Abstract
The related proteins D1 and D2 together build up the photosystem II reaction center. Synthesis of D1 (PsbA) is highly regulated in all photosynthetic organisms. The mechanisms and specific protein factors involved in controlled expression of the psbA gene in higher plants are highly elusive. Here, we report on the identification of a chloroplast-located protein, HCF244 (for high chlorophyll fluorescence244), which is essentially required for translational initiation of the psbA messenger RNA in Arabidopsis (Arabidopsis thaliana). The factor is highly conserved between land plants, algae, and cyanobacteria. HCF244 was identified by coexpression analysis of HCF173, which encodes a protein that is also necessary for psbA translational initiation and in addition for stabilization of this messenger RNA. Phenotypic characterization of the mutants hcf244 and hcf173 suggests that the corresponding proteins operate cooperatively during psbA translation. Immunolocalization studies detected the majority of the two proteins at the thylakoid membrane. Both HCF244 and HCF173 are members of the atypical short-chain dehydrogenase/reductase superfamily, a modified group, which has lost enzyme activity but acquires new functions in the metabolism of the cell.
PSII is a multiprotein complex catalyzing the light-driven electron transfer from water to plastoquinone in oxygen-evolving photosynthetic organisms (Baena-González and Aro, 2002). The heart of this thylakoid membrane complex consists of the D1/D2 heterodimer, which binds most of the redox-active cofactors involved in PSII electron transfer. On either side, the chlorophyll a-binding proteins CP47 and CP43 flank the internal heterodimer. In addition, many low-molecular-mass proteins surround this PSII core complex. At the lumenal side of the membrane, the subunits PsbO, PsbP, and PsbQ are attached and build up the water-splitting complex.
In consequence of the evolutionary gene transfer from the cyanobacterial ancestor to the host genome, PSII forms a genetic mosaic, consisting of nucleus- and plastid-encoded subunits. These circumstances require a well-orchestrated regulation of gene expression in both genetic compartments during biogenesis of the membrane complex. Nucleus-encoded proteins operate as essential or supportive factors during the expression of plastid-encoded PSII genes. They are prime candidates for the regulation of plastid gene expression during developmental processes and in response to environmental cues. Thus, their identification is a prerequisite to understand the gene regulatory network between the nucleus and chloroplasts.
While the basic structure of PSII is highly conserved during the evolution of chloroplasts, cyanobacteria and photosynthetic eukaryotes show marked differences with regard to the regulation of PSII gene expression. In cyanobacteria, PSII biogenesis is predominantly regulated at the transcriptional level. In contrast, in green algae and land plants, plastid-encoded PSII genes are mainly regulated at the posttranscriptional level (Rochaix, 2006; Marín-Navarro et al., 2007).
The expression of psbA, encoding the PSII reaction center protein D1, reflects this differential regulation. In cyanobacteria, a small psbA gene family consisting of two to six genes encodes for distinct D1 isoforms (Mulo et al., 2009). According to the environmental conditions, the transcriptional activity of these genes is up- or down-regulated, generating variable mRNA amounts of the different D1 isoforms. Some regulatory cis-elements and trans-acting factors involved in psbA transcription have been identified (Mulo et al., 2012). The psbA mRNA in cyanobacteria is rather unstable, with a half-life of only 10 to 20 min (Mohamed and Jansson, 1991; Constant et al., 1997). Ribosomes immediately associate with the psbA RNA even in darkness (Tyystjärvi et al., 2004). Moreover, it was suggested that membrane targeting of the D1 nascent complex and translational elongation are important regulatory steps in expression of the D1 genes in Synechocystis sp. PCC6803 (referred to as Synechocystis herein) and Synechococcus sp. PCC7942 (Mulo et al., 2012).
In higher plants and green algae, D1 is encoded by a single gene. In contrast to cyanobacteria, psbA is more or less constitutively transcribed and the psbA mRNA stability is considerably increased, facilitating a shift of main regulatory steps to posttranscriptional processes (Klaff and Gruissem, 1991). In Chlamydomonas reinhardtii, a model is proposed in which translational initiation of the psbA mRNA demands the presence of a multisubunit complex (Danon and Mayfield, 1991; Yohn et al., 1996). In the light, this complex is believed to bind to the 5′ untranslated region (UTR) of psbA, thereby enhancing the efficiency of translation initiation (Fong et al., 2000). In addition, the synthesis rate of D1 in C. reinhardtii directly depends on the availability of the interacting partner, D2. This regulatory relationship was found also for core subunits of the other photosynthetic membrane complexes and was termed control by epistasy of synthesis (Choquet and Wollman, 2002; Minai et al., 2006).
The existence of comparable mechanisms for the regulation of D1 translation in higher plants is still unclear. Nevertheless, cis-acting elements and trans-acting factors determining D1 translational initiation were identified. In the psbA 5′ UTR of tobacco (Nicotiana tabacum), two potential ribosome-binding sites (RBS1 and RBS2) flank an adenine uracil box, which is a putative target of at least one trans-acting factor (Hirose and Sugiura, 1996). Proteins that bind to the psbA 5′ UTR were also found in spinach (Spinacia oleracea; Alexander et al., 1998). The 43-kD RNA-binding protein of spinach is a homolog of the ribosomal protein S1 and binds to uracil-rich single-stranded RNA in response to light (Klaff and Gruissem, 1995; Alexander et al., 1998). Detailed information about the second identified factor, a 47-kD protein, are not available. UV cross-linking experiments with the psbA 5′ UTR of Arabidopsis (Arabidopsis thaliana) established the presence of 43- and 30-kD proteins, both of which bind in a redox-dependent manner but have not been identified yet (Shen et al., 2001). Altogether, these findings suggest that the 5′ UTR of the psbA mRNA from higher plants is very likely also a primary target of control mechanisms for D1 synthesis.
Previously, we identified HCF173 (for high chlorophyll fluorescence173), an essential factor involved in early processes of D1 synthesis in Arabidopsis (Schult et al., 2007). The loss of HCF173 causes a specific destabilization of the psbA transcript together with impaired translation initiation. HCF173 shows low similarity to members of the classical short-chain dehydrogenase/reductase (SDR) superfamily. SDRs represent a large group of oxidoreductases, including epimerases, lyases, and isomerases, that depend on NAD+ or NADP+ or their reduced forms [NAD(P)(H)] as cofactors. These enzymes have a wide substrate spectrum and are involved in a variety of key metabolic processes (Filling et al., 2002; Kallberg et al., 2002). Classical SDR proteins contain two motifs. The N-terminal Gly-rich motif TGXXXGXG mediates the binding of the dinucleotide cofactor and is arranged within a conserved domain of alternating α-helices and β-sheets designated the Rossmann fold. The enzymatic activity of SDR proteins is associated with a catalytic tetrad (N-S-Y-K) consisting of the YXXXK motif in cooperation with conserved Asn (N) and Ser (S) residues (Kavanagh et al., 2008; Persson et al., 2009).
HCF173 belongs to the subfamily of atypical SDRs. Compared with classical SDRs, this subfamily has an altered Gly-rich NAD(P)(H)-binding motif and partly or completely lacks the signature catalytic tetrad; however, the three-dimensional architecture of the SDR fold is conserved (Persson et al., 2009). Atypical SDRs examined so far have obviously lost their enzymatic activity, and some of them gained new functions in RNA metabolism or as regulatory proteins. One of the well-characterized atypical SDRs is NmrA, a transcriptional repressor in fungi able to inactivate the transcription factor AreA in response to the nitrogen status of the cell (Stammers et al., 2001). A long-known chloroplast-located atypical SDR protein is CSP41 (for chloroplast stem loop-binding protein41; Yang et al., 1996; Bollenbach et al., 2003). Many different roles in the RNA metabolism of chloroplasts have been assigned to CSP41. The first reports detected endoribonuclease activity in spinach chloroplasts (Yang et al., 1996). In Arabidopsis, which encodes two copies of this gene, roles for CSP41 in ribosomal RNA (rRNA) metabolism and in transcription were described (Beligni and Mayfield, 2008), and recently, a role in RNA stabilization was demonstrated (Qi et al., 2012).
The factor HCF173 is also directly involved in the RNA metabolism of the chloroplast. Affinity purification revealed its association with the psbA mRNA in a high-Mr thylakoid membrane complex (Schult et al., 2007). Because a significant amount of HCF173 was also detected in the stroma, we discussed a role as an RNA-binding factor involved in the membrane targeting of nascent D1 ribosomal complex.
To identify additional components required during regulated expression of the D1 protein, we analyzed the coexpression profile of HCF173. In this context, we identified the new D1 biogenesis factor HCF244. Its knockout resulted in a very similar phenotype to that described previously for HCF173 inactivation. As in hcf173, the analysis of polysomal association revealed drastically impaired initiation of translation of the psbA mRNA in hcf244. This results in a severe reduction of D1 protein accumulation coupled with destabilization of all other PSII core subunits. By comparison with hcf173, only a slight reduction of the psbA mRNA level was detectable in hcf244. Interestingly, the factor HCF244 also belongs to the atypical SDR subfamily and possesses the modified Rossmann fold dinucleotide-binding site of this subgroup, together with an altered catalytic motif. HCF244 is associated with an approximately 300-kD thylakoid membrane complex. The hcf173hcf244 double mutant shows a more pronounced phenotype than the single mutants, suggesting that photosynthetic activity is further improved due to a lesser extent of D1 synthesis.
RESULTS
Identification of the SDR Protein HCF244
In a forward genetic approach, we identified the nucleus-encoded factor HCF173 (At1g16720), which is a chloroplast-located SDR protein associated with the psbA mRNA and required for translational initiation of this transcript (Schult et al., 2007). To explore the functional and biological network of At1g16720 in Arabidopsis, we analyzed condition-specific coexpression data sets using the Bio-Array Resource (Toufighi et al., 2005). All positively correlated genes (r > 0.8) have been analyzed concerning their function and the localization of the gene product. Interestingly, four genes with known functional relationships to PSII biogenesis and five SDR or SDR-like proteins were found (Supplemental Table S1). The first nine highly correlated genes encoding for unknown or less characterized products with a predicted or experimentally shown chloroplast localization were selected, and corresponding T-DNA insertion lines from the SALK (http://signal.salk.edu/index.html) and GABI-Kat (http://www.gabi-kat.de/) collections were analyzed (Supplemental Table S2). We screened the progeny of these lines for seedlings showing an hcf phenotype, indicative for an impaired photosynthetic capacity. Under our growth conditions (60–80 μmol m−2 s−1, 22°C), the mutant line N408380 (GABI-Kat) segregated an hcf phenotype and mutant seedlings were unable to survive under photoautotrophic growth conditions, revealing the relevance of the inactivated nuclear gene for an efficient photosynthesis rate. According to the observed phenotype, the mutant was designated hcf244. The hcf244 T-DNA line possesses an insertion in the nuclear gene At4g35250 (Fig. 1A), which has the second best correlation to HCF173 expression. To localize the position of the T-DNA, we amplified the region around the insertion point by gene- and T-DNA-specific primers. Sequencing of the PCR products monitored the incorporation of the T-DNA at position +186 in the first of seven exons of At4g35250. As a consequence of the T-DNA insertion, the HCF244 transcript is missing in the mutant (Fig. 1B), resulting in the absence of the HCF244 protein (Fig. 1C).
Figure 1.
