Abstract
Legumes overcome nitrogen shortage by developing root nodules in which symbiotic bacteria fix atmospheric nitrogen in exchange for host-derived carbohydrates and mineral nutrients. Nodule development involves the distinct processes of nodule organogenesis, bacterial infection, and the onset of nitrogen fixation. These entail profound, dynamic gene expression changes, notably contributed to by microRNAs (miRNAs). Here, we used deep-sequencing, candidate-based expression studies and a selection of Lotus japonicus mutants uncoupling different symbiosis stages to identify miRNAs involved in symbiotic nitrogen fixation. Induction of a noncanonical miR171 isoform, which targets the key nodulation transcription factor Nodulation Signaling Pathway2, correlates with bacterial infection in nodules. A second candidate, miR397, is systemically induced in the presence of active, nitrogen-fixing nodules but not in that of noninfected or inactive nodule organs. It is involved in nitrogen fixation-related copper homeostasis and targets a member of the laccase copper protein family. These findings thus identify two miRNAs specifically responding to symbiotic infection and nodule function in legumes.
Legume plants such as bean (Phaseolus vulgaris), soybean (Glycine max), and pea (Pisum sativum) can overcome a dependence on soil nitrogen sources by forming nitrogen-fixing symbiosis with rhizobial bacteria. The rhizobia reside in special root organs, the nodules, where reduction of atmospheric dinitrogen and nutrient exchange between bacterial and host cells takes place. Specific recognition of rhizobial symbionts relies primarily on chitooligosaccharide signaling molecules, the Nod factors. In the model legume Lotus japonicus, these are perceived by the LysM receptor kinases Nod Factor Receptor1 (NFR1) and NFR5 (Madsen et al., 2003; Radutoiu et al., 2003). Recognition of compatible rhizobia induces curling of root hair tips, and bacteriae form microcolonies in the resulting cavities. The associated root hair cells develop tube-like infection threads that mediate the internalization of bacteria into the host epidermis and cortex.
Epidermal infection is paralleled by the induction of nodule organogenesis in the root cortex, and cells of emerging nodule primordia are infected with bacteria through infection threads. Upon release into the host cell cytoplasm, from which they are separated by the plant-derived symbiosome membrane, bacteria develop into bacteroids metabolically adapted to nitrogen fixation and survival in the plant cell environment. With no access to resources outside the host cell, bacteroids fully depend on the plant for nutrient supplies, including carbohydrates and mineral nutrients such as sulfur and phosphorus required to maintain their metabolism and nitrogen-fixing ability (Krusell et al., 2005; Delmotte et al., 2010). Transport across the symbiosome membrane, therefore, is crucial for nutrient exchange between the host cell and bacteroid, as illustrated with the sulfate transporter Symbiotic Sulfate Transporter1 (SST1; Krusell et al., 2005), which is thought to provide sulfate required for the bacterial nitrogenase activity. In the absence of SST1, nitrogen reduction cannot take place, and the fully infected, yet inefficient, nodules forming on sst1 loss-of-function mutants are subject to premature senescence (Krusell et al., 2005).
Legume nodules can be categorized into two major groups based on the persistence of their apical meristem. The “determinate” type, as found in the important crop legume soybean or the model plant L. japonicus, has a transient meristem that eventually ceases activity, resulting in globular organs going through a succession of developmental steps. By contrast, “indeterminate” types of nodules, such as those of pea and Medicago truncatula, maintain an active meristem (Oldroyd and Downie, 2008). A determinate organogenetic pattern implies that different stages of symbiotic maturity are spatiotemporally separated, allowing for symbiosis progression to be monitored in a stepwise manner. The extensive collection of symbiotic mutants isolated in the model legume L. japonicus has helped to dissect the signaling pathways involved in the distinct steps of bacterial infection and nodule organogenesis (Madsen et al., 2010). A series of transcription factors, including Nodule Inception (Schauser et al., 1999) and the two GRAS-type proteins Nodulation Signaling Pathway1 (NSP1) and NSP2 (Heckmann et al., 2006), are notably required for both epidermal bacterial infection and nodule organogenesis, suggesting a tight link between these processes. Identification of a class of mutants generating spontaneous nodules in the absence of bacterial infection (Tirichine et al., 2006b) demonstrated, however, that nodule organogenesis and endosymbiotic infection can be uncoupled. L. japonicus plants encoding autoactive versions of the Ca2+-calmodulin kinase (spontaneous nodule formation1 [snf1]; Tirichine et al., 2006a) or the cytokinin receptor Lotus Histidine Kinase1 (LHK1; snf2; Tirichine et al., 2007) induce nodule organogenesis independently of compatible rhizobia. The resulting nodule organs show the same morphology and tissue composition as symbiotic nodules, but they contain a center of uninfected, parenchymatic cells instead of the bacteria-filled infected tissue found under normal symbiotic conditions (Tirichine et al., 2006a, 2007).
The dissection of signaling pathways involved in legume nodulation symbiosis has been largely focused on a combination of mutant (Madsen et al., 2010) and transcriptome (Høgslund et al., 2009) analyses. More recently, however, small RNAs (sRNAs) that orchestrate RNA silencing have emerged as potential contributors to the complex and dynamic gene regulation changes that accompany the nodulation process. Plant silencing sRNAs fall into several distinct classes, among which microRNAs (miRNAs) have received the most attention in the field so far. Plant miRNAs are processed by the Dicer-like1 endoribonuclease from stem-loop precursor RNAs derived from polyadenylated, longer primary microRNA transcripts (Voinnet, 2009). Mature miRNAs are 20 to 24 nucleotides long and mediate posttranscriptional regulation of target transcripts displaying local sequence complementarity (Voinnet, 2009). In both monocot and dicot plants, the majority of miRNAs associates with the ARGONAUTE (AGO) effector protein AGO1, although other AGO proteins might be recruited in some cases (Mi et al., 2008; Vaucheret, 2008; Czech and Hannon, 2011). AGOs are guided to target mRNA transcripts by the incorporated miRNA strand in a sequence-dependent manner. A high degree of sequence complementarity between miRNA and target appears to trigger AGO-mediated endonucleolytic cleavage or “slicing” at or near the center of miRNA/target duplexes. Alternative or additional control through translational inhibition, commonly observed in metazoans, seems to be widespread in plants as well (Brodersen et al., 2008; Mallory and Bouché, 2008; Brodersen and Voinnet, 2009).
To date, a large number of miRNA families have been described in legumes, many of which are conserved beyond this lineage and also occur in Arabidopsis (Arabidopsis thaliana), as evidenced in the miRBase registry (www.mirbase.org; Subramanian et al., 2008; Jagadeeswaran et al., 2009; Lelandais-Brière et al., 2009; Joshi et al., 2010; Chi et al., 2011; Devers et al., 2011; Li et al., 2011; Peláez et al., 2012; Turner et al., 2012). Furthermore, symbiosis-responsive sRNAs have also recently been identified at different stages of nodule development (Subramanian et al., 2008; Jagadeeswaran et al., 2009; Lelandais-Brière et al., 2009; Li et al., 2010; Kulcheski et al., 2011), including several miRNAs found to be highly expressed in nodules (Lelandais-Brière et al., 2009; Wang et al., 2009). Nonetheless, evidence for functional involvement in nodulation symbiosis has been obtained for only a small number of candidates, including miR166 and miR169 in M. truncatula. miR166 isoforms regulate meristem activity and vascular differentiation in both roots and nodules by controlling HD-ZIPIII targets (Boualem et al., 2008), while miR169 regulates the spatial distribution of the transcription factor HAP2-1 involved in meristem maintenance and bacterial release in nodules (Combier et al., 2006). While both miR166 (Boualem et al., 2008) and miR169 (Combier et al., 2006) are thus likely to be involved in nodule organogenesis, no miRNA has so far been linked to nitrogen fixation activity in nodules. In parallel, miRNAs involved in arbuscular mycorrhizal symbiosis have also been assayed in recent years (Branscheid et al., 2011; Devers et al., 2011; Lauressergues et al., 2012).
In this study, we used deep sequencing to identify novel and symbiosis-responsive miRNAs from determinate nodules of the model legume L. japonicus, in which the existence of miRNAs has only been inferred from in silico approaches so far (miRBase registry; Sunkar and Jagadeeswaran, 2008; Hsieh et al., 2009; Wang et al., 2009). Using a series of mutants that uncouple nodule organ formation, infection, and function, we identify two miRNAs specifically linked to bacterial infection and nodule function rather than organogenesis.
RESULTS
Identification of sRNAs from L. japonicus Roots and Nodules
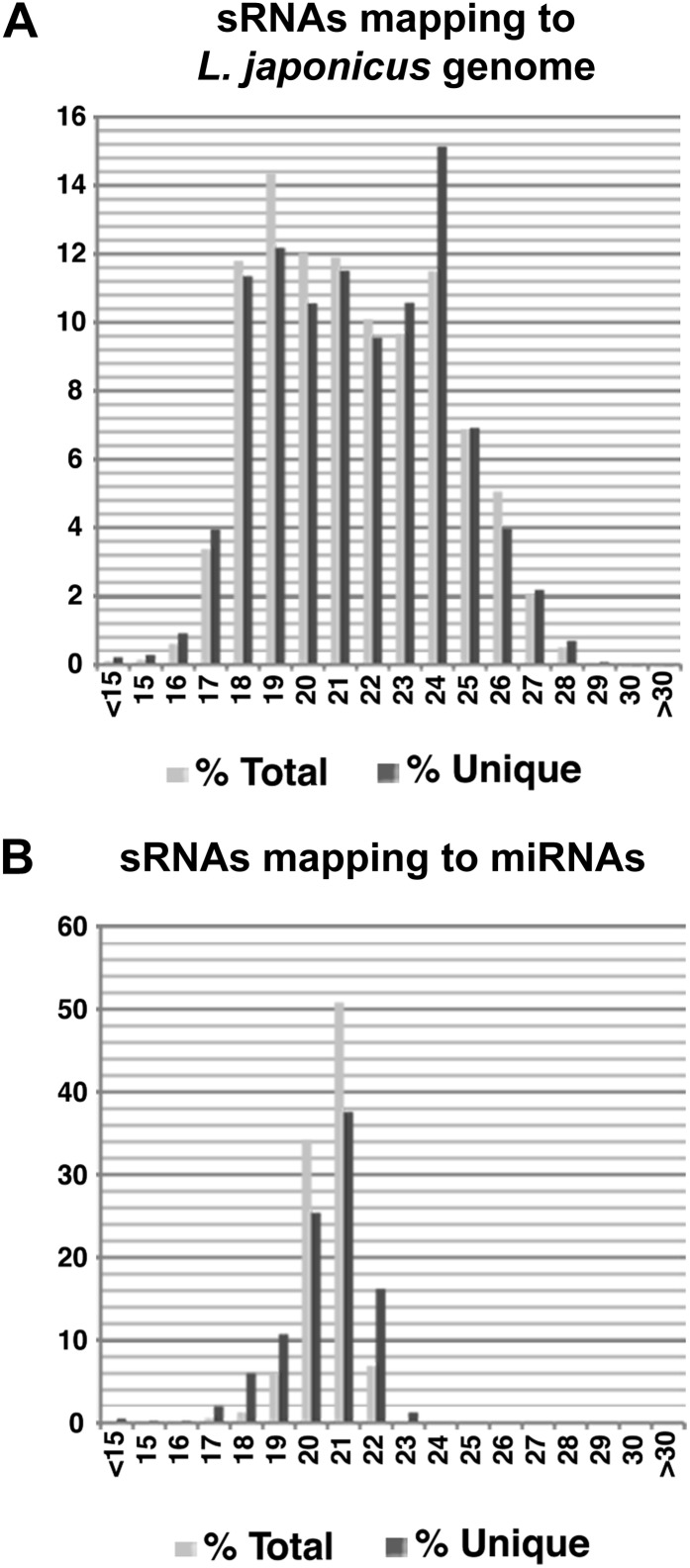
To identify miRNAs involved in L. japonicus root nodulation symbiosis, entire roots were harvested at 3 h post inoculation (hpi) or 3 d post inoculation (dpi) with the rhizobial symbiont Mesorhizobium loti or upon control treatments with medium (mock). Nodules harvested at 3 weeks post inoculation (wpi) were also included in the analysis. sRNAs were isolated from these tissues and sequenced using the 454 technology (Roche), yielding a total of about 300,000 quality reads corresponding to 147,692 unique sequences (Table I). A total of 71.2% of unique sRNA sequences from the combined four root libraries mapped to the L. japonicus genome (Sato et al., 2008; version 2.5), compared with 46.2% from the 3-wpi nodule library, which contained nearly 40% of unique sequences mapping to the genome of M. loti (Table I). About one-quarter of unique sequences from the combined libraries could not be mapped to either of these sources and probably correspond to as yet unsequenced regions of the L. japonicus genome (Table I). Of the unique sequences mapping to the L. japonicus genome, the 24-nucleotide fraction was the most abundant in all five samples (Fig. 1).
Table I. sRNA sequencing output and mapping to genomic sequences.
