Abstract
Dehydration inhibits petal expansion resulting in abnormal flower opening and results in quality loss during the marketing of cut flowers. We constructed a suppression subtractive hybridization library from rose (Rosa hybrida) flowers containing 3,513 unique expressed sequence tags and analyzed their expression profiles during cycles of dehydration. We found that 54 genes were up-regulated by the first dehydration, restored or even down-regulated by rehydration, and once again up-regulated by the second dehydration. Among them, we identified a putative NAC family transcription factor (RhNAC2). With transactivation activity of its carboxyl-terminal domain in yeast (Saccharomyces cerevisiae) cell and Arabidopsis (Arabidopsis thaliana) protoplast, RhNAC2 belongs to the NAC transcription factor clade related to plant development in Arabidopsis. A putative expansin gene named RhEXPA4 was also dramatically up-regulated by dehydration. Silencing RhNAC2 or RhEXPA4 in rose petals by virus-induced gene silencing significantly decreased the recovery of intact petals and petal discs during rehydration. Overexpression of RhNAC2 or RhEXPA4 in Arabidopsis conferred strong drought tolerance in the transgenic plants. RhEXPA4 expression was repressed in RhNAC2-silenced rose petals, and the amino-terminal binding domain of RhNAC2 bound to the RhEXPA4 promoter. Twenty cell wall-related genes, including seven expansin family members, were up-regulated in Arabidopsis plants overexpressing RhNAC2. These data indicate that RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals and that RhEXPA4 expression may be regulated by RhNAC2.
Drought or dehydration results in water deficit stress (WDS), one of the most important abiotic stresses to which plants are subjected. WDS can cause numerous morphological and developmental changes in plants, including reduced longevity, growth inhibition, enhancement of root growth, and early flowering (Xiong and Zhu, 2002). Roses (Rosa spp.) are the most important ornamental plants worldwide, and cut roses account for about 21% and 31% of all cut flowers traded in China and at the European auctions, respectively (Heinrichs, 2008). Commercially, roses are usually harvested at the open bud stage and are extremely susceptible to WDS during postharvest handling; WDS causes abnormal flower opening, flower wilting, wilting of the pedicel (bent neck), and failure to open (Jin et al., 2006; Xue et al., 2008). The effect of dehydration on flower opening and senescence in cut roses, therefore, has been studied at the physiological and biochemical levels for several decades (Kumar et al., 2008).
The effect of WDS on the transcriptome has been studied, using microarray technologies, in different plant tissues and species, including seedlings, leaves, and roots of Arabidopsis (Arabidopsis thaliana; Seki et al., 2002), seedlings, leaves, roots, and panicles of rice (Oryza sativa; Rabbani et al., 2003; Wang et al., 2011), leaves and roots of poplar (Populus spp.; Cohen et al., 2010), seedlings and leaves of barley (Hordeum vulgare; Talamè et al., 2007; Tommasini et al., 2008), and leaves of tomato (Solanum lycopersicum) and potato (Solanum tuberosum; Schafleitner et al., 2007; Gong et al., 2010). The number of ESTs analyzed ranges from 1,000 in barley (Talamè et al., 2007) to tens of thousands in Arabidopsis and rice (Kawaguchi et al., 2004; Zhou et al., 2007). To date, several hundred genes have been reported to respond to drought stress and are classified into two major groups. The first group comprises regulatory proteins, including transcription factors and protein kinases (Seki et al., 2002; Shinozaki and Yamaguchi-Shinozaki, 2007). The second group comprises functional proteins, which are downstream effectors in the stress response pathway. WDS-induced transcriptomes differ significantly depending on species, developmental stage, and organs and tissues analyzed, but they also show strong overlapping of gene function, including many orthologs.
Transcription factors are important regulatory proteins controlling the differential expression of physiological and biochemical processes during plant growth and development. They function by binding to cis-elements in the promoters or enhancers of their target genes. There are about 1,600 genes encoding for independent transcription factors in the genome of Arabidopsis (Riechmann et al., 2000; Melzer and Theissen, 2011). Based on structural features or conserved domains, they are grouped into families, such as basic leucine zipper, WRKY, MYB, APETALA2/ethylene responsive element binding protein (AP2/EREBP), NAM, ATAF, and CUC (NAC), zinc finger, MADS, and heat shock factor family proteins (Zhou et al., 2007; Nakashima et al., 2009; Qiu and Yu, 2009; Seo et al., 2009; Jeong et al., 2010). Many transcription factor families have been implicated in the regulation of WDS responses in plants (Hussain et al., 2011).
Gene Ontology (GO) enrichment analysis has been a useful tool for identifying biological processes that may be involved in the response to WDS. An overview of functional groups involved in WDS responses has been obtained from several plant species, including Arabidopsis (Wilkins et al., 2010), poplar (Street et al., 2006; Cohen et al., 2010), and chickpea (Cicer arietinum; Molina et al., 2008). Biological functions and metabolic processes that are indicated by the GO analysis vary, depending on the organ or plant species exposed to WDS. To date, studies of changes in gene expression profiles during WDS have mostly focused on vegetative organs, including seedlings, leaves, and roots, of model and crop plants. Less information is available on floral organs, apart from a global transcriptomics study of the response to WDS in Alstroemeria aurantiaca (Wagstaff et al., 2010) and a suppression subtractive hybridization (SSH) study of desiccation in lily (Lilium spp.) anthers (Hsu et al., 2007). No such study has been conducted on roses, a multipetaled and dicotyledonous model flower that is also the most important commercial cut flower.
Plant-specific NAC (for NAM, ATAF, and CUC) transcription factors are characterized by highly conserved DNA-binding NAC domains located in the N-terminal region and a variable C terminus (Ooka et al., 2003; Jeong et al., 2010). Arabidopsis and rice contain approximately 117 and 151 NAC members, respectively (Nuruzzaman et al., 2010). There are at least five different NAC subgroups based on their target DNA-binding sites in Arabidopsis (Le et al., 2011). NAC transcription factors play important roles in regulating diverse biological processes, including development, senescence, growth, cell division, and responses to environmental stress stimuli (Olsen et al., 2005).
The involvement of NACs in the regulation of the Arabidopsis WDS response was suggested by the identification and functional analysis of the AtNAC2, ANAC019, ANAC055, ANAC072, and ANAC102 genes under drought conditions (Tran et al., 2004; He et al., 2005; Christianson et al., 2009). Genome-wide transcriptomic analysis indicates that 20% to 25% of NAC genes in the whole genome of Arabidopsis and rice are responsive to drought stress (Tran et al., 2010). There are also a number of studies on abiotic stress-related NAC transcription factors in other plant species, such as that on GmNACs from soybean (Glycine max; Tran et al., 2009), TaNAC69 from wheat (Triticum aestivum; Xue et al., 2011), and AhNAC2 from groundnut (Arachis hypogaea; Liu et al., 2011). There has been no study, so far, of the possible role of NAC family genes in the response to WDS during petal expansion of flowers.
Expansins are plant cell wall-loosening proteins grouped into four subfamilies, designated as α-expansin, β-expansin, expansin-like A, and expansin-like B (Sampedro and Cosgrove, 2005). Expansin genes were first isolated from cucumber (Cucumis sativus) hypocotyls and play a primary role in cell wall extension (McQueen-Mason et al., 1992). Expansin proteins have been identified in widely different species and are involved in a variety of developmental processes requiring cell wall modification (McQueen-Mason and Cosgrove, 1995; Cosgrove, 2000). For example, they function in leaf and root growth (Wu et al., 2001; Shin et al., 2005), internode elongation (Choi et al., 2003), leaf development (Fleming et al., 1997; Pien et al., 2001), and floral development (Zenoni et al., 2004).
There is increasing evidence of a role for expansins in plant stress responses. Expansins are strongly regulated at the transcriptional level by WDS in maize (Zea mays; Wu et al., 2001; Zhu et al., 2007) and in a resurrection plant, Craterostigma plantagineum (Jones and McQueen-Mason, 2004). Heat tolerance, shade stress, low oxygen, and pathogens also up-regulate the expression of expansins in plants (Colmer et al., 2004; Xu et al., 2007; Ding et al., 2008; Fudali et al., 2008; Sasidharan et al., 2008). Overexpression of TaEXPB23, a wheat expansin gene, improves drought tolerance in tobacco (Nicotiana tabacum; Li et al., 2011). The transgenic plants showed improved tissue integrity during WDS, suggesting that expansins increase the flexibility of the cell wall (Li et al., 2011).
In ornamental plants, α- and β-expansin were identified from opening and senescing flowers of Mirabilis jalapa (Gookin et al., 2003). Two expansin genes, DcEXPA1 and DcEXPA2, are associated with petal growth and development during carnation (Dianthus caryophyllus) flower opening (Harada et al., 2011). In roses, RhEXPA, one of three α-expansin genes (RhEXPA1–RhEXPA3), is mainly involved in petal expansion (Takahashi et al., 2007; Yamada et al., 2009). However, information is very limited on the role of expansins in flower opening under WDS conditions.
Our previous work has indicated that dehydration can inhibit petal expansion, resulting in abnormal flower opening in roses. It is known that petal expansion depends on three important processes: cell wall metabolism, changes in cell turgor pressure, and restructuring of the cytoskeleton (Cosgrove, 2005; Smith and Oppenheimer, 2005; Zonia and Munnik, 2007). Nevertheless, the molecular mechanism of the effect of dehydration on petal expansion is still unclear. In this work, we focused on RhNAC2, a transcription factor identified in a dehydration-response SSH library from rose (Rosa hybrida) flowers. Functional analysis of this gene suggested that it is involved in the regulation of dehydration tolerance during petal expansion. An up-regulated expansin gene, RhEXPA4, may be directly regulated by RhNAC2. Our findings indicate that RhNAC2 and RhEXPA4 play an important role in flower opening under water stress conditions.
RESULTS
Isolation of Unique ESTs in Response to Dehydration in Rose Flowers
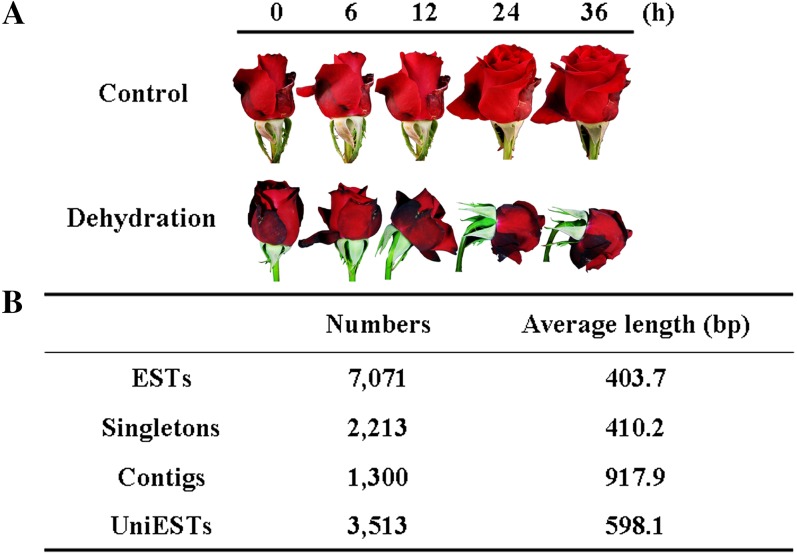
To analyze changes in the transcriptome resulting from dehydration on flower opening in roses, we first constructed a subtracted complementary DNA (cDNA) library by SSH. For the dehydration treatment, we held the cut flowers without water for 36 h, by which time the flowers showed considerable petal wilting and bent neck (Fig. 1A). We combined RNA samples isolated from the flowers at 6, 12, 24, and 36 h of dehydration as tester and from the mixture of RNA samples from the control flowers at the corresponding time points as driver. The forward subtractive PCR products were ligated into the pGEM T-Easy vector (Promega) and then transferred into Escherichia coli. We obtained 7,071 high-quality ESTs with an average length of 403 bp through random sequencing of bacterial colonies. A total of 3,513 uniESTs, including 1,300 contigs and 2,213 singletons, were identified by clustering and assembly analysis. Among the unique ESTs (uniESTs), 527 were putative full-length cDNAs based on the BLAST analysis using GenBank. The uniESTs had an average length of 598 bp (Fig. 1B). Ninety percent of the ESTs and the uniESTs were between 200 and 800 bp in length; the number of ESTs over 800 bp in length was greater for the contigs than for the singletons (Supplemental Fig. S1).
Figure 1.
Subtracted cDNA library of rose flowers under dehydration. A, Phenotypes of rose flowers under dehydration. Dehydrating flowers were held in air (25°C, 40%–50% relative humidity) under continuous light (140 μmol m−2 s−1), and control flowers remained in water under the same conditions. Both were sampled at 6, 12, 24, and 36 h. B, Rose flower ESTs in the subtracted cDNA library prepared from the combined samples. cDNA prepared from flowers under dehydration was used as tester, and cDNA prepared from the control flowers was used as driver.
Classification of Genes Responding to Dehydration in Rose Petals
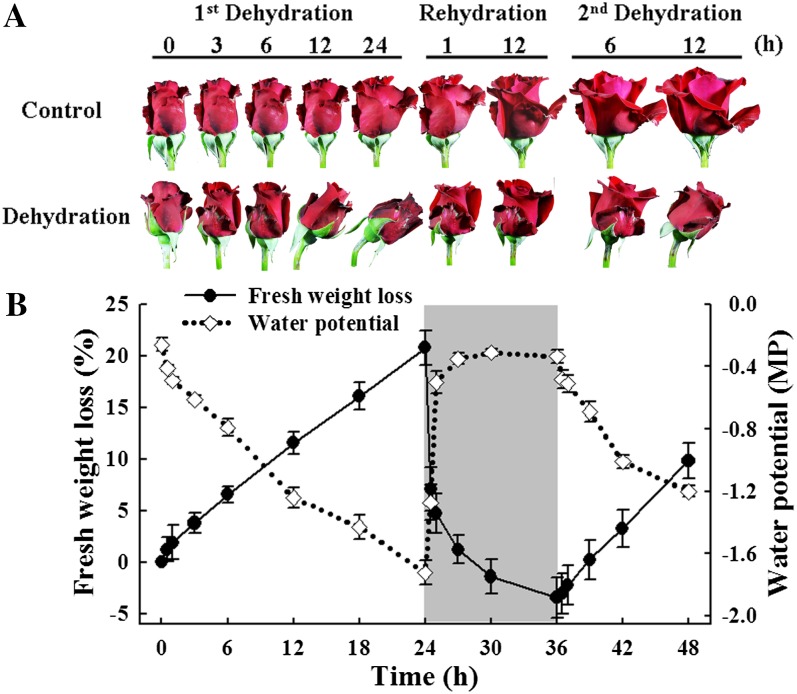
To evaluate the expression profiles of dehydration-regulated genes in rose petals, we amplified all uniESTs from the SSH library by PCR and spotted them on amino-silaned glass slides to make a cDNA microarray. Flowers were exposed to cyclic dehydration, namely dehydration for 24 h (first dehydration), rehydration for 12 h in water, then dehydration for 12 h (second dehydration). We recorded the phenotypes, water potential, and fresh weight of flowers during the treatment. During the first dehydration, flowers started to wilt after 12 h, and neck bending occurred at 24 h. The necks recovered after 1 h of rehydration (Fig. 2A). In the second dehydration, flowers wilted after 6 h and recovered after 12 h of rehydration (data not shown), indicating that the second dehydration resulted in greater stress. During the first dehydration, flowers lost fresh weight linearly, reaching approximately 20% loss after 36 h. Water uptake was very rapid in the first 1 h of rehydration, and the flowers reached their original fresh weight within 3 h. Changes in water potential reflected the changes in flower fresh weight (Fig. 2B).
