Abstract
The truncated light-harvesting antenna size3 (tla3) DNA insertional transformant of Chlamydomonas reinhardtii is a chlorophyll-deficient mutant with a lighter green phenotype, a lower chlorophyll (Chl) per cell content, and higher Chl a/b ratio than corresponding wild-type strains. Functional analyses revealed a higher intensity for the saturation of photosynthesis and greater light-saturated photosynthetic activity in the tla3 mutant than in the wild type and a Chl antenna size of the photosystems that was only about 40% of that in the wild type. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and western-blot analyses showed that the tla3 strain was deficient in the Chl a/b light-harvesting complex. Molecular and genetic analyses revealed a single plasmid insertion in chromosome 4 of the tla3 nuclear genome, causing deletion of predicted gene g5047 and plasmid insertion within the fourth intron of downstream-predicted gene g5046. Complementation studies defined that gene g5047 alone was necessary and sufficient to rescue the tla3 mutation. Gene g5047 encodes a C. reinhardtii homolog of the chloroplast-localized SRP43 signal recognition particle, whose occurrence and function in green microalgae has not hitherto been investigated. Biochemical analysis showed that the nucleus-encoded and chloroplast-localized CrCpSRP43 protein specifically operates in the assembly of the peripheral components of the Chl a/b light-harvesting antenna. This work demonstrates that cpsrp43 deletion in green microalgae can be employed to generate tla mutants with a substantially diminished Chl antenna size. The latter exhibit improved solar energy conversion efficiency and photosynthetic productivity under mass culture and bright sunlight conditions.
There is current interest and ongoing efforts to renewably generate fuel and chemical products for human consumption through the process of microalgal photosynthesis. Such bioproducts include H2 (Hankamer et al., 2007; Melis, 2007), biofuel and chemical molecules (Hu et al., 2008; Greenwell et al., 2010; Mata et al., 2010; Melis, 2012), antigens (Dauvillée et al., 2010; Michelet et al., 2011), and high-value biopharmaceuticals (Mayfield et al., 2007). For this effort, sunlight energy conversion in photosynthesis must take place with the utmost efficiency, as this would help to make renewable fuel and chemical processes economically feasible. In plants and algae, the solar energy conversion efficiency of photosynthesis is thus a most critical factor for the economic viability of renewable fuel and chemical production (Melis, 2009, 2012).
Green microalgae and other photosynthetic systems tend to develop large arrays of light-harvesting complexes, especially when cultivated under high-density mass culture conditions. This physiological response of the cells reflects an effort to absorb as much sunlight as possible as they compete in a light-limited environment (Kirk, 1994). However, in mass culture with cells possessing large chlorophyll (Chl) antennae, cells at the surface of the reactor would absorb incident sunlight (intensity of 2,500 μmol photons m−2 s−1) with rates that far exceed the capacity of the photosynthetic apparatus to utilize them (light saturation of photosynthesis occurs at less than 500 μmol photons m−2 s−1). The excess absorbed sunlight energy is dissipated via a process of nonphotochemical quenching to prevent photodamage and photoinhibition phenomena at the thylakoid membrane level (for review, see Müller et al., 2001).
It has been shown that high-density cultures of microalgae with a truncated Chl antenna size are photosynthetically more productive under bright sunlight due to the elimination of overabsorption and wasteful dissipation of excess energy (Nakajima and Ueda, 1997, 1999; Melis et al., 1999; Polle et al., 2002, 2003; Melis, 2009). Identification of genes that confer a permanently truncated light-harvesting antenna size phenotype in plants and algae is thus of interest, as they could be applied in efforts to improve solar-to-product conversion efficiencies (Mitra and Melis, 2008; Melis, 2009; Ort et al., 2011). To this end, and to better understand the genetic mechanism that defines the size of the light-harvesting antenna in green microalgae, and also in an effort to generate truncated light-harvesting antenna size (tla) mutants, we generated and screened a library of Chlamydomonas reinhardtii DNA insertional mutagenesis strains. This work presents a molecular, genetic, and physiological analysis of one of these mutants, termed tla3, which exhibited a stably truncated light-harvesting Chl antenna size. The corresponding TLA3 gene was cloned and found to encode a homolog of the chloroplast signal recognition particle protein CpSRP43. Detailed functional analysis revealed that the phenotype of the tla3-ΔCpSRP43 mutant in C. reinhardtii entailed substantial reductions of the light-harvesting Chl antenna size. Accordingly, the cpsrp43 mutant phenotype and the CrCpSRP43 gene can be employed in C. reinhardtii, and possibly other green microalgae and plants, as a tool by which to truncate the Chl antenna size without affecting the function of the photosystems or thylakoid membrane electron transport properties of the chloroplast.
RESULTS
Pigmentation and Composition of the tla3 Mutant
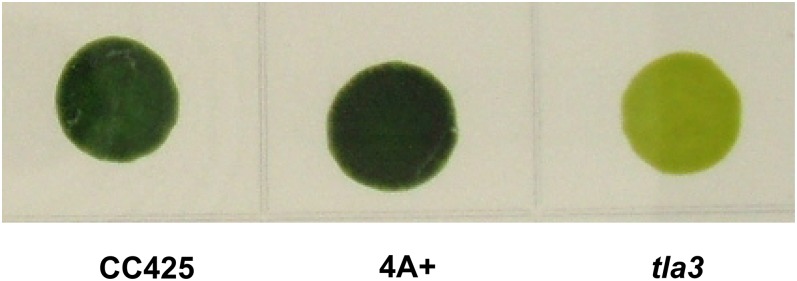
The C. reinhardtii tla3 strain was generated by rescue of an Arg-requiring mutant (arg7) upon transformation with the cloned wild-type ARG7 gene contained in plasmid pJD67. Such rescue is the result of random integration of the pJD67 plasmid DNA into the nuclear genome, causing mutation(s) to other genes. When cultivated on agar, the C. reinhardtii tla3 strain displayed a yellow-green coloration, compared with the dark green of wild-type strains (Fig. 1). Measurements of pigment content showed that cells of the tla3 mutant contained only about 15% of the Chl present in wild-type strains (Table I). While the Chl a content in the tla3 mutant is about 30% of the wild type, Chl b content dropped to about 5%. This disproportional loss of Chl b compared with Chl a changed the Chl a/Chl b ratio in the tla3 mutant to about 13:1, while this ratio in the wild types normally ranges between 2.7:1 and 3.0:1. An elevated Chl a/Chl b ratio suggests that the peripheral Chl a/b light-harvesting antenna complexes are substantially depleted in abundance (i.e. they do not assemble to form large arrays of antennae like those found in the wild type). Accordingly, this strain is a putative tla mutant. The total carotenoid per cell in the tla3 mutant was also lowered to about 35% of that in the wild type.
Figure 1.
Single-cell colonies of the C. reinhardtii wild type and the tla3 mutant grown on agar. Note the dark coloration of the wild-type strains as compared with the pale coloration of the tla3 mutant. [See online article for color version of this figure.]