Unique sequences from sRNA libraries were mapped using an in-house sRNA bioinformatic platform, allowing one mismatch.
| Genome sRNA Mapping | Roots | Nodules | All Libraries |
|---|---|---|---|
| L. japonicus genome v2.5 | 74,116 (71.2%) | 16,467 (46.2%) | |
| M. loti genomea | 795 (0.76%) | 20,063 (41.1%) | |
| Unknown | 29,140 (28%) | 12,270 (25.1%) | |
| Sequencing Output | |||
| Unique sequences | 104,051 | 48,800 | 147,692 |
| Total reads | 234,060 | 71,802 | 305,862 |
Sequences mapping to the L. japonicus genome were removed prior to analysis of the M. loti genome.
Figure 1.
Size fractionation of L. japonicus sRNAs sequenced using Roche 454 pyrosequencing. Data sets from all root and nodule libraries combined were mapped to the L. japonicus genome version 2.5 (A) and the miRBase registry (B). Values are expressed in percentages of total (light gray) or unique (dark gray) read counts.
To annotate sRNA populations obtained from our libraries and identify conserved plant miRNAs or repeat-associated sRNAs present in L. japonicus, we mapped the sequences to publicly available databases (Table II; Supplemental Fig. S1). Between 0.3% and 0.6% of unique sequences mapped to members of 45 families of proposed plant miRNAs, as deposited in the miRBase registry (Tables II and III; www.mirbase.org). Predicted miRNA* sequences, representing the passenger strands of miRNA duplexes upon release from their stem-loop precursors, were cloned at low frequencies for members of six of these families (Table III). Most L. japonicus sequences mapping to conserved miRNAs were 20 to 21 nucleotides long (Fig. 1). In both root and nodule libraries, about 25% of total sequences mapping to the L. japonicus genome showed high homology to ribosomal RNAs (rRNAs), tRNAs, small nuclear (snRNAs), or small nucleolar RNAs, collectively referred to here as “miscellaneous” RNAs (miscRNAs). Another 0.3% to 0.6% mapped to conserved repetitive DNA sequences including transposable elements (Table II).
Table II. Annotation of sRNA libraries from L. japonicus roots and nodules.
Unique sequences were used for this analysis. For parameters and database references, see “Materials and Methods.”
| sRNA Annotation | 3-h Mock | 3 hpi | 3-d Mock | 3 dpi | Total Roots | 3-wpi Nodules |
|---|---|---|---|---|---|---|
| miscRNAa | 2,382 (25.1%) | 9,953 (25.5%) | 9,716 (26.7%) | 11,761 (28.3%) | 26,279 (25.3%) | 6,439 (13.2%) |
| Known miRNAb | 61 (0.6%) | 196 (0.5%) | 197 (0.5%) | 207 (0.5%) | 457 (0.4%) | 162 (0.3%) |
| DNA repeats | 57 (0.6%) | 178 (0.5%) | 139 (0.4%) | 132 (0.3%) | 490 (0.5%) | 143 (0.3%) |
| Sequencing output | ||||||
| Unique sequences | 9,500 | 39,031 | 36,363 | 41,598 | 104,051 | 48,800 |
| Total reads | 11,932 | 72,129 | 66,945 | 83,054 | 234,060 | 71,802 |
Table III. Identification of known miRNA families in L. japonicus roots and nodules.
| Known miRNA Familya | Total Readsb | Normalized Total Rootsc | Normalized 3-d Mockd | Normalized 3-wpi Nodulee | miRNA*f | miRCatg |
|---|---|---|---|---|---|---|
| miR1120 | 1 | 0.0 | 0.0 | 1.7 | – | – |
| miR1310 | 1 | 0.7 | 2.3 | 0.0 | – | – |
| miR1426, miR172 | 1 | 0.0 | 0.0 | 1.7 | – | – |
| miR1509 | 2 | 0.0 | 0.0 | 3.4 | – | – |
| miR1510 | 4 | 2.7 | 4.7 | 0.0 | – | – |
| miR1511 | 75 | 48.4 | 37.2 | 3.4 | – | Yes |
| miR156 | 41 | 22.6 | 23.3 | 11.8 | – | Yes |
| miR156, miR157 | 30 | 11.3 | 9.3 | 21.9 | – | – |
| miR159 | 886 | 524.6 | 762.7 | 159.9 | – | – |
| miR159, miR319 | 4 | 2.0 | 4.7 | 1.7 | – | – |
| miR160 | 4 | 2.7 | 2.3 | 0.0 | – | Yes |
| miR162 | 18 | 10.6 | 9.3 | 3.4 | – | – |
| miR164 | 25 | 15.3 | 25.6 | 3.4 | – | Yes |
| miR165 | 2 | 1.3 | 2.3 | 0.0 | – | – |
| miR166 | 950 | 590.9 | 581.3 | 99.3 | Yes | Yes |
| miR167 | 41 | 9.9 | 7.0 | 43.8 | – | Yes |
| miR168 | 60 | 39.8 | 55.8 | 0.0 | – | Yes |
| miR169 | 4 | 2.7 | 2.3 | 0.0 | – | Yes |
| miR171 | 24 | 11.9 | 18.6 | 10.1 | Yes | Yes |
| miR172 | 67 | 0.7 | 0.0 | 111.1 | – | Yes |
| miR1875 | 1 | 0.0 | 0.0 | 1.7 | – | – |
| miR2109 | 1 | 0.7 | 0.0 | 0.0 | – | – |
| miR2111 | 6 | 4.0 | 9.3 | 0.0 | – | – |
| miR2119 | 4 | 2.7 | 9.3 | 0.0 | – | – |
| miR2199 | 8 | 5.3 | 4.7 | 0.0 | – | – |
| miR2916 | 1 | 0.7 | 0.0 | 0.0 | – | – |
| miR319 | 127 | 82.2 | 102.3 | 5.1 | – | Yes |
| miR3447 | 1 | 0.7 | 0.0 | 0.0 | – | – |
| miR390 | 16 | 2.0 | 0.0 | 21.9 | Yes | Yes |
| miR393 | 120 | 75.6 | 39.5 | 10.1 | Yes | Yes |
| miR394 | 2 | 1.3 | 0.0 | 0.0 | – | – |
| miR396 | 173 | 109.4 | 123.2 | 13.5 | Yes | Yes |
| miR397 | 46 | 11.3 | 9.3 | 48.8 | – | Yes |
| miR398 | 6 | 4.0 | 4.7 | 0.0 | – | – |
| miR399 | 1 | 0.7 | 0.0 | 0.0 | – | – |
| miR408 | 8 | 3.3 | 2.3 | 5.1 | – | Yes |
| miR414 | 1 | 0.7 | 0.0 | 0.0 | – | – |
| miR419 | 1 | 0.0 | 0.0 | 1.7 | – | – |
| miR477 | 2 | 1.3 | 0.0 | 0.0 | – | Yes |
| miR482 | 18 | 11.3 | 9.3 | 1.7 | – | – |
| miR5054 | 3 | 2.0 | 2.3 | 0.0 | – | – |
| miR5059 | 4 | 2.0 | 2.3 | 1.7 | – | – |
| miR5072 | 20 | 11.3 | 7.0 | 5.1 | – | – |
| miR5077, miR894 | 201 | 120.0 | 93.0 | 33.7 | – | – |
| miR894 | 20 | 11.3 | 16.3 | 5.1 | – | – |
miRNA families from the miRBase registry to which sRNA reads were assigned through sequence homology (two or fewer mismatches) using miRProf default parameters for miRNA family grouping. Reads that matched to registered miRNA* sequences were not considered. Some reads matched to several miRNA families simultaneously, as indicated.
Total reads present in all libraries combined two or fewer mismatches.
Normalized read counts per 100,000 non rRNA/tRNA reads in all root libraries combined.
Normalized read counts per 100,000 non rRNA/tRNA reads in 3-d noninoculated roots.
Normalized read counts per 100,000 non rRNA/tRNA reads in 3-wpi nodule libraries. miRNA* sequences that are present in our dataset.
Families whose precursor sequences were predicted as bona-fide miRNA precursors by miRCat.
Prediction and Validation of L. japonicus-Specific miRNAs
Different studies have shown that, in addition to a set of evolutionarily conserved miRNAs found across phyla, plants including legumes possess lineage- or species-specific miRNAs of more recent evolutionary origin (He et al., 2008; Szittya et al., 2008; Lelandais-Brière et al., 2009; Cuperus et al., 2011; Peláez et al., 2012; Turner et al., 2012). To identify new miRNAs in L. japonicus, we used the plant de novo miRNA prediction algorithm miRCat (Moxon et al., 2008; Supplemental Fig. S2). We isolated 32 novel candidate miRNAs mapping to 45 genomic loci and not homologous (more than two mismatches) to plant miRNAs already found in the miRBase registry. The predicted precursor RNAs form stable hairpin structures as typically found in plant MIR gene transcripts (Meyers et al., 2008; Supplemental Fig. S3). The sequencing frequencies of these nonconserved miRNAs were consistently lower than those of conserved plant miRNAs predicted on the basis of sequence homology (Tables III and IV). This is consistent with observations from previous studies in both legume and nonlegume plants showing that evolutionarily young miRNAs tend to be less expressed than conserved ones (Szittya et al., 2008; Lelandais-Brière et al., 2009; Fahlgren et al., 2010; Cuperus et al., 2011; Li et al., 2011), a possible adaptation reducing potential undesirable off-targeting effects (Voinnet, 2009). An important criterion in the validation of novel miRNAs is the availability of the corresponding, less stable miRNA* sequence, indicating precise excision of a discrete miRNA/miRNA* duplex from the stem-loop precursor (Meyers et al., 2008). Our data set contained miRNA* sequences for one of the 32 novel miRNA candidates (Table IV; Supplemental Fig. S3), and higher-coverage sequencing approaches will be necessary to verify miRNA* sequences of the remaining candidates.
Table IV. Novel miRNAs identified in L. japonicus.
| miRNA Identifier | Sequence (5´–3´) | Nucleotides | Readsa | miRNA*b | Loci No.c | Detectiond |
| lja-miR7516 | UAGCGGGUGUCUUCGCCUCUGA | 22 | 55 | Yes (5) | 1 | N |
| lja-miR1507 | UCUUCCAUCCAUACAUCAUCU | 21 | 53 | – | 2 | N |
| lja-miR7517 | AUAUGGUAAAGGUUAGGGACC | 21 | 19 | – | 1 | – |
| lja-miR7518 | UUGCGCACUGAGCAAGGACAGG | 22 | 16 | – | 1 | – |
| lja-miR7519 | CAAAUUUUCUAAGUGGGCUAGC | 22 | 3 | – | 1 | – |
| lja-miR7520 | GAGGGGGAAGGUGAUGACAUC | 21 | 3 | – | 1 | – |
| lja-miR7521 | UCAUGGGUGGGGUGUUAAACC | 21 | 3 | – | 1 | – |
| lja-miR7522 | AACUGCGGACAGGUUUUAUGAC | 22 | 2 | – | 1 | – |
| lja-miR7523 | ACCACCGGGCUCGAGGAUCAGC | 22 | 2 | – | 2 | – |
| lja-miR7524 | ACCAGUGAGUCAUUGGGCGGA | 21 | 2 | – | 1 | – |
| lja-miR7525 | AGGGCGUUUUGGUACAUGACU | 21 | 2 | – | 1 | – |
| lja-miR7526 | AUCAAGGUAGCUGUAACUUCC | 21 | 2 | – | 8 | – |
| lja-miR7527 | CAUGGCGUGCAAAACCCCACGC | 22 | 2 | – | 1 | – |
| lja-miR7528 | CCGAAAUGCUAAUCUGAAGCUU | 22 | 2 | – | 1 | – |
| lja-miR7529 | CCGUAGCAUCAAUUUAUCCGA | 21 | 2 | – | 1 | – |
| lja-miR7530 | CCUUCCUCUCUUCACUAUCUUC | 22 | 2 | – | 1 | – |
| lja-miR7531 | CGUGUUUUCUUUCAUUCCCCA | 21 | 2 | – | 1 | – |
| lja-miR7532 | GAAGCUGCCUCUGGUCGUGGU | 21 | 2 | – | 2 | – |
| lja-miR7533 | GAGGGGAUGGAGAGAAGCUGG | 21 | 2 | – | 2 | – |
| lja-miR7534 | GCAACUUGACUACAGUUUGAC | 21 | 2 | – | 1 | – |
| lja-miR7535 | GGGAAAAUGUGGGUGUGGU | 19 | 2 | – | 1 | – |
| lja-miR7536 | UAAGACAUGCUCAAGAGUG | 19 | 2 | – | 2 | – |
| lja-miR7537 | UAGGAAAUACGCCUGCGGUUCC | 22 | 2 | – | 1 | – |
| lja-miR7538 | UCAACGGAGAGCUUGCUGUC | 20 | 2 | – | 1 | – |
| lja-miR7539 | UCGAGAGAGAGAGCGACGAGG | 21 | 2 | – | 1 | – |
| lja-miR7540 | UGAUAUGAUAAGUGAUGUGA | 20 | 2 | – | 2 | – |
| lja-miR7541 | UGCAUUCUCUUUUGGUGGCCC | 21 | 2 | – | 1 | – |
| lja-miR7542 | UGCUUGCUUAUAGAUGGUG | 19 | 2 | – | 1 | – |
| lja-miR7543 | UUAAUGAUACAUGUUUGACU | 20 | 2 | – | 1 | – |
| lja-miR7544 | UUAGAAAGAAAAUGUUGUUAGC | 22 | 2 | – | 1 | – |
| lja-miR7545 | UUGGGAAAGCUAGAGUGCU | 19 | 2 | – | 1 | – |
| lja-miR7546 | UUGGUGACCGACAGGCGCGUGC | 22 | 2 | – | 1 | – |
To validate the specific accumulation of novel miRNAs, we performed systematic northern hybridization analyses. The two most sequenced candidates, lja-miR7516 and lja-miR1507, accumulated as discrete species in different L. japonicus tissues and were absent in RNA samples from Arabidopsis seedlings tested alongside (Fig. 2), supporting the in silico finding that no Arabidopsis homologs of these sequences exist. This suggests that lja-miR7516 and lja-miR1507 are specific to L. japonicus or its lineage. It is noteworthy, however, that mature lja-miR7516 presents a certain degree of similarity to the recently identified gma-miR2109 family found in the genus Glycine (www.mirbase.org). Sequence similarity between the corresponding miRNA precursor sequences also suggests possible homology (Supplemental Fig. S4). Similarly, lja-miR1507, considered novel based on the absence of apparent homologs at the mature sequence level, is predicted to originate from two precursor genes with possible homology to MIR1507 family members found in several other legume species based on precursor similarity. It is therefore named lja-miR1507 in this publication and has been registered as such in the miRBase registry. The remaining candidate miRNAs could not be traced using northern analysis, in line with their low frequencies in our sequencing data sets, suggesting that their abundance levels are below the detection limit for this method. Further analysis will thus be necessary to confirm if these miRNA candidates accumulate as distinct species. In addition to the 32 novel miRNA candidates, miRCat predicted miRNA identity for members of 19 additional known miRNA families, representing less than half of the 45 conserved miRNAs identified in L. japonicus on the basis of sequence similarity (Table III). This low overlap likely reflects the high stringency of the miRCat prediction algorithm, which has been specifically designed for miRNA prediction in plants using 454 pyrosequencing data (Moxon et al., 2008).
