Abstract
Plant resistance to necrotrophic fungi is regulated by a complex set of signaling pathways that includes those mediated by the hormones salicylic acid (SA), ethylene (ET), jasmonic acid (JA), and abscisic acid (ABA). The role of ABA in plant resistance remains controversial, as positive and negative regulatory functions have been described depending on the plant-pathogen interaction analyzed. Here, we show that ABA signaling negatively regulates Arabidopsis (Arabidopsis thaliana) resistance to the necrotrophic fungus Plectosphaerella cucumerina. Arabidopsis plants impaired in ABA biosynthesis, such as the aba1-6 mutant, or in ABA signaling, like the quadruple pyr/pyl mutant (pyr1pyl1pyl2pyl4), were more resistant to P. cucumerina than wild-type plants. In contrast, the hab1-1abi1-2abi2-2 mutant impaired in three phosphatases that negatively regulate ABA signaling displayed an enhanced susceptibility phenotype to this fungus. Comparative transcriptomic analyses of aba1-6 and wild-type plants revealed that the ABA pathway negatively regulates defense genes, many of which are controlled by the SA, JA, or ET pathway. In line with these data, we found that aba1-6 resistance to P. cucumerina was partially compromised when the SA, JA, or ET pathway was disrupted in this mutant. Additionally, in the aba1-6 plants, some genes encoding cell wall-related proteins were misregulated. Fourier transform infrared spectroscopy and biochemical analyses of cell walls from aba1-6 and wild-type plants revealed significant differences in their Fourier transform infrared spectratypes and uronic acid and cellulose contents. All these data suggest that ABA signaling has a complex function in Arabidopsis basal resistance, negatively regulating SA/JA/ET-mediated resistance to necrotrophic fungi.
Plants are exposed in their natural environments to biotic and abiotic stresses. Under these conditions, plant survival depends on their ability to detect stress-associated signals and to respond to these stimuli quickly and efficiently (Bari and Jones, 2009). The mechanisms involved in regulating the activation of defensive responses upon biotic and abiotic stresses are not fully understood. Intricate networks involving different signaling pathways are required for the activation of specific plant responses to a particular stress, but how these pathways interact to balance the final output response is largely unknown (Fujita et al., 2006; Robert-Seilaniantz et al., 2007; Spoel and Dong, 2008; Bari and Jones, 2009). Plant resistance to pathogens depends on the interplay of different signaling mechanisms, such as those mediated by the hormones salicylic acid (SA), jasmonic acid (JA), and ethylene (ET; Thomma et al., 1998; Glazebrook, 2005). In addition to these well-characterized pathways, other plant hormones, such as abscisic acid (ABA), brassinosteroids, gibberellins, and auxins, are emerging as important coregulators of plant resistance to pathogens, including necrotrophic fungi (Bari and Jones, 2009).
ABA regulates many aspects of plant development, such as seed dormancy and germination, and also controls plant responses to abiotic and biotic stress (Fujita et al., 2006; Wasilewska et al., 2008). The mechanism of action of at least one arm of the ABA perception/signaling pathway has been recently characterized at the molecular level (Raghavendra et al., 2010). The activated ABA receptor was demonstrated to be a heteromeric complex formed by an ABA-binding RCAR/PYR1/PYL family member (for Regulatory Component of ABA Receptor/Pyrabactin Resistance1/PYR1-Like) and a “clade A” type 2C protein phosphatase (PP2C; Schweighofer et al., 2004) such as Abscisic Acid Insensitive1 (ABI1), ABI2, or Hypersensitive to ABA1; Ma et al., 2009; Park et al., 2009). In the presence of ABA, the receptor blocks the phosphatase activity of PP2Cs; consequently, protein kinases, such as OST1 and SnRKs, are no longer inhibited, and they phosphorylate key targets of the ABA signaling pathway (Umezawa et al., 2009; Vlad et al., 2009).
The role of ABA signaling in the regulation of plant basal resistance to pathogens is complex and not completely understood. For example, in plant resistance to necrotrophic pathogens, positive and negative actions have been described for ABA depending on the pathosystem studied, the plant developmental stage tested, or the environmental conditions used for plant growth (Mauch-Mani and Mauch, 2005; Ton et al., 2009; García-Andrade et al., 2011; Robert-Seilaniantz et al., 2011). In tomato (Solanum lycopersicum), a negative regulatory role of ABA in resistance to necrotrophic pathogens, such as the fungus Botrytis cinerea and the bacterium Dickeia dadantii, has been demonstrated. These pathogens are less virulent on the tomato sitiens mutant, in which ABA biosynthesis is disrupted, than in wild-type plants (Audenaert et al., 2002; Asselbergh et al., 2007, 2008a, 2008b). Similarly, Arabidopsis (Arabidopsis thaliana) ABA-deficient mutants (e.g. aba2 and aao3) are more resistant than wild-type plants to the necrotroph B. cinerea and to the vascular fungal pathogen Fusarium oxysporum (Audenaert et al., 2002; L’Haridon et al., 2011). Several molecular mechanisms were proposed to explain the enhanced resistance phenotype of Arabidopsis and tomato ABA-deficient mutants. For example, in tomato sitiens plants, enhanced up-regulation of defense-related transcripts (e.g. PR-1), accumulation of reactive oxygen species (ROS), and increase in cuticle permeability were suggested to enhance resistance to B. cinerea and D. dadantii (Audenaert et al., 2002; Asselbergh et al., 2007; Curvers et al., 2010). Similarly, enhanced resistance of Arabidopsis ABA-deficient mutants (aba2) to B. cinerea was linked to increased cuticle permeability, which was also predicted to account for the stronger and faster accumulation of ROS seen following fungal infection (L’Haridon et al., 2011).
Exogenous application of ABA to Arabidopsis wild-type plants resulted in an enhanced susceptibility to Pseudomonas syringae pv tomato DC3000 that was correlated with inhibition of lignin biosynthesis, reduced SA accumulation, and suppression of transcripts of several defense-related genes, including some regulated by SA and JA/ET signaling pathways (Anderson et al., 2004; Mohr and Cahill, 2007). Treatment of Arabidopsis with ABA also blocked the activation of systemic acquired resistance (SAR) both upstream and downstream of SA biosynthesis (Yasuda et al., 2008). Congruently, SAR activation disrupted the up-regulation of ABA biosynthesis and signaling in plants exposed to salt stress, further indicating a negative cross talk between SAR and ABA signaling pathways (Yasuda et al., 2008). Plant pathogens also utilize ABA-mediated suppression of plant defensive responses to colonize plant tissues. The AvrPtoB effector of P. syringae pv tomato DC3000 induces the accumulation of ABA in Arabidopsis to suppress the induction of basal defense by down-regulating SA biosynthesis and signaling pathways (de Torres-Zabala et al., 2007, 2009). Further support for an important role for ABA-SA cross talk in plant defense comes from a recent study demonstrating that activation of EDS1/PAD4-dependent immune responses rapidly disrupts ABA signaling transduction (Kim et al., 2011).
ABA also has a positive role in the regulation of Arabidopsis basal resistance to certain pathogens. Increased susceptibility to the necrotrophic fungus Alternaria brassicicola, the vascular oomycete Pythium irregulare, and the necrotrophic fungus Plectosphaerella cucumerina was observed in ABA-deficient mutants (Adie et al., 2007; Flors et al., 2008; García-Andrade et al., 2011). ABA is required for JA biosynthesis and induction of the expression of JA-regulated defense genes, two essential processes for full Arabidopsis resistance to P. irregulare (Adie et al., 2007). ABA triggers callose deposition upon A. brassicicola infection, a defense mechanism associated with restriction of colonization by the fungus. A reduction of ABA levels in plants infected with A. brassicicola may represent a virulence defense mechanism evolved to favor colonization (Flors et al., 2008). A role for ABA in mimicking β-aminobutyric acid-primed callose deposition and some defense responses against the necrotrophic pathogens A. brassicicola and P. cucumerina has also been described (Ton and Mauch-Mani, 2004; Flors et al., 2005; García-Andrade et al., 2011). Moreover, the enhanced and constitutive accumulation of ABA of the Arabidopsis irx1-6 mutant, impaired in a secondary cell wall cellulose synthase (AtCESA8), and of the ocp3 mutant, defective in a transcriptional factor of the homeodomain family, has been suggested to contribute to increased resistance to several pathogens (Hernández-Blanco et al., 2007; García-Andrade et al., 2011).
In this work, we show that disruption of ABA biosynthesis or signaling leads to a constitutive activation of Arabidopsis defensive response. We provide genetic evidence that supports a negative function of ABA in the regulation of SA/ET/JA-dependent immune responses, which are essential for Arabidopsis resistance to the virulent, necrotrophic fungal strain P. cucumerina Brigitte Mauch-Mani (PcBMM). Moreover, we show that the aba1-6 mutant displays alterations in its cell wall structure and composition.