Gene structure and expression levels of At4g35250 in wild-type and mutant seedlings. A, Structure of the gene HCF244. The HCF244 open reading frame (1,869 bp) consists of seven exons separated by six introns. The mutant allele hcf244 contains a T-DNA insertion at position +186, which is schematically marked by a triangle. Exons are shown in dark gray. B, Transcript analysis of HCF244. Five micrograms of total leaf RNA or the indicated dilutions, isolated from 2- to 3-week-old wild-type (WT), hcf244 mutant, and complemented hcf244 plants, were analyzed by RNA gel-blot hybridization using a radiolabeled probe against the HCF244 cDNA. The distribution of rRNAs was visualized by methylene blue staining. The degradation product of the 23S rRNA is indicated by an asterisk. C, Immunoblot analysis of the HCF244 protein. Thylakoid membrane proteins were isolated from wild-type and hcf244 leaves and separated by SDS-PAGE. Thirty micrograms of protein was loaded on the SDS gel. The detection of HCF244 was performed with the specific HCF244 antibody, whereas cytochrome f detection resulted from its heme-associated peroxidase activity. The position of cytochrome f is indicated by an asterisk.
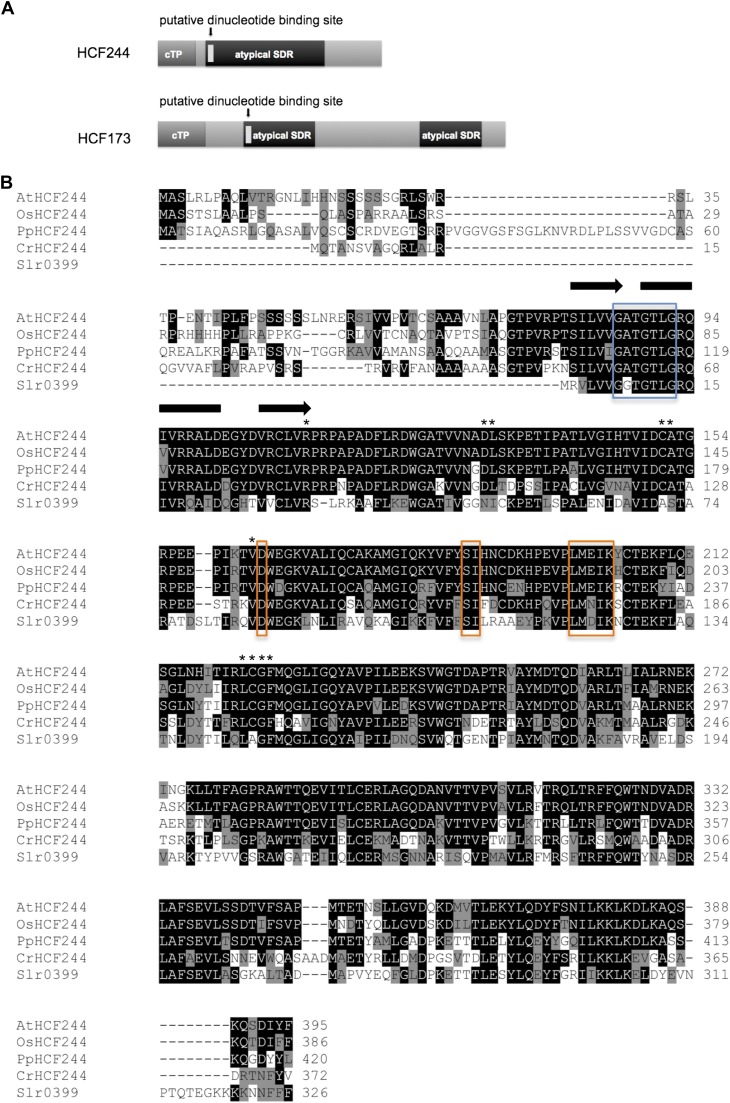
HCF244 encodes a protein consisting of 395 amino acids with a predicted Mr of 43.7 kD. The first 63 amino acids show characteristics of a chloroplast transit peptide, based on ChloroP prediction (Emanuelsson et al., 1999). Referring to the conserved domain database (Marchler-Bauer et al., 2011), the region of amino acids 81 to 284 is annotated as an atypical group (subgroup 5) of the SDR superfamily (Fig. 2A). The NAD(P)(H)-binding domain of HCF244 shows the conserved sequence GXXGXXG, which is similar to that of extended SDRs (Kavanagh et al., 2008). In contrast, a poorly conserved active site tetrad is present, consisting of only the Lys residue in the YXXXK motif. A Ser is located right beside its conserved position, and Asp is present instead of Asn (Fig. 2B). This results in the modified tetrad D-(S)-L-K. With regard to the stringent requirement of Tyr for dehydrogenase/reductase activity (Jörnvall et al., 1995), the altered composition of this motif suggests the loss of enzymatic activity in HCF244.
Figure 2.
Domain structure of HCF244 and similarities to other homologous proteins. A, Domain composition of HCF244 in comparison with HCF173. Both nucleus-encoded factors contain a putative chloroplast transit peptide, indicated in dark gray. The conserved atypical SDR domain, including a putative dinucleotide binding site, is indicated in black and light gray, respectively. B, Multiple amino acid sequence alignment of Arabidopsis HCF244 and homologs from rice (Oryza sativa; OsHCF244), moss (Physcomitrella spp.; PpHCF244), green algae (C. reinhardtii; CrHCF244), and cyanobacteria (Slr0399). Identical amino acids are marked white on black; similar amino acids are black on gray. The predicted secondary structure of the Rossmann fold dinucleotide binding site is schematically indicated above the sequence: black arrow for β-strand; black bar for α-helix. The position of the conserved Gly-rich motif (GXXGXXG) is indicated by a blue box. The conserved and altered residues of the catalytic tetrad (N-S-YXXXK) are marked by orange boxes. Further conserved amino acids are indicated by asterisks. The multiple amino acid sequence alignment was generated with ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). For secondary structure prediction, the Web tool Jpred (http://www.compbio.dundee.ac.uk/www-jpred/) was used.
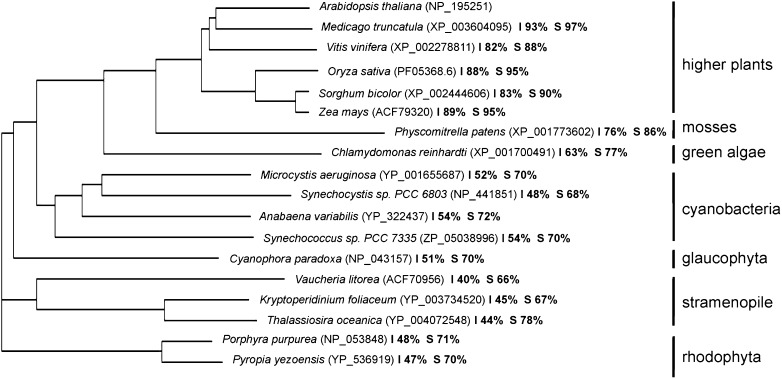
BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Phytozome (http://www.phytozome.net/) searches revealed a number of orthologs ranging from higher plants and mosses to green algae, glaucophytes, red algae, stramenopiles, and cyanobacteria (Fig. 3). Phylogenetic analyses of the representative sequences revealed very close evolutionary relationships among the HCF244 homologs of higher plants, mosses, and green algae, with protein identities in the range of 63% to 93%. Moreover, strong relationships were observed between orthologs of plants and cyanobacteria, revealing protein identities with AtHCF244 around 50%. In addition, orthologs of HCF244 were also found in the stramenopiles (dinoflagellata and heterokontophyta) and in rhodophyta. The phylogenetic analysis of HCF244 suggested that the protein is encoded by an ancient gene, which originated from a cyanobacterial progenitor.
Figure 3.
Phylogenetic analysis of HCF244. The phylogenetic tree was constructed using the ClustalW2 tool for multiple sequence alignment. The values of identity (I) and similarity (S) compared with the Arabidopsis HCF244 protein (NP_195251) are listed in percentages. Accession numbers are bracketed.
The Mutant hcf244 Is Deficient in PSII Function
To assess the function of HCF244, we explored the photosynthetic capacity and the accumulation of the corresponding thylakoid membrane complexes in the selected knockout mutant. Under autotrophic growth conditions, hcf244 mutants merely produced cotyledons and died in the seedling stage. On Suc-supplemented medium, mutants could be cultivated under low-light conditions (60–80 µmol m−2 s−1), but compared with the wild type, these mutant seedlings showed an impaired growth rate and pale green leaf color (Fig. 4). In addition, fertile flowers were not generated by the mutant plants under these conditions.
Figure 4.
Presentation of 3- to 4-week-old hcf mutants and wild-type (WT) plants. Plants were grown under a 16-h-light/8-h-dark period at a PFD of 60 to 80 μmol m−2 s−1 and a constant temperature of 22°C. A, Comparison of the wild type, hcf173 and hcf244 single mutants, and the hcf173hcf244 double mutant. B, Closeup of the hcf173hcf244 double mutant.
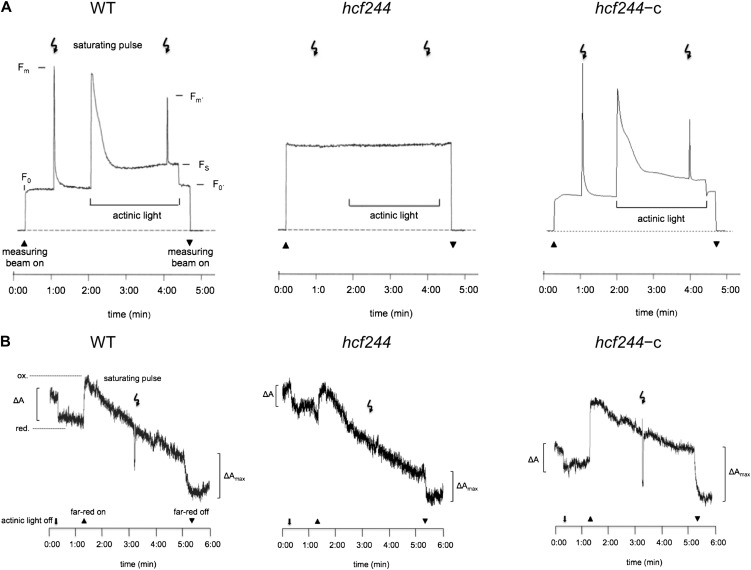
To gain insights into the functional state of PSII and the photosynthetic electron transport chain, chlorophyll a fluorescence was recorded. In dark-adapted wild-type seedlings, a short saturating light pulse induces a characteristic chlorophyll a emission reflecting high efficiency of PSII photochemistry (Fig. 5A). The ratio of variable to maximal fluorescence reached the usual value of 0.78 ± 0.013. In contrast, a saturating light pulse did not induce an increased fluorescence signal in hcf244, indicating a severe defect in PSII photochemistry. To complement these measurements, we analyzed P700 redox reactions in the reaction center of PSI via its absorbance change at 830 nm (Fig. 5B). In the wild type, actinic light partially oxidized P700 whereas far-red illumination induced a maximum oxidation. In far-red background light, P700 was rereduced by a saturating light pulse due to the induction of electron transport from PSII. In contrast, actinic light already results in complete P700 oxidation in hcf244, and the application of a saturating light pulse failed to rereduce P700. This observation indicates a functional PSI reaction center in the mutant but insufficient electron transport rate or absence of electron production in PSII.