Figure 2.
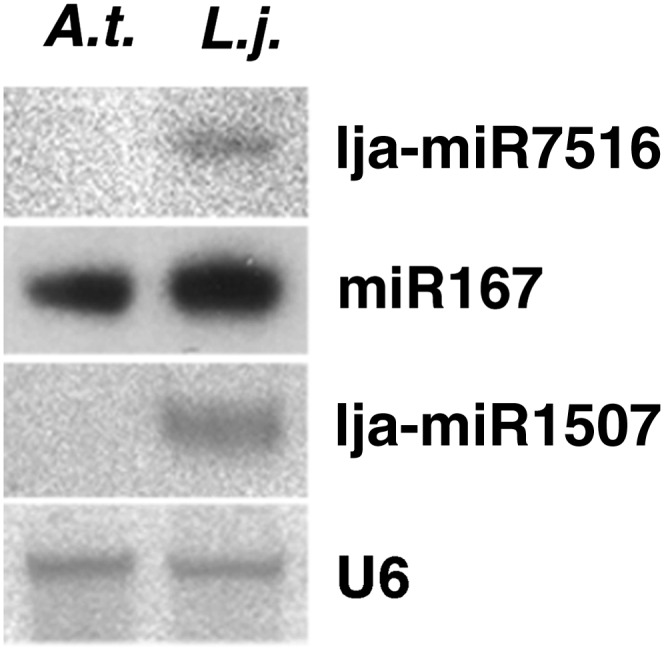
Experimental validation of novel miRNAs lja-miR7516 and lja-miR1507 by northern analysis using RNA extracted from L. japonicus (L.j.) and Arabidopsis (A.t.) seedlings. Blots correspond to independent assays, where miR167 or U6 was used as the loading control, respectively.
L. japonicus Nodules Express a Specific Set of miRNAs
Having established a repertoire of L. japonicus root and nodule sRNAs including miRNAs, we then aimed to isolate miRNAs specifically involved in bacterial infection and/or the establishment of symbiosis by comparing the abundance profiles of conserved and predicted novel miRNAs between the root and nodule libraries. At 3 hpi, measurable physiological responses of L. japonicus roots to M. loti include an influx of Ca2+ ions into developing root hair cells, followed by distinct Ca2+ spikes in and around the nucleus. These responses, along with an alkalinization of the extracellular space, are initiated within minutes of the application of Nod factors or compatible bacteria (Miwa et al., 2006), but morphological root or root hair responses to the symbiont are not generally observed at this stage. Subsequently, at 3 dpi, infection threads are emerging at a time when cross signaling to inner cortical cells initiating primordium formation has commenced. None of the miRNAs contained in our libraries, however, showed significantly altered abundance between M. loti-infected and mock-treated root samples at either of these early time points preceding organogenesis. We thus focused on the analysis of later symbiotic stages.
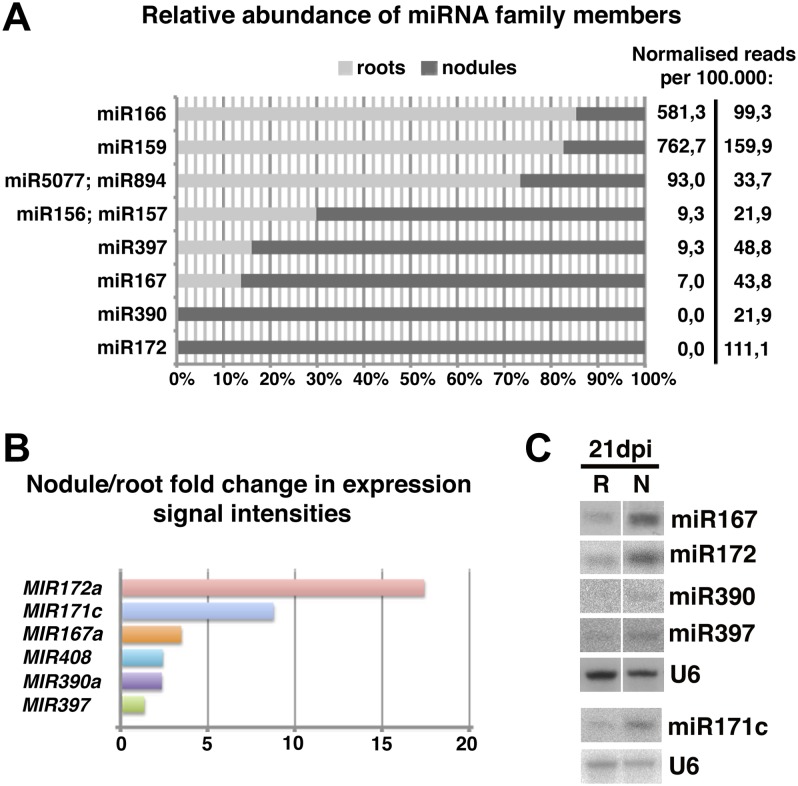
Comparative expression analysis from root and mature nodule samples has been successfully employed to predict symbiosis-related genes (Colebatch et al., 2004; El Yahyaoui et al., 2004; Høgslund et al., 2009; Lelandais-Brière et al., 2009). This led us to analyze sequencing data from nodule and root libraries in a comparative profiling approach. Using the miRProf algorithm (Moxon et al., 2008), we selected miRNAs represented by a minimum of 10 read counts in nodules and a minimum 3-fold higher read abundance in nodule versus mock-inoculated root libraries (Supplemental Fig. S5). Members of four conserved miRNA families, miR167, miR172, miR390, and miR397, followed these criteria, showing a markedly higher relative abundance in mature nodule as compared with root libraries (Fig. 3A; Table III). Some miRNA families, such as MIR159 and MIR166, were found in nodules at high accumulation levels (Table III), but their relative accumulation in root tissue was even stronger (Fig. 3A). These miRNAs were not considered in further analyses, because specific involvement in the symbiotic process was considered unlikely on the basis of the observed abundance patterns. In an independent approach, we analyzed expression profiles of MIR genes spotted on the L. japonicus Affymetrix GeneChip (Høgslund et al., 2009) to explore these results at the level of primary MIRNA transcript abundance. Out of the 65 predicted miRNA precursor transcripts represented on the chip, three were significantly more abundant in mature nodules compared with noninoculated roots showing 3-fold or higher expression levels (Fig. 3B). These include MIR167a, MIR172a, and a member of the MIR171 gene family referred to here as MIR171c. Out of five MIR171 paralogs represented on the L. japonicus Affymetrix GeneChip, only MIR171c showed differential regulation in nodules versus roots, while all other loci remained unresponsive in nodules (Supplemental Fig. S6; Høgslund et al., 2009), suggesting a specific role for miR171c in symbiosis or nodule development. The Affymetrix-based monitoring also showed an up-regulation, although modest, of members of the MIR390, MIR397, and MIR408 families (Fig. 3B). Expression data of miRNA primary transcripts is thus in line with the 454 sequencing results and further suggests that miR167, miR171c, miR172, miR390, miR397, and miR408 could potentially undergo nodule-specific transcriptional up-regulation as opposed to being translocated from cell to cell from other parts of roots, as has been reported for some miRNAs (Lin et al., 2008; Pant et al., 2008; Carlsbecker et al., 2010). For further validation, we performed northern hybridization from root and nodule tissue extracts. While miR408 could not be detected under our experimental conditions, we confirmed a higher accumulation of miR167, miR171c, and miR172 in nodules as compared with roots and a modest induction of miR390 and miR397 (Fig. 3C).
Figure 3.
Identification of miRNAs that are differentially expressed in nodules and roots. A, Abundance of members of known miRNA families between 3-wpi nodule and noninoculated root libraries, expressed as percentages. Normalized counts per 100,000 reads filtered for rRNAs/tRNAs are listed on the right, for roots (left column) and nodules (right column). Only miRNA families present in nodule libraries with 10 or more reads were considered. B, Relative expression signal intensities of PRI-MIRNA genes spotted on the L. japonicus Affymetrix GeneChip (for data and statistical analysis, see Høgslund et al., 2009). The fold change was calculated as the ratio of normalized average expression values from wild-type 21-dpi nodules as compared with noninoculated roots (three independent biological replicates). From all PRI-MIRNA genes spotted on the chip, only those with a consistently observed statistically significant differential expression are shown (false discovery rate-corrected P ≤ 0.05). C, miRNA accumulation in L. japonicus roots and nodules at 21 dpi. Root (R) samples comprise remaining tissue after nodule (N) excision. The top and bottom panels in indicate different hybridization experiments, where U6 served as a loading control.
We conclude that a set of specific miRNAs overaccumulate in mature determinate L. japonicus nodules, showing different levels of nodule/root specificity of these miRNAs. Therefore, they constitute strong candidates as possible riboregulators of various stages of root nodulation symbiosis in L. japonicus.