Table I. Chl and carotenoid pigment contents in femtomoles (fmol) and ratios for the wild type, the tla3 mutant, and tla3 complemented strains of C. reinhardtii (n = 3–5; values shown are means ± sd).
| Strain | Chl per Cell | Chl a per Cell | Chl b per Cell | Chl a/Chl b Ratio | Carotenoid per Cell | Carotenoid per Chl |
|---|---|---|---|---|---|---|
| fmol | fmol | fmol | fmol | |||
| 4A+ | 2.57 ± 0.43 | 1.84 ± 0.32 | 0.73 ± 0.11 | 2.72 ± 0.07 | 1.07 ± 0.17 | 0.42 ± 0.00 |
| CC125 | 2.66 ± 0.13 | 1.95 ± 0.10 | 0.71 ± 0.03 | 3.00 ± 0.03 | 1.11 ± 0.06 | 0.42 ± 0.00 |
| CC504 | 2.36 ± 0.05 | 1.69 ± 0.04 | 0.67 ± 0.01 | 2.73 ± 0.05 | 0.93 ± 0.04 | 0.39 ± 0.01 |
| CC425 | 2.33 ± 0.10 | 1.69 ± 0.07 | 0.64 ± 0.03 | 2.86 ± 0.04 | 0.95 ± 0.04 | 0.41 ± 0.01 |
| tla3 | 0.38 ± 0.03 | 0.34 ± 0.01 | 0.04 ± 0.00 | 12.70 ± 1.72 | 0.37 ± 0.01 | 0.97 ± 0.01 |
| C1 | 1.26 ± 0.35 | 0.91 ± 0.25 | 0.36 ± 0.10 | 2.77 ± 0.02 | 0.70 ± 0.15 | 0.56 ± 0.04 |
| C2 | 1.59 ± 0.10 | 1.17 ± 0.05 | 0.42 ± 0.05 | 3.08 ± 0.22 | 0.67 ± 0.02 | 0.42 ± 0.01 |
| C3 | 0.81 ± 0.02 | 0.61 ± 0.02 | 0.19 ± 0.00 | 3.59 ± 0.03 | 0.54 ± 0.04 | 0.67 ± 0.03 |
| C4 | 0.45 ± 0.02 | 0.38 ± 0.02 | 0.07 ± 0.00 | 6.64 ± 0.07 | 0.40 ± 0.01 | 0.89 ± 0.03 |
Photosynthetic Activity and Functional Chl Antenna Size in the tla3 Mutant and the Wild Type
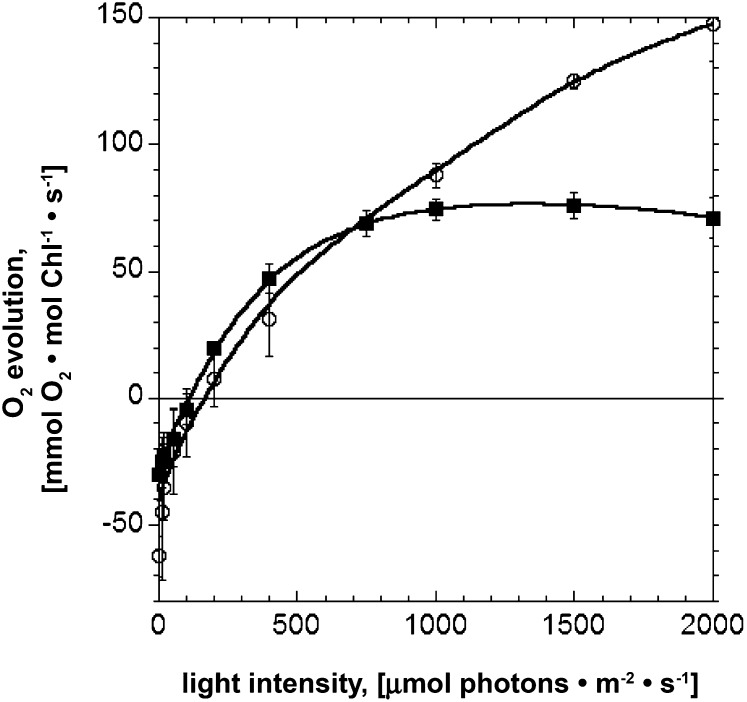
To investigate the functional properties of the photosynthetic apparatus in the tla3 mutant relative to that in the wild type, the light-saturation curve of photosynthesis was measured under in vivo conditions with photoautotrophically grown cells (Fig. 2). On a per Chl basis, the dark respiration rate of the cells, measured at 0 light intensity, was approximately 30 mmol oxygen (mol Chl)−1 s−1 in the wild type and 62 mmol oxygen (mol Chl)−1 s−1 in the tla3 mutant (Table II).
Figure 2.
Light-saturation curves of photosynthesis obtained with the C. reinhardtii wild type (closed squares) and the tla3 mutant (open circles). The initial slopes of both curves are similar, suggesting equal quantum yield of photosynthesis. The light-saturated Pmax was 2-fold higher in the tla3 mutant than in the wild type, suggesting a greater productivity on a per Chl basis in tla3 than in the wild type.
Table II. Photosynthesis, respiration, and photochemical apparatus characteristics of the wild type and the tla3 mutant of C. reinhardtii grown photoautotrophically under medium-light (450 μmol photons m−2 s−1) conditions.
Photosystem Chl antenna size and reaction center concentrations were measured spectrophotometrically (Melis, 1989; n = 3; values shown are means ± sd).
| Parameter Measured | Wild Type | tla3 |
|---|---|---|
| Respiration [mmol oxygen (mol Chl)−1 s−1] | 30.2 ± 11.9 | 62.2 ± 8.0 |
| Pmax [mmol oxygen (mol Chl)−1 s−1] | 106.3 ± 12.8 | 209.7 ± 16.7 |
| Quantum yield of photosynthesis (relative units) | 100 ± 25 | 102 ± 20 |
| Half-saturation intensity (μmol photons m−2 s−1) | 210 | >1,000 |
| Respiration (amol oxygen cell−1 s−1) | 55.8 ± 26.3 | 20.6 ± 5.4 |
| Pmax (amol oxygen cell−1 s−1) | 196.2 ± 46.2 | 69.3 ± 6.6 |
| Pmax/respiration (relative units) | 3.5 ± 1.9 | 3.4 ± 0.7 |
| Amount of PSII per cell (amol cell−1) | 6.49 ± 1.81 | 1.73 ± 0.18 |
| Amount of PSI per cell (amol cell−1) | 4.64 ± 0.24 | 1.85 ± 0.14 |
| Functional PSIIα Chl antenna size | 333 ± 27 | – |
| Functional PSIIβ Chl antenna size | 119 ± 31 | 94 ± 5 |
| Proportion of PSIIβ (%) | 40 ±1 | – |
| Average PSII Chl antenna size | 245 ± 30 | 94 ± 5 |
| Functional PSI Chl antenna size | 230 ± 26 | 120 ± 10 |
In the light intensity range of 0 to 400 μmol photons m−2 s−1, photosynthetic activity increased as a function of irradiance. The increase was linear for tla3 and the wild type, specifically in the region between 20 and 300 μmol photons m−2 s−1 light intensity, where there is a linear regression for both strains (Fig. 2). The slope of these linear regressions was measured to be 0.56 and 0.57 in relative units for tla3 and the wild type, respectively. Identical initial slopes in the light-saturation curves indicated that the quantum yield of photosynthesis for tla3 and the wild type is essentially the same. It may be concluded that the mutation in tla3 and the putative truncated Chl antenna size did not interfere with the quantum yield of the photosynthetic apparatus (Table II).
Photosynthetic activity in the wild type saturated at about 500 μmol photons m−2 s−1. Higher light intensities did not bring about any further increase in the rate of photosynthesis (Fig. 2). The light-saturated rate of photosynthesis minus respiration is defined as Pmax, equal to about 106 mmol oxygen (mol Chl)−1 s−1 in the wild type (Table II). The photosynthetic activity of the tla3 mutant continued to increase beyond the 500 μmol photons m−2 s−1 intensity and did not quite saturate, even at light intensities corresponding to full sunlight at about 2,000 μmol photons m−2 s−1. For the purposes of this work, Pmax in the tla3 mutant was defined as the rate of oxygen evolution achieved at 2,000 μmol photons m−2 s−1 [Pmax = 210 mmol oxygen (mol Chl)−1 s−1]. The rate of photosynthesis in the light-saturation measurement is based on the Chl content of the samples. Hence, Pmax provides a measurement of the productivity of Chl in the two strains. On this basis, tla3 outperforms the wild type by about 100% [i.e. 210 compared with 106 mmol oxygen (mol Chl)−1 s−1 for tla3 and the wild type, respectively; Table II].
The half-saturation intensity for the wild type was determined to be about 200 μmol photons m−2 s−1, whereas the half-saturation intensity for tla3 was estimated to be greater than 600 μmol photons m−2 s−1. Because of the reciprocal relationship between the half-saturation intensity and the size of the light-harvesting antenna of both photosystems, it can be estimated that the size of the light-harvesting antenna of the photosystems in the tla3 mutant is about one-third of that in the wild type. This observation suggests that there is a substantially smaller Chl antenna size of the photosystems in the mutant.