Figure 5.
Spectroscopic analyses of wild type (WT), hcf244 mutant, and complemented hcf244 (hcf244-c) plants. Measurements of chlorophyll fluorescence induction (A) and P700 absorbance kinetics (B) were performed with intact 2- to 3-week-old plants.
HCF244 Is Specifically Necessary for the Accumulation of PSII Complex Proteins
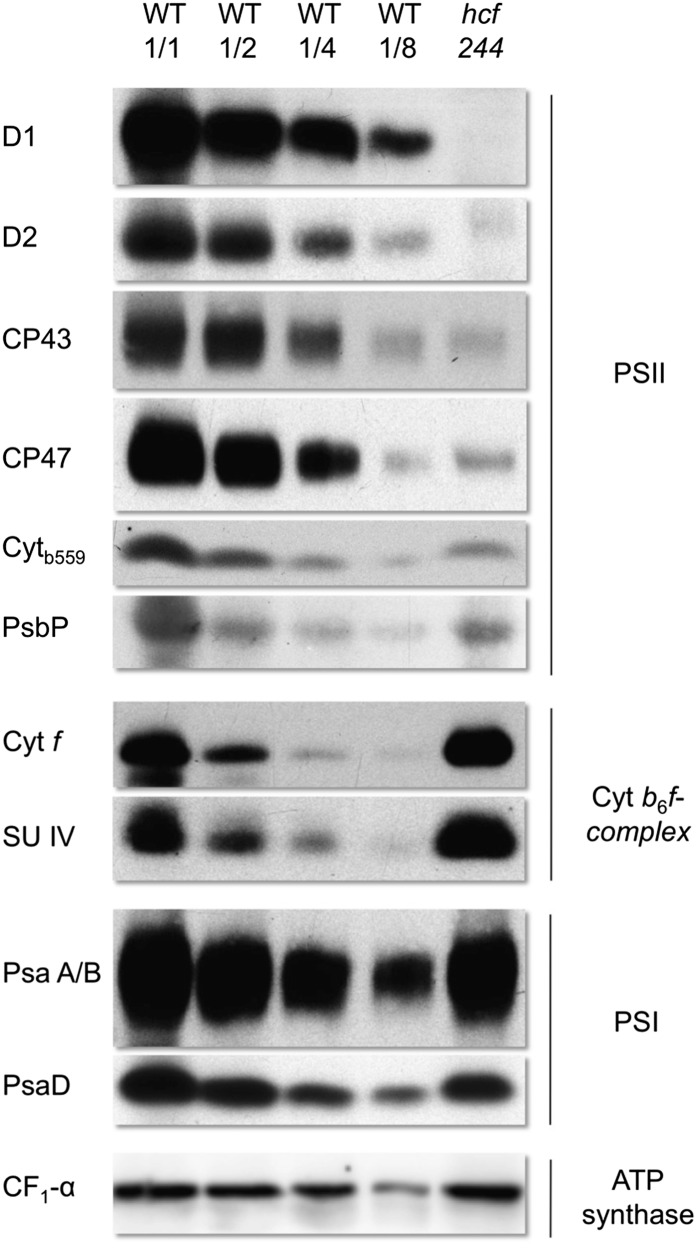
To verify the results of the spectroscopic measurements, we analyzed the accumulation of distinct subunits of the photosynthetic electron transport chain by immunoblot analyses (Fig. 6). The levels of the reaction center proteins D1 and D2 of PSII were decreased below 12.5% and hardly detectable in the mutant. The PSII inner antenna polypeptides CP43, CP47, and cytochrome b559 accumulated to about 12.5% to 25% of wild-type levels, whereas PsbP, a nucleus-encoded subunit of the oxygen-evolving complex, was found to be reduced to 50% in the mutant.
Figure 6.
Protein composition of photosynthetic membranes of 2- to 3-week-old hcf244 and wild-type (WT) plants. Membrane proteins were separated by SDS-PAGE and subsequently transferred onto nitrocellulose membranes. Lanes were loaded with 20 μg (WT 1/1 and hcf244), 10 μg (WT 1/2), 5 μg (WT 1/4), and 2.5 μg (WT 1/8) of protein.
In parallel, we analyzed representative subunits of PSI (PsaD and PsaA/B), the cytochrome b6f complex (Cytf and SUIV), and ATP synthase (CF1-α-SU). In hcf244, 50% to 80% of PsaD and PsaA/B were present. The accumulation of the cytochrome b6f complex and ATP synthase was not impaired. Modification of the light conditions to a lower intensity led to wild-type levels of PSI (data not shown), suggesting that the reduced PSI level is a secondary effect of the mutation, like those previously observed in hcf136 (Meurer et al., 1998) and hcf173 (Schult et al., 2007). Taken together, the immunoblot analysis data suggest that hcf244 is characterized by a specific deficiency in PSII accumulation, whereas other complexes of the photosynthetic electron transport chain are not affected by the mutation.
To confirm that the hcf244 phenotype is based on the single T-DNA insertion in the gene At4g35250, we transformed heterozygous hcf244 plants with the At4g35250 complementary DNA (cDNA) under the control of the 35S promoter employing the Agrobacterium tumefaciens-mediated floral dip method (Clough and Bent, 1998). Transgenic plants, which were homozygous for the hcf244 mutation, were able to grow on soil under photoautotrophic conditions. In addition, spectroscopic measurements (Fig. 5) and immunoblot analysis (data not shown) confirmed the presence of a fully functional PSII. Thus, the mutant phenotype was rescued by the introduced cDNA, indicating that the single T-DNA insertion in the gene At4g35250 was exclusively responsible for the mutant defect.
In addition, we tried to complement hcf244 with several C-terminally tagged protein versions. As the smallest tag, we used the 27-amino acid triple hemagglutinin epitope (Sato and Wada, 1997) or this tag in combination with the 28-amino acid StrepIII tag (Junttila et al., 2005) and the protein A tag (IgG-binding domain of Staphylococcus aureus protein A). However, none of these fusion proteins was able to complement the mutant phenotype, indicating that the C terminus of the protein does not tolerate modifications of this type.
Translational Initiation of the psbA mRNA Is Drastically Impaired in the hcf244 Mutant
To address the question whether the PSII defect in hcf244 is caused by reduced synthesis rates of chloroplast-encoded PSII proteins, we performed in vivo labeling studies with [35S]Met in intact leaves. Newly synthesized proteins were electrophoretically separated to analyze specific translation products. To simplify the complex labeling pattern, cytoplasmic translation was blocked by cycloheximide.
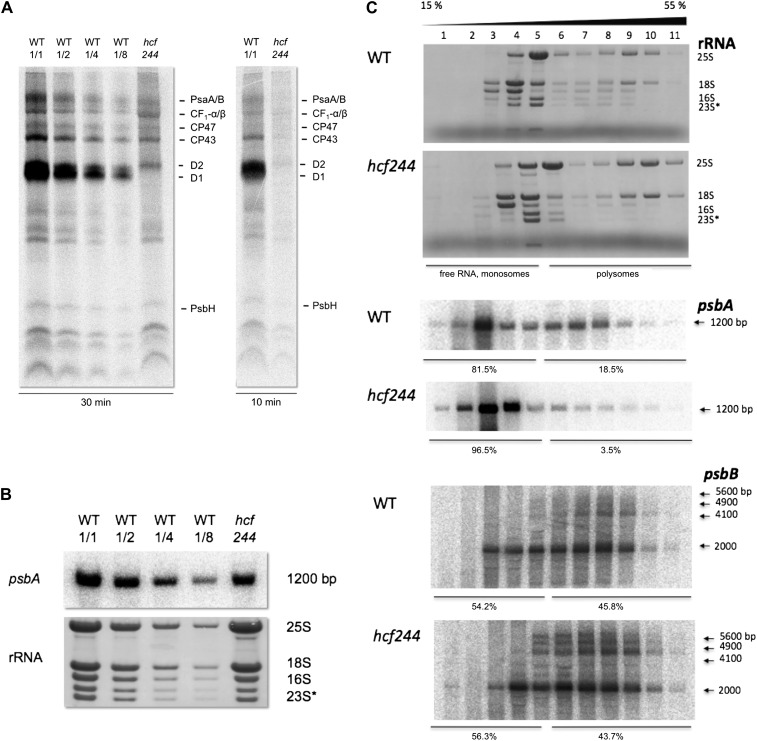
Comparison of the whole labeling pattern between wild-type and mutant leaves revealed that protein labeling is reduced by about 50% in hcf244, indicating that protein synthesis capacity is generally impaired in mutant chloroplasts (Fig. 7A). After a pulse of 30 min, we were able to visualize reaction center proteins A/B of PSI, the α/β-subunits of ATP synthase, and CP47, CP43, D2, and PsbH of PSII in the wild type and mutant. However, the protein with the strongest labeling intensity in wild-type leaves, the D1 protein, was not detectable in the mutant (Fig. 7A). To assess whether this drastically impaired labeling is caused by a decreased half-life, we allowed 35S incorporation only for 10 min. This short pulse period should facilitate D1 detection, even if D1 is degraded very fast. However, our results did not differ from the 30-min pulse, suggesting that the reduced D1 level results from an impaired D1 synthesis rate and not from increased protein degradation.
Figure 7.
Gene expression of psbA in wild-type (WT) and hcf244 mutant seedlings. A, In vivo labeling of newly synthesized membrane proteins by radioactive [35S]Met. Primary leaves of 2-week-old wild-type and hcf244 mutant plants were radiolabeled for 10 and 30 min. Subsequently, membrane proteins were isolated, separated by SDS-PAGE, and visualized by autoradiography. Lanes were loaded with an equivalent amount to 100,000 cpm (WT 1/1 and hcf244), 50,000 cpm (WT 1/2), 25,000 cpm (WT 1/4), and 12,500 cpm (WT 1/8). B, RNA gel-blot analyses of the psbA transcript from wild-type and hcf244 mutant plants. Total leaf RNA was isolated from 2- to 3-week-old plants. Lanes were loaded with 5 μg of total RNA or the indicated dilutions. The degradation product of the 23S rRNA is indicated by an asterisk. C, Polysome association studies in the wild type and the hcf244 mutant. Whole cell extracts of wild-type and hcf244 mutant plants were fractionated by linear 15% to 55% Suc gradients. Afterward, 11 equal fractions were harvested and extracted, and RNA was analyzed by RNA gel-blot hybridization. The distribution of rRNAs was visualized by methylene blue staining. The RNA amounts of psbA and psbB were detected by a specific probe and quantified by phosphor imaging.
To determine if an altered transcript level caused the low synthesis rate of photosynthetic proteins, we measured the amount of numerous plastid- and nucleus-encoded transcripts by RNA gel-blot hybridization. These analyses showed that the psbA mRNA is decreased to 60% of the wild-type level in hcf244 (Fig. 7B). All other analyzed transcripts of PSII (psbB, psbC, psbD, psbE/F, psbO, psbR, psbP, psbH, psbK, and psbN), PSI (psaA and psaC), the cytochrome b6/f complex (petA), and ATP synthase (atpA) accumulated in size and abundance comparable to the wild type (data not shown), indicating that plastid RNA accumulation is not generally impaired by the mutation.