Figure 2.
Morphological changes of rose flowers during cyclic dehydration. A, Flower phenotypes. Flowers were held dry in air for 24 h (first dehydration), returned to water for 12 h (rehydration), and then held dry in air for a further 12 h (second dehydration). Control flowers were held continuously in water. The flowers were held at 25°C and 40% to 50% relative humidity in continuous light (140 μmol m−2 s−1). B, Fresh weight loss (solid line) and water potential (dotted line) for the flowers. Vertical lines represent sd; n = 30 for fresh weight loss and n = 10 for water potential.
Petal total RNA extracts were hybridized to the microarray chips; spot intensities were quantified using the Axon GenePix pro 6.0 software, and the Lowess strategy was applied to normalize the ratio values for each array using the Bioconductor marray R package (Yang et al., 2002). We first compared differentially expressed genes identified by SSH in rose flower with genes that were up-regulated (more than 2.0-fold change, corrected P < 0.05) during the first dehydration. We found up-regulation of approximately 54% of the uniESTs present in more than 10 copies in the SSH library. In contrast, only 5.5% of the singleton uniESTs in the SSH library were found to be up-regulated in the microarray assay (Supplemental Fig. S2).
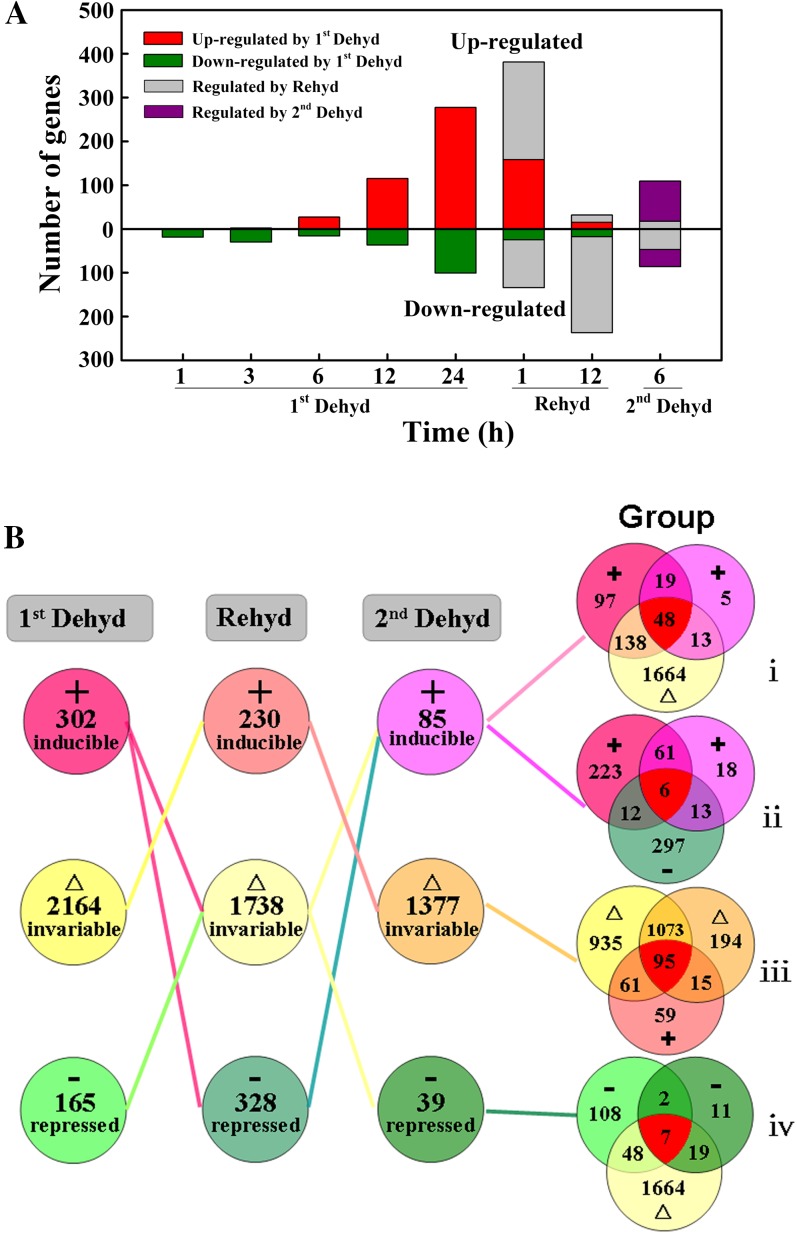
We considered genes with an expression ratio of 2 or greater or 0.5 or less and corrected P < 0.05 at any time-course point in the cyclic dehydration treatment as dehydration-regulated genes. During the first dehydration, the number of such genes increased rapidly after 12 h of dehydration. After rehydration for 12 h, transcript abundance for these genes, especially those up-regulated by the first dehydration, was restored to control levels; a number of genes were concomitantly down-regulated. By 6 h of the second dehydration, the number of genes responding to the stress was similar to that seen in the first dehydration at 12 h (Fig. 3A).
Figure 3.
Differentially expressed genes in rose petals during cyclic dehydration. Flowers were held dry for 36 h (first dehydration), placed in water for 12 h (rehydration), and then held dry again for 8 h (second dehydration). Experiments were conducted under controlled conditions of 25°C, 40% to 50% relative humidity, and continuous light (140 μmol m−2 s−1). Genes were considered induced or repressed by petal dehydration (Dehyd) and rehydration (Rehyd) when the ratio of their transcript abundance to that in control petals was 2 or greater or 0.5 or less, respectively (P < 0.05); other genes were considered invariable. A, Number of differentially expressed genes. B, Selected gene groups classified by their expression pattern during dehydration and rehydration. Groups are as follows: i, first Dehyd-induced, Rehyd-invariable, second Dehyd-induced genes; ii, first Dehyd-invariable, Rehyd-repressed, second Dehyd-invariable genes; iii, first Dehyd-invariable, Rehyd-induced, second Dehyd-invariable genes; iv, first Dehyd-repressed, Rehyd-invariable, second Dehyd-repressed genes. [See online article for color version of this figure.]
To classify the genes on the basis of expression patterns in the cyclic dehydration treatment, we regarded genes with an expression ratio of 2 or greater and corrected P < 0.05 at any time-course point as induced genes, genes with an expression ratio of greater than 0.5 to less than 2 as invariable genes, and genes with an expression ratio of 0.5 or less and corrected P < 0.05 as repressed genes. Genes that were induced, invariable, and repressed during the cyclic dehydration treatment (Fig. 3B) are listed in Supplemental Table S1. Using Venn diagram analysis, we analyzed the specificity and cross talk in gene expression in the responses to dehydration and rehydration. We classified them into 27 groups based on their gene expression patterns (Supplemental Fig. S3), and four of the 27 groups are shown in Figure 3B. We postulated that the genes most closely related to dehydration tolerance would be those whose transcript abundance was up-regulated during the first dehydration, restored to control abundance or unaffected during rehydration, and up-regulated again during the second dehydration. Groups i (induced-invariable-induced genes) and ii (induced-repressed-induced genes) meet these criteria (Fig. 3B). Group i comprises 48 genes and group ii comprises six genes, including genes known to be associated with drought stress (dehydrin, Suc synthase), transcription factors (NAC protein, zinc ion-binding protein), and cell wall-related genes (expansin, pectate lyase; Supplemental Table S2).
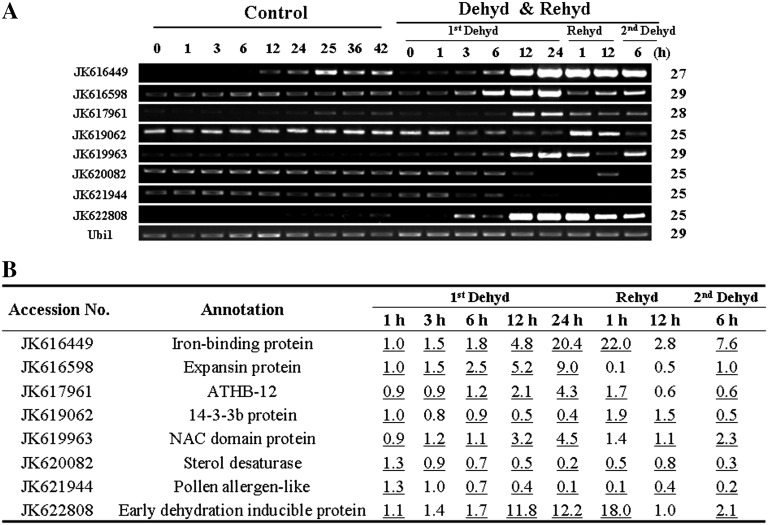
To confirm the microarray results, we conducted reverse transcription (RT)-PCR of eight genes selected from the dehydration-responsive genes identified in microarray analysis (Fig. 4A). Results from semiquantitative RT-PCR are in agreement with the microarray data for 55 out of 64 (86%) data points (Fig. 4B).
Figure 4.
Verification of microarray results by RT-PCR. A, RT-PCR analysis of selected genes. Ubi1 was used as the internal control. Numbers of PCR cycles are listed on the right. Dehyd, Dehydration; Rehyd, rehydration. B, Expression ratios of the selected genes derived from the microarray analysis. Data matching the RT-PCR results are underlined.
Transcription Factors Responding to Dehydration in Rose Petals
To understand transcriptional regulation during dehydration of rose petals, we first analyzed all transcription factors that were responsive to dehydration. From 101 annotated transcription factor unigenes in our microarray, we observed 41 unigene homologs that responded to dehydration and rehydration, comprising members of at least 17 transcription factor families (Supplemental Table S3). We focused on transcription factors up-regulated by dehydration, especially those from four major families: NAC, Homeodomain, Broad-Complex, Tramtrack, and Bric-a-Brac/Transcriptional Adaptor Zinc finger (BTB/TAZ), and AP2 (Table I). Some NAC family transcription factors were strongly up-regulated by dehydration; transcripts of a unigene, JK619963, increased 4.5-fold after 24 h of the first dehydration, decreased to 1.4-fold after 1 h of rehydration, and then increased to 2.3-fold after 6 h of the second dehydration. JK619963 is in group i of the Venn diagram (Fig. 3B; Supplemental Table S2).
Table I. Transcription factors up-regulated by dehydration in rose petals.
Significant differences (corrected P < 0.05 and expression ratio ≥ 2) in relative levels are shown in boldface.
| First Dehydration |
Rehydration |
Second Dehydration |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Accession No. | Annotation | 1 h | 3 h | 6 h | 12 h | 24 h | 1 h | 12 h | 6 h |
| NAC family | |||||||||
| JK616545 | NAC domain-containing protein | 1.0 | 0.9 | 0.9 | 2.0 | 4.2 | 3.0 | 1.7 | 1.5 |
| JK617046 | NAC domain protein | 0.8 | 0.9 | 1.3 | 2.1 | 3.2 | 3.5 | 1.1 | 1.7 |
| JK617768 | NAC domain protein | 1.1 | 1.2 | 0.9 | 3.0 | 3.6 | 3.0 | 2.1 | 1.2 |
| JK619963 | NAC domain protein NAC2 | 0.9 | 1.2 | 1.1 | 3.2 | 4.5 | 1.4 | 1.1 | 2.3 |
| JK620479 | NAC domain transcription factor | 1.0 | 1.0 | 1.1 | 2.8 | 4.0 | 2.7 | 1.4 | 1.5 |
| Homeodomain family | |||||||||
| JK616983 | HD domain transcription factor | 0.9 | 1.1 | 1.3 | 2.5 | 2.6 | 1.3 | 1.1 | 1.9 |
| JK617961 | ATHB-12 | 0.9 | 0.9 | 1.2 | 2.1 | 4.3 | 1.7 | 0.6 | 0.6 |
| BTB/TAZ family | |||||||||
| JK619064 | BTB and TAZ domain protein4 | 0.9 | 0.8 | 0.8 | 1.6 | 2.7 | 3.6 | 1.2 | 1.0 |
| JK622967 | BTB and TAZ domain protein1 | 1.0 | 0.9 | 1.0 | 1.4 | 2.4 | 4.4 | 1.1 | 0.8 |
| AP2 family | |||||||||
| JK619081 | EREB protein | 1.1 | 1.3 | 1.0 | 1.2 | 2.2 | 1.3 | 1.7 | 1.1 |
| JK622912 | ABR1 (ABA REPRESSOR1) | 0.9 | 0.9 | 0.8 | 1.7 | 3.7 | 16.7 | 0.5 | 0.9 |
| Other transcription factors | |||||||||
| JK617262 | SET domain-containing protein | 0.9 | 1.1 | 1.6 | 2.5 | 3.1 | 3.0 | 0.9 | 2.7 |
| JK617756 | Zinc-binding family protein | 0.9 | 1.1 | 1.0 | 1.8 | 3.4 | 0.9 | 1.3 | 1.5 |
| JK619813 | DNA-binding protein S1FA | 1.1 | 1.2 | 1.0 | 2.1 | 2.4 | 1.6 | 2.7 | 1.2 |
| JK621262 | WRKY65 transcription factor | 1.0 | 1.1 | 1.4 | 1.6 | 2.4 | 0.3 | 1.1 | 1.6 |
| JK622333 | MYB transcription factor | 1.1 | 1.5 | 1.7 | 1.7 | 2.0 | 0.5 | 1.4 | 1.8 |
Biological Processes in the Dehydration Response of Rose Petals
To understand the functional significance of the dehydration-responsive genes in rose petals, we classified differentially regulated genes into different categories using a set of plant-specific GO slims (http://www.geneontology.org/GO.slims.shtml). This analysis suggested that cyclic dehydration caused significant changes in a range of biological processes, including drought, defense, and stimulus responses (Supplemental Tables S4 and S5). Processes such as Pro biosynthesis, carbohydrate metabolism, and polysaccharide catabolism relate to osmotic adjustment. This suggests that the synthesis and breakdown of osmotic components is tightly linked to the dehydration response. Cytokinin metabolism was also very responsive to dehydration; two genes encoding cytokinin oxidase/dehydrogenases were highly up-regulated during dehydration, suggesting that endogenous cytokinin content of petals may be reduced during dehydration.
