Abstract
The soybean (Glycine max) genome contains 18 members of the 14-3-3 protein family, but little is known about their association with specific phenotypes. Here, we report that the Glyma0529080 Soybean G-box Factor 14-3-3c (SGF14c) and Glyma08g12220 (SGF14l) genes, encoding 14-3-3 proteins, appear to play essential roles in soybean nodulation. Quantitative reverse transcription-polymerase chain reaction and western-immunoblot analyses showed that SGF14c mRNA and protein levels were specifically increased in abundance in nodulated soybean roots 10, 12, 16, and 20 d after inoculation with Bradyrhizobium japonicum. To investigate the role of SGF14c during soybean nodulation, RNA interference was employed to silence SGF14c expression in soybean roots using Agrobacterium rhizogenes-mediated root transformation. Due to the paleopolyploid nature of soybean, designing a specific RNA interference sequence that exclusively targeted SGF14c was not possible. Therefore, two highly similar paralogs (SGF14c and SGF14l) that have been shown to function as dimers were silenced. Transcriptomic and proteomic analyses showed that mRNA and protein levels were significantly reduced in the SGF14c/SGF14l-silenced roots, and these roots exhibited reduced numbers of mature nodules. In addition, SGF14c/SGF14l-silenced roots contained large numbers of arrested nodule primordia following B. japonicum inoculation. Transmission electron microscopy further revealed that the host cytoplasm and membranes, except the symbiosome membrane, were severely degraded in the failed nodules. Altogether, transcriptomic, proteomic, and cytological data suggest a critical role of one or both of these 14-3-3 proteins in early development stages of soybean nodules.
Protein phosphorylation is required for many biochemical functions of a cell, such as signal transduction, and 14-3-3 proteins are often involved in these reactions. The name 14-3-3 is based on the discovery of this group of proteins that were found in the 14th fraction of an ion-exchange column and the subsequent fraction 3.3 of a starch gel (Moore and Perez, 1967), but they were subsequently shown to be phospho-Ser/phospho-Thr-binding proteins present in most tissues examined. They function mainly as homodimers and heterodimers (Yaffe et al., 1997). Binding of 14-3-3s generally requires phosphorylation of a target protein with the 14-3-3-binding motif. Three major binding motifs have been identified (Bachmann et al., 1996; Fuglsang et al., 1999), with additional putative 14-3-3-binding motifs proposed more recently (Ottmann et al., 2007; Chan et al., 2011). The consequences of 14-3-3/target interactions were summarized by Darling et al. (2005) and Gökirmak et al., (2010) and include such effects as conformational changes that affect target protein activity, scaffolding, and movement. Performing these activities, 14-3-3 proteins function as regulators of important biological processes in plants, such as development, metabolism, transcription, organellar protein trafficking, and stress responses (Bunney et al., 2001; Roberts et al., 2002; Sehnke et al., 2002; Roberts, 2003; Shi et al., 2007). In legume species such as Medicago truncatula, plastid-located Gln synthetase (GS) is regulated by phosphorylation catalyzed by a calcium-dependent protein kinase and a 14-3-3 interaction when GS phosphorylation is mediated by nitrogen fixation in root nodules (Lima et al., 2006a, 2006b).
The full genome sequencing of numerous plant species has allowed for the reliable identification of 14-3-3 genes in plants by sequence homology. For example, a total of 15 14-3-3 genes, called General Regulatory Factor1 to GRF15, were identified in Arabidopsis (Arabidopsis thaliana), whereas only eight 14-3-3 genes were identified in the rice (Oryza sativa) genome (DeLille et al., 2001; Rosenquist et al., 2001; Chen et al., 2006; Yao et al., 2007). In the recently sequenced genome of soybean (Glycine max; Schmutz et al., 2010), 18 14-3-3 genes were identified and named Soybean G-box Factor 14-3-3a (SGF14a) to SGF14r (Li and Dhaubhadel, 2011).
There is evidence that 14-3-3 proteins play a role in plant-microbe interactions. For example, the expression of several 14-3-3 genes was altered by Pseudomonas syringae inoculation of soybean (Seehaus and Tenhaken, 1998; Finnie et al., 2002; Zou et al., 2005) as well as tomato (Solanum lycopersicum; Oh et al., 2010). Reaction to powdery mildew (Blumeria graminis) pathogens was shown to involve 14-3-3 proteins for both barley (Hordeum vulgare; Finnie et al., 2002) and Arabidopsis (Yang et al., 2009). In barley, 14-3-3 proteins were suggested to form a complex with an H+-ATPase to trigger defense, whereas in Arabidopsis, a 14-3-3 protein was reported to interact with the plant resistance protein RPW8.2 to affect the host response. A survey involving 10 14-3-3 genes from tomato showed that three were induced by the Cf9-mediated hypersensitive defense response (Roberts and Bowles, 1999). A final example is that of 14-3-3 proteins suggested in Agrobacterium rhizogenes to bind RolB and direct it to the nucleus (Moriuchi et al., 2004).
Although implicated in several plant-pathogen interactions, there are no published reports detailing the importance of 14-3-3 proteins in the symbiotic interaction involving rhizobia or the establishment of nodulation. A microarray study (Brechenmacher et al., 2008) identified a 14-3-3 gene, SGF14c, whose mRNA levels significantly increased in response to inoculation, suggesting a potential role for this gene during nodulation. The aim of this work was to use functional genomic tools to examine the role of SGF14c and its isoform and dimerization partner, SGF14l, during Bradyrhizobium japonicum-induced nodulation in soybean. In this study, we show that RNA interference (RNAi)-mediated silencing of SGF14c/SGF14l resulted in a severe reduction of nodule primordium development and maturation. In addition, transmission electron microscopy (TEM) showed that the cells within the nodule primordia were undergoing host cytoplasm degradation early in nodule development, when SGF14c/SGF14l are lacking. Together, these results reflect a critical role of 14-3-3 proteins during the establishment of mature soybean nodules.
RESULTS
Soybean 14-3-3 Gene Family
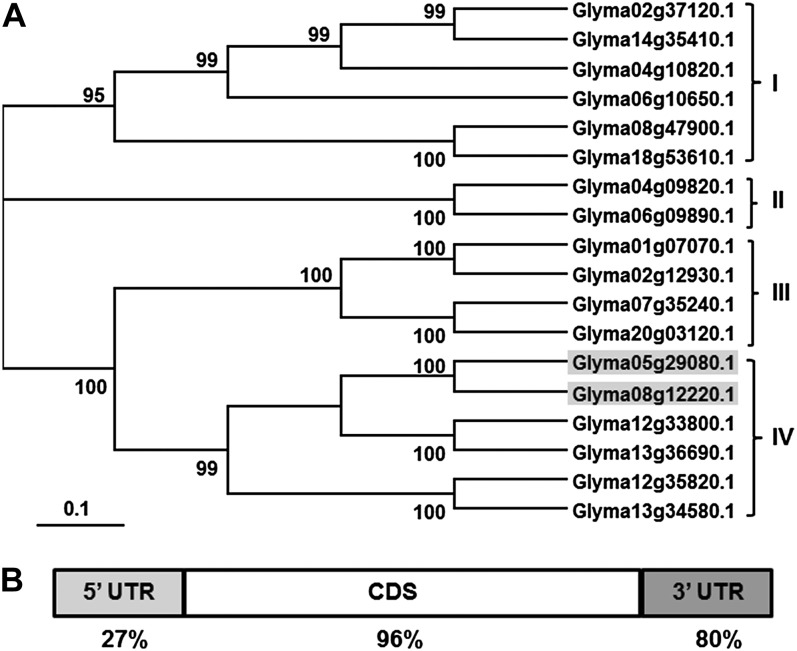
The two soybean 14-3-3 isoforms examined in this study were previously named SGF14c and SGF14l (Supplemental Table S1; Li and Dhaubhadel, 2011). DNA identity among soybean 14-3-3 genes (SGF14a–SGF14r) ranges from 45% to 97%, while the protein similarity ranges from 46% to 99% (Supplemental Fig. S1). A phylogenetic tree of these soybean genes (Fig. 1A) divides them into four clades (I–IV), supported by high bootstrap values ranging from 95 to 100. Clade I includes six genes with DNA identity ranging from 57% to 96%, while clade II includes two genes with DNA identity of 96%. Clade III includes four genes with DNA identity ranging from 84% to 95%, while clade IV includes six genes with DNA identity ranging from 61% to 96%. Alignment of soybean, Medicago truncatula, Lotus japonicus, and Arabidopsis 14-3-3 protein sequences (Supplemental Fig. S2) showed high similarity between the different members. The highest protein similarity of 98% was shown between MT5g046960 (M. truncatula) and Glyma01g07070 (soybean). A phylogenetic tree of 14-3-3 proteins from the four species (Supplemental Fig. S3) divided them into nine clades, with three of the nine clades containing members from the four species. Only one clade contained 14-3-3 protein members from soybean, M. truncatula, and L. japonicus. Two clades contained only 14-3-3 members belonging to Arabidopsis, and three other clades contained 14-3-3 proteins belonging to soybean with proteins either from M. truncatula or L. japonicus, reflecting the high level of protein similarity across the legume species.
Figure 1.
Phylogenetic tree of soybean 14-3-3 DNA sequences, and DNA sequence identity between SGF14c and SGF14l. A, The phylogenetic tree divided into four clades (I–IV). The numbers shown on branches are the percentages of bootstrap replications supporting the clades. Gene identifiers for SGF14c (Glyma05g29080) and SGF14l (Glyma08g12220) are highlighted in clade IV. Phylogenetic analysis was conducted using MEGA 3.1. B, DNA sequence identity of the 5′ UTR (27%), coding DNA sequence (CDS) region (96%), and the 3′ UTR (80%) between SGF14c and SGF14l.
