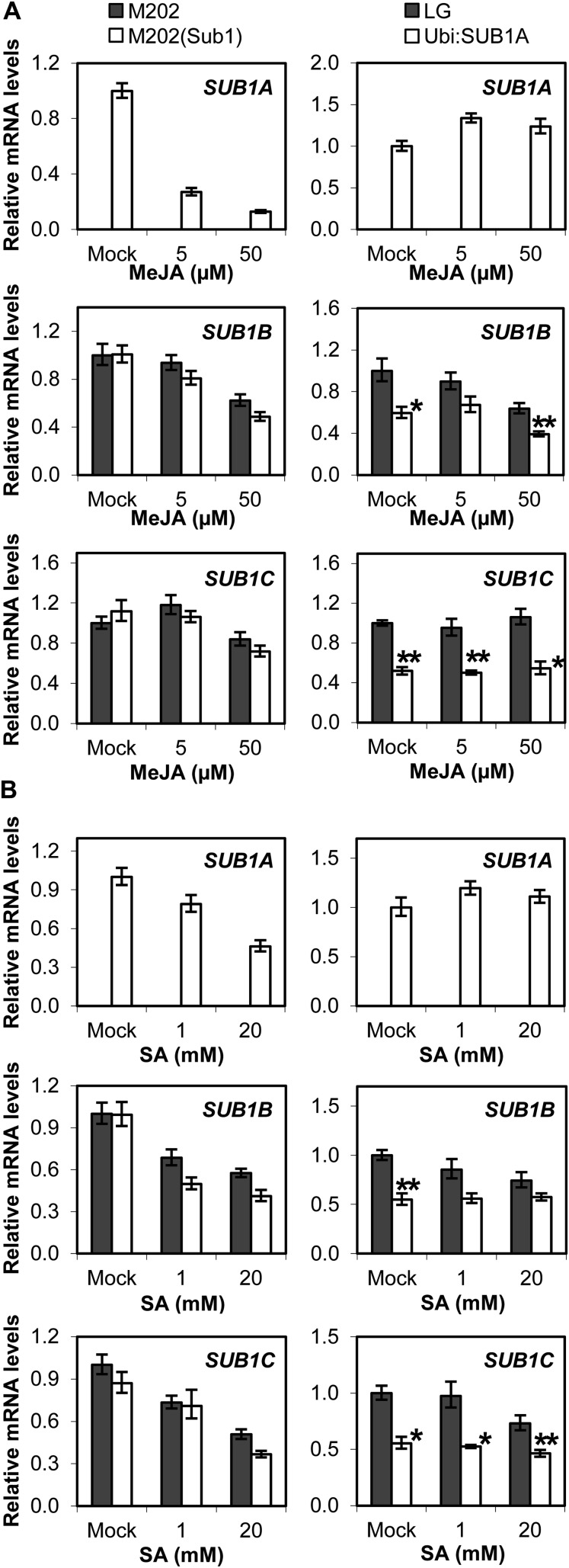
JA and SA are key phytoregulators of molecular and biochemical processes of leaf senescence (Lim et al., 2007; Balbi and Devoto, 2008; Pauwels et al., 2009). Application of a derivative of JA, MeJA, significantly decreased the basal level of SUB1A mRNA in aerial tissue of M202(Sub1) plants grown under control conditions (Fig. 6A). By contrast, the level of ectopically expressed SUB1A mRNA was minimally affected by MeJA in Ubi:SUB1A. MeJA treatment slightly repressed the basal level of SUB1B transcript, whereas there was little effect of the hormone on SUB1C mRNA in all genotypes. Similar trends in SUB1 gene accumulation were observed in SA-treated plants (Fig. 6B).
Figure 6.
Relative transcript levels of SUB1 genes following MeJA or SA treatment. The levels of SUB1A, SUB1B, and SUB1C mRNAs were compared between M202 versus M202(Sub1) (A) and LG versus Ubi:SUB1A (B) plants treated with MeJA or SA by qRT-PCR. The relative level of each mRNA in aerial tissue was calculated by comparison with the nonstressed control [M202(Sub1) or Ubi:SUB1A at day 0 for SUB1A; M202 or LG at day 0 for SUB1B and SUB1C]. The data represent means ± se from three biological replicates. Asterisks indicate significant differences between the two genotypes (*P < 0.05, **P < 0.01).
