Abstract
Adventitious rooting is an essential but sometimes rate-limiting step in the clonal multiplication of elite tree germplasm, because the ability to form roots declines rapidly with age in mature adult plant tissues. In spite of the importance of adventitious rooting, the mechanism behind this developmental process remains poorly understood. We have described the transcriptional profiles that are associated with the developmental stages of adventitious root formation in the model tree poplar (Populus trichocarpa). Transcriptome analyses indicate a highly specific temporal induction of the AINTEGUMENTA LIKE1 (PtAIL1) transcription factor of the AP2 family during adventitious root formation. Transgenic poplar samples that overexpressed PtAIL1 were able to grow an increased number of adventitious roots, whereas RNA interference mediated the down-expression of PtAIL1 expression, which led to a delay in adventitious root formation. Microarray analysis showed that the expression of 15 genes, including the transcription factors AGAMOUS-Like6 and MYB36, was overexpressed in the stem tissues that generated root primordia in PtAIL1-overexpressing plants, whereas their expression was reduced in the RNA interference lines. These results demonstrate that PtAIL1 is a positive regulator of poplar rooting that acts early in the development of adventitious roots.
The ability to rapidly form numerous adventitious roots provides a selective advantage for plant species that propagate in a vegetative manner. The ability of a plant species to root is also economically important for forest trees, because this capability creates the ability to rapidly amplify millions of cuttings from elite clones for plantations. The Populus genus is a typical example of a woody plant that is propagated by directly planting its stem cuttings in the field (Dickmann, 2006). While representing approximately 30 species that are widely distributed throughout the Northern Hemisphere (Eckenwalder, 1996), poplars are fast-growing plants with high adaptability to marginal soils that make them the plants of choice for use in timber, pulp, and bioenergy-related applications. However, one major limitation to clonal propagation of the elite germplasm of poplar and several other tree species is their rapid loss of or highly reduced ability to form adventitious roots. Therefore, the identification of molecular mechanisms that cause adventitious rooting should create avenues for improving this trait in economically important plants for which clonal propagation is a requirement.
Adventitious roots are distinct from lateral roots in that they form from any tissue that is not a root, such as leaves and stems. Various molecular and genetic approaches have been used to study adventitious root development in Arabidopsis (Arabidopsis thaliana) and other plants (Geiss et al., 2009). For example, Gutierrez et al. (2009) have shown that a balance of AUXIN RESPONSE FACTOR transcripts can control adventitious root initiation in the Arabidopsis hypocotyl. Our knowledge of adventitious rooting in trees is limited. It has been shown that the expression of SCARECROW-like genes is induced during rooting (Sánchez et al., 2007), whereas the expression of a cytokinin type B response regulator (PtRR13) is negatively modulated during the early steps of adventitious root formation in poplar (Populus trichocarpa) stem cuttings (Ramírez-Carvajal et al., 2009). Moreover, the PtRR13-dependent pathway interfered with auxin transport and inhibited the transcription of two APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factor genes. The AP2/ERF proteins belong to a family of transcription factors that are unique to plants (Riechmann and Meyerowitz, 1998), and all of them contain the conserved AP2 DNA-binding domain (Jofuku et al., 1994). This superfamily consists of 145 members in Arabidopsis (Sakuma et al., 2002; Zhuang et al., 2008), 157 in rice (Oryza sativa; Nakano et al., 2006), and 210 in poplar (according to the database of transcriptional factors in poplar (http://planttfdb.cbi.pku.edu.cn/). AtAP2 subfamily members such as PLETHORA1 (AtPLT1) and AtPLT2, BABY BOOM (AtBBM), AINTEGUMENTA (AtANT), and AINTEGUMENTA-like (AtAIL) genes regulate a number of developmental processes (Krizek et al., 2000; Mizukami and Fischer, 2000; Mizukami, 2001; Nole-Wilson et al., 2005; Imin et al., 2007). ANT is part of a pathway that regulates floral organ initiation and growth (Krizek et al., 2000), and it also contributes to the specification of organ polarity by interacting with other genes (Nole-Wilson and Krizek, 2000). AtANT also plays a role in the regulation of shoot development during organ primordium initiation and growth (Elliott et al., 1996) as well as in maintaining cell meristematic competence during shoot organogenesis (Mizukami and Fischer, 2000). A putative function of the AtANT gene in root formation and development has not yet been identified. However, it was recently shown that crown rootless5 is essential for crown root formation in rice. Crown rootless5 encodes a member of the AP2/ERF transcription factor family that shares 50% of its structure with AtANT (Kitomi et al., 2011). PtAIL1 was previously noted as a Populus spp. homolog of the Arabidopsis gene ANT and has been shown to be involved in the modulation of D-type cyclin gene expression (Elliott et al., 1996; Karlberg et al., 2011).
Here, we demonstrate that PtAIL1 is a key endogenous regulator of adventitious rooting in poplar. Global transcript profiling that was performed during poplar adventitious root development showed an increasing level of the PtAIL1 transcription factor transcript during the early stages of adventitious rooting. Transgenic poplar lines that overexpressed PtAIL1 were able to exhibit an increased number of adventitious roots, while RNA interference (RNAi) lines with a reduced level of PtAIL1 transcripts had fewer adventitious roots. Microarray analysis of PtAIL1 overexpressors and RNAi lines from samples that were taken during adventitious rooting led to the identification of potential downstream regulators of the adventitious rooting process. Taken together, our data have identified a key role for PtAIL1 in the control of adventitious rooting in poplars.