Interestingly, we found that the abundance of transcripts for enzymes associated with plant cell wall organization changed significantly during cyclic dehydration. These genes were up-regulated at the first 6, 12, and 24 h of dehydration (Supplemental Table S4), down-regulated immediately after 1 h of rehydration (Supplemental Table S5), and up-regulated during the second 6 h of dehydration (Supplemental Table S4). These findings suggest that the assembly, disassembly, and rearrangement of constituent parts of the cellulose- and pectin-containing cell wall are important components of the dehydration response in rose petals.
We analyzed the effects of cyclic dehydration on all 41 of the cell wall-associated unigenes on the rose petal microarray. We found that 13 out of 41 unigenes were involved, comprising homologs to eight expansins, one cellulase, one cellulose synthase, one Pro-rich cell wall protein, one pectin methylesterase, and one pectate lyase (Table II). One of the 13 unigenes, JK616598, was rapidly and strongly up-regulated during the first dehydration and then was quickly down-regulated during rehydration. This unigene, therefore, was further evaluated to determine its role in rose flower opening during the cyclic dehydration treatment.
Table II. Genes involved in the biological process of plant-type cell wall organization responsive to dehydration.
Significant differences (corrected P < 0.05 and expression ratio ≥ 2 or ≤ 0.5) in relative levels are shown in boldface.
| Dehydration |
Rehydration |
||||
|---|---|---|---|---|---|
| Accession No. | Annotation | 6 h | 12 h | 24 h | 1 h |
| JK616598 | ATEXPA10 | 2.5 | 5.2 | 9.0 | 0.1 |
| JK617470 | Pro-rich cell wall protein | 1.1 | 0.6 | 0.3 | 0.3 |
| JK617691 | ATEXPA9 | 1.6 | 1.2 | 0.9 | 0.02 |
| JK617882 | ATEXPA1 | 1.7 | 1.2 | 0.9 | 0.04 |
| JK618171 | Cellulase | 1.6 | 3.1 | 5.0 | 3.7 |
| JK618328 | ATEXPA1 | 2.4 | 3.1 | 2.5 | 0.1 |
| JK618813 | Pectate lyase | 1.4 | 1.0 | 0.3 | 0.1 |
| JK619324 | ATEXPA11 | 0.9 | 1.8 | 4.4 | 1.1 |
| JK619325 | ATEXPA8 | 1.3 | 1.2 | 1.2 | 0.1 |
| JK619868 | Cellulose synthase | 0.7 | 0.6 | 0.4 | 0.8 |
| JK620968 | ATEXPA4 | 1.2 | 1.6 | 1.8 | 2.3 |
| JK621413 | ATEXPA1 | 2.4 | 3.2 | 2.1 | 0.1 |
| JK622421 | Pectin methylesterase31 | 1.5 | 1.8 | 1.2 | 2.3 |
Characterization of RhNAC2 and RhEXPA4
To characterize the functional role of JK619963 and JK616598 in dehydration tolerance of rose petals, we first cloned the two full-length cDNAs. JK619963 is 1,395 bp in length with a 227-bp 5′-untranslated region (UTR) and a 103-bp 3′-UTR; the gene encodes a polypeptide of 355 amino acids. Analysis of the deduced amino acid sequence indicates that the protein shares homology to known NAC family proteins and contains five conserved subdomains (A–E). In our previous work, a unigene induced by ethylene was identified and designated as RhNAC1 (data not shown); therefore, JK619963 is designated as RhNAC2 (Supplemental Fig. S4, A and B). Phylogenetic comparison with NAC proteins from Arabidopsis indicates that RhNAC2 is a close homolog of embryo development-related genes CUC1 and CUC2 and also of secondary cell wall NACs VND6 and VND7 (Supplemental Fig. S4C).
JK616598 is 1,298 bp in length with a 43-bp 5′-UTR and a 499-bp 3′-UTR; the gene encodes a polypeptide of 252 amino acids. Phylogenetic analysis indicates that JK616598 is a homolog of α-type expansin proteins in Arabidopsis and is designated as RhEXPA4 (Supplemental Fig. S5), since three other expansin genes (RhEXPA1, AB370116; RhEXPA2, AB370117; RhEXPA3, AB370118) have been described previously (Takahashi et al., 2007; Yamada et al., 2009).
We examined the expression patterns of the two genes during cyclic dehydration of rose petals using RT-PCR and found that they mirrored the pattern previously determined using the microarray. We also measured the expression levels of RhNAC2 and RhEXPA4 in different floral organs and leaves. During dehydration, both genes were strongly induced in petals, and RhNAC2 was also clearly induced in other organs, but RhEXPA4 expression showed no obvious changes in other floral organs or leaves (Supplemental Fig. S6). To determine subcellular localization of RhNAC2, we transiently expressed p35S::GFP-RhNAC2 in onion (Allium cepa) epidermal cells. GFP fluorescence in the onion cells was detected exclusively in the nucleus, indicating that the gene encodes a nucleus-localized protein (Supplemental Fig. S7).
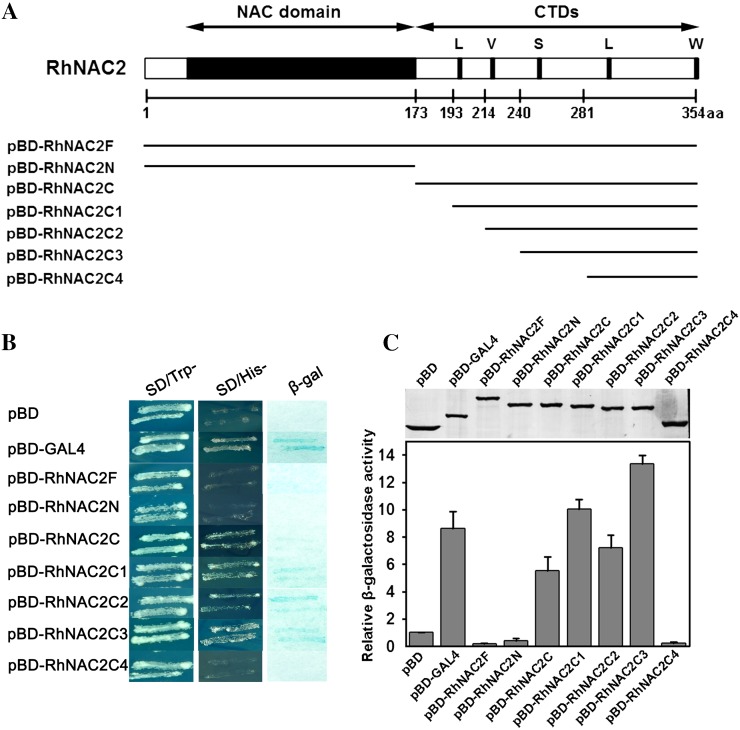
To examine the transactivation potential and the function of the different conserved motifs of RhNAC2, we performed a yeast (Saccharomyces cerevisiae) one-hybrid assay. We truncated and fused different portions of RhNAC2 to the GAL4 DNA-binding domain in the pBD vector (Fig. 5A). We tested the growth on synthetic dextrose (SD)-His plates of yeast bearing four different C-terminal domain truncations of RhNAC2. In comparison with the positive control (pBD-GAL4), the full length of RhNAC2 (pBD-RhNAC2F) and the NAC domain (pBD-RhNAC2N) showed no obvious β-galactosidase activity, while the C-terminal domain (pBD-RhNAC2C) retained 0.64-fold activity. One, pBD-RhNAC2C4, failed to grow; the other three grew well and showed higher relative β-galactosidase activity than colonies bearing the full C-terminal domain (Fig. 5, B and C). The abilities of RhNAC2C and RhNAC2C3 to activate transcription were also examined using the GAL4 transient expression system in Arabidopsis protoplasts. Compared with the GAL4-DBD negative control, both fusion proteins strongly activated the reporter GUS gene, and the relative GUS activities of RhNAC2C and RhNAC2C3 were 2.11- and 3.91-fold higher than the negative control, respectively (Supplemental Fig. S8). The results indicate that the 240 to 281 region of the polypeptide, with a putative S motif, may play an important role in the transactivation activity of RhNAC2.
Figure 5.
Truncation analysis of transactivation activity of RhNAC2 in yeast. A, Constructs used in the transactivation analysis. The regions shown as lines below the RhNAC2 diagrams were inserted into the expression vector pBD-GAL4 to create expression vectors comprising partial RhNAC2 proteins fused with the GAL4 DNA-binding domain in yeast. The numbers below the line indicate the positions of each truncated portion of RhNAC2. aa, Amino acids; CTDs, C-terminal domains; L, V, S, and W indicate highly conserved motifs common to NAC proteins. B, Transactivation activity of RhNAC2 in yeast. Expression vectors (pBD-X) were introduced into yeast YRG-2; pBD-GAL4 was used as the positive control and pBD vector was used as the negative control. The transformants were streaked on SD-Trp medium (left panel) and SD-His medium (middle panel) for examination of growth. C, β-Galactosidase activity was measured in the transformed yeast cells (bar graph). The top panel shows the expression of pBD-X proteins. Rabbit anti-GAL4 DNA-BD antibody was used western-blot analysis. Each data point represents the mean ± sd (n = 6). [See online article for color version of this figure.]
Dehydration Tolerance in RhNAC2- or RhEXPA4-Silenced Rose Petals
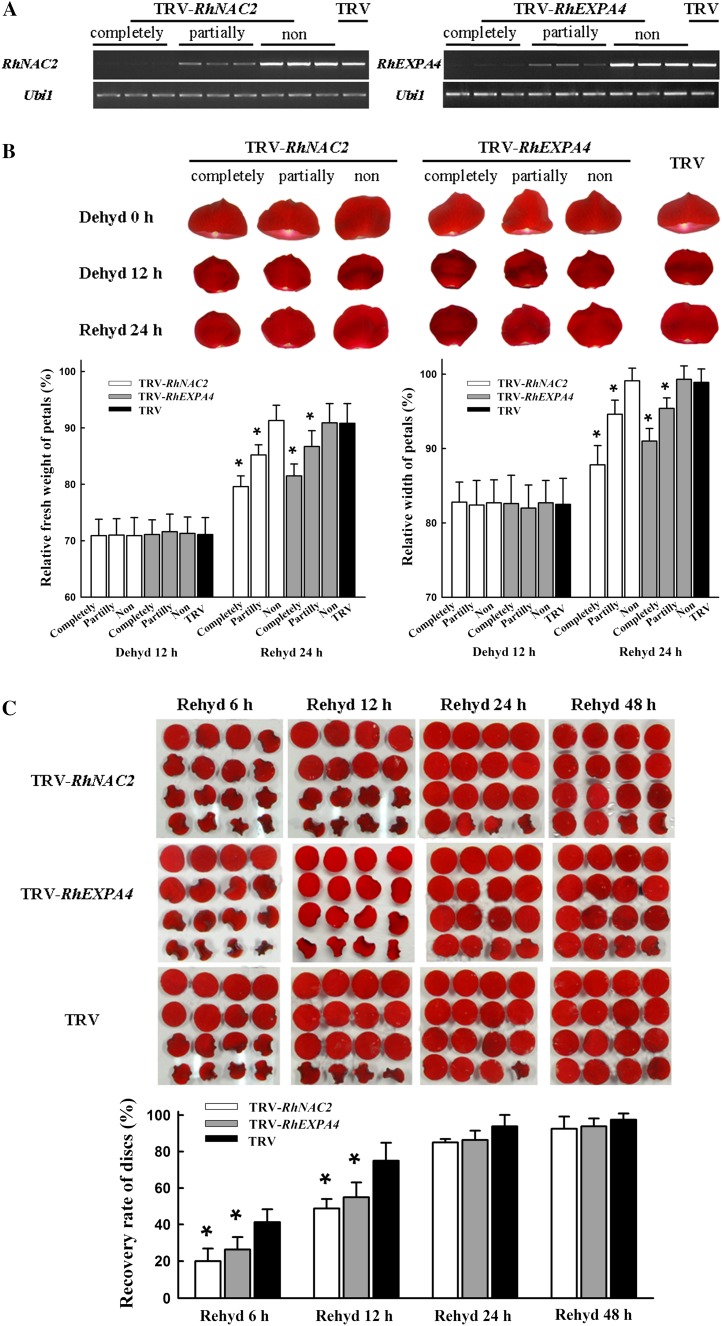
To test the possible role of RhNAC2 or RhEXPA4 in dehydration tolerance in rose petals, we silenced each gene in rose petals using a virus-induced gene silencing (VIGS) approach. By infiltrating entire petals or petal discs with Agrobacterium tumefaciens containing tobacco rattle virus (TRV)-RhNAC2, we were able to partially or completely silence the RhNAC2 gene (Fig. 6A). After 12 h of dehydration, the fresh weight of all the petals was about 70% of its initial value and petal width was about 83% of its initial value. During 24 h of rehydration, TRV control petals returned to 91% of their initial fresh weight, but it was still only 79% in the completely RhNAC2-silenced petals. Similarly, petal width returned to near the initial levels in the TRV controls but was still only 88% of the initial width in the completely RhNAC2-silenced petals (Fig. 6B; Supplemental Table S6).
Figure 6.