Because the observed reduction of the psbA transcript alone is unlikely to be responsible for the absence of D1, we addressed the question of whether the efficiency of psbA translation is affected in the mutant. To this end, we analyzed ribosomal loading of this transcript by fractionation of leaf RNA in a Suc gradient and subsequent gel-blot hybridization of RNA fractions (Fig. 7C). Efficiently translated RNAs are located in polyribosomes, which migrate into the high-density fractions of a gradient; in contrast, monosomes and free RNA are located in the low-density range. Chelating agents like EDTA disrupt ribosomes, and once they are added in the RNA preparation, the typical distribution pattern of a polysomal gradient is shifted drastically to the low-density fractions, thereby confirming the position of free RNAs and monosomes in the gradient (Schult et al., 2007).
Staining of rRNAs reflected a comparable distribution in both the gradient of the wild type and the mutant hcf244, revealing that there is no general difference in the distribution of ribosomes between the mutant and the wild type. Detection of the psbA mRNA by phosphor imaging and subsequent quantification of the results showed that 19% of wild-type psbA mRNA is located in polysomal fractions. Accordingly, 81% is located in monosomes or represents free RNA. In contrast, in the hcf244 mutant, only 4% of the psbA mRNA is associated with polysomes, implying that the translational activity of this RNA is very low. A subsequent detection of polycistronic psbB transcripts (Westhoff and Herrmann, 1988) displayed a similar distribution in the wild type and the hcf244 mutant.
Taken together, the results of the in vivo labeling assays and the analysis of polysomal distribution of the psbA mRNA indicated that the synthesis rate of the D1 protein is specifically impaired in hcf244 due to reduced translational initiation. This leads to drastically reduced amounts of the D1 protein, with the consequence that no stable PSII core complex could be assembled and mutants are photosynthetically inactive.
HCF244 Is a Peripheral Thylakoid Membrane Protein
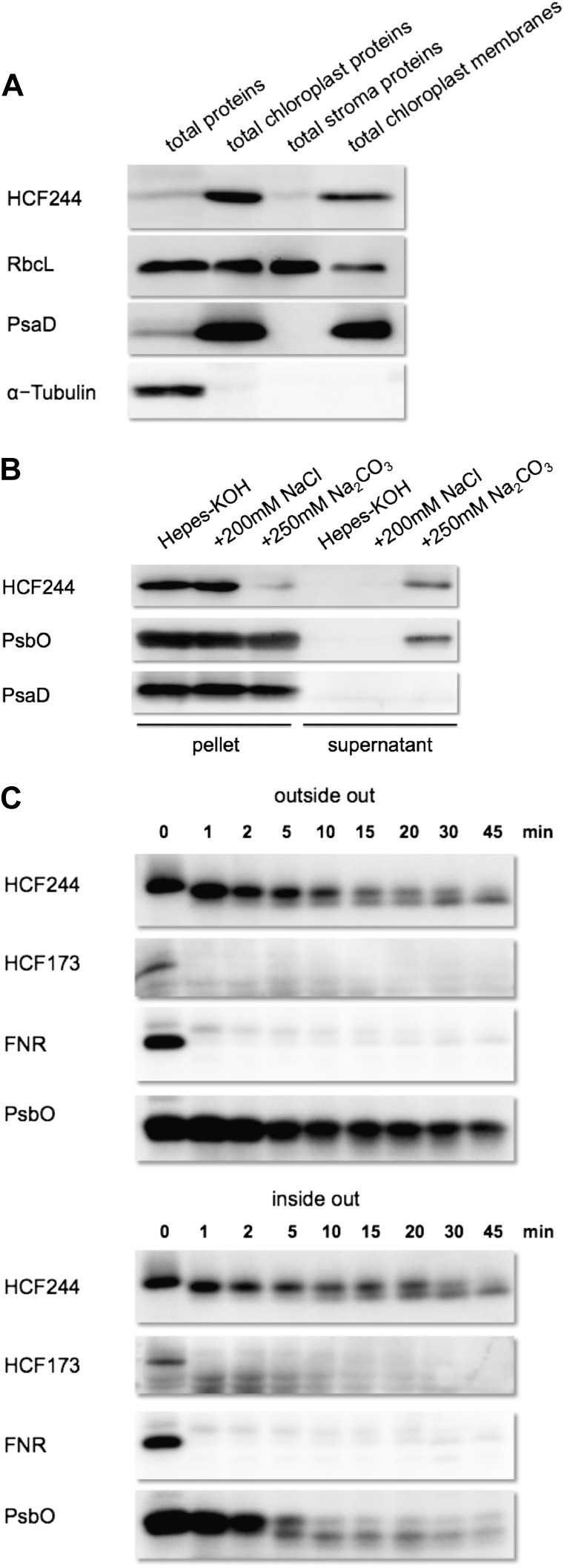
The database entry of the Plant Proteome Database (Sun et al., 2009) defines HCF244 to be a thylakoid-associated protein. To verify this localization and to precisely determine the partitioning of HCF244 in the chloroplast compartments, we prepared a specific antibody against the recombinant protein. In the first immunoblot analyses, the HCF244 antibody detected one clear signal around 38 kD in wild-type membrane protein extracts (Fig. 1C). Subsequently, intact chloroplasts were isolated from wild-type plants and further fractionated into stroma and membrane fractions (Fig. 8A). Compared with total leaf extracts, HCF244 is highly enriched in the chloroplast fraction. The stroma and membrane fractions both revealed a signal of HCF244, while the vast majority of the protein was bound to the membrane fraction. Defined chloroplast proteins like the soluble RbcL and the membrane integral PsaD corroborate the purity of the organelle fractions, whereas α-tubulin as a cytosolic marker confirmed the complete removal of the cytosol. These findings verify that HCF244 is targeted into chloroplasts, where it mainly associates with the membrane fraction. The relative mass of HCF244 differs from the predicted value of 44 kD for the full-length product, suggesting that the import process of HCF244 is accompanied by cleavage of the predicted chloroplast transit peptide.
Figure 8.
Immunolocalization of HCF244. A, Chloroplast localization of HCF244. Intact chloroplasts were isolated from 5- to 6-week-old wild-type plants by Percoll step gradient centrifugation and fractionated into chloroplast membranes and stroma proteins. An equivalent of 10 μg of chlorophyll was loaded on the gel, together with a total leaf extract containing the same protein amount as the total chloroplast extract. B, Salt washing of thylakoid membranes. Thylakoids were sonificated in the indicated buffers, then afterward separated into membrane and soluble proteins by centrifugation and analyzed by SDS-PAGE. C, Protease protection assay. Intact and sonificated thylakoids were treated with thermolysin (final concentration, 0.1 mg mL−1). To stop the reaction, EDTA was added to a final concentration of 50 mm. A total of 10 μg of chlorophyll was loaded in each lane.
To explore the intensity of membrane binding of HCF244, sonificated thylakoids were treated with different salt washings (Fig. 8B). In the presence of sodium chloride, HCF244 remained attached to the membrane, whereas sodium carbonate released the majority of HCF244 protein into the supernatant. The same result was detected for the peripheral oxygen-evolving complex protein PsbO. As expected, the integral PSI protein PsaD was not affected by these treatments.
To address the question of whether HCF244 is located at the stromal or luminal side of the thylakoid membrane, we performed thermolysin protection assays. When intact thylakoids were treated with thermolysin, HCF244 was truncated by approximately 1 kD, indicating that HCF244 is accessible for this protease, but the main part of the protein remained protected (Fig. 8C). A second distinct HCF244 degradation product with a relative size of around 35 kD accumulated with increasing incubation time. In contrast, HCF173 was degraded within 1 min, comparable to the stromal ferredoxin NADP oxidoreductase. This result suggested that HCF244 might be located at the luminal side of the thylakoid membrane, where it is protected against proteolytic degradation. To prove this assumption, we investigated the luminal peripheral proteins in a preparation of thylakoids that were disrupted by sonification. While the luminal PsbO was degraded rapidly, the behavior of HCF244 proteolysis was not altered. Accordingly, the protection of HCF244 is very likely not the result of luminal localization but seems to be based on a highly protected stroma-exposed position.
Together, these data indicate that HCF244 is a peripheral membrane protein, which is tightly attached to the thylakoids. Only a very small domain of HCF244 is directly exposed to the stroma, whereas the residual part is protected against protease activity.
HCF244 Is Part of a Protein Complex, Which Does Not Stably Bind RNA
Recent explorations of accessory proteins involved in thylakoid membrane biogenesis often revealed the assembly of these factors with high-Mr complexes containing both proteins and RNA (Vaistij et al., 2000; Dauvillée et al., 2003; Schwarz et al., 2007; Boulouis et al., 2011). Our studies of HCF173 also showed that this factor is associated in a high-Mr complex of the chloroplast membrane fraction. In addition, an affinity-purified protein fraction of HCF173 preferentially contained the psbA mRNA, indicating specific binding of this transcript (Schult et al., 2007). Hence, the question arises whether HCF244 is also part of a high-Mr complex and, if so, whether the complex includes RNA.
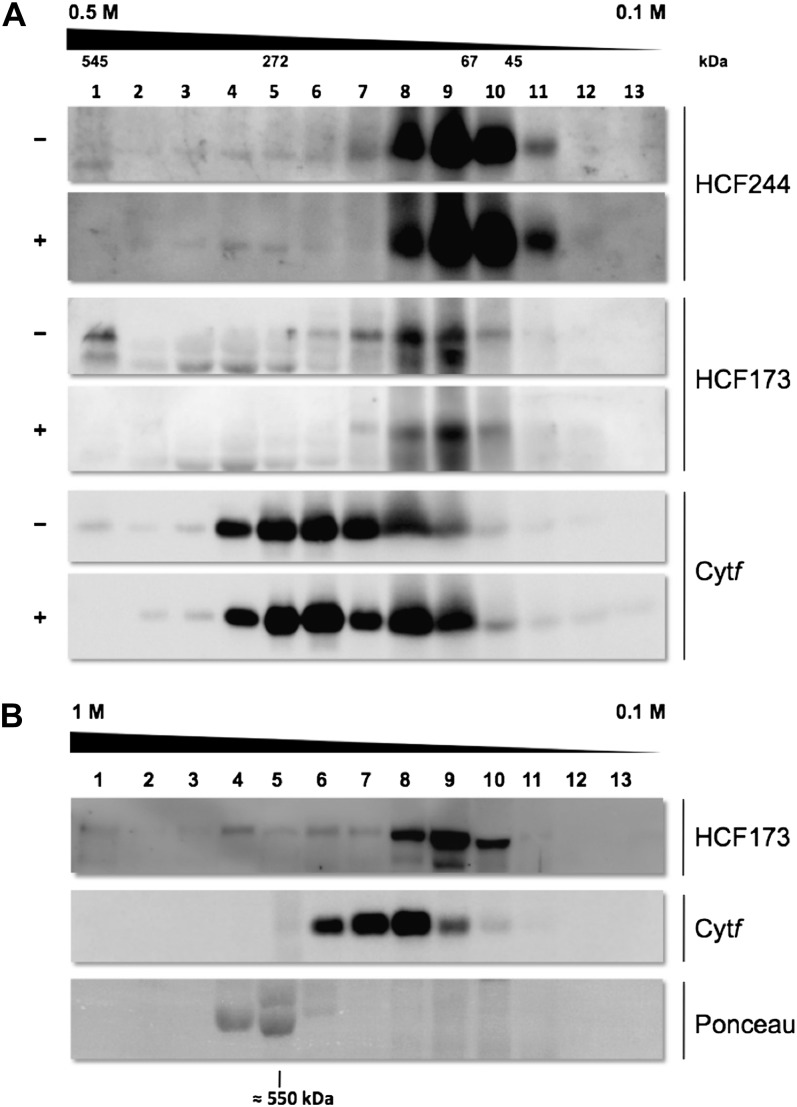
To investigate the accumulation of higher order protein complexes of HCF244, dodecyl-maltoside-solubilized chloroplast membrane extracts of the wild type were separated by Suc density gradient centrifugation. In parallel, we examined the ability of RNA to bind to the HCF244 complex by digestion of membrane protein extract with RNase A and subsequent monitoring of the fractionation behavior of HCF244 and HCF173 (Fig. 9A). Immunodetection of HCF244 in the fractions of the gradient showed that the majority of the protein was detectable in fractions 8 to 10, corresponding to an Mr between 40 and 80 kD; however, a faint signal of HCF244 was detectable up to fraction 2. These results indicate that HCF244 accumulates mainly in a small form. Besides this, the Suc gradient verified the existence of larger HCF244 complexes predominantly separating in a range between 200 and 400 kD. A related behavior could be observed for HCF173, which was enriched in fractions 7 to 10, but significant amounts of the protein were also detectable in the bottom fraction 1 that contained complexes of at least 500 kD. Since fraction 1 contained the complete protein pelleted at the bottom of the gradient, even larger complexes could be present in this fraction. To prove this assumption, we analyzed the migration behavior of HCF173 in a gradient with higher resolution. Figure 9B shows that parts of HCF173 can be detected far beyond the size of the Rubisco enzyme, which migrates at about 550 kD.