RESULTS AND DISCUSSION
Adventitious Root Formation in Poplar Involves Complex Cellular Events
Stem cuttings from poplar (from clone 101-74) were rooted in liquid medium without growth regulators (in basal medium). The first macroscopic evidence of root initiation was the appearance of bulges at the stem surface after 3 to 4 d had passed since their transfer to basal medium (Fig. 1A). Cross sections showed numerous dividing cells that formed root primordia (Fig. 1D). One to 2 d later, the bark split (Fig. 1B), and the organized sequence of cell division and differentiation steps in the primordium led to the establishment of the main root tissues as well as the vascular connections from the incipient roots to the preexisting stem vasculature (Fig. 1, E and G). The adventitious root subsequently grew out and emerged (Fig. 1, C and F). We refer to the dormant cutting as stage 0, the organizing primordium as stage 1 (Fig. 1D), the primordium differentiation as stage 2 (Fig. 1E), and the elongating roots as stage 3 (Fig. 1F). Stage 2 clearly showed that the vascular cells have differentiated from the cambium cells and their immediate derivatives (Fig. 1G), suggesting that these cells could be the initial cells of adventitious roots. Interestingly, it has been shown that adventitious roots arise directly from cambial tissues in easy-to-root species such as poplar (Ginzburg, 1967; Zhou et al., 1992). Contrarily, in difficult-to-root species such as Pinus spp., adventitious rooting initiation starts with a previous callus stage (Rasmussen at al., 2009). The first emerging roots were observed 6 d after the start of the cutting cultures (Fig. 1H). The highest average root number per cutting (10 ± 2 roots per cutting) was obtained after 14 d.
Figure 1.
Adventitious root development in poplar. A to F, Images of the first visible stages with representative cross sections of the stem. In stage 1, intensely dividing cells that form the root primordium are visible (A and D). Stage 2 is characterized by the establishment of the main root tissues as well as the vascular connections of the incipient root to the preexisting stem vasculature (B, E, and G). At stage 3, the outgrowth and emergence of the adventitious roots are visible (C and F). D to F represent harvested samples that were used for transcriptomic analyses. G, Observation at the base of a new adventitious root in stage 2. Arrowheads illustrate the vascular connections. H, Number of adventitious roots formed by poplar dormant cuttings that were transferred to liquid medium. Means and se are indicated (n = 8). I, Rooted dormant stem after 14 d of subirrigation. The frames in A to C indicate the harvest regions for microarrays and real-time PCR analysis. C, Cambium; P, root primordium; Pa, parenchyma tissues; V, vascular connections. Bars = 1 mm in D to F and 100 μm in G.
Adventitious Root Primordium Formation Remodels the Poplar Transcriptome
To identify the molecular processes that are involved in poplar root primordium formation and activation, oligoarray-based transcription profiles of stage 1 and stage 2 were generated from poplar cuttings. The organization of the adventitious root primordium (stage 1) was accompanied by the differential expression of 5,781 genes (fold change > 5, Benjamini and Hochberg-corrected ANOVA P < 0.01; Supplemental Table S1) in comparison with the dormant stage. At stage 2, 6,538 genes were found to be differentially expressed when compared with the dormant cuttings. Between stages 1 and 2, 1,146 genes were overexpressed or repressed, suggesting their possible role in the activation of the primordium and root meristem formation. A total of 7,107 transcript levels corresponding to 13% of the predicted gene models (55,970 gene models predicted) were changed during these early stages of adventitious root development, reflecting a profound cellular and metabolic reorganization during the early stages of root primordia initiation and activation. Among the most highly overexpressed transcripts in stage 1 were genes coding for proteins involved in cell wall remodeling, such as several glycoside hydrolases (GH1, GH3, GH9, GH16, GH17, and GH28), pectate lyases, pectin esterases and expansins, auxin-, gibberellin-, or ethylene-responsive genes, as well as genes that have been implicated in signaling such as the Ser/Thr protein kinases (Supplemental Table S1). Because the surrounding tissue was analyzed together with the root primordium, the cell wall remodeling action could take place in the adjacent zone that would be partially destroyed by the developing root. Given the transcripts that show significant expression changes in stage 1, 289 genes (5%) encoded putative transcription factors that belonged to 35 transcription factor families (http://planttfdb.cbi.pku.edu.cn; Fig. 2). Among these genes, we identified several transcription factors with a clear link to root primordia initiation, including lateral root primordium (lrp1), which is an SRS-type transcription factor and is known to be involved in lateral and adventitious root primordium formation (Smith and Fedoroff, 1995). We also identified members of the GRAS family, such as SCARECROW. The transcriptional factors with significant expression changes during stage 1 mostly belonged to the AP2/ERF, MYB, NAC, WRKY, and bHLH families, with 42, 36, 21, 19, and 19 members, respectively (Fig. 2A). In stage 2, MYB family expression levels changed the most, followed by AP2/ERF transcription factors (Fig. 2A; Supplemental Table S2). However, during the adventitious root formation in poplar, the most highly modulated transcription factor group is the AP2/ERF family (Fig. 2B).
Figure 2.
Differentially expressed transcription factor (TF) families during poplar adventitious root formation. A, Venn diagram illustrating differentially expressed transcription factor families at stages 1 and 2 of adventitious root formation in poplar. The number of regulated transcription factor members is indicated for each family. Genes were considered to be differentially expressed when they met the following criteria: fold change > 5 and P < 0.01. Asterisks indicate Myb and Myb-related transcription factors. B, Number of AP2-ERF, Myb/Myb-related, bHLH, NAC, bZIP, and WRKY transcription factors presented as a percentage of the total number of transcription factors in the genome (dark gray bars) and as a percentage of transcription factors with significant transcript level changes at stage 1 or 2 (light gray bars). At the bottom, the percentage of transcription factors among the genes that are present on the array, as well as the percentage of transcription factors among the genes with changes in transcript levels during adventitious root formation, are given. [See online article for color version of this figure.]
Among these families, several genes, such as PLT, SCARECROW-like6, and PISTILLATA, which are all expressed in the Arabidopsis root quiescent center, have been shown to be implicated in root development (Nawy et al., 2005). SCARECROW genes in the GRAS family are known to be involved in the earliest stages of adventitious root formation. Interestingly, the expression of these genes is induced by exogenous auxin in the rooting-competent cuttings of two distantly related forest species, Pinus radiata and Castanea sativa, during the early stage leading to adventitious root formation (Sánchez et al., 2007). In the WRKY family, one of the genes that is most modulated during primordium formation is an ortholog of WRKY75, which has been found to be involved in root development (Devaiah et al., 2007). The AP2 subfamily is known to be involved in various aspects of plant growth and development (Jofuku et al., 1994; Mizukami and Fischer, 2000; Aida et al., 2004). The AIL members of the AP2 subfamily are expressed in young tissues and may be involved in maintaining the cells in a meristematic and/or division-competent state (Nole-Wilson et al., 2005).
