Abstract
Self-incompatibility RNases (S-RNases) are an allelic series of style glycoproteins associated with rejection of self-pollen in solanaceous plants. The nucleotide sequences of S-RNase alleles from several genera have been determined, but the structure of the gene products has only been described for those from Nicotiana alata. We report on the N-glycan structures and the disulfide bonding of the S3-RNase from wild tomato (Lycopersicon peruvianum) and use this and other information to construct a model of this molecule. The S3-RNase has a single N-glycosylation site (Asn-28) to which one of three N-glycans is attached. S3-RNase has seven Cys residues; six are involved in disulfide linkages (Cys-16-Cys-21, Cys-46-Cys-91, and Cys-166-Cys-177), and one has a free thiol group (Cys-150). The disulfide-bonding pattern is consistent with that observed in RNase Rh, a related RNase for which radiographic-crystallographic information is available. A molecular model of the S3-RNase shows that four of the most variable regions of the S-RNases are clustered on one surface of the molecule. This is discussed in the context of recent experiments that set out to determine the regions of the S-RNase important for recognition during the self-incompatibility response.
Self-incompatibility is a prezygotic barrier that prevents a plant from being fertilized by its own pollen (de Nettancourt, 1977). In solanaceous plants such as wild tomato (Lycopersicon peruvianum), self-incompatibility is controlled by a single locus (the S-locus) with multiple alleles (Clarke and Newbigin, 1993). Fertilization is prevented if one of the S-alleles of the diploid female reproductive tissue, the pistil, matches the S-allele expressed by the haploid pollen. The only product known to be encoded by the S-locus of solanaceous plants is an allelic series of extracellular stylar glycoproteins with RNase activity, known as the S-RNases (McClure et al., 1989). S-RNases are presumed to interact in an allele-specific manner with the unknown product of the S-locus in pollen to initiate a response that discriminates between “self” and “nonself” pollen.
Amino acid sequence identity between S-RNase alleles can be as low as 40%, although alignment of the sequence of different S-RNase alleles reveals a common structure comprised of five regions of highly conserved sequence and two regions of a sequence highly variable between alleles (hypervariable regions) (Tsai et al., 1992). The amino acid residues of the hypervariable regions are typically hydrophilic and are thus predicted to be exposed on the surface of the molecule, where they presumably interact with the pollen component of the S-locus (Anderson et al., 1989). Presence of the hypervariable region on the surface depends on the overall structure of the S-RNase, which is in part determined by disulfide bonding. The number of Cys residues in different S-RNases varies between 7 and 10, although the position of these residues in the molecule is conserved (Tsai et al., 1992). This suggests that despite their widely different primary sequences, the S-RNases produced by different S-alleles have similar three-dimensional structures.
Carbohydrates are also expected to be a dominant feature on the S-RNase surface. S-RNases from another solanaceous plant, Nicotiana alata, are N-glycosylated (Woodward et al., 1989, 1992; Oxley et al., 1996). Most S-RNases have several potential sites for the attachment of N-glycans, with one site toward the N terminus present in almost all of the S-RNases sequenced to date (Oxley et al., 1996). The structure of the N-linked oligosaccharides of several S-RNases from N. alata is known (Woodward et al., 1992; Oxley et al., 1996).
To further understand the overall structure of the solanaceous S-RNases, we established the disulfide bonds and the N-linked glycans of S3-RNase from L. peruvianum. The disulfide bonds, together with the primary sequence of the protein gained from the cDNA sequence (Royo et al., 1994) and the crystal structure of the related fungal RNase Rh (Kurihara et al., 1992), allowed us to construct a molecular model of the S3-RNase. This model has provided insights into the spatial arrangements of the variable regions of the S-RNases.
MATERIALS AND METHODS
Wild tomato (Lycopersicon peruvianum L. Mill.) plants with the self-incompatibility genotype S3S3 were grown and maintained as described previously (Mau et al., 1986).
Purification of S3-RNase from L. peruvianum
S3-RNase was prepared by HIC, as described previously (Parry et al., 1997). For experiments requiring native protein, the fraction from the HIC column containing S3-RNase (where S3-RNase comprises 80% of total protein by weight) was used. After treatment, the S3-RNase was purified to homogeneity by RP-HPLC (see below). For experiments not requiring native protein, RP-HPLC-purified S3-RNase was used.