RESULTS
ABA-Deficient and Signaling Mutants Are More Resistant Than Wild-Type Plants to the Necrotrophic Fungus P. cucumerina BMM
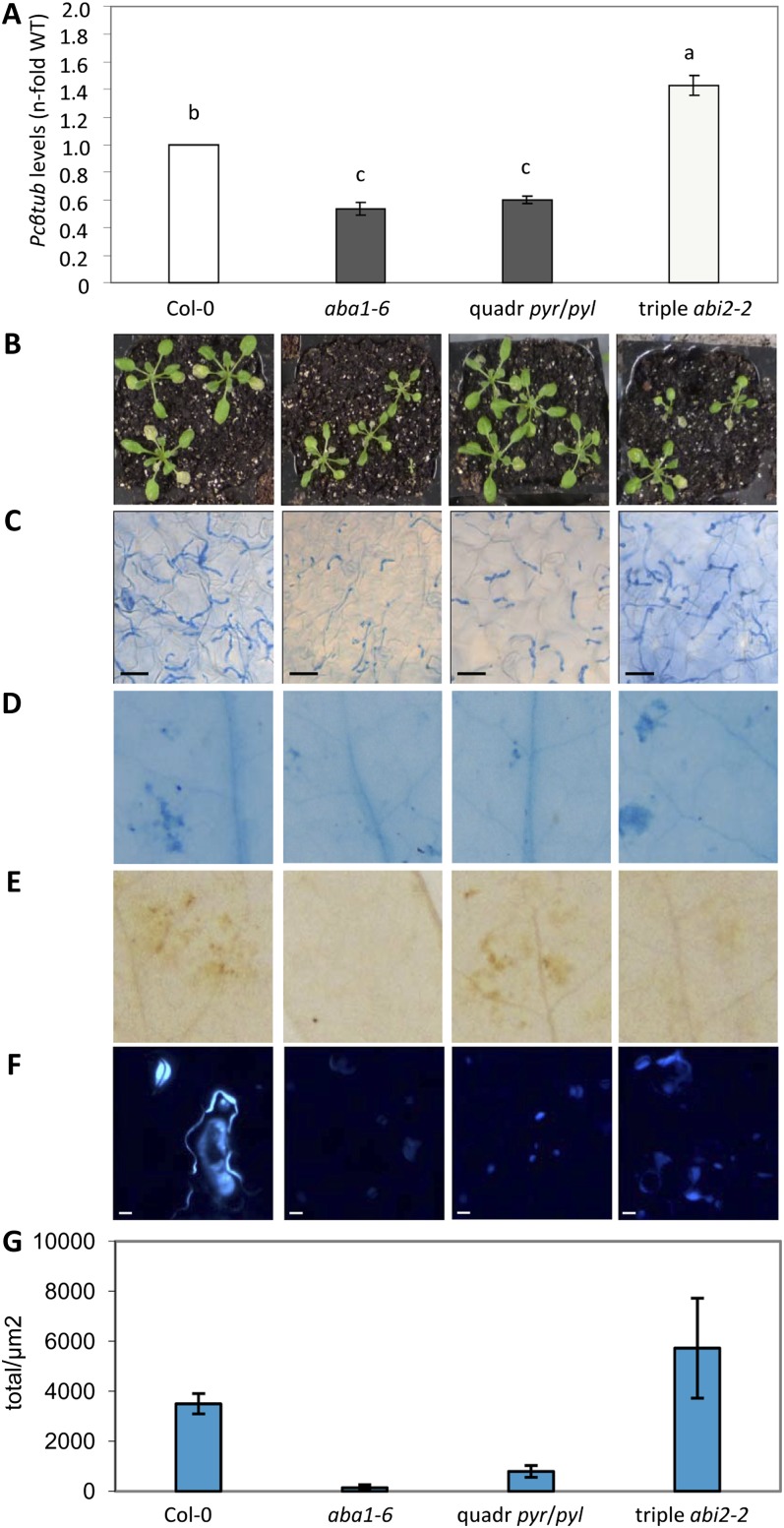
To investigate the role of ABA in the regulation of Arabidopsis resistance to necrotrophic fungal pathogens, we tested the susceptibility to PcBMM of plant mutants impaired in ABA biosynthesis, like aba1-6 (Niyogi et al., 1998), in ABA signaling, such as in quadruple pyr/pyl (pyr1pyl1pyl2pyl4; Park et al., 2009), or displaying a partial constitutive activation of the ABA pathway, as occurs in the triple abi2-2 mutant hab1-1abi1-2abi2-2 (Rubio et al., 2009). We inoculated 3-week-old ecotype Columbia (Col-0) wild-type plants and the ABA mutants with a spore suspension (4 × 106 spores mL−1) of the PcBMM isolate that causes disease in a broad range of Arabidopsis accessions (Llorente et al., 2005; Sánchez-Vallet et al., 2010). Progression of the infection was examined at different hours/days post inoculation (hpi/dpi) by determining fungal growth and plant cell death, using trypan blue (TB) staining of inoculated leaves, the fungal biomass using quantitative PCR (qPCR) of the PcBMM β-Tubulin gene, and by macroscopic evaluation of disease rating (DR; Sánchez-Vallet et al., 2010). As shown in Figure 1, aba1-6 and quadruple pyr/pyl mutant supported lower fungal growth than wild-type plants, as revealed by TB staining of inoculated leaves 1 dpi and by fungal biomass quantification 3 dpi (Fig. 1, A and C). In contrast, fungal growth in the triple abi2-2 mutant was higher than that determined in Col-0 plants (Fig. 1, A and C). These results were corroborated by confocal microscopy analyses of Col-0 and aba1-6 plants inoculated with PcBMM-GFP, a fungal transformant constitutively expressing GFP (Supplemental Fig. S1; Ramos et al., 2012). A positive correlation was found between in planta fungal biomass, plant cell death, and macroscopic disease symptoms (Fig. 1, A, B, and D; data not shown). Thus, the level of plant cell death, as determined by TB staining, was lower in aba1-6 and quadruple pyr/pyl mutants than in wild-type plants, whereas it was enhanced in the triple abi2-2 mutant (Fig. 1D). Accordingly, the aba1-6 mutant, which has a dwarf phenotype (Barrero et al., 2005), and the quadruple pyr/pyl mutant displayed similar, less severe macroscopic symptoms than those observed in inoculated wild-type plants, whereas the opposite was true for the triple abi2-2 mutant (Fig. 1B; data not shown). These data strongly supported a negative function of the cytosolic ABA perception and signaling pathways in the regulation of Arabidopsis resistance to PcBMM and further corroborated the relevant function of the ABA pathway in the control of Arabidopsis resistance to necrotrophic fungal pathogens.
Figure 1.
Resistance of ABA-deficient and signaling mutants to the necrotrophic fungus PcBMM. Wild-type plants (Col-0; WT) and the aba1-6, quadruple pyr1pyl1pyl2pyl4 (quadr pyr/pyl), and triple abi1-2abi2-2hab1-1 mutants were inoculated with a spore suspension (4 × 106 spores mL−1) of PcBMM. A, qPCR quantification of fungal biomass at 3 dpi. Specific primers of P. cucumerina β-Tubulin and Arabidopsis UBIQUITIN21 were used. Values ± se were normalized to Arabidopsis UBIQUITIN21 and are represented as averages of the fold increased expression compared with the corresponding wild-type plants. Data represent average values of two replicates from one out of three independent experiments performed, which gave similar results. The letters indicate different statistically significant groups (ANOVA, P ≤ 0.05, Bonferroni test). B, Macroscopic disease symptoms of inoculated plants at 7 dpi. C and D, Fungal hyphae (C) and plant cell death (D) were visualized in the inoculated leaves by lactophenol TB 1 dpi. E, Hydrogen peroxide accumulation in inoculated leaves was determined by DAB staining at 1 dpi. F, Callose deposition in inoculated leaves (1 dpi) was visualized using aniline blue, and the number of total pixels of these depositions per μm2 was quantified (G). At least four leaves from each genotype were used for pixel quantification in the two experiments performed that gave similar results. Bars = 10 μm.
ROS regulates Arabidopsis responses to biotic and abiotic stresses (Torres et al., 2005). ABA has been suggested to negatively regulate ROS accumulation in Arabidopsis and tomato in resistance responses to the necrotrophic fungus B. cinerea (Asselbergh et al., 2007; Curvers et al., 2010; L’Haridon et al., 2011). Therefore, we used 3,3′-diaminobenzidine (DAB) staining to determine ROS production in PcBMM-inoculated wild-type plants and in the three ABA mutants, but no significant differences were macroscopically observed in ROS production among the genotypes tested (Fig. 1E), further indicating that ABA-mediated regulation of Arabidopsis resistance to this fungus is independent of ROS production. ABA has also been described to trigger callose deposition upon pathogen infection, and ABA-deficient mutants were shown to be impaired in callose accumulation after inoculation with some fungal pathogens (Ton and Mauch-Mani, 2004; García-Andrade et al., 2011). However, PcBMM-infected plants stained with aniline blue displayed lower callose content in the aba1-6 and quadruple pyr/pyl mutants than in wild-type plants (Fig. 1, F and G), indicating that callose deposition is not determining the enhanced resistance phenotype of these mutants. In line with these results, we found that the pmr4 mutant, impaired in callose deposition (Nishimura et al., 2003), was as susceptible to PcBMM as wild-type plants (Supplemental Fig. S2). This pmr4 phenotype was not due to the previously described enhanced SA content of this mutant (Nishimura et al., 2003), as the susceptibility to PcBMM of the pmr4 sid2-1 double mutant, defective in SA biosynthesis, was similar to that of susceptible sid2-1 plants (Supplemental Fig. S2). These results further corroborate that, under our inoculation conditions, callose deposition is not an essential defensive barrier against the necrotrophic fungus PcBMM and that additional defense mechanisms determine the enhanced resistance phenotype of aba1-6 and quadruple pyr/pyl mutants.
Comparative Transcriptomic Analyses Reveal Constitutive Activation of a Complex Defensive Network in the aba1-6 Mutant
To further characterize the molecular mechanisms regulating ABA-mediated resistance to necrotrophs, we selected the well-characterized aba1-6 mutant (Niyogi et al., 1998; Barrero et al., 2005) and performed a comparative transcriptomic analysis of Col-0 and aba1-6 plants prior to inoculation (time zero) and at 1 d after water treatment (mock) or inoculation with a spore suspension (4 × 106 spores mL−1) of PcBMM. Out of the 22,000 probe sets tested in these transcriptomic analyses, 8,687 showed statistically significant differential abundance according to a two-way ANOVA and a Benjamini and Hochberg multiple testing correction (P ≤ 0.01; GeneSpring 7.2 software). From these probe sets, 213 were differentially regulated in untreated (time zero) aba1-6 compared with untreated wild-type plants (Supplemental Table S1). In PcBMM-inoculated aba1-6 plants, 497 probe sets were differentially expressed compared with mock-treated plants, whereas 1,474 probe sets were found differentially expressed between PcBMM- and mock-inoculated wild-type plants (Supplemental Tables S2 and S3).
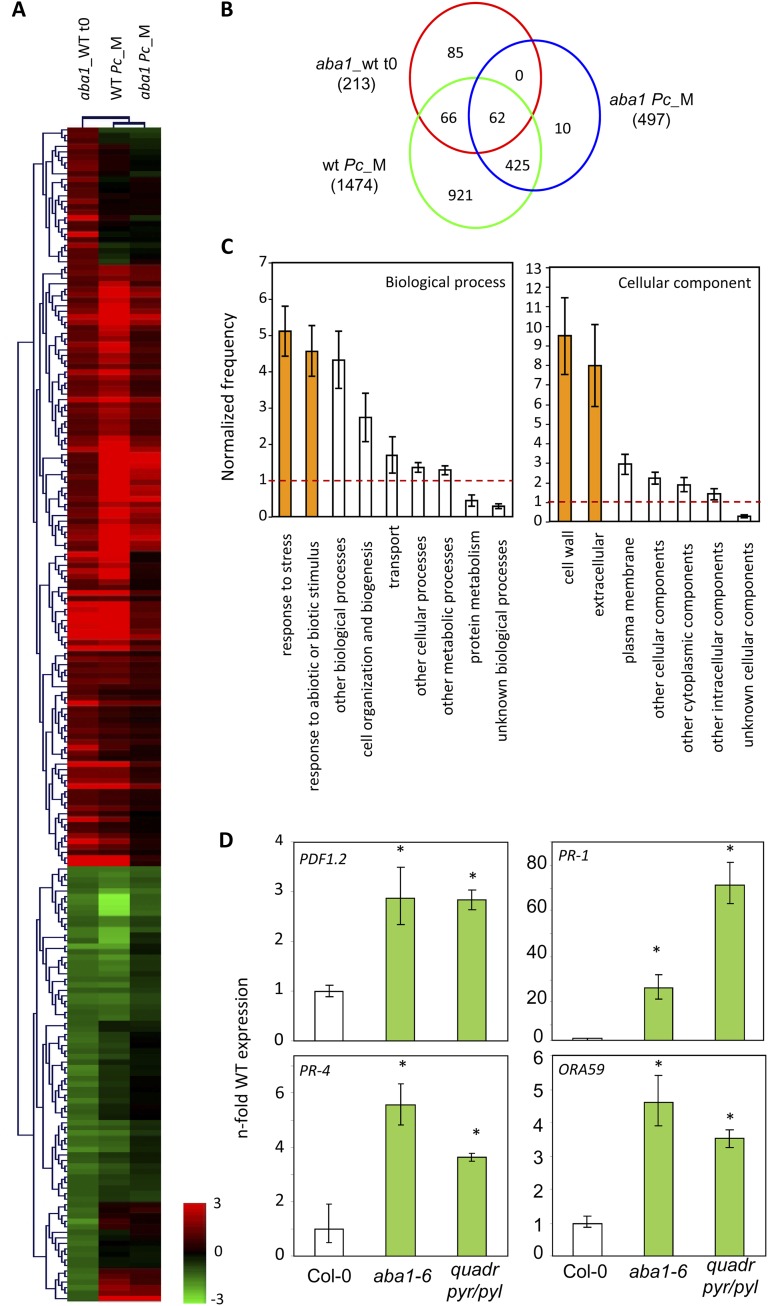
The 213 probe sets differentially regulated in the untreated (time zero) aba1-6 plants (Supplemental Table S1) were analyzed by hierarchical clustering. As shown in Figure 2A, a significant overlap was found between the transcriptional profiles of untreated aba1-6 and PcBMM-inoculated wild-type plants. In agreement, 128 from the 213 probe sets (60%) that were constitutively up- or down-regulated in untreated aba1-6 plants were found to be also differentially regulated in wild-type plants upon PcBMM inoculation (Fig. 2B; Supplemental Table S4). Thus, 67% (90 genes) of the genes constitutively up-regulated (134 genes) in aba1-6 mutant plants were also induced in wild-type plants upon PcBMM infection (Fig. 2B) and might be considered as constitutively up-regulated defensive genes. Meta-analysis, using Genevestigator tools (Zimmermann et al., 2004), demonstrated that the expression of a high number of these up-regulated genes was also induced after Arabidopsis infection with other necrotrophic fungi such as B. cinerea (60 genes) and with other pathogens like Phytophthora infestans (70 genes), Erysiphe orontii (27 genes), Erysiphe cichoracearum (31 genes), or P. syringae (19 genes); in contrast, a reduced number of these genes were found to be differentially regulated after insect or nematode attack (eight and five genes, respectively). About 63% (57 genes) of the genes up-regulated both in aba1-6 untreated plants and in PcBMM-inoculated wild-type plants were also up-regulated by JA, SA, or ET treatment. These data indicated that an extensive cross talk existed among these pathways and ABA signaling.