The Induction of PtAIL1 Expression Is Associated with Adventitious Root Primordia Development
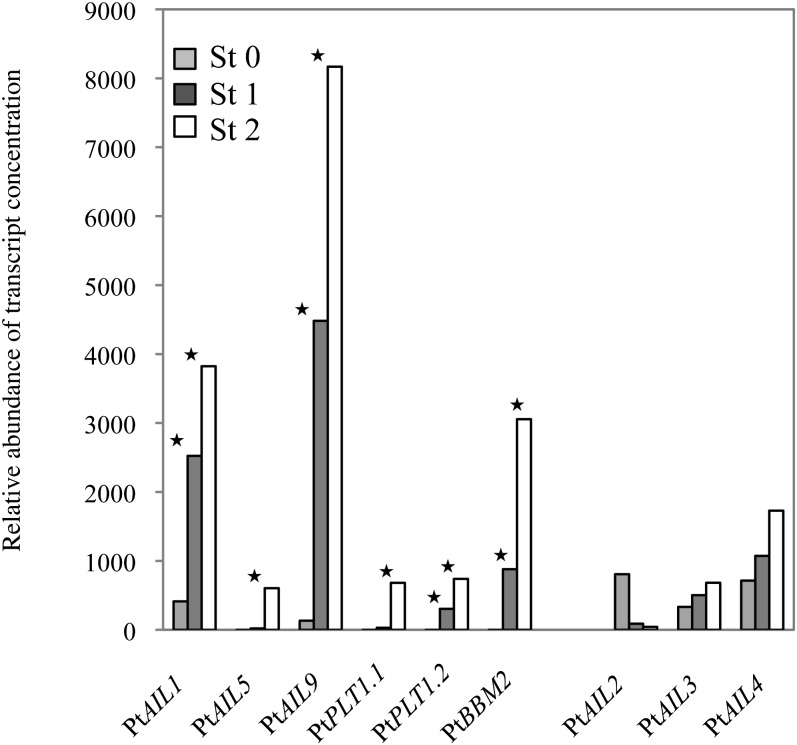
Because genes from the AIL subgroup of the AP2/ERF subfamily (Table I) play a key role in cell division activity (Mizukami and Fischer, 2000; Krizek, 2009) and cell differentiation (Nole-Wilson et al., 2005), we monitored the expression of this subgroup during stages 1 and 2 (Fig. 3) using microarray data as described above. Among the 13 ANT-like genes in poplar (Karlberg et al., 2011; Table I), we found differential abundance in nine of them. Thus, the organization of the root primordium (stage 1) was accompanied by increased levels of PtAIL1, PtAIL9, PtPLT1.2, and PtBBM2 transcripts. The differentiation of the root primordium (stage 2) showed increased levels of PtAIL1, PtAIL5, PtAIL9, PtPLT1.1, PtPLT1.2, and PtBBM2 transcripts. In contrast, the abundance of PtAIL2, PtAIL3, and PtAIL4 transcripts was not significantly modified during these developmental stages (Fig. 3). Quantitative real-time PCR analysis confirmed the above results and showed that at stage 1, the abundance of PtAIL1, PtPLT1.1, and PtAIL9 transcripts was increased by 20-, 30-, and 7-fold on a linear scale, respectively, in comparison with dormant cuttings (Fig. 4). In contrast, PtAIL2 expression was not significantly modified during the two stages that were studied. We also analyzed the relative transcript abundance of PtPLT1.1, PtAIL1, PtAIL2, and PtAIL9 in mature roots (Fig. 4). In contrast to the others, PtAIL1 transcript levels were low in the entire root, indicating a very specific mRNA accumulation during adventitious root primordium activation and formation. PtPLT1.1 expression increased during the organization and differentiation stages in the adventitious root primordia, but it was also predominantly expressed in the root tip (Z1) containing the root apical meristem (Fig. 4). Thus, PtAIL1 became particularly interesting because of its distinct regulatory pattern. These data suggested that PtAIL1 might be involved in the organization and function of adventitious root primordia.
Table I. The AIL gene family in poplar and Arabidopsis.
Gene model identifiers as well as gene names are given. For poplar, the names used in this study are given according to the sequence similarity with Arabidopsis genes. The gene model names of both version 1.1 and Phytozome 8.0 were added.
| Arabidopsis Gene Identifier | Joint Genome Institute P. trichocarpa Version 1.1. Gene Model | Phytozome 8.0 Identifier | Name Used in This Study |
|---|---|---|---|
| At1g51190 (PLT2) and At3g20840 (PLT1) | fgenesh4_pg.C_LG_III001621 | POPTR_0003s20470.1 | PtPLT1.2 |
| gw1.I.7266.1 | POPTR_0001s05580.1 | PtPLT1.1 | |
| At1g72570.1 (AIL1) | fgenesh4_pg.C_LG_I001053 | POPTR_0001s16960.1 | PtAIL5 |
| gw1.III.223.1 | POPTR_0003s06330.1 | PtAIL6 | |
| At4g37750.1 (ANT) | gw1.II.4141.1 | POPTR_0002s11550.1 | PtAIL1 |
| gw1.VII.75.1 | POPTR_0007s14690.1 | PtAIL3 | |
| gw1.XIV.508.1 | POPTR_0014s01260.1 | PtAIL4 | |
| estExt_Genewise1_v1.C_LG_V4387/gw1.V.4418.1 | POPTR_0005s19220.1 | PtAIL2 | |
| At5g57390 (AIL5) | gw1.121.61.1 | POPTR_0006s18330.1 | PtAIL7 |
| gw1.XVIII.2738.1 | POPTR_0018s09900.1 | PtAIL8 | |
| At5g65510.1 (AIL7) | gw1.VII.3975.1 | POPTR_0007s14210.1 | PtAIL9 |
| At5g10510 (AIL6) | |||
| At5g17430 (BBM) | gw1.X.1523.1 | POPTR_0010s18840.1 | PtBBM1 |
| gw1.VIII.931.1 | POPTR_0008s07610.1 | PtBBM2 |
Figure 3.