RP-HPLC
RP-HPLC was performed on a system comprised of a solvent delivery system and a diode array detector (model 126 and model 168, respectively, System Gold HPLC, Beckman). RP separations were performed on C-8 columns (RP-300, Brownlee Aquapore, Santa Clara, CA; 4.6 × 100 mm or 2.1 × 100 mm). Solvent A was 0.1% (v/v) aqueous TFA, and solvent B was 0.089% TFA in 60% aqueous acetonitrile (v/v). The columns were developed over 60 min at flow rates of 1 mL/min (4.6-mm column) or 0.4 mL/min (2.1-mm column), with a linear gradient of 0 to 100% solvent B. The effluent from the HIC and the RP-HPLC columns was continuously monitored at 215 and 280 nm, and fractions containing the S3-RNase were collected.
Alkylation of Thiol Groups
S3-RNase was alkylated under either nonreducing or reducing conditions. Under nonreducing conditions, native S3-RNase (100 μg) was exchanged into 50 mm sodium acetate, pH 5.0, using a PD-10 column (Pharmacia). After the volume of this solution was reduced to approximately 100 μL using a spin column (Centricon 10, Amicon), the S3-RNase was treated with 20 mm 4-vinylpyridine, 10 mm EDTA, and 6 m guanidinium chloride for 2.5 h at room temperature. Under reducing conditions, S3-RNase (200 μg) purified by RP-HPLC was reduced for 30 min at 50°C in 6 m guanidinium chloride, 0.1 m (NH4)HCO3, pH 8.6, 10 mm EDTA, and 20 mm DTT (final reaction volume, 500 μL), and then alkylated with 100 mm 4-vinylpyridine for 30 min at room temperature. In both cases the alkylation reaction was diluted (4×) with water and fractionated on the 4.6- × 100-mm RP-HPLC column, and the collected fraction was dried by vacuum centrifugation.
Trypsin Digestion
Alkylated S3-RNase was dissolved in 0.1 m (NH4)HCO3, pH 8.6, containing 20% (v/v) propan-2-ol (Welinder, 1988), and treated with sequencing-grade trypsin (Sigma) at an enzyme:substrate ratio of approximately 1:50 (w/w) at 22°C for 18 h. Tryptic peptides were fractionated by RP-HPLC on a 2.1- × 100-mm column and stored at −20°C.
ESI-MS
ESI-MS was performed on a double-focusing, two-sector mass spectrometer (MAT 95, Finnigan MAT, Bremen, Germany) equipped with an electrospray source (Finnigan). A constant stream of acetonitrile:0.02% aqueous acetic acid (50:50, v/v) was pumped into the source at 3 μL/min using a syringe pump (Harvard Apparatus, Inc., South Natick, MA). Samples dissolved in the same solvent were introduced into the source using an injector (Rheodyne, Cotati, CA). Spectra were acquired by scanning from 400 to 2000 D at 10 s/decade.
N-Terminal Peptide Sequencing
Peptides were sequenced on an automated protein sequencer (model LF3400, Beckman) according to the manufacturer's instructions.
Enzymatic Deglycosylation of S3-RNase
S3-RNase purified by RP-HPLC was dried by vacuum centrifugation and deglycosylated with PNGase F (Boehringer Mannheim), as described previously (Oxley et al., 1996). Deglycosylated S3-RNase was precipitated with methanol (4 volumes) at −20°C for 40 min, and pelleted by centrifugation at 15,800g for 5 min at 4°C. The supernatant was dried to approximately 200 μL by vacuum centrifugation, applied to prewashed 3MM paper (Whatman), and run at room temperature for 24 h in a descending chromatography system in butanol:ethanol:water (4:1:1, v/v) to remove contaminants (Takasaki et al., 1982). N-glycans were eluted from the paper with water, dried in vacuo, and fractionated by high-pH anion-exchange HPLC on a column (4 × 250 mm, CarboPac PA1, Dionex, Sunnyvale, CA) coupled to an HPLC system (Dionex), essentially as described previously (Oxley et al., 1996). Chromatography was performed at 1 mL/min using a 30-min linear gradient of 0 to 50 mm sodium acetate in 0.1 m sodium hydroxide.