Figure 2.
Transcriptomic profiles of untreated aba1-6 mutant and PcBMM-inoculated wild-type plants strongly overlapped. A, Hierarchical clustering of the 213 differentially expressed probe sets in untreated aba1-6 mutant plants compared with wild-type plants (aba1_WT t0). Genes (rows) and experiments (columns) were clustered with the Multiexperiment Viewer (The Institute for Genomic Research) using Pearson uncentered distance and average linkage. The columns corresponded to untreated aba1-6 compared with wild-type plants (aba1_WT t0) and PcBMM-inoculated wild-type and aba1-6 plants at 1 dpi compared with their respective mock samples (WT Pc_M and aba1 Pc_M, respectively). B, Venn diagram showing overlapping of the differentially regulated probe sets (P ≤ 0.01, ANOVA, Benjamini and Hochberg multiple testing correction) in the untreated aba1-6 mutant line (red circle), in PcBMM-inoculated wild-type plants (green circle), and in PcBMM-inoculated aba1-6 plants (blue circle). The numbers of differentially regulated probe sets in each treatment and of common genes among these treatments are indicated. C, The 128 probe sets that were common to untreated aba1-6 plants and PcBMM-inoculated wild-type plants were classified using tools of the Bio-Array Resources for Plant Biology (http://bar.utoronto.ca/). The normalized score value for each functional class is represented. Normalized score values greater than 1 indicate overrepresented classes in comparison with the whole genome. D, Validation of differentially expressed genes in aba1-6 mutants. The expression of PDF1.2, PR-1, PR-4, and ORA59 genes was determined by qPCR in untreated wild-type plants (WT; Col-0), aba1-6, and quadruple pyr1pyl1pyl2pyl4 (quadr pyr/pyl) mutants. Values were normalized to Arabidopsis UBIQUITIN21 expression levels and, for a better comparison with transcriptomic data, were represented as relative fold expression to the one in wild-type plants. Bars represent average values ± sd of two technical replicates. Asterisks indicate differences statistically significant from those of Col-0 plants (P ≤ 0.05, ANOVA, lsd test). Data are from one out of three independent experiments, which gave similar results.
In addition, 33 (42%) out of the 79 probe sets down-regulated in untreated aba1-6 were also down-regulated after infection of wild-type plants with PcBMM. Interestingly, expression of a high percentage of these genes was also down-regulated after infection with B. cinerea (67%) and P. infestans (48%), as revealed by meta-analysis (data not shown). The expression of 30% of these aba1-6 and PcBMM down-regulated genes was found to be repressed either by SA or ET treatment, further supporting the existence of a negative interplay among these signaling pathways in response to pathogen infection.
Functional classification of the proteins encoded by the genes differentially regulated in untreated aba1-6 mutant and in PcBMM-inoculated wild-type plants was performed using the Classification SuperViewer tool (Bio-Array Resource; http://bar.utoronto.ca/; Toufighi et al., 2005). Notably, the most overrepresented cellular component categories were “cell wall” (P = 7 × 10−15) and “extracellular” (P = 3 × 10−9), and the most overrepresented biological process categories were “response to stress” (P = 2 × 10−17) and “response to abiotic and biotic stimulus” (P = 2 × 10−12; Fig. 2C). Among the proteins in the “biotic stress” functional category, there were some previously described to be involved in defense against necrotrophs (Table I): (1) PDF1.2, PR-1, and PR-4, defensive markers of the JA/ET, SA, and ET pathways, respectively; (2) the respiratory burst oxidase homolog D (RbohD), an enzymatic subunit of the plant NADPH oxidase involved in the generation of hydrogen peroxide; (3) the transcription factors WRKY33 and ORA59; and (4) several proteins, such as CYP81F2, PAD3, and PEN3, involved in the synthesis and delivery of Trp-derived secondary metabolites, like camalexin and indole glucosinolates. Similarly, some genes encoding cell wall-related proteins were found to be misregulated in the aba1-6 mutant compared with wild-type plants (Supplemental Table S5). Remarkably, cell wall architecture/alteration has been recently linked to the regulation of tomato and Arabidopsis resistance to necrotrophic pathogens, such B. cinerea and PcBMM, respectively (Asselbergh et al., 2007; Sánchez-Rodríguez et al., 2009; Delgado-Cerezo et al., 2012).
Table I. Defense-related genes constitutively up-regulated in the aba1-6 mutant and induced in wild-type plants after PcBMM challenge.
| Gene Description | Locus | Fold Change | Reference | |
|---|---|---|---|---|
|
aba1/Wild Typea |
Wild Type (Pc/M)b |
|||
| PR-4/HEL (hevein-like) | AT3G04720 | 3.43 | 3.16 | Hejgaard et al. (1992); Potter et al. (1993) |
| PDR8 (pleiotropic drug resistance8)/PEN3 | AT1G59870 | 1.83 | 1.07 | Stein et al. (2006) |
| PR-1 (pathogenesis-related1) | AT2G14610 | 4.10 | 3.15 | Van Loon and Van Strien (1999) |
| PDF1.2B (plant defensin1.2B) | AT2G26020 | 5.81 | 7.78 | Thomma et al. (2002) |
| PLP2 (patatin-like protein) | AT2G26560 | 4.12 | 3.33 | La Camera et al. (2005) |
| WRKY33 | AT2G38470 | 3.74 | 1.15 | Zheng et al. (2006) |
| ATTI1 (trypsin inhibitor protein1) | AT2G43510 | 2.44 | 1.36 | Silverstein et al. (2005) |
| CHI-B; PR-3 (basic chitinase) | AT3G12500 | 4.13 | 3.39 | Verburg and Huynh (1991) |
| PRX33 (peroxidase33) | AT3G49110 | 2.02 | 1.20 | Bindschedler et al. (2006); O’Brien et al. (2012) |
| OSM34 (osmotin34) | AT4G11650 | 3.74 | 2.41 | Monteiro et al. (2003) |
| Blue copper-binding protein | AT5G20230 | 4.03 | 3.05 | Mishina and Zeier (2007) |
| PDF1.2A (plant defensin1.2A) | AT5G44420 | 6.20 | 8.90 | Manners et al. (1998) |
| Thomma et al. (2002) | ||||
| RBOHD (respiratory burst oxidase protein D) | AT5G47910 | 2.26 | 1.56 | Torres et al. (2005) |
| PAD3 (phytoalexin deficient3; CYP71B15) | AT3G26830 | 6.39 | 1.16 | Glazebrook and Ausubel (1994); Zhou et al. (1999) |
| CYP81F2 (cytochrome P450) | AT5G57220 | 4.36 | 1.50 | Bednarek et al. (2009); Clay et al. (2009) |
| ORA59 (octadecanoid-responsive Arabidopsis AP2/ERF59) | AT1G06160 | 3.20 | 2.88 | Pré et al. (2008) |
| PNP-A (plant natriuretic peptide A) | AT2G18660 | 1.13 | 5.05 | Meier et al. (2008) |
| ATGRXS13 (glutaredoxin13) | AT1G03850 | 1.55 | 3.25 | La Camera et al. (2011) |
| WRKY26 | AT5G07100 | 1.20 | 1.88 | Eulgem and Somssich (2007) |
Normalized, average fold ratios (log2) of aba1-6 versus wild-type plants at time zero.
Normalized, average fold ratios (log2) of PcBMM-inoculated versus mock-treated wild-type plants. Genes were selected using values of P ≤ 0.01 (see “Materials and Methods”).
To validate the transcriptomic data, the steady-state expression of some defensive genes (PDF1.2, PR-1, PR-4, and ORA59), which were up-regulated in aba1-6 plants, was determined in untreated wild-type plants and the ABA-resistant mutants (aba1-6 and quadruple pyr/pyl). qPCR analysis demonstrated that the expression of these genes was higher in both ABA-deficient and ABA signaling mutants than in wild-type plants (Fig. 2D). The role of one of these differentially expressed genes in Arabidopsis resistance to PcBMM was validated by demonstrating the enhanced susceptibility to this fungus of the wrky33-1 mutant, which is defective in the WRKY33 transcriptional factor previously shown to control resistance to the necrotroph B. cinerea (Supplemental Fig. S3; Zheng et al., 2006). These data further corroborated the relevance in plant immunity of constitutively up-regulated genes in the Arabidopsis aba1-6 and quadruple pyr/pyl mutants.
Hormonal Regulation of the Genes Differentially Expressed in aba1-6 Plants
To gather additional information on the processes regulated by genes differentially regulated in aba1-6, meta-analysis was performed on the transcriptomic data of the untreated aba1-6 mutant (Supplemental Table S1), wild-type plants inoculated with PcBMM (Supplemental Table S2), and publicly available transcriptomic data of Arabidopsis wild-type plants treated with the hormones SA, 1-aminocyclopropane-1-carboxylic acid (ACC), an ET precursor, or methyl jasmonate (Goda et al., 2008). Using Pearson uncentered hierarchical clustering on genes differentially regulated in aba1-6 compared with wild-type plants upon inoculation with PcBMM, we found that the transcriptomic profiles of the untreated aba1-6 mutant and PcBMM-inoculated wild-type plants clustered together and separately from the transcriptomic profiles of plants treated with the different hormones (Fig. 3), indicating that the overall transcriptomic profile of the aba1-6 mutant could not be linked specifically to any of these hormones. Pearson uncentered hierarchical clustering differentiated seven statistically significant gene clusters using a distance threshold of 0.7 (Fig. 3; data not shown). The largest clusters (I–IV) were further analyzed: clusters I to III corresponded to genes up-regulated in both untreated aba1-6 and PcBMM-inoculated wild-type plants, whereas cluster IV contained genes down-regulated in both PcBMM-inoculated wild-type plants and untreated aba1-6 plants (Supplemental Table S6).
Figure 3.
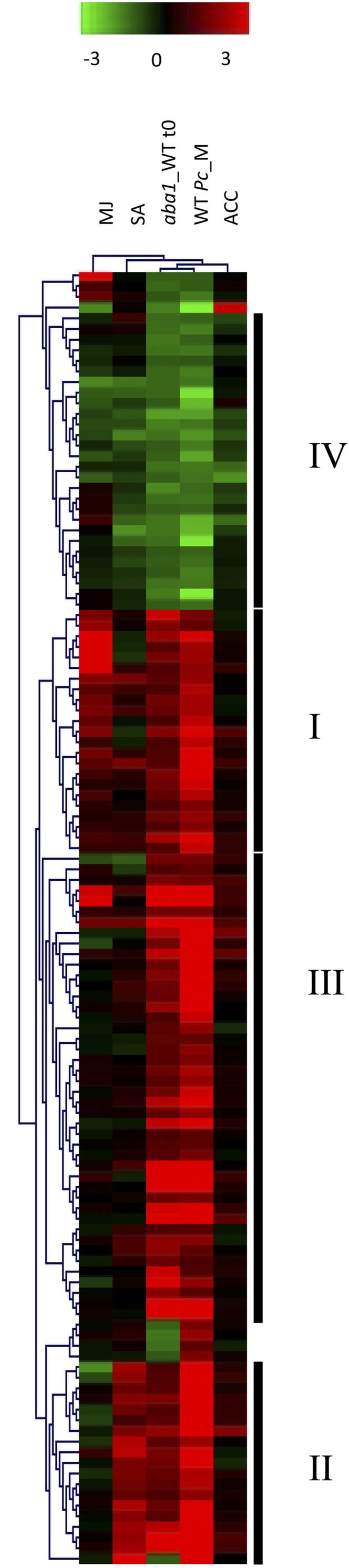
Hierarchical clustering of genes differentially regulated in untreated aba1-6 and in PcBMM-inoculated wild-type plants. Hierarchical clustering is shown for the 128 probe sets differentially expressed in both aba1-6 untreated plants (aba1_WT t0) and in PcBMM-inoculated wild-type plants (WT Pc_M) and those from treatments of Arabidopsis with different hormones (SA, ACC, and methyl jasmonate [MJ]; http://www.arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp). All data sets were normalized with the robust multiarray average method using GeneSpring software. Genes (rows) and samples (columns) were clustered with the Multiexperiment Viewer (The Institute for Genomic Research) using Pearson uncentered distance and average linkage (threshold = 0.7). The resulting clusters (I–IV), which are indicated by vertical bars, were further analyzed, and the results are shown in Supplemental Table S5.