14-3-3 Transcript Levels Increase during Soybean-B. japonicum Interaction
Microarray analysis (Brechenmacher et al., 2008) revealed that the 14-3-3 gene, SGF14c (Glyma05g29080; microarray clone Gm-r1021-944), increased in transcript abundance following B. japonicum inoculation. The SGF14c gene shares high homology with SGF14l (Glyma08g12220, with 96% identity in the coding sequence, 27% in the 5′ untranslated region [UTR], and 80% in the 3′ UTR; Fig. 1B). The syntenic relationship between SGF14c and SGF14l (Supplemental Fig. S4) indicates that these two genes are paralogs and likely shared a common ancestor before soybean genome duplication. Li and Dhaubhadel (2011) conducted bimolecular fluorescence complementation analyses demonstrating that SGF14c and SCF14l can form homodimers and heterodimers and were localized to both the cytoplasm and nucleus, suggesting that these proteins likely function together.
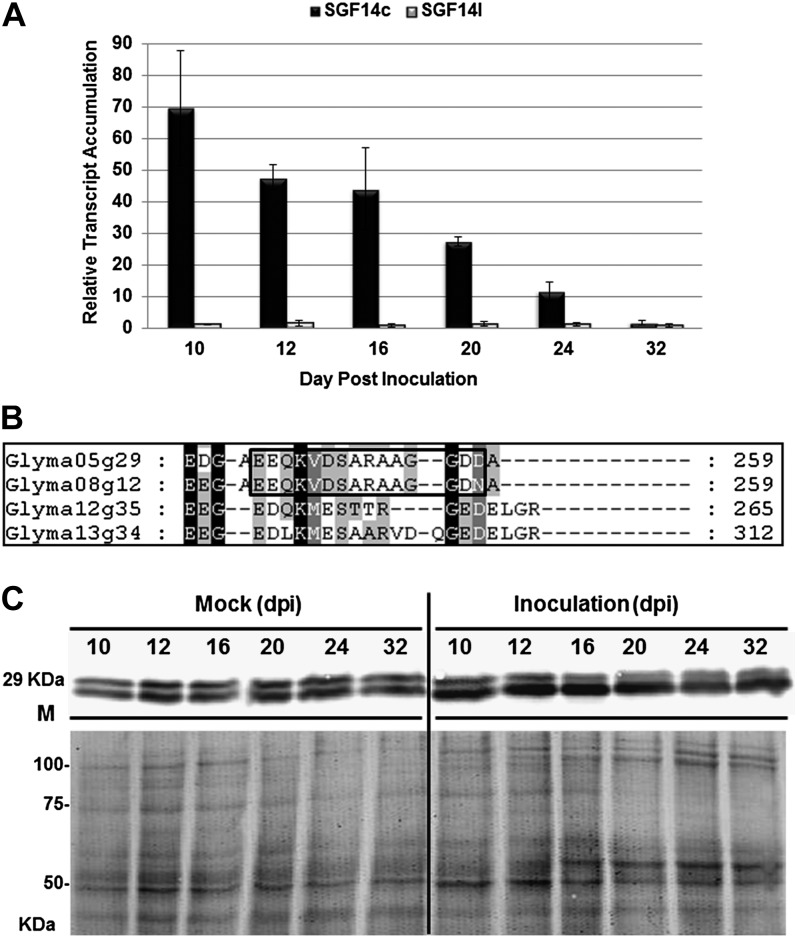
To measure the transcript accumulation of each gene, we employed gene-specific quantitative reverse transcription (qRT)-PCR using primers that differentiate between SGF14c and SGF14l (Supplemental Table S2). The qRT-PCR analysis was performed with total RNA isolated from mock-inoculated roots and nodules collected at 10, 12, 16, 20, 24, and 32 d post inoculation (dpi). The expression levels of these two genes were normalized against the geometric mean of two stably expressed reference genes: cons4, encoding an ATP-binding cassette transporter (Libault et al., 2008), and β-Actin (Zou et al., 2005). The qRT-PCR results (Fig. 2A) showed that the SGF14c transcript increased in response to B. japonicum inoculation at 10, 12, 16, and 20 dpi, with the highest increase at 10 dpi (a change of 69.5-fold). No such transcript accumulation was found for SGF14l, which reflects a potential isoform-specific regulation of SGF14c during the B. japonicum-soybean interaction and reveals that only SGF14c, and not SGF14l, is induced during early stages of nodule development.
Figure 2.
Transcriptomic and proteomic profiles of SGF14c (Glyma05g29080) and SGF14l (Glyma08g12220) upon inoculation with B. japonicum. A, qRT-PCR results showing the relative transcript accumulation of each gene after normalization using the geometric mean of Actin and cons4 reference genes. In each histogram, the means of three biological replicates and three technical replicates are represented ± se (n = 3). The expression levels of each gene were measured in nodules and mock-inoculated roots at 10, 12, 16, 20, 24, and 32 dpi. The mock-inoculated roots were treated with water. Bars are as follows: black bars, SGF14c (Glyma05g29080); gray bars, SGF14l (Glyma08g12220). B, Specific antibodies (anti-SGF14cl) derived from SGF14c (Glyma05g29080) and SGF14l (Glyma08g12220) protein sequences (the polypeptide of the antibody is boxed). C, Western blot using anti-SGF14cl against proteins extracted from nodules and mock-inoculated roots at 10, 12, 16, 20, 24, and 32 dpi and the corresponding Coomassie blue-stained gel showing equal loading. The bottom band on the western blot belongs to SGF14c (Glyma05g29080), while the top band belongs to SGF14l (Glyma08g12220).
Both SGF14c and SGF14l Are Expressed Constitutively in Different Soybean Tissues
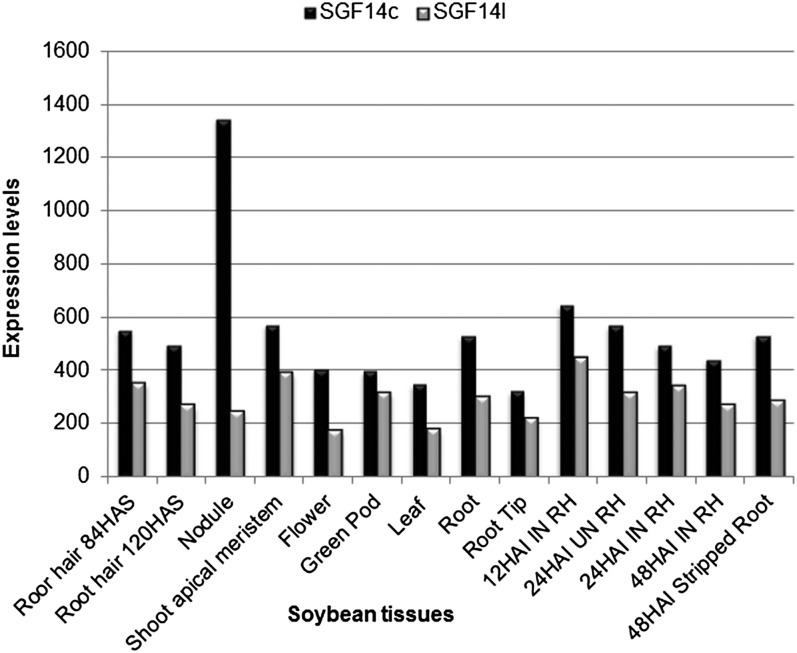
The expression of SGF14c and SGF14l was quantified as part of an RNA-seq analysis involving nine different soybean tissues: root hair, nodule, shoot apical meristem, flower, green pod, leaf, root, root tip, and stripped, hairless root (Libault et al., 2010a). Mining those gene expression results (Fig. 3) showed that both genes were expressed constitutively in these different soybean tissues, but SGF14c mRNA levels were higher (1.26- to 2.35-fold) than SGF14l in all tissues tested. Additionally, the transcription of SGF14c, but not SGF14l, was nearly 6-fold higher in nodules, consistent with our qRT-PCR results.
Figure 3.
Gene expression levels of SGF14c (Glyma05g29080; black bars) and SGF14l (Glyma08g12220; gray bars) from different soybean tissues. The expression of genes was quantified by ultra-high-throughput sequencing after normalization of the read number by the total number of soybean reads (Libault et al., 2010a). Nine different soybean tissues (root hair, nodule, shoot apical meristem, flower, green pod, leaf, root, root tip, and stripped root) were used (Libault et al., 2010a). HAS, Hours after sowing; RH, root hairs inoculated (IN) and mock-inoculated (UN) with B. japonicum at 12, 24, and 48 h after inoculation.
14-3-3 Protein Levels Increase during Soybean-B. japonicum Interaction
To measure the protein levels of SGF14c and SGF14l in soybean tissue extracts, we produced a peptide antibody that targets a unique C-terminal sequence of SGF14c and SGF14l (Fig. 2B). This anti-SGF14cl antibody cross-reacted with two bands of approximately 29 kD on immunoblots (Fig. 2C; Supplemental Figure S5). The abundance of the lower protein band increased considerably following inoculation with B. japonicum at 10, 12, 16, and 20 dpi, consistent with the increase in transcript levels of SGF14c detected by qRT-PCR (Fig. 2A).