Silencing of RhNAC2 or RhEXPA4 in rose petals and discs by VIGS. Rose petals or discs were infiltrated with A. tumefaciens containing TRV control (TRV, pTRV1 + pTRV2), TRV carrying a RhNAC2 fragment (TRV-RhNAC2, pTRV1 + pTRV2-RhNAC2), or TRV carrying a RhEXPA4 fragment (TRV-RhEXPA4, pTRV1 + pTRV2-RhEXPA4). After the VIGS procedure, petals were dehydrated for 12 h and then rehydrated for 48 h to observe the recovery ability. Finally, the petals were sampled after 12 h of redehydration for the determination of VIGS efficiency by RT-PCR. Based on the expression level of VIGS-targeted genes, the samples were classified into three groups: completely silenced petals, partially silenced petals, and nonsilenced petals. A, RT-PCR analysis of RhNAC2 or RhEXPA4 expression in silenced petals. A total of 31, 27, and 29 PCR cycles were used for RhNAC2, RhEXPA4, and Ubi1, respectively. B, The phenotype of RhNAC2- or RhEXPA4-silenced petals and petal recovery in terms of width and fresh weight relative to initial values. Values are means ± sd (n = 30 for both groups of completely silenced petals and n = 60 for other groups; *P < 0.05). C, Phenotype and recovery of RhNAC2- or RhEXPA4-silenced petal discs. After the VIGS procedure, petal discs were dehydrated for 9 h and then examined at intervals during 48 h of rehydration. Discs were scored as recovered if they were fully expanded. Values are means ± sd (n = 5, with 90 petal discs per repeat; *P < 0.05). Dehyd, Dehydration; Rehyd, rehydration.
The effects of gene silencing and dehydration were also tested in petal discs; the percentage of fully expanded discs provided a convenient rehydration assay. Discs were dehydrated for 9 h and then rehydrated for 48 h. By 6 h of rehydration, 41% of the discs in the TRV controls had recovered compared with only 20% in the RhNAC2-silenced discs (Fig. 6C). The difference between TRV controls and RhNAC2-silenced discs was still significant at 12 h of rehydration but disappeared after 24 h. These results are consistent with those obtained from intact petals and indicate that RhNAC2, a transcription factor gene, is involved in dehydration tolerance of rose petals.
We used TRV-RhEXPA4 to silence the RhEXPA4 gene both in entire petals and in petal discs. After 24 h of rehydration, the fresh weight and petal width of RhEXPA4-silenced petals were significantly less than in the TRV controls (Fig. 6B; Supplemental Table S6). Recovery of petal discs after a 9-h dehydration confirmed this response. RhNAC2-silenced discs showed significantly higher recovery than TRV controls at 6 and 12 h of rehydration (Fig. 6C). These results indicate that RhEXPA4, a functional gene, is involved in dehydration tolerance of rose petals, but it has less effect on the tolerance than RhNAC2.
Drought Tolerance of Arabidopsis Plants Overexpressing RhNAC2 or RhEXPA4
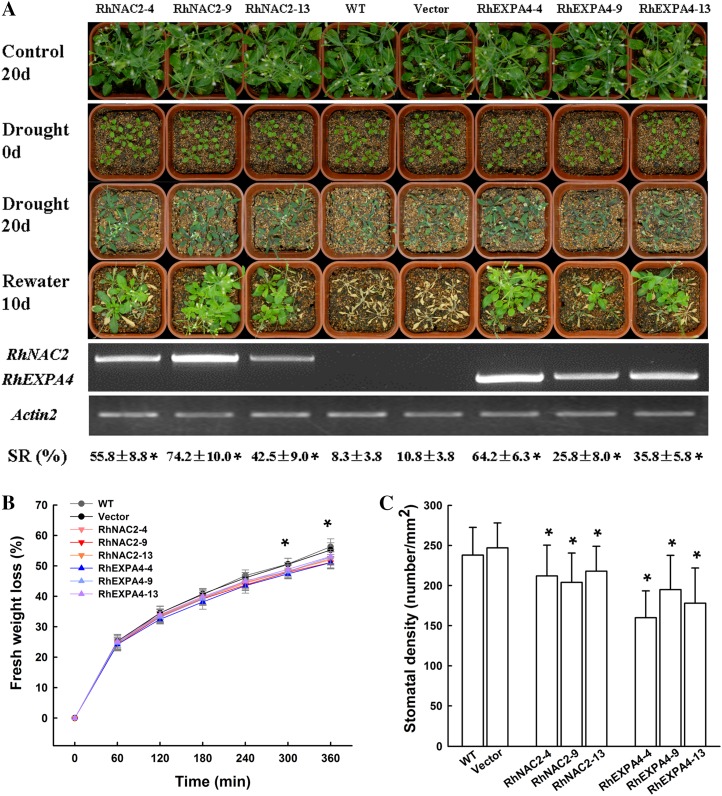
To further examine the function of RhNAC2 and RhEXPA4 in dehydration tolerance, we overexpressed RhNAC2 and RhEXPA4 in Arabidopsis. Based on the expression level of the transgene, we chose three each from 14 35S::RhNAC2 transgenic lines and 12 35S::RhEXPA4 transgenic lines to test their drought tolerance. Overexpression of both transgenes resulted in reduced plant height (Supplemental Fig. S9), but there were no changes in leaf shape, bolting time, silique size, root number, and architecture (Supplemental Fig. S10).
After 20 d without water, control plants (wild type and vector only) were severely wilted, while plants overexpressing RhNAC2 or RhEXPA4 were only slightly wilted (Fig. 7A). Ten days after watering resumed, over 60% of the transgenic plants had recovered, compared with only about 10% of the control plants (Fig. 7A). Improvement in survival rate was closely related to RhNAC2 or RhEXPA4 expression levels in the overexpressing lines (Fig. 7A). For a given level of gene expression, plants overexpressing RhNAC2 had a higher survival rate than those overexpressing RhEXPA4.
Figure 7.
Tolerance of RhNAC2- and RhEXPA4-overexpressing Arabidopsis to water deficit stress. A, Drought tolerance and gene expression of RhNAC2 and RhEXPA4 overexpressors. T2 homozygous transformants were used in this experiment. Control 20d, Twenty-day-old plants growing under normal watering conditions; Drought 0d, 10-d-old well-watered plants; Drought 20d, 20 d after withholding water; Rewater 10d, 10 d after rewatering; SR, survival rate; RhNAC2-4, RhNAC2-9, and RhNAC2-11, three independent RhNAC2-overexpressing lines; RhEXPA4-4, RhEXPA4-9, and RhEXPA4-13, three independent RhEXPA4-overexpressing lines; WT, wild type. RT-PCR was conducted on fully expanded leaves of control plants. Actin2 was used as the internal control. The RhNAC2, RhEXPA4, and Actin2 genes were amplified for 28, 28, and 22 PCR cycles, respectively. Survival rate was calculated from three independent experiments (40 plants per line in one experiment; *P < 0.05). B, Water loss assay of aerial parts during dehydration. Four-week-old plants were derooted and held in air. Asterisks denote significant differences (P < 0.05). Vertical lines represent sd (n = 15). C, Leaf stomatal density. Vertical lines represent sd (n = 20).
Rosette leaves of RhNAC2- and RhEXPA4-overexpressing Arabidopsis plants lost water more slowly than those of wild-type plants (Fig. 7B). This was associated with a lower leaf stomatal density (Fig. 7C), but there was no obvious difference in stomatal aperture under normal growth conditions or in response to abscisic acid (ABA) treatment (Supplemental Fig. S11). It was reported that higher drought tolerance was also caused by lower stomatal density in an Arabidopsis mutant (Yoo et al., 2010). These data suggest that drought tolerance in the Arabidopsis RhNAC2 or RhEXPA4 overexpressors may be partially due to lower stomatal density.
Role of RhNAC2 in the Regulation of Expansin Gene Expression in Rose Petals and Arabidopsis
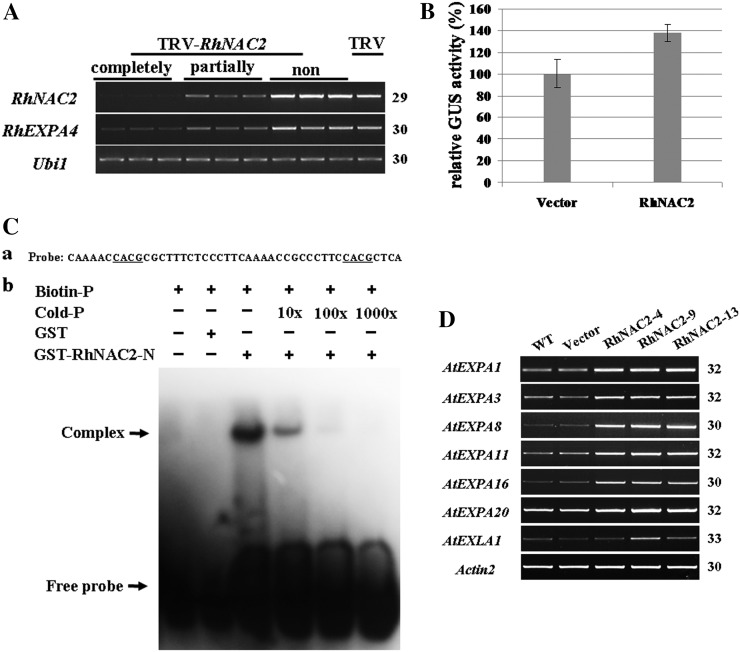
Our results showed that RhNAC2 and RhEXPA4 exhibited similar expression patterns under dehydration, similar phenotypes when silenced in petals or petal discs, and similar effects in extending drought tolerance when overexpressed in Arabidopsis. To test the hypothesis that RhEXPA4 might be a candidate downstream gene of RhNAC2 in rose petals, we first determined the effect of silencing RhNAC2 on the expression of RhEXPA4. The expression of RhEXPA4 was obviously repressed in RhNAC2-silenced petals, and the degree of repression was correlated with the level of silencing of RhNAC2 (Fig. 8A).
Figure 8.
Analysis of the interaction between RhNAC2 and expansins in rose and Arabidopsis. A, RhEXPA4 expression in RhNAC2-silenced rose petals. The first-strand cDNA was generated from 1 µg of total RNA from petals where RhNAC2 was completely, partially, or not silenced. B, Transactivation of ProRhEXPA4::GUS gene expression by RhNAC2 in Arabidopsis protoplasts. A pUC19-RhNAC2::GFP construct (RhNAC2) and pUC19-GFP (Vector) as a control were transformed into Arabidopsis mesophyll protoplasts isolated from 4-week-old ProRhEXPA4::GUS transgenic leaves. Nine independent transformation experiments were performed. C, Gel-shift analysis of RhNAC2 N-terminal binding to the RhEXPA4 promoter. a, Oligonucleotide sequence of the −216 to −171 region of the RhEXPA4 promoter. Underlined letters indicate the core sequences of NAC protein-targeted promoters. b, Interaction between GST-RhNAC2-N1–173 and biotin-labeled probe on native PAGE. Purified protein (2 µg) was incubated with 0.2 pmol of biotin probe. For the competition test, nonlabeled probe at different concentrations (from 10 to 1,000 times) was added to the reaction mixture. D, Transcript abundance of AtEXPA genes is enhanced in RhNAC2 overexpressors. The first-strand cDNAs from 4-week-old transgenic Arabidopsis leaves were used for RT-PCR with Actin2 as the internal control. WT, Wild type. PCR cycles are listed on the right side in A and D.
To test the possibility that the RhNAC2 protein activates the RhEXPA4 promoter, we transformed Arabidopsis plants with a construct comprising the promoter for RhEXPA4 driving the gene for glucuronidase (ProRhEXPA4::GUS). When mesophyll protoplasts isolated from 4-week-old ProRhEXPA4::GUS transgenic leaves were transformed with a construct inducing transient expression of RhNAC2 (pUC19-RhNAC2::GFP), GUS activity in the protoplasts was nearly 40% higher than in protoplasts transformed with pUC19-GFP, the empty vector (Fig. 8B).
We also used electrophoresis mobility shift assay (EMSA) to investigate whether the RhNAC2 protein binds to the RhEXPA4 promoter. We selected a fragment from the −216 to −171 region of the RhEXPA4 promoter containing two core sequences common to NAC protein-targeted promoters (Olsen et al., 2005) as probe and the DNA-binding domain of RhNAC2 labeled with glutathione S-transferase (GST; GST-RhNAC2-N1-173) as target. DNA-binding bands were detected when GST-RhNAC2-N1-173 was combined with the labeled probe but not in the GST control (Fig. 8C). When unlabeled probe was added to the reaction mixture, the DNA-binding band was abolished (Fig. 8C). These data suggest that the N-terminal binding domain of RhNAC2 binds to the promoter of RhEXPA4 and support the hypothesis that RhEXPA4 may be a direct downstream gene of RhNAC2.
To investigate the potential gene network related to cell expansion regulated by RhNAC2, we performed a microarray experiment with the GeneChip ATH1 Arabidopsis genome array (Affymetrix; http://www.affymetrix.com/index.affx) using aerial parts of transgenic 35S::RhNAC and vector control Arabidopsis plants. A total of 108 genes were up-regulated at least 2-fold by RhNAC2 overexpression; among them, there are 20 cell wall-related genes (Supplemental Table S7). Seven expansin family genes, especially AtEXPA1, a close homolog of RhEXPA4, were up-regulated by RhNAC2. The microarray data were confirmed using semiquantitative RT-PCR (Fig. 8D).
DISCUSSION
Expression Profiles of Genes in Response to Dehydration in Rose Petals
Water deficit is recognized as one of the main environmental constraints restricting natural and agricultural ecosystem productivity. The genetic basis of drought tolerance has been extensively studied by targeting relevant genes through transcriptomic techniques (Seki et al., 2007). In rose, a model for multipetaled flowers, several small-scale EST sequencing projects have resulted in a total of approximately 9,300 ESTs available in public databases, mainly genes involved in volatile biosynthesis and flowering control (Debener and Linde, 2009). Interestingly, around 30% of rose transcripts did not match any annotated sequence in the common databases (Channelière et al., 2002; Guterman et al., 2002), indicating that a number of rose petal genes are either unique or too divergent from the genes from other taxa to be identified by sequence homology. Our study comprehensively explored the temporal expression patterns of dehydration-induced genes in rose petals through SSH and cDNA microarray technology. We obtained a total of 3,513 uniESTs and detected 1,006 transcripts differentially expressed under the cyclic dehydration treatment, accounting for around 30% of total transcripts on the cDNA microarray.