Expression levels of PtAIL genes at stage 0 (St 0), stage 1 (St 1), and stage 2 (St 2) of poplar adventitious root formation that resulted from microarray data. Asterisks indicate significance in comparison with stage 0 as determined by Benjamini and Hochberg-corrected ANOVA, with P < 0.01. Unlike the group of genes that is indicated on the left, the group of genes indicated on the right is not significantly modulated, including PtAIL2, -3, and -4. PtAIL6, PtAIL7, PtAIL8, and PtBBM1, for which all three probes are at risk of cross hybridizing with other transcripts, are not shown.
Figure 4.
Expression of AIL genes during adventitious root formation and in fully developed poplar roots. Relative expression is shown for PtAIL1, PtAIL2, PtPLT1.1, and PtAIL9 as measured by quantitative reverse transcription-PCR at stage 1 (St 1) and stage 2 (St 2) of adventitious root formation, in the mature root (R), and in two different zones of the mature root, root cap and meristem zone (Z1) and elongation and maturation zones (Z2). Expression was normalized to the reference genes and scaled to stage 0 expression for each gene. We used elongation factor 1B and the putative protein A as reference genes, because their expression was not significantly modulated according to the microarray data. Averages and se values are derived from three biological replicates. Student’s t test and Fisher’s test were performed by comparing all results with stage 0 (left part of each graph) or between Z1 and Z2 (right part). Asterisks indicate significance at P < 0.05.
Interestingly, Arabidopsis orthologs of the same genes were expressed in the roots (Nole-Wilson et al., 2005) and were also necessary for the formation of root stem cell niches (Imin et al., 2007). AtPLT1, AtPLT2, AtAIL6, and AtBBM are required for root development (Aida et al., 2004; Galinha et al., 2007). Specifically, AtPLT1 and AtPLT2 are involved in maintaining the stem cell specification of the root meristem. Following the high expression of PtPLT1.1 in the root tip, we could hypothesize a similar role in poplar for PtPLT1.1.
The Arabidopsis ortholog of PtAIL1 (AtANT) is known to promote growth within floral meristems and plays a role in organ primordium initiation throughout the shoot’s development (Elliott et al., 1996; Mizukami and Fischer, 2000). Furthermore, OsCR5 shares 50% homology with PtANT and is involved in rice crown root initiation (Kitomi et al., 2011). The expression of PtAIL1 is consistent with prior results in Arabidopsis and rice. This transcription factor is expressed at the early stages in developing organs, and our transcriptional analysis strongly suggests a putative regulatory role for PtAIL1 in poplar adventitious root formation.
PtAIL1 Is a Positive Regulator of Adventitious Root Formation
The highly specific expression of PtAIL1 during adventitious root development prompted us to study its function in this process. Therefore, we analyzed the rooting capacity of transgenic Populus spp. lines in which the transcript level of PtAIL1 was perturbed. We first analyzed the ability of transgenic hybrid aspen (Populus tremula × Populus tremuloides; clone T89) lines to form adventitious roots that constitutively overexpressed PtAIL1 (35S:AIL1) or that had reduced PtAIL1 expression due to the expression of the RNAi construct (Karlberg et al., 2011). Second, the P. tremula × Populus alba hybrid (clone 717-1B4) was transformed with the 35S:AIL1 construct (Supplemental Fig. S1). As shown in Figure 5, A and B, the transformed lines that overexpressed PtAIL1 showed an earlier root emergence in comparison with the wild type as well as a significant increase in the average root number per cutting (Fig. 5, C–F). In contrast, the RNAi line showed a delay in adventitious root formation and a significant decrease in the number of total roots per cutting (Fig. 5, A, C, and E). These findings suggest that PtAIL1 could be a positive regulator of adventitious root formation in several genotypes of Populus spp.
Figure 5.
Transgenic manipulation of AIL1 affects adventitious root development. A, Percentage of rooted cuttings of P. tremula × P. tremuloides T89 (wild type), 35S:AIL1, and RNAi PtAIL1 lines. B, Percentage of rooted cuttings of P. tremula × P. alba 717-1B4 (wild type) and 35S:AIL transgenic lines (K4-5, K4-8, and K4-13). C, Number of adventitious roots per cutting in P. tremula × P. tremuloides T89 cuttings (wild type), 35S:AIL1, and RNAi line. D, Number of adventitious roots per cutting in P. tremula × P. alba 717-1B4 (wild type) and 35S:AIL transgenic lines (K4-5, K4-8, and K4-13). The curves show means and se. Student’s t and Fisher tests (P < 0.05) were used for analysis. Single asterisks indicate the day on which the differences became statistically significant in transgenic lines compared with wild-type plants, and double asterisks indicate a significant difference in the 35S:AIL1 K4-13 line compared with the 35S:AIL1 K4-8 line. E, Representative images of the adventitious root systems of T89 (wild type), 35S:AIL1, and RNAi line after 6 d of transfer to rooting induction medium. Bars = 1 cm. F, Representative poplar stem cross sections from 717-1B4 (wild type) and 35S:AIL1 (K4-8). Bars = 100 μm.
It was previously demonstrated that PtAIL1/PtANT1 is expressed in the cambial zone within cells that undergo intensive cell proliferation (Schrader et al., 2004). Recent findings in hybrid aspen trees and Arabidopsis highlight that PtAIL1 has been shown to bind to the D-type cyclin promoter (Karlberg et al., 2011). Moreover, the overexpression of ANT (an ortholog of PtAIL1) inside fully differentiated organs results in neoplastic activity and the production of calli and adventitious roots and shoots. These data suggest that ANT could be involved in organ growth and cell proliferation, indicating its role in the control of cell division activity (Mizukami and Fischer, 2000).
A large majority of these data suggest that PtAIL1 is most likely a positive cell proliferation regulator. Its positive effect on adventitious rooting is mediated by either promoting the reentry of differentiated cells into the cell cycle and/or by increasing the rate of initiation and differentiation of root primordia from undifferentiated cells such as those that can be found in the cambial zone and in areas where PtANT1 was previously expressed (Schrader et al., 2004). These data are in accordance with our anatomical observation, suggesting that cambial cells could be the initials of adventitious root development.