miR397 and miR171c Accumulation in L. japonicus Nodules Is Linked to Infection But Not Nodule Development
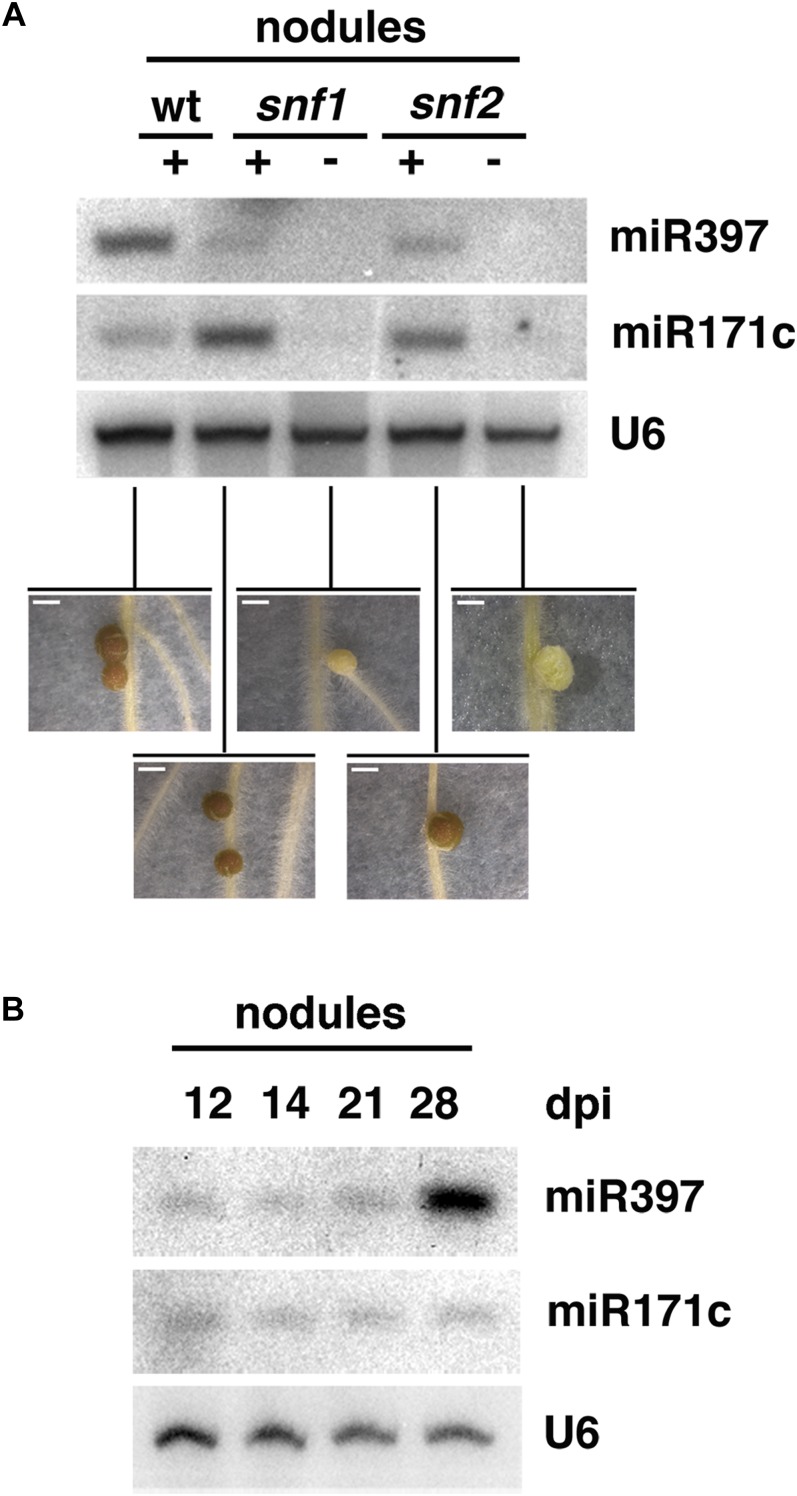
A high accumulation of miRNAs in symbiotic nodules could indicate their involvement in morphological processes such as induction and progression of nodule organogenesis or, alternatively, in the establishment/maintenance of an efficient endosymbiotic interaction. To identify candidates specifically linked to bacterial infection and nodule symbiotic functionality rather than organogenesis, we exploited the availability of snf mutants. snf1 and snf2 produce autoactive versions of the Ca2+-calmodulin-activated kinase and the cytokinin receptor LHK1, respectively. As a result, both lines form nodule organs in the absence of the bacterial symbiont, but they also develop fully infected functional nodules upon M. loti inoculation (Tirichine et al., 2006a, 2006b, 2007). Our candidate miRNAs were tested using northern analysis, and two of them, miR397 and miR171c, showed a consistently higher accumulation in infected nodules of snf mutants compared with noninfected, spontaneous nodules (Fig. 4A). This indicates that miR397 and miR171c induction in wild-type nodules versus roots is linked to bacterial infection and/or to nodule functionality rather than organogenesis per se. By contrast, the remaining miRNAs tested showed similar abundance in infected and bacteria-free snf nodules or were below the detection limit (Supplemental Fig. S7).
Figure 4.
miR397 and miR171c are specifically up-regulated in infected nodules. A, miRNA accumulation in infected wild-type nodules (wt) and infected or spontaneous nodules in snf1-1 and snf2 mutant backgrounds. Tissues were harvested at 6 wpi. The bottom panels present nodule phenotypes at the time of harvest. Bars = 1 mm. +, Plants were inoculated with M. loti; −, plants were mock inoculated. B, Accumulation of miR171c and miR397 at different stages of nodule development. U6 served as a loading control.
We then sought to analyze whether the induction of miR171c and miR397 expression in M. loti-infected nodules is implicated in the establishment/maintenance of endosymbiotic infection or in processes related to N2 fixation and symbiosis functionality. We first tested if the expression levels of these miRNAs depend on the nodule developmental stage. In determinate nodules like those formed by L. japonicus, meristematic activity ceases once the nodule has reached a certain size, such that infection of nodule cells, bacteroid maturation, nitrogen fixation, and nodule senescence occur sequentially thereafter. Under our conditions, nodules start fixing nitrogen at 7 to 14 dpi, reaching full maturity at 21 to 28 dpi, followed by senescence. Northern hybridization analyses revealed that, while miR171c abundance was constant in infected nodules of different ages, miR397 levels were drastically enhanced at 28 dpi compared with younger nodules (Fig. 4B). This result suggests a link between miR397 expression and the presence or maintenance of an efficient symbiosis or, alternatively, the initiation of nodule senescence. By contrast, the elevated miR171c expression levels in nodules compared with roots seem to be linked to the infected stage per se, as expression levels remain stable as the nodule matures and enters nitrogen fixation activity.
L. japonicus miR171c Accumulates Specifically in Nodules and Targets NSP2, a Transcription Factor Required for Root Nodule Symbiosis
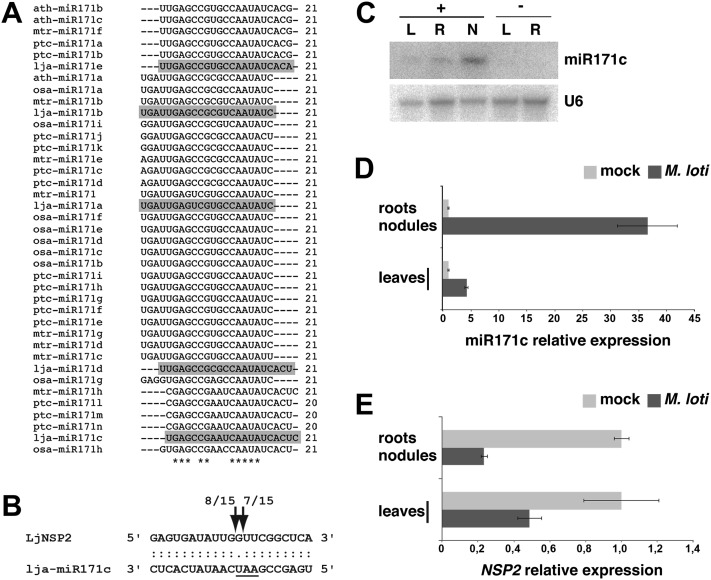
Using a combination of cloning and sequence homology searches, we identified a total of five different miR171 family variants in L. japonicus (Fig. 5A). Interestingly, the sequence of lja-miR171c aligns only partially with that of other plant miR171 family members, clustering with a group of miR171 isoforms that are shifted by several nucleotides along the precursor RNA backbone and contain a central two- to three-nucleotide polymorphism that sets them apart from the canonical miR171 sequences (Fig. 5A). In silico target predictions using the Target Finder algorithm (Allen et al., 2005; Fahlgren et al., 2007) suggested that lja-miR171c might specifically regulate the L. japonicus GRAS transcription factor NSP2, which is essential for nodulation in legumes (Kaló et al., 2005; Heckmann et al., 2006; Murakami et al., 2006). 5′ RACE analysis confirmed endonucleolytic cleavage of the L. japonicus NSP2 mRNA within the miR171c complementary site and, more specifically, within the central three-nucleotide polymorphism observed (Fig. 5B), indicating that NSP2 is a direct target of this miRNA. As for the remaining members of the L. japonicus miR171 family, similar target-prediction analysis suggested that three of them, but not miR171c, could target members of the Scarecrow-like homology group of GRAS-type transcription factors. This is consistent with a widely conserved miRNA-mediated regulation mechanism initially observed in Arabidopsis (Llave et al., 2002). 5′ RACE analysis confirmed that the mRNA of a L. japonicus homolog of Scarecrow-like6, LjSCL6, is indeed targeted by one or more members of the miR171 family (Supplemental Fig. S8). Moreover, lja-miR171c is homologous to mtr-miR171h and other miRNAs belonging to a recently identified group that is part of the conserved MIR171 family, but it has so far exclusively been found in endosymbiotic plant species (Fig. 5A; Lauressergues et al., 2012). Taken together, these observations demonstrate the conservation of both canonical and noncanonical functions of the MIR171 family in the regulation of GRAS-type transcription factors in L. japonicus.
Figure 5.
Lja-miR171c targets NSP2 and is up-regulated in L. japonicus nodule tissue. A, Alignment of mature miR171 isoforms from L. japonicus and several plant species. L. japonicus miR171 members, marked in gray, were identified in this study by cloning (lja-miR171b to lja-miR171e) or homology mapping (lja-miR171a). Rice, poplar, Arabidopsis, and M. truncatula sequences were retrieved from the miRBase registry. Sequences were aligned using Clustal version 2.1; ath, Arabidopsis; lja, L. japonicus; mtr, M. truncatula; osa, rice; ptc, poplar. B, Validation of NSP2 as a miR171c target in L. japonicus. Cleavage sites were determined by 5′ RACE analysis. The number of sequenced fragments ending at a respective site (out of total clones sequenced) is indicated. Predicted sites of endonucleolytic cleavage are indicated by arrows. The nucleotide triplet that is different from the canonical miR171 family members is underlined. C, miR171c accumulation in L. japonicus leaves (L), roots (R), and nodules (N) from M. loti-inoculated (+) or mock-treated (−) plants. Tissues were collected at 21 dpi. Root (R) samples from M. loti-inoculated plants (+) comprise remaining root tissue after nodule excision. D and E, Relative expression levels of mature miR171c (D) and NSP2 (E) in nodules compared with mock-treated roots and leaves from inoculated compared with mock-treated plants. Light gray bars indicate tissues harvested from mock-treated plants, and dark gray bars indicate tissues harvested from inoculated plants. All tissues were harvested at 21 dpi. Three biological and two technical replicates were analyzed. Error bars show the se.
Affymetrix data initially pinpointed a strong up-regulation of MIR171c in L. japonicus nodules compared with roots (Høgslund et al., 2009; Fig. 3B). We performed northern analysis and saw a similar tendency for the mature miR171c, along with modestly increased abundance levels of this miRNA in leaves from M. loti-inoculated plants versus mock-inoculated controls (Fig. 5C). On the other hand, NSP2 is known to be strongly down-regulated in nodule organs from L. japonicus (Murakami et al., 2006; Høgslund et al., 2009; Supplemental Fig. S9). To analyze the behavior of miR171c and its target gene in more detail, we performed quantitative PCR followed by reverse transcription (qRT)-PCR analysis from different plant tissues. When compared with roots, nodules showed a striking greater than 30-fold higher abundance of miR171c, which coincided with a reduced NSP2 signal (Fig. 5, D and E). A complementary expression pattern of miR171c and NSP2 was also present in leaves (Fig. 5, D and E). However, the relative NSP2 expression values in aerial tissue were substantially lower than in nodules or roots (Supplemental Fig. S10), a fact similarly observed using northern analysis (Fig. 5C). In light of these findings, we performed 35S promoter-mediated complementation of nsp2 mutants using an NSP2 version carrying several silent mutations in the miR171c complementary site. However, this construct fully restored the formation of infection threads and symbiotically active nodules, and nodules were indistinguishable from those of plants complemented with a 35S-driven wild-type NSP2 gene (Supplemental Fig. S11).