Among the differentially expressed genes responsive to WDS, genes encoding transcription factors are considered to be the most interesting category because of their potential involvement in regulating the expression of downstream genes (Trewavas and Malho, 1997; Bray, 2004). It is known that there are more differences than similarities in transcription factor families based on their classification among plant species. The role of different transcription factor families in plant growth and development differs widely among different plant species. The most highly conserved are the changes observed in AP2/EREBP transcription factors responding to WDS in Arabidopsis, rice, and chickpea. However, even in this family, different members show different expression patterns in response to WDS (Molina et al., 2008; Narsai et al., 2010; Wang et al., 2011). Types and numbers of transcription factors responding to WDS vary from species to species (Tommasini et al., 2008), and some also show tissue- or development stage-specific features (Zhou et al., 2007; Wang et al., 2011).
We have identified 41 transcription factor genes that are differentially regulated by cyclic dehydration treatment in rose petals, accounting for approximately 40% of the total transcription factor transcripts on our cDNA microarray. The ratio is much higher than the 5% seen in genome-wide gene expression profiling in Arabidopsis or rice with Affymetrix microarrays (Narsai et al., 2010), because our rose EST library was derived by subtractive hybridization of ESTs from dehydrated petals. The differentially regulated transcription factor genes belong to a diverse range of transcription factor families, including NAC, Homeobox, AP2, and BTB/TAZ (Table I; Supplemental Table S3), and most have been reported as being responsive to drought stress in other plant species such as Arabidopsis and wheat (Olsen et al., 2005; Ergen et al., 2009). It seems that transcriptional regulation of the dehydration response in rose petals shares features of that in other plants.
GO functional categories are useful tools for analyzing changes in the transcriptome during growth and development and in response to stress in model and crop plants (Street et al., 2006; Rodriguez et al., 2010; Wilkins et al., 2010; Wang et al., 2011). Genes grouped in the functional categories of “response to stresses” and “response to biotic and abiotic stimuli” were identified in the Arabidopsis response to drought (Wilkins et al., 2010). In rice, WDS caused tissue-specific down-regulation of genes in distinct categories, including photosynthesis-related genes in leaves and genes involved in cell membrane biogenesis and cell wall modification in the roots and in the young panicle (Wang et al., 2011). In poplar, genes involved in the maintenance of cellular homeostasis and in GA response and signaling were up-regulated by drought (Street et al., 2006). In C. plantagineum, GO analysis of the 500 most variable unigenes revealed metabolic processes that are hallmarks of the stages of dehydration and rehydration (Rodriguez et al., 2010).
GO analysis of the cyclic dehydration response in rose petals indicated a primary role for three processes: osmotic adjustment, cytokinin metabolism, and cell wall metabolism (Supplemental Tables S4 and S5). Osmotic adjustment can help to maintain cell turgor and stabilize cell proteins under stress conditions (Seki et al., 2007). Recent reports have described the involvement of cytokinins in stress responses of vegetative organs of plants (Rivero et al., 2007; Tran et al., 2007; Werner et al., 2010; Nishiyama et al., 2011). However, there is little information on the role of cytokinins in the dehydration response of reproductive organs, particularly petals. In rose petals, higher expression levels of cytokinin oxidase/dehydrogenase genes accompanied cyclic dehydration (Supplemental Table S4), which may lead to a decrease in endogenous cytokinin levels and, therefore, better adaptation to stresses. Our results in rose share the common feature with other plants that reduction of active cytokinin content by stimulation of cytokinin degradation contributes to increased drought stress tolerance (for review, see Ha et al., 2012). Changes in cell wall organization are also strongly implicated in the dehydration response of rose petals. This confirms results from other studies; for example, the processes of cell wall modification, cell wall loosening, cell wall organization and biogenesis, and plant-type cell wall organization are associated with the drought responses of poplar (Street et al., 2006), C. plantagineum (Rodriguez et al., 2010), and chickpea (Molina et al., 2008).
Our analyses suggest that rose petal dehydration is accompanied by a number of other biological processes commonly reported as plant responses to WDS, including response to water, defense response, and response to stimulus (Supplemental Tables S4 and S5). It appears that dehydration may trigger defense reactions that enhance petal cells’ ability to restrict damage and tolerate stress. Our data suggest that roses not only share similar biological processes with model and crop plants responsive to the WDS but also have species-, stage-, or tissue-specific GO categories.
RhNAC2 and RhEXPA4 Increase Tolerance to Dehydration in Rose Petals
The indicators of WDS tolerance reported so far include the survival rate of seedlings (Tran et al., 2004; Lu et al., 2007), wilting degree (Gao et al., 2010), maintenance of green leaves (Huang et al., 2011), biomass of roots and leaves (Nelson et al., 2007; Yu et al., 2008), transpiration rate (Yoo et al., 2010), and photosynthesis rate (Nelson et al., 2007). For cut flowers, a consumer quality requirement is that the flowers open normally, gradually, and sufficiently from the harvested bud stage after postharvest handling. In this study, dehydration tolerance of rose flowers is defined as the ability of the dehydrated flowers to open fully during rehydration. Since flower opening depends mainly on petal expansion, we tested the dehydration tolerance of rose petals by examining the expansion of intact petals or petal discs after rehydration. In VIGS experiments, we used the width and length of petals as tolerance indicators for intact petals and the disc expansion as a tolerance indicator for petal discs.
NAC proteins, one of the largest families of plant-specific transcription factors, have been identified and functionally characterized in Arabidopsis, rice, soybean, and wheat (Tran et al., 2010). Based on sequence alignments and phylogenetic analyses, NACs can be classified into subgroups with different functions in plant growth, development, and senescence (Tran et al., 2004; Olsen et al., 2005; Le et al., 2011). Previous reports have provided a close match between amino acid sequence and function for a range of NAC proteins (Fang et al., 2008; Zhang et al., 2008), so phylogenetic analysis of amino acid sequences can be used systematically to predict the functions for new NAC members (Le et al., 2011).
One group of NACs targets a DNA-binding site termed a drought-responsive NAC recognition sequence, which contains a core CACG motif. This group clusters in the stress-responsive clades identified by phylogenetic analysis. Group members responsive to drought or dehydration were identified in Arabidopsis (Tran et al., 2004), rice (Hu et al., 2006, 2008; Nakashima et al., 2007), cotton (Gossypium hirsutum; Meng et al., 2009), and soybean (Le et al., 2011). Overexpression of WDS-responsive NACs conferred drought tolerance and resulted in morphological, physiological, and molecular adjustments to stress conditions in transgenic Arabidopsis, rice, and wheat (Tran et al., 2004; Hu et al., 2006; Zheng et al., 2009; Jeong et al., 2010; Xue et al., 2011). Our study indicates a similar role for RhNAC2 in the dehydration response of rose petals (Fig. 4). Silencing this gene resulted in a significant increase in sensitivity to WDS and reduced petal expansion (Fig. 6); overexpressing it in Arabidopsis conferred significant drought tolerance (Fig. 7). Unlike other NACs involved in stress responses (Le et al., 2011), RhNAC2 appears to be a member of a development-related clade of the phylogenetic tree. Members of this clade and homologs of RhNAC2, such as CUC in Arabidopsis, NAM in petunia (Petunia hybrida), CUP in snapdragon (Antirrhinum majus), and NAM/CUC in tomato, play an evolutionarily conserved role mainly in shoot apical meristem function and organ separation (for review, see Hasson et al., 2011).
Genome-wide analysis of direct targets of a VASCULAR-RELATED NAC DOMAIN transcription factor, VND7, reveals that this protein directly activates the expression of a number of genes involved in secondary wall biosynthesis and cell wall modification (Zhong et al., 2010). Based on this relationship between dehydration tolerance and cell wall development, we hypothesize that RhNAC2 is involved in dehydration tolerance as one of key regulators in the process of cell wall metabolism under stress conditions.
Expansins are cell wall proteins that induce pH-dependent cell wall extension and stress relaxation in a characteristic and unique manner by disrupting hydrogen bonds between cellulose microfibrils and matrix polymers, thus increasing the flexibility of the cell wall (Sampedro and Cosgrove, 2005). Expansin proteins play a role in cell enlargement, pollen tube invasion of the stigma, cell wall disassembly during fruit ripening and softening, organ abscission, leaf organogenesis, and response to environmental stimuli (Cosgrove, 2000; Li et al., 2003). In ornamental crops, expansins have been reported to be involved in development-related processes, such as opening and senescence of flowers of M. jalapa (Gookin et al., 2003), expansion of all floral tissues and leaves in gladiolus (Gladiolus gandavensis; Azeez et al., 2010), and petal growth and development during carnation flower opening (Harada et al., 2011). Three α-expansin genes (RhEXPA1–RhEXPA3) in roses are reported to be mainly involved in expansion growth of rose petals (Takahashi et al., 2007; Yamada et al., 2009).
Increasing evidence suggests the involvement of expansins in WDS tolerance in plants such as maize (Wu et al., 2001; Zhu et al., 2007), C. plantagineum (Jones and McQueen-Mason, 2004), and wheat (Li et al., 2011). Generally, WDS does harm to the cells through increasing the rigidity of the cell wall shrinkage in plants. Under drought conditions, cell wall extensibility in Arabidopsis leaves increased. This was a response that was thought to prevent damage that might result from the folding of rigid cell walls as cell volume decreases (Jones and McQueen-Mason, 2004). Expansins, as well as glucanase and pectin-modifying enzymes, confer this capacity to remodel cell wall composition and to maintain cell wall flexibility during WDS (Lu and Neumann, 1998; Rodriguez et al., 2010; Li et al., 2011). We identified eight expansin genes up-regulated during cyclic dehydration of rose petals (Table II). The results of silencing and overexpression experiments indicate that RhEXPA4, like RhNAC2, is involved in dehydration tolerance of rose petals.
Since RhNAC2 is a homolog of SND1 and VND7, which are the master regulator genes of secondary wall biosynthesis involved in cell wall modifications (Zhong et al., 2010), we wondered if RhNAC2 was a regulator of cell wall-related genes in rose petals. We noted that RhNAC2 was strongly up-regulated (after 12–24 h of dehydration) and that RhEXPA4 expression levels also increased quickly (Fig. 4). Although RhEXPA4 expression increased earlier than that of RhNAC2, this may be attributed to other putative regulators of RhEXPA4, whose promoter (accession no. JN903506) contains other cis-elements in addition to the NAC protein-targeted core sequence. Silencing RhNAC2 in rose petals reduced the expression of RhEXPA4 (Fig. 8A). RhEXPA4 is a close homolog of AtEXPA1 and AtEXPA10 in Arabidopsis (Supplemental Fig. S5), and RhNAC2 overexpression in Arabidopsis enhanced the expression of cell wall-related genes, including seven expansin family genes. We obtained additional evidence of the link between RhNAC2 and RhEXPA4 through an assay of GUS activity in protoplasts and EMSA (Fig. 8). Taken together, these data are strong evidence that RhNAC2 and RhEXPA4 are involved in petal cell expansion and confer the dehydration tolerance in rose petals and that RhNAC2 acts as a potential regulator of RhEXPA4.
Concerning functional mechanisms of RhNAC2 and RhEXPA4 in rose leaf, RhNAC2 probably does not regulate RhEXPA4 expression under dehydration, because the RhNAC2 gene was induced by dehydration in rose leaf but RhEXPA4 was not (Supplemental Fig. S6). However, 35S:RhEXPA4 transgenic Arabidopsis showed enhanced drought tolerance at the vegetative stage (Fig. 7), suggesting that RhEXPA4 may play roles in drought stress response in Arabidopsis young seedlings. Thus, our data here are not sufficient to clearly illustrate the roles of RhNAC2 and RhEXPA4 in rose leaf under dehydration.
EXPANSIN proteins could disrupt hydrogen bonds between cellulose microfibrils and matrix polymers (Sampedro and Cosgrove, 2005). Thus, overexpression of EXPANSIN may break the elaborate microtubule arrays, cellulose depositions, and cell wall thickenings that are required for the development of stomatal pores and their adjacent cells during stomatal morphogenesis (for review, see Nadeau and Sack, 2002). RhEXPA4 perhaps influenced stomatal development through affecting cell wall structures. Our results showed that the constitutive RhEXPA4 overexpression contributed to lower stomatal density in the 35S:RhEXPA4 transgenic plants than 35S:RhNAC2 plants (Fig. 7C). At present, it is not clear that the lower stomatal density found in 35S:RhEXPA4 transgenic plants may be contributed by the different expression levels of EXPANSIN in these two transgenic plants or may be due to stronger potency of RhEXPA4 in 35S:RhEXPA4 transgenic plants than its Arabidopsis counterparts in the 35S:RhNAC2 plants. We also could not rule out the possibility that this may be caused by another mechanism independently of RhNAC2.
We also noticed that RhNAC2 overexpressors with higher stomatal density showed stronger tolerance than RhEXPA4 overexpressors (Fig. 7). This may be because RhNAC2, as a transcription factor, could regulate some stress-related genes (Supplemental Table S7). These genes may contribute to the higher drought tolerance in RhNAC2 overexpressors from a broad range of morphological and physiological mechanisms. For the analysis reported here, we selected RhNAC2 and RhEXPA4 from a number of NAC and EXPANSIN genes that were up-regulated during rose petal dehydration (Fig. 3B). There may be functional redundancy among NAC family members (Taoka et al., 2004; Ko et al., 2007; Zhong et al., 2007; Yoon et al., 2008) and expansins (Choi et al., 2006). On the other hand, mutant phenotypes sometimes result from the loss of function of key NAC genes (Kim et al., 2006; Mitsuda et al., 2007) or expansins (Cho and Cosgrove, 2000). It would be worthwhile in future studies to examine the contributions of the other NAC and EXPANSIN genes we identified in the response of rose petals to dehydration.