When measured to a per cell basis, wild-type cells respired about 56 attomoles (amol) of oxygen per second, while the tla3 mutant consumed only 21 amol of oxygen per second. This phenomenon of a lower respiration rate per cell for tla-type mutants has been observed previously with the tla2 ΔCpFTSY strain (Kirst et al., 2012) and may be explained as a consequence of the slower supply of respiratory substrate in these mutants. Consistent with this interpretation is the light-saturated photosynthetic activity of tla3, which was only about 35% of that in the wild type (Pcell = 69 amol oxygen cell−1 s−1 for tla3 compared with 196 amol oxygen cell−1 s−1 for the wild type). This observation suggests fewer thylakoid membranes and/or photosynthetic electron transport chains in the mutant. An independent and more direct measurement of the abundance of electron transport chains was obtained from the quantification of PSII (QA) and PSI (P700) in the wild type and the tla3 mutant (Table II). The amount of PSII was lowered from 6.5 in the wild type to 1.7 amol per cell in the mutant. Similarly, the amount of PSI was lowered from 4.6 amol per cell in the wild type to 1.8 amol per cell in the mutant. These measurements clearly show that tla3 cells possess only about 35% of photosynthetic electron transport chains when compared with the wild type.
To measure precisely the Chl molecules functionally associated with each photosystem, the kinetic-spectrophotometric method of Melis (1989) was applied. The number of Chl molecules associated with the reaction center of PSI and PSII was measured in cells grown photoautotrophically under medium light intensity (450 μmol photons m−2 s−1). The wild type showed biphasic PSII conversion kinetics attributed to the two PSII populations, PSIIα and PSIIβ (Melis and Homann, 1976; Melis and Duysens, 1979), having a large and a smaller light-harvesting Chl antenna size, respectively. The Chl (a and b) molecules associated with these two forms of PSII were measured to be 333 and 119, respectively (Table II). It was further determined that the ratio of PSIIα to PSIIβ was about 60:40 in the wild type. Thus, the average number of Chl molecules associated with PSII in the wild type was calculated to be 245, consistent with earlier measurements from this laboratory (Polle et al., 2003). The PSII conversion kinetics of the tla3 strain showed a single QA reduction phase; thus, only one type of PSII unit is present in the tla3 mutant. The functional light-harvesting Chl antenna size of PSII in the tla3 strain was measured to be 95 Chl molecules, which is about 38% of the wild-type Chl antenna size. The light-harvesting antenna of PSI contained 230 and 120 Chl molecules for the wild type and the tla3 mutant, respectively, a reduction to about 50% of the wild-type antenna (Table II).
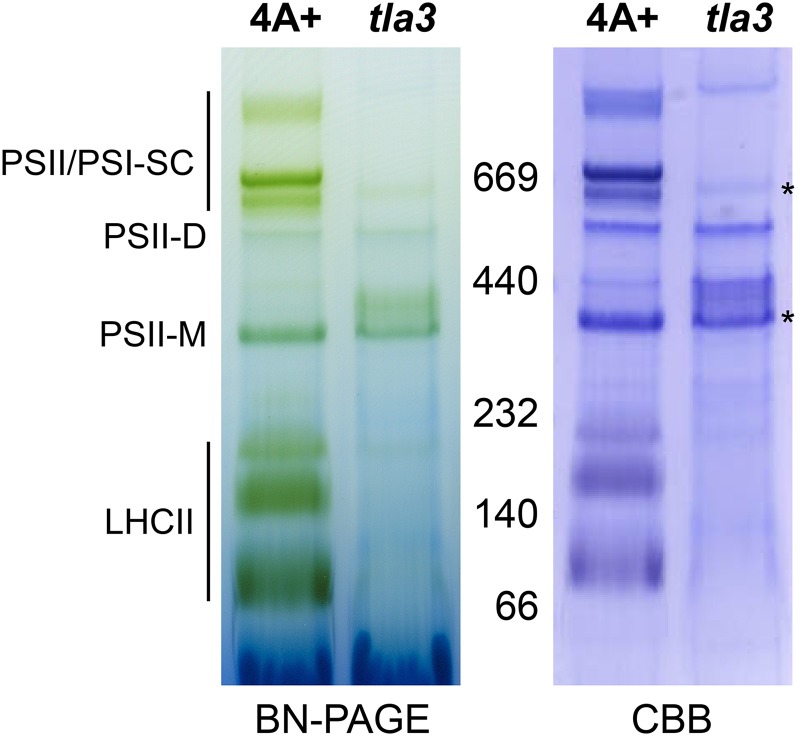
A comparative thylakoid membrane protein profile analysis of the wild type and the tla3 mutant was undertaken, aiming to identify biochemical and functional differences between the two strains. For an initial resolution of the integral and hydrophobic components of the thylakoid membrane, holocomplexes were separated via nondenaturing PAGE (Fig. 3). Comparative analysis of the green profile of the wild type and the tla3 mutant (Fig. 3, left panels) showed a substantially diminished amount of the Chl a/b proteins and some reduction in the amount of the PSI complexes. The same conclusion was drawn from the Coomassie blue-stained blue-native gel of wild-type thylakoid membrane proteins compared with those of the tla3 mutant (Fig. 3, right panels).
Figure 3.
Analysis of the C. reinhardtii wild type and tla3 mutant thylakoid membrane protein complexes by blue-native (BN) PAGE and Coomassie Brilliant Blue (CBB) staining. Solubilization of thylakoid membranes was performed in the presence of 1% n-dodecyl-β-d-maltoside and a Chl concentration of 1 or 0.25 mg mL−1 for the wild type or the tla3 mutant, respectively. Identification of the thylakoid membrane protein complexes was done according to Dewez et al. (2009). PSII/PSI-SC, PSII-LHCII and PSI-LHCI supercomplexes; PSII-D, PSII dimer; PSII-M, PSII monomer; LHCII, Chl a/b light-harvesting complex. Note the absence of peripheral Chl-containing light-harvesting antenna from isolated tla3 mutant thylakoids, whereas reaction center (PSII and PSI) complexes were clearly present (marked by asterisks). [See online article for color version of this figure.]
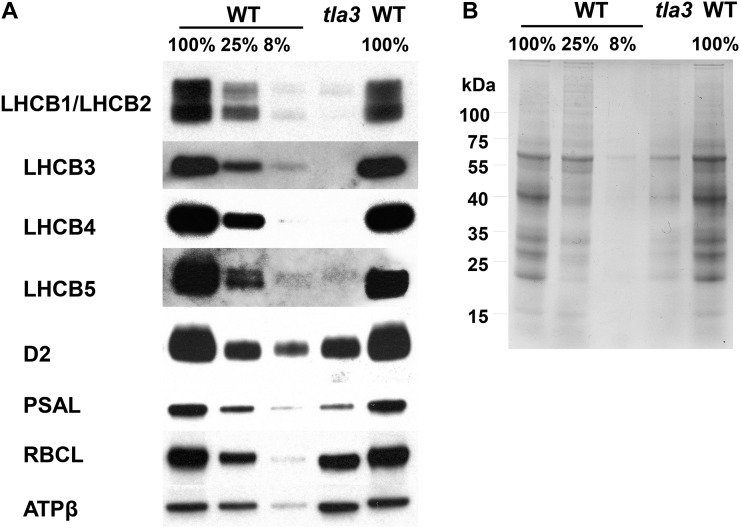
To further investigate the abundance of the proteins associated with the Chl a/b light-harvesting complex in the wild type and the tla3 mutant, western-blot analysis of total cell protein extracts was undertaken. C. reinhardtii proteins were probed with specific polyclonal antibodies raised against the Arabidopsis (Arabidopsis thaliana) light-harvesting proteins 1 to 5 (Fig. 4). For better comparison of the results, the wild-type lanes were loaded in three different concentrations, equal cell basis (100%), 25% cells (about equal reaction center basis), and 8% cells. Antibodies against the major light-harvesting proteins LHCB1 and LHCB2 cross-reacted with corresponding proteins in C. reinhardtii. The abundance of the latter was lower to about 5% to 10% in the tla3 mutant as compared with the wild type. The protein band cross-reacting with anti-LHCB3 was not detectable in the tla3 mutant. The protein bands cross-reacting with the minor light-harvesting antenna proteins LHCB4 and LHCB5 were both reduced to about 5% or less in the tla3 mutant. The psbD/D2 PSII reaction center protein was lowered to about 35% in the mutant relative to the wild type. The PSI reaction center protein psaL was also lower in abundance to about 20%. These results clearly showed that loss of the light-harvesting proteins is proportionally greater than the loss of reaction centers in the mutant, confirming the truncated light-harvesting antenna size phenotype at the protein level. The abundance of the large subunit of Rubisco and the β-subunit of ATP synthase (ATPβ) were not affected by the mutation (Fig. 4).