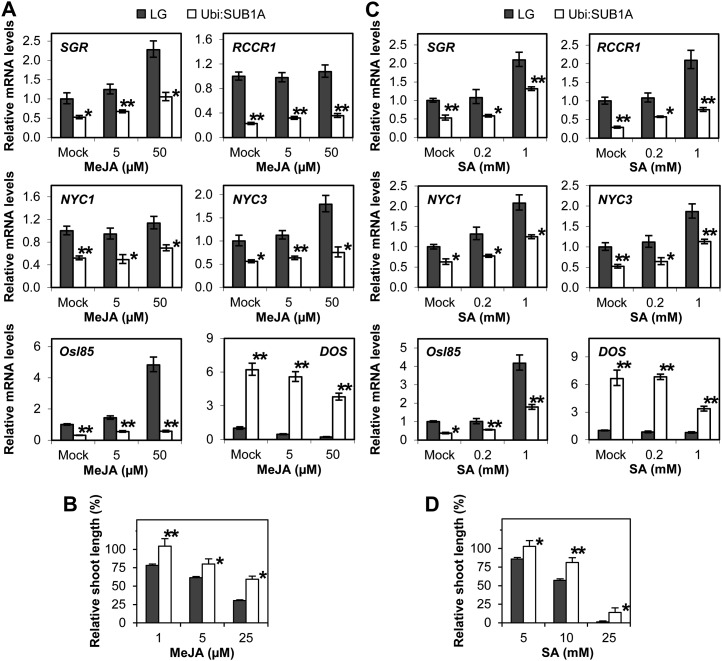
Since MeJA and SA reduce basal levels of SUB1A transcript in M202(Sub1), we focused our attention on the influence of SUB1A on mRNA accumulation of representative senescence-regulating genes in MeJA- or SA-treated Ubi:SUB1A and LG plants. SGR, NYC3, and Osl85 were induced by the application of MeJA (Fig. 7A). However, overexpression of SUB1A significantly restricted the accumulation of these transcripts regardless of the hormone treatment. The abundance of NYC1 and RCCR1 was constitutively repressed in Ubi:SUB1A, although the two genes were not influenced by MeJA. DOS mRNA was highly accumulated in mock- and MeJA-treated Ubi:SUB1A. To discern whether SUB1A modulates the responsiveness to MeJA, inhibition of shoot elongation by the hormone was assayed at the seedling stage (Fig. 7B). Application of MeJA repressed elongation growth of seedling shoots in LG and Ubi:SUB1A, but overexpression of SUB1A significantly dampened the inhibition mediated by MeJA. The effect of SUB1A on transcript accumulation of senescence-regulating genes was also evaluated in plants treated with SA (Fig. 7C). SGR, RCCR1, NYC1, NYC3, and Osl85 were responsive to SA in LG and Ubi:SUB1A. However, overexpression of SUB1A significantly restrained the mRNA abundance of these genes in mock- and SA-treated plants. Conversely, DOS mRNA constitutively accumulated in Ubi:SUB1A. As seen with MeJA, shoot elongation was repressed by SA treatment in LG and Ubi:SUB1A (Fig. 7D), but the growth inhibition was significantly lower in the SUB1A overexpression line. Together, these data indicate that constitutive overexpression of SUB1A diminishes the responsiveness to MeJA and SA.
Figure 7.
Constitutive expression of SUB1A diminishes responsiveness to MeJA and SA. A and C, Relative mRNA levels of senescence-regulated genes following MeJA (A) or SA (C) treatment. The transcript levels of representative genes were monitored in aerial tissue of plants treated with MeJA or SA by qRT-PCR. The relative level of each mRNA was calculated by comparison with nonstressed LG. The data represent means ± se from three biological replicates. Asterisks indicate significant differences between the two genotypes (*P < 0.05, **P < 0.01). B and D, MeJA-inhibited (B) or SA-inhibited (D) shoot elongation in germinating seeds. Seeds were incubated on wet filter paper containing a series of MeJA or SA solutions for 6 d. Relative shoot elongation was calculated by comparison with the nontreated seeds of individual genotypes. The data represent means ± se (n = 25). Asterisks indicate significant differences between the two genotypes (*P < 0.05, **P < 0.01).
Ethylene Promotes Dark- and JA-Induced Senescence, But SUB1A Restricts Ethylene Accumulation during Prolonged Darkness
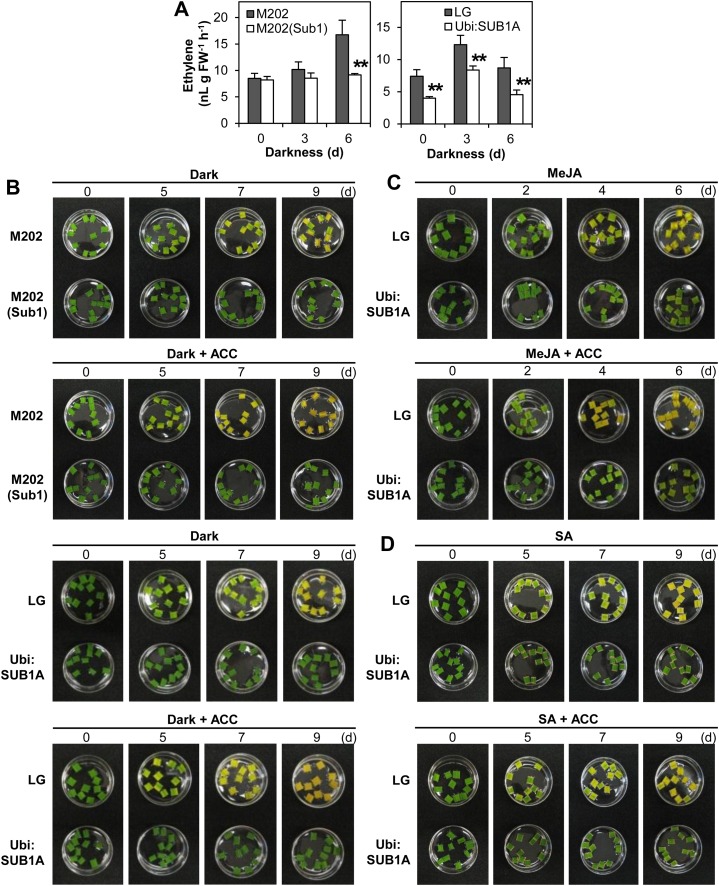
Ethylene is an endogenous regulator of leaf senescence (Bleecker and Kende, 2000; Lim et al., 2007). Previously, we reported that SUB1A mRNA accumulation was significantly up-regulated by ethylene, although SUB1A repressed ethylene production under submergence (Fukao et al., 2006). To investigate whether this SUB1A-mediated restriction occurs during prolonged darkness, the rate of ethylene evolution was quantified in plants exposed to the stress (Fig. 8A). No difference in ethylene production was evident in M202 or M202(Sub1) at the 0- and 3-d time points. However, 6 d of darkness clearly increased the level of ethylene evolved by M202 but not M202(Sub1). LG also significantly increased ethylene production within 3 d of darkness (P < 0.001). However, the overexpression of SUB1A restricted ethylene evolution under dark conditions and also during standard growth conditions. These data indicate that SUB1A down-regulates ethylene production during constant darkness, as documented for submergence (Fukao et al., 2006).