Figure 9.
Analysis of HCF complexes by discontinuous Suc gradient centrifugation. A, RNA association study. Wild-type thylakoid membrane proteins were solubilized with 1% (w/v) n-dodecyl-β-d-maltoside and treated with (+) or without (−) RNase A. An equivalent of 500 μg of chlorophyll was loaded on each gradient (0.5–0.1 m Suc). After centrifugation, 13 fractions were harvested. Proteins were precipitated with TCA and analyzed by SDS-PAGE. Exposure of immunoblots incubated with HCF244 antibody was increased to detect minor protein levels. B, Analysis of HCF173 complexes. Wild-type thylakoid membrane proteins were solubilized with 1% (w/v) digitonin and 0.5% n-dodecyl-β-d-maltoside. An equivalent of 500 μg of chlorophyll was loaded on the gradient (1–0.1 m Suc). The position of Rubisco is indicated by the Ponceau red staining of RbcL.
The RNase treatment does not alter the separation pattern of HCF244, indicating that RNA is not part of the detected protein assemblies. In contrast, the portion of HCF173 associated with high-Mr complexes disappeared after RNase treatment, and only signals in the range between 40 and 200 kD were still detectable (Fig. 9A).
Together, these findings support the assumption that HCF244 associates with other binding partners; however, this association seems to be rather weak or is especially instable under our conditions of membrane protein extraction. In addition, RNA is not stably associated with the HCF244 complex. In contrast, the data confirm the existence of high-Mr complexes of HCF173 that stably bind RNA. The size of the HCF173 complex presumably extends into the megadalton region, suggesting that HCF173 and a variety of proteins and RNA associate in a functional assembly.
Analysis of the hcf173hcf244 Double Mutant
The detailed characterization of hcf173 and hcf244 knockout mutants discloses many parallels between the phenotypes of these mutants: both mutants are seedling lethal, show a substantially reduced ratio of variable to maximal fluorescence, and, according to current knowledge, the primary lesion in both is the drastically impaired translational initiation of the psbA mRNA, resulting in strong reduction of D1 accumulation. In addition, HCF173 and HCF244 are directly or indirectly necessary for stabilization of the psbA transcript, but not to the same extent. Altogether, these parallels provide evidence that both HCF proteins are players of the same or related steps of PSII biogenesis.
In order to analyze the relationship genetically, we crossed the mutants and examined the phenotype of the double mutant. While hcf173 and hcf244 single mutants grown on Suc-supplemented medium were pale green and reached nearly one-half of the wild-type size with eight rosette leaves after 3 weeks, the hcf173hcf244 double mutant showed a substantially reduced size and produced only cotyledons and two small rosette leaves with yellow-green color (Fig. 4). The extreme small size of these seedlings prevented physiological measurements and further analyses; however, it suggests that D1 expression is further improved and PSII activity is close to zero in the double mutant.
To further verify the relationship between both proteins, the stability of HCF244 was measured in hcf173 mutants and vice versa. Three independent experiments showed varying accumulation of HCF173 in hcf244 and nearly no alterations for HCF244 in hcf173 (data not shown). Thus, a mutual relationship between the accumulation behaviors of both factors could not be distinguished.
DISCUSSION
Coexpression Analysis of HCF173 Identifies the Highly Conserved SDR Protein HCF244
Coexpression analyses using the psbA translation factor HCF173 as bait revealed around 50 highly correlated genes. Among them, we identified several genes that encode for proteins involved in PSII biogenesis. We found HCF136, a factor essential for the assembly of PSII reaction center proteins (Meurer et al., 1998; Plücken et al., 2002), and the PSII repair proteins LQY1 (Lu et al., 2011) as well as the luminal PPL1 (Ishihara et al., 2007) and Psb27 (Chen et al., 2006). This result shows that functionally related genes cluster with the SDR protein HCF173, implying that there was a reasonable chance to find new PSII biogenesis factors among the unknown, coexpressed genes.
In a reverse genetic approach, we selected the first nine correlated genes with unknown or less characterized function and analyzed the phenotype of the corresponding T-DNA insertion mutants. The knockout of At4g35250 resulted in phenotypical alterations visible under our low-light screening conditions during 2 to 3 weeks of seedling growth. It is possible that the collection of nine candidates comprises further hcf mutants, which have a weaker phenotype and thus are not detectable under our screening conditions.
The isolated T-DNA insertion line, hcf244, is seedling lethal and possesses specifically impaired PSII activity and protein levels. Like the bait gene HCF173, HCF244 encodes a protein with similarity to the SDR superfamily. HCF244 is highly conserved in higher plants and mosses (Physcomitrella spp.), with identities of 75% to 95%. However, even between land plants and algae as well as cyanobacteria, identities remain 45% to 65% among HCF244 and different orthologs. Between higher plants and cyanobacteria, high conservation is found for the N terminus comprising the dinucleotide-binding domain. Thus, it can be suggested that one functional role of these atypical SDR proteins is coupled to the Rossmann fold cofactor-binding domain and is conserved in all these photosynthetic organisms.
Knockout and Suppressor Mutants of Synechocystis HCF244 Reveal a Chaperone Function for the Cyanobacterial Protein
The ortholog of HCF244 from Synechocystis is designated Slr0399 and reveals 48% protein identity. It is similar to the hypothetical protein Ycf39 encoded in the chloroplast genomes of stramenopiles and rhodophyta and the cyanelle genome of Cyanophora paradoxa (Ermakova-Gerdes and Vermaas, 1999). The functional role of Slr0399 has been analyzed in suppressor studies of a Synechocystis photosynthesis mutant with point mutations in the D2 subunit of PSII. The mutant is unable to grow photoautotrophically due to a modified binding site of the primary electron-accepting plastoquinone of PSII (Ermakova-Gerdes and Vermaas, 1998). The isolation of pseudorevertants of this phenotype revealed a secondary mutation in the reading frame of slr0399 (Ermakova-Gerdes and Vermaas, 1999). The mutations in slr0399 represent two different point mutations or a large deletion of around 60%, suggesting that even inactivation of slr0399 leads to the restoration of photosynthetic growth. Knockout of slr0399 in wild-type Synechocystis has no effect on photosynthetic growth but induces a decrease in thermotolerance of the mutant line. Due to these observations, Ermakova-Gerdes and Vermaas (1999) speculate that Slr0399 has a chaperone-like function that is not essential but advantageous during quinone binding to D2 and possibly other quinone-binding complexes.
While knockout of slr0399 in Synechocystis has no significant phenotypic effects under moderate temperatures, inactivation of the orthologous gene HCF244 in Arabidopsis has tremendous consequences for the plant. The hcf244 mutant is unable to grow under autotrophic conditions due to drastically impaired accumulation of PSII proteins. PSII core polypeptides (CP47, CP43, D1, and D2) reach only about 10% to 20% of the wild-type levels. Determination of plastid protein synthesis rates indicates that D1 synthesis is specifically decreased; however, the corresponding psbA mRNA accumulates more or less normally, indicating that a reduced psbA mRNA level cannot account for impaired D1 synthesis. Instead, the analysis of ribosomal loading showed that only a minority of the psbA mRNA in hcf244 is associated with polysomes. This demonstrates that impaired D1 accumulation is not the result of increased protein turnover but rather a consequence of a reduced synthesis rate.
Direct comparison of the knockout phenotypes in Synechocystis and Arabidopsis is currently difficult because only growth rates under different temperatures have been analyzed for the slr0399− mutant. However, marked discrepancies observed for knockout mutants of orthologous genes from cyanobacteria and higher plants are not unusual and were also found for mutants of HCF136/Ycf48 (Meurer et al., 1998; Komenda et al., 2008) and PAM68/Sll0933 (Armbruster et al., 2010). On the other hand, it cannot be ruled out that the proteins HCF244 and Slr0399 have evolved different functions in cyanobacteria and higher plants. To allow convincing conclusions, the phenotypes of hcf244 and slr0399− mutants have to be analyzed in detail in a direct comparison.
HCF173 and HCF244 Mutational Analyses Reveal Their Direct Functions in Translational Initiation of psbA
Phenotypic characterization of hcf244 reveals nearly the same phenotypic properties as we described previously for the hcf173 mutant (Schult et al., 2007). In this photosynthesis mutant, we also found specifically reduced synthesis of D1 coupled with impaired polysomal association of the psbA mRNA, indicating that translational initiation of the psbA mRNA is impaired in both mutants. This deficiency could be initiated directly or indirectly. An indirect effect is conceivable as a feedback regulation in consequence of the impaired D1 assembly. However, a feedback regulation is not supported by the known D1 assembly mutants of Arabidopsis, lpa1, pam68, and hcf243 (Peng et al., 2006; Armbruster et al., 2010; Zhang et al., 2011). For all these mutants, a reduced labeling of the D1 protein was observed, but none of them showed impaired ribosomal loading for the psbA mRNA. Thus, a defective D1 assembly predominantly causes increased D1 degradation, but translational initiation of psbA is unaltered in these mutants. Decreased D1 synthesis was also reported for the Arabidopsis mutant soldat10 (Meskauskiene et al., 2009). Phenotypic examination of the mutant revealed that decreased levels of plastid-specific rRNAs led to a disproportionate reduction of protein synthesis for proteins with high turnover, like D1. However, control panels for the mutants hcf244 and hcf173 and the wild type with methylene blue-stained rRNAs indicate that these RNAs accumulate normally in leaves of both mutants. Hence, we conclude that the identified chloroplast-located proteins HCF244 and HCF173 are both directly involved in the process of translational initiation of the D1-encoding psbA mRNA.