Figure 4.
Western-blot analysis of the light-harvesting antenna proteins of PSII in the wild type (WT) and the tla3 mutant. A, Immunodetection of proteins with specific polyclonal antibodies against the light-harvesting proteins LHCB1/LHCB2, LHCB3, LHCB4, and LHCB5, the PSII reaction center protein D2, the PSI reaction center protein PSAL, Rubisco (RBCL), and the β-subunit of ATP synthase (ATPβ). B, Coomassie blue-stained SDS-PAGE analysis of the samples shown in A.
Collectively, the above analysis showed that the tla3 strain has a bona fide truncated Chl antenna size, raising the question of the gene(s) interrupted in this phenotype. Accordingly, a gene-cloning analysis was undertaken.
Flanking Sequence Analysis of Plasmid Insertion in the tla3 Mutant
Southern-blot analysis of the tla3 genomic DNA showed that a single pJD67 plasmid was inserted into the genomic DNA of the tla3 mutant (Supplemental Fig. S1). For cloning the flanking sequence of the insertion, the structure and integrity of the inserted plasmid was mapped by PCR to an accuracy of 100 bp (Supplemental Fig. S2). This analysis revealed a break point in the pJD67 plasmid, which occurred 650 to 750 bp within the ARG7 gene promoter. On the basis of the above findings, thermal asymmetric interlaced (TAIL)-PCR was applied with a beginning point located about 100 bp upstream of the break point in the ARG7 promoter of the inserted plasmid. In this approach, a mix of two products is expected, one from the endogenous ARG7 promoter and one from the pJD67 sequence. After cloning the PCR products into Escherichia coli, 50 clones were sequenced to determine if any of them contained C. reinhardtii genomic DNA sequence flanking the pJD67 insertion. However, all clones sequenced contained exclusively sequences flanking the endogenous ARG7 promoter but not that of the insertion. This negative result could be attributed to a greater efficiency of amplifying the flanking sequence of the endogenous ARG7 promoter but not that of the insert. To increase the chances of cloning the flanking sequence of the plasmid insertion site, the DNA adapter method was employed (Supplemental Fig. S3). The insertion site was thus localized on chromosome 4, occurring within the fourth intron of predicted gene g5046 (Fig. 5). PCR analysis was employed to screen the insertion site for possible deletions. This analysis revealed that a predicted gene immediately upstream, namely g5047, was deleted in the tla3 mutant. Bioinformatic analysis showed that predicted gene g5047 has homology to the CpSRP43 gene in higher plants. Similarly, gene g5046 was putatively assigned oxidation/reduction properties and might be involved in cell redox homeostasis.
Figure 5.
Map of the insertion site in the genomic DNA of the tla3 mutant. The gray areas are deleted in the tla3 mutant, including the g5047 CrCpSRP43 homolog and a portion of the g5046 gene. [See online article for color version of this figure.]
Complementation of the tla3 Mutant
The complementary DNA (cDNA) of genes g5046 and g5047 was cloned into the C. reinhardtii overexpression vector pSL18. After KpnI linearization, the constructs were transformed separately into the tla3 strain. The pSL18-g5046 construct failed to rescue the mutation, as it did not generate any complemented strains. The pSL18-g5047 construct yielded several complemented strains with various degrees of recovery of the dark-green coloration, ranging anywhere from dark-green wild-type coloration to pale-green tla3-type coloration (Fig. 6). On the basis of the successful complementation of the tla3 mutant with the g5047 gene, it was concluded that the tla3 mutant phenotype was caused by a knockout of the C. reinhardtii homolog of the CpSRP43 gene known from higher plants (Amin et al., 1999; Klimyuk et al., 1999). Four complemented tla3 lines were selected randomly to measure pigment content and composition properties. All complemented lines, namely C1 through C4, showed various degrees of restoration of the Chl a/Chl b ratio, as they also showed a significantly greater amount of pigmentation per cell than the uncomplemented tla3 mutant (Table I). However, none of the four selected complemented lines reached as high of a Chl content and as low of a Chl a/Chl b ratio as the wild type. The most promising of the complemented strains, C2, accumulated Chl on a per cell basis equal to only about 60% of that in the wild type.
Figure 6.
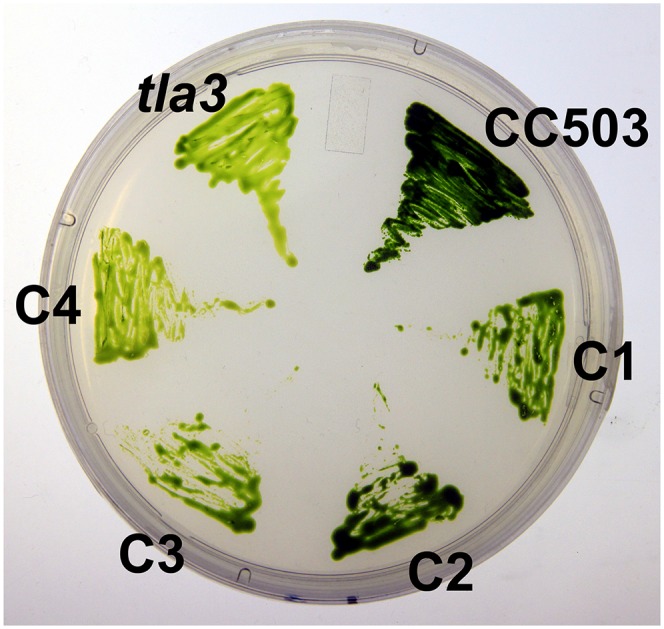
Colonies of C. reinhardtii wild-type CC503, the tla3 mutant, and the CrCpSRP43 complemented lines C1 through C4 grown on agar. Note the dark coloration of the wild type, the pale coloration of the tla3 mutant, and the intermediate phenotypes of the complemented strains. [See online article for color version of this figure.]
Western-blot analyses of the wild type, the tla3 mutant, and complemented strains were conducted with specific polyclonal antibodies raised against a synthetic peptide of the TLA3-CpSRP43 protein. A strong cross-reaction was observed between antibodies and a protein band migrating to about 43 kD in the wild type (Fig. 7). This cross-reaction was missing from the protein extracts of the tla3 mutant (Fig. 7, tla3) but was recovered to various degrees in the complemented strains (Fig. 7, C2). These results clearly show that the TLA3-CpSRP43 protein is responsible for the tla3 phenotype.
Figure 7.
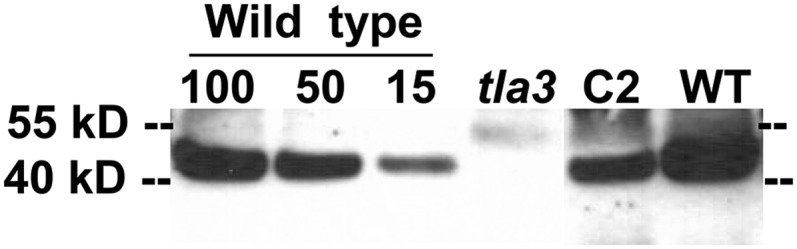
Western-blot analysis of total protein extracts from the C. reinhardtii wild type (WT), the tla3 mutant, and the CrCpSRP43 complemented line C2. For quantitative analysis, different amounts of wild-type protein were loaded (100%, 50%, and 15%). The corresponding loadings of the tla3 and C2 proteins were equivalent to 100% loading.