PtAIL1 Expression Modulates Target Gene Expression
To gain more mechanistic insights into the role of PtAIL1 in adventitious rooting, we compared global gene expression profiles between the wild type, PtAIL1 overexpressors, and RNAi suppression lines. Line 3B (for 35S:AIL1) and line 255-6 of RNAi PtAIL1 were chosen because they showed the strongest rooting phenotypes in P. tremula × P. tremuloides (T89; data not shown). Total RNA was isolated from 2-mm sections of the basal stem region at 24 and 72 h after stem excision and transfer to the medium. One hundred twenty-six genes were found to be differentially expressed at 24 h between at least two lines (fold change > 5, Benjamini and Hochberg-corrected ANOVA P < 0.01; Supplemental Table S3). This set of 126 transcripts was then searched for either overexpression in 35S:AIL1 transgenic plants compared with T89 and less expressed in RNAi PtAIL1 transgenic plants compared with T89 or vice versa (fold change > 5, Benjamini and Hochberg-corrected Student’s t test P < 0.05). A total of 15 genes were selected for their ability to maintain this antagonistic expression pattern at 72 h after excision (Table II). These included genes that code for phosphorylases, a Gly-rich protein, a cytochrome P450 CYP86B1 lipase with a GDSL-like motif, a tandem zinc knuckle/PLU3 domain-encoding gene, as well as transcription factors such as MYB36, the AGAMOUS-like AGL16, and MINI ZINC FINGER MIF2 (Table II). In comparing trends in expression among the three genotypes, the majority (10) showed an increase of expression in the overexpressed genotypes and down-expression in the suppressed transgenic lines (Table II). The preponderance of up-modulated transcripts is consistent with the presumed transcriptional activation function of AtANT (Krizek and Sulli, 2006) and suggests that the direct target(s) are likely among these genes.
Table II. List of genes that showed opposite expression patterns between 35S:AIL1 and RNAi PtAIL1 transgenic plants.
Genes with increased transcript abundance in PtAIL1-overexpressing plantlets and, in contrast, reduced transcript level in the PtAIL1 RNAi lines were compared with the wild-type P. tremula × P. tremuloides T89 line or vice versa. Data were searched for transcripts with more than 5-fold changes at 24 h between T89 and 35S:AIL1 and RNAi PtAIL1 transgenic plants using Benjamini and Hochberg-corrected ANOVA P < 0.01. In addition, pairwise comparisons using Student’s t tests (Benjamini and Hochberg-corrected Student’s t test P < 0.01) were applied to filter out 126 transcripts that were significantly modulated in a minimum of one of the mutants compared with T89. The 126 remaining transcripts were then searched to detect either overexpression in 35S:AIL1 transgenic plants compared with T89 and down-expression in RNAi PtAIL1 transgenic plants compared with T89 or vice versa (fold change > 5, Benjamini and Hochberg-corrected Student’s t test P < 0.05). Transcripts that maintained the antagonist expression pattern at 72 h after excision were selected.
| Arabidopsis Defline | Fold Change in Expression |
|||||
|---|---|---|---|---|---|---|
| 24 h of Cutting Excision |
72 h of Cutting Excision |
|||||
| 35S:AIL1 versus the Wild Type | RNAi PtAIL1 versus the Wild Type | 35S:AIL1 versus RNAi PtAIL1 | 35S:AIL1 versus the Wild Type | RNAi PtAIL1 versus the Wild Type | 35S:AIL1 versus RNAi PtAIL1 | |
| MYB36 | 105.250 | 0.256 | 411.353 | 25.343 | 1.158 | 21.880 |
| Gly-rich protein | 77.363 | 0.392 | 197.483 | 5.338 | 1.418 | 3.765 |
| Phosphorylase family protein | 12.206 | 0.883 | 13.830 | 2.936 | 0.003 | 1131.083 |
| Phosphorylase family protein | 7.548 | 0.618 | 12.211 | 3.163 | 0.187 | 16.886 |
| Unknown protein | 6.620 | 0.842 | 7.861 | 8.568 | 1.789 | 4.790 |
| AGAMOUS-LIKE16 | 5.770 | 0.479 | 12.051 | 7.111 | 0.972 | 7.318 |
| Unknown protein | 5.227 | 0.774 | 6.754 | 3.194 | 0.929 | 3.436 |
| Unknown protein | 3.180 | 0.053 | 60.345 | 18.249 | 0.082 | 221.604 |
| Unknown protein | 2.422 | 0.055 | 44.198 | 12.607 | 0.118 | 107.008 |
| Unknown protein | 2.398 | 0.007 | 339.816 | 59.817 | 1.000 | 59.817 |
| Cytochrome P450 CYP86B1 | 1.000 | 102.286 | 0.010 | 0.164 | 8.183 | 0.020 |
| GDSL-motif lipase | 0.129 | 1.762 | 0.073 | 0.633 | 4.128 | 0.153 |
| GDSL-motif lipase | 0.101 | 1.759 | 0.058 | 0.572 | 4.109 | 0.139 |
| MINI ZINC FINGER2 | 0.082 | 1.408 | 0.058 | 0.184 | 1.993 | 0.092 |
| Tandem zinc knuckle/PLU3 domain (TZP) | 0.011 | 1.609 | 0.007 | 0.187 | 3.478 | 0.054 |
We further examined transcript patterns for the above 15 genes during the three stages of adventitious root formation in poplar (Supplemental Table S1; Table III). The majority of these genes were found to be differentially expressed (Table III). Seven differentially modulated genes showed a positive and increasing relation of their expression pattern to that of PtAIL1, further supporting an activator function for the PtAIL1-encoded protein. Among the seven genes that showed positive relationships with PtAIL1 expression, more than one-half (four) were of unknown function, suggesting a significant lack of mechanistic knowledge on how PtAIL1 regulates adventitious root formation and possible other processes in plants. Two of the genes that encoded transcription factors for the AGL and MYB families have been linked to the regulation of root formation. Previous studies have revealed that AGL16 is expressed at relatively high levels in the root quiescent center in Arabidopsis (Nawy et al., 2005). Interestingly, it was reported that AGAMOUS is a possible target of AtANT (Krizek et al., 2000; Nole-Wilson and Krizek, 2000). A number of different MYBs are also almost exclusively expressed in the root quiescent center of Arabidopsis (Nawy et al., 2005). Müller et al. (2006) have shown that AtMYB36 was almost exclusively found in the root.