Figure 8.
SUB1A limits dark- and MeJA-induced senescence promoted by ethylene. A, Ethylene evolution during dark treatment. Developmentally matched 9-day-old [M202, M202(Sub1), and LG] or 12-d-old (Ubi:SUB1A) plants grown in test tubes were exposed to complete darkness for up to 6 d. Following treatment, ethylene accumulated in the test tube was quantified by gas chromatography. The data represent means ± sd (n = 5). Asterisks indicate significant differences between the two genotypes (**P < 0.01). FW, Fresh weight. B to D, Photographs of leaf segments treated with prolonged darkness (B), MeJA (C), or SA (D) with and without ACC. Leaf segments of uppermost leaves in 14-d-old [M202, M202(Sub1), and LG] and 21-d-old (Ubi:SUB1A) plants were incubated on one-half-strength MS medium containing hormone solution with and without ACC (100 µm) for up to 9 d (n = 10).
To evaluate the effect of ethylene on dark-induced senescence, leaf segments were incubated on one-half-strength Murashige and Skoog (MS) medium containing the immediate precursor of ethylene, 1-aminocyclopropane-1-carboxylic-acid (ACC), in the dark (Fig. 8B). In the absence of ACC, the greenness of leaves was gradually lost over the 9 d of dark treatment in M202 and LG. In the genotypes carrying SUB1A, leaf color was unchanged after 9 d, consistent with the observation at the whole-plant level (Fig. 2A). The addition of ACC accelerated dark-induced senescence in M202 and LG but had no visible effect on M202(Sub1) or Ubi:SUB1A. Interaction of ethylene with MeJA and SA in leaf senescence was also evaluated in the same experimental system using LG and Ubi:SUB1A. M202 and M202(Sub1) were not tested because the two hormones reduced rather than increased basal SUB1A mRNA levels (Fig. 6). MeJA induced a leaf color change in both genotypes, but the loss of greenness was slower in the SUB1A genotype (Fig. 8C). In LG, MeJA-mediated senescence was further promoted by the addition of ACC. In both genotypes, SA promoted cell death along the cut edge of the leaf segments, consistent with a functional hypersensitive response. SA stimulated leaf senescence more severely in control genotypes than in SUB1A-containing genotypes (Fig. 8D). However, ACC did not influence SA-mediated senescence. Overall, it appears that SUB1A delays dark- and JA-induced senescence through the restriction of an ethylene response pathway.
DISCUSSION
Complete submergence imposes multiple environmental stimuli due to a 10,000-fold reduction in the diffusion rate of oxygen, CO2, and ethylene as well as a restriction of light and nutrient availability. A combination of these environmental factors induces a reduction in photosynthesis and aerobic respiration and an increase in catabolism of energy reserves, including carbohydrates, proteins, and lipids, resulting in carbohydrate starvation, chlorophyll degradation, and leaf senescence. Previously, our studies demonstrated that SUB1A restricts carbohydrate consumption and chlorophyll breakdown through the regulation of ethylene production and gibberellin responsiveness during submergence (Fukao et al., 2006; Fukao and Bailey-Serres, 2008). To further dissect the role of SUB1A in adaptation to the complex stress, we investigated physiological and molecular responses of vegetative-stage plants to prolonged darkness, which also provokes common environmental alterations to plants.