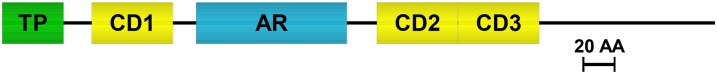
The genomic DNA of the CpSRP43 gene in C. reinhardtii from start to stop codon was found to be 4,988 bp long and to contain 12 exons. The gene encodes for a protein of 430 amino acids, of which the first 37 amino acids are predicted, on the basis of ChloroP (http://www.cbs.dtu.dk/services/ChloroP/; Emanuelsson et al., 1999) and also on the basis of TargetP (http://www.cbs.dtu.dk/services/TargetP/; Emanuelsson et al., 2007), to serve as a chloroplast-targeting sequence and transit peptide (Fig. 8). The mature protein of 393 amino acids contains four protein domains, determined by Pfam (http://pfam.sanger.ac.uk), three chromodomains, CD1 (amino acids 27–76), CD2 (amino acids 214–266), and CD3 (amino acids 305–324), and an ankyrin-repeat domain of three repeats (amino acids 80–171). It is of interest that Predalgo (http://giavap-genomes.ibpc.fr/predalgo; Tardif et al., 2012), a more recently described transit sequence-defining software, also predicted the TLA3 protein to be localized in the chloroplast but assigned a slightly longer transit peptide of 42 amino acids.
Figure 8.
The C. reinhardtii CpSRP43 protein domain structure. AA, Amino acids; AR, ankyrin domain; CD1, CD2, and CD3, chromodomains; TP, chloroplast transit peptide. [See online article for color version of this figure.]
DISCUSSION
The chloroplast signal recognition particle (SRP) is defined as a collection of four proteins that work together: CpSRP54, CpSRP43, CpFTSY, and ALB3 (Aldridge et al., 2009). The CpSRP component proteins are postulated to be involved in the proper folding of light-harvesting proteins and targeting of these proteins to the thylakoid membrane, thereby facilitating the biogenesis and assembly of the photosystem holocomplexes (Pilgrim et al., 1998; Klimyuk et al., 1999). In a temporal sequence of events, it is postulated that CpSRP43 primarily, and CpSRP54 secondarily, operate in the chloroplast stroma, where they bind to hydrophobic domains of imported LHC proteins, thereby preventing them from misfolding and/or precipitating from the aqueous phase. The receptor CpFTSY protein recognizes the CpSRP54-CpSRP43-LHC protein complex and guides the complex to the integral thylakoid membrane protein ALB3. The latter facilitates incorporation of the transmembrane domains of the LHC protein into the thylakoid membrane. The chloroplast signal recognition particle proteins CpSRP43 and CpSRP54 are predicted to exist in green algae based on DNA sequence analysis, but they have not yet been experimentally verified or their function determined. The corresponding receptor protein CpFTSY was recently identified in C. reinhardtii and its unique effect on the assembly of the Chl a/b light-harvesting complex defined (Kirst et al., 2012).
This work showed that the tla3 locus of C. reinhardtii encodes for the chloroplast-localized light-harvesting chaperone protein known as CpSRP43. This annotation for the tla3 locus was based on the successful complementation of the tla3 mutant with the cDNA construct of the CrCpSRP43 gene. This work provides, to our knowledge for the first time, experimental evidence for the occurrence and functional role of the CpSRP43 protein in green microalgae. The CpSRP43 protein of C. reinhardtii shares a sequence identity of 27.6% and a similarity of 38.3% with the Arabidopsis AtCpSRP43 protein. Even though these numbers are relatively high, the phenotype of the CpSRP43-null mutant differed between C. reinhardtii and Arabidopsis, at least in terms of the pigments accumulating in the chloroplast. The Chl content dropped to about 15% of that of the wild type in C. reinhardtii, while Arabidopsis retained 50% of its Chl content (Amin et al., 1999; Klimyuk et al., 1999). Consequently, the CpSRP43-null mutation in microalgae appears to have a more severe effect on the assembly of the light-harvesting proteins, which are nearly absent in the tla3 mutant, while their abundance is not so adversely affected in the corresponding Arabidopsis mutant (Amin et al., 1999; Klimyuk et al., 1999).
The CrCpSRP43 protein has a similar domain configuration to that of the AtCpSRP43 protein. Both proteins contain an ankyrin-repeat domain and three chromodomains, one in front of the ankyrin domain and two immediately after it. Ankyrin repeats are among the most common protein motifs and function in protein-protein interactions (Mosavi et al., 2004). The ankyrin repeats are responsible for binding the L18 motif of light-harvesting apoproteins (DeLille et al., 2000; Tu et al., 2000; Stengel et al., 2008), thereby preventing a misfolding or precipitation of the hydrophoblic LHCB and LHCA apoproteins in the stroma phase (Falk and Sinning, 2010). Denaturation of the LHC proteins would be a likely outcome in the absence of this molecular chaperone and could occur as the nascent LHCB and LHCA apoproteins are imported into the aqueous stroma phase of the chloroplast. The chromodomains are commonly found in proteins that function to manipulate or remodel chromatin. However, Goforth et al. (2004) and Funke et al. (2005) showed that CD2 is involved in the binding of CpSRP43 to CpSRP54 to form the signal recognition particle. Due to the high similarity of the AtCpSRP43 and CrCpSRP43 proteins, and their similar domain structure, it can be inferred that they perform similar functions.
It is of interest that rates of cellular respiration in the tla3 mutant were substantially lower, on a per cell basis down to about 30%, of those measured in the wild type. A truncated Chl antenna in the photosynthetic apparatus should not a priori affect the cell’s respiration capacity (Polle et al., 2000, 2003). Rather, loss of respiration fitness in the tla3 mutant could be attributed to the loss or modification of genes flanking the pJD67 insertion site(s) in the genomic DNA (Kirst et al., 2012). The possibility could not be excluded that one of the affected genes in the tla3 mutant adversely impacted properties of respiration, PSII and PSI content, and/or cell size. These anomalies may contribute to the slower cellular respiration rate. An additional consideration is that tla strains, by necessity due to their diminished ability to absorb enough light, cannot generate photosynthate in sufficient quantities to saturate the respiratory rate of the cells, thus attenuating the respiratory capacity of the mutants.
The lower photosystem content of tla3 compared with the wild type is an indication of overall lower thylakoid membrane development and abundance in the chloroplast of the mutants. Inability to assemble the imported light-harvesting proteins in these tla mutants may trigger a feedback inhibition in light-harvesting protein and Chl biosynthesis, indirectly affecting the Chl supply and lowering the chloroplast ability to assemble the full complement of PSII and PSI units. Another possible consideration is that a deficiency of light-harvesting proteins may have had an adverse effect on thylakoid morphology. It has been proposed that the light-harvesting proteins are needed to form the grana stacking in the appressed thylakoid membranes (for review, see Allen and Forsberg, 2001). Absence of the light-harvesting proteins in the tla3 mutants would thus prevent grana stacking, substantially lowering the thylakoid membrane surface area in which PSII can be placed. The amount of PSI is by consequence negatively adjusted to meet the needs of the chloroplast in terms of an efficient and coordinated electron transport through the thylakoid membranes.
In spite of the total absence of the CpSRP43, the tla3 mutant retained some LHC assembly activity for the photosystem inner light-harvesting proteins. It is possible that CpSRP54 alone is sufficient to form a complex with imported LHC apoproteins and further interact with the CpFTSY and ALB3 translocase (Tzvetkova-Chevolleau et al., 2007; Bals et al., 2010), explaining the observation that some light-harvesting antenna proteins are incorporated into the thylakoid membrane of the tla3 strain, albeit with a much lower efficiency. In this respect, Moore et al. (2003) showed that CpSRP43 is not required for the interaction of CpSRP54-CpFTSY with the insertase protein ALB3. This observation could explain how some light-harvesting complexes are integrated into the thylakoid membrane.