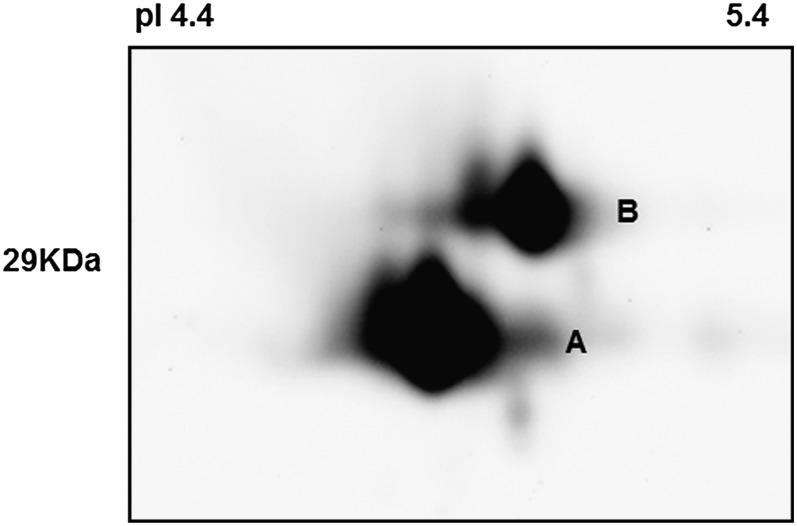
Considering the close migration of the two protein bands on one-dimensional gels, we used isoelectric focusing and two-dimensional electrophoresis (IEF-2DE) to further separate the two proteins by pI. The IEF-2DE results indicated that both upper and lower bands were composed of a single major protein (Fig. 4). The protein with the lower molecular mass and pI was termed spot A, while the protein with the higher molecular mass and pI was termed spot B. According to the protein sequence, isoform SGF14c is estimated to be 29.11 kD with a pI of 4.8, while isoform SGF14l is estimated to be 29.24 kD and pI 4.9. Thus, it is likely that spot A is SGF14c while spot B is SGF14l. To confirm the identities of the two 14-3-3 isoforms, we excised the corresponding spots from the IEF-2DE Coomassie blue-stained gel and sequenced them by liquid chromatography-tandem mass spectrometry. As shown in Table I, the mass spectrometry analysis unequivocally identified spot A as SGF14c with 36% protein coverage and two discriminative peptides (K.ERENFVYVAK.L and K.AYQLASTTAEAELASTHPIR.L). Spot B was unambiguously identified as SGF14l with the aid of 17% coverage and one discriminatory peptide (K.ERENFVYTAK.L). The mass spectrum identification (Supplemental Fig. S6) further confirmed our interpretation of the one-dimensional immunoblot results, with the levels of SGF14c protein increasing during soybean nodule development while SGF14l protein levels remained unchanged (Fig. 2C).
Figure 4.
IEF-2DE immunoblots and mass spectrometry unequivocally identified the bottom band (spot A) as SGF14c (Glyma05g29080; Table I) and the top band (spot B) as SGF14l (Glyma08g12220; Table I). Proteins extracted from nodules were collected at 24 dpi.
Table I. Protein identification of SGF14c (Glyma05g29080.1) and SGF14l (Glyma08g12220.1) from two-dimensional gel and mass spectrometry.
Boldface sequences are discriminative peptides for SGF14c and SGF14l. (ox)M, Oxidized Met; Mr(expt), relative molecular mass (experimental); Mr(calc), relative molecular mass (calculated); Δ, difference between experimental and calculated relative molecular masses.
| Start-End | Mr(expt) | Mr(calc) | Δ | Sequence | Mascot Score |
|---|---|---|---|---|---|
| Spot A Identificationa | |||||
| 6-15 | 1,253.69 | 1,253.64 | 0.0494 | K.ERENFVYVAK.L | 35 |
| 23-31 | 1,160.524 | 1,160.47 | 0.0513 | R.YEEMVEAMK.N | 44 |
| 36-45 | 1,200.682 | 1,200.64 | 0.0469 | K.LNVELTVEER.N | 70 |
| 46-53 | 892.5384 | 892.502 | 0.0366 | R.NLLSVGYK.N | 44 |
| 65-72 | 916.56 | 916.523 | 0.0371 | R.ILSSIEQK.E | 41 |
| 154-173 | 2,129.168 | 2,129.08 | 0.0928 | K.AYQLASTTAEAELASTHPIR.L | 137 |
| 200-218 | 2,174.091 | 2,173.99 | 0.1012 | K.QAFDEAISELDTLSEESYK.D | 110 |
| 219-228 | 1,188.705 | 1,188.65 | 0.0516 | K.DSTLIMQLLR.D | 69 |
| 219-228 | 1,204.698 | 1,204.65 | 0.0497 | K.DSTLI(ox)MQLLR.D | 61 |
| Spot B Identificationb | |||||
| 6-15 | 1,255.672 | 1,255.62 | 0.0528 | K.ERENFVYTAK.L | 48 |
| 36-45 | 1,200.671 | 1,200.64 | 0.0363 | K.LNVELTVEER.N | 52 |
| 46-53 | 892.5282 | 892.502 | 0.0264 | R.NLLSVGYK.N | 19 |
| 65-72 | 916.5614 | 916.523 | 0.0385 | R.ILSSIEQK.E | 31 |
| 219-228 | 1,204.68 | 1,204.65 | 0.0319 | K.DSTLI(ox)MQLLR.D | 46 |
Gene locus: Glyma05g29080.1; Mr: 29,160; pI: 4.8; coverage: 36%; annotation: Soybean 14-3-3 SGF14c.
Gene locus: Glyma08g12220.1; Mr: 29,475; pI: 4.9; coverage: 17%; annotation: Soybean 14-3-3 SGF14l.
Reduction in SGF14c/SGF14l Transcript Levels Affects Nodulation
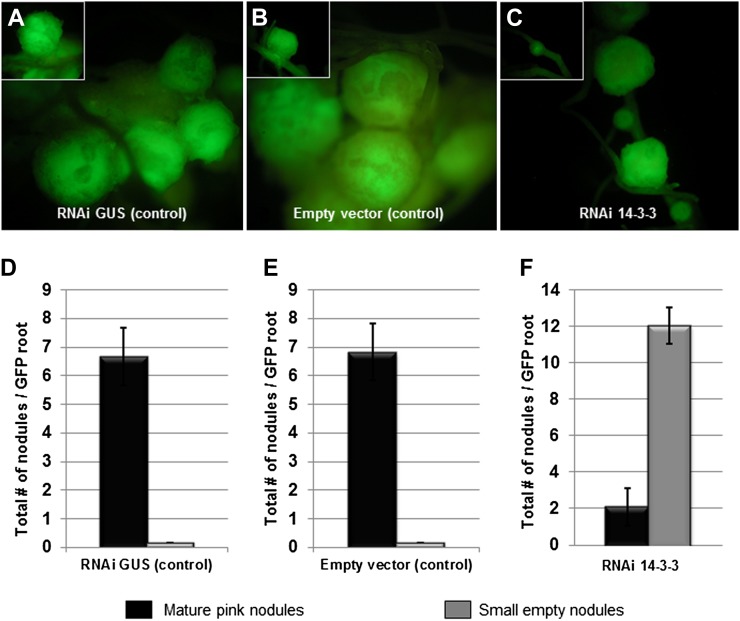
In order to examine the function and role of SGF14c/SGF14l during nodulation, we silenced both SGF14c and SGF14l using RNAi in soybean roots transformed with A. rhizogenes. Composite plants (Collier et al., 2005; Govindarajulu et al., 2008, 2009), possessing transformed roots and nontransformed shoots, were used to express the RNAi constructs under control of the strong, constitutive figwort mosaic virus promoter. An empty vector and an RNAi GUS construct were used as negative controls. GFP driven from the superubiquitin promoter was used as the marker for identifying transformed roots (Collier et al., 2005; Govindarajulu et al., 2008, 2009). Abnormal nodule morphology was the only obvious alteration of RNAi SGF14c/SGF14l composite plants, while no abnormalities were observed in the morphology of leaves, stems, or roots (data not shown). The nodule number per transgenic root varied according to the examined construct, including the empty vector control, RNAi GUS control, and RNAi SGF14c/SGF14l fragment.
Roots transformed with the empty vector control produced 6.8 ± 1.68 pink, fully formed nodules per transgenic root (Fig. 5, A and D). The RNAi GUS control formed 6.7 ± 0.48 mature nodules per transgenic root (Fig. 5, B and E). Roots expressing the RNAi SGF14c/SGF14l fragment showed a dramatic decrease in the number of mature pink nodules, forming only 2.1 ± 0.83 mature nodules per transgenic root (Fig. 5, C and F). In the SGF14c/SGF14l-silenced roots, we also observed numerous arrested nodule primordia and the formation of small transparent bumps emerging on the root surface from the root cortex that can be referred to as either “small empty” or medium sized (Govindarajulu et al., 2009). These small empty and medium nodules in the SGF14c/SGF14l-silenced transgenic roots were translucent and failed to develop into mature pink nodules (12.1 ± 1.93 small/medium nonpink nodules formed per transgenic root; Fig. 5, C and F), even after extended incubation following the initial B. japonicum inoculation. The decrease in mature nodules may be due to the inhibition of nodule development after nodule primordia are formed (Govindarajulu et al., 2009).
Figure 5.
Nodulation response in transgenic soybean 14-3-3-silenced roots after inoculation with B. japonicum. A to C, Five-week-old GFP-expressing root nodule phenotypes for empty vector control (A), RNAi GUS control (B), and RNAi 14-3-3 (C) in soybean hairy roots. The RNAi 14-3-3 roots exhibited poor nodulation (as small bumps or empty nodules), whereas the controls showed many fully mature nodules. D to F, Quantitation of nodulation from empty vector control (D), RNAi GUS control (E), and RNAi 14-3-3 (F) soybean hairy roots. Empty nodules (gray bars) were counted as small bumps just emerging on the root surface from the root cortex, while mature nodules (black bars) were counted as full sized with a pink internal color. The data represent average values of 24 individual plants (per biological replicate) containing transgenic nodulated roots. se bars are shown for total number of mature nodules and small empty nodules of three independent biological replicates.