The presence of conserved putative cis-regulatory elements was determined in the genes included in clusters I to IV with Transplanta software (http://bioinfogp.cnb.csic.es/transplanta_dev/). Subsequently, TOMTOM (http://meme.nbcr.net/meme/cgi-bin/tomtom.cgi; Gupta et al., 2007) and PLACE (for Plant cis-acting Regulatory DNA Elements; http://www.dna.affrc.go.jp/PLACE/; Higo et al., 1999) databases were used to identify the statistically overrepresented regulatory elements in the promoters of the genes from each cluster. We observed that cluster I contained genes (14 out of 23) predicted to be predominantly up-regulated by JA treatment, some of which encoded proteins (RbohD, PEN3, ATKTI1, and ATTI1) involved in plant basal resistance (Supplemental Table S6). Notably, the cis-regulatory elements of these JA-dependent genes were enriched in W-box sequences, suggesting a transcriptional regulation by WRKY transcription factors (Rushton et al., 1996). Cluster II contained genes (18 out of 19) associated with SA-mediated induction, such as defense-related genes like PR-1, WRKY33, PNP-A (encoding the plant natriuretic peptide A), ATGRXS13, and a Receptor-Like Kinase (Table I). Among these genes there were no cis-regulatory elements statistically overrepresented. The 44 genes in cluster III, which were not predominantly regulated by any of the hormones analyzed, encoded proteins involved in biotic and abiotic stress responses, such as CHIB, PR-4, PRX33, and PRX34 (Table I), cell wall-related proteins, and two transcription factors (WRKY26 and ORA59). The W-box cis-regulatory element was also overrepresented in the promoters of cluster III genes. Finally, nearly all the genes down-regulated in untreated aba1-6 plants were contained in cluster IV, and they were found to be negatively regulated by either ACC or SA treatment and, to a lesser extent, by the JA signaling pathway. Among the genes found in this cluster, several encoded cell wall-related proteins (Fig. 3; Supplemental Table S6). There were no cis-regulatory elements statistically overrepresented in cluster IV genes.
The cluster analyses suggested that a significant number of the genes differentially regulated in aba1-6 plants responded to different hormones, in particular to SA and JA. Moreover, a significant number of the differentially expressed genes in PcBMM-inoculated wild-type plants corresponded to genes that are up-regulated (110 probe sets) or down-regulated (87 probe sets) by ABA. Accordingly, we measured the concentration of these hormones in ABA-deficient and ABA signaling mutants and in wild-type plants to evaluate if the misregulation of these genes could be linked to alterations in hormone levels. SA content was not altered in untreated aba1-6 and quadruple pyr/pyl, while it was lower in the triple abi2-2 mutant plants (Supplemental Fig. S4A). The concentration of JA was lower in the three mutants than in wild-type plants (Supplemental Fig. S4A). As expected, the aba1-6 mutant displayed lower ABA content than wild-type plants and quadruple pyr/pyl and triple abi2-2 mutants, which are not impaired in ABA biosynthesis (Supplemental Fig. S4A). Inoculation of wild-type plants with PcBMM (1 dpi) induced an increase in the concentration of JA and SA (Supplemental Fig. S4B), which was consistent with the previously described positive roles of SA and JA in the regulation of Arabidopsis resistance to this fungal pathogen (Berrocal-Lobo et al., 2002). Interestingly, the elevation of JA and SA contents in the PcBMM-inoculated ABA mutants did not exceed that in wild-type plants (Supplemental Fig. S4B). Notably, significant differences in the levels of ABA were not found among PcBMM-inoculated and mock-treated wild-type plants (Supplemental Fig. S4B).
SA, JA, and ET Signaling Pathways Are Essential for aba1-Mediated Resistance to PcBMM
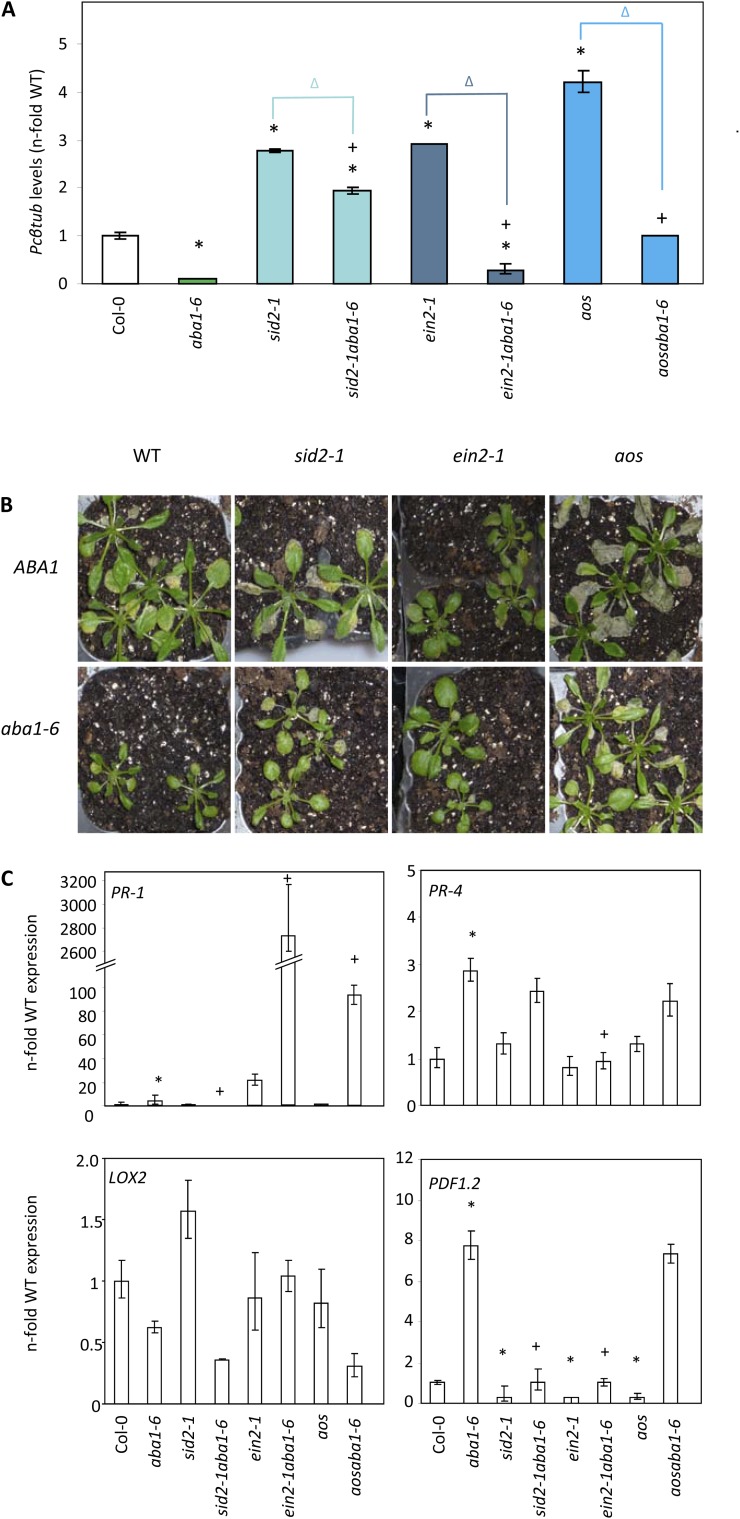
To further determine if the enhanced resistance of aba1-6 plants to PcBMM was dependent on JA, SA, and ET signaling, we genetically disrupted these pathways in the aba1-6 mutant by generating the sid2-1 aba1-6, ein2-1 aba1-6, aos aba1-6, and coi1-1 aba1-6 double mutants (Fig. 4; data not shown), which were impaired, respectively, in SA biosynthesis, ET signaling, and JA biosynthesis and signaling. All these double mutants were viable and showed developmental phenotypes that in some cases were a combination of those observed in their corresponding parental lines (Supplemental Fig. S5). Susceptibility of these double mutants to PcBMM was analyzed by spraying them alongside their parental lines and wild-type plants with a spore suspension (4 × 106 spores mL−1) of the fungus. Notably, fungal biomass and DR were significantly higher in sid2-1 aba1-6, ein2-1 aba1-6, aos aba1-6, and coi1-1 aba1-6 double mutants than in the aba1-6 single mutant (Fig. 4, A and B; data not shown), indicating that the SA, ET, and JA signaling pathways are essential for the enhanced resistance of aba1-6 to the necrotrophic fungus PcBMM. However, none of the double mutants tested was as susceptible as their corresponding parental lines sid2-1, ein2-1, or aos (Fig. 4, A and B). These data indicate that all these signaling pathways partially contributed to aba1-mediated resistance, as any of them on its own was sufficient to explain the enhanced resistance phenotype of aba1-6.
Figure 4.
SA, ET, and JA are essential for aba1-mediated resistance to PcBMM. Wild-type plants (WT; Col-0), aba1-6, sid2-1, ein2-1, and aos single mutants, and sid2-1 aba1, ein2-1 aba1-6, and aos aba1-6 double mutants were inoculated with a spore suspension (4 × 106 spores mL−1) of PcBMM. A, Fungal biomass in the inoculated plants (3 dpi) was determined by qPCR using specific primers for P. cucumerina β-Tubulin. Values were normalized to Arabidopsis UBIQUITIN21 expression levels and represented as fold increased expression compared with wild-type plants. Data represent average values of two technical replicates ± se from one of the three experiments performed, which gave similar results. Asterisks indicate genotypes that show significant differences in fungal biomass compared with that in the wild-type; plus signs indicate double mutants that show significant differences in fungal biomass compared with that in aba1-6 plants; and triangles indicate statistical differences in fungal biomass between the double mutants and the corresponding parental line (P ≤ 0.05, ANOVA, Bonferroni test). B, Macroscopic disease symptoms of PcBMM-inoculated plants at 7 dpi. C, Expression of PR-1 (AT2G14610), PR-4 (AT3G04720), LOX2 (AT3G45140), and PDF1.2 (AT5G44420), marker genes of the SA, ET, JA, and ET+JA pathways, respectively, was determined by qPCR on untreated 3-week-old plants. Values are represented as fold increased expression compared with the wild-type. Data correspond to average values ± sd of two replicates from one representative experiment out of the three performed. Asterisks indicate single mutants showing a significant differential expression of the gene compared with the wild-type, whereas plus signs indicate double mutants showing significant differential expression of the gene compared with aba1-6 plants (P ≤ 0.05, ANOVA, lsd test).
The impact of hormone imbalance in the resistance phenotype observed in the sid2-1 aba1-6, ein2-1 aba1-6, and aos aba1-6 double mutants was studied by quantifying the expression levels of PR-1, PR-4, LOX2, and PDF1.2 (marker genes of SA, ET, JA, and ET/JA signaling, respectively). The steady-state expression of PR-1, which was higher in the aba1-6 mutant than in wild-type plants (Fig. 2D), was further enhanced in the ein2-1 aba1-6 and aos aba1-6 double mutants (Fig. 4C). As expected, PR-1 expression was not detected in the sid2-1 and sid2-1 aba1-6 mutants impaired in SA biosynthesis (Fig. 4C). These data suggested a negative regulation of PR-1 expression by ABA, ET, and JA signaling pathways. The enhanced expression of PR-4 in the aba1-6 mutant was disrupted in the ein2-1 aba1-6 plants but not in the sid2-1 aba1-6 and aos aba1-6 double mutants, further corroborating the ET-dependent regulation of the PR-4 gene in the aba1-6 background (Fig. 4C). No significant changes were observed in the expression levels of LOX2, a marker gene for the JA signaling pathways, in all the genotypes tested (Fig. 4C). The enhanced expression of the PDF1.2 gene in the aba1-6 mutant (Figs. 2D and 4C) was partially blocked in the sid2-1 aba1-6 and ein2-1 aba1-6 double mutants, whereas it was not affected in the aos aba1-6 genotype (Fig. 4C). All together, these data illustrate the complex interplay between hormone signaling pathways in the regulation of defensive genes in Arabidopsis resistance responses.