Enzymatic Deglycosylation of Glycopeptides
Glycopeptides, generated by tryptic digestion and purified by RP-HPLC, were dried by vacuum centrifugation and deglycosylated using PNGase F in the presence of 50 mm (NH4)HCO3 buffer (35°C for 18 h). The reaction was loaded onto a 2.1-mm RP-HPLC column, and the released N-glycans present in the unbound fraction were collected and dried.
Methylation Analysis
Methylation analyses were performed as described previously (Oxley et al., 1996) except that the mass spectrometer was operated in selected-ion monitoring mode for the ions of m/z 101, 117, 118, 129, 131, 145, 159, 161, 162, 175, 189, 190, 203, 205, 261, 262, and 305.
Determination of Three-Dimensional Structure
The homology model of S3-RNase from L. peruvianum was constructed using the program INSIGHT (Biosym Technologies, San Diego, CA) based on the radiographic coordinates of RNase Rh (Kurihara et al., 1992). Sequences were aligned using a proprietary algorithm within INSIGHT, and then adjusted manually to preserve as much of the secondary structure as possible. The coordinates for residues of S3-RNase in the conserved regions were then derived from those of RNase Rh. The coordinates for nonconserved loop regions were generated from a search for sequences of similar size and termini spacing from the PDB database, except the loop between strands β1 and β2, which was generated de novo. The loops were then grafted onto the coordinates of the conserved regions of secondary structure. The complete set of coordinates was then energy minimized and subjected to restrained molecular dynamics to produce the final homology model.
RESULTS
Structure of N-Linked Glycans on S3-RNase
The cDNA sequence of S3-RNase from L. peruvianum has been determined (Royo et al., 1994) and the predicted amino acid sequence contains a single potential N-glycosylation site (consensus sequence Asn-28-Phe-Thr; Fig. 1). Carbohydrate is attached to this site since treatment with PNGase F caused a decrease in the molecular mass of S3-RNase when analyzed by SDS-PAGE (data not shown). S3-RNase was incubated with the alkylating agent 4-vinylpyridine and then digested with trypsin. The resulting peptides were separated by RP-HPLC and analyzed by ESI-MS (Fig. 2; Table I). ESI-MS showed that fractions 3 and 4 (both containing Asn-28) had molecular masses that were 893 and 1096 D larger than those calculated from their amino acid sequences. When corrected for the loss of 18 D (water) in the formation of the N-glycosidic bond, these increases in molecular mass correspond to the glycan compositions Hex3HexNAc2 and Hex3HexNAc3, respectively (where Hex is hexose and HexNAc is N-acetyl-hexosamine) (Table II). Extensive fragmentation of the glycan moiety of the glycopeptide was observed in the ESI-MS of fractions 3 and 4. Thus, in addition to the intact glycopeptides (peptide+Hex3HexNAc2 and peptide+Hex3HexNAc3), ions corresponding to peptide, peptide+HexNAc, peptide+HexNAc2, peptide+Hex1HexNAc2, and peptide+Hex2HexNAc2 were also observed.
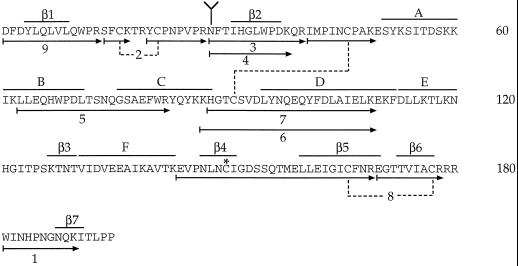
Figure 1.
Deduced amino acid sequence from the cDNA sequence of S3-RNase from L. peruvianum (Royo et al., 1994). Arrows with numbers indicate peptides generated from trypsin digestion of S-pyridylethylated S3-RNase characterized in Table I. Predicted structural motifs (α-helices A–F and β-sheets 1–7) are indicated above the amino acid sequence. Branch structure designates the N-glycosylation site. The asterisk represents the site of S-pyridylethylation. Dotted lines show positions of disulfide bonds determined in this study.