Light Microscopy and TEM Reveal Altered Nodule Morphology in SGF14c/SGF14l-Silenced Roots
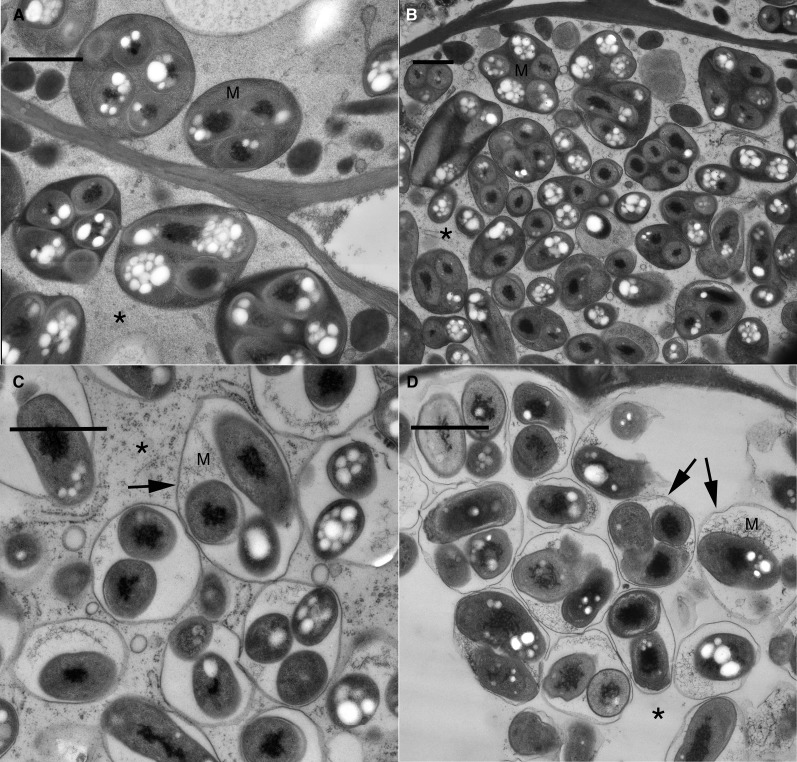
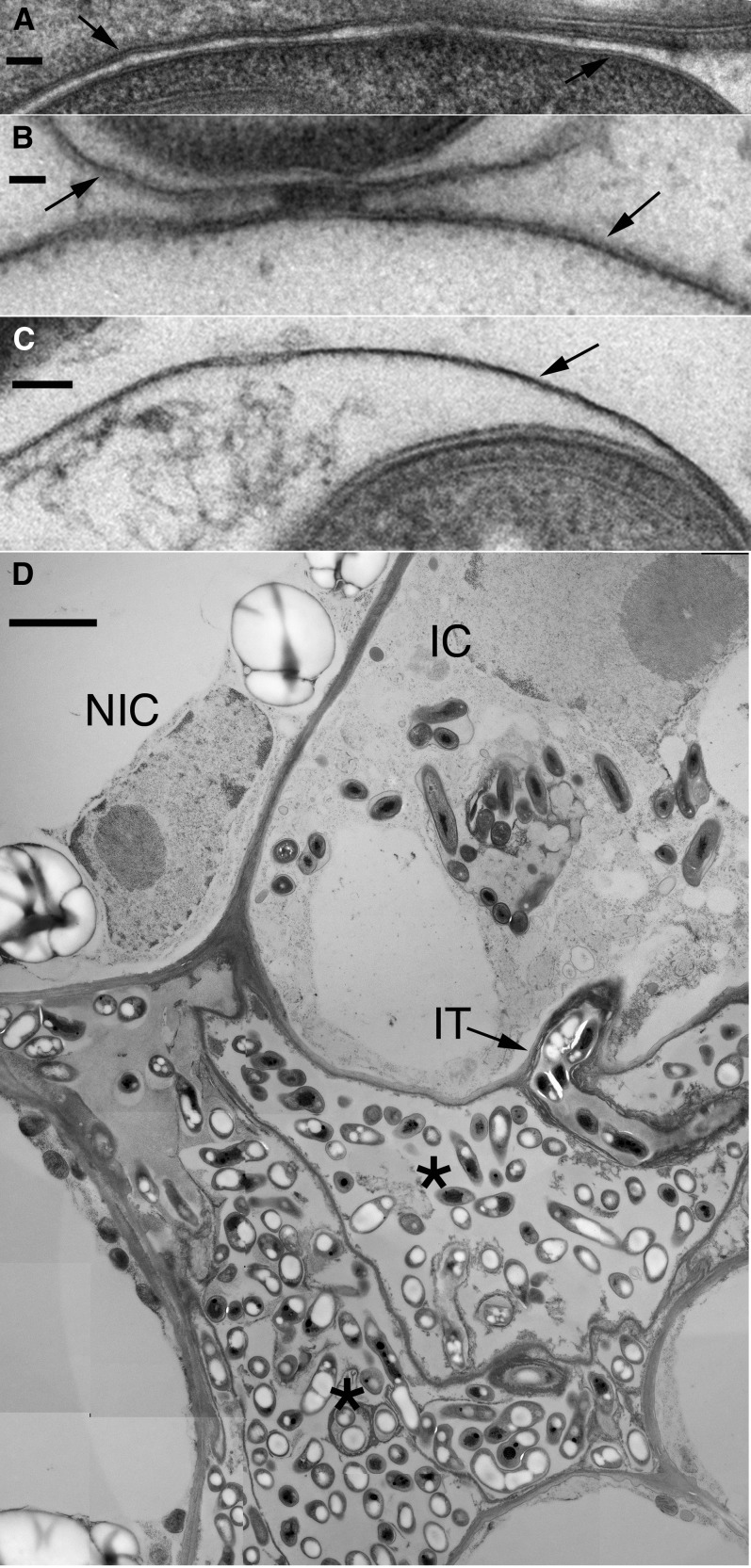
To understand the role of SGF14c/SGF14l in nodule development, nodules formed from roots transformed with the empty vector control, RNAi GUS control, and RNAi SGF14c/SGF14l were analyzed using light microscopy and TEM. Thin sections of mature pink nodules from the controls and various-sized nodules from the RNAi SGF14c/SGF14l were cytologically examined. In the vector and RNAi GUS controls, the host cells of nodules were filled with mature bacteroids within symbiosomes (Fig. 6, A and B). In these controls, the symbiosomes contained a normal-looking host matrix and the host cytoplasm remained intact. The mature nodules that formed on the RNAi SGF14c/SGF14l roots differed from the controls in that there was apparent degradation of the symbiosome matrix (Fig. 6C), whereas the cytoplasm remained intact, in comparison with the empty nodules (Fig. 6D). The RNAi SGF14c/SGF14l empty nodules were severely affected and only partially developed and/or were undergoing degradation. The empty nodules contained relatively few infected cells that had bacteroids within a symbiosome (Fig. 6D), and these cells exhibited loss of host cytoplasm, organelles, and membranes. The symbiosome matrix of the empty nodules also showed apparent degradation of the host cell matrix. Although the symbiosome membrane was not completely degraded, and thus was the most resistant to degradation of the host cell membranes, it was still adversely affected in the empty nodules. The symbiosome membrane in these empty nodules had reduced thickness, with the membranes of medium-sized nodules (10.17 ± 0.79 nm [se]) being reduced by about 10% and the membranes of small-sized nodules (5.67 ± 0.56 nm) being reduced about 50% compared with the thickness of the symbiosome membranes (11.00 ± 0.63 nm) of the controls (Fig. 7, A–C). A typical observation was that the RNAi SGF14c/SGF14l small empty nodules did not contain symbiosomes and had relatively few infected cells, these being arrested in development. For example, the infected cell in Figure 7D had a few bacteroids that were released by the infection thread, but further development of symbiosomes had not occurred. The low contrast of the cytoplasm in this cell and the adjacent noninfected cell is likely due to a general degradation process that eventually will lead to cell death and collapse, which occurred in the lowermost two cells (Fig. 7D). Electron micrographs suggest that silencing of SGF14c/SGF14l led to malformation and/or degradation of nodular cell components, which reflects a general loss in plant cell viability expected with the silencing of a key protein regulating essential plant cell signaling systems.
Figure 6.
TEM micrographs showing the cytoplasm and symbiosome matrix from mature (A and B), medium-sized (C), and small empty (D) nodules. A, Empty vector. Typical symbiosomes with matrix (M) and host cytoplasm intact (asterisk) are seen. B, GUS control (as in A). C, 14-3-3 medium-sized nodule. Cytoplasm is intact (asterisk), but matrix (M) is degraded. D, 14-3-3 small empty nodule. Cytoplasm (asterisk) and matrix (M) are degraded; the symbiosome membrane remains (arrows). These micrographs are derived from one representative experiment of three independent biological replications. Bars = 1 μm.
Figure 7.
TEM micrographs showing the symbiosome membrane from different types of nodules and the general degradation observed in empty nodules. A, Symbiosome membranes (arrows) for GUS control. Both leaflets are visible. Average thickness is 11.00 ± 0.63 nm. B, Symbiosome membranes (arrows) for 14-3-3 medium-sized nodule. Both leaflets are visible. Average thickness is 10.17 ± 0.79 nm. C, Symbiosome membrane (arrow) for 14-3-3 small empty nodule. Only one leaflet is apparent. Average thickness is 5.67 ± 0.56 nm. D, 14-3-3 small empty nodule. A few infected cells are present. A recently infected cell (IC) has an infection thread (IT) and bacteroids. The cytoplasm and organelles of this cell, and the adjacent noninfected cell (NIC), have a loss of electron density likely due to early degradation of these components. Two infected cells (asterisks) have completely lost the host cell components and have collapsed, leaving remnant bacteria. These micrographs are derived from one representative experiment of three independent biological replications. Bars = 50 nm for A to C and 2 μm for D.