D1 synthesis and membrane insertion occur in a concerted manner at the thylakoid membrane (Zhang et al., 2000) under the assistance of membrane chaperones like ALB3 and LPA1 (Ossenbühl et al., 2004; Peng et al., 2006). Accordingly, we detected the majority of the translation initiation factor HCF244 at the thylakoid membrane. Sodium chloride washes were not sufficient to remove it, but sodium carbonate treatment extracted most of the protein, indicating that HCF244 resembles extrinsic polypeptides at the thylakoid membrane. This observation corresponds to the highly hydrophilic nature of the HCF244 protein sequence, which lacks any predicted membrane-spanning helices. Interestingly, this peripheral membrane association is a further parallel to HCF173, which behaves similarly when treated with both salts (Schult et al., 2007).
While membrane binding of HCF244 and HCF173 is obviously mediated by comparable interactions, both proteins are differently exposed to the stromal compartment. A thermolysin treatment of intact thylakoid membranes resulted in immediate degradation of HCF173, implying an exposed position of the factor. HCF244 was slightly reduced in size by this treatment, but this truncated form was stable for a considerable time period, independent of whether the lumenal or stromal side of the thylakoid membrane was exposed. Obviously, HCF244 is protected by other proteins, which have a shielding function and retard the complete degradation by the protease.
HCF173 Is Part of a High-Mr Complex That Binds and Protects psbA mRNA
Many of the recently identified accessory factors involved in biogenesis of the photosynthetic membrane complexes interact with other partners in a stable manner and can be detected as high-Mr complexes (Sane et al., 2005; Schwarz et al., 2007). In very small quantities, HCF244 could be detected in a membrane-bound complex of around 200 to 400 kD. A considerable amount of HCF244 accumulated in the low-molecular-mass range, between 40 and 80 kD, and one can suppose that the vast majority of the larger assemblies do not survive the isolation procedure. HCF173 was found in different high-molecular-mass membrane complexes, reaching from 150 kD far beyond 550 kD (Schult et al., 2007, and refs. therein). Based on the predicted role of HCF173 in translation initiation, it can be assumed that ribosomal subunits contribute to the largest entities. Interestingly, in an independent proteomic approach, HCF173 was detected in a megadalton fraction of the chloroplast stroma (Olinares et al., 2010). Mass spectrometric analyses of this fraction revealed a high enrichment with ribosomal subunits and accessory and regulatory proteins of plastid gene expression. This observation corroborates our formerly stated hypothesis that HCF173 operates in nascent D1 ribosomal complexes, which assemble in the stroma and subsequently have to be attached to the thylakoids, where translation continues and membrane insertion of D1 commences.
The large complexes of HCF173, detected after Suc gradient fractionation, disappeared when extracted leaf proteins were RNase treated prior to fractionation. This clearly indicates that RNA binds to the HCF173 high-Mr complexes. In a previous work, we were able to show that immunoaffinity-purified HCF173-hemagglutinin or its complex specifically binds the psbA mRNA (Schult et al., 2007). Together, these data provide strong evidence that the psbA mRNA is the specific RNA component of the high-Mr complex of HCF173.
The psbA transcript accumulated to a substantial amount (60%) in hcf244 mutants. In contrast, this RNA is highly unstable in hcf173, resulting in a drastically reduced level of only 10% (Schult et al., 2007). On the other hand, both mutants show the same marked decrease in psbA polysome association. With regard to this observation, we conclude that psbA mRNA destabilization in hcf173 is not a secondary effect of impaired ribosomal loading, since the same effect has to be expected in hcf244. In fact, we assume that impaired psbA accumulation is a specific consequence of the missing HCF173 protein. Therefore, it is highly suggestive that, in addition to a role in translation initiation, HCF173 or its complex binds to the psbA mRNA to protect it against degrading RNases.
An RNA binding and stabilizing function was recently also shown for the atypical chloroplast-located SDR protein CSP41b (Qi et al., 2012). It is proposed that the factor stabilizes nontranslated RNAs coding for photosynthetic proteins and precursors of the 23S and 16S rRNA during the night, when translational activity is low. CSP41b, together with its relative CSP41a, acts as a component of a high-Mr RNA-protein complex (greater than 1 MD), whose formation is very likely redox regulated.
The NAD(P)(H)-Binding Fold of SDRs Can Evolve RNA-Binding Capacity
The precise molecular mechanisms of HCF244 and HCF173 functions are still elusive. Both proteins are members of the atypical SDR family. As typically found in this group, HCF244 possesses the characteristic Rossmann fold dinucleotide-binding domain of atypical SDRs at the N terminus and a partly conserved catalytic domain. The absence of the conserved Tyr in the catalytic tetrad, whose hydroxyl group delivers or accepts protons to or from the substrates (Kavanagh et al., 2008), indicates that HCF244 has very likely lost its enzyme activity. In HCF173, the NAD(P)(H)-binding site shows a slightly altered sequence, and the catalytic center is fragmentary (Schult et al., 2007). The conservation of the dinucleotide-binding site in many atypical SDRs suggests that this motif has retained its function. In most SDR proteins, this domain binds NAD(H) or NADP(H). Moreover, this motif evolved the capability to bind RNA (Hentze, 1994). This was demonstrated for the NAD+-binding fold of glyceraldehyde-3-P dehydrogenase, which specifically binds to AU-rich RNA sequences (Nagy et al., 2000). This RNA binding interferes with the glycolytic activity of the enzyme. Likewise, the Rossmann fold dinucleotide-binding site of lactate dehydrogenase was identified to bind the AU-rich element of granulocyte macrophage colony-stimulating factor RNA (Pioli et al., 2002). Lactate dehydrogenase was identified in translationally active fractions of a polysomal gradient, indicating a role in posttranscriptional gene expression. It is conceivable, therefore, that HCF244 and/or HCF173 act as RNA-binding proteins, which allow translational initiation of psbA mRNA.
Furthermore, the SDR scaffold can also operate as a redox sensor. This property has been demonstrated for the atypical SDR NmrA, a fungal transcriptional regulator able to bind oxidized dinucleotide cofactor, thus mediating the redox status of the cell to the activity of a transcription factor (Lamb et al., 2003). A similar regulatory role has been identified for the factor CC3, a protein with proapoptotic and antimetastatic properties (El Omari et al., 2005). CC3 contains an SDR fold and binds NAD(P)(H); it is suggested that this binding modifies the interaction between CC3 and other proteins (e.g. the importins, which affect nuclear transport, or transcription factors such as c-myc/CIA). Many studies in plants and C. reinhardtii revealed that the synthesis of D1 is regulated by the generation of reducing equivalents from photosynthetic electron transport (Mulo et al., 2012). Thus, redox-sensor proteins are a prerequisite for this regulatory network. Both HCF244 and HCF173 are putative candidates for this function; they possibly could directly sense the redox state of the chloroplast by binding NAD(P)(H) to adapt the rate of D1 synthesis.
The nuclear gene HCF244 was selected by coexpression analysis of HCF173, indicating that both gene products act in a related or even similar biological process. The phenotypic characterization of the single mutants hcf244 and hcf173 indicates that both proteins closely cooperate during the process of translational initiation of psbA. However, the precise relationship is still unclear. So far, the first coimmunoprecipitation experiments and yeast (Saccharomyces cerevisiae) split-ubiquitin analyses do not show a direct interaction between both proteins (data not shown). However, it is conceivable that they act together in one complex, which is involved in the controlled expression of D1 in higher plants, reminiscent of the situation found in the chloroplast of C. reinhardtii. The coexpression analysis revealed that further SDR or SDR-like proteins cluster with HCF173. Thus, we assume that Rossmann fold-containing proteins not only recruit enzymes of the chloroplast but might also acquire further important roles in the process of D1 synthesis or other steps of plastid gene expression.
MATERIALS AND METHODS
Growth Conditions
Mutant and wild-type seeds of Arabidopsis (Arabidopsis thaliana) were plated on Suc-supplemented Murashige and Skoog medium (Murashige and Skoog, 1962) containing 0.3% (w/v) gerite. Seedlings were illuminated at a photon flux density (PFD) of approximately 60 to 80 μmol m−2 s−1 and a 16-h photoperiod (long-day conditions). The selection of mutant plants exhibiting the high-chlorophyll phenotype was performed under UV light according to Meurer et al. (1996).
For blue-native PAGE, Suc density centrifugation, and thermolysin protease protection assay, wild-type seeds were sown on Floraton I-soil (Florogard; http://www.floragard.de/) and cultivated in a growth chamber under short-day conditions (8 h of light/16 h of darkness), constant temperature of 21°C, and a PFD of 60 to 80 μmol m−2 s−1.
Mutant Isolation, Determination of T-DNA Insertion, and Crossing of the Mutants hcf244 and hcf173
The hcf phenotype was screened in the progeny of T-DNA insertion lines for genes coexpressed with HCF173. The hcf244 mutant (mutant line N408380) was recovered from the GABI-Kat collection of T-DNA insertion lines. To determine the position of T-DNA incorporation, the region around the insertion was amplified using the oligonucleotides 8409 (5′-ATATTGACCATCATACTCATTGC-3′) and 250R1 (5′-CTGCATATTGACCAATAAGACC-3′) and subsequently sequenced.
To generate hcf173hcf244 double mutants, heterozygous hcf173-2 (Schult et al., 2007) was crossed with heterozygous hcf244. The genotype of the F1 seedlings was determined by PCR using the oligonucleotides 16720-R10 (5′-CGGTTGTGTCCTGAAAGTAAGTTCC-3′) and pPC161LB (5′-CCCATTTGGACGTGAATGTAGACAC-3′) for the hcf173 mutant T-DNA allele and 250R1 and 8409 for the hcf244 mutant T-DNA allele.
Homozygous hcf173hcf244 double mutants were selected from the F2 generation. The T-DNA insertions and the wild-type genes were verified using the primers described above as well as primer pairs 16720-H4 (5′-GTGGGATTAGGACGACGATCGC-3′)/16720-R10 for HCF173 and 2505UTRupstream (5′-CGTGGCAGCAAATGCAAG-3′)/250R1 for HCF244.
Complementation of the Mutant hcf244
For complementation of the hcf244 mutant phenotype, a construct consisting of the At4g35250 open reading frame and the 5′ UTR under the control of a 2x35S cauliflower mosaic virus promoter was created using Gateway technology (Hartley et al., 2000).
The HCF244 cDNA was amplified by PCR with the primers 250attB15UTR (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGTCAAAACACCAGCTGACG-3; the attB1 site is underlined and the beginning of the 5′ UTR is shown in italics) and 250attB2stop (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGTCAGAAGTAGATGTCTGATTGCTTGG-3; the attB2 site is underlined and the stop codon is shown in italics), employing wild-type cDNA as template. To create the entry clone, a pENTRY-HCF244 BP clonase reaction (Invitrogen; http://www.invitrogen.com/) between the HCF244 PCR product and donor vector pDONR221 was performed according to the Gateway manual. pENTRY-HCF244 was transformed into Escherichia coli strain DH5α and subsequently subjected to sequence analysis. To introduce the HCF244 gene under the control of the 2x35S cauliflower mosaic virus promoter, LR clonase reaction (Invitrogen) was accomplished between pENTRY-HCF244 and the binary Ti destination vector pMDC32 (Curtis and Grossniklaus, 2003). The created expression clone pMDC32-HCF244 was transferred to heterozygous hcf244 plants by Agrobacterium tumefaciens-mediated transformation according to the floral dip method (Clough and Bent, 1998). Complemented transgenic plants were named HCF244-c.