An early physiological response to constant darkness is a reduction in carbohydrate reserves of aerial tissue. We observed that the abundance of starch, Glc, Fru, and Suc quickly decreased after 1 d of dark treatment (Fig. 4), followed by chlorophyll degradation and anthocyanin accumulation after 6 d of stress (Fig. 3) in all four genotypes. However, the decline in carbohydrate reserves and chlorophylls was significantly dampened in genotypes carrying SUB1A as compared with non-SUB1A lines (Figs. 3A and 4), consistent with the observations during submergence (Fukao et al., 2006; Barding et al., 2012). Notably, the major soluble carbohydrates were constitutively higher in aerial tissue of Ubi:SUB1A plants even under normal growth conditions, presumably due to SUB1A-mediated restriction of carbohydrate catabolism, since CO2 assimilation by photosynthesis was not altered by the overexpression of SUB1A (Figs. 3C and 4). Photosynthesis and carbohydrate catabolism are regulated by light/dark transitions and the circadian clock (Lu et al., 2005; Farré and Weise, 2012). In Arabidopsis, levels of starch are most abundant at the end of the light cycle and decline significantly over the course of each night to fuel rapid growth (Graf et al., 2010). If the night is prolonged or the central oscillator of the circadian clock (CIRCADIAN CLOCK ASSOCIATED1/LATE ELONGATED HYPOCOTYL4) is disrupted, starch is consumed more rapidly during the night, to the detriment of the overall biomass of the plant. Previously, we monitored the level of SUB1A mRNA in aerial tissue of M202(Sub1) every 3 h for 24 h, but its abundance was unaltered by the light/dark transition (Peña-Castro et al., 2011), indicating that SUB1A may not be regulated by the depletion of carbohydrate reserves during the anticipated nighttime. However, SUB1A mRNA levels rose dramatically (more than 90-fold) by 24 h of darkness initiated at midday, correlating with a rapid decline in carbohydrate reserves (Figs. 1 and 4). This pronounced but transient accumulation of SUB1A mRNA could be due to the decline of carbohydrate reserves beyond that which occurs overnight. Overall, our results indicate that dark-induced SUB1A enables more conservative carbon use to prolong the maintenance of cellular homeostasis under conditions of prolonged darkness or submergence.
The catabolism of chlorophyll and chloroplast proteins is actively regulated during natural leaf senescence, which remobilizes nutrients or energy resources to storage organs or seeds (Lim et al., 2007). It is feasible that this genetically coordinated process is also part of the acclimation response to prolonged darkness and submergence. However, excessive breakdown of chlorophyll and chloroplast proteins prevents the recommencement of photosynthesis during recovery. Here, we show that the induction of two chlorophyll catabolic enzyme genes, NYC1 and RCCR1, is significantly reduced by SUB1A during constant darkness, especially at later time points (Fig. 5). Recently, it was shown that direct or indirect interaction of SGR with NYC1 and RCCR is necessary for recruitment of these enzymes into senescing chloroplasts (Sakuraba et al., 2012). It seems that SUB1A-dependent regulation of the key regulators for chlorophyll degradation limits the catabolism of chlorophyll and chloroplast proteins under stress conditions. Consistent with this, our previous 1H-NMR spectrometry study of metabolites demonstrated that SUB1A represses the accumulation of nine amino acids, which were elevated during submergence (Barding et al., 2012). Here, we found that genotypes carrying SUB1A better retained chlorophyll content during constant darkness and recovered photosynthetic activity more quickly after reillumination (Fig. 3C). Despite the impediment of gas exchange and light availability, the degree of underwater photosynthesis influences the survival of terrestrial wetland plants, including rice (Colmer et al., 2011). It follows that the maintenance of chlorophylls in SUB1A genotypes may benefit photosynthetic energy production during submergence stress and upon recovery.
We demonstrated that transcript accumulation of representative SAGs was significantly restricted during constant darkness in genotypes carrying SUB1A, indicating that SUB1A functions as a repressor of these senescence-inducible genes (Fig. 5). By contrast, a negative regulator of leaf senescence, DOS, was further induced in the presence of SUB1A during prolonged darkness. This gene was also induced by submergence, with a higher level in M202(Sub1) than in M202 (Jung et al., 2010; Mustroph et al., 2010). DOS encodes a nucleus-localized CCCH-type zinc finger protein that regulates the expression of a subset of genes associated with JA biosynthesis and signaling in rice (Kong et al., 2006). Genetic analysis revealed that responsiveness to JA is elevated in DOS knockdown lines but repressed in DOS overexpression lines. Thus, DOS acts upstream of the JA pathway to restrain JA-dependent leaf senescence. In accordance with the observations in DOS overexpression lines, SUB1A-mediated constitutive accumulation of DOS mRNA displayed diminished responsiveness to JA as well as postponed dark- and JA-induced leaf senescence (Figs. 7B and 8). It seems that SUB1A potentiates DOS up-regulation to dampen responsiveness to JA, thereby limiting JA-promoted senescence. Further investigation will be required to elucidate the regulatory mechanism of DOS mRNA accumulation governed by SUB1A.
In Arabidopsis, SA is specifically involved in age-induced but not dark- or starvation-induced senescence. Microarray analysis revealed that a subset of genes down-regulated in a SA-defective transgenic line are induced in age-dependent senescence but not in dark- and starvation-induced senescence in Arabidopsis (Buchanan-Wollaston et al., 2005). Additionally, age-dependent senescence was significantly delayed in the SA-defective line, but dark-induced senescence occurred similarly in the wild type and the transgenic line. Based on our results, SA may be important in dark-induced senescence in rice. Here, we show that constitutive expression of SUB1A restricts the accumulation of mRNAs induced by SA and represses SA-mediated inhibition of shoot elongation (Fig. 7C), indicating that responsiveness to SA is down-regulated by constitutive expression of SUB1A. Previously, we reported a marked delay of seed maturation as well as vegetative growth and flowering in the two independent SUB1A overexpression lines (Fukao and Bailey-Serres, 2008). Thus, SUB1A-dependent reduction of SA responsiveness may also delay seed maturation, a developmental senescence process.