miR397 Correlates with Functional Symbiotic Infection in L. japonicus
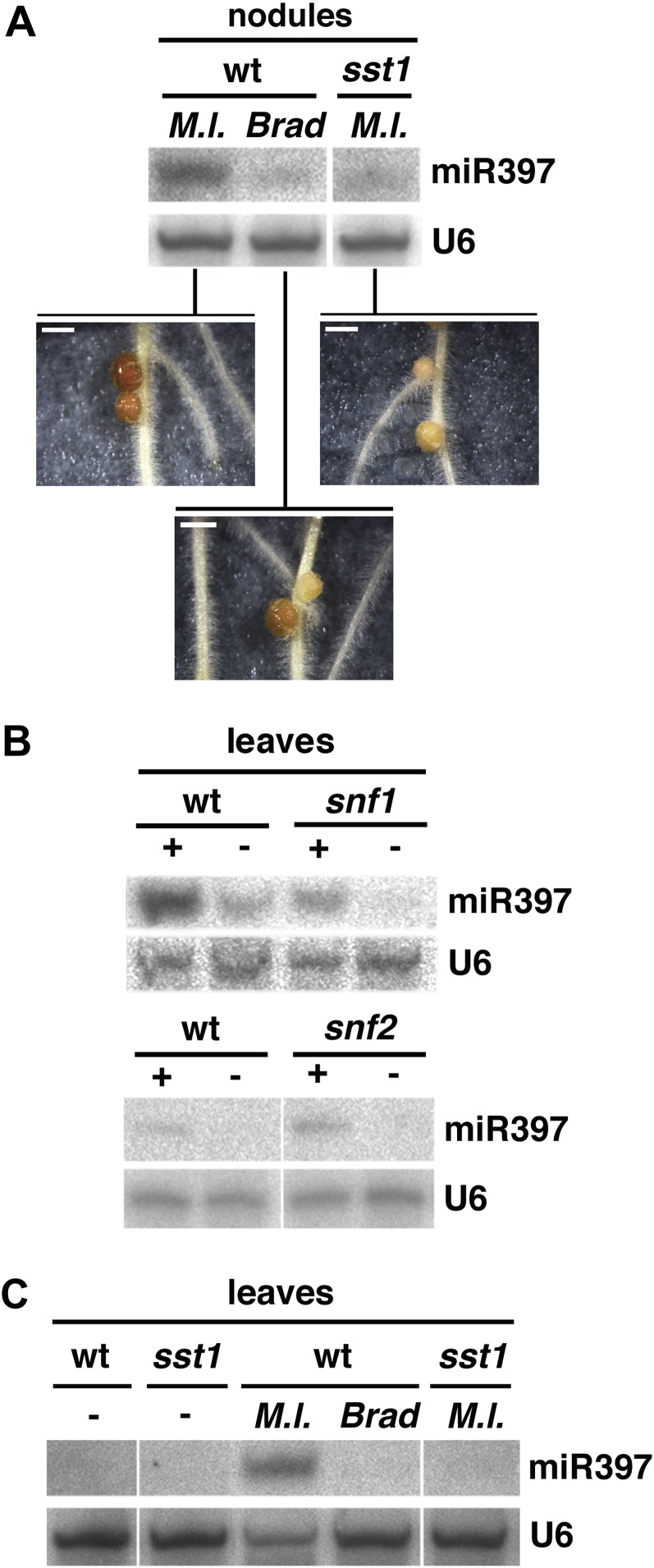
To investigate whether the observed differential abundance of miRNA levels in bacteria-containing nodules as opposed to empty spontaneous nodules (Fig. 4A) was related to nodule functionality, we analyzed miRNA expression during interactions where bacterial infection progresses normally but nitrogen fixation is compromised. To that aim, we first exploited Bradyrhizobium sp. (Lotus) strain NZP2309, an alternative symbiont that efficiently infects some members of the Lotus genus but induces nodules that are either inefficient or go through a very short period of efficient nitrogen fixation on L. japonicus, despite normal endosymbiotic infection (Bek et al., 2010; Fig. 6A, bottom panel). Nodules infected with Bradyrhizobium sp. showed basal expression levels of miR397, much lower than those observed with effective M. loti-infected nodules of the same age (Fig. 6A). We further analyzed nodules formed with the natural symbiont M. loti on L. japonicus sst1 mutant plants, which lack a functional SST1 sulfate transporter in the symbiosome membrane and consequently develop fully infected, but non-nitrogen-fixing, nodules (Krusell et al., 2005). Although to a lesser extent than Bradyrhizobium sp.-infected nodules on wild-type plants, sst1 nodules also showed reduced miR397 abundance compared with wild-type M. loti-infected nodules (Fig. 6A). By contrast, no difference in miR171c levels was found (data not shown). Collectively, these results indicate that the accumulation of miR397 to enhanced levels in mature nodules depends on their nitrogen-fixing ability and on the presence of a fully compatible microsymbiont. At the same time, young but fixing nodules or infection with compatible M. loti bacteria in the absence of nitrogen fixation, as in the case of sst1 nodules, do not lead to elevated miR397 levels, implying that either factor alone is insufficient to trigger elevated miR397 abundance levels.
Figure 6.
Systemic lja-miR397 accumulation correlates with the presence of functional nodules. A, miR397 expression in nodules (6 wpi) representing inefficient symbiotic interactions. Wild-type (wt) nodules inoculated with M. loti served as a fully functional reference. Inocula are as follows: Brad, Bradyrhizobium sp. (Lotus); M.l., M. loti strain MAFF303099. The top panels correspond to the same hybridization experiment, and the bottom panels present nodule phenotypes at the time of harvest. Bars = 1 mm. B and C, miR397 is up-regulated systemically in the presence of functional nodules. B, Northern blotting was done using RNA extracted from leaf tissues of the same plants used in Figure 4A. C, miR397 levels in leaves from the same nodulated plants shown in A. In all panels, U6 served as a loading control. +, Plants were inoculated with M. loti; −, plants were mock inoculated. Harvest was at 6 wpi. Panels in A, B, and C correspond to the same hybridization experiment.
Functional symbiosis requires nutrient exchange between root and shoot, and the de novo development of symbiotic nodules is regulated systemically in a nitrogen-dependent manner (Krusell et al., 2005; Oka-Kira and Kawaguchi, 2006). The fact that miR397 belongs to a series of miRNAs involved in systemic nutrient regulation in Arabidopsis and other species (Abdel-Ghany and Pilon, 2008; Jagadeeswaran et al., 2009; Yamasaki et al., 2009; Lu et al., 2011) thus prompted us to investigate miR397 expression at the systemic level. We found that miR397 abundance was strongly enhanced in leaves from wild-type, snf1, and snf2 plants bearing infected nodules as compared with mock-inoculated plants, although systemic miR397 levels in infected snf1 mutants were slightly lower than in infected wild-type plants (Fig. 6B). Remarkably, systemically elevated miR397 levels were neither observed in wild-type plants infected with Bradyrhizobium sp. nor in sst mutant plants infected with M. loti (Fig. 6C), none of which develop functional, nitrogen-fixing nodules. Preliminary analysis showed a similar absence of systemic induction and low miR397 expression in nonfixing nodules of another L. japonicus mutant, sen1 (Supplemental Fig. S12). Like sst1, sen1 lacks nitrogen fixation capacities despite a wild-type-like infection pattern (Suganuma et al., 2003). These combined data, therefore, demonstrate that miR397 specifically accumulates in efficient, nitrogen-fixing nodules and in leaves of plants bearing such nodules on their root systems. Therefore, miR397 represents a novel sRNA marker that signals a compatible bacterial infection and the presence of symbiotically functional nodules at both the local and systemic levels.
miR397 Is Related to Copper Homeostasis in L. japonicus and Targets a Copper-Containing Laccase Gene
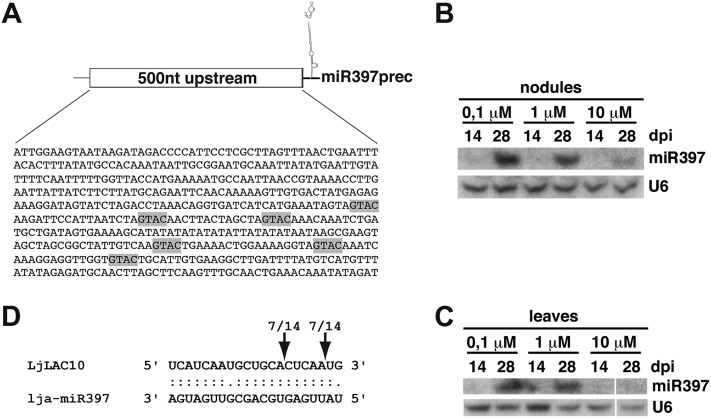
miR397 belongs to an endogenous miRNA network regulating copper homeostasis at the systemic level and is conserved across different plant species (Abdel-Ghany and Pilon, 2008; Jagadeeswaran et al., 2009; Yamasaki et al., 2009; Lu et al., 2011). While Arabidopsis, rice (Oryza sativa), and poplar (Populus trichocarpa) contain two, two, and three MIR397 loci, respectively (miRBase registry), we could only identify a single MIR397 locus in L. japonicus. The 600-nucleotide upstream promoter region of Arabidopsis MIR397a contains several copper-response (CuRE) GTAC motifs, which are essential for transcriptional activation upon Cu2+ shortage (Quinn et al., 2000; Yamasaki et al., 2009). Similar promoter features are observed in poplar MIR397a, MIR397b, and MIR397c, where three to five CuRE motifs are found (Lu et al., 2011). In silico analysis of the 500-nucleotide sequence fragment upstream of the predicted L. japonicus pre-MIR397 revealed the presence of six of these motifs (Fig. 7A). By contrast, CuRE sequences were absent (lja-MIR166 and lja-MIR171c) or present only once (lja-MIR167a and lja-390a) in corresponding upstream regions of other MIRNAs that we found to be abundant in nodules or enriched in nodules compared with roots (Supplemental Fig. S13).
Figure 7.
miR397 is related to copper homeostasis in L. japonicus and targets a copper-containing laccase. A, Predicted copper-responsive transcriptional activation motifs present in the 500-nucleotide (500nt) upstream genomic sequence of the lja-miR397 precursor. B and C, miR397 accumulation in nodules (B) and leaves (C) from plants grown at the indicated CuSO4 concentrations. U6 served as a loading control. Panels in B and C correspond to the same hybridization experiment. D, Validation of LjLACCASE10 as a miR397 target. Cleavage sites were determined by 5′ RACE analysis. The number of sequenced fragments ending at a respective site (out of total clones sequenced) is indicated. Predicted sites of endonucleolytic cleavage are indicated by arrows.
To test whether miR397 expression is dependent on copper availability in L. japonicus, we supplemented the growth medium with Cu2+ to yield final concentrations ranging from 0.1 to 10 μm (Fig. 7, B and C). The standard Cu2+ concentration of Fahraeus Plant (FP; Fahraeus, 1957) or B&D (Broughton and Dilworth, 1971) media routinely used for L. japonicus sterile culture is close to 0.1 μm, which is within the copper deprivation range in Arabidopsis (Yamasaki et al., 2007). We found that the abundance of miR397 in L. japonicus was dependent on Cu2+ concentration in both nodules and leaves (Fig. 7, B and C). At low Cu2+ levels (0.1 μm), miR397 was strongly up-regulated in nodules and leaves harvested at 28 dpi, corresponding to the highest miR397 levels previously observed in standard in vitro growth (Fig. 4B). This induction was significantly reduced at a concentration of 1 μm Cu2+ and almost absent under 10 μm Cu2+ growth conditions (Fig. 7, B and C). Strikingly, 14-dpi nodule and leaf samples did not show this induction when compared with corresponding 28-dpi tissues. These observations indicate that L. japonicus miR397 expression not only correlates with available Cu2+ levels but also with the symbiotic stage of the plant.
To further investigate the possible function of this miRNA in nodulation symbiosis, we predicted putative mRNA targets using the Target Finder algorithm (Allen et al., 2005; Fahlgren et al., 2007). Several Cu2+-containing LACCASE-like genes were retrieved as target candidates, and 5′ RACE analysis in nodulated roots confirmed that a L. japonicus homolog of Arabidopsis LACCASE10 indeed undergoes endonucleolytic cleavage within the miR397 complementary site (Fig. 7D). Collectively, these results strongly suggest that miR397 expression, which correlates with functional symbiosis, is linked to copper homeostasis at the nodule organ and systemic levels, at least in part via posttranscriptional regulation of the LACCASE10 homolog in L. japonicus.
DISCUSSION
The objective of this study was the identification and characterization of miRNAs potentially involved in nitrogen-fixing nodulation symbiosis in a determinate nodule-type legume species. We have identified a group of conserved and young miRNA families in the model legume L. japonicus and further investigated two candidates, miR171c and miR397, that are regulated specifically in the context of symbiotic infection.
While some of the miRNAs identified in this study have been bioinformatically predicted before (Sunkar and Jagadeeswaran, 2008; Hsieh et al., 2009; Wang et al., 2009), L. japonicus miRNAs have so far not been confirmed by sequencing. Our data expand on initiatives from recent years to decipher the small RNAome of other legume plants (Subramanian et al., 2008; Jagadeeswaran et al., 2009; Lelandais-Brière et al., 2009; Joshi et al., 2010; Chi et al., 2011; Devers et al., 2011; Li et al., 2011; Peláez et al., 2012; Turner et al., 2012).
In our approach, no sRNAs with significantly differential expression between infected and control libraries could be observed at early stages of infection, namely 3 hpi and 3 dpi. Using soybean roots, Szittya et al. (2008) similarly detected only small variation between miRNA abundances in infected versus mock-treated roots at 3 hpi. It is surprising that at 3 dpi, when endocytotic uptake of bacteria and plant developmental responses to the presence of compatible rhizobia have commenced, little variation in expression levels was observed in our libraries. We applied stringent cutoffs for the detection of differential abundances to avoid high numbers of false positives, which may contribute to this observation. Besides, in a whole-root cloning approach like ours, the expression of certain miRNAs in specific cell types or infected cells could be masked or diluted by miRNAs strongly accumulated in other root regions, including the highly active root tips.