Spectroscopic Measurements
Chlorophyll fluorescence and P700 absorbance measurements were performed as described by Meurer et al. (1996) with the following modifications: saturating light pulses had a length of 1 s, and actinic light had a PFD of 200 μmol m−2 s−1. The P700 absorbance kinetic was measured with the DualPAM100 (Walz; http://www.walz.com/).
Thylakoid Membrane Extraction
Crude leaf proteins from 2- to 3-week-old wild type and mutant plants were isolated as described by Meurer et al. (1996).
For blue-native PAGE, Suc density gradient centrifugation, and protease protection assay, thylakoid membrane proteins were isolated from 4- to 6-week-old wild-type plants grown on soil under short-day conditions. Plant material was homogenized in 10 volumes of lysis buffer (10 mm HEPES/KOH, pH 7.8, 10 mm MgCl2, and 25 mm KCl) per gram fresh weight, filtered through two layers of Miracloth, and centrifuged at 5,900g and 4°C for 2 min. Subsequently, pelleted membranes were dissolved in the appropriate buffers.
SDS-PAGE and Immunoblotting
Proteins were separated by SDS-PAGE according to Schägger and von Jagow (1987) or Laemmli (1970) and transferred onto nitrocellulose membranes (BA79; 0.45 μm; Whatman; http://www.whatman.com/) or polyvinylidene difluoride membranes (0.45 μm; GE Healthcare; http://www.ge.com/). HCF244 and HCF173 proteins were immunodecorated with the specific antibodies described below for antiserum production. Other proteins were detected with specific antibodies as listed by Meurer et al. (1996) and visualized by enhanced chemiluminescence (GE Healthcare).
Antiserum Production
The nucleotide sequence encoding amino acids 55 to 395 of the HCF244 protein was fused to the sequence of an N-terminal T7 tag and a C-terminal hexa-His tag using the expression vector pET-21d(+) (Novagen; http://www.novagen.com/). The construct was transformed into E. coli strain Rosetta2 (DE3) (Novagen). Three hours after the induction of HCF244 expression with 1 mm isopropyl-β-d-thiogalactopyranoside, the 41-kD fusion protein was isolated under denaturing conditions according to the QIAexpressionist manual (Qiagen; http://www.qiagen.com/) and purified on a nickel-nitrilotriacetic acid agarose column (Qiagen). The eluate was separated by SDS-PAGE. The Coomassie blue-stained HCF244 fusion protein was excised from the gel and used to generate polyclonal antibody in rabbits (Agrisera; http://www.agrisera.com/). In parallel, the same procedure was performed to produce a specific antibody against the HCF173 protein. The recombinant protein product comprised amino acids 80 to 598 of HCF173.
Both HCF antibodies detected several unspecific signals in addition to the HCF protein. Therefore, we decided to purify the polyclonal HCF antibodies. The affinity-purified HCF fusion protein was loaded on a SDS gel and transferred onto a polyvinylidene difluoride membrane. To excise the region of HCF protein, the membrane was stained with 1% (w/v) Ponceau red in 2% (w/v) TCA. The antiserum was diluted 1:4 in Tris-buffered saline (20 mm Tris-HCl, 137 mm NaCl, and 0.1% [v/v] Tween 20, pH 7.6) and incubated with the blotted HCF fusion protein overnight. Unbound components of the sera were removed by washing the membrane three times in Tris-buffered saline at 4°C. To elute the HCF antibody, 1.125 volumes of elution buffer (0.1 m Gly, 0.5 m NaCl, and 0.05% [v/v] Tween 20, pH 2.6; related to the amount of antiserum used) was added to the membrane, vortexed for 90 s, and 1 m Tris-HCl, pH 8.0, was added to a final concentration of 100 mm Tris-HCl. For long-term storage of the purified antibody, 0.1 mg mL−1 bovine serum albumin and 50% (v/v) glycerin (final concentration) were added.
Membrane-bound HCF proteins were visualized by chemiluminescence using the Supersignal West femto maximum sensitivity substrate (Thermo Scientific; http://www.thermoscientific.com/).
RNase Treatment and Suc Density Gradient Centrifugation of Protein Complexes
Membranes were isolated as described above, adjusted to a chlorophyll concentration of 1 mg mL−1 containing 1% (w/v) n-dodecyl-β-d-maltoside, and solubilized for 30 min at 4°C. Unsolubilized material was removed by centrifugation for 20 min at 4°C and 15,000g. Two units of RNase A (Qiagen) was added to 500 μL of supernatant and incubated for 1 h at 4°C. In parallel, samples were incubated without RNase. A total of 500 μL of the supernatant was loaded on a linear Suc density gradient (12.5 mL) ranging from 0.5 to 0.1 m Suc in lysis buffer supplemented with 0.06% (w/v) n-dodecyl-β-d-maltoside. Gradients were centrifuged at 4°C for 16 to 17 h at 180,000g (SW40Ti). One-milliliter fractions were collected using a gradient fractionator (Beckman Coulter; http://www.beckmancoulter.de/), precipitated with 15% (w/v) TCA, and analyzed by SDS-PAGE.
Immunolocalization Studies
To isolate total leaf extracts, wild-type plants were homogenized in a small volume of ice-cold lysis buffer and filtrated through glass wool to discard cell debris.
For the isolation of intact chloroplasts, wild-type plants were grown under short-day conditions for 4 to 6 weeks. According to Kunst (1998), the plant material was homogenized in 10 volumes of homogenization buffer (450 mm sorbitol, 20 mm Tricine/KOH, pH 8.4, 10 mm EDTA, 10 mm NaHCO3, and 0.1% [w/v] bovine serum albumin) per gram fresh weight and filtrated through two layers of Miracloth. The chloroplast suspension was centrifuged at 4°C for 4 min and 300g. The pellet was resuspended in 0.04 volume of resuspension buffer (0.3 m sorbitol, 20 mm Tricine/KOH, pH 8.4, 2.5 mm EDTA, and 5 mm MgCl2), loaded on a Percoll step gradient consisting of 80% and 40% Percoll in resuspension buffer, and centrifuged for 20 min at 4°C and 6,500g. Intact chloroplasts were collected, washed in 3 volumes of resuspension buffer three times, and lysed in ice-cold lysis buffer for 10 min. Stroma and membrane proteins were separated by 1 h of centrifugation at 17,000g at 4°C. Total leaf, total chloroplast, chloroplast membrane, and stroma proteins were separated by SDS-PAGE, blotted, and analyzed by immunoblot experiments.
The strength of the membrane association of HCF244 was analyzed by resuspending pelleted membranes in lysis buffer or lysis buffer supplemented with 200 mm NaCl or 250 mm Na2CO3. Membranes were centrifuged for 1 h at 100,000g and 4°C. Supernatants were precipitated with 15% (w/v) TCA. All samples were analyzed by SDS-PAGE and immunodetected with specific antibodies.
The intraorganellar location and topology of HCF244 were examined further by protease protection assay according to Meurer et al. (1998) with the following modifications: thylakoid membranes were isolated as above, and the output of the sonifier (Branson Sonifier B12; Hielscher; http://www.hielscher.com) was set to 5.
In Vivo Labeling of Chloroplast Proteins
The in vivo labeling was performed as described (Lennartz et al., 2001) with the variation that the labeling occurred for 30 and 10 min and the membrane pellet was resuspended in gel-loading buffer (Schägger and von Jagow, 1987). After SDS-PAGE, radiolabeled proteins were analyzed by autoradiography.
RNA Isolation and Gel-Blot Analysis
Isolation of total leaf RNA, electrophoresis, gel-blot analysis, and hybridization were performed according to Westhoff et al. (1991). Specific RNA probes listed by Meurer et al. (1998) were applied for hybridization. Gene-specific probes for At4g35250 were obtained by first-strand cDNA synthesis with total leaf RNA from wild-type plants and subsequent amplification by PCR with the following primer pair: 2505UTRupstream and 250R1 (see above).
Polysome Purification
Polysome fractions were prepared as described before (Barkan, 1993). After gel-blot analysis and hybridization with radiolabeled probes specific for psbA and psbB RNA, the observed signals were quantified using a Bioimager FLA3000 (http://www.fujifilm.com/).
The accession number of the protein that is encoded by the HCF244 gene is NP_195251. Accession numbers of other proteins described in the text are as follows: XP_003604095 (MtHCF244), XP_002278811 (VvHCF244), PF05368.6 (OsHCF244), XP_002444606 (SbHCF244), ACF79320 (ZmHCF244), XP_001773602 (PpHCF244), XP_001700491 (CrHCF244), YP_001655687 (MaHCF244), NP_441851 (Slr0399), YP_322437 (AvHCF244), ZP_05038996 (SsHCF244), NP_043157 (CpHCF244), ACF70956 (VlHCF244), YP_003734520 (KfHCF244), YP_004072548 (ToHCF244), NP_053848 (PpuHCF244), and YP_536919 (PppHCF244).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. List of genes positively coexpressed with HCF173 revealing a relationship to PSII biogenesis or the SDR superfamily.
Supplemental Table S2. List of the first nine unknown or less characterized genes showing high correlation to HCF173 expression.