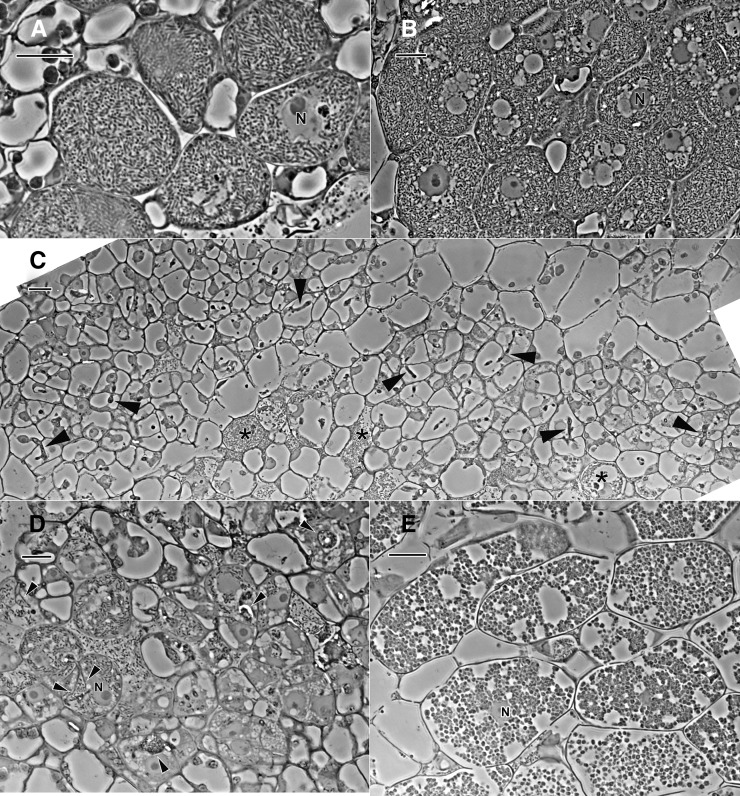
The difference in ultrastructure between the GUS control and medium-size nodules of the 14-3-3-silenced roots discussed above (Figs. 6 and 7) was not apparent in phase-contrast micrographs (Fig. 8, A and B). However, the more severe ultrastructural changes in small nodules of the 14-3-3-silenced roots were manifested in a variety of phenotypes on the tissue level, as shown in the phase micrographs of small nodules (Fig. 8, C–E). In the nodule in Figure 8C, there were only a few cells with bacteroids, and they appeared senescent. There were a large number of cells with infection threads, indicating that bacteria are adequately delivered to the cells in this tissue. The infected cells in the small nodule in Figure 8D were more developed, but still they did not have the full population of rhizobia seen in the controls, and the infection threads of these cells were distended with large numbers of bacteria, consistent with a phenotype of an interruption in the normal host cell endocytosis of bacteria. In the small nodule of Figure 8E, the infected cells have developed atypical, spherical symbiosomes (compared with the rod-shaped bacteria in symbiosomes in the control tissue), and the only remnant of host cytoplasm in these senescent cells was the nucleus. These cells are similar to those shown in the TEM image of Figure 6D.
Figure 8.
Phase-contrast micrographs of 1-µm-thick resin sections of freeze-substituted nodules. A, GUS control. B, 14-3-3-silenced roots, medium-size nodule. C to E, 14-3-3-silenced roots, small-size nodule. N, Nucleus. The GUS control (A) and medium nodules (B) are similar in structure and are filled with typical symbiosomes. Small nodules contain cells that are arrested in development. In C, there are a few senescent host cells that contain rhizobia (asterisks), but most cells that have infection threads (arrowheads) are devoid of internalized rhizobia. There are more fully developed infected cells in the small nodule in D, but they are not as fully developed as those in medium nodules (B), and the infection threads (arrowheads) are distended with large numbers of rhizobia. Infected cells in the small nodule in E are filled with atypical symbiosomes that are spherical, and the host cytoplasm is degraded. Bars = 20 µm.
Decreased SGF14c/SGF14l mRNA and Protein Levels in RNAi-Silenced Tissues
The mRNA and protein levels of the SGF14c/SGF14l-silenced tissues after B. japonicum inoculation were measured by qRT-PCR and immunoblotting. The 186-bp RNAi fragment exhibited 100% and 91% DNA identity with SGF14c and SGF14l, respectively (Supplemental Fig. S7), reflecting a high probability to silence both paralogs. To discriminate transcript abundance between SGF14c and SGF14l, we designed discrete qRT-PCR primer pairs for each of these genes, in addition to a PCR primer pair that amplified a fragment from both genes. Figure 9 shows the mRNA abundance levels of SGF14c, SGF14l, both genes, and GUS measured by qRT-PCR from total RNA isolated from empty vector control, RNAi GUS control, and SGF14c/SGF14l-silenced roots after inoculation with B. japonicum. The geometric mean of two stably expressed reference genes was used to normalize the mRNA expression levels. The transcript levels of SGF14c were significantly reduced (approximately 3.9-fold) in the SGF14c/SGF14l-silenced roots, whereas SGF14c expression in RNAi GUS control roots remained unaffected when compared with the empty vector control (Fig. 9A). Moreover, SGF14l expression was also altered (approximately 3-fold) in SGF14c/SGF14l-silenced roots compared with the transcript levels in the empty vector control (Fig. 9A). The concomitant silencing of both SGF14c and SGF14l is expected, given the high level of sequence identity (i.e. identity in the coding region sequence is 96%) between these two genes. In order to verify that the silencing constructs do not affect the expression of other 14-3-3 genes in addition to the expression of SGF14c and SGF14l, we also designed specific primers (Supplemental Table S2) for each gene located in clade IV of the soybean 14-3-3 gene family (Fig. 1), since this clade contains the genes of highest DNA identity with SGF14c (61%–77%). qRT-PCR (data not shown) indicated that the mRNAs of these other 14-3-3 genes were not altered in the silenced infected roots, supporting that only the expression of the genes SGF14c and SGF14l was silenced.
Figure 9.
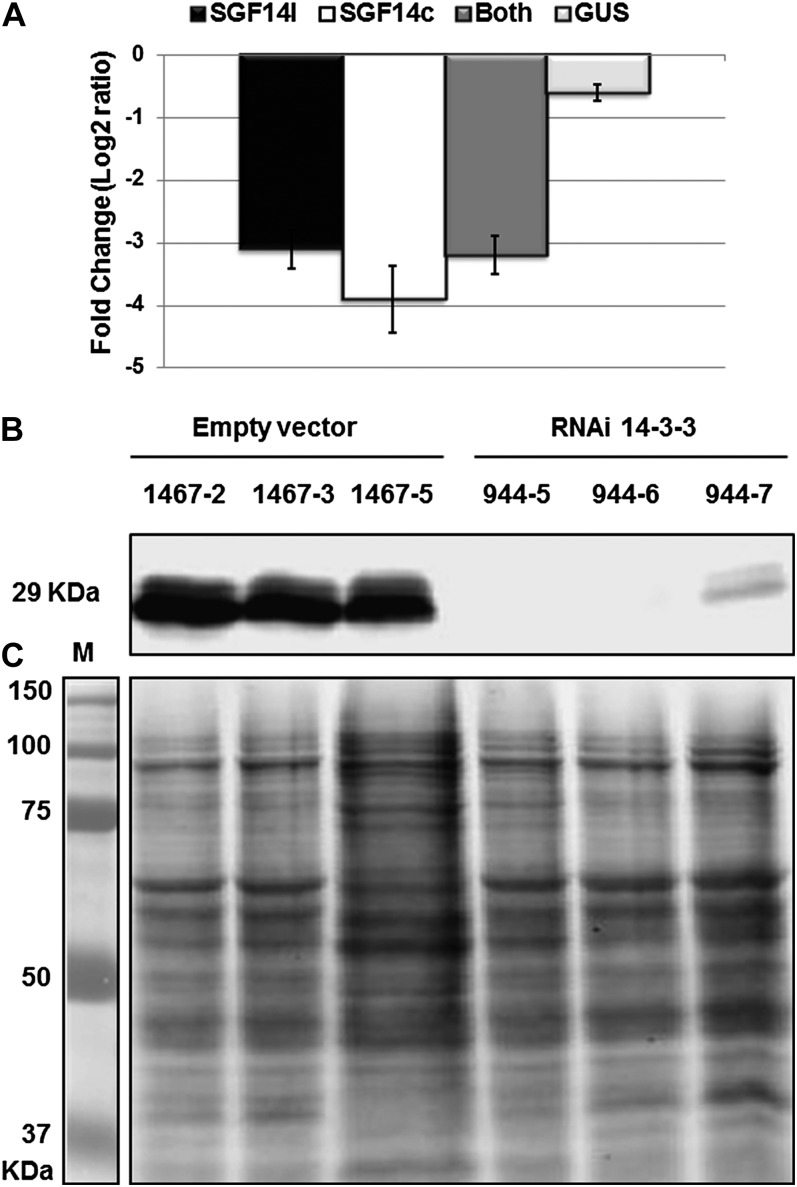
mRNA and protein reduction in soybean silenced tissues after inoculation with B. japonicum. A, qRT-PCR results showing the relative transcript accumulation of different genes after normalization using the geometric mean of cons4 and Actin reference genes. Bars are as follows: light gray bar, RNAi GUS; dark gray bar, SGF14c (Glyma05g29080); black bar, SGF14l (Glyma08g12220); white bar, both genes. In each histogram, the means of three biological replicates are represented ± se (n = 3). B, Western blot using soybean-specific antibody (anti-SGF14cl) against soybean protein extracted from nodules collected at 16 dpi from three independent RNAi 14-3-3 lines (944-5, 944-6, and 944-7) and three independent empty vector lines (1467-2, 1467-3, and 1467-5). C, Partnered Coomassie blue-stained gels are shown as evidence of equal loading.