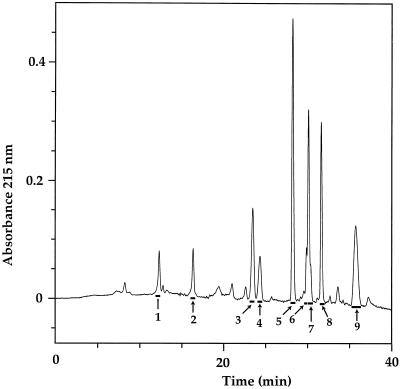
Figure 2.
RP-HPLC chromatogram of the trypsin digestion products of S-pyridylethylated S3-RNase. Numbered bars indicate the fractions that were collected and analyzed.
Table I.
ESI-MS and N-terminal sequencing data of peptides generated from a tryptic digest of S-pyridylethylated S3-RNase (see Fig. 2)
| Fractiona | Molecular Massb
|
N-Terminal Sequence | Assignment | |
|---|---|---|---|---|
| Observed | Calculatedc | |||
| D | ||||
| 1 | 1207.7 | 1207.3 | Trp-181-Lys-190 | |
| 2 | 1426.1 | 1426.7 | Ser-Phe-Xaa-Xaad | Ser-14-Lys-17-SS-Tyr-20-Arg-27 |
| Tyr-Xaa-Pro-Asn | ||||
| 3 | 2504.6, 2707.0 | 1611.8 | Asn-28-Arg-40+glycan | |
| 4 | 2220.0, 2424.0 | 1327.5 | Asn-28-Lys-38+glycan | |
| 5 | 2415.4 | 2414.6 | Leu-63-Arg-82 | |
| 6 | 3699.3 | 3699.3 | Ile-Met-Pro-Ile | Ile-41-Lys-49-SS-Lys-87-Lys-109 |
| Lys-His-Gly-Thr | ||||
| 7 | 3571.2 | 3571.2 | Ile-Met-Pro-Ile-Asn-Xaa | Ile-41-Lys-49-SS-His-88-Lys-109 |
| His-Gly-Thr-Xaa-Ser-Val | ||||
| 8 | 3948.0 | 3948.6 | Glu-Val-Pro-Asn-Leu-Asn-peCyse | Glu-144-Arg-169-SS-Glu-170-Arg-178 |
| Glu-Gly-Thr-Thr-Val-Ile-Ala | ||||
| 9 | 1692.3 | 1692.9 | Asp-1-Arg-13 | |
Collected fractions from Figure 2.
Molecular masses determined by ESI-MS. Multiple molecular masses of some fractions are due to microheterogeneity of N-glycosylation at Asn-28.
Molecular mass of tryptic peptides calculated from the S3-RNase amino acid sequence (see Fig. 1).
Xaa represents a blank cycle.
peCys, Pyridylethylcysteine.
Table II.
ESI-MS data for N-glycans of S3-RNase from L. peruvianum
| Glycan | Molecular
Mass
|
Composition | |||
|---|---|---|---|---|---|
| Native
Glycana
|
Per-O-Methylated Glycan
|
||||
| Measured | Calculated | Calculated | Measuredb | ||
| D | |||||
| S3a | 910.2 | 910.8 | 1149.3 | 1149.3 (60) | Hex3HexNAc2 |
| S3b | 1113.4 | 1114.0 | 1394.5 | 1394.2 (35) | Hex3HexNAc3 |
| S3c | NDc | – | 1554.7 | 1555.4 (5) | Hex3Pent1HexNAc3 |
Molecular masses of N-glycans were deduced from the molecular masses of the glycopeptides.
Numbers in parentheses indicate the percentage of individual pseudomolecular ions as estimated by ESI-MS.
ND, Not detected.
Some plant N-glycans have been shown to have a truncated core structure (Bouwstra et al., 1990), so it is possible that some of these putative fragment ions were actually pseudomolecular ions of peptides bearing truncated glycans. To confirm that the smaller pseudomolecular ions observed in the mass spectra of the glycopeptides were due to fragmentation of the glycans and did not reflect the natural state of glycosylation, fractions 3 and 4 were enzymatically deglycosylated by PNGase F and, after per-O-methylation, the mixture of N-glycans was analyzed by ESI-MS (Table II). Two major components with molecular masses of 1149.3 and 1394.2 D corresponded to the glycans Hex3HexNAc2 and Hex3HexNAc3, respectively (Table II). These glycans constituted 60 and 35% of the total N-glycans, respectively. A glycan not previously detected on the glycopeptides, and with a molecular mass of 1555.4 D after per-O-methylation, represented the remaining 5%, and corresponded to a glycan with the composition Hex3Pent1HexNAc3 (where Pent is pentose). No glycans with molecular masses smaller than 1149.3 D were detected.