Previously, we reported that submergence-induced SUB1A down-regulates underwater ethylene production and the accumulation of ethylene-inducible genes that are associated with elongation and anaerobic metabolism (Fukao et al., 2006). We found that SUB1A also diminishes the induction of ethylene production during constant darkness in both M202(Sub1) and Ubi:SUB1A (Fig. 8A). Overexpression of SUB1A also repressed dark- and JA-induced senescence promoted by the application of ACC (Fig. 8, B and C). Thus, ethylene modulates GA-mediated processes during submergence (Fukao and Bailey-Serres, 2008), and ethylene stimulates JA-mediated processes that promote stress-induced senescence. Both of these ethylene-induced responses promote the consumption of energy reserves and are inhibited by SUB1A. Identification of direct targets of SUB1A and functional characterization of the downstream ramifications will aid elucidation of the integrated molecular mechanisms underlying SUB1A-mediated hormonal regulation conferring multiple stress tolerance to rice.
CONCLUSION
Plants encounter similar physiological alterations under submergence and constant darkness, such as extensive catabolism of carbohydrate reserves, breakdown of chlorophyll, and recycling of chloroplast proteins. These acclimation responses provide nutrient resources under conditions where energy production through photosynthesis is limited or not available. However, prolonged exposure to the stress eventually exhausts energy resources and causes death. This study demonstrates that the key regulator of submergence tolerance, SUB1A, coordinates physiological and molecular responses to prolonged darkness, resulting in enhanced survival of the stress. Prolonged darkness places constraints on carbohydrate availability for growth and stimulates the accumulation of ethylene and JA (Lim et al., 2007; Seltmann et al., 2010). Ethylene accelerates leaf senescence mediated by JA. In the genotype with an endogenous SUB1A, ethylene induces SUB1A mRNA accumulation, however, which restricts ethylene production. As a result, JA-mediated senescence responses are restrained, enhancing survival of prolonged darkness. Stress-induced senescence occurs as a consequence of prolonged exposure to a variety of biotic and abiotic stresses, including submergence and drought. Further investigation of the regulatory mechanism underlying stress-induced senescence may provide additional strategies to improve the resilience of crops to the extremes in weather associated with climate change. A question raised by these studies is whether the influence of SUB1A on JA and SA responsiveness associated with senescence may conditionally regulate innate immunity to pathogens. To date, there is no evidence that Sub1 rice varieties grown in farmers’ fields are altered in pathogen resistance relative to near-isogenic non-Sub1 control lines (D. Mackill, personal communication).
MATERIALS AND METHODS
Plant Materials
Rice (Oryza sativa) lines cv M202 (SUB1B-2, SUB1C-2), cv LG (SUB1B-2, SUB1C-2), the SUB1 introgression line M202(Sub1) (SUB1A-1, SUB1B-1, SUB1C-1), and the SUB1A overexpression line Ubi:SUB1A-3 (SUB1A-1, SUB1B-2, SUB1C-2) were used in this study (Xu et al., 2006). Ubi:SUB1A-3 is one of two well-characterized representative transgenic lines with constitutively expressed SUB1A-1 in the LG background, referred as to Ubi:SUB1A (Fukao and Bailey-Serres, 2008; Fukao et al., 2011). Sterilized seeds were placed on wet filter paper for 3 d at 25°C in the light (50 µmol m−2 s−1), and germinated seeds were transplanted into soil-containing plastic pots (10 × 10 × 10 cm). Plants were grown in a greenhouse (30°C day, 20°C night) for 14 d [M202, M202(Sub1), and LG] or 21 d (Ubi:SUB1A) under natural light conditions. Ubi:SUB1A exhibits a semidwarf phenotype, but all four genotypes are developmentally matched at the ages used in these analyses (Fukao and Bailey-Serres, 2008). The plants have three fully opened leaves under our growth conditions. All experiments were performed at this developmental age unless otherwise indicated.
Dark and Hormone Treatments
All dark and hormone treatments were replicated in at least three independent experiments. For dark treatment, 14- or 21-d-old plants were transferred to a growth chamber at midday and subjected to complete darkness at 25°C for up to 18 d. Aerial tissue was harvested at midday on the day specified under green light in the darkroom to avoid a light response. For MeJA treatment, the entire aerial tissue was excised at the base of the stem and immediately placed into 20 mL of mock (0.1% [v/v] dimethyl sulfoxide) or MeJA (5 or 50 µm in 1% [v/v] dimethyl sulfoxide) solution in a 250-mL glass beaker for 24 h in the light (50 µmol m−2 s−1). For SA treatment, deionized water and SA (1 or 20 mm) were used as mock and hormone solution, respectively. After each treatment, collected tissue was immediately frozen in liquid nitrogen and stored at −80°C until use. To observe the effect of darkness and hormones on leaf senescence, the fully expanded uppermost leaves were cut into pieces (8 mm in length), and the leaf segments were floated on one-half-strength MS medium in the dark or containing hormone solution in the light (50 µmol m−2 s−1) at 25°C for up to 9 d. To monitor hormonal effects on seedling growth, sterilized seeds were incubated on wet filter paper containing MeJA (1, 5, and 25 µm) or SA (5, 10, and 25 mm) at 25°C in the light (50 µmol m−2 s−1), and the length of each shoot was recorded after 6 d.