To determine whether the protein levels of SGF14c and SGF14l were also decreased as a result of mRNA silencing, we analyzed protein extracted from SGF14c/SGF14l-silenced nodules as well as the various controls. As shown in Figure 9B, immunoblotting revealed two bands with an approximate size of 29 kD. These two bands were both significantly decreased in the SGF14c/SGF14l-silenced plants (i.e. lines 944-5, 944-6, and 944-7), while the intensity of these two proteins was not affected in the empty vector lines (1467-2, 1467-3, and 1467-5). Coomassie staining of the gels indicated equal loading (Fig. 9C). Therefore, the specific reduction of both mRNA and protein levels of both SGF14c and SGF14l supports the notion that they play an important role in nodulation.
DISCUSSION
In a soybean transcriptome expression study focused on B. japonicum-induced nodulation, Brechenmacher et al. (2008) reported that the mRNA abundance of approximately 5,000 genes changed significantly within 16 dpi in young roots. Some of the differentially expressed genes identified from that study were subsequently shown to play critical roles in soybean nodulation, such as FW2.2-like1 (Libault et al., 2010b), GS52 ecto-apyrase (Govindarajulu et al., 2009; Tanaka et al., 2011), and CND (for control of nodule development), encoding a Myb transcription factor (Libault et al., 2009). Additionally, in a recent study, Nguyen et al. (2012) conducted experiments on quantitative phosphoproteomics where 273 phosphopeptides corresponding to 240 phosphoproteins were found to be significantly regulated in soybean root hairs in response to B. japonicum inoculation, reflecting a critical role of phosphorylation events during the initiation of the B. japonicum infection process. Here, we report another nodulation-induced gene identified from that microarray study as being critical for establishing healthy nodules, the 14-3-3 gene SGF14c.
This study verified that transcript levels of the 14-3-3 SGF14c gene increased during nodule development. Silencing of this gene led to greatly reduced nodulation (functional nodule formation) in soybean. Using qRT-PCR and by mining the previously published, high-throughput RNA-seq data (Libault et al., 2010a), we confirmed the results published by Brechenmacher et al. (2008) that SGF14c transcript abundance increased in response to B. japonicum. We also showed that this induction in transcript level correlated with an increase in SGF14c protein abundance. To determine the importance of this 14-3-3 gene in nodule development, we silenced SGF14c by transforming soybean hairy roots with an RNAi hairpin/loop construct. As genome data became available, we realized that our RNAi construct would also silence paralog SGF14l, and data presented here verified that, indeed, both SGF14c and SGF14l were silenced in our RNAi experiments. It should be noted that the expression level of SGF14l is not affected by nodulation (Libault et al., 2010a; this study); therefore, although the protein SGF14l may be partnered with SGF14c, only SGF14c and its protein product appear to be nodulation inducible. SGF14c/SGF14l-silenced plants showed a significant reduction in the formation of mature, pink nodules and instead formed immature or empty nodules, consistent with an arrest in nodule development. Li and Dhaubhadel (2011) showed that SGF14c and SGF14l can form homodimers or heterodimers. Therefore, it may be that the disruption of a SGF14c/SGF14l protein complex, with loss of either 14-3-3 protein, leads to abnormal nodulation.
TEM and phase-contrast micrographs showed that silencing of SGF14c/SGF14l led to poorly developed or degraded plant cell components, reflecting a general loss in plant cell viability. It is possible that the silencing inhibits processes downstream from termination of the infection thread in the host cell, from bacterium endocytosis onward, and that the medium nodules have managed to progress much more than the small nodules. These severe ultrastructure defects reinforce the notion that 14-3-3 proteins are a critical component of the nodule development regulatory pathway, and this dramatic phenotype would be expected with the silencing of a protein regulating a key plant cell signaling system or other vital function. It should also be noted that this study shows a very clear phenotypic effect, which is not the case for most reported 14-3-3 knockouts or knockdowns (Roberts et al., 2002; Roberts, 2003).
The 14-3-3 proteins act to bind phosphorylated proteins and therefore play important roles in numerous cellular events involving phosphorylation and/or signal transduction as well as changes in cellular localization. A recent study of soybean 14-3-3 proteins (Li and Dhaubhadel, 2011) reported that the SGF14s are localized in different subcellular compartments, with the specific localizations determined by each isoform. SGF14c homodimers, as well as SGF14c/SGF14l heterodimers, localized to the cytoplasm and nucleus in tobacco (Nicotiana tabacum) leaf assays (Li and Dhaubhadel, 2011). The presence of a large number of 14-3-3 isoforms in the soybean genome means that many 14-3-3-interacting combinations are possible, but it also leaves open the possibility that isoform-specific interactions with specific targets are also playing critical roles. A very recent study (Li et al., 2012) suggested that the SGF14l protein regulates the intracellular localization of soybean transcription factor GmMYB176 and that the interaction between SGF14l and its client (GmMYB176) affects isoflavonoid synthesis in soybean.
Critical functions of a cell may be controlled by 14-3-3 proteins, as so many essential functions involve phosphorylation. For example, in Arabidopsis, it was shown that a 14-3-3 may play a role in cytoskeleton organization through the regulation of Endosperm Defective1, a microtubule-associating protein that is essential for Arabidopsis cell division (Pignocchi and Doonan, 2011). Plant 14-3-3 proteins were also shown to interact with numerous transcription factors and signaling proteins during development and in response to various stresses (Camoni et al., 1998; Pan et al., 1999; Cotelle et al., 2000; Ikeda et al., 2000; Roberts, 2003). Basic metabolic processes are also controlled by 14-3-3s, such as fundamental nitrogen and carbon assimilation pathways (Finnie et al., 1999; Vance, 2008). In this study, we assume that SGF14c or SGF14c/SGF14l interacts with target proteins that are involved in essential cell functions. Silencing of 14-3-3 caused a loss of function of these target proteins, leading to the severe phenotypic and ultrastructure defects that were observed (Figs. 5–8). Interestingly, the dramatic effect on cellular degradation was only seen in the cells of the developing nodules, the tissue where SGF14c is induced. The cells of the root tissue neighboring the arrested nodules looked normal and healthy. Since the biological function of 14-3-3 proteins involves interaction with phosphorylated targets (Obsil et al., 2001; Roberts, 2003; Chevalier et al., 2009), further work to identify possible interacting proteins of SGF14l, or SGF14c/SGF14l, will need to be performed.
MATERIALS AND METHODS
Plant and Bacteria Materials
Soybean (Glycine max ‘Williams 82’) was used in this study. For isolating nodulated roots, sterilized soybean seeds were placed between humidified Whatman paper and kept under dark conditions (80% humidity at 25°C). Three-day-old germinated seedlings were placed in vermiculite:perlite mix (3:1), and each seedling was inoculated with 10 mL of Bradyrhizobium japonicum USDA110 suspension (optical density at 600 nm = 0.08), while control seedlings were mock inoculated with 10 mL of water. The nodulated roots were harvested at 10, 12, 16, 20, 24, and 32 dpi and stored at −80°C for RNA extraction. Three independent biological replicates were performed and analyzed. Escherichia coli, Agrobacterium rhizogenes (K599), and B. japonicum (USDA110 strain) were grown and handled as described by Govindarajulu et al. (2009).
RNAi Plasmid Construction
The RNAi gene constructs were made according to Collier et al. (2005). In brief, a 186-bp gene fragment for the RNAi construct was amplified by PCR from soybean cDNA using SGF14c-specific primers as indicated in Supplemental Table S2. The amplified PCR fragment was cloned into the entry vector CGT11050 by TA cloning using two AhdI sites (Libault et al., 2010b). This RNAi entry vector was recombined with the destination vector CGT11017A (Libault et al., 2009) using the LR clonase reaction, creating the 14-3-3-RNAi vector. The RNAi empty vector control and RNAi GUS constructs were described previously by Govindarajulu et al. (2009). The clones were verified by sequencing, electroporated into A. rhizogenes strain K599, and used for composite plant production.
Soybean Composite Plant Production and Nodulation Analysis
Soybean composite plants were produced according to Govindarajulu et al. (2008). After growing sterilized soybean seeds for 2 weeks in vermiculite/perlite, 10 mL of B. japonicum suspended in nitrogen-free plant nutrient solution (optical density at 600 nm = 0.08) was added to the root area of each plant. Control plants (noninoculated) were treated with 10 mL of nitrogen-free plant nutrient solution. Roots expressing the RNAi 14-3-3, RNAi GUS, or the empty vector control, pCGT6419A (Govindarajulu et al., 2009), were analyzed 5 weeks after inoculation. Transgenic roots expressing GFP were isolated, and nodule numbers were counted using an Olympus stereomicroscope equipped with epifluorescence excitation and a GFP long-pass filter (http://www.olympusamerica.com), according to Libault et al. (2010b) and Govindarajulu et al. (2009). Twenty-four individual plants were scored for GFP roots containing nodules for each RNAi construct. Twenty-four root tissues with nodules were frozen in liquid nitrogen after nodule counting and stored at −80°C for RNA extractions. For each RNAi construct, experiments were repeated three times as three independent biological replicates.