To gain information on the linkages of the carbohydrate residues, N-glycans were released from intact S3-RNase by PNGase F and fractionated by high-pH anion-exchange HPLC. Each of the collected peaks was methylated and then analyzed by ESI-MS. Only two peaks (S3a and S3b) contained carbohydrate. Glycans S3a and S3b yielded pseudomolecular ions ([M+Na]+) at m/z 1172.3 and 1417.2, corresponding to Hex3HexNAc2 and Hex3HexNAc3, respectively. No pseudomolecular ions corresponding to Hex3Pent1HexNAc3 were detected at this level of loading.
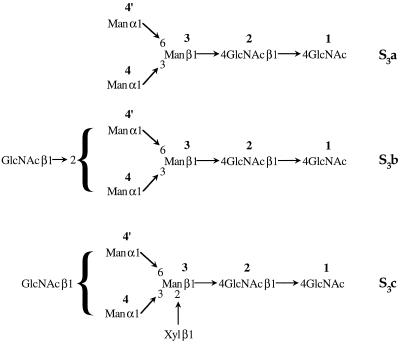
The per-O-methylated glycans S3a and S3b were hydrolyzed, reduced, acetylated, and then analyzed by GC-MS. Glycan S3a yielded the derivatives of t-Manp, 3,6-Manp, and 4-GlcpNAc residues. Glycan S3b yielded these same derivatives, as well as derivatives of 2-Manp and t-GlcpNAc residues. Based on this linkage information and the ESI-MS data, the structures of S3a and S3b were deduced (Fig. 3). It was not possible to ascertain whether the terminal, nonreducing GlcNAc of S3b was linked to Man-4 or Man-4′. However, in N-glycans from N. alata S-RNases, this GlcNAc is always linked to Man-4 (Woodward et al., 1992; Oxley and Bacic, 1995; Oxley et al., 1996), and it is likely to be the same in the L. peruvianum S-RNases. Similarly, based on the known N-glycan structures found on N. alata S-RNases, S3c is probably based on the S3b core (Man3GlcNAc3) with a Xylp residue linked at O-2 of Man-3 (Fig. 3).
Figure 3.
Proposed structure of the N-glycans on S3-RNase from L. peruvianum.
Identification of Disulfide Bonds in S3-RNase
There are seven Cys residues in the amino acid sequence of S3-RNase from L. peruvianum (Royo et al., 1994; Fig. 1). Therefore, at least one of the Cys residues cannot be involved in an intramolecular disulfide linkage. To prevent disulfide bond interchange, native S3-RNase was treated with the alkylating agent 4-vinylpyridine. Under nondenaturing conditions no alkylation of thiol groups was observed, so the reaction was performed under denaturing conditions but at low pH to favor the alkylation of free thiol groups over disulfide interchange (Friedman et al., 1970). After 30 min an absorbance band characteristic of a pyridylethyl group was detected on the S3-RNase. The alkylated S3-RNase was digested with trypsin, and the products were separated by RP-HPLC (Fig. 2).
The molecular mass of the tryptic peptide(s) in each collected fraction was determined by ESI-MS and the results are summarized in Table I. Peptides were assigned by comparing the observed molecular masses with those calculated for every possible tryptic peptide of nonreduced S3-RNase. Four fractions (2, 6, 7, and 8) contained peptides linked by disulfide bonds (Fig. 1; Table I). Fraction 2 was identified by ESI-MS as Ser-14-Lys-17-S-S-Tyr-20-Arg-27, indicating a disulfide bond between Cys-16 and Cys-21. Edman sequencing of this fraction yielded two N-terminal sequences (Ser-Phe-Xaa-Xaa and Tyr-Xaa-Pro-Asn), which is consistent with this assignment (Table I). The pseudomolecular ion ([M+H]+) at m/z 3572.2 observed in the mass spectrum of fraction 7 corresponded to the peptide Ile-41-Lys-49-S-S-His-88-Lys-109, indicating a disulfide bond between Cys-46 and Cys-91. Again, the N-terminal sequences of fraction 7 (Ile-Met-Pro-Ile-Asn-Xaa and His-Gly-Thr-Xaa-Ser-Val) confirmed this assignment. Fraction 6, which was a shoulder on the leading edge of fraction 7, was identical to fraction 7 except that tryptic cleavage occurred after Lys-86 rather than Lys-87.