A Specific Set of miRNAs Is Highly Expressed in Nodules, Two of Them Linked Specifically with Bacterial Infection
Using a combination of high-throughput sequencing, northern blotting, and primary microRNA transcript expression analysis, we have identified members of five MIRNA families in L. japonicus with strong expression in symbiotic nodules: miR167, miR172, miR390, miR397, and the miR171 isoform lja-miR171c. To dissect whether these miRNAs are potentially involved in processes related to either symbiotic bacterial infection or nodule organogenesis, we made use of the availability of L. japonicus snf mutants spontaneously producing uninfected nodule organs but generating wild-type-like symbiotic nodules in the presence of M. loti. A comparison of symbiotic and uninfected snf mutant nodules showed that, although strongly expressed in infected nodules, levels of miR171c and miR397 were low in nodule organs devoid of the symbiont M. loti. As spontaneous and infected nodules are indistinguishable in terms of developmental and morphological characteristics (Tirichine et al., 2006b), these results strongly suggest that enhanced expression of both miR171c and miR397 in nodules is specifically linked to the presence of the bacterial symbiont.
For the remaining miRNAs, no consistent differences in expression levels were detected between functional infected and nonfunctional or uninfected nodules. miR167 and miR172 may thus play roles in differentiation and maintenance of the nodule as an independent organ, rather than in infection or nodule functionality. Both miRNAs have been previously found to be expressed in soybean nodules (Wang et al., 2009), which, like L. japonicus nodules, show determinate development. In indeterminate M. truncatula nodules, northern hybridization analyses showed that members of the miR167 and miR172 families were more abundant in nodules than in root tips, while no nodule-specific up-regulation was detected for homologs of miR171c, miR390, or miR397 in this species (Lelandais-Brière et al., 2009). While differences in sampling and sequencing depth preclude direct comparisons between these data sets, these combined observations indicate a potential key role for miR167 and miR172 in the nodulation process in both determinate and indeterminate nodulators. In summary, the use of spontaneous nodulation mutants in the determinate nodulator L. japonicus has allowed the identification of miR167 and miR172 as miRNAs potentially involved in the context of nodule organogenesis or maintenance, on the one hand, and of lja-miR171c and lja-miR397 as two miRNAs specifically linked to bacterial infection during symbiotic root nodulation, on the other.
Infection-Related Abundance Levels of the NSP2-Targeting miR171c Isoform Are Enhanced in Determinate L. japonicus Nodules
Our work shows different lines of evidence supporting a role for miR171c in determinate-type nodulation in L. japonicus. First, lja-miR171c is highly accumulated in L. japonicus nodules, and gene expression analyses indicate that this is due to elevated tissue-specific expression at the precursor level. Next, although the high levels of this miRNA in nodules are constant across different nodule developmental stages, the analysis of spontaneous nodulation mutants shows that this accumulation is strictly dependent on the presence of the symbiotic partner M. loti. Furthermore, the establishment of a successful infection in nodules also leads to altered systemic lja-miR171c levels, as evident by enhanced expression levels in shoot tissues upon nodulation. We also show that lja-miR171c targets the NSP2 gene encoding a GRAS-type transcription factor essential for nodulation symbiosis (Kaló et al., 2005; Heckmann et al., 2006; Murakami et al., 2006), which coincides with inversely correlated expression patterns of miR171c and NSP2 in nodules and roots and, to a lesser extent, in shoot tissues. The observed cleavage of NSP2 is in line with reports on M. truncatula miR171h targeting a NSP2 homolog in this species (Branscheid et al., 2011; Devers et al., 2011; Lauressergues et al., 2012). M. truncatula NSP2 acts in concert with NSP1, another GRAS-type transcription factor, to regulate the expression of downstream symbiosis genes (Hirsch et al., 2009).
The combined evidence suggests a role for miR171c-mediated NSP2 regulation in the establishment or maintenance of a successful symbiosis. Strikingly, an NSP2 version blocking miR171c-mediated cleavage was fully competent to restore infection thread formation as well as nodule development and colonization in nsp2 mutant plants. One possible explanation might be that, although NSP2 is essential for bacterial symbiosis in L. japonicus, its miRNA171c-mediated regulation is not required for this process, at least under laboratory growth conditions. While it certainly deserves further investigation, this situation would not be unprecedented: several (plants) or indeed many (metazoans) miRNAs are known to act redundantly with other sRNA-independent regulatory mechanisms at the pretranscriptional or posttranscriptional level or the protein level. Only the prior inactivation or environmental perturbation of these additional regulatory layers (thereby creating “sensitized” backgrounds) would reveal the importance of miRNA function in conferring robustness to the system (for review, see Voinnet, 2009; Ambros, 2010). Alternatively, it is possible that the biological role of the observed infection-related miR171c expression in L. japonicus is not linked to NSP2 regulation in the nodulation symbiotic process but, instead, to the regulation of additional target genes.
Lja-miR171c and mtr-miR171h are members of a miR171 subfamily that seems to have coevolved with NSP2 genes, such that the miRNA target region in NSP2 has been conserved exclusively in plants undergoing endosymbiosis (Lauressergues et al., 2012). In that study, the authors showed that mtr-171h-mediated regulation of NSP2 has been established as a mediator of arbuscular mycorrhizal symbiosis in M. truncatula. The authors performed a parallel analysis of bacterial symbiosis and showed that, in contrast to fungal symbiosis, expression of a NSP2 version resistant to mtr-171h cleavage did not impair bacterial colonization or nodule development. However, intriguingly, mtr-171h did not present a nodule-specific expression pattern resembling the responses we reproducibly observed in L. japonicus for lja-miR171c. Discrepancies found in both model legumes are not restricted to lja-miR171c/mtr-171h expression patterns. Indeed, while in M. truncatula NSP2 shows a gradual increase in expression levels in roots inoculated with Sinorhizobium meliloti from 1 to 7 dpi (Kaló et al., 2005), M. loti-treated L. japonicus roots show a basal high NSP2 expression at 1 dpi, which is progressively reduced and peaks again at 8 dpi (Heckmann et al., 2006). Considering that both species undergo distinct types of nodulation, it thus seems possible that lja-miR171c and mtr-miR171h play distinct roles in gene regulation underlying symbiotic events. Further investigation will thus be needed to reveal whether there is a specific role for miR171c in determinate nodulation, considering that the nodule-specific regulation of this miRNA is tightly linked to the presence of bacteria in the symbiotic organs.
miR397 Correlates with Both Functional Symbiotic Nitrogen Fixation and Copper Deprivation
miR397, the second miRNA found in this work to be specifically enhanced in abundance in infected nodules, shows an elevated expression in late nodule developmental stages (28 dpi). This is in contrast to miR171c, which retains stable expression levels as nodules undergo maturation, indicating a possible link between nodule functionality, or even the onset of nodule senescence, and miR397 expression. The analysis of inefficient interactions of L. japonicus mutant sst1 (Krusell et al., 2005) and wild-type plants infected with the inefficient bacterial symbiont Bradyrhizobium sp. (Bek et al., 2010) confirmed that the accumulation of miR397 in both nodules and aerial plant parts correlates with the progression of a functional N2-fixing symbiosis. miR397 thus serves as a systemic marker for the presence of functional nodules in L. japonicus.
We further demonstrate that miR397 targets a LACCASE-like gene in L. japonicus. Laccases are Cu2+-containing polyphenol oxidoreductases playing diverse roles in different organisms such as bacteria, fungi, plants, and insects. LACCASE-like genes have also been confirmed as targets of miR397 in Arabidopsis (Abdel-Ghany and Pilon, 2008) and poplar (Lu et al., 2011), where these Cu2+-containing genes are down-regulated upon Cu2+ deficiency, thereby regulating systemic allocation of this important micronutrient.
In Arabidopsis, miR397 responds to Cu2+ availability through the action of SQUAMOSA Promoter-Binding Protein-Like7 (SPL7), a transcription factor that binds to specific GATC motifs present in its miRNA target promoters. Upon Cu2+ deficiency, a set of SPL7-responsive miRNAs (i.e. miR397, miR398, miR408, and miR857) are systemically activated. As a result, their Cu2+-containing targets, including laccases, plantacyanin, or superoxide dismutases, are down-regulated, so that copper is made more available to essential, copper-demanding cellular processes (Abdel-Ghany and Pilon, 2008; Pilon et al., 2009; Yamasaki et al., 2009). In fact, these miRNAs are popularly referred to as a “Cu-miRNA” (Burkhead et al., 2009). Enhanced miR397 levels in response to copper deprivation have also been observed in poplar (Lu et al., 2011) and, under nonsymbiotic conditions, in M. truncatula, where targeting of two LACCASE-like genes by this miRNA has been demonstrated (Jagadeeswaran et al., 2009). Therefore, a laccase-mediated Cu2+-related regulatory activity of miR397 is conserved in many eurosid angiosperm lineages.
Our results suggest that similar Cu2+-homeostatic mechanisms involving miR397 exist in the context of symbiotic N2 fixation. Root nodule symbiosis requires an intensive exchange of macronutrients and micronutrients between the plant and bacterial partners (Krusell et al., 2005; Delmotte et al., 2010), and evidence for a need for Cu2+ transport into bacteroids exists (Preisig et al., 1996b). At the biochemical level, nitrogen fixation requires low concentrations of free oxygen, which could otherwise inactivate the oxygen-sensitive nitrogenase enzymatic complex. On the other hand, a high respiratory rate is needed in nodules to meet the high demands in ATP, which is satisfied through strategies that include the action of the oxygen carrier leghemoglobin and the use of a specialized respiratory electron transport chain terminated by a Cu2+-containing bb3-type cytochrome oxidase (cbb3) with high affinity for oxygen (Preisig et al., 1996b; Delgado et al., 1998; Ott et al., 2005; Arunothayanan et al., 2010). Cu2+ availability seems crucial to the functionality of this complex, as deletion of the fixGHIS genes in the soybean symbiont B. japonicum, encoding for the copper-transporting ATPase FixI, results in defective nitrogen fixation (Preisig et al., 1996a). The same effect is found with B. japonicum carrying mutations in the bll4880 gene, encoding a metallochaperone proposed to transfer Cu2+ to cbb3, further emphasizing the need for Cu2+ trafficking from the plant to the nodule and bacteroids (Arunothayanan et al., 2010). Moreover, a nodule-specific expression pattern has been reported for a putative plant copper transporter in M. truncatula (Fedorova et al., 2002). In line with these findings, we show here that L. japonicus plants harboring mature functional N2-fixing nodules accumulate miR397 in both nodule and leaf tissues, a process reverted when higher Cu2+ levels are supplied in the growth medium. Based on these collective data, it seems likely that nodules become major Cu2+ sinks in the plant at the onset of bacterial N2 fixation. Subsequent regulation of nutrient allocation and prioritization would then rely on the host molecular machinery, including miR397, which could thus play an important role in ensuring N2 fixation and nodule functionality. This would explain the codependence of L. japonicus miR397 induction on functional symbiosis and Cu2+ availability. Indeed, plants harboring 28-dpi fully N2-fixing nodules show strongly enhanced miR397 levels when compared with the earlier symbiotic stages, where N2 fixation-associated Cu2+ demands are predicted to be less pronounced. Accordingly, elevated Cu2+ levels in the growth medium are sufficient for a marked reduction in the local and systemic accumulation of miR397 at the later nodule stages. Nevertheless, we cannot exclude the possibility that it is the onset of nodule senescence, setting in at 4 to 6 wpi under our conditions, that activates the Cu2+-deprivation miR397 systemic network through an unknown molecular mechanism.
CONCLUSION
In this work, we have taken advantage of a unique set of L. japonicus mutants that uncouple nodule organ formation, infection, and function, leading to the identification of two miRNAs that accumulate specifically when infected with rhizobial bacteria. miR171c targets NSP2, an essential gene for nodulation, and shows a symbiosis-dependent expression in determinate L. japonicus nodules and shoot tissues, which, to our knowledge, has not been observed in other legume species. Systemic up-regulation of miR397 is strictly linked to the presence of efficient, N2-fixing nodules but could be suppressed by providing excess Cu2+ levels in the growth medium. As a mediator of Cu2+ homeostasis, miR397 up-regulation would then signal nutrient reallocation as a consequence of nodulation, thereby reflecting the importance of balanced micronutrient levels for efficient agricultural legume production. Besides, given moderate copper availability levels, miR397 qualifies as a reliable marker reflecting the presence of an efficient symbiotic interaction. Nodulation with inefficient rhizobial strains that evade plant defense systems or induce nodules failing to deliver nitrogen derivatives continues to cause significant yield losses. A simple systemic marker for symbiotic efficiency, such as miR397, could thus represent a valuable diagnostic tool.
MATERIALS AND METHODS
Biological Material
Lotus japonicus ecotypes ‘Gifu’ B-129 (Handberg and Stougaard, 1992) and ‘Miyakojima’ MG-20 (Kawaguchi et al., 2001) were used in this study. In addition to wild-type plants, the following mutants were used (ecotype Gifu): snf1-1 (Tirichine et al., 2006a), snf2 (Tirichine et al., 2007), sst1-1 (Krusell et al., 2005), sen1 (sym11; Schauser et al., 1998; Suganuma et al., 2003; Sandal et al., 2006; Hakoyama et al., 2012). Bacterial strains Mesorhizobium loti MAFF303099 and Bradyrhizobium sp. (Lotus) strain NZP2309 (Scott et al., 1985; Bek et al., 2010) were used for plant inoculation. Transgenic (hairy) roots were induced using Agrobacterium rhizogenes AR1193 (Stougaard et al., 1987).