Table III. Transcript levels during adventitious root formation in poplar of candidate genes showing an opposite pattern of expression between 35S:AIL1 and RNAi PtAIL1 transgenic plants.
The transcript levels of these genes are indicated at stages 0, 1, and 2 of adventitious root formation in poplar. Data were obtained from microarray analyses. Expression values in italics indicate significance at P < 0.05, those in boldface indicate significance at P < 0.01 for the Benjamini and Hochberg-corrected ANOVA P, and underlined values are significantly different from stage 0 using Tukey’s honest significant difference pairwise post hoc tests between stages with P < 0.05.
| Joint Genome Institute Version 1.1 | Phytozome Version 8.0 | Name | Stage 0 | Stage 1 | Stage 2 |
|---|---|---|---|---|---|
| gw1.II.4141.1 | POPTR_0002s11550.1 | PtAIL1 | 413 | 2,524 | 3,824 |
| gw1.XVIII.2879.1 | POPTR_0018s10390.1 | PtMYB36 | 1 | 32 | 608 |
| fgenesh4_pg.C_LG_IV000635 | POPTR_0004s09190.1 | Unknown protein | 1 | 2,951 | 3,554 |
| fgenesh4_pg.C_LG_II001012 | POPTR_0002s11030.1 | PtAGL16 | 24 | 140 | 1,514 |
| estExt_fgenesh4_pg.C_LG_XVI1239 | POPTR_0016s14020.1 | Unknown protein | 1 | 1,793 | 5,609 |
| eugene3.00640195 | POPTR_0017s04950.1 | Unknown protein | 69 | 745 | 24,781 |
| eugene3.00640196 | POPTR_1962s00200.1 | Unknown protein | 1 | 848 | 29,904 |
| fgenesh4_pg.C_LG_I000707 | POPTR_0001s11670.1 | Unknown protein | 1 | 1 | 97 |
| gw1.V.3321.1 | POPTR_0005s09500.1 | PtCYP86B1 cytochrome P450 | 1 | 1 | 1 |
| estExt_Genewise1_v1.C_LG_XVI3628 | POPTR_0016s12310.1 | PtGDSL-motif lipase | 3,433 | 17,561 | 17,670 |
| estExt_Genewise1_v1.C_LG_XVII1072 | POPTR_0017s11900.1 | PtMIF2 (MINI ZINC FINGER2) | 3,279 | 5747 | 3,544 |
| fgenesh4_pg.C_LG_VIII001446 | POPTR_0008s16200.1 | PtTZP (Tandem zinc knuckle/PLU3 domain) | 2,283 | 146 | 81 |
| genesh4_pg.C_LG_VIII001448 | POPTR_0008s16200.1 | 2,769 | 164 | 90 |
Together, these data suggest that PtAIL1, PtAGL16, and PtMYB36 could be part of a regulatory network that controls adventitious root formation in poplars.
CONCLUSION
A global transcriptional analysis that was conducted during the organization and differentiation stages of adventitious root formation in poplar clearly highlighted the significant modulation of AP2/ERF transcription factors. Our study shows that several members of the ANT-AIL group, including PtAIL1, PtPLT1.1, PtAIL9, and PtBBM, are highly expressed in stage 1 and even more so in stage 2 of adventitious root development. Although several members of this subfamily may be of particular interest in relation to adventitious root formation, PtAIL1 became a strong candidate because of its distinct expression pattern. Functional analysis clearly indicated that perturbing PtAIL1 expression affected adventitious rooting. Therefore, the sum of our studies revealed that PtAIL1 is a positive regulator of adventitious rooting that acts by promoting the formation of root primordia.
MATERIALS AND METHODS
Plant Material and Growth Conditions
To induce rooting within a subirrigation system, the dormant hardwood stems from poplar (Populus trichocarpa; clone 101-74) were collected from stool beds in January 2010. Dormant stems were then cut into 25-cm cuttings in which each had six to seven nodes. These cuttings were sealed in polyethylene bags and kept in cold storage (0°C) until they were removed for rooting. The experiments in which these cuttings were used were then performed during the following year. Selected cuttings had a range of diameters from 0.8 to 1.1 cm. During rooting and growth, cuttings were subirrigated using a hydroponic culture system that has been described previously (Merret et al., 2010). In vitro rooting experiments were performed with the wild type and transgenic lines of Populus tremula × Populus alba Institut National de la Recherche Agronomique 717-1B4 and of P. tremula × Populus tremuloides Umeä Plant Science Center clone T89 as described previously (Felten et al., 2009; Karlberg et al., 2011). The number of roots per rooted cutting or micropropagated plantlet was estimated throughout the rooting period. For adventitious root quantification, eight to 40 individual plants of poplar, P. tremula × P. alba, and/or P. tremula × P. tremuloides were observed for each set of experimental conditions on a daily basis. Adventitious roots were counted, and images were taken using a Canon camera (EOS 350 digital) and a Discovery V.8 stereomicroscope (Zeiss). For the poplar transformation, we used the Agrobacterium tumefaciens-mediated transformation procedure as described previously (Han et al., 2000; Karlberg et al., 2011).
Histological Analysis
Samples were fixed for 4 h at 4°C in 4% (w/v) paraformaldehyde in 0.1 m phosphate-buffered saline (pH 7). After washing them in 0.1 m phosphate-buffered saline buffer, tissue samples were sequentially dehydrated in a graded series of ethanol (45 min each on ice at 75%, 80%, and 90% [v/v] and three times in absolute ethanol), followed by an ethanol:Otix+ (Microm Microtech) series (45 min each on ice at rates of 75:25, 50:50, and 25:75 [v/v] and three times in 100% Otix+). After Otix+ removal, samples were embedded in Diawax (Microm Microtech), sectioned into 7-µm-thick sections using a rotary microtome (Microm Microtech), and then stained with toluidine blue.