The UV spectrum of fraction 8 contained a strong absorbance band at 254 nm, indicative of a pyridylethyl group. The molecular mass of fraction 8 was greater than that calculated for the peptide Glu-144-Arg-169-S-S-Glu-170-Arg-178 by 105.1 D, an amount corresponding to a pyridylethyl group. N-terminal sequence analysis (Glu-Val-Pro-Asn-Leu-Asn-peCys [where pe is pyridylethyl] and Glu-Gly-Thr-Thr-Val-Ile-Ala) confirmed the assignment of fraction 8 and identified Cys-150 as the pyridylethyl-labeled residue. The disulfide bond linking Glu-144-Arg-169 to Glu-170-Arg-178 must therefore be between Cys-166 and Cys-177.
Three-Dimensional Structure of S3-RNase
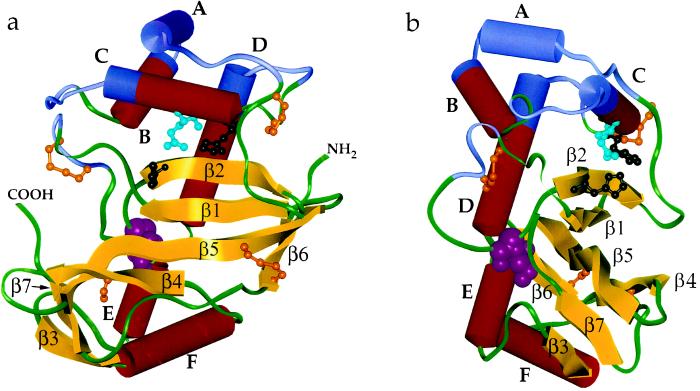
A homology model for the three-dimensional structure of S3-RNase from L. peruvianum was constructed from the coordinates of RNase Rh (Kurihara et al., 1992) and this is shown in Figure 4. A sequence alignment of the two proteins shows that most of the differences occur in loop regions rather than in regions of secondary structure. The model thus incorporates all of the elements of the secondary structure of RNase Rh. These include seven β-strands, β1 (4–9), β2 (31–37), β3 (127–130), β4 (147–151), β5 (160–170), β6 (173–177), and β7 (188–191); and six helices, A (51–60), B, (60–71), C (76–88), D (95–112), E (113–120), and F (131–143) (Fig. 1). Differences between the two proteins include the fact that the N-terminal 22 residues of RNase Rh are absent in S3-RNase and the loop between strand β2 and helix A is significantly shorter in S3-RNase. In contrast, the loop between strands β1 and β2 is extended by eight residues, and the loop between strands β4 and β5 is extended by three residues in S3-RNase relative to RNase Rh. It is interesting that most of the residues that are absent in S3-RNase originate from a contiguous surface patch near the N terminus of RNase Rh (upper right side of Fig. 4a), whereas most of the extra residues in S3-RNase form a new surface patch near the C terminus (left side of Fig. 4a).
Figure 4.
a and b, Two views (rotated by 90° about a vertical axis) of a homology model of the S3-RNase from L. peruvianum. The model is based on the three-dimensional crystal structure of RNase Rh from Rhizopus niveus (Kurihara et al., 1992), which has 32% sequence identity with S3-RNase. α-Helices (A–F) and β-sheets (β1–β7) are labeled (see Fig. 1 for an alignment between the amino acid sequence and the structural motifs). The conformation of the loop between strands β1 and β2 is arbitrary, as it is of substantially different length and has no sequence identity to the corresponding loop in RNase Rh. Ball and stick structures of the three disulfide bonds, the free Cys residue (brown), and the active site residues (His-32 and His-88, black; Gln-84, light blue) are shown. The N-glycosylation site (Asn-28, pink) is shown as a space-filled structure. The four hypervariable regions are shown in blue. The N and C termini of S3-RNase are labeled in a.