Glossary
- UTR
untranslated region
- SDR
short-chain dehydrogenase/reductase
- PFD
photon flux density
- rRNA
ribosomal RNA
- cDNA
complementary DNA
References
- Alexander C, Faber N, Klaff P. (1998) Characterization of protein-binding to the spinach chloroplast psbA mRNA 5′ untranslated region. Nucleic Acids Res 26: 2265–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U, Zühlke J, Rengstl B, Kreller R, Makarenko E, Rühle T, Schünemann D, Jahns P, Weisshaar B, Nickelsen J, et al. (2010) The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Aro E-M. (2002) Biogenesis, assembly and turnover of photosystem II units. Philos Trans R Soc Lond B Biol Sci 357: 1451–1459, discussion 1459–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1993) Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beligni MV, Mayfield SP. (2008) Arabidopsis thaliana mutants reveal a role for CSP41a and CSP41b, two ribosome-associated endonucleases, in chloroplast ribosomal RNA metabolism. Plant Mol Biol 67: 389–401 [DOI] [PubMed] [Google Scholar]
- Bollenbach TJ, Tatman DA, Stern DB. (2003) CSP41a, a multifunctional RNA-binding protein, initiates mRNA turnover in tobacco chloroplasts. Plant J 36: 842–852 [DOI] [PubMed] [Google Scholar]
- Boulouis A, Raynaud C, Bujaldon S, Aznar A, Wollman FA, Choquet Y. (2011) The nucleus-encoded trans-acting factor MCA1 plays a critical role in the regulation of cytochrome f synthesis in Chlamydomonas chloroplasts. Plant Cell 23: 333–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang D, Guo J, Wu H, Jin M, Lu Q, Lu C, Zhang L. (2006) A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol Biol 61: 567–575 [DOI] [PubMed] [Google Scholar]
- Choquet Y, Wollman FA. (2002) Translational regulations as specific traits of chloroplast gene expression. FEBS Lett 529: 39–42 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Constant S, Perewoska I, Alfonso M, Kirilovsky D. (1997) Expression of the psbA gene during photoinhibition and recovery in Synechocystis PCC 6714: inhibition and damage of transcriptional and translational machinery prevent the restoration of photosystem II activity. Plant Mol Biol 34: 1–13 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Mayfield SPY. (1991) Light regulated translational activators: identification of chloroplast gene specific mRNA binding proteins. EMBO J 10: 3993–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée D, Stampacchia O, Girard-Bascou J, Rochaix JD. (2003) Tab2 is a novel conserved RNA binding protein required for translation of the chloroplast psaB mRNA. EMBO J 22: 6378–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Omari K, Bird LE, Nichols CE, Ren J, Stammers DK. (2005) Crystal structure of CC3 (TIP30): implications for its role as a tumor suppressor. J Biol Chem 280: 18229–18236 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermakova-Gerdes S, Vermaas W. (1998) Mobility of the primary electron-accepting plastoquinone QA of photosystem II in a Synechocystis sp. PCC 6803 strain carrying mutations in the D2 protein. Biochemistry 37: 11569–11578 [DOI] [PubMed] [Google Scholar]
- Ermakova-Gerdes S, Vermaas W. (1999) Inactivation of the open reading frame slr0399 in Synechocystis sp. PCC 6803 functionally complements mutations near the Q(A) niche of photosystem II: a possible role of Slr0399 as a chaperone for quinone binding. J Biol Chem 274: 30540–30549 [DOI] [PubMed] [Google Scholar]
- Filling C, Berndt KD, Benach J, Knapp S, Prozorovski T, Nordling E, Ladenstein R, Jörnvall H, Oppermann U. (2002) Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J Biol Chem 277: 25677–25684 [DOI] [PubMed] [Google Scholar]
- Fong CL, Lentz A, Mayfield SP. (2000) Disulfide bond formation between RNA binding domains is used to regulate mRNA binding activity of the chloroplast poly(A)-binding protein. J Biol Chem 275: 8275–8278 [DOI] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. (2000) DNA cloning using in vitro site-specific recombination. Genome Res 10: 1788–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW. (1994) Enzymes as RNA-binding proteins: a role for (di)nucleotide-binding domains? Trends Biochem Sci 19: 101–103 [DOI] [PubMed] [Google Scholar]
- Hirose T, Sugiura M. (1996) Cis-acting elements and trans-acting factors for accurate translation of chloroplast psbA mRNAs: development of an in vitro translation system from tobacco chloroplasts. EMBO J 15: 1687–1695 [PMC free article] [PubMed] [Google Scholar]
- Ishihara S, Takabayashi A, Ido K, Endo T, Ifuku K, Sato F. (2007) Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiol 145: 668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörnvall H, Persson B, Krook M, Atrian S, Gonzàlez-Duarte R, Jeffery J, Ghosh D. (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry 34: 6003–6013 [DOI] [PubMed] [Google Scholar]
- Junttila MR, Saarinen S, Schmidt T, Kast J, Westermarck J. (2005) Single-step Strep-tag purification for the isolation and identification of protein complexes from mammalian cells. Proteomics 5: 1199–1203 [DOI] [PubMed] [Google Scholar]
- Kallberg Y, Oppermann U, Jörnvall H, Persson B. (2002) Short-chain dehydrogenase/reductase (SDR) relationships: a large family with eight clusters common to human, animal, and plant genomes. Protein Sci 11: 636–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh KL, Jörnvall H, Persson B, Oppermann U. (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily. Functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci 65: 3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaff P, Gruissem W. (1991) Changes in chloroplast mRNA stability during leaf development. Plant Cell 3: 517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaff P, Gruissem W. (1995) A 43 kD light-regulated chloroplast RNA-binding protein interacts with the psbA 5′ non-translated leader RNA. Photosynth Res 46: 235–248 [DOI] [PubMed] [Google Scholar]
- Komenda J, Nickelsen J, Tichý M, Prásil O, Eichacker LA, Nixon PJ. (2008) The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J Biol Chem 283: 22390–22399 [DOI] [PubMed] [Google Scholar]
- Kunst L. (1998) Preparation of physiologically active chloroplasts from Arabidopsis. Methods Mol Biol 82: 43–48 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lamb HK, Leslie K, Dodds AL, Nutley M, Cooper A, Johnson C, Thompson P, Stammers DK, Hawkins AR. (2003) The negative transcriptional regulator NmrA discriminates between oxidized and reduced dinucleotides. J Biol Chem 278: 32107–32114 [DOI] [PubMed] [Google Scholar]
- Lennartz K, Plücken H, Seidler A, Westhoff P, Bechtold N, Meierhoff K. (2001) HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b(6)f complex in Arabidopsis. Plant Cell 13: 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Hall DA, Last RL. (2011) A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23: 1861–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Navarro J, Manuell AL, Wu J, P Mayfield S. (2007) Chloroplast translation regulation. Photosynth Res 94: 359–374 [DOI] [PubMed] [Google Scholar]
- Meskauskiene R, Würsch M, Laloi C, Vidi PA, Coll NS, Kessler F, Baruah A, Kim C, Apel K. (2009) A mutation in the Arabidopsis mTERF-related plastid protein SOLDAT10 activates retrograde signaling and suppresses (1)O(2)-induced cell death. Plant J 60: 399–410 [DOI] [PubMed] [Google Scholar]
- Meurer J, Meierhoff K, Westhoff P. (1996) Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198: 385–396 [DOI] [PubMed] [Google Scholar]
- Meurer J, Plücken H, Kowallik KV, Westhoff P. (1998) A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J 17: 5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minai L, Wostrikoff K, Wollman FA, Choquet Y. (2006) Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell 18: 159–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed A, Jansson C. (1991) Photosynthetic electron transport controls degradation but not production of psbA transcripts in the cyanobacterium Synechocystis 6803. Plant Mol Biol 16: 891–897 [DOI] [PubMed] [Google Scholar]
- Mulo P, Sakurai I, Aro EM. (2012) Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim Biophys Acta 1817: 247–257 [DOI] [PubMed] [Google Scholar]
- Mulo P, Sicora C, Aro EM. (2009) Cyanobacterial psbA gene family: optimization of oxygenic photosynthesis. Cell Mol Life Sci 66: 3697–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nagy E, Henics T, Eckert M, Miseta A, Lightowlers RN, Kellermayer M. (2000) Identification of the NAD(+)-binding fold of glyceraldehyde-3-phosphate dehydrogenase as a novel RNA-binding domain. Biochem Biophys Res Commun 275: 253–260 [DOI] [PubMed] [Google Scholar]
- Olinares PD, Ponnala L, van Wijk KJ. (2010) Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol Cell Proteomics 9: 1594–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenbühl F, Göhre V, Meurer J, Krieger-Liszkay A, Rochaix JD, Eichacker LA. (2004) Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. Plant Cell 16: 1790–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Ma J, Chi W, Guo J, Zhu S, Lu Q, Lu C, Zhang L. (2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson B, Kallberg Y, Bray JE, Bruford E, Dellaporta SL, Favia AD, Duarte RG, Jörnvall H, Kavanagh KL, Kedishvili N, et al. (2009) The SDR (short-chain dehydrogenase/reductase and related enzymes) nomenclature initiative. Chem Biol Interact 178: 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioli PA, Hamilton BJ, Connolly JE, Brewer G, Rigby WF. (2002) Lactate dehydrogenase is an AU-rich element-binding protein that directly interacts with AUF1. J Biol Chem 277: 35738–35745 [DOI] [PubMed] [Google Scholar]
- Plücken H, Müller B, Grohmann D, Westhoff P, Eichacker LA. (2002) The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett 532: 85–90 [DOI] [PubMed] [Google Scholar]
- Qi Y, Armbruster U, Schmitz-Linneweber C, Delannoy E, de Longevialle AF, Rühle T, Small I, Jahns P, Leister D. (2012) Arabidopsis CSP41 proteins form multimeric complexes that bind and stabilize distinct plastid transcripts. J Exp Bot 63: 1251–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix JD. (2006) The role of nucleus- and chloroplast-encoded factors in the synthesis of the photosynthetic apparatus. In RR Wise, JK Hoober, eds, Advances in Photosynthesis and Respiration. Vol 23. The Structure and Function of Plastids. Springer, Dordrecht, The Netherlands, pp 145–165
- Sane AP, Stein B, Westhoff P. (2005) The nuclear gene HCF107 encodes a membrane-associated R-TPR (RNA tetratricopeptide repeat)-containing protein involved in expression of the plastidial psbH gene in Arabidopsis. Plant J 42: 720–730 [DOI] [PubMed] [Google Scholar]
- Sato MH, Wada Y. (1997) Universal template plasmid for introduction of the triple-HA epitope sequence into cloned genes. Biotechniques 23: 254–256 [DOI] [PubMed] [Google Scholar]
- Schägger H, Cramer WA, von Jagow G. (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217: 220–230 [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Schult K, Meierhoff K, Paradies S, Töller T, Wolff P, Westhoff P. (2007) The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell 19: 1329–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz C, Elles I, Kortmann J, Piotrowski M, Nickelsen J. (2007) Synthesis of the D2 protein of photosystem II in Chlamydomonas is controlled by a high molecular mass complex containing the RNA stabilization factor Nac2 and the translational activator RBP40. Plant Cell 19: 3627–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Danon A, Christopher DA. (2001) RNA binding-proteins interact specifically with the Arabidopsis chloroplast psbA mRNA 5′ untranslated region in a redox-dependent manner. Plant Cell Physiol 42: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Stammers DK, Ren J, Leslie K, Nichols CE, Lamb HK, Cocklin S, Dodds A, Hawkins AR. (2001) The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases. EMBO J 20: 6619–6626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ. (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. (2005) The Botany Array Resource: e-Northern, Expression Angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Tyystjärvi T, Sirpiö S, Aro EM. (2004) Post-transcriptional regulation of the psbA gene family in the cyanobacterium Synechococcus sp. PCC 7942. FEBS Lett 576: 211–215 [DOI] [PubMed] [Google Scholar]
- Vaistij FE, Boudreau E, Lemaire SD, Goldschmidt-Clermont M, Rochaix JD. (2000) Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 97: 14813–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff P, Herrmann RG. (1988) Complex RNA maturation in chloroplasts: the psbB operon from spinach. Eur J Biochem 171: 551–564 [DOI] [PubMed] [Google Scholar]
- Westhoff P, Offermann-Steinhard K, Höfer M, Eskins K, Oswald A, Streubel M. (1991) Differential accumulation of plastid transcripts encoding photosystem II components in the mesophyll and bundle-sheath cells of monocotyledonous NADP-malic enzyme-type C4 plants. Planta 184: 377–388 [DOI] [PubMed] [Google Scholar]
- Yang J, Schuster G, Stern DB. (1996) CSP41, a sequence-specific chloroplast mRNA binding protein, is an endoribonuclease. Plant Cell 8: 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn CB, Cohen A, Danon A, Mayfield SP. (1996) Altered mRNA binding activity and decreased translational initiation in a nuclear mutant lacking translation of the chloroplast psbA mRNA. Mol Cell Biol 16: 3560–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhou G, Liu B, Kong Y, Chen N, Qiu Q, Yin H, An J, Zhang F, Chen F. (2011) HCF243 encodes a chloroplast-localized protein involved in the D1 protein stability of the Arabidopsis photosystem II complex. Plant Physiol 157: 608–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LX, Paakkarinen V, van Wijk KJ, Aro EM. (2000) Biogenesis of the chloroplast-encoded D1 protein: regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell 12: 1769–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]