Preparation of Transgenic Nodules for Light Microscopy and TEM
Light microscopy was used to examine longitudinal cross-sections of SGF14c/l-silenced nodules, empty vector control, and RNAi GUS control. For TEM examination, isolated nodules from GFP-expressing RNAi 14-3-3 transgenic roots were prepared according to Govindarajulu et al. (2009). In brief, fresh nodules were frozen in a Bal-Tec high-pressure freezer (Danforth Plant Science Center) and freeze substituted in acetone containing 2% osmium tetroxide and 0.1% uranyl acetate for 5 d at −85°C, 24 h at −20°C, and 1 h at 0°C (on ice). Subsequently, the samples were embedded with Spurr’s resin (Electron Microscopy Sciences; http://www.emsdiasum.com) for 7 d (Hess, 2007). Thin sections were stained in uranyl and lead salts and observed using a LEO 912 energy filter transmission electron microscope (Carl Zeiss; http://www.zeiss.com/nts/). Digital images were analyzed manually. For phase micrograph sections, thick resin sections (1 µm) from TEM blocks were stained with toluidine blue and digitally imaged with phase-contrast optics, using a 60× Plan Apo, 1.4 numerical aperture, phase objective and a Nikon Eclipse 800 microscope. Montages allowing large fields of view were stitched together using the photomerge function in Photoshop version CS4.
Sample Collection and RNA Extraction
Soybean nodules were collected at 10, 12, 16, 20, 24, and 32 d after B. japonicum inoculation, with mock-inoculated roots collected at the same time points. Each collected sample consisted of pooled nodules (B. japonicum inoculation) or roots (mock inoculation) from seven to 10 different plants. The entire experiment was repeated three times on different dates using new preparations of B. japonicum inoculum to obtain three independent biological replications. The three biological replicates showed consistency in nodulation development. Total RNA was isolated using TRIzol reagent (Invitrogen) and verified for quality as described previously in detail (Zou et al., 2005).
Reverse Transcription-PCR and qRT-PCR
Reverse transcription-PCR and qRT-PCR were performed according to Radwan et al. (2011). In brief, RNA was treated with DNase I (Invitrogen) to remove genomic DNA contamination. Two micrograms of DNase-treated RNA was reverse transcribed using the SuperScript III First-Strand Synthesis System for reverse transcription-PCR (Invitrogen). A “minus” reverse transcription-PCR, in which no reverse transcriptase enzyme was added during the cDNA synthesis reaction, was used to test each mRNA sample for genomic DNA contamination. The transcriptional expression of each gene was analyzed using qRT-PCR. cDNA was diluted to a final concentration of 30 ng µL−1, and then 2 µL was used in a 20-µL PCR containing 8.9 µL of Brilliant QPCR Master Mix (catalog no. 600549; Stratagene) and 0.2 μm of each primer. PCR was carried out under the following conditions: an initial denaturation step at 95°C for 10 min, followed by 40 cycles of 15 s at 95°C, 20 s at the approximate melting temperature for primers and target °C (Supplemental Table S2), and 30 s at 72°C. After each run, a dissociation curve was checked for amplification of a single product. In addition, PCR products were sequenced to verify that the correct amplicon was produced. qRT-PCR data were analyzed using the δ-δ method (McMaugh and Lyon, 2003). Two constitutive controls were selected for data normalization: a soybean β-actin (GB AI507761) and the recently reported soybean constitutive gene (Libault et al., 2008), cons4 (GB BU578186), where the geometric mean of both reference genes was used for normalization. Three biological replicates with three technical replicates of each treatment were performed, and the data were calculated as the average of three biological replicates.
Protein Extraction and Western-Blot Analysis
Soybean nodules and root tissues were ground into powder in liquid nitrogen and then quickly mixed with SDS direct extraction buffer (60 mm Tris-HCl, 0.7 m β-mercaptoethanol, 2% SDS, 1 m urea, 1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, 1 mm Na3VO4, 2 mm EDTA, 10% glycerol, and 0.005% bromophenol blue) in a 3:1 (w/v) ratio. The extraction mixture was boiled at 95°C for 5 min and vortexed for 1 min. Total soluble proteins were collected as the supernatant after centrifugation at 16,000g for 10 min at room temperature. The concentration of the total proteins was quantified by Bradford assay, and equal amounts of total proteins from different treatments were analyzed by 12% SDS-PAGE and immunoblotting.
The soybean SB14-3-3 antigen (CEEQKVDSARAAGGD) was synthesized by GenScript and used at a 1:5,000 dilution ratio. An Alexa Fluor 680-labeled anti-rabbit tagged secondary antibody (Invitrogen) was used at a 1:20,000 dilution ratio. The fluorescence signal was quantified using an Odyssey imaging system (LI-COR Biotechnology).
IEF-2DE Analysis and Protein Identification
The IEF-2DE proteins were prepared as described by Wu et al. (2011). Briefly, protein extracts were extracted in saturated Tris-phenol (pH 8.0) and precipitated in 0.1 m ammonium acetate in methanol overnight at −20°C. The pellets were washed three times with ice-cold 0.1 m ammonium acetate in methanol and once with ice-cold 100% ethanol before dissolving in IEF sample buffer (7 m urea, 2 m thiourea, 4% CHAPS, 2% immobilized pH gradient buffer, 65 mm dithiothreitol, and 0.002% [v/v] bromphenol blue). The proteins were resolved in pI 4 to 7 (GE) and 12% SDS-PAGE. The IEF-2DE gel was transferred to a polyvinylidene difluoride membrane and analyzed by anti-SGF14cl antibodies. The corresponding spots of 14-3-3 proteins on the Coomassie blue-stained gel were excised and digested with trypsin (Promega). The digested peptides were analyzed by liquid chromatography-tandem mass spectrometry (Wu et al., 2011).
Sequence and Phylogeny Analyses
DNA and protein sequences of all soybean 14-3-3 genes were obtained from the soybean genome database (http://www.phytozome.net/soybean). The sequences were aligned using ClustalX software with default options (Thompson et al., 1997), and the resulting alignments were shaded using GENEDOC software (Nicholas et al., 1997). A neighbor-joining tree of soybean 14-3-3 genes and a tree of soybean and Arabidopsis (Arabidopsis thaliana) 14-3-3 genes were produced for analysis using bootstrap resampling (k = 1,000 permutations). Phylogenetic trees were made using MEGA 3.1 (http://www.megasoftware.net). To study the syntenic relationship between SGF14c and SGF14l, 50 kb surrounding both genes was isolated from the soybean genome database (http://www.phytozome.net/soybean), and gene content and order were compared.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of soybean 14-3-3 proteins.
Supplemental Figure S2. Alignment of soybean, M. truncatula, L. japonicus, and Arabidopsis 14-3-3 proteins.
Supplemental Figure S3. Phylogenetic tree of soybean, M. truncatula, L. japonicus, and Arabidopsis 14-3-3 proteins.
Supplemental Figure S4. Syntenic relationship between SGF14c and SGF14l genes.
Supplemental Figure S5. Entire western blot viewed in Figure 2C.
Supplemental Figure S6. Mass spectrum results of SGF14c and SGF14l.
Supplemental Figure S7. SGF14c/l RNAi sequence.
Supplemental Table S1. Listing of soybean 14-3-3 proteins.
Supplemental Table S2. PCR primers used in the study.
Acknowledgments
We thank Wan (Aline) Wu, Jeff Hinchman, and Andreina Chiu for assistance with RNA extractions.