All disulfide bonds determined experimentally in the current study are indicated on the model in Figure 4 and are consistent with the radiographic structure of RNase Rh. One of them, Cys-46-Cys-91, is homologous with a related disulfide bond in RNase Rh (Cys-63-Cys-112), despite being located in a region of the sequence that is not highly conserved and is heavily truncated in S3-RNase relative to RNase Rh. This disulfide bond appears to play a crucial role in tethering two loops, one that joins strand β2 to helix A and another that joins helix C to helix D, as can been seen from Figure 4. These elements of secondary structure are crucial to the orientation of the active site residues, so the high degree of conservation of Cys residues in the connecting loops is not surprising. The disulfide bonds Cys-16-Cys-21 and Cys-166-Cys-177 are not present in RNase Rh, but in the case of the latter bond, the corresponding residues in RNase Rh (Tyr-193 and Thr-205) are adjacent based on the sequence alignment used in building the homology model. The other disulfide bond, Cys-16-Cys-21, occurs across a new extended loop between strands β1 and β2, and thus has no homology with RNase Rh. It presumably serves to conformationally restrict what might otherwise be a relatively flexible loop.
The highest sequence conservation between RNase Rh and S3-RNase occurs in strand β2, which is identical in the two proteins except for a Leu-Ile substitution. This strand contains one of two histidines (His-32, using S3-RNase numbering) implicated at the active site of RNase Rh. The other active-site His (His-88, using S3-RNase numbering) is located at the terminus of helix C, in another highly conserved region. In RNase Rh, Glu-105 is hydrogen bonded to the two active-site His residues. In S3-RNase the corresponding Glu is replaced by Gln-84. In RNase Rh a Trp residue was predicted from chemical modification studies (Sanda and Irie, 1980) to be near the active site. In the radiographic structure, this is hydrogen bonded to the carboxyl group of Glu-105 and stacked with the imidazole ring of His-109. It was proposed that the function of the indole ring of Trp is to fix the orientation of the side chains of these two groups (Kurihara et al., 1992). The high sequence conservation and predicted structural similarity suggests a similar role for this conserved Trp-35 in S3-RNase.
DISCUSSION
The microheterogeneity of the N-glycans from S3-RNase of L. peruvianum is consistent with that observed in the S-RNases from N. alata (Woodward et al., 1992; Oxley and Bacic, 1995; Oxley et al., 1996), although, overall, the glycans from L. peruvianum are smaller than those found in N. alata. The single potential N-glycosylation site in the amino acid sequence of S3-RNase from L. peruvianum is conserved in almost all S-RNases sequenced to date (Oxley et al., 1996). Although the glycans attached to this site may have some biological role, they do not appear to be involved in determining the self-incompatibility phenotype. Transgenic petunia plants expressing an S-RNase lacking this site still rejected pollen carrying the corresponding S-allele (Karunanandaa et al., 1994). Furthermore, the structural similarities of the glycans in N. alata suggest that the N-glycans do not encode allelic specificity (Woodward et al., 1992), although the possibility that these oligosaccharides participate in a nonallelic recognition event cannot be excluded. Alternatively, it is possible that the N-glycans assist in stabilizing the conformation of the native molecule in solution.
The position of each of the three disulfide bonds in S3-RNase from L. peruvianum is the same as that in S2-RNase from N. alata (Oxley and Bacic, 1996), although S2-RNase has an additional disulfide bond in the C-terminal region of the molecule. The extra disulfide bond in N. alata S2-RNase is between the N. alata equivalent of Cys-150 and a Cys residue that is absent in the L. peruvianum S3-RNase. Since S3-RNase shares a similar disulfide bond pattern with both S2-RNase and the fungal RNase, RNase Rh (Kurihara et al., 1992), it is likely that all of these molecules have a similar three-dimensional structure.