An important consideration is the thylakoid membrane domain, where such assembly takes place. We propose that the CpSRP complex is specifically localized and functions in the “polar” regions of the chloroplast, where thylakoid membrane biogenesis takes place. This suggestion is based on the observation that photosystem complexes and their antenna size cannot be altered once assembly has occurred and once transmembrane complexes become components of the developed thylakoid membrane (Melis, 1991; Kim et al., 1993). According to this concept, it is in the polar regions of the chloroplast that the simultaneous assembly of the photosystems, their antenna size, and formation of the thylakoid membrane take place. The nascent thylakoid membrane receives via the ALB3 protein the CpSRP43-CpSRP54-CpFTSY-LHC complex and guides the transmembrane domains of the LHC in the developing thylakoid membrane lipid bilayer. Upon GTP hydrolysis, the LHC protein is integrated into the thylakoid membrane (Tu et al., 1999). The CpSRP proteins CpSRP54 and ALB3 are apparently needed for the proper integration of other transmembrane proteins, as is evident by the pleiotropic phenotype generated in the corresponding knockout mutants (Amin et al., 1999; Bellafiore et al., 2002).
MATERIALS AND METHODS
Cell Cultivation
Chlamydomonas reinhardtii strains CC-503 cw92 mt+, CC-425 arg2 cw15 sr-u-2-60 mt+, and CC-125 wild type mt+ 137C, obtained from the Chlamydomonas Center (http://www.chlamy.org/), and laboratory strains 4A+ and tla3 were maintained under orbital shaking in 100-mL liquid cultures in Erlenmeyer flasks at 25°C under continuous illumination at low light (50 μmol photons m−2 s−1). Irradiance was provided by balanced cool-white and warm-white fluorescent lamps. Cells were grown photoheterotrophically in Tris-acetate phosphate medium (Gorman and Levine, 1965) or photoautotrophically in high-salt medium (Harris, 1989) under a combination of cool-white, warm-white fluorescent, and incandescent irradiance at a light intensity of 450 μmol photons m−2 s−1. For physiological measurements, cultures were harvested during the logarithmic growth phase (approximately 1–3 × 106 cells mL−1).
Cell Count and Pigment Quantification
Cell density was measured using an improved Neubauer ultraplane hemacytometer and a BH-2 light microscope (Olympus). Pigments from intact cells or thylakoid membranes were extracted in 80% acetone, and cell debris was removed by centrifugation at 20,000g for 5 min. The absorbance of the supernatant was measured with a Shimadzu UV-1800 spectrophotometer, and the Chl concentration of the samples was determined according to Arnon (1949), with equations corrected as described by Melis et al. (1987). Total carotenoid content was determined according to the method of Lichtenthaler (1987).
Mutagenesis and Screening Protocols
Mutants of C. reinhardtii were obtained upon DNA insertional mutagenesis and transformation with plasmid DNA by the glass-bead method, as described in Debuchy et al. (1989). Parental strain CC-425, an Arg auxotroph, was transformed with 1 μg of HindIII-linearized plasmid pJD67, containing the structural gene (ARG7) of the argininosuccinate lyase to complement the Arg-requiring phenotype of the CC-425 strain (Davies et al., 1996). Wild-type strain CC-503 was transformed with 0.5 μg of KpnI-linearized pBC1 plasmid (a gift from the laboratory of Dr. Kris Niyogi) conferring paromomycin resistance. Transformants were selected on Tris-acetate phosphate-only medium and initially screened upon measurement of the Chl a/b ratio of the strains, following extraction of Chl with 80% acetone. A BioTek Epoch spectrophotometer equipped with a 96-well plate reader was used in these measurements.
Nucleic Acid Extractions
C. reinhardtii genomic DNA was isolated for PCR analysis using Qiagen’s Plant DNA purification kit. For Southern-blot analysis, genomic DNA was isolated by harvesting cells from a 50-mL aliquot of the culture upon centrifugation at 5,000g for 5 min, followed by resuspension of the pellet in 500 μL of sterile water. Cells were lysed upon addition of 500 μL of lysis buffer containing 2% SDS, 400 mm NaCl, 40 mm EDTA, and 100 mm Tris-HCl (pH 8.0) and upon incubation for 2 h at 65°C. To this mix, 170 μL of 5 m NaCl solution was added. SDS and carbohydrates were precipitated upon addition of 135 μL of 10% cetyl-trimethyl-ammonium bromide in 0.7 m NaCl and incubation for 10 min at 65°C. They were extracted by mixing with chloroform:isoamylalcohol (24:1) followed by centrifugation at 20,000g for 5 min. Proteins were removed by extraction with phenol:chloroform:isoamylalcohol (25:24:1) solution. RNA in the water phase was digested using 10 ng mL−1 RNase A and upon incubation at 37°C for 20 min. RNase A was removed by extraction with phenol:chloroform:isoamylalcohol (25:24:1) solution. Desalting of the DNA solution was achieved upon precipitation with isopropanol and resuspension in 10 mm Tris-HCl (pH 8.0).
Southern-Blot Analysis
Approximately 5 μg of genomic DNA was digested by various restriction enzymes (New England Biolabs) in a 500-μL volume at 37°C with overnight incubation (16 h). The digested DNA was precipitated with isopropanol, washed in 70% ethanol, and resuspended in 20 μL of buffer containing 5 mm Tris, pH 8.0. DNA fragments were separated on a 0.6% agarose gel, transferred on a positively charged nylon membrane (Hybond-N+; Amersham), and UV cross linked. Probes were obtained upon PCR using specific primers (Supplemental Table S1) and the pJD67 plasmid as template DNA and labeled with alkaline phosphatase using the Gene Images AlkPhos Direct Labeling and Detection System kit (Amersham). The manufacturer’s protocol was used for labeling, hybridization, washing, and signal detection with the following modifications: hybridization temperature and primary washing buffer temperature were maintained at 72°C.
Measurements of Photosynthetic Activity
The oxygen evolution activity of the cultures was measured at 25°C with a Clark-type oxygen electrode illuminated with light from a halogen lamp projector. A Corning 3-69 filter (510-nm cutoff filter) defined the yellow actinic excitation via which photosynthesis measurements were made. Samples of 5-mL cell suspension containing 1.3 μm Chl were loaded into the oxygen electrode chamber. Sodium bicarbonate (100 μL of 0.5 m solution, pH 7.4) was added to the cell suspension prior to the oxygen evolution measurements to ensure that oxygen evolution was not limited by the carbon supply available to the cells. After registration of the rate of dark respiration by the cells, samples were illuminated with gradually increasing light intensities. The rate of oxygen exchange (uptake or evolution) under each of these irradiance conditions was recorded continuously for a period of about 5 min.
Isolation of Thylakoid Membranes
Cells were harvested by centrifugation at 1,000g for 3 min at 4°C. Samples were resuspended with ice-cold sonication buffer containing 50 mm Tricine (pH 7.8), 10 mm NaCl, 5 mm MgCl2, 0.2% polyvinylpyrrolidone 40, 0.2% sodium ascorbate, 1 mm aminocaproic acid, 1 mm aminobenzamidine, and 100 μm phenylmethylsulfonyl fluoride. Cells were broken by sonication in a Branson 250 Cell Disrupter operated at 4°C three times for 30 s each time (pulse mode, 50% duty cycle, output power of 5) with 30-s cooling intervals on ice. Unbroken cells and starch grains were removed by centrifugation at 3,000g for 4 min at 4°C. Thylakoid membranes were collected by centrifugation of the first supernatant at 75,000g for 30 min at 4°C. The thylakoid membrane pellet was resuspended in a buffer containing 50 mm Tricine (pH 7.8), 10 mm NaCl, and 5 mm MgCl2 for spectrophotometric measurements or 250 mm Tris-HCl (pH 6.8), 20% glycerol, 7% SDS, and 2 m urea for protein analysis.