Chlorophyll and Anthocyanin Assays
Chlorophyll a and b contents were quantified from 50 mg of tissue in 5 mL of 100% methanol as described by Porra (2002). After centrifugation at 4°C for 20 min at 16,800g, the absorbance of the supernatant was measured at 652.0 and 665.2 nm with a spectrophotometer (DU800; Beckman). Anthocyanin content was assayed following the method of Jeong et al. (2010). Aerial tissue (50 mg) was homogenized in 600 µL of 1% (v/v) HCl in methanol on ice and then incubated for 16 h at 4°C in the dark with gentle shaking. After incubation, the extract was mixed with 400 µL of water and 400 µL of chloroform. Following centrifugation at 4°C for 2 min at 16,800g, A530 and A657 were measured.
Carbohydrate Assays
Glc, Fru, and Suc contents were measured by the method of Guglielminetti et al. (1995). Aerial tissue (50 mg) was homogenized in 1 mL of 80% (v/v) ethanol and incubated at 80°C for 20 min. Following centrifugation for 10 min at 16,800g, the supernatant was collected and the extraction process was repeated twice, with the three tissue extracts combined and dried under a vacuum. After rehydration in 0.5 mL of water, the samples were subjected to coupled enzymatic methods with a spectrophotometer. Glc was assayed in a reaction mixture (1 mL) containing 50 µL of extract, 100 mm Tris-HCl (pH 7.6), 8 mm MgCl2, 1 mm ATP, 1.5 mm NADP, 1 unit of hexokinase (Sigma-Aldrich), and 1 unit of Glc-6-P dehydrogenase (Sigma-Aldrich). The mixture was incubated at 37°C for 30 min, and the increase in A340 was measured. For Fru, 6 units of phosphoglucose isomerase (Sigma-Aldrich) was added to the Glc assay mixture. For Suc, a reaction mixture (100 µL) containing the extract (50 µL), 15 mm sodium acetate (pH 4.6), and 40 units of invertase (Sigma-Aldrich) was incubated at 37°C for 15 min, and the resulting Glc was quantified as described above. Starch content was measured following the method of Fukao et al. (2006). The pellet obtained after ethanol extraction was washed with water, resuspended in 1 mL of water containing 10 units of heat-resistant α-amylase (Sigma-Aldrich), and incubated at 95°C for 15 min. After cooling, the suspension was adjusted to 25 mm sodium citrate (pH 4.8), and 5 units of amyloglucosidase (Sigma-Aldrich) was added. Following incubation at 55°C for 30 min, the reaction mixture was centrifuged at 16,800g for 30 min, and the supernatant was subjected to Glc assay as described above. The reaction efficiency of each method was validated by analyzing known amounts of each carbohydrate.
Photosynthetic Activity Measurement
To analyze photosynthetic light response, maximum rates of net CO2 assimilation were quantified with a portable photosynthesis analysis system (model 6400; Li-Cor) equipped with a red-blue light source (model 6400-02B, no. SI-710; Li-Cor) as described by Santiago (2007). Uppermost expanded leaves of eight plants were measured at 370 µmol mol−1 CO2 and 1,200 µmol m−2 s−1 irradiance at noon. Since plants treated with prolonged darkness required recovery in the light to produce a detectable amount of CO2, plants were placed in the light (50 µmol m−2 s−1) at 25°C for 2 to 24 h. Following photosynthetic measurements, leaf area was recorded and used for standardization.
qRT-PCR
Total RNA was extracted from frozen aerial tissue using the RNeasy Plant mini kit (Qiagen). Genomic DNA was removed by the on-column digestion method described in the manufacturer’s protocol. complementaryDNA was synthesized from 2 µg of total RNA following the method of Fukao et al. (2006). Real-time PCR was performed in a 20-µL reaction using iQ SYBR Green Supermix (Bio-Rad) in the CFX96 real-time PCR detection system (Bio-Rad). PCR efficiency (95%–105%) was verified as described by Schmittgen and Livak (2008). Amplification specificity was validated by melt-curve analysis at the end of each PCR cycle. Relative transcript abundance was calculated using the comparative cycle threshold method (Livak and Schmittgen, 2001). ACTIN1 or α-TUBULIN was used as a normalization control. Sequences and annealing temperatures of primer pairs are listed in Supplemental Table S1.
Ethylene Measurement
The rate of ethylene production was quantified as described by Larsen and Cancel (2004). Dehulled seeds were sterilized in 70% (v/v) ethanol for 10 min and in 2.5% (v/v) sodium hypochlorite solution for 20 min. After rinsing with sterilized water thoroughly, each seed was cultured on one-half-strength MS medium in a test tube for 9 d [M202, M202(Sub1), and LG] or 12 d (Ubi:SUB1A; 16 h of light/8 h of dark; light level, 100 µmol m−2 s−1). For dark treatment, the test tubes were placed in the dark at 25°C for up to 6 d. Following stress treatment, each tube was tightly closed with a rubber serum stopper and incubated in the dark for 2 h. The accumulated gas sample (0.9 mL) was withdrawn from each tube with a 1-mL syringe and assayed by a gas chromatograph (6850 Series; Hewlett-Packard) equipped with an alumina-based capillary column (Agilent Technologies).