JA, SA, and ET Signaling Pathways Do Not Interfere with ABA Function in the Regulation of Arabidopsis Resistance to Abiotic Stresses
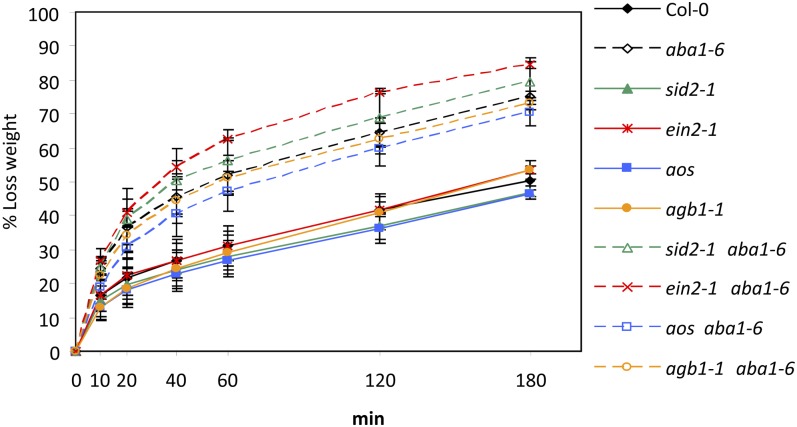
ABA signaling also has a role in the regulation of plant responses to abiotic stress, such as drought and osmotic stress. Thus, the aba1 mutant was shown to be impaired in ABA-mediated resistance to desiccation (Xiong et al., 2002). Taking advantage of the double mutants generated in this work, we decided to assess whether the SA, ET, and JA signaling pathways might interfere with the regulatory function of ABA signaling in abiotic stresses. We determined plant water loss (represented as percentage of lost weight) at different time points (0–3 h) in wild-type plants and the single and double mutants. As shown in Figure 5, water loss was similar in Col-0, sid2-1, ein2-1, and aos genotypes, which harbored the wild-type ABA1 allele. Interestingly, the sid2-1 aba1-6, ein2-1 aba1-6, and aos aba1-6 double mutants were as sensitive to this abiotic stress as the single mutant aba1-6, which showed an enhanced and faster water loss than that determined in Col-0 plants (Fig. 5). These data indicated that ET, SA, and JA signaling did not play a major role in the regulation of Arabidopsis responses to this abiotic stress and that they did not interfere with ABA function in the regulation of this process.
Figure 5.
SA, ET, and JA pathways do not affect the transpiration rates of aba1-6 plants. The sensitivity of wild-type plants (Col-0), aba1-6, sid2-1, ein2-1, and aos single mutants, and sid2-1 aba1, ein2-1 aba1-6, and aos aba1-6 double mutants to water desiccation is shown. Transpiration rates were determined as percentage lost weight at different time points (0–180 min). Each data point represents the mean of three replicates ± se. This experiment was repeated three times and gave similar results.
ABA Modulates Arabidopsis Cell Wall Composition
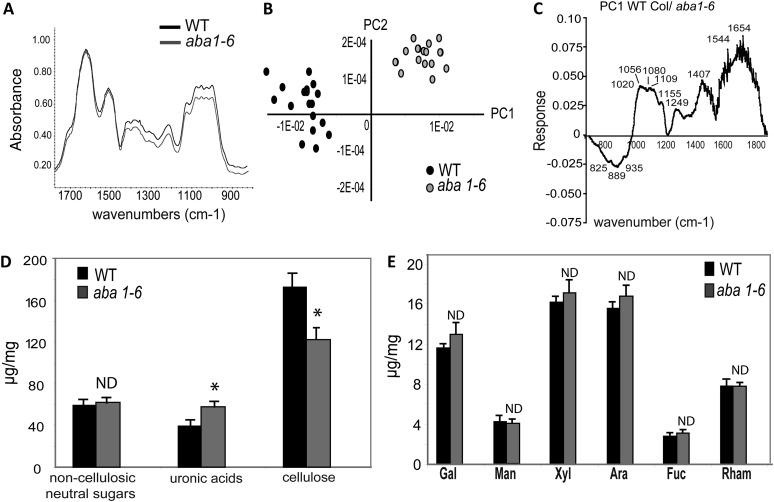
Arabidopsis resistance to necrotrophic fungi, such as PcBMM, can be regulated by remodeling plant cell wall structure and composition (Hernández-Blanco et al., 2007; Cantu et al., 2008; Sánchez-Rodríguez et al., 2009; Ramírez et al., 2011; Delgado-Cerezo et al., 2012). Ontology analyses of the genes differentially regulated in untreated aba1-6 plants identified cell wall-related genes misregulated in the mutant compared with wild-type plants (P = 7 × 10−15; Supplemental Table S5). Thus, we determined cell wall structure/composition of the aba1-6 mutant and compared it with that of wild-type plants. Cell walls from noninoculated leaves of 3-week-old aba1-6 and Col-0 plants were subjected to Fourier transform infrared (FTIR) spectroscopy to obtain qualitative spectratypes (i.e. cell wall phenotypes). The comparison of averaged FTIR spectra (n = 15) obtained between aba1-6 and Col-0 showed significant differences (Fig. 6A). Much of the total sample variation (86%) was explained by principal component 1 (Fig. 6B). Wave numbers at 1,056, 1,080, and 1,109 cm−1, which are common in the fingerprint for several cell wall polysaccharides (Kauráková et al., 2000), and 1,544 and 1,654 cm−1, which are associated with proteins, were relatively more abundant in Col-0 than in aba1-6 cell walls (Fig. 6C). On the contrary, bands at 825, 889, and 935 cm−1 were more intense in aba1-6 than in wild-type plants (Fig. 6C). Due to the complexity of infrared spectra even after principal component analysis, it was difficult to assign infrared band differences to specific cell wall polysaccharides.
Figure 6.
FTIR and biochemical analyses of the cell walls from aba1-6 mutant and wild-type (WT) plants. A, FTIR average spectra (n = 15) from 3-week-old rosettes of Col-0 wild-type plants and the aba1-6 mutant. B, Biplot analysis showing a clear separation of aba1-6 and wild-type plants. C, Mid-infrared spectra were analyzed by the covariance-matrix approach for principal component 1 (PC1). D and E, Quantification of cellulose, total uronic acid content (g mg−1 dry weight; D), and individual neutral sugars (mol %) from the noncellulosic carbohydrate fraction (E) from the cell walls of wild-type plants (black bars) and the aba1-6 mutant (gray bars). Data represent average values ± se of three replicates. Asterisks indicate values statistically different from those of mutant plants (Student’s t test); ND indicates no significant differences (P ≤ 0.05).
To obtain a more quantitative analysis of the cell wall changes in aba1-6 plants, we determined the noncellulosic neutral monosaccharide composition of leaves from noninoculated 3-week-old aba1-6 and Col-0 plants (Blakeney et al., 1983; Reiter et al., 1993) as well as cellulose and uronic acid contents of their walls (Fig. 6, D and E). We found that the amount of cellulose was much lower in aba1-6 samples than in wild-type plants, whereas the content of uronic acid was much higher in the mutant than in the wild-type plants (Fig. 6D). No significant changes were observed in noncellulosic neutral sugar between aba1-6 samples and wild-type plants (Fig. 6E). To further correlate these cell wall alterations in the aba1-6 mutant with its resistance phenotype, we determined the cellulose and uronic acid contents, as well as the noncellulosic neutral monosaccharide composition, in leaves from noninoculated 3-week-old quadruple pyr/pyl and triple abi2-2 mutants. As shown in Supplemental Figure S6, no significant alterations in the contents of uronic acid and cellulose were found between these mutants and wild-type plants, whereas a subtle reduction in the content of noncellulosic neutral sugars was found in the triple abi2-2 mutant compared with that in wild-type plants. All together, these results revealed that disruption of ABA biosynthesis, but not ABA signaling, leads to a modification of cell wall structure and composition. However, the lack of correlation between altered cell wall chemistry and basal resistance in the pyr/pyl mutant and the triple abi2-1 mutant suggests that these cell wall modifications have no significant contribution to ABA-regulated resistance.
DISCUSSION
ABA has multiple and diverse functions in plants, such as the regulation of development-associated processes, the response to abiotic stresses, and the outcome of pathogen infections (Fujita et al., 2006; Wasilewska et al., 2008). For the latter, positive and negative functions have been described for ABA depending on the specific plant-pathogen interaction studied (Mauch-Mani and Mauch, 2005; Ton et al., 2009). Here, we show that ABA plays a negative regulatory function in balancing Arabidopsis resistance response to PcBMM, a necrotrophic fungal pathogen that naturally colonizes a broad range of Arabidopsis accessions (Llorente et al., 2005; Sánchez-Vallet et al., 2010). The aba1-6 mutant defective in ABA biosynthesis and the quadruple pyr/pyl mutant impaired in four ABA receptors (Niyogi et al., 1998; Park et al., 2009) were more resistant to PcBMM than wild-type plants, whereas the triple hab1-1abi1-2abi2-1 mutant, which has a constitutive activation of ABA signaling due to the impairment of three PP2C phosphatases (from clade A) that negatively regulate ABA signaling (Rubio et al., 2009), displayed an enhanced susceptibility to this fungal pathogen (Fig. 1). Thus, in addition to ABA biosynthesis, we show here that the ABA signaling pathway is essential for the suppression of Arabidopsis resistance to necrotrophic fungi, as impairment or constitutive activation of the pathway led to enhanced or reduced resistance responses, respectively. In line with these data, treatment of the ABA-deficient mutant aba1-1 (Landsberg erecta background) with ABA (50 μm) reverted its enhanced resistance to PcBMM to the susceptibility levels observed in wild-type plants (Supplemental Table S7). The data presented here are consistent with the negative regulatory role of ABA seen on resistance in tomato to the necrotrophic pathogens B. cinerea and D. dadantii and in Arabidopsis to the fungi F. oxysporum and B. cinerea (Audenaert et al., 2002; Anderson et al., 2004; Asselbergh et al., 2008a; L’Haridon et al., 2011). In contrast, ABA plays a positive regulatory role in Arabidopsis basal resistance to the necrotrophic fungus A. brassicicola or the vascular oomycete P. irregulare and in the modulation of ocp3 resistance to necrotrophic fungi (Adie et al., 2007; Flors et al., 2008; García-Andrade et al., 2011). Strikingly, a positive role of ABA in resistance to P. cucumerina has been described previously (e.g. the aba2 mutant was shown to be hypersusceptible to the fungus), and this function has been associated with the regulation of callose deposition upon infection (García-Andrade et al., 2011). However, in these experiments, the plants used for inoculation were older (5 weeks old) than those used here (3 weeks old), and drop inoculation instead of spray inoculation of expanded leaves was performed (García-Andrade et al., 2011). Moreover, under our experimental conditions, callose deposition does not seem to be essential for resistance to PcBMM, as the susceptibility of the callose-deficient pmr4 plants to the fungus was similar to that of wild-type plants (Supplemental Fig. S2). The data presented here reinforce the different roles that ABA plays in plant basal resistance depending on the plant-pathogen interaction analyzed and also suggest a putative effect of plant developmental stage and growth environment conditions on ABA regulatory function. This last conclusion is in line with the recently described effect of humidity on the effectiveness of Arabidopsis wound-induced resistance to B. cinerea (L’Haridon et al., 2011).