RNA Extraction and Complementary DNA Synthesis
For microarray experiments performed with poplar, a region that included the developing adventitious root at different stages and the surrounding stem tissues was harvested and immediately frozen in liquid nitrogen as indicated in Figure 1, A to C. Time-zero samples were excised from a comparable stem region of dormant cuttings at the moment of submersion. We used the same regions for quantitative reverse transcription-PCR analyses as for the microarrays along with one additional stage, the mature adventitious root (the first 1.5 cm including the root tip). Two parts of mature adventitious root were also analyzed: the apical zone (the first 5 mm) was called Z1 and corresponded to the root cap and meristem zone, and the basal zone (the 2 cm behind zone 1) was called Z2 and corresponded to the elongation and maturation zones.
At each time point for the microarray and quantitative reverse transcription-PCR analyses, three pools of approximately 50 mg of material were harvested from several plants, frozen in liquid nitrogen, and kept at −80°C until RNA extraction. Samples from three cuttings were harvested for each root tip sample. For experiments that were performed with hybrids, P. tremula × P. tremuloides (T89) and P. tremula × P. alba (Institut National de la Recherche Agronomique 717-1B4), and their transgenic plants, 2 mm of the stem from the base of four excised shoots were collected.
Total RNA was extracted from poplar samples (except zone Z1) using the RNeasy Plant Mini Kit (Qiagen) following the manufacturer’s instructions. An in-column digestion step with DNAse I (Qiagen) was included in the extraction. For the zone Z1 and T89 samples (with harvested material less than 50 mg), we used the RNeasy Micro Kit (Qiagen), which was first preceded by a cleanup step with QIAshredder Mini Spin Columns (Qiagen). An in-column digestion step with DNAse I (Qiagen) was also part of the extraction. RNA quality and integrity were checked prior to complementary DNA (cDNA) synthesis using Experion StdSens or HighSens capillary gels (Bio-Rad). The cDNAs that were needed for the NimbleGen microarrays were synthesized using the SMART or SMARTer PCR cDNA Synthesis Kit (Clontech) according to the manufacturer’s instructions. The cDNA that was needed for real-time PCR was synthesized from RNA using the iScript Kit (Bio-Rad).
Quantitative Real-Time PCR
All primer sequences in this study are noted in Supplemental Table S4. We used elongation factor 1B (Brunner et al., 2004) and putative protein A (Gutierrez et al., 2008) as reference genes (Supplemental Table S4) because their expression was not significantly modified according to the microarray data. To design specific primer pairs for selected genes, we used QuantPrime (Arvidsson et al., 2008) and Primer3 software. We designed primers using the annotations from the Joint Genome Institute Populus trichocarpa version 1.1 data bank and double checked them with Phytozome version 5.0 (http://www.phytozome.net/). Quantitative real-time PCR was performed using SYBR Green Supermix (Bio-Rad) following the manufacturer’s instructions, a Chromo4 Light Cycler, and OpticonMonitor software (Bio-Rad). These analyses were performed in three biological replicates and were independent from microarray samples, with each two technical replicates using 2.5 ng of cDNA. The cycle number at which the fluorescence crosses the threshold is called Ct value. The relative gene expression for each stage was based on ΔCt calculations using the mean of the two reference gene expressions, according to Pfaffl (2001). The ΔCt values were scaled to the average of the stage 0 expression for the gene of interest and transformed to log2 afterward. The means of the three expression values are presented as histograms with se values. Student’s t test associated with a Fisher test was performed.
NimbleGen Microarray Transcript Profiling
During this study, the genome version of poplar changed from genome assembly version 1.1 to version 2. Therefore, two whole-genome expression arrays that were manufactured by NimbleGen Systems were used for the experiments, one based on first poplar assembly, GPL2699, and a second that was designed in 2010 and based on version 2 and Phytozome 5.0 of the poplar genome, which is called GPL13485. GPL2699 contained three independent, nonidentical, 60-mer probes in duplicates for 55,970 gene models that were predicted for the poplar genome sequence version 1.0, while GPL13485 contained three independent, nonidentical, 60-mer probes from 43,929 annotated gene models of the Populus trichocarpa genome version 2 (Phytozome 5.0).
Transcriptome analyses were performed on the first visible step of adventitious root formation in poplar (GPL2699) and on the stem bases (zone of emergence) at two culture times (24 and 72 h) for the P. tremula × P. tremuloides wild type and transgenic hybrids (GPL13485).
Single-dye labeling of samples, the hybridization procedures, and data acquisition were all performed at the NimbleGen facilities in Reykjavik, Iceland, following their standard protocol. Three biological replicates were performed for each condition. Microarray probe intensities were normalized across chips using ArrayStar software (DNASTAR). Natural log-transformed data were calculated and subjected to the CyberT statistical framework (http://cybert.ics.uci.edu/; Baldi and Long, 2001) using the one-way ANOVA multiple condition data module and the data module with the standard Student’s t test unpaired two conditions. ANOVA was followed by Tukey’s honestly significant difference pairwise post hoc tests. Benjamini and Hochberg multiple-hypothesis testing corrections with false discovery rate were used for both the ANOVA and the Student’s t test. Transcripts with a q value (Bayesian posterior P value; Storey, 2003) of less than 0.01 for the Benjamini and Hochberg test were considered to be significantly differentially expressed within the two sets of microarray analyses (http://cybert.ics.uci.edu/). Randomly designed probes were used on the array to estimate unspecific hybridization. Three times the mean intensity of the random probes was subtracted from each value, and the negative values were set to 1. In addition, we filtered our data for very-low-concentration transcripts to avoid artifacts. We only kept transcripts with an expression level of 300 or greater in at least one condition (Supplemental Tables S1–S3). The poplar genome contains a large number of duplicated genes or gene families. In some cases, it is impossible to define array probes that do not hybridize with at least one more transcript than the transcript for which they were designed. Therefore, we added a column in Supplemental Tables S1 to S3 to indicate the genes in which all three probes are at risk of cross hybridizing with other transcripts. In both data sets, we focused on genes with a fold change ≥ 5 in transcript level.
The two complete transcriptome data sets are available as a series (GSE34096 and GSE34162) at the Gene Expression Omnibus at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcript level of AIL1 in P. tremula × P. alba (717-1B4) and in the 35S:AIL1 transgenic lines determined by semi-quantitative RT-PCR from apices.