Sequence data from this article can be found in the Michigan State University Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu) under the following accession numbers: Actin1 (LOC_Os03g50890), SUB1A (DQ011598b [GenBank/EMBL accession number; this gene is absent from Nipponbare and therefore has no LOC number]), α-TUBULIN (LOC_Os07g38730) SUB1B (LOC_Os09g11480), SUB1C (LOC_Os09g11460), SGR (LOC_Os09g36200), RCCR1 (LOC_Os10g25030), NYC1 (LOC_Os01g12710), NYC3 (LOC_Os06g24730), Osl85 (LOC_Os07g34520), and DOS (LOC_Os01g09620). To obtain GenBank sequences, which are linked to Rice Annotation Project identification numbers, see http://rapdb.dna.affrc.go.jp/tools/converter to convert from Michigan State University identification numbers (LOC_Os00g00000) to Rice Annotation Project identification numbers (Os00g0000000).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Primers and PCR conditions used for quantitative RT-PCR.
Acknowledgments
We thank David Mackill and Abdelbagi Ismail for critical review and helpful comments on the manuscript. We are grateful to Lois Santiago and Paul Larsen for technical assistance with photosynthesis activity measurement and ethylene quantification, respectively.
Glossary
- MeJA
methyl jasmonate
- JA
jasmonate
- SA
salicylic acid
- LG
Liaogeng
- qRT
quantitative reverse transcription
- ACC
1-aminocyclopropane-1-carboxylic-acid
- MS
Murashige and Skoog
References
- Akman M, Bhikharie AV, McLean EH, Boonman A, Visser EJ, Schranz ME, van Tienderen PH. (2012) Wait or escape? Contrasting submergence tolerance strategies of Rorippa amphibia, Rorippa sylvestris and their hybrid. Ann Bot (Lond) 109: 1263–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbi V, Devoto A. (2008) Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytol 177: 301–318 [DOI] [PubMed] [Google Scholar]
- Barding GA, Jr, Fukao T, Béni S, Bailey-Serres J, Larive CK. (2012) Differential metabolic regulation governed by the rice SUB1A gene during submergence stress and identification of alanylglycine by 1H NMR spectroscopy. J Proteome Res 11: 320–330 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Kende H. (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16: 1–18 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Chen X, Pierik R, Peeters AJ, Poorter H, Visser EJ, Huber H, de Kroon H, Voesenek LA. (2010) Endogenous abscisic acid as a key switch for natural variation in flooding-induced shoot elongation. Plant Physiol 154: 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmer TD, Winkel A, Pedersen O. (2011) A perspective on underwater photosynthesis in submerged terrestrial wetland plants. AoB Plants 2011: plr030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré EM, Weise SE. (2012) The interactions between the circadian clock and primary metabolism. Curr Opin Plant Biol 15: 293–300 [DOI] [PubMed] [Google Scholar]
- Fukao T, Bailey-Serres J. (2008) Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc Natl Acad Sci USA 105: 16814–16819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J. (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18: 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Yeung E, Bailey-Serres J. (2011) The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23: 412–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepstein S, Sabehi G, Carp M-J, Hajouj T, Nesher MFO, Yariv I, Dor C, Bassani M. (2003) Large-scale identification of leaf senescence-associated genes. Plant J 36: 629–642 [DOI] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, Smith AM. (2010) Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci USA 107: 9458–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić V, Bleecker AB. (1995) Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J 8: 595–602 [Google Scholar]
- Guglielminetti L, Perata P, Alpi A. (1995) Effect of anoxia on carbohydrate metabolism in rice seedlings. Plant Physiol 108: 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Fukushige H, Hildebrand DF, Gan S. (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Tang W, Swain JD, Green AL, Jack TP, Gan S. (2001) Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiol 126: 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S. (2009) Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci 14: 155–162 [DOI] [PubMed] [Google Scholar]
- Jeong SW, Das PK, Jeoung SC, Song JY, Lee HK, Kim YK, Kim WJ, Park YI, Yoo SD, Choi SB, et al. (2010) Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis. Plant Physiol 154: 1514–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Li M, Liang N, Yan H, Wei Y, Xu X, Liu J, Xu Z, Chen F, Wu G. (2007) Molecular cloning and function analysis of the stay green gene in rice. Plant J 52: 197–209 [DOI] [PubMed] [Google Scholar]
- Jing HC, Sturre MJ, Hille J, Dijkwel PP. (2002) Arabidopsis onset of leaf death mutants identify a regulatory pathway controlling leaf senescence. Plant J 32: 51–63 [DOI] [PubMed] [Google Scholar]
- Jung KH, Seo YS, Walia H, Cao P, Fukao T, Canlas PE, Amonpant F, Bailey-Serres J, Ronald PC. (2010) The submergence tolerance regulator Sub1A mediates stress-responsive expression of AP2/ERF transcription factors. Plant Physiol 152: 1674–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Z, Li M, Yang W, Xu W, Xue Y. (2006) A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol 141: 1376–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, et al. (2007) Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19: 1362–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PB, Cancel JD. (2004) A recessive mutation in the RUB1-conjugating enzyme, RCE1, reveals a requirement for RUB modification for control of ethylene biosynthesis and proper induction of basic chitinase and PDF1.2 in Arabidopsis. Plant J 38: 626–638 [DOI] [PubMed] [Google Scholar]