Comparative transcriptomic analysis of the aba1-6 mutant and wild-type plants demonstrated that in aba1-6 there is a constitutive activation of resistance responses, as the expression of a significant subset of defense-related genes was up-regulated in the mutant compared with wild-type plants (Table I). Furthermore, we found a significant overlap (60% of the genes) between the transcriptomic profiles of untreated aba1-6 mutant and P. cucumerina-infected wild-type plants 1 dpi. This is a clear indication that the Arabidopsis defensive responses activated upon fungal infection were constitutively up-regulated in aba1-6 plants and, therefore, that ABA negatively regulates these responses against this fungal pathogen. Among the defensive proteins encoded by constitutively up-regulated genes in aba1-6 were antimicrobial proteins (e.g. PDF1.2, PR-1, and LTP3), enzymes involved in the biosynthesis and target delivery of Trp-derived secondary metabolites, such as indole glucosinolates and camalexin (e.g. PEN3, PAD3, and CYP81F2), and the ATRBOHD and PRX33/PRX34 proteins that are involved in ROS generation (Fig. 2). The enhanced accumulation of antimicrobial proteins and secondary metabolites in Arabidopsis has been shown to confer broad-spectrum resistance to pathogens, including the necrotroph PcBMM (García-Olmedo et al., 2001; Stein et al., 2006; Hernández-Blanco et al., 2007; Bednarek et al., 2009; Sánchez-Vallet et al., 2010). ROS accumulation has been linked to the enhanced resistance phenotype of some ABA mutants (Asselbergh et al., 2007; L’Haridon et al., 2011), but a significant increase in ROS production upon PcBMM infection was not observed in the ABA mutants in comparison with wild-type plants (Fig. 1E). Notably, some transcriptional factors, such as WRKY33 and ORA59, that regulate Arabidopsis immune responses to pathogens, including necrotrophic fungi (Supplemental Fig. S3; Zheng et al., 2006; Pré et al., 2008), were also up-regulated in unchallenged aba1-6 and quadruple pyr/pyl mutants (Fig. 2; data not shown). Interestingly, the cis-regulatory elements of the aba1-6 up-regulated genes were enriched in W-box and GCC-box sequences that bind transcriptional factors such as WRKY33 and ORA59, respectively (Rushton et al., 1996; Zarei et al., 2011). These data further corroborate the link between ABA signaling and the Arabidopsis resistance response to the necrotrophic fungus PcBMM.
Hierarchical clustering of genes differentially regulated in the aba1-6 mutant identified four statistically significant clusters (I–IV; Fig. 3; Supplemental Table S6). The functions of the genes contained in these clusters suggested an extensive and complex cross talk among different signaling pathways. For example, two of the aba1-6 clusters contained up-regulated genes whose expression was positively modulated by either JA or SA treatment (Fig. 3). Genetic disruption of these pathways by generating the coi1-1 aba1-6, aos aba1-6, and sid2-1 aba1-6 double mutants led to a partial impairment of the aba1-6 resistance phenotype (Fig. 4; data not shown). Similarly, the cluster analysis suggested an ET function in aba1-6 resistance, which was further corroborated by showing that the ein2-1 aba1-6 double mutant was more susceptible to PcBMM than aba1-6 plants (Fig. 4). These data are in accordance with previous reports describing the antagonistic interaction between ABA and the JA, ET, and SA signaling pathways in the regulation of plant resistance to pathogens. With a few exceptions, these antagonistic interactions were hypothesized based on comparative gene expression analysis (Anderson et al., 2004; Adie et al., 2007; de Torres-Zabala et al., 2007; Asselbergh et al., 2008b; Flors et al., 2008). Here, we have genetically validated some of the interactions of this complex network and also found additional, previously uncharacterized ones (Fig. 7). Notably, the interaction between ABA and the JA and SA hormones in balancing the resistance response of aba1-6 to PcBMM seems to take place at the signaling level, as the content of these hormones in aba1-6 and quadruple pyr/pyl mutants (untreated or PcBMM inoculated) was not higher than those found in wild-type plants. Moreover, ABA content was not increased in PcBMM-inoculated wild-type plants (Supplemental Fig. S4), and exogenous application of ABA (50 μm) did not have any effect on the resistance of wild-type plants, whereas it restored the resistance of the aba1 mutant to the susceptible levels of wild-type plants (Supplemental Table S7). In contrast to their genetic interaction in the regulation of Arabidopsis innate immunity, JA, ET, and SA signaling do not interfere with ABA function in the regulation of Arabidopsis responses to abiotic stress, such as drought (Fig. 5).
Figure 7.
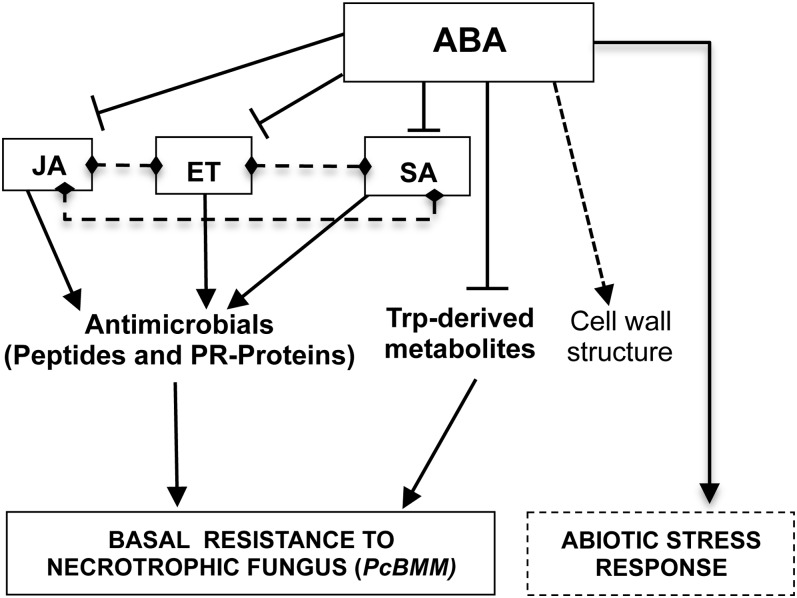
The complex regulatory function of ABA in Arabidopsis resistance to the necrotrophic fungus PcBMM. Upon Arabidopsis infection with the fungus, the activation of ET, SA, and JA pathways and the biosynthesis of Trp-derived metabolites occur, positively regulating (arrows) the basal resistance response. ABA signaling negatively regulates (lines) these pathways; therefore, inactivation of ABA signaling (e.g. aba1-6 or quadruple pyr/pyl) leads to their constitutive activation and an enhanced resistance to the fungus. Previously described interactions among ET, SA, and JA signaling are indicated by dotted lines (for additional details, see text). The putative function of ABA in regulating wall architecture is indicated by the dotted arrow. The regulation of abiotic stress responses by ABA seems to be independent of SA, ET, and JA.
The intermediate resistance phenotype observed in the double mutants in comparison with that of aba1-6 plants suggests that ABA, JA, ET, and SA signaling pathways cooperatively contribute to Arabidopsis resistance to PcBMM. How these pathways interact at the molecular level to regulate this resistance needs further characterization, but it seems to be more complex than initially anticipated (Tsuda et al., 2009). For example, the constitutive overexpression of the SA marker gene PR-1 in the ein2-1 aba1-6 and aos aba1-6 double mutants (Fig. 4C) was not linked to an enhanced accumulation of SA in these double mutants (data not shown). Moreover, the ET/JA marker gene PDF1.2 was similarly up-regulated in the aba1-6 plants and in the aos aba1-6 double mutant defective in JA biosynthesis (Fig. 4C). Therefore, the results presented here indicate that an intricate and complex regulatory network among these pathways occurs upon Arabidopsis challenge with necrotrophic pathogens (Fig. 7). It cannot be discarded that additional hormones such as auxin, gibberellin, cytokinin, and brassinosteroids might be involved in this multifaceted regulatory network conferring resistance to necrotrophic fungi (Bari and Jones, 2009).
Comparative transcriptomic analyses and genetic data revealed that in addition to the defensive responses regulated by hormones, other mechanisms might contribute to explain the resistance phenotypes of ABA mutants. For example, a significant number of the aba1-6 differently regulated genes encoded proteins that were related to cell wall biosynthesis/modification (Fig. 2C). In line with these data, we found that aba1-6 plants, which have a dwarf phenotype, showed alterations in the structure/composition of their walls compared with that of wild-type plants (Fig. 6), further suggesting that aba1-6 resistance can be partially dependent on these wall alterations. Our data are in line with the described cell wall alterations found in the tomato ABA-deficient sitiens mutant (Asselbergh et al., 2007) and with the hypothesized modulation of cell wall architecture by ABA signaling (Gimeno-Gilles et al., 2009). The relevance of the cell wall in directly mediating the resistance of Arabidopsis to different pathogens, including PcBMM, has been reported previously (Hernández-Blanco et al., 2007; Denoux et al., 2008; Sánchez-Rodríguez et al., 2009; Ramírez et al., 2011; Delgado-Cerezo et al., 2012). However, we did not find similar wall modifications in the PcBMM-resistant quadruple pyr/pyl mutant; therefore, it is tentative to speculate about the putative contribution of aba1-6 wall alterations to PcBMM resistance. Also, a decrease in cuticle permeability and the alteration in callose deposition at the cell wall have been suggested to explain the resistance phenotypes of Arabidopsis and tomato ABA-deficient mutants (Ton and Mauch-Mani, 2004; Asselbergh et al., 2007a; García-Andrade et al., 2011; L’Haridon et al., 2011). However, in our plant growth and inoculation conditions, callose does not seem to contribute to the enhanced resistance to PcBMM of the aba1-6 and quadruple pyr/pyl mutants (Fig. 1E; Supplemental Fig. S2). In conclusion, with this work, we elucidate the complex role that phytohormones, in particular ABA, play in balancing plant resistance to pathogens.
MATERIALS AND METHODS
Biological Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in phytochambers on a sterilized mixture of soil and vermiculite (3:1) with a 10-h-day/14-h-night photoperiod, a temperature of 22°C day/20°C night, and 50% relative humidity (Hernández-Blanco et al., 2007). Light intensity was fixed to 120 to 150 µmol m−2 s−1, according to Weigel and Glazebrook (2002). The following lines, in the Col-0 background, were used: aba1-6 (Niyogi et al., 1998), pyr1pyl1pyl2pyl4 (Park et al., 2009), hab1-1abi1-2abi2-2 (Rubio et al., 2009), sid2-1 (Nawrath and Métraux, 1999), ein2-1 (Guzmán and Ecker, 1990), agb1-1 (Lease et al., 2001), coi1-1 (Xie et al., 1998), wrky33 (salk_006603), and pmr4 and pmr4 sid2-1 (Nishimura et al., 2003). The aos mutant is in the Col-6 background (gl1; Park et al., 2002), and the aba1-1 allele is in the Landsberg erecta background (Koornneef et al., 1982). The Plectosphaerella cucumerina strain BMM was provided by Dr. B. Mauch-Mani (University of Neuchatel).
Double mutants were generated by standard genetic crosses followed by identification of the mutant alleles. Genotyping of the aba1-6 mutation was confirmed by sequencing the PCR product of 5′-GAATCGGAGGATTGGTGTTTG-3′ and 5′-CTCCAGAATCTTCAAAATCAAC-3′. The aos mutation was confirmed by detection of the T-DNA insert using the flanking primers (5′-GTACGATCAAAGCCGTTGAAATTCCGAGTTTT-3′ and 5′-TGGAACAAGAAAACAGATGGACTACACAGGT-3′) and the T-DNA internal primer (5′-CGGGCCTAACTTTTGGTGTGATGATGCT-3′; Park et al., 2002). The sid2-1 mutation was detected by selective digestion with MunI of the product of 5′-TGTCTGCAGTGAAGCTTTGG-3′ and 5′-CACAAACAGCTGGAGTTGGA-3′. The ein2-1 mutation was selected as reported (Guzmán and Ecker, 1990). The coi1-1 mutant was selected by determining plant insensitivity to JA (Xie et al., 1998).