Spectrophotometric and Kinetic Analyses
The concentration of the photosystems in thylakoid membranes was measured spectrophotometrically from the amplitude of the light-minus-dark absorbance difference signal at 700 nm (P700) for PSI and 320 nm (QA) for PSII (Melis and Brown, 1980; Melis, 1989; Smith et al., 1990). The functional light-harvesting Chl antenna size of PSI and PSII was measured from the kinetics of P700 photooxidation and QA photoreduction, respectively (Melis, 1989).
5′ and 3′ RACE Analysis
Total RNA was isolated from CC-503 cells in the early log phase of growth (0.5 × 106 cells mL−1) using the Trizol reagent (Invitrogen). Genomic DNA in these samples was digested according to the protocol provided by the Turbo DNA-free kit (Ambion). The 5′ and 3′ RACE analyses were done with the FirstChoice RLM-RACE kit (Ambion) using primers as indicated (Supplemental Table S1). The manufacturer’s protocol was followed in all procedures.
Transformation of C. reinhardtii
CpSRP43 cDNA was cloned into pSL18 and incorporated into the genomic DNA of the tla3 mutant using the conventional glass-bead transformation protocol (Kindle, 1990). pSL18 contains a paromomycin resistance gene (selectable marker) operated under the control of the C. reinhardtii Hsp70A and RbcS2 promoters (Sizova et al., 2001) and linked to the PsaD promoter and terminator that was used to express the CpSRP43 gene. Transformants were isolated upon screening independent cell lines on the basis of coloration and the Chl a/Chl b ratio of the cells.
Analysis of Genomic DNA Flanking the Plasmid Insert Site
In the effort to isolate C. reinhardtii genomic DNA flanking the plasmid insertion site, a TAIL-PCR protocol was used (Liu et al., 1995), optimized for C. reinhardtii genomic DNA, as described recently (Dent et al., 2005). Primers used for the TAIL-PCR are listed in Supplemental Table S1. Briefly, flanking genomic DNA was amplified by PCR from the region adjacent to the inserted pJD67 plasmid that was used for DNA insertional mutagenesis. Specific primers for primary, secondary, and tertiary reactions were designed (Supplemental Table S1). Two arbitrary degenerate primers were tested for amplification, HK030 and HK031, as described previously (Dent et al., 2005). The general TAIL-PCR protocol of Liu et al. (1995) was used with minor modifications for the various PCR amplification reactions. Nucleotide sequences of the resulting PCR products were obtained via an ABI3100 sequence analyzer. The adapter method was used to isolate C. reinhardtii genomic DNA flanking the plasmid insertion. tla3 was digested by XmaI (New England Biolabs), and fragments between 3 and 5 kb were purified after gel electrophoresis using the QIAquick Gel Extraction Kit (Qiagen) following the manufacturer’s protocol. The adapter (Supplemental Data S1) was ligated using a T4 DNA ligase (New England Biolabs). Nested PCR (Supplemental Data S1) was used to specifically amplify the sequence flanking the plasmid insertion. C. reinhardtii genomic DNA sequence information was obtained from the Chlamydomonas Genome Project Web site: http://phyto5.phytozome.net/chlamy.php.
Western-Blot Analysis
Specific polyclonal antibodies were generated against a synthetic peptide from the CpSRP43 protein (ProSci). The following amino acid sequence served as antigen: CERRFKIRWSDGYPTSWEPEE. Western-blot analyses were performed with total protein from cell extracts, resolved in precast SDS-PAGE Any KD (Bio-Rad). Loading of samples was based on equal cell count. SDS-PAGE-resolved proteins were transferred to a polyvinylidene difluoride membrane (Immobilon-FL 0.45 µm; Millipore) by a tank transfer system. Specific polyclonal antibodies raised against LHCB(1-5), and PSAL from Arabidopsis (Arabidopsis thaliana) and D2, Rubisco, and ATPβ from C. reinhardtii were visualized by the Supersignal West Pico Chemiluminescent substrate detection system (Thermo Scientific). NIH Image 1.62 software was employed for the deconvolution and quantification of the western-blot bands.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number XP_001703704.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Southern blot analysis to define copy number and integrity of inserted pJD67 plasmid into the genomic DNA of the tla3 C. reinhardtii insertional transformant.
Supplemental Figure S2. Genomic DNA PCR analysis to map the plasmid insertion site in the tla3 genomic DNA.
Supplemental Figure S3. Strategy for the cloning of the genomic DNA sequence flanking the tla3 insertion site using the adapter method.
Supplemental Table S1. Primers used in this work
Supplemental Data S1. Flanking sequence analysis of the insertion in the tla3 mutant.
Acknowledgments
We thank Dr. Kris Niyogi for the AG1x3.24 (arg2) strain of C. reinhardtii and for providing the pBC1 plasmid employed in this work, Dr. Jürgen Polle and Dr. Sarada Kanakagiri, as well as Mr. Ian McRae and Ms. Carine Marshall, all of whom contributed to the screening of putative tla strains in the Melis laboratory.
Glossary
- Chl
chlorophyll
- TAIL
thermal asymmetric interlaced
- cDNA
complementary DNA
- amol
attamoles
References
- Aldridge C, Cain P, Robinson C. (2009) Protein transport in organelles: protein transport into and across the thylakoid membrane. FEBS J 276: 1177–1186 [DOI] [PubMed] [Google Scholar]
- Allen JF, Forsberg J. (2001) Molecular recognition in thylakoid structure and function. Trends Plant Sci 6: 317–326 [DOI] [PubMed] [Google Scholar]
- Amin P, Sy DAC, Pilgrim ML, Parry DH, Nussaume L, Hoffman NE. (1999) Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol 121: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. (1949) Copper enzymes in isolated chloroplasts: polyphenol oxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bals T, Dünschede B, Funke S, Schünemann D. (2010) Interplay between the cpSRP pathway components, the substrate LHCP and the translocase Alb3: an in vivo and in vitro study. FEBS Lett 584: 4138–4144 [DOI] [PubMed] [Google Scholar]
- Bellafiore S, Ferris P, Naver H, Göhre V, Rochaix JD. (2002) Loss of Albino3 leads to the specific depletion of the light-harvesting system. Plant Cell 14: 2303–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée D, Delhaye S, Gruyer S, Slomianny C, Moretz SE, d’Hulst C, Long CA, Ball SG, Tomavo S. (2010) Engineering the chloroplast targeted malarial vaccine antigens in Chlamydomonas starch granules. PLoS ONE 5: e15424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Yildiz FH, Grossman AR. (1996) Sac1, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO J 15: 2150–2159 [PMC free article] [PubMed] [Google Scholar]
- Debuchy R, Purton S, Rochaix JD. (1989) The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J 8: 2803–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLille J, Peterson EC, Johnson T, Moore M, Kight A, Henry R. (2000) A novel precursor recognition element facilitates posttranslational binding to the signal recognition particle in chloroplasts. Proc Natl Acad Sci USA 97: 1926–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK. (2005) Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol 137: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewez D, Park S, García-Cerdán JG, Lindberg P, Melis A. (2009) Mechanism of REP27 protein action in the D1 protein turnover and photosystem II repair from photodamage. Plant Physiol 151: 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S, Sinning I. (2010) cpSRP43 is a novel chaperone specific for light-harvesting chlorophyll a,b-binding proteins. J Biol Chem 285: 21655–21661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke S, Knechten T, Ollesch J, Schünemann D. (2005) A unique sequence motif in the 54-kDa subunit of the chloroplast signal recognition particle mediates binding to the 43-kDa subunit. J Biol Chem 280: 8912–8917 [DOI] [PubMed] [Google Scholar]