Plant Growth, Inoculation, and Transformation
L. japonicus seeds were scarified using concentrated H2SO4 and sterilized with a bleach-SDS solution. Upon imbibition, seeds were kept overnight at 4°C to synchronize germination, germinated in the dark at 23°C for 3 d, and then transferred to a 16-/8-h day/night cycle for 3 d. Seedlings were then transferred to plates containing FP (Fahraeus, 1957) or one-quarter-strength B&D (Broughton and Dilworth, 1971) medium and grown until inoculation. For the Cu2+-response assay, FP medium was supplemented with CuSO4 to obtain final concentrations of 0.1, 1, or 10 μm Cu2+. Arabidopsis (Arabidopsis thaliana) plants used as controls in this study were grown under greenhouse conditions as indicated above.
For in vitro inoculation, M. loti was grown on tryptone yeast medium for 48 to 72 h at 28°C. The resulting culture was pelleted twice at 3,000g, resuspended to optical density at 600 nm (OD600) = 0.1, and applied directly to roots. For the assay of N2 fixation-deficient nodules, Bradyrhizobium sp. (Lotus) were grown on yeast mannitol broth medium supplemented with tetracycline (2.5 mg mL−1) for 4 d. Bacterial cultures were pelleted twice, resuspended (OD600 = 0.01), and applied to roots. Plant growth plates were covered and grown at a 21°C constant, 16-h-light/8-h-dark regime.
For sRNA library generation, a large-scale flood inoculation procedure was followed. Open culture trays were filled with autoclaved clay granule substrate and soaked with sterile FP medium. One-week-old L. japonicus seedlings pregrown on agar petri dishes were transferred to the culture trays and grown for 2 more weeks. M. loti bacteria were grown at 28°C for 48 h in liquid tryptone yeast medium, and the resulting culture was pelleted and resuspended in FP medium to an OD600 of 0.01. Culture trays were soaked with the resulting solution, and excess medium was removed. Control plant cultures were treated similarly using sterile FP medium alone. Hairy root induction using A. rhizogenes was performed as described (Stougaard, 1995).
For quantitative PCR (qPCR) assays, seeds of L. japonicus wild type were surface scarified and imbibed overnight at 4°C, then germinated for 3 d at 21°C in darkness. Seedlings were grown on square plastic dishes with wedged one-half-strength B&D medium (Broughton and Dilworth, 1971) supplemented with 1 mm KNO3 at 21°C (16 h of light, 8 h of dark). Roots were shielded from light access. Plants were inoculated at day 7 post germination. For inoculation, liquid cultures of M. loti MAFF303099 expressing DsRED (Maekawa et al., 2009) were grown for 2 d and harvested by centrifuging for 10 min at 3,000g. The bacterial pellet was washed twice in one-half-strength B&D medium (1 mm KNO3) and diluted to OD600 of 0.01. A total of 100 µL of bacterial suspension was applied directly to each root. Mock roots were treated with an equal amount of sterile medium. Roots were harvested and shock frozen in nitrogen 10 d post germination.
Microscopy
Plant phenotypes were monitored and photographs were taken using a Leica M165FC stereomicroscope. Fluorescence was visualized using a DsRED filter (Leica 10447412).
Vectors and Cloning Procedures
An NSP2 miR171c-resistant version was generated using primers 5′-tTTaAGgCGcACgAggATaACaCGAGCCAGTTCaCGGT-3′ (reverse) and 5′-TtATccTcGTgCGcCTtAAaGAGTTGGTGTCCCACACCGAC-3′ (forward) carrying silent mutations (lowercase letters) in and near the miR171c complementary region of the coding sequence with primers 5′-caccCTCAGGCATGGAAATGGA-3′ (forward) and 5′-TTCTGTTTTCGGAAGGTCAA-3′ (reverse), respectively, to PCR amplify mutated 5′ and 3′ fragments of the gene from L. japonicus leaf DNA (ecotype Gifu). A full-length version was obtained from these using an overlapping PCR strategy with primers 5′-caccCTCAGGCATGGAAATGGA-3′ (forward) and 5′-TTCTGTTTTCGGAAGGTCAA-3′ (reverse). The same primers and template were used to isolate full-length wild-type NSP2. Both fragments were subsequently cloned into pENTR/D/TOPO (Invitrogen) and transferred to modified pIV10 vector (Stougaard et al.,1987) equipped with a Gateway destination cassette (Invitrogen) preceded by a 405-nucleotide cauliflower mosaic virus 35S promoter fragment (5′-CGTACCCCTACTCCAAAAATG…TTCATTTGGAGAGGACAGCCC-3′). As a control, the Gateway destination cassette was eliminated from the pIV10 plasmid carrying the 35S promoter fragment. The resulting pIV10-based constructs were introduced into A. rhizogenes AR1193 via triparental mating (Stougaard et al., 1987) and used for hairy root induction.
Cloning of sRNAs
Isolation and cloning of sRNAs from L. japonicus roots and nodules were carried out as described previously (Pfeffer, 2007). Each of the five libraries was PCR labeled using distinct four-nucleotide tags, pooled, and subjected to two rounds of 454 pyrosequencing.
RNA Analyses
Total RNA from L. japonicus tissue was extracted by adapting the hot borate buffer protocol (Wan and Wilkins, 1994). Briefly, plant tissue was ground under liquid nitrogen and homogenized with an extraction buffer (2,000 mm sodium tetraborate decahydrate, 30 mm EGTA, 5 mm EDTA, 1% SDS, 1% sodium deoxycholate; adding before use 100 µL of dithiothreitol, 35 µL of β-mercaptoethanol, and 0.1 g of polyvinylpyrrolidone-40 per 5 mL of buffer). Samples were Proteinase K digested and treated with TRIzol-LS reagent (Invitrogen) following the manufacturer’s instructions. RNA from Arabidopsis was extracted with Tri-Reagent (Sigma) according to the manufacturer’s instructions.
RNA gel-blot analysis of low-molecular-weight RNAs was done as described previously (Dunoyer et al., 2004). To detect highly expressed miRNAs, the sRNA-containing membrane was UV cross linked in Stratalinker. To detect low expressed miRNAs, RNAs were chemically cross linked to the membrane with a fixing solution including N-ethyl-N′-3-dimethylaminopropyl carbodiimide hydrochloride and 1-methylimidazole as described previously (Pall and Hamilton, 2008). For miRNA detection, complementary DNA (cDNA) oligonucleotides were radiolabeled with [γ-32P]ATP using T4 PNK enzyme (New England Biolabs). The U6 snRNA signal was used as an internal loading control. The following DNA oligonucleotides were used for miRNA detection: TCAGAGGCGAAGACACCCGCTA (lja-A1), AGATGATGTATGGATGGAAGA (lja-A2), AGATCATGCTGGCAGCTTCA (lja-miR167), GAGTGATATTGATTCGGCTCA, (lja-miR171c), CTGCAGCATCATCAAGATTCT (lja-miR172a), and TCATCAACGCTGCACTCAATA (lja-miR397).
Bioinformatic Analyses
For sRNA library annotation and analysis, following 454 sequencing, adaptor sequences were removed and high-quality sequences were annotated with BLAST (word size = 7/no filter) using an in-house analysis platform (bioimage.u-strasbg.fr/bioinfo/) and the following databases as references: plant miRNA sequences were analyzed using miRBase (version 17; Viridiplantae); tRNA, rRNA, and other noncoding RNA sequences were extracted from GenBank (release 180.0; plants only); M. loti genomic sequence was retrieved at Rhizobase (version 1.0); DNA repeat sequences were obtained from Repbase (version 16.01; L. japonicus and ancestral loci); L. japonicus genomic sequences were retrieved at the Kazusa Institute (version 2.5). The results were filtered to authorize two mismatches for all databases.
Novel miRNA identification was carried out using miRCat, selecting default parameters except for read abundance (2 or greater; Moxon et al., 2008), and predicted miRNA precursors were folded using RNAfold from the Vienna RNA package (Hofacker, 2003). The novelty of miRNAs was confirmed by sequence alignment of individual species to the miRBase registry (version 18) using BLASTN and authorizing a maximum of two mismatches. miRNA abundance profiling was done using the miRProf algorithm, allowing two mismatches and/or overhangs for the sequence search and selecting default values for output grouping (Moxon et al., 2008). miRNA target predictions were carried out with the Target Finder algorithm (Allen et al., 2005; Fahlgren et al., 2007). miR171 family sequence alignments were performed using Clustal 2.1 and sequences from the miRBase registry (version 18). Promoter analyses were carried out through the Plant Cis-Acting Regulatory DNA Elements database (www.dna.affrc.go.jp/PLACE/; Higo et al., 1998).
Analysis of Affymetrix Transcriptome Data
Normalized and pairwise-compared expression data from the L. japonicus Affymetrix GeneChip were kindly provided by S. Radutoiu (Department of Molecular Biology, Aarhus University; M, FDR, where M is the log2 ratio of average signal intensity values from any two conditions [three biological replicates each] and FDR is the false discovery rate-corrected P value; Høgslund et al., 2009). Nodule expression values as compared with those from uninoculated root samples of the same genotype were analyzed. A significance criterion of FDR-corrected P ≤ 0.05 was applied (Høgslund et al., 2009). Fold change values were calculated as |M|2.
qRT-PCR Analysis
Total RNA was extracted from 10 to 15 independent root systems per sample using miRVana (Ambion) kit materials and following the supplier’s recommendations. RNA was eluted in diethyl pyrocarbonate-treated water. cDNA for sRNA qPCR assays was prepared using RevertAid reverse transcriptase (Fermentas) following a protocol by Varkonyi-Gasic et al. (2007; modified). Stem-loop primers for reverse transcription of sRNAs (Supplemental Table S1) were designed such that the 6 bp at the 5′ end of the stem-loop primer were complementary to six nucleotides at the 3′ end of the sRNA. In addition to one or more stem-loop primers, each cDNA reaction contained oligo(dT) primers (Supplemental Table S1). Pulsed reverse transcription conditions were 60× 16°C for 30 min, 30°C for 30 s, and 42°C for 30 s, then 50°C for 1 s, and 85°C for 5 min. Primers for qPCR amplification are listed in Supplemental Table S1. Reference genes were L. japonicus homologs of ATP synthase, protein phosphatase2, and the nuclear RNA U6. Due to disproportionally high expression levels of U6, which is used as a standard reference in low-Mr northern blots, the presented graphs were calculated using only ATP synthase and protein phosphatase2A for normalization. While introducing slightly more variation, standardization with U6 supported all conclusions drawn.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Bioinformatic pipeline for sRNA bioinformatic mapping and annotation of L. japonicus cloned sRNAs.
Supplemental Figure S2. Prediction of new miRNAs in L. japonicus.
Supplemental Figure S3. Secondary structures of precursor sequences from novel miRNAs identified in L. japonicus.
Supplemental Figure S4. Sequence alignment of lja-A1, gma-miR2109, and gso-miR2109 precursor sequences.
Supplemental Figure S5. Pipeline for nodule/root miRNA profiling used by the miRProf algorithm.
Supplemental Figure S6. Relative expression signal intensities from all five PRI-MIR genes from the miR171 family represented on the L. japonicus Affymetrix GeneChip.
Supplemental Figure S7. Accumulation of other miRNAs in rhizobia-free spontaneous nodules from snf1 and snf2 mutants.
Supplemental Figure S8. Targeting of SCL6 genes by canonical miR171s is conserved in L. japonicus.
Supplemental Figure S9. Relative expression signal intensities of PRI-MIR171c and NSP2 on the L. japonicus Affymetrix GeneChip.
Supplemental Figure S10. Relative expression levels determined by qRT-PCR of mature miR171c and NSP2 in nodules or leaves normalized to mock-inoculated roots.
Supplemental Figure S11. Complementation of nsp2 mutant plants with NSP2 wild type and miR171c-resistant versions.
Supplemental Figure S12. Preliminary analysis of miR397 accumulation in sen1 mutants.
Supplemental Figure S13. Upstream sequences of MIRNA genes searched for GTAC transcriptional activation motifs.
Supplemental Table S1. Oligonucleotide sequences used in reverse transcription and in qPCR.
Acknowledgments
We are grateful to Simon Moxon and Frank Schwach (University of East Anglia) for adapting miRNA prediction softwares to the L. japonicus genome. We also thank Shusei Sato and Satoshi Tabata (Kazusa Institute) for access to unpublished L. japonicus sequence data.