Biological Assays
Three-week-old Arabidopsis plants were spray inoculated with a spore suspension (4 × 106 spores mL−1) of PcBMM (0.6 mL per pot containing five plants). After inoculation, plants were transferred to a growth chamber with a 10-h-day/14-h-night photoperiod, a temperature of 24°C day/22°C night, and light intensity was maintained at 120 to 150 µmol m−2 s−1. Inoculated plants were kept under high humidity in covered trays. Disease progression was estimated by determining the average DR (0–5; 0, no infection; 5, dead plant), TB staining, and relative quantification of fungal DNA by qPCR as described previously (Sánchez-Vallet et al., 2010). At least three independent experiments were performed, and statistically significant differences among the inoculated Arabidopsis genotypes were determined by one-way ANOVA and Bonferroni post hoc test, as reported previously (Sánchez-Rodríguez et al., 2009). For ABA treatment, aba1-1 and wild-type plants were sprayed with ABA (50 μm) 24 and 48 h prior to inoculation with PcBMM, and then the DR was determined at different dpi and expressed as a percentage of disease susceptibility in wild-type plants (100%). Water loss assays were performed as described by Saez et al. (2006). Three samples containing four rosettes of 3-week-old plants were excised and weighed separately at different time points, and the average values (±se) were determined. This experiment was performed three times.
Plant Tissue Staining
TB staining was performed on mock- and PcBMM-inoculated leaves at 20 hpi (for hyphal growth analysis) and 48 hpi (for cell death detection; Sánchez-Vallet et al., 2010). To determine the production of hydrogen peroxide, DAB (Sigma) staining was performed on at least six leaves collected 24 hpi and vacuum infiltrated with the DAB solution (1 mg mL−1), placed in a plastic box under high humidity until brown precipitate was observed (3 h), and then fixed with 3:1:1 ethanol:lactic acid:glycerol (Torres et al., 2002). Callose deposition was determined by aniline blue staining in leaves harvested 24 hpi, which were incubated in 95% ethanol and then washed with a solution of 0.07 m phosphate buffer (pH 7) prior to their staining with aniline blue (0.02% in 0.07 m phosphate buffer, pH 7; Luna et al., 2011). Leaves were mounted on slides with glycerol and observed with an epifluorescence microscope using UV filters. Pixel quantification of callose was performed using Adobe Photoshop (Luna et al., 2011). At least four leaves per genotype were used for pixel quantification in the experiments performed.
Gene Expression Analyses
RNA extractions from PcBMM-inoculated or mock-treated plants were done as described (Llorente et al., 2005). Real-time qPCR analyses were performed as reported previously (Sánchez-Vallet et al., 2010). The oligonucleotide sequences, designed using Primer Express version 2.0 (Applied Biosystems), used for qPCR have been described previously (Sánchez-Vallet et al., 2010), except those of ORA59 (5′-GCAGCCTCGCAGTACTCAATTT-3′ and 5′-TCAAGGCTATCACCGGAGACTC-3′) and LOX2 (5′-ATCAACAAGCCCCAATGGAA-3′ and 5′-CGGCGTCATGAGAGATAGCAT-3′). qPCR results are mean values (±sd) from two technical replicates. Differences in expression ratios among the samples were analyzed by ANOVA (lsd test) using StatGraphics (StatPoint Technologies). Experiments for qPCR were performed at least three times.
For the microarray experiment, approximately 25 rosettes from 3-week-old noninoculated, mock-treated (1 dpi) or P. cucumerina-inoculated Col-0 and aba1-6 plants (1 dpi) were collected (four biological replicates). Plant total RNA was extracted as described previously (Berrocal-Lobo et al., 2002) and further purified using the RNeasy Kit (Qiagen). RNA quality was tested by using a Bioanalyzer 2100 (Agilent Technologies). Three of the four biological replicates were independently hybridized for each transcriptomic comparison. Biotinylated complementary RNA (20 μg) was prepared, and the resulting complementary RNA was used to hybridize ATH1 Arabidopsis GeneChips (Affymetrix) using the manufacturer’s protocols at the Genomic Unit of the Centro Nacional de Biotecnología-Consejo Superior de Investigaciones Científicas. Array images were analyzed with a GenePix 400B scanner (Molecular Devices) at 10-mm resolution. The images were quantified with GenePixPro 5.1. Gene expression levels were analyzed with GeneSpring 7.2 software (Silicon Genetics), and the chip-to-chip signal variation was minimized by normalizing signal intensities to the averaged intensity values of wild-type plants using the expression levels of the top 20th percentile of probe sets. Differentially expressed genes in untreated mutants relative to wild-type samples or in PcBMM-inoculated relative to the mock-treated samples were identified using two-way ANOVA and a Benjamini and Hochberg multiple testing correction (GeneSpring 7.2), as described previously (Stein et al., 2006). Genes were considered differentially expressed at P ≤ 0.01. Up- and down-regulated genes were selected using normalized values higher than 2-fold or lower than 0.5-fold relative to control plants.
The Classification SuperViewer Tool (Bio-Array Resource, University of Toronto; http://bbc.botany.utoronto.ca/; Provart et al., 2003) was used to generate an overview of the Gene Ontology classification of the list of differentially regulated genes in aba1-6 and wild-type plants. The Genevestigator Meta-Analyzer tool (www.genevestigator.ethz.ch/; Zimmermann et al., 2004) was used to analyze expression profiles of differentially regulated genes in the microarray database. Hierarchical clustering of differentially expressed probe sets in aba1-6 and wild-type plants was performed using Pearson uncentered distance, average linkage, and the Multiexperiment Viewer application from TM4 software (http://www.tm4.org/; Saeed et al., 2003).
The presence of conserved putative cis-regulatory elements in the selected genes was determined with Transplanta software (http://bioinfogp.cnb.csic.es/transplanta_dev/), which integrates a suite of bioinformatic tools (Bioprospector [http://ai.stanford.edu/∼xsliu/BioProspector/], DME [http://rulai.cshl.edu/dme/], MEME [http://meme.nbcr.net/meme/], and Motif Sampler [http://bayesweb.wadsworth.org/gibbs/gibbs.html]). Subsequently, TOMTOM (http://meme.nbcr.net/meme/cgi-bin/tomtom.cgi; Gupta et al., 2007) and PLACE (http://www.dna.affrc.go.jp/PLACE/; Higo et al., 1999) were used to identify the statistically overrepresented regulatory elements in the promoters of the genes from each cluster.
Quantification of Plant Hormones
Three-week-old plants were collected before and after treatment (1 dpi) with water or a spore suspension of PcBMM. They were frozen immediately in liquid nitrogen and subsequently freeze dried. Tissue was then ground to a powder in a tissue lyser (Qiagen; http://www.qiagen.com/). Hormone extractions were performed on 10 mg of freeze-dried tissue as described by Forcat et al. (2008). Data are averages of three independent replicates ± se. Statistical differences among Arabidopsis genotypes were determined by one-way ANOVA and lsd post hoc test.
Cell Wall Analysis
For FTIR, precleaned (by solvent extraction) dried leaves from at least 30 3-week-old individual plants per genotype were pooled and homogenized by ball milling. The powder was dried and mixed with potassium bromide and then pressed into 13-mm pellets. For each plant genotype, 15 FTIR spectra were collected on a Thermo Nicolet Nexus 470 spectrometer (ThermoElectric) over a range of 4,000 to 400 cm–1. For each spectrum, 32 scans were coadded at a resolution of 4 cm–1 for Fourier transform processing and absorbance spectrum calculation using OMNIC software (Thermo Nicolet). Using win-das software (Wiley), spectra were baseline corrected, normalized, and analyzed using the principal component analysis covariance matrix method (Kemsley et al., 1996). Cell wall monosaccharides were assayed as alditol acetate derivatives (Stevenson and Furneaux, 1991) by gas chromatography performed on an Agilent 6890N gas chromatograph, and the results obtained were validated with the method described previously by Gibeaut and Carpita (1991). Myoinositol (Sigma-Aldrich) was added as an internal standard. Cellulose was determined on the fraction resistant to extraction with 2 m trifluoroacetic acid (n = 4) by phenol-sulfuric assay using Glc equivalents as standard (DuBois et al., 1956). Uronic acids were quantified using the soluble 2 m trifluoroacetic acid fraction (n = 5; Filisetti-Cozzi and Carpita, 1991). The data were analyzed using two-way Student’s t test (P = 0.05). All statistical analyses were performed using the statistical software package SPSS 13.0.
Expression data from this article was deposited according to Minimum Information About a Microarray Experiment standards at ArrayExpress (EMBL-European Bioinformatics Institute) with the accession number E-MEXP-3733.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Infection of aba1-6 and wild-type plants with the fungal transformant PcBMM-GFP.
Supplemental Figure S2. Callose deposition is not essential for Arabidopsis resistance to PcBMM.
Supplemental Figure S3. Susceptibility of the wkry33-1 mutant to PcBMM.
Supplemental Figure S4. Determination of ABA, JA, and SA concentrations in leaves of wild-type plants and ABA-defective mutants.
Supplemental Figure S5. Development-associated phenotypes of 3-week-old single and double mutants defective in different hormone pathways.
Supplemental Figure S6. Biochemical analyses of the cell walls from quadruple pyr/pyl and triple abi2-2 mutants and wild-type plants.
Supplemental Table S1. Genes differentially regulated in untreated (time zero) aba1-6 plants compared with wild-type plants.
Supplemental Table S2. Genes differentially regulated in wild-type plants inoculated with PcBMM (1 dpi) compared with mock-treated plants.
Supplemental Table S3. Genes differentially regulated in aba1-6 inoculated with PcBMM (1 dpi) compared with mock-treated plants.
Supplemental Table S4. Genes differentially regulated in untreated (time zero) aba1-6 plants compared with wild-type plants and in wild-type plants inoculated with PcBMM (1 dpi) compared with mock-treated plants
Supplemental Table S5. Genes contained in the “cell wall” Gene Ontology category.
Supplemental Table S6. Genes included in clusters I to IV described in Figure 3.
Supplemental Table S7. Effect of ABA treatment on aba1-1 resistance to PcBMM.
Acknowledgments
We thank Dr. Roberto Solano and the Unidad de Genómica from the Centro Nacional de Biotecnología-Consejo Superior de Investigaciones Científicas for technical assistance with transcriptomic analyses as well as Dr. Marta de Torres-Zabala (College of Life and Environmental Sciences, University of Exeter) for technical assistance with hormone quantification and critical reading of the manuscript.