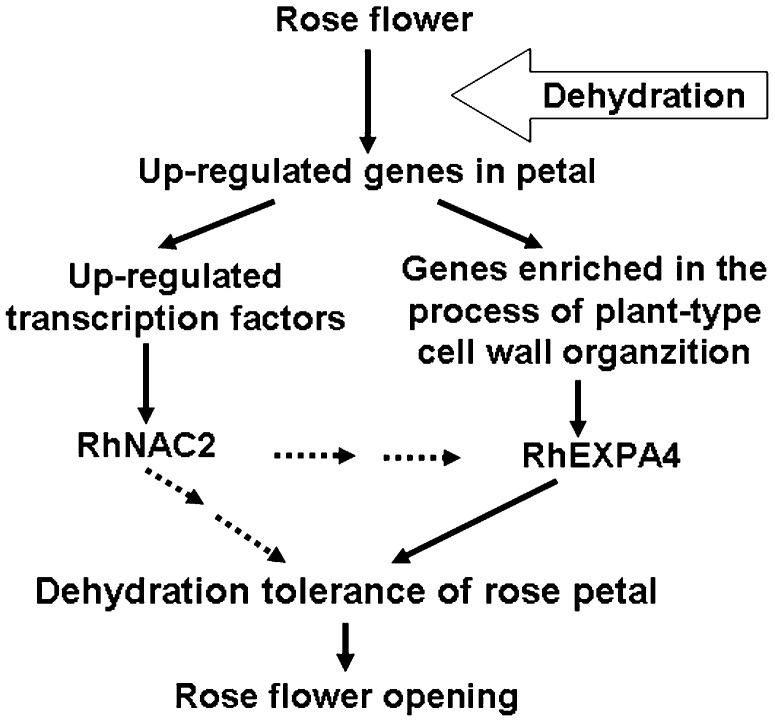
Based on the results described above, we propose a hypothetical model for the functions of RhNAC2 and RhEXPA4 during dehydration of expanding rose petals (Fig. 9). Dehydration inhibits petal expansion and results in abnormal flower opening. Rose petals adapt to dehydration through regulating the expression of dehydration-responsive genes. Overexpression of RhNAC2, one of the up-regulated transcription factors, improves petal expansion. RhEXPA4 is closely involved in petal expansion, and RhNAC2 may function as one of the regulators of RhEXPA4 gene expression in the dehydration response.
Figure 9.
Schematic model describing the involvement of RhNAC2 and RhEXPA4 in dehydration tolerance of rose petals. The up-regulated genes in rose petals under dehydration are obtained by SSH and microarray hybridization. Dehydration causes the inhibition of petal expansion and results in abnormal flower opening. Rose petals adapt and tolerate dehydration through regulating the dehydration-related gene expression and biological processes. Among the up-regulated transcription factor genes, increased RhNAC2 expression improves petal expansion. A biological process, plant type cell wall organization, is closely involved in petal expansion, and RhEXPA4 plays a key role in this process. RhNAC2 may function as a potential regulator gene of RhEXPA4 in response to dehydration during petal expansion. In a word, two development-related proteins, RhNAC2 and RhEXPA4, are involved in dehydration tolerance of rose petal during flower opening.
MATERIALS AND METHODS
Plant Materials
Cut rose flowers (Rosa hybrida ‘Samantha’) were harvested at opening stage 2 of flower from a local commercial greenhouse and placed immediately in water. Flower-opening stages were defined as described by Ma et al. (2005). The flowers were delivered to the laboratory within 1 h of harvest. Their stems were recut to 25 cm under water, and the flowers were held in deionized water until needed.
Dehydration Treatment of Rose Flowers
Flowers were placed horizontally on the bench for 6, 12, 24, or 36 h in a climate-controlled room at 25°C, 40% to 50% relative humidity, and a continuous light with intensity of 140 µmol m−2 s−1. The sepals, petals, stamens, gynoecia, and receptacle of flowers were taken separately and frozen into −80°C for RNA isolation and construction of the cDNA library.
For microarray hybridization, the flowers were subjected to a cyclic dehydration treatment in the same climate-controlled room. The flowers were held in air for 24 h (first dehydration), recut, removing 1 cm from the base under water, and placed in water for 12 h (rehydration), then removed from the water and held in air for a further 12 h (second dehydration). Control flowers remained in water throughout the experiment. The outermost whorl of petals was collected and frozen to −80°C for RNA isolation.
Measurement of Fresh Weight and Water Potential
The weight of each harvested flower was measured at the start and at intervals throughout the cyclic dehydration experiment. The water potential of cut flowers was measured at intervals using a pressure chamber as described by Jin et al. (2006).
Construction of a Subtracted cDNA Library
To construct the subtracted cDNA library, we used RNA from dehydrated flowers as “tester” and the control flowers as “driver.” Flowers were separated into five distinct parts: sepals, petals, stamens, gynoecia, and receptacles. Total RNA of sepals, petals, stamens, and receptacles was extracted using the hot borate method described by Ma et al. (2005), and total RNA from gynoecia was extracted using the hot phenol method described by Xue et al. (2008). We purified mRNA from total RNA using an Oligotex mRNA Purification Kit (Qiagen).
The subtracted cDNA library was constructed using the PCR-Select cDNA Subtraction Kit (Clontech) following the manufacturer’s instructions. The tester cDNAs were reverse transcribed from 2 µg of mRNA mixed from five distinct parts of dehydrated rose flowers in the ratio of 4 (petal):1 (sepal):1 (receptacle):2 (gynoecia):2 (stamen). The RNAs of different parts of rose flower were mixed equally from 6-, 12-, 24-, and 36-h dehydration samples. The driver cDNAs were obtained from control flowers using the same method. Both tester and driver cDNAs were digested with RsaI, and the tester cDNAs were then ligated to the adapters for forward subtraction. Two rounds of hybridization and PCR amplification were conducted to normalize and enrich the library with differentially expressed cDNAs. Products of the secondary PCR were directly inserted into pGEM T-Easy Vector (Promega) and transformed into Escherichia coli DH5α cells. Recombinant colonies were collected to establish the forward subtracted cDNA library. Colonies were randomly picked from the subtractive cDNA library for sequence analysis. A total of 7,071 independent clones were successfully sequenced. All 7,071 sequences have been submitted to GenBank with accession numbers JK616121 to JK623191. EST assembly was performed to obtain uniESTs using the iAssembler program (http://bioinfo.bti.cornell.edu/tool/iAssembler) with a minimum overlap of 40 bp and a minimum identity of 97% compared with the National Center for Biotechnology Information and 454 sequence libraries (J. Gao, N. Ma, and H. Pei, unpublished data).
Preparation of cDNA Microarray Slides
Bacterial clones containing all uniESTs from the subtracted cDNA library were selected and distributed individually onto 96-well plates for cDNA amplification by PCR. The nested PCR primer 5′-TCGAGCGGCCGCCCGGGCAGGT-3′ was used as the forward primer and 5′-AGCGTGGTCGCGGCCGAGGT-3′ was used as the reverse primer. The 50-µL reaction contained 34 µL of sterile water, 5 µL of 10× Taq buffer, 2 µL of each primer (10 µm each), 4 µL of deoxyribonucleotide triphosphate mix (2.5 mm each), 5 units of Taq polymerase (TaKaRa), and 1 µL of bacterial culture as template. PCR was performed as follows: at 95°C for 5 min, then 35 cycles at 95°C for 30 s, 68°C for 30 s, and 72°C for 1 min, then a final extension at 72°C for 10 min. PCR products were precipitated by the addition of 100 µL of anhydrous ethanol and resuspended in 40 µL of sterile water for preparation of the array. One-microliter aliquots of the resuspended products were analyzed by electrophoresis on 1% agarose gels to verify the quality and quantity of the cDNA. The remaining PCR products were spotted onto amino-silaned glass slides (CapitalBio). All uniESTs were spotted two times onto the microarray in different subgrids.
Preparation of Labeled DNA, Hybridization, and Scanning
The petal RNA samples were used to generate Cy3- and Cy5-labeled cDNA probes using the SuperScript direct cDNA Labeling System (Invitrogen) following the manufacturer’s instructions. Cy3 and Cy5 probes were mixed in equal quantities and purified on spin columns (Minipore) to a final volume of 20 µL. Prehybridization, hybridization, and posthybridization washes were carried out according to the UltraGAPS coated slides instruction manual (Corning). After the washes, the slides were scanned using an Axon Genepix 4000B scanner (Molecular Devices) at a resolution of 10 μm. Laser and photomultiplier tube voltages were adjusted manually to minimize background and the number of spots that had saturated signal values. For biological repeats, the RNA samples used for preparations of labeled cDNAs were prepared from six replicate untreated and treated petal samples.
Data Processing and Analysis
Spot intensities were quantified using GenePix pro 6.0 (Axon Instruments). Spots with mean signal intensities less than local background intensities plus 2 sd from the local background in both channels were regarded as empty spots and not included in the downstream statistical analysis. A Print-tip Lowess Normalization strategy was applied to normalize the ratio values for each array using the Bioconductor marray R package (Yang et al., 2002). The significance of gene expression changes between stressed and control plants was identified using the Patterns from Gene Expression package (Grant et al., 2005). Genes with corrected P < 0.05 and fold change no less than 2 were considered to be differentially expressed genes. Identification of highly enriched GO terms and functional classification of differentially expressed genes were performed using the Plant MetGenMAP system (http://bioinfo.bti.cornell.edu/cgi-bin/MetGenMAP/home.cgi; Joung et al., 2009). All microarray data were deposited at the Gene Expression Omnibus-National Center for Biotechnology Information with the following accession numbers: GSE33217 and GPL14787.
Silencing of RhNAC2 and RhEXPA4 in Rose Petals by VIGS
The silencing of RhNAC2 and RhEXPA4 in petals by VIGS was performed according to the procedures described by Ma et al. (2008) with some modifications. For the construction of the RhNAC2 VIGS vector, we used a 351-bp fusion fragment combining 249 bp from the 5′ end of the gene with 102 bp from its 3′ end. For the construction of the RhEXPA4 VIGS vector, we used a 302-bp fragment from the 3′ end of the gene. All primers used are listed in Supplemental Table S8. The two resulting products were cloned into pTRV2 to form the pTRV2-RhNAC2 and pTRV2-RhEXPA4 constructs, respectively. The two constructs, as well as the pTRV1 and pTRV2 vectors, were transformed into Agrobacterium tumefaciens GV3101 by electroporation. The transformed A. tumefaciens lines were grown at 28°C in Luria-Bertani medium supplemented with 10 mm MES, 20 mm acetosyringone, and 50 mg L−1 kanamycin for about 24 h. A. tumefaciens cells were harvested and suspended in the infiltration buffer (10 mm MgCl2, 200 mm acetosyringone, and 10 mm MES, pH 5.6) to a final optical density at 600 nm of around 1.5. A mixture of A. tumefaciens cultures containing empty or silencing pTRV1 and pTRV2 vectors in a ratio of 1:1 (v/v) was placed at room temperature for 4 h before vacuum infiltration into petal tissues.
Rose petals were collected from the outermost whorl of the flowers at flower stage 2. One-centimeter-diameter discs were taken from the center of the petals with a hole punch. For vacuum infiltration, rose petals or discs were placed into the bacterial suspension solution and infiltrated under vacuum at 0.5 MPa for 15 s. After release of the vacuum, petals and discs were washed in deionized water and kept in deionized water for 3 d at 8°C, then at 23°C for 1 d. Petals were dehydrated for 12 h and then placed in deionized water for rehydration for 48 h. The width, length, and fresh weight of all petals were determined at intervals. The petals were sampled after the 12-h dehydration to determine the VIGS efficiency using RT-PCR. We classified VIGS-silenced petals into three groups, completely silenced petals, partially silenced petals, and nonsilenced petals, based on the expression level of the VIGS-targeted gene determined by RT-PCR. Widths and fresh weights of petals in each group were determined according to image records or measurements of individual petals at intervals.
After the VIGS procedure, petal discs were dehydrated for 9 h and then rehydrated for 48 h. The discs were monitored by using a digital camera (Nikon D200). The discs were regarded as recovered when they expanded fully. The percentage of recovered discs was determined at intervals. Silencing experiments with petal discs were repeated five times using at least 90 discs per gene in each repetition. Student’s t test (P < 0.05) was used for statistical analysis of the recovery data.
Semiquantitative RT-PCR Analysis
To verify microarray results, eight dehydration-responsive genes were selected and subjected to semiquantitative RT-PCR analysis. Total RNAs were isolated from rose petals under the cyclic dehydration process as well as under control conditions following the procedures described by Xue et al. (2008). cDNAs were synthesized from 1 µg of total RNA using Moloney murine leukemia virus reverse transcriptase (Promega). The rose Ubiquitin1 gene (Ubi1; GenBank accession no. JK622648) was used as the internal control. PCRs were performed with 25 to 29 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min, using 1 µL of cDNA as the template. PCR products were analyzed on 1.0% agarose gels and imaged using the AlphaImager 2200 (α Innotech).
To determine the expression levels of RhNAC2 and RhEXPA4 in silenced petals, we isolated total RNAs from the petals using the hot borate method (Wan and Wilkins, 1994). cDNA synthesis and RT-PCR were performed as described above. The primers were designed outside the region used for VIGS to avoid amplification of RNA from the silencing vectors (Supplemental Table S8). The rose Ubi1 gene was used as the internal standard.
To determine the expression levels of RhNAC2, RhEXPA4, and Arabidopsis (Arabidopsis thaliana) expansin genes in transgenic Arabidopsis plants, we extracted total RNA from leaves collected from 4-week-old RhNAC2- and RhEXPA4-overexpressing transgenic plants (T2 generation) and wild type (Columbia ecotype) plants using the Trizol agent (Invitrogen). cDNA synthesis and RT-PCR were performed as described above. The Arabidopsis Actin2 gene (GenBank accession no. NM_112764) was used as the internal control. PCR amplification of the RhNAC2, RhEXPA4, and Actin2 genes was performed using 28, 28, and 22 cycles, respectively. All primers are listed in Supplemental Table S8.
Transactivation Assay of RhNAC2 in Yeast Cells
For transactivation assay in yeast (Saccharomyces cerevisiae), different portions of RhNAC2 to be examined were PCR amplified using forward primers with the SalI site at the 5′ end and reverse primers with the PstI site at the 5′ end. The amplified fragments were digested with SalI and PstI and inserted in frame into the SalI and PstI sites of pBD (Clontech) to make expression vectors. The proteins fused with pBD-GAL4 are as follows: pBD-RhNAC2F (1–354 of RhNAC2), pBD-RhNAC2N (1–173 of RhNAC2), pBD-RhNAC2C (173–354 of RhNAC2), pBD-RhNAC2C1 (193–354 of RhNAC2), pBD-RhNAC2C2 (214–354 of RhNAC2), pBD-RhNAC2C3 (240–354 of RhNAC2), and pBD-RhNAC2C4 (281–354 of RhNAC2). The primers are listed in Supplemental Table S8.
According to the protocol of the manufacturer (Stratagene), expression vectors (pBD-X), with pBD-GAL4 as the positive control or pBD vector as the negative control, were introduced into the yeast host strain YRG-2, containing the His-3 and LacZ reporter genes with three and four GAL4-binding elements in the promoters, respectively.