Supplemental Table S1. List of genes where transcript level changed during adventitious root formation in P. trichocarpa. The first data set includes all genes significantly regulated (Benjamini and Hochberg-corrected ANOVA P value < 0.01). The second set includes regulated genes with fold change > 5 (Benjamini and Hochberg-corrected ANOVA P value < 0.01).
Supplemental Table S2. List of AP2/ERF (first data set) and MYB (second data set) genes with changes in transcript level during adventitious root formation in P. trichocarpa.
Supplemental Table S3. List of 126 genes with changes in transcript level in transgenic plants at 24 h compared with P. tremula × P. tremuloides T89 (wild type) and between the two transgenic plants at 24 h.
Supplemental Table S4. Primer sequences used for qPCR analysis.
Acknowledgments
We thank P. Vion, J.P. Lecler, C. Delaruelle, and E. Morin for technical assistance, F. Elegbede, I. Hummel, and C. Fourrey for helpful discussions, and C. Bastien (Institut National de la Recherche Agronomique-Orléans) for providing the cuttings.
Glossary
- RNAi
RNA interference
- cDNA
complementary DNA
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B. (2008) QuantPrime: a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P, Long AD. (2001) A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509–519 [DOI] [PubMed] [Google Scholar]
- Brunner AM, Yakovlev IA, Strauss SH. (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG. (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143: 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickmann DI. (2006) Silviculture and biology of short-rotation woody crops in temperate regions: then and now. Biomass Bioenergy 30: 696–705 [Google Scholar]
- Eckenwalder JE. (1996) Systematics and evolution of Populus. In RF Stettler, HD Bradshaw Jr, PE Heilman, TM Hinckley, eds, Biology of Populus and Its Implications for Management and Conservation. NRC Research Press, National Research Council of Canada, Ottawa, pp 7–32
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR. (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felten J, Kohler A, Morin E, Bhalerao RP, Palme K, Martin F, Ditengou FA, Legué V. (2009) The ectomycorrhizal fungus Laccaria bicolor stimulates lateral root formation in poplar and Arabidopsis through auxin transport and signaling. Plant Physiol 151: 1991–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Geiss G, Gutierrez L, Bellini C. (2009) Adventitious root formation: new insights and perspectives. In T Beeckman, ed, Annual Plant Reviews: Root Development, Vol 37. Wiley-Blackwell, Oxford, pp 127–156
- Ginzburg C. (1967) Organization of the adventitious root apex in Tamarix aphylla. Am J Bot 54: 4–8 [Google Scholar]
- Gutierrez L, Bussell JD, Pacurar DI, Schwambach J, Pacurar M, Bellini C. (2009) Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, et al. (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6: 609–618 [DOI] [PubMed] [Google Scholar]
- Han KH, Meilan R, Ma C, Strauss SH. (2000) An Agrobacterium tumefaciens transformation protocol effective on a variety of cottonwood hybrids (genus Populus). Plant Cell Rep 19: 315–320 [DOI] [PubMed] [Google Scholar]
- Imin N, Nizamidin M, Wu T, Rolfe BG. (2007) Factors involved in root formation in Medicago truncatula. J Exp Bot 58: 439–451 [DOI] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlberg A, Bako L, Bhalerao RP. (2011) Short day-mediated cessation of growth requires the downregulation of AINTEGUMENTALIKE1 transcription factor in hybrid aspen. PLoS Genet 7: e1002361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitomi Y, Ito H, Hobo T, Aya K, Kitano H, Inukai Y. (2011) The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J 67: 472–484 [DOI] [PubMed] [Google Scholar]
- Krizek BA. (2009) Making bigger plants: key regulators of final organ size. Curr Opin Plant Biol 12: 17–22 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Prost V, Macias A. (2000) AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. Plant Cell 12: 1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Sulli C. (2006) Mapping sequences required for nuclear localization and the transcriptional activation function of the Arabidopsis protein AINTEGUMENTA. Planta 224: 612–621 [DOI] [PubMed] [Google Scholar]
- Merret R, Moulia B, Hummel I, Cohen D, Dreyer E, Bogeat-Triboulot MB. (2010) Monitoring the regulation of gene expression in a growing organ using a fluid mechanics formalism. BMC Biol 8: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y. (2001) A matter of size: developmental control of organ size in plants. Curr Opin Plant Biol 4: 533–539 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D, Schmitz G, Theres K. (2006) Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. (2005) Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17: 1908–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S, Krizek BA. (2000) DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Res 28: 4076–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S, Tranby TL, Krizek BA. (2005) AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol Biol 57: 613–628 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: 2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Carvajal GA, Morse AM, Dervinis C, Davis JM. (2009) The cytokinin type-B response regulator PtRR13 is a negative regulator of adventitious root development in Populus. Plant Physiol 150: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A, Smith TE, Hunt MA. (2009) Cellular stage of root formation, root system quality and survival of Pinus elliottii var. elliottii × P. caribaea var. hondurensis cuttings in different temperature environments. New For 38: 285–294 [Google Scholar]
- Riechmann JL, Meyerowitz EM. (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379: 633–646 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Sánchez C, Vielba JM, Ferro E, Covelo G, Solé A, Abarca D, de Mier BS, Díaz-Sala C. (2007) Two SCARECROW-LIKE genes are induced in response to exogenous auxin in rooting-competent cuttings of distantly related forest species. Tree Physiol 27: 1459–1470 [DOI] [PubMed] [Google Scholar]
- Schrader J, Nilsson J, Mellerowicz E, Berglund A, Nilsson P, Hertzberg M, Sandberg G. (2004) A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16: 2278–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Fedoroff NV. (1995) LRP1, a gene expressed in lateral and adventitious root primordia of Arabidopsis. Plant Cell 7: 735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. (2003) The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Stat 31: 2013–2035 [Google Scholar]
- Zhou J, Wu H, Collet GF. (1992) Histological study of initiation and development in vitro of adventitious roots in mini cutting of apple rootstocks of M26 and EMLA 9. Physiol Plant 84: 433–440 [Google Scholar]
- Zhuang J, Cai B, Peng RH, Zhu B, Jin XF, Xue Y, Gao F, Fu XY, Tian YS, Zhao W, et al. (2008) Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem Biophys Res Commun 371: 468–474 [DOI] [PubMed] [Google Scholar]