Glossary
- qRT
quantitative reverse transcription
- RNAi
RNA interference
- UTR
untranslated region
- dpi
days post inoculation
- IEF-2DE
two-dimensional electrophoresis
- TEM
transmission electron microscopy
- IEF-2DE
isoelectric focusing and two dimensional electrophoresis
References
- Bachmann M, Huber JL, Athwal GS, Wu K, Ferl RJ, Huber SC. (1996) 14-3-3 proteins associate with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Lett 398: 26–30 [DOI] [PubMed] [Google Scholar]
- Brechenmacher L, Kim MY, Benitez M, Li M, Joshi T, Calla B, Lee MP, Libault M, Vodkin LO, Xu D, et al. (2008) Transcription profiling of soybean nodulation by Bradyrhizobium japonicum. Mol Plant Microbe Interact 21: 631–645 [DOI] [PubMed] [Google Scholar]
- Bunney TD, van Walraven HS, de Boer AH. (2001) 14-3-3 protein is a regulator of the mitochondrial and chloroplast ATP synthase. Proc Natl Acad Sci USA 98: 4249–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camoni L, Harper JF, Palmgren MG. (1998) 14-3-3 proteins activate a plant calcium-dependent protein kinase (CDPK). FEBS Lett 430: 381–384 [DOI] [PubMed] [Google Scholar]
- Chan PM, Ng YW, Manser E. (2011) A robust protocol to map binding sites of the 14-3-3 interactome: Cdc25C requires phosphorylation of both S216 and S263 to bind 14-3-3. Mol Cell Proteomics 10: M110.005157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Li Q, Sun L, He Z. (2006) The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res 13: 53–63 [DOI] [PubMed] [Google Scholar]
- Chevalier D, Morris ER, Walker JC. (2009) 14-3-3 and FHA domains mediate phosphoprotein interactions. Annu Rev Plant Biol 60: 67–91 [DOI] [PubMed] [Google Scholar]
- Collier R, Fuchs B, Walter N, Lutke WK, Taylor CG. (2005) Ex vitro composite plants: an inexpensive, rapid method for root biology. Plant J 43: 449–457 [DOI] [PubMed] [Google Scholar]
- Cotelle V, Meek SEM, Provan F, Milne FC, Morrice N, MacKintosh C. (2000) 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. EMBO J 19: 2869–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling DL, Yingling J, Wynshaw-Boris A. (2005) Role of 14-3-3 proteins in eukaryotic signaling and development. Curr Top Dev Biol 68: 281–315 [DOI] [PubMed] [Google Scholar]
- DeLille JM, Sehnke PC, Ferl RJ. (2001) The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol 126: 35–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnie C, Andersen CH, Borch J, Gjetting S, Christensen AB, de Boer AH, Thordal-Christensen H, Collinge DB. (2002) Do 14-3-3 proteins and plasma membrane H+-ATPases interact in the barley epidermis in response to the barley powdery mildew fungus? Plant Mol Biol 49: 137–147 [DOI] [PubMed] [Google Scholar]
- Finnie C, Borch J, Collinge DB. (1999) 14-3-3 proteins: eukaryotic regulatory proteins with many functions. Plant Mol Biol 40: 545–554 [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Visconti S, Drumm K, Jahn T, Stensballe A, Mattei B, Jensen ON, Aducci P, Palmgren MG. (1999) Binding of 14-3-3 protein to the plasma membrane H(+)-ATPase AHA2 involves the three C-terminal residues Tyr(946)-Thr-Val and requires phosphorylation of Thr(947). J Biol Chem 274: 36774–36780 [DOI] [PubMed] [Google Scholar]
- Gökirmak T, Paul AL, Ferl RJ. (2010) Plant phosphopeptide-binding proteins as signaling mediators. Curr Opin Plant Biol 13: 527–532 [DOI] [PubMed] [Google Scholar]
- Govindarajulu M, Elmore JM, Fester T, Taylor CG. (2008) Evaluation of constitutive viral promoters in transgenic soybean roots and nodules. Mol Plant Microbe Interact 21: 1027–1035 [DOI] [PubMed] [Google Scholar]
- Govindarajulu M, Kim S-Y, Libault M, Berg RH, Tanaka K, Stacey G, Taylor CG. (2009) GS52 ecto-apyrase plays a critical role during soybean nodulation. Plant Physiol 149: 994–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess MW. (2007) Cryopreparation methodology for plant cell biology. Methods Cell Biol 79: 57–100 [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Koizumi N, Kusano T, Sano H. (2000) Specific binding of a 14-3-3 protein to autophosphorylated WPK4, an SNF1-related wheat protein kinase, and to WPK4-phosphorylated nitrate reductase. J Biol Chem 275: 31695–31700 [DOI] [PubMed] [Google Scholar]
- Li X, Chen L, Dhaubhadel S. (2012) 14-3-3 proteins regulate the intracellular localization of the transcriptional activator GmMYB176 and affect isoflavonoid synthesis in soybean. Plant J 71: 239–250 [DOI] [PubMed] [Google Scholar]
- Li X, Dhaubhadel S. (2011) Soybean 14-3-3 gene family: identification and molecular characterization. Planta 233: 569–582 [DOI] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. (2010a) An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J 63: 86–99 [DOI] [PubMed] [Google Scholar]
- Libault M, Joshi T, Takahashi K, Hurley-Sommer A, Puricelli K, Blake S, Finger RE, Taylor CG, Xu D, Nguyen HT, et al. (2009) Large-scale analysis of putative soybean regulatory gene expression identifies a Myb gene involved in soybean nodule development. Plant Physiol 151: 1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Thibivilliers S, Bilgin DD, Radwan O, Benitez M, Clough SJ, Stacey G. (2008) Identification of four soybean reference genes for gene expression normalization. Plant Genome 1: 44–54 [Google Scholar]
- Libault M, Zhang X-C, Govindarajulu M, Qiu J, Ong YT, Brechenmacher L, Berg RH, Hurley-Sommer A, Taylor CG, Stacey G. (2010b) A member of the highly conserved FWL (tomato FW2.2-like) gene family is essential for soybean nodule organogenesis. Plant J 62: 852–864 [DOI] [PubMed] [Google Scholar]
- Lima L, Seabra A, Melo P, Cullimore J, Carvalho H. (2006a) Phosphorylation and subsequent interaction with 14-3-3 proteins regulate plastid glutamine synthetase in Medicago truncatula. Planta 223: 558–567 [DOI] [PubMed] [Google Scholar]
- Lima L, Seabra A, Melo P, Cullimore J, Carvalho H. (2006b) Post-translational regulation of cytosolic glutamine synthetase of Medicago truncatula. J Exp Bot 57: 2751–2761 [DOI] [PubMed] [Google Scholar]
- McMaugh SJ, Lyon BR. (2003) Real-time quantitative RT-PCR assay of gene expression in plant roots during fungal pathogenesis. Biotechniques 34: 982–986 [DOI] [PubMed] [Google Scholar]
- Moore BW, Perez VJ. (1967) Specific acidic proteins of the nervous system. Carlson FD, , Physiological and Biochemical Aspects of Nervous Integration. Prentice-Hall/The Marine Biological Laboratory, Woods Hole, MA, 343–359 [Google Scholar]
- Moriuchi H, Okamoto C, Nishihama R, Yamashita I, Machida Y, Tanaka N. (2004) Nuclear localization and interaction of RolB with plant 14-3-3 proteins correlates with induction of adventitious roots by the oncogene rolB. Plant J 38: 260–275 [DOI] [PubMed] [Google Scholar]
- Nguyen TH, Brechenmacher L, Aldrich J, Clauss T, Gritsenko M, Hixson K, Libault M, Tanaka K, Yang F, Yao Q, et al. (2012) Quantitative phosphoproteomic analysis of soybean root hairs inoculated with Bradyrhizobium japonicum. Mol Cell Proteomics (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Deerfield DW. (1997) GeneDoc: analysis and visualization of genetic variation. EMBnet News 4: 1–4 [Google Scholar]
- Obsil T, Ghirlando R, Klein DC, Ganguly S, Dyda F. (2001) Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex: a role for scaffolding in enzyme regulation. Cell 105: 257–267 [DOI] [PubMed] [Google Scholar]
- Oh CS, Pedley KF, Martin GB. (2010) Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKKα. Plant Cell 22: 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann C, Yasmin L, Weyand M, Veesenmeyer JL, Diaz MH, Palmer RH, Francis MS, Hauser AR, Wittinghofer A, Hallberg B. (2007) Phosphorylation-independent interaction between 14-3-3 and exoenzyme S: from structure to pathogenesis. EMBO J 26: 902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S, Sehnke PC, Ferl RJ, Gurley WB. (1999) Specific interactions with TBP and TFIIB in vitro suggest that 14-3-3 proteins may participate in the regulation of transcription when part of a DNA binding complex. Plant Cell 11: 1591–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignocchi C, Doonan JH. (2011) Interaction of a 14-3-3 protein with the plant microtubule-associated protein EDE1. Ann Bot (Lond) 107: 1103–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwan O, Liu Y, Clough SJ. (2011) Transcriptional analysis of soybean root response to Fusarium virguliforme, the causal agent of sudden death syndrome. Mol Plant Microbe Interact 24: 958–972 [DOI] [PubMed] [Google Scholar]
- Roberts MR. (2003) 14-3-3 proteins find new partners in plant cell signalling. Trends Plant Sci 8: 218–223 [DOI] [PubMed] [Google Scholar]
- Roberts MR, Bowles DJ. (1999) Fusicoccin, 14-3-3 proteins, and defense responses in tomato plants. Plant Physiol 119: 1243–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MR, Salinas J, Collinge DB. (2002) 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol Biol 50: 1031–1039 [DOI] [PubMed] [Google Scholar]
- Rosenquist M, Alsterfjord M, Larsson C, Sommarin M. (2001) Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes: expression is demonstrated for two out of five novel genes. Plant Physiol 127: 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Seehaus K, Tenhaken R. (1998) Cloning of genes by mRNA differential display induced during the hypersensitive reaction of soybean after inoculation with Pseudomonas syringae pv. glycinea. Plant Mol Biol 38: 1225–1234 [DOI] [PubMed] [Google Scholar]
- Sehnke PC, DeLille JM, Ferl RJ. (2002) Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell (Suppl) 14: S339–S354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Li D, Tang W, Wang H, Xu W, Li X. (2007) Molecular characterization of cotton 14-3-3L gene preferentially expressed during fiber elongation. J Genet Genomics 34: 151–159 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nguyen CT, Libault M, Cheng J, Stacey G. (2011) Enzymatic activity of the soybean ecto-apyrase GS52 is essential for stimulation of nodulation. Plant Physiol 155: 1988–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance CP. (2008) Carbon and nitrogen metabolism in legume nodules. Dilworth MJ, James EK, Sprent JI, Newton WE, , Nitrogen-Fixing Leguminous Symbioses. Springer-Verlag, New York, 293–320 [Google Scholar]
- Wu X, Oh MH, Schwarz EM, Larue CT, Sivaguru M, Imai BS, Yau PM, Ort DR, Huber SC. (2011) Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol 155: 1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91: 961–971 [DOI] [PubMed] [Google Scholar]
- Yang X, Wang W, Coleman M, Orgil U, Feng J, Ma X, Ferl RJ, Turner JG, Xiao S. (2009) Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J 60: 539–550 [DOI] [PubMed] [Google Scholar]
- Yao Y, Du Y, Jiang L, Liu JY. (2007) Molecular analysis and expression patterns of the 14-3-3 gene family from Oryza sativa. J Biochem Mol Biol 40: 349–357 [DOI] [PubMed] [Google Scholar]
- Zou J, Rodriguez-Zas S, Aldea M, Li M, Zhu J, Gonzalez DO, Vodkin LO, DeLucia E, Clough SJ. (2005) Expression profiling soybean response to Pseudomonas syringae reveals new defense-related genes and rapid HR-specific downregulation of photosynthesis. Mol Plant Microbe Interact 18: 1161–1174 [DOI] [PubMed] [Google Scholar]