- Goforth RL, Peterson EC, Yuan J, Moore MJ, Kight AD, Lohse MB, Sakon J, Henry RL. (2004) Regulation of the GTPase cycle in post-translational signal recognition particle-based protein targeting involves cpSRP43. J Biol Chem 279: 43077–43084 [DOI] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. (1965) Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 54: 1665–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell HC, Laurens LML, Shields RJ, Lovitt RW, Flynn KJ. (2010) Placing microalgae on the biofuels priority list: a review of the technological challenges. J R Soc Interface 7: 703–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankamer B, Lehr F, Rupprecht J, Mussgnug JH, Posten C, Kruse O. (2007) Photosynthetic biomass and H2 production by green algae: from bioengineering to bioreactor scale-up. Physiol Plant 131: 10–21 [DOI] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego
- Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A. (2008) Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J 54: 621–639 [DOI] [PubMed] [Google Scholar]
- Kim JH, Nemson JA, Melis A. (1993) Photosystem II reaction center damage and repair in the green alga Dunaliella salina: analysis under physiological and adverse irradiance conditions. Plant Physiol 103: 181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL. (1990) High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 87: 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk JTO. (1994) Light and Photosynthesis in Aquatic Ecosystems, Ed 2. Cambridge University Press, Cambridge, UK
- Kirst H, García-Cerdán JG, Zurbriggen A, Melis A. (2012) Assembly of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii requires expression of the TLA2-CpFTSY gene. Plant Physiol 158: 930–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimyuk VI, Persello-Cartieaux F, Havaux M, Contard-David P, Schuenemann D, Meiherhoff K, Gouet P, Jones JD, Hoffman NE, Nussaume L. (1999) A chromodomain protein encoded by the Arabidopsis CAO gene is a plant-specific component of the chloroplast signal recognition particle pathway that is involved in LHCP targeting. Plant Cell 11: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Mata TM, Martins AA, Caetano NS. (2010) Microalgae for biodiesel production and other applications: a review. Renew Sustain Energy Rev 14: 217–232 [Google Scholar]
- Mayfield SP, Manuell AL, Chen S, Wu J, Tran M, Siefker D, Muto M, Marin-Navarro J. (2007) Chlamydomonas reinhardtii chloroplasts as protein factories. Curr Opin Biotechnol 18: 126–133 [DOI] [PubMed] [Google Scholar]
- Melis A. (1989) Spectroscopic methods in photosynthesis: photosystem stoichiometry and chlorophyll antenna size. Philos Trans R Soc Lond B 323: 397–409 [Google Scholar]
- Melis A. (1991) Dynamics of photosynthetic membrane composition and function. Biochim Biophys Acta 1058: 87–106 [Google Scholar]
- Melis A. (2007) Photosynthetic H2 metabolism in Chlamydomonas reinhardtii (unicellular green algae). Planta 226: 1075–1086 [DOI] [PubMed] [Google Scholar]
- Melis A. (2009) Solar energy conversion efficiencies in photosynthesis: minimizing the chlorophyll antennae to maximize efficiency. Plant Sci 177: 272–280 [Google Scholar]
- Melis A. (2012) Photosynthesis-to-fuels: from sunlight to hydrogen, isoprene, and botryococcene production. Energy Environ Sci 5: 5531–5539 [Google Scholar]
- Melis A, Brown JS. (1980) Stoichiometry of system I and system II reaction centers and of plastoquinone in different photosynthetic membranes. Proc Natl Acad Sci USA 77: 4712–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A, Duysens LNM. (1979) Biphasic energy conversion kinetics and absorbance difference spectra of PS II of chloroplasts: evidence for two different PS II reaction centers. Photochem Photobiol 29: 373–382 [Google Scholar]
- Melis A, Homann PH. (1976) Heterogeneity of the photochemical centers in system II of chloroplasts. Photochem Photobiol 23: 343–350 [DOI] [PubMed] [Google Scholar]
- Melis A, Neidhardt J, Benemann JR. (1999) Dunaliella salina (Chlorophyta) with small chlorophyll antenna sizes exhibit higher photosynthetic productivities and photon use efficiencies than normally pigmented cells. J Appl Phycol 10: 515–525 [Google Scholar]
- Melis A, Spangfort M, Andersson B. (1987) Light-absorption and electron transport balance between photosystem-II and photosystem-I in spinach chloroplasts. Photochem Photobiol 45: 129–136 [Google Scholar]
- Michelet L, Lefebvre-Legendre L, Burr SE, Rochaix JD, Goldschmidt-Clermont M. (2011) Enhanced chloroplast transgene expression in a nuclear mutant of Chlamydomonas. Plant Biotechnol J 9: 565–574 [DOI] [PubMed] [Google Scholar]
- Mitra M, Melis A. (2008) Optical properties of microalgae for enhanced biofuels production. Opt Express 16: 21807–21820 [DOI] [PubMed] [Google Scholar]
- Moore M, Goforth RL, Mori H, Henry R. (2003) Functional interaction of chloroplast SRP/FtsY with the ALB3 translocase in thylakoids: substrate not required. J Cell Biol 162: 1245–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. (2004) The ankyrin repeat as molecular architecture for protein recognition. Protein Sci 13: 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Li XP, Niyogi KK. (2001) Non-photochemical quenching: a response to excess light energy. Plant Physiol 125: 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y, Ueda R. (1997) Improvement of photosynthesis in dense microalgal suspension by reduction of light harvesting pigments. J Appl Phycol 9: 503–510 [Google Scholar]
- Nakajima Y, Ueda R. (1999) Improvement of microalgal photosynthetic productivity by reducing the content of light harvesting pigments. J Appl Phycol 11: 195–201 [Google Scholar]
- Ort DR, Zhu X, Melis A. (2011) Optimizing antenna size to maximize photosynthetic efficiency. Plant Physiol 155: 79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim ML, van Wijk KJ, Parry DH, Sy DA, Hoffman NE. (1998) Expression of a dominant negative form of cpSRP54 inhibits chloroplast biogenesis in Arabidopsis. Plant J 13: 177–186 [DOI] [PubMed] [Google Scholar]
- Polle JEW, Benemann JR, Tanaka A, Melis A. (2000) Photosynthetic apparatus organization and function in the wild type and a chlorophyll b-less mutant of Chlamydomonas reinhardtii: dependence on carbon source. Planta 211: 335–344 [DOI] [PubMed] [Google Scholar]
- Polle JEW, Kanakagiri S, Jin E, Masuda T, Melis A. (2002) Truncated chlorophyll antenna size of the photosystems: a practical method to improve microalgal productivity and hydrogen production in mass culture. Int J Hydrogen Energy 27: 1257–1264 [Google Scholar]
- Polle JEW, Kanakagiri SD, Melis A. (2003) tla1, a DNA insertional transformant of the green alga Chlamydomonas reinhardtii with a truncated light-harvesting chlorophyll antenna size. Planta 217: 49–59 [DOI] [PubMed] [Google Scholar]
- Sizova IA, Fuhrmann M, Hegemann P. (2001) A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277: 221–229 [DOI] [PubMed] [Google Scholar]
- Smith BM, Morrissey PJ, Guenther JE, Nemson JA, Harrison MA, Allen JF, Melis A. (1990) Response of the photosynthetic apparatus in Dunaliella salina (green algae) to irradiance stress. Plant Physiol 93: 1433–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel KF, Holdermann I, Cain P, Robinson C, Wild K, Sinning I. (2008) Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43. Science 321: 253–256 [DOI] [PubMed] [Google Scholar]
- Tardif M, Atteia A, Specht M, Cogne G, Rolland N, Brugière S, Hippler M, Ferro M, Bruley C, Peltier G, et al. (2012) PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol Biol Evol (in press) [DOI] [PubMed] [Google Scholar]
- Tu CJ, Peterson EC, Henry R, Hoffman NE. (2000) The L18 domain of light-harvesting chlorophyll proteins binds to chloroplast signal recognition particle 43. J Biol Chem 275: 13187–13190 [DOI] [PubMed] [Google Scholar]
- Tu CJ, Schuenemann D, Hoffman NE. (1999) Chloroplast FtsY, chloroplast signal recognition particle, and GTP are required to reconstitute the soluble phase of light-harvesting chlorophyll protein transport into thylakoid membranes. J Biol Chem 274: 27219–27224 [DOI] [PubMed] [Google Scholar]
- Tzvetkova-Chevolleau T, Hutin C, Noël LD, Goforth R, Carde JP, Caffarri S, Sinning I, Groves M, Teulon JM, Hoffman NE, et al. (2007) Canonical signal recognition particle components can be bypassed for posttranslational protein targeting in chloroplasts. Plant Cell 19: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]