- Lee RH, Wang CH, Huang LT, Chen SC. (2001) Leaf senescence in rice plants: cloning and characterization of senescence up-regulated genes. J Exp Bot 52: 1117–1121 [DOI] [PubMed] [Google Scholar]
- Lee SC, Mustroph A, Sasidharan R, Vashisht D, Pedersen O, Oosumi T, Voesenek LA, Bailey-Serres J. (2011) Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol 190: 457–471 [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Lin JF, Wu SH. (2004) Molecular events in senescing Arabidopsis leaves. Plant J 39: 612–628 [DOI] [PubMed] [Google Scholar]
- Liu L, Zhou Y, Zhou G, Ye R, Zhao L, Li X, Lin Y. (2008) Identification of early senescence-associated genes in rice flag leaves. Plant Mol Biol 67: 37–55 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu Y, Gehan JP, Sharkey TD. (2005) Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol 138: 2280–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R, Sato Y, Masuda Y, Nishimura M, Kusaba M. (2009) Defect in non-yellow coloring 3, an alpha/beta hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J 59: 940–952 [DOI] [PubMed] [Google Scholar]
- Morquecho-Contreras A, Méndez-Bravo A, Pelagio-Flores R, Raya-González J, Ortíz-Castro R, López-Bucio J. (2010) Characterization of drr1, an alkamide-resistant mutant of Arabidopsis, reveals an important role for small lipid amides in lateral root development and plant senescence. Plant Physiol 152: 1659–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K, MacKerness SA, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V. (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23: 677–685 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, Yaghoubi-Masihi A, Fukao T, Bailey-Serres J. (2010) Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 152: 1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Liu S, Tang D. (2012) HPR1, a component of the THO/TREX complex, plays an important role in disease resistance and senescence in Arabidopsis. Plant J 69: 831–843 [DOI] [PubMed] [Google Scholar]
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al. (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19: 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Inzé D, Goossens A. (2009) Jasmonate-inducible gene: what does it mean? Trends Plant Sci 14: 87–91 [DOI] [PubMed] [Google Scholar]
- Peña-Castro JM, van Zanten M, Lee SC, Patel MR, Voesenek LA, Fukao T, Bailey-Serres J. (2011) Expression of rice SUB1A and SUB1C transcription factors in Arabidopsis uncovers flowering inhibition as a submergence tolerance mechanism. Plant J 67: 434–446 [DOI] [PubMed] [Google Scholar]
- Porra RJ. (2002) The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res 73: 149–156 [DOI] [PubMed] [Google Scholar]
- Pruzinská A, Anders I, Aubry S, Schenk N, Tapernoux-Lüthi E, Müller T, Kräutler B, Hörtensteiner S. (2007) In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19: 369–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Schelbert S, Park SY, Han SH, Lee BD, Andrès CB, Kessler F, Hörtensteiner S, Paek NC. (2012) STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 24: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago LS. (2007) Extending the leaf economics spectrum to decomposition: evidence from a tropical forest. Ecology 88: 1126–1131 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97: 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: 1991–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltmann MA, Stingl NE, Lautenschlaeger JK, Krischke M, Mueller MJ, Berger S. (2010) Differential impact of lipoxygenase 2 and jasmonates on natural and stress-induced senescence in Arabidopsis. Plant Physiol 152: 1940–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Li M, Chen Y, Wu P, Wu G, Jiang H. (2011) Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. J Plant Physiol 168: 1952–1959 [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge UI, Kunze R. (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashisht D, Hesselink A, Pierik R, Ammerlaan JM, Bailey-Serres J, Visser EJ, Pedersen O, van Zanten M, Vreugdenhil D, Jamar DC, et al. (2011) Natural variation of submergence tolerance among Arabidopsis thaliana accessions. New Phytol 190: 299–310 [DOI] [PubMed] [Google Scholar]
- Vogelmann K, Drechsel G, Bergler J, Subert C, Philippar K, Soll J, Engelmann JC, Engelsdorf T, Voll LM, Hoth S. (2012) Early senescence and cell death in Arabidopsis saul1 mutants involves the PAD4-dependent salicylic acid pathway. Plant Physiol 159: 1477–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HJ, Wan AR, Hsu CM, Lee KW, Yu SM, Jauh GY. (2007) Transcriptomic adaptations in rice suspension cells under sucrose starvation. Plant Mol Biol 63: 441–463 [DOI] [PubMed] [Google Scholar]
- Wawrzynska A, Christiansen KM, Lan Y, Rodibaugh NL, Innes RW. (2008) Powdery mildew resistance conferred by loss of the ENHANCED DISEASE RESISTANCE1 protein kinase is suppressed by a missense mutation in KEEP ON GOING, a regulator of abscisic acid signaling. Plant Physiol 148: 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Zacarias L, Reid MS. (1990) Role of growth regulators in the senescence of Arabidopsis thaliana leaves. Physiol Plant 80: 549–554 [Google Scholar]
- Zhou C, Cai Z, Guo Y, Gan S. (2009) An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol 150: 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]