All transformants were streaked on SD-Trp and SD-His plates. The transcriptional activation activities were evaluated by growth of the colonies. Transfected yeast cells were also transferred onto filter paper and incubated at 30°C for 0.5 to 8 h in the presence of 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside acid to check the β-galactosidase activity by monitoring the generation of blue color. Quantitation was performed using o-nitrophenyl β-d-galactopyranoside to test β-galactosidase activity as described in the yeast protocols handbook (PT3024-1; Clontech). Three transformants from each transfection were selected randomly and subjected to the o-nitrophenyl β-d-galactopyranoside assay. To monitor fusion protein production in yeast, total protein (0.3 mL of yeast culture equivalent) was extracted and size fractionated by 10% SDS-PAGE. Electrophoresed protein samples were transferred to nitrocellulose membrane and blocked with 3% (w/v) nonfat dry milk before incubation with rabbit anti-GAL4 DNA-BD antibody (Sigma-Aldrich) to detect pBD-X fusion proteins. Anti-rabbit IgG alkaline phosphatase (Sigma) was used as the secondary antibody and detected using alkaline phosphatase substrate.
Arabidopsis Transformation and Drought Tolerance Determination
To transform Arabidopsis with the RhNAC2 and RhEXPA4 genes, we first amplified the open reading frames of RhNAC2 and RhEXPA4 using gene-specific primers providing XbaI and SacI sites (Supplemental Table S8). The PCR products were digested with XbaI and SacI and inserted into the pBI121 vector. The transformed vectors were introduced into A. tumefaciens strain GV3101. Arabidopsis plants were transformed using the flower dip method described by Clough and Bent (1998).
Seeds were surface sterilized using 5% NaClO for 10 min, washed off and stratified at 4°C for 3 d, and then sown on Murashige and Skoog (MS) medium to allow germination. Eight-day-old seedlings were then transplanted into 7-cm pots filled with a 1:1 mixture of peat and vermiculite in a controlled room at 23°C ± 1°C, 16/8-h day/night period, and 80 to 100 µmol m−2 s−1 illumination. The plants were watered every 4 d. The T0 seeds collected from transformed plants were germinated on MS medium containing 50 mg L−1 kanamycin to screen for kanamycin resistance. T1 seeds obtained from individual T0 plants were planted to obtain T2 seeds. To ensure homozygous T2 seedlings, the T2 seeds obtained from individual T1 seedlings were further sown on the MS medium containing 50 mg L−1 kanamycin, and only lines that germinated completely and grew normally were chosen. The homozygous T2 seeds were stored for further use. Based on the overexpression levels of RhNAC2 and RhEXPA4 in Arabidopsis leaves, we chose three lines each for further analysis. The morphological phenotypes of RhNAC2- or RhEXPA4-overexpressing plants were observed under normal growth conditions. Primary root length and lateral root number of 10-d-old seedlings were determined on MS medium. Water loss assay of aerial parts of 4-week-old RhNAC2- and RhEXPA4-overexpressed Arabidopsis was conducted on the laboratory bench at 23°C to 25°C, 30% to 40% relative humidity, and 40 µmol m−2 s−1 light intensity. The percentage loss of fresh weight was calculated on the basis of the initial weight of the plants. Leaf stomatal density was observed with a light microscope using the leaf surface imprint method (Yoo et al., 2010).
For the evaluation of drought tolerance, 8-d-old T2 transgenic and control Arabidopsis seedlings were transplanted into 7-cm pots filled with 100 g of substrate and five seedlings per pot and grown under the conditions described above. The 10-d-old plants were watered fully, then water was withheld for 20 d. The plants were then rewatered to determine recovery. The phenotype of the plants was monitored by digital photography (Nikon), and survival of the plants was scored 10 d after rewatering. All experiments were repeated at least three times, and more than 40 plants from at least three lines were used in each comparison.
Purification of Recombinant Protein and EMSA
The N terminus (N1–173) of RhNAC2, which contains the DNA-binding domain, was fused in frame with GST and expressed in E. coli BL21. The fusion protein was induced by 0.2 mm isopropyl β-d-1-thiogalactopyranoside, and the cultures were incubated at 28°C for 6 h. The recombinant protein was purified by GST-agarose affinity chromatography. The EMSA was carried out using the LightShift Chemiluminescent EMSA Kit (Pierce; 20148) according to the manufacturer’s instructions. The fragment derived from the −216 to −171 region of the RhEXPA4 promoter, 5′-CAAAACCACGCGCTTTCTCCCTTCAAAACCGCCCTTCCACGCTCA-3′, was biotin labeled as a probe; it contains two (underlined) target binding sequences for NAC proteins (Olsen et al., 2005). The same unlabeled DNA fragment was used as a competitor in the assay. The probes were incubated with the fusion protein at room temperature for 25 min in a binding buffer (10× concentration: 100 mm Tris, 500 mm KCl, and 10 mm dithiothreitol, pH 7.5). Each 20-µL binding reaction contained 0.2 pmol of biotin probe and 2 µg of fusion protein; the reaction was supplemented with 1 µg of poly(dI·dC) to minimize nonspecific interactions. The reaction products were analyzed by 5% native PAGE. Electrophoresis was performed at 120 V for approximately 35 min at 4°C in 0.5× Tris-borate/EDTA buffer. DNA fragments were transferred from the gel to nitrocellulose membrane with 0.5× Tris-borate/EDTA at 380 mA (approximately 100 V) for 30 min at 4°C. After cross linking, the membrane was incubated in the blocking buffer for 15 min with gently shaking and then transferred to conjugate/blocking buffer by mixing 16.75 µL of stabilized streptavidin-horseradish peroxidase conjugate with 5 mL of blocking buffer. The membrane was washed four times, each for 5 min, with a 1× washing buffer. Biotin-labeled DNA was detected by the chemiluminescence method according to the manufacturer’s protocol.
Construction of the RhEXPA4 Promoter-GUS Fusion and Transient Expression Assays
The 1.6-kb promoter fragment (accession no. JN903506) upstream of the RhEXPA4 coding region was amplified from rose genomic DNA and cloned into a modified pBI121 binary vector to replace the cauliflower mosaic virus 35S promoter. Arabidopsis mesophyll protoplasts were transformed with the resulting vector (pBI121-ProRhEXPA4::GUS). The open reading frame of RhNAC2 was amplified and cloned into a modified pUC19 containing GUS. The primers used are listed in Supplemental Table S8. The resulting pUC19-RhNAC2::GUS was used as an effector plasmid, and the pUC19-GUS plasmid was used as a control. Isolation of Arabidopsis mesophyll protoplasts from pBI121-ProRhEXPA4::GUS-transformed plants, transformation of protoplasts, and GUS activity assays were carried out as described previously (Jefferson, 1987; Sheen, 2001).
ATH1 Microarray Analysis
Total RNAs were prepared from the aerial portion of 3-week-old Arabidopsis plants of two overexpressing lines, RhNAC2-4 and RhNAC2-9, using the Trizol agent (Invitrogen). The vector-only plant was used as a control. Twenty micrograms of total RNA was used for microarray analysis. We used the Affymetrix ATH1 arrays (Affymetrix), and probe labeling, hybridization, and scanning for microarray analysis were conducted according to the manufacturer’s instructions (Affymetrix; http://www.affymetrix.com). The raw microarray data were normalized (GeneChip Robust Multiarray Averaging) and analyzed using the Genespring GX package (Agilent Technologies). Seven expansin genes were confirmed by RT-PCR: AtEXPA1 (The Arabidopsis Information Resource [TAIR] accession no. AT1G69530), AtEXPA3 (TAIR accession no. AT2G37640), AtEXPA8 (TAIR accession no. AT2G40610), AtEXPA11 (TAIR accession no. AT1G20190), AtEXPA16 (TAIR accession no. AT3G55500), AtEXPA20 (TAIR accession no. AT4G38210), and AtEXLA1 (TAIR accession no. AT3G45970).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JN857363 (RhNAC2) and JN857364 (RhEXPA4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Length distributions of rose flower ESTs and assembled sequences.
Supplemental Figure S2. Differentially expressed genes identified by SSH and microarray analysis.
Supplemental Figure S3. All 27 groups of genes classified by their expression pattern during dehydration and rehydration.
Supplemental Figure S4. Analysis of the RhNAC2 protein sequence.
Supplemental Figure S5. Analysis of the RhEXPA4 protein sequence.
Supplemental Figure S6. Expression of RhNAC2 and RhEXPA4 in different floral organs and leaves.
Supplemental Figure S7. Nuclear localization assay of RhNAC2.
Supplemental Figure S8. Transcriptional activation of RhNAC2 fragments in Arabidopsis protoplast.
Supplemental Figure S9. Morphological phenotypes of Arabidopsis plants overexpressing RhNAC2 or RhEXPA4.
Supplemental Figure S10. Root growth in MS medium of Arabidopsis plants overexpressing RhNAC2 or RhEXPA4.
Supplemental Figure S11. Effect of different concentrations of ABA on stomatal aperture of wild-type and transgenic Arabidopsis plants overexpressing RhNAC2 and RhEXPA4.
Supplemental Table S1. List of genes regulated by dehydration and rehydration in rose petals.
Supplemental Table S2. Genes comprising the induced-repressed-induced and induced-invariable-induced groups responding to cyclic dehydration.
Supplemental Table S3. Transcription factors regulated by dehydration in rose petals.
Supplemental Table S4. GO terms significantly overrepresented for UniProt identifiers under biological processes with up-regulated genes in response to dehydration in rose petals.
Supplemental Table S5. GO terms significantly overrepresented for UniProt identifiers under biological processes with down-regulated genes in response to dehydration in rose petals.
Supplemental Table S6. Petal recovery ability in width and fresh weight of RhNAC2- or RhEXPA4-silencing petals.
Supplemental Table S7. Up-regulated genes in Arabidopsis transgenic plants overexpressing RhNAC2.
Supplemental Table S8. Primers used in RT-PCR analysis and vector construction.
Acknowledgments
We are grateful to the laboratory of Dr. Zhangjun Fei of Boyce Thompson Institute for Plant Research, Cornell University, for kind help with bioinformatic analysis and to the laboratory of Dr. Zhizhong Gong of College of Biological Sciences, China Agricultural University, for technical support in the EMSA experiments. We also thank Dr. Cai-Zhong Jiang and Dr. Michael Reid of the University of California, Davis, for critical review of the manuscript.