Glossary
- miRNA
microRNA
- hpi
hours post inoculation
- dpi
days post inoculation
- wpi
weeks post inoculation
- snRNA
small nuclear RNA
- miscRNA
miscellaneous RNA
- miRNA*
passenger strands of miRNA duplexes upon release from their stem-loop precursors
- qRT
quantitative reverse transcription
- CuRE
copper-response
- FP
Fahraeus Plant
- OD600
optical density at 600 nm
- qPCR
quantitative PCR
- rRNA
ribosomal RNA
- sRNA
to be defined
- sRNA
small RNA
- cDNA
complementary DNA
References
- Abdel-Ghany SE, Pilon M. (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem 283: 15932–15945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Ambros V. (2010). MicroRNAs: genetically sensitized worms reveal new secrets. Curr Biol 20: R598–R600 [DOI] [PubMed] [Google Scholar]
- Arunothayanan H, Nomura M, Hamaguchi R, Itakura M, Minamisawa K, Tajima S. (2010) Copper metallochaperones are required for the assembly of bacteroid cytochrome c oxidase which is functioning for nitrogen fixation in soybean nodules. Plant Cell Physiol 51: 1242–1246 [DOI] [PubMed] [Google Scholar]
- Bek AS, Sauer J, Thygesen MB, Duus JO, Petersen BO, Thirup S, James E, Jensen KJ, Stougaard J, Radutoiu S. (2010) Improved characterization of nod factors and genetically based variation in LysM receptor domains identify amino acids expendable for nod factor recognition in Lotus spp. Mol Plant Microbe Interact 23: 58–66 [DOI] [PubMed] [Google Scholar]
- Boualem A, Laporte P, Jovanovic M, Laffont C, Plet J, Combier JP, Niebel A, Crespi M, Frugier F. (2008) MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J 54: 876–887 [DOI] [PubMed] [Google Scholar]
- Branscheid A, Devers EA, May P, Krajinski F. (2011) Distribution pattern of small RNA and degradome reads provides information on miRNA gene structure and regulation. Plant Signal Behav 6: 1609–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O. (2009) Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol 10: 141–148 [DOI] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ. (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhead JL, Reynolds KA, Abdel-Ghany SE, Cohu CM, Pilon M. (2009) Copper homeostasis. New Phytol 182: 799–816 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. (2010) Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X, Yang Q, Chen X, Wang J, Pan L, Chen M, Yang Z, He Y, Liang X, Yu S. (2011) Identification and characterization of microRNAs from peanut (Arachis hypogaea L.) by high-throughput sequencing. PLoS ONE 6: e27530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebatch G, Desbrosses G, Ott T, Krusell L, Montanari O, Kloska S, Kopka J, Udvardi MK. (2004) Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J 39: 487–512 [DOI] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernié T, Ott T, Gamas P, Crespi M, et al. (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20: 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC. (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. (2011) Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 12: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado MJ, Bedmar EJ, Downie JA. (1998) Genes involved in the formation and assembly of rhizobial cytochromes and their role in symbiotic nitrogen fixation. Adv Microb Physiol 40: 191–231 [DOI] [PubMed] [Google Scholar]
- Delmotte N, Ahrens CH, Knief C, Qeli E, Koch M, Fischer HM, Vorholt JA, Hennecke H, Pessi G. (2010) An integrated proteomics and transcriptomics reference data set provides new insights into the Bradyrhizobium japonicum bacteroid metabolism in soybean root nodules. Proteomics 10: 1391–1400 [DOI] [PubMed] [Google Scholar]
- Devers EA, Branscheid A, May P, Krajinski F. (2011) Stars and symbiosis: microRNA- and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol 156: 1990–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O. (2004) Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16: 1235–1250 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- El Yahyaoui F, Küster H, Ben Amor B, Hohnjec N, Pühler A, Becker A, Gouzy J, Vernié T, Gough C, Niebel A, et al. (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136: 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Jogdeo S, Kasschau KD, Sullivan CM, Chapman EJ, Laubinger S, Smith LM, Dasenko M, Givan SA, Weigel D, et al. (2010) MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22: 1074–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahraeus G. (1957) The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol 16: 374–381 [DOI] [PubMed] [Google Scholar]
- Fedorova M, van de Mortel J, Matsumoto PA, Cho J, Town CD, VandenBosch KA, Gantt JS, Vance CP. (2002) Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol 130: 519–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakoyama T, Niimi K, Yamamoto T, Isobe S, Sato S, Nakamura Y, Tabata S, Kumagai H, Umehara Y, Brossuleit K, et al. (2012) The integral membrane protein SEN1 is required for symbiotic nitrogen fixation in Lotus japonicus nodules. Plant Cell Physiol 53: 225–236 [DOI] [PubMed] [Google Scholar]
- Handberg K, Stougaard J. (1992) Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J 2: 487–496 [Google Scholar]
- He XF, Fang YY, Feng L, Guo HS. (2008) Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett 582: 2445–2452 [DOI] [PubMed] [Google Scholar]
- Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA. (2006) Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol 142: 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Higo H. (1998) PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res 26: 358–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Kim J, Muñoz A, Heckmann AB, Downie JA, Oldroyd GE. (2009) GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. (2003) Vienna RNA secondary structure server. Nucleic Acids Res 31: 3429–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høgslund N, Radutoiu S, Krusell L, Voroshilova V, Hannah MA, Goffard N, Sanchez DH, Lippold F, Ott T, Sato S, et al. (2009) Dissection of symbiosis and organ development by integrated transcriptome analysis of Lotus japonicus mutant and wild-type plants. PLoS ONE 4: e6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ. (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151: 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeswaran G, Zheng Y, Li YF, Shukla LI, Matts J, Hoyt P, Macmil SL, Wiley GB, Roe BA, Zhang W, et al. (2009) Cloning and characterization of small RNAs from Medicago truncatula reveals four novel legume-specific microRNA families. New Phytol 184: 85–98 [DOI] [PubMed] [Google Scholar]
- Joshi T, Yan Z, Libault M, Jeong DH, Park S, Green PJ, Sherrier DJ, Farmer A, May G, Meyers BC, et al. (2010) Prediction of novel miRNAs and associated target genes in Glycine max. BMC Bioinformatics (Suppl 1) 11: S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Motomura T, Imaizumi-Anraku H, Akao S, Kawasaki S. (2001) Providing the basis for genomics in Lotus japonicus: the accessions Miyakojima and Gifu are appropriate crossing partners for genetic analyses. Mol Genet Genomics 266: 157–166 [DOI] [PubMed] [Google Scholar]
- Krusell L, Krause K, Ott T, Desbrosses G, Krämer U, Sato S, Nakamura Y, Tabata S, James EK, Sandal N, et al. (2005) The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17: 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulcheski FR, de Oliveira LF, Molina LG, Almerão MP, Rodrigues FA, Marcolino J, Barbosa JF, Stolf-Moreira R, Nepomuceno AL, Marcelino-Guimarães FC, et al. (2011) Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics 12: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D, Delaux PM, Formey D, Lelandais-Briere C, Fort S, Cottaz S, Becard G, Niebel A, Roux C, Combier JP. (2012). The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J 72: 512–522 [DOI] [PubMed] [Google Scholar]
- Lelandais-Brière C, Naya L, Sallet E, Calenge F, Frugier F, Hartmann C, Gouzy J, Crespi M. (2009) Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell 21: 2780–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Deng Y, Wu T, Subramanian S, Yu O. (2010) Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol 153: 1759–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Dong Y, Yin H, Wang N, Yang J, Liu X, Wang Y, Wu J, Li X. (2011) Characterization of the stress associated microRNAs in Glycine max by deep sequencing. BMC Plant Biol 11: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ. (2008) Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056 [DOI] [PubMed] [Google Scholar]
- Lu S, Yang C, Chiang VL. (2011) Conservation and diversity of microRNA-associated copper-regulatory networks in Populus trichocarpa. J Integr Plant Biol 53: 879–891 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. (2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Maekawa-Yoshikawa M, Takeda N, Imaizumi-Anraku H, Murooka Y, Hayashi M. (2009) Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J 58: 183–194 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bouché N. (2008) MicroRNA-directed regulation: to cleave or not to cleave. Trends Plant Sci 13: 359–367 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, et al. (2008) Criteria for annotation of plant microRNAs. Plant Cell 20: 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H, Sun J, Oldroyd GE, Downie JA. (2006) Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol Plant Microbe Interact 19: 914–923 [DOI] [PubMed] [Google Scholar]
- Moxon S, Schwach F, Dalmay T, Maclean D, Studholme DJ, Moulton V. (2008) A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics 24: 2252–2253 [DOI] [PubMed] [Google Scholar]
- Murakami Y, Miwa H, Imaizumi-Anraku H, Kouchi H, Downie JA, Kawaguchi M, Kawasaki S. (2006). Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res 13: 255–265 [DOI] [PubMed] [Google Scholar]
- Oka-Kira E, Kawaguchi M. (2006) Long-distance signaling to control root nodule number. Curr Opin Plant Biol 9: 496–502 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Ott T, van Dongen JT, Günther C, Krusell L, Desbrosses G, Vigeolas H, Bock V, Czechowski T, Geigenberger P, Udvardi MK. (2005) Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr Biol 15: 531–535 [DOI] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ. (2008) Improved northern blot method for enhanced detection of small RNA. Nat Protoc 3: 1077–1084 [DOI] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR. (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peláez P, Trejo MS, Iñiguez LP, Estrada-Navarrete G, Covarrubias AA, Reyes JL, Sanchez F. (2012) Identification and characterization of microRNAs in Phaseolus vulgaris by high-throughput sequencing. BMC Genomics 13: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S. (2007) Identification of virally encoded microRNAs. Methods Enzymol 427: 51–63 [DOI] [PubMed] [Google Scholar]
- Pilon M, Cohu CM, Ravet K, Abdel-Ghany SE, Gaymard F. (2009) Essential transition metal homeostasis in plants. Curr Opin Plant Biol 12: 347–357 [DOI] [PubMed] [Google Scholar]
- Preisig O, Zufferey R, Hennecke H. (1996a) The Bradyrhizobium japonicum fixGHIS genes are required for the formation of the high-affinity cbb3-type cytochrome oxidase. Arch Microbiol 165: 297–305 [DOI] [PubMed] [Google Scholar]
- Preisig O, Zufferey R, Thöny-Meyer L, Appleby CA, Hennecke H. (1996b) A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J Bacteriol 178: 1532–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JM, Barraco P, Eriksson M, Merchant S. (2000) Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J Biol Chem 275: 6080–6089 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Sandal N, Petersen TR, Murray J, Umehara Y, Karas B, Yano K, Kumagai H, Yoshikawa M, Saito K, Hayashi M, et al. (2006) Genetics of symbiosis in Lotus japonicus: recombinant inbred lines, comparative genetic maps, and map position of 35 symbiotic loci. Mol Plant Microbe Interact 19: 80–91 [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. (2008) Genome structure of the legume, Lotus japonicus. DNA Res 15: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Handberg K, Sandal N, Stiller J, Thykjaer T, Pajuelo E, Nielsen A, Stougaard J. (1998) Symbiotic mutants deficient in nodule establishment identified after T-DNA transformation of Lotus japonicus. Mol Gen Genet 259: 414–423 [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Scott DB, Chua KY, Jarvis BDW, Pankhurst CE. (1985) Molecular cloning of a nodulation gene from fast- and slow-growing strains of Lotus rhizobia. Mol Gen Genet 201: 43–50 [Google Scholar]
- Stougaard J. (1995) Agrobacterium rhizogenes as a vector for transforming higher plants: application in Lotus corniculatus transformation. Methods Mol Biol 49: 49–61 [DOI] [PubMed] [Google Scholar]
- Stougaard J, Abildsten D, Marcker KA. (1987) The Agrobacterium rhizogenes pRi TL-DNA segment as a gene vector system for transformation of plants. Mol Gen Genet 207: 251–255 [Google Scholar]
- Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Yu O. (2008) Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics 9: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma N, Nakamura Y, Yamamoto M, Ohta T, Koiwa H, Akao S, Kawaguchi M. (2003) The Lotus japonicus Sen1 gene controls rhizobial differentiation into nitrogen-fixing bacteroids in nodules. Mol Genet Genomics 269: 312–320 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Jagadeeswaran G. (2008) In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol 8: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, Moulton V, Dalmay T. (2008) High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics 9: 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L, Imaizumi-Anraku H, Yoshida S, Murakami Y, Madsen LH, Miwa H, Nakagawa T, Sandal N, Albrektsen AS, Kawaguchi M, et al. (2006a) Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156 [DOI] [PubMed] [Google Scholar]
- Tirichine L, James EK, Sandal N, Stougaard J. (2006b) Spontaneous root-nodule formation in the model legume Lotus japonicus: a novel class of mutants nodulates in the absence of rhizobia. Mol Plant Microbe Interact 19: 373–382 [DOI] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J. (2007) A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107 [DOI] [PubMed] [Google Scholar]
- Turner M, Yu O, Subramanian S. (2012) Genome organization and characteristics of soybean microRNAs. BMC Genomics 13: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. (2008) Plant ARGONAUTES. Trends Plant Sci 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA. (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223: 7–12 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li P, Cao X, Wang X, Zhang A, Li X. (2009) Identification and expression analysis of miRNAs from nitrogen-fixing soybean nodules. Biochem Biophys Res Commun 378: 799–803 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M. (2007) Regulation of copper homeostasis by micro-RNA in Arabidopsis. J Biol Chem 282: 16369–16378 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T. (2009) SQUAMOSA Promoter Binding Protein-Like7 Is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]