Glossary
- SA
salicylic acid
- ET
ethylene
- JA
jasmonic acid
- ABA
abscisic acid
- ROS
reactive oxygen species
- SAR
systemic acquired resistance
- Col-0
ecotype Columbia
- hpi
hours post inoculation
- dpi
days post inoculation
- TB
trypan blue
- qPCR
quantitative PCR
- DR
disease rating
- DAB
3,3′-diaminobenzidine
- ACC
1-aminocyclopropane-1-carboxylic acid
- FTIR
Fourier transform infrared
References
- Adie BAT, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sánchez-Serrano J-J, Schmelz EA, Solano R. (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K. (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, Achuo AE, Höfte M, Van Gijsegem F. (2008a) Abscisic acid deficiency leads to rapid activation of tomato defence responses upon infection with Erwinia chrysanthemi. Mol Plant Pathol 9: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, Curvers K, Franca SC, Audenaert K, Vuylsteke M, Van Breusegem F, Höfte M. (2007) Resistance to Botrytis cinerea in sitiens, an abscisic acid-deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiol 144: 1863–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, De Vleesschauwer D, Höfte M. (2008b) Global switches and fine-tuning-ABA modulates plant pathogen defense. Mol Plant Microbe Interact 21: 709–719 [DOI] [PubMed] [Google Scholar]
- Audenaert K, De Meyer GB, Höfte MM. (2002) Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol 128: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Jones JD. (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Barrero JM, Piqueras P, González-Guzmán M, Serrano R, Rodríguez PL, Ponce MR, Micol JL. (2005) A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. J Exp Bot 56: 2071–2083 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R. (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Bindschedler LV, Dewdney J, Blee KA, Stone JM, Asai T, Plotnikov J, Denoux C, Hayes T, Gerrish C, Davies DR, et al. (2006) Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J 47: 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeney NB, Harris PJ, Henry RJ, Stone BA. (1983) A simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr Res 182: 291–299 [Google Scholar]
- Cantu D, Vicente AR, Greve LC, Dewey FM, Bennett AB, Labavitch JM, Powell AL. (2008) The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc Natl Acad Sci USA 105: 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curvers K, Seifi H, Mouille G, de Rycke R, Asselbergh B, Van Hecke A, Vanderschaeghe D, Höfte H, Callewaert N, Van Breusegem F, et al. , et al (2010) Abscisic acid deficiency causes changes in cuticle permeability and pectin composition that influence tomato resistance to Botrytis cinerea. Plant Physiol 154: 847–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Cerezo M, Sánchez-Rodríguez C, Escudero V, Miedes E, Fernández PV, Jordá L, Hernández-Blanco C, Sánchez-Vallet A, Bednarek P, Schulze-Lefert P, et al. (2012) Arabidopsis heterotrimeric G-protein regulates cell wall defense and resistance to necrotrophic fungi. Mol Plant 5: 98–114 [DOI] [PubMed] [Google Scholar]
- Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, De Lorenzo G, Ferrari S, Ausubel FM, Dewdney J. (2008) Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol Plant 1: 423–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres-Zabala M, Bennett MH, Truman WH, Grant MR. (2009) Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J 59: 375–386 [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bögre L, Grant M. (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J 26: 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28: 350–356 [Google Scholar]
- Eulgem T, Somssich IE. (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Filisetti-Cozzi TMCC, Carpita NC. (1991) Measurement of uronic acids without interference from neutral sugars. Anal Biochem 197: 157–162 [DOI] [PubMed] [Google Scholar]
- Flors V, Ton J, Jakab G, Mauch-Mani B. (2005) Abscisic acid and callose: team players in defence against pathogens? J Phytopathol 153: 377–383 [Google Scholar]
- Flors V, Ton J, van Doorn R, Jakab G, García-Agustín P, Mauch-Mani B. (2008) Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J 54: 81–92 [DOI] [PubMed] [Google Scholar]
- Forcat S, Bennett MH, Mansfield JW, Grant MR. (2008) A rapid and robust method for simultaneously measuring changes in the phytohormones ABA, JA and SA in plants following biotic and abiotic stress. Plant Methods 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9: 436–442 [DOI] [PubMed] [Google Scholar]
- García-Andrade J, Ramírez V, Flors V, Vera P. (2011) Arabidopsis ocp3 mutant reveals a mechanism linking ABA and JA to pathogen-induced callose deposition. Plant J 67: 783–794 [DOI] [PubMed] [Google Scholar]
- García-Olmedo F, Rodríguez-Palenzuela P, Molina A, Alamillo JM, López-Solanilla E, Berrocal-Lobo M, Poza-Carrión C. (2001) Antibiotic activities of peptides, hydrogen peroxide and peroxynitrite in plant defence. FEBS Lett 498: 219–222 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Carpita NC. (1991) Tracing cell wall biogenesis in intact cells and plants: selective turnover and alteration of soluble and cell wall polysaccharides in grasses. Plant Physiol 97: 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno-Gilles C, Lelièvre E, Viau L, Malik-Ghulam M, Ricoult C, Niebel A, Leduc N, Limami AM. (2009) ABA-mediated inhibition of germination is related to the inhibition of genes encoding cell-wall biosynthetic and architecture-modifying enzymes and structural proteins in Medicago truncatula embryo axis. Mol Plant 2: 108–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Ausubel FM. (1994) Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA 91: 8955–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al. (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. (2007) Quantifying similarity between motifs. Genome Biol 8: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR. (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejgaard J, Jacobsen S, Bjørn SE, Kragh KM. (1992) Antifungal activity of chitin-binding PR-4 type proteins from barley grain and stressed leaf. FEBS Lett 307: 389–392 [DOI] [PubMed] [Google Scholar]
- Hernández-Blanco C, Feng DX, Hu J, Sánchez-Vallet A, Deslandes L, Llorente F, Berrocal-Lobo M, Keller H, Barlet X, Sánchez-Rodríguez C, et al. (2007) Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19: 890–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauráková M, Capek P, Sasinková V, Wellner N, Ebringerová A. (2000) FT-IR study of plant cell wall model compounds: pectic polysaccharides and hemicelluloses. Carbohydr Polym 43: 195–203 [Google Scholar]
- Kemsley EK, Holland JK, Defernez M, Wilson RH. (1996) Detection of adulteration of raspberry purees using infrared spectroscopy and chemometrics. J Agric Food Chem 44: 3864–3870 [Google Scholar]
- Kim TH, Hauser F, Ha T, Xue S, Böhmer M, Nishimura N, Munemasa S, Hubbard K, Peine N, Lee BH, et al. (2011) Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr Biol 21: 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Jorna ML, Brinkhorst-Van der Swan DLC, Karssen CM. (1982) The isolation of abscisic acid (ABA)-deficient mutants by selection of induced revertants in non-germinating gibberellin-sensitive lines of Arabidopsis thaliana. Theor Appl Genet 61: 385–393 [DOI] [PubMed] [Google Scholar]
- La Camera S, Geoffroy P, Samaha H, Ndiaye A, Rahim G, Legrand M, Heitz T. (2005) A pathogen-inducible patatin-like lipid acyl hydrolase facilitates fungal and bacterial host colonization in Arabidopsis. Plant J 44: 810–825 [DOI] [PubMed] [Google Scholar]
- La Camera S, L’Haridon F, Astier J, Zander M, Abou-Mansour E, Page G, Thurow C, Wendehenne D, Gatz C, Métraux JP, et al. (2011) The glutaredoxin ATGRXS13 is required to facilitate Botrytis cinerea infection of Arabidopsis thaliana plants. Plant J 68: 507–519 [DOI] [PubMed] [Google Scholar]
- Lease KA, Wen J, Li J, Doke JT, Liscum E, Walker JC. (2001) A mutant Arabidopsis heterotrimeric G-protein beta subunit affects leaf, flower, and fruit development. Plant Cell 13: 2631–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Haridon F, Besson-Bard A, Binda M, Serrano M, Abou-Mansour E, Balet F, Schoonbeek HJ, Hess S, Mir R, Léon J, et al. (2011) A permeable cuticle is associated with the release of reactive oxygen species and induction of innate immunity. PLoS Pathog 7: e1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente F, Alonso-Blanco C, Sánchez-Rodriguez C, Jorda L, Molina A. (2005) ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J 43: 165–180 [DOI] [PubMed] [Google Scholar]
- Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, Ton J. (2011) Callose deposition: a multifaceted plant defense response. Mol Plant Microbe Interact 24: 183–193 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Manners JM, Penninckx IA, Vermaere K, Kazan K, Brown RL, Morgan A, Maclean DJ, Curtis MD, Cammue BP, Broekaert WF. (1998) The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol Biol 38: 1071–1080 [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Mauch F. (2005) The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol 8: 409–414 [DOI] [PubMed] [Google Scholar]
- Meier S, Bastian R, Donaldson L, Murray S, Bajic V, Gehring C. (2008) Co-expression and promoter content analyses assign a role in biotic and abiotic stress responses to plant natriuretic peptides. BMC Plant Biol 8: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J. (2007) Bacterial non-host resistance: interactions of Arabidopsis with non-adapted Pseudomonas syringae strains. Physiol Plant 131: 448–461 [DOI] [PubMed] [Google Scholar]
- Mohr PG, Cahill DM. (2007) Suppression by ABA of salicylic acid and lignin accumulation and the expression of multiple genes, in Arabidopsis infected with Pseudomonas syringae pv. tomato. Funct Integr Genomics 7: 181–191 [DOI] [PubMed] [Google Scholar]
- Monteiro S, Barakat M, Piçarra-Pereira MA, Teixeira AR, Ferreira RB. (2003) Osmotin and thaumatin from grape: a putative general defense mechanism against pathogenic fungi. Phytopathology 93: 1505–1512 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP. (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC. (2003) Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301: 969–972 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Björkman O. (1998) Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10: 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien JA, Daudi A, Finch P, Butt VS, Whitelegge JP, Souda P, Ausubel FM, Bolwell GP. (2012) A peroxidase-dependent apoplastic oxidative burst in cultured Arabidopsis cells functions in MAMP-elicited defense. Plant Physiol 158: 2013–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-H, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R. (2002) A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Park S-Y, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S, Uknes S, Lawton K, Winter AM, Chandler D, DiMaio J, Novitzky R, Ward E, Ryals J. (1993) Regulation of a hevein-like gene in Arabidopsis. Mol Plant Microbe Interact 6: 680–685 [DOI] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CMJ, Memelink J. (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provart NJ, Gil P, Chen W, Han B, Chang HS, Wang X, Zhu T. (2003) Gene expression phenotypes of Arabidopsis associated with sensitivity to low temperatures. Plant Physiol 132: 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Ramírez V, García-Andrade J, Vera P. (2011) Enhanced disease resistance to Botrytis cinerea in myb46 Arabidopsis plants is associated to an early down-regulation of CesA genes. Plant Signal Behav 6: 911–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos B, Gonzalez-Melendi P, Sánchez-Vallet A, Sánchez-Rodríguez C, López G, Molina A. (2012) Functional genomic tools to decipher the pathogenicity mechanisms of the necrotrophic fungus Plectosphaerella cucumerina in Arabidopsis thaliana. Mol Plant Pathol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter WD, Chapple CC, Somerville CR. (1993) Altered growth and cell wall a fucose-deficient mutant of Arabidopsis. Science 261: 1032–1035 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JDG. (2007) Pathological hormone imbalances. Curr Opin Plant Biol 10: 372–379 [DOI] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL. (2009) Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol 150: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15: 5690–5700 [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Saez A, Robert N, Maktabi MH, Schroeder JI, Serrano R, Rodriguez PL. (2006) Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol 141: 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C, Estévez JM, Llorente F, Hernández-Blanco C, Jordá L, Pagán I, Berrocal M, Marco Y, Somerville S, Molina A. (2009) The ERECTA receptor-like kinase regulates cell wall mediated resistance to pathogens in Arabidopsis thaliana. Mol Plant Microbe Interact 22: 953–963 [DOI] [PubMed] [Google Scholar]
- Sánchez-Vallet A, Ramos B, Bednarek P, López G, Piślewska-Bednarek M, Schulze-Lefert P, Molina A. (2010) Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. Plant J 63: 115–127 [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9: 236–243 [DOI] [PubMed] [Google Scholar]
- Silverstein KA, Graham MA, Paape TD, VandenBosch KA. (2005) Genome organization of more than 300 defensin-like genes in Arabidopsis. Plant Physiol 138: 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X. (2008) Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3: 348–351 [DOI] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S. (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson TT, Furneaux RH. (1991) Chemical methods for the analysis of sulphated galactans from red algae. Carbohydr Res 210: 277–298 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Cammue BP, Thevissen K. (2002) Plant defensins. Planta 216: 193–202 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BP, Broekaert WF. (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95: 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Flors V, Mauch-Mani B. (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci 14: 310–317 [DOI] [PubMed] [Google Scholar]
- Ton J, Mauch-Mani B. (2004) Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J 38: 119–130 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JD. (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL. (2005) Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet 37: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon LC, Van Strien EA. (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55: 85–97 [Google Scholar]
- Verburg JG, Huynh QK. (1991) Purification and characterization of an antifungal chitinase from Arabidopsis thaliana. Plant Physiol 95: 450–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S. (2009) Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frei dit Frey N, Leung J. (2008) An update on abscisic acid signaling in plants and more.... Mol Plant 1: 198–217 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Xie D-X, Feys BF, James S, Nieto-Rostro M, Turner JG. (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Zhu JK. (2002) Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J Biol Chem 277: 8588–8596 [DOI] [PubMed] [Google Scholar]
- Yasuda M, Ishikawa A, Jikumaru Y, Seki M, Umezawa T, Asami T, Maruyama-Nakashita A, Kudo T, Shinozaki K, Yoshida S, et al. (2008) Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 20: 1678–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei A, Körbes AP, Younessi P, Montiel G, Champion A, Memelink J. (2011) Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol Biol 75: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T. (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48: 592–605 [DOI] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Glazebrook J. (1999) Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11: 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]