Glossary
- WDS
water deficit stress
- GO
Gene Ontology
- SSH
suppression subtractive hybridization
- RT
reverse transcription
- UTR
untranslated region
- SD
synthetic dextrose
- VIGS
virus-induced gene silencing
- ABA
abscisic acid
- EMSA
electrophoresis mobility shift assay
- GST
glutathione S-transferase
- MS
Murashige and Skoog
- TAIR
The Arabidopsis Information Resource
- cDNA
complementary DNA
- uniEST
unique EST
- TRV
tobacco rattle virus
References
- Azeez A, Sane AP, Tripathi SK, Bhatnagar D, Nath P. (2010) The gladiolus GgEXPA1 is a GA-responsive alpha-expansin gene expressed ubiquitously during expansion of all floral tissues and leaves but repressed during organ senescence. Postharvest Biol Technol 58: 48–56 [Google Scholar]
- Bray EA. (2004) Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J Exp Bot 55: 2331–2341 [DOI] [PubMed] [Google Scholar]
- Channelière S, Rivière S, Scalliet G, Szecsi J, Jullien F, Dolle C, Vergne P, Dumas C, Bendahmane M, Hugueney P, et al. (2002) Analysis of gene expression in rose petals using expressed sequence tags. FEBS Lett 515: 35–38 [DOI] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. (2000) Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 9783–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D, Cho HT, Lee Y. (2006) Expansins: expanding importance in plant growth and development. Physiol Plant 126: 511–518 [Google Scholar]
- Choi D, Lee Y, Cho HT, Kende H. (2003) Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell 15: 1386–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JA, Wilson IW, Llewellyn DJ, Dennis ES. (2009) The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol 149: 1724–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cohen D, Bogeat-Triboulot MB, Tisserant E, Balzergue S, Martin-Magniette ML, Lelandais G, Ningre N, Renou JP, Tamby JP, Le Thiec D, et al. (2010) Comparative transcriptomics of drought responses in Populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genomics 11: 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmer TD, Peeters AJM, Wagemaker CAM, Vriezen WH, Ammerlaan A, Voesenek LACJ. (2004) Expression of α-expansin genes during root acclimations to O2 deficiency in Rumex palustris. Plant Mol Biol 56: 423–437 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2000) Loosening of plant cell walls by expansins. Nature 407: 321–326 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Debener T, Linde M. (2009) Exploring complex ornamental genomes: the rose as a model plant. Crit Rev Plant Sci 28: 267–280 [Google Scholar]
- Ding XH, Cao YL, Huang LL, Zhao J, Xu CG, Li XH, Wang SP. (2008) Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20: 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergen NZ, Thimmapuram J, Bohnert HJ, Budak H. (2009) Transcriptome pathways unique to dehydration tolerant relatives of modern wheat. Funct Integr Genomics 9: 377–396 [DOI] [PubMed] [Google Scholar]
- Fang YJ, You J, Xie KB, Xie WB, Xiong LZ. (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics 280: 547–563 [DOI] [PubMed] [Google Scholar]
- Fleming AJ, McQueen-Mason S, Mandel T, Kuhlemeier C. (1997) Induction of leaf primordia by the cell wall protein expansin. Science 276: 1415–1418 [Google Scholar]
- Fudali S, Janakowski S, Sobczak M, Griesser M, Grundler FMW, Golinowski W. (2008) Two tomato alpha-expansins show distinct spatial and temporal expression patterns during development of nematode-induced syncytia. Physiol Plant 132: 370–383 [DOI] [PubMed] [Google Scholar]
- Gao F, Xiong AS, Peng RH, Jin XF, Xu J, Zhu B, Chen JM, Yao QH. (2010) OsNAC52, a rice NAC transcription factor, potentially responds to ABA and confers drought tolerance in transgenic plants. Plant Cell Tissue Organ Cult 100: 255–262 [Google Scholar]
- Gong PJ, Zhang JH, Li HX, Yang CX, Zhang CJ, Zhang XH, Khurram Z, Zhang YY, Wang TT, Fei ZJ, et al. (2010) Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J Exp Bot 61: 3563–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gookin TE, Hunter DA, Reid MS. (2003) Temporal analysis of alpha- and beta-expansin expression during floral opening and senescence. Plant Sci 164: 769–781 [Google Scholar]
- Grant GR, Liu J, Stoeckert CJJ., Jr (2005) A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics 21: 2684–2690 [DOI] [PubMed] [Google Scholar]
- Guterman I, Shalit M, Menda N, Piestun D, Dafny-Yelin M, Shalev G, Bar E, Davydov O, Ovadis M, Emanuel M, et al. (2002) Rose scent: genomics approach to discovering novel floral fragrance-related genes. Plant Cell 14: 2325–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. (2012) Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci 17: 172–179 [DOI] [PubMed] [Google Scholar]
- Harada T, Torii Y, Morita S, Onodera R, Hara Y, Yokoyama R, Nishitani K, Satoh S. (2011) Cloning, characterization, and expression of xyloglucan endotransglucosylase/hydrolase and expansin genes associated with petal growth and development during carnation flower opening. J Exp Bot 62: 815–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson A, Plessis A, Blein T, Adroher B, Grigg S, Tsiantis M, Boudaoud A, Damerval C, Laufs P. (2011) Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell 23: 54–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY. (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44: 903–916 [DOI] [PubMed] [Google Scholar]
- Heinrichs F. (2008) International statistics flowers and plants. AIPH/Union Fleurs 56: 16–90 [Google Scholar]
- Hsu YF, Wang CS, Raja R. (2007) Gene expression pattern at desiccation in the anther of Lilium longiflorum. Planta 226: 311–322 [DOI] [PubMed] [Google Scholar]
- Hu HH, Dai MQ, Yao JL, Xiao BZ, Li XH, Zhang QF, Xiong LZ. (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103: 12987–12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HH, You J, Fang YJ, Zhu XY, Qi ZY, Xiong LZ. (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67: 169–181 [DOI] [PubMed] [Google Scholar]
- Huang XS, Luo T, Fu XZ, Fan QJ, Liu JH. (2011) Cloning and molecular characterization of a mitogen-activated protein kinase gene from Poncirus trifoliata whose ectopic expression confers dehydration/drought tolerance in transgenic tobacco. J Exp Bot 62: 5191–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain SS, Kayani MA, Amjad M. (2011) Transcription factors as tools to engineer enhanced drought stress tolerance in plants. Biotechnol Prog 27: 297–306 [DOI] [PubMed] [Google Scholar]
- Jefferson RA. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK. (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JS, Shan NW, Ma N, Bai JH, Gao JP. (2006) Regulation of ascorbate peroxidase at the transcript level is involved in tolerance to postharvest water deficit stress in the cut rose (Rosa hybrida L.) cv. Samantha. Postharvest Biol Technol 40: 236–243 [Google Scholar]
- Jones L, McQueen-Mason S. (2004) A role for expansins in dehydration and rehydration of the resurrection plant Craterostigma plantagineum. FEBS Lett 559: 61–65 [DOI] [PubMed] [Google Scholar]
- Joung JG, Corbett AM, Fellman SM, Tieman DM, Klee HJ, Giovannoni JJ, Fei ZJ. (2009) Plant MetGenMAP: an integrative analysis system for plant systems biology. Plant Physiol 151: 1758–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Girke T, Bray EA, Bailey-Serres J. (2004) Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Park JE, Park HY, Lim MH, Chua NH, Park CM. (2006) A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18: 3132–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Yang SH, Park AH, Lerouxel O, Han KH. (2007) ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J 50: 1035–1048 [DOI] [PubMed] [Google Scholar]
- Kumar N, Srivastava GC, Dixit K. (2008) Flower bud opening and senescence in roses (Rosa hybrida L.). Plant Growth Regul 55: 81–99 [Google Scholar]
- Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. (2011) Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res 18: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Xing SC, Guo QF, Zhao MR, Zhang J, Gao Q, Wang GP, Wang W. (2011) Drought tolerance through over-expression of the expansin gene TaEXPB23 in transgenic tobacco. J Plant Physiol 168: 960–966 [DOI] [PubMed] [Google Scholar]
- Li Y, Jones L, McQueen-Mason S. (2003) Expansins and cell growth. Curr Opin Plant Biol 6: 603–610 [DOI] [PubMed] [Google Scholar]
- Liu X, Hong L, Li XY, Yao Y, Hu B, Li L. (2011) Improved drought and salt tolerance in transgenic Arabidopsis overexpressing a NAC transcriptional factor from Arachis hypogaea. Biosci Biotechnol Biochem 75: 443–450 [DOI] [PubMed] [Google Scholar]
- Lu PL, Chen NZ, An R, Su Z, Qi BS, Ren F, Chen J, Wang XC. (2007) A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol 63: 289–305 [DOI] [PubMed] [Google Scholar]
- Lu ZJ, Neumann PM. (1998) Water-stressed maize, barley, and rice seedlings show species diversity in mechanisms of leaf growth inhibition. J Exp Bot 49: 1945–1952 [Google Scholar]
- Ma N, Cai L, Lu WJ, Tan H, Gao JP. (2005) Exogenous ethylene influences flower opening of cut roses (Rosa hybrida) by regulating the genes encoding ethylene biosynthesis enzymes. Sci China C Life Sci 48: 434–444 [DOI] [PubMed] [Google Scholar]
- Ma N, Xue JQ, Li YH, Liu XJ, Dai FW, Jia WS, Luo YB, Gao JP. (2008) Rh-PIP2;1, a rose aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiol 148: 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. (1992) Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4: 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Cosgrove DJ. (1995) Expansin mode of action on cell walls: analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol 107: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer R, Theissen G. (2011) MADS and more: transcription factors that shape the plant. Methods Mol Biol 754: 3–18 [DOI] [PubMed] [Google Scholar]
- Meng CM, Cai CP, Zhang TZ, Guo WZ. (2009) Characterization of six novel NAC genes and their responses to abiotic stresses in Gossypium hirsutum L. Plant Sci 176: 352–359 [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M. (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina C, Rotter B, Horres R, Udupa SM, Besser B, Bellarmino L, Baum M, Matsumura H, Terauchi R, Kahl G, et al. (2008) SuperSAGE: the drought stress-responsive transcriptome of chickpea roots. BMC Genomics 9: 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD. (2002) Stomatal development in Arabidopsis. The Arabidopsis Book 1: e0066, doi/10.1199/tab.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149: 88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51: 617–630 [DOI] [PubMed] [Google Scholar]
- Narsai R, Castleden I, Whelan J. (2010) Common and distinct organ and stress responsive transcriptomic patterns in Oryza sativa and Arabidopsis thaliana. BMC Plant Biol 10: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu JR, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, et al. (2007) Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA 104: 16450–16455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M, Werner T, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Kakimoto T, et al. (2011) Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23: 2169–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S. (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465: 30–44 [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K. (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Pien S, Wyrzykowska J, McQueen-Mason S, Smart C, Fleming A. (2001) Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc Natl Acad Sci USA 98: 11812–11817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu YP, Yu DQ. (2009) Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ Exp Bot 65: 35–47 [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E. (2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci USA 104: 19631–19636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MCS, Edsgärd D, Hussain SS, Alquezar D, Rasmussen M, Gilbert T, Nielsen BH, Bartels D, Mundy J. (2010) Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. Plant J 63: 212–228 [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. (2005) The expansin superfamily. Genome Biol 6: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R, Chinnappa CC, Voesenek LACJ, Pierik R. (2008) The regulation of cell wall extensibility during shade avoidance: a study using two contrasting ecotypes of Stellaria longipes. Plant Physiol 148: 1557–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafleitner R, Gutierrez Rosales RO, Gaudin A, Alvarado Aliaga CA, Martinez GN, Tincopa Marca LR, Bolivar LA, Delgado FM, Simon R, Bonierbale M. (2007) Capturing candidate drought tolerance traits in two native Andean potato clones by transcription profiling of field grown plants under water stress. Plant Physiol Biochem 45: 673–690 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Seki M, Umezawa T, Urano K, Shinozaki K. (2007) Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol 10: 296–302 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Xiang FN, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM. (2009) The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol 151: 275–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Jeong DH, Park MC, An G. (2005) Characterization and transcriptional expression of the α-expansin gene family in rice. Mol Cells 20: 210–218 [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Smith LG, Oppenheimer DG. (2005) Spatial control of cell expansion by the plant cytoskeleton. Annu Rev Cell Dev Biol 21: 271–295 [DOI] [PubMed] [Google Scholar]
- Street NR, Skogström O, Sjödin A, Tucker J, Rodríguez-Acosta M, Nilsson P, Jansson S, Taylor G. (2006) The genetics and genomics of the drought response in Populus. Plant J 48: 321–341 [DOI] [PubMed] [Google Scholar]
- Takahashi R, Fujitani C, Yamaki S, Yamada K. (2007) Analysis of the cell wall loosening proteins during rose flower opening. Acta Hortic 755: 483–488 [Google Scholar]
- Talamè V, Ozturk NZ, Bohnert HJ, Tuberosa R. (2007) Barley transcript profiles under dehydration shock and drought stress treatments: a comparative analysis. J Exp Bot 58: 229–240 [DOI] [PubMed] [Google Scholar]
- Taoka K, Yanagimoto Y, Daimon Y, Hibara K, Aida M, Tasaka M. (2004) The NAC domain mediates functional specificity of CUP-SHAPED COTYLEDON proteins. Plant J 40: 462–473 [DOI] [PubMed] [Google Scholar]
- Tommasini L, Svensson JT, Rodriguez EM, Wahid A, Malatrasi M, Kato K, Wanamaker S, Resnik J, Close TJ. (2008) Dehydrin gene expression provides an indicator of low temperature and drought stress: transcriptome-based analysis of barley (Hordeum vulgare L.). Funct Integr Genomics 8: 387–405 [DOI] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nishiyama R, Yamaguchi-Shinozaki K, Shinozaki K. (2010) Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 1: 32–39 [DOI] [PubMed] [Google Scholar]
- Tran LS, Quach TN, Guttikonda SK, Aldrich DL, Kumar R, Neelakandan A, Valliyodan B, Nguyen HT. (2009) Molecular characterization of stress-inducible GmNAC genes in soybean. Mol Genet Genomics 281: 647–664 [DOI] [PubMed] [Google Scholar]
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA 104: 20623–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas AJ, Malho R. (1997) Signal perception and transduction: the origin of the phenotype. Plant Cell 9: 1181–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff C, Bramke I, Breeze E, Thornber S, Harrison E, Thomas B, Buchanan-Wollaston V, Stead T, Rogers H. (2010) A specific group of genes respond to cold dehydration stress in cut Alstroemeria flowers whereas ambient dehydration stress accelerates developmental senescence expression patterns. J Exp Bot 61: 2905–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA. (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223: 7–12 [DOI] [PubMed] [Google Scholar]
- Wang D, Pan YJ, Zhao XQ, Zhu LH, Fu BY, Li ZK. (2011) Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genomics 12: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Nehnevajova E, Köllmer I, Novák O, Strnad M, Krämer U, Schmülling T. (2010) Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22: 3905–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins O, Bräutigam K, Campbell MM. (2010) Time of day shapes Arabidopsis drought transcriptomes. Plant J 63: 715–727 [DOI] [PubMed] [Google Scholar]
- Wu YJ, Thorne ET, Sharp RE, Cosgrove DJ. (2001) Modification of expansin transcript levels in the maize primary root at low water potentials. Plant Physiol 126: 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu JK. (2002) Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ 25: 131–139 [DOI] [PubMed] [Google Scholar]
- Xu JC, Tian J, Belanger FC, Huang BR. (2007) Identification and characterization of an expansin gene AsEXP1 associated with heat tolerance in C3 Agrostis grass species. J Exp Bot 58: 3789–3796 [DOI] [PubMed] [Google Scholar]
- Xue GP, Way HM, Richardson T, Drenth J, Joyce PA, McIntyre CL. (2011) Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant 4: 697–712 [DOI] [PubMed] [Google Scholar]
- Xue JQ, Li YH, Tan H, Yang F, Ma N, Gao JP. (2008) Expression of ethylene biosynthetic and receptor genes in rose floral tissues during ethylene-enhanced flower opening. J Exp Bot 59: 2161–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Takahashi R, Fujitani C, Mishima K, Yoshida M, Joyce DC, Yamaki S. (2009) Cell wall extensibility and effect of cell-wall-loosening proteins during rose flower opening. J Jpn Soc Hortic Sci 78: 242–251 [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CY, Pence HE, Jin JB, Miura K, Gosney MJ, Hasegawa PM, Mickelbart MV. (2010) The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 22: 4128–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HK, Kim SG, Kim SY, Park CM. (2008) Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol Cells 25: 438–445 [PubMed] [Google Scholar]
- Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB. (2008) Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenoni S, Reale L, Tornielli GB, Lanfaloni L, Porceddu A, Ferrarini A, Moretti C, Zamboni A, Speghini A, Ferranti F, et al. (2004) Downregulation of the Petunia hybrida α-expansin gene PhEXP1 reduces the amount of crystalline cellulose in cell walls and leads to phenotypic changes in petal limbs. Plant Cell 16: 295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GY, Chen M, Chen XP, Xu ZS, Guan S, Li LC, Li AL, Guo JM, Mao L, Ma YZ. (2008) Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J Exp Bot 59: 4095–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XN, Chen B, Lu GJ, Han B. (2009) Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun 379: 985–989 [DOI] [PubMed] [Google Scholar]
- Zhong RQ, Lee C, Ye ZH. (2010) Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant 3: 1087–1103 [DOI] [PubMed] [Google Scholar]
- Zhong RQ, Richardson EA, Ye ZH. (2007) The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19: 2776–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JL, Wang XF, Jiao YL, Qin YH, Liu XG, He K, Chen C, Ma LG, Wang J, Xiong L, et al. (2007) Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol Biol 63: 591–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JM, Alvarez S, Marsh EL, Lenoble ME, Cho IJ, Sivaguru M, Chen SX, Nguyen HT, Wu YJ, Schachtman DP, et al. (2007) Cell wall proteome in the maize primary root elongation zone. II. Region-specific changes in water soluble and lightly ionically bound proteins under water deficit. Plant Physiol 145: 1533–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonia L, Munnik T. (2007) Life under pressure: hydrostatic pressure in cell growth and function. Trends Plant Sci 12: 90–97 [DOI] [PubMed] [Google Scholar]