Cys-150 has a free sulfydryl group in S3-RNase, in contrast to RNase Rh, in which the corresponding Cys-182 is paired in a disulfide bond. Since Cys-150 did not react with 4-vinylpyridine, the free thiol group must be at least partially inaccessible to solvent, probably because of the steric hindrance of neighboring residues. At least one free Cys residue has been found in all of the S-RNases examined, including those with an even number of Cys residues, although the location of the free Cys residue is not conserved. For example, in the L. peruvianum S3-RNase Cys-150 is free, whereas in the N. alata S6-RNase, which has 10 Cys residues, both Cys-76 and Cys-92 (numbers relative to L. peruvianum S3-RNase) are free (Ishimizu et al., 1995). Interactions between S-RNases and other molecules, particularly those in pollen, are important for determining the self-incompatibility response. The presence of a free thiol group on S-RNase may play a role in these interactions; for example, under appropriate conditions it can lead to self-association of S-RNase molecules (Oxley and Bacic, 1996).
In an alignment of S-RNase sequences from different solanaceous genera, there are two major stretches of sequence (Hva and Hvb) with very little sequence identity (Tsai et al., 1992). In the model presented here, these regions form a continuous surface on one side of the S-RNase (top surface, Fig. 4), although these sequences are separated from each other in the primary structure of the molecule. This surface also includes two smaller regions, one between Lys-17 and Pro-22, and one between Val-93 and Gln-100, identified as variable between different S-RNase alleles (Haring et al., 1990). Together, these four variable regions may constitute an important part of the allele-specific binding site for the pollen product of the S-locus.
Recent studies in which sequence domains were exchanged between two N. alata S-RNases concluded that recognition was not localized to a specific domain on the S-RNase but was dispersed over the entire molecule (Zurek et al., 1997). Chimeric proteins containing most of the sequence from one S-RNase (including the four variable domains identified by Haring et al. [1990]) and a small amount of sequence from another did not cause transgenic plants to reject pollen in an allele-specific manner (Zurek et al., 1997). It is possible that the pollen component of the S-locus binds to a more extended region of the S-RNase, perhaps including some C-terminal regions, or that the chimeric S-RNases did not present the allelic recognition domains in their native conformation. Differences in the amount of carbohydrate attached to the various chimeric S-RNases (which have two to four potential N-glycosylation sites) may also contribute to perturbations in folding. The model of L. peruvianum S3-RNase presented here may help to predict which regions are likely to be exposed and hence could interact with the pollen component of the S-locus in future experiments with chimeric S-RNases.
ACKNOWLEDGMENTS
Thanks to Dr. G. Currie and Ms. K. Dunse for assistance with the ESI-MS and peptide sequencing, respectively. We also thank Prof. A.E. Clarke for her support in this project and for helpful discussions.
Abbreviations:
- ESI
electrospray ionization
- HIC
hydrophobic-interaction chromatography
- RP
reversed-phase
- S-RNase
self-incompatibility RNase
- TFA
trifluoroacetic acid
Footnotes
This work was supported by a Special Research Centre grant from the Australian Research Council. We acknowledge support for the purchase of the electrospray-ionization mass spectrometer from the Clive and Vera Ramaciotti Foundations, the Ian Potter Foundation, The Australian Research Council, and the University of Melbourne. S.P. is the recipient of a Melbourne University Scholarship from the Faculty of Science and School of Botany.
LITERATURE CITED
- Anderson MA, McFadden GI, Bernatzky R, Atkinson A, Orpin T, Dedman H, Tregear G, Fernley R, Clarke AE. Sequence variability of three alleles of the self-incompatibility gene of Nicotiana alata. Plant Cell. 1989;1:483–491. doi: 10.1105/tpc.1.5.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwstra JB, Spoelstra EC, de Waard P, Leeflang BR, Kamerling JP, Vliegenthart JFG. Conformational studies on the N-linked carbohydrate chain of bromelain. Eur J Biochem. 1990;190:113–122. doi: 10.1111/j.1432-1033.1990.tb15553.x. [DOI] [PubMed] [Google Scholar]
- Clarke AE, Newbigin E. Molecular aspects of self-incompatibility in flowering plants. Annu Rev Genet. 1993;27:257–279. doi: 10.1146/annurev.ge.27.120193.001353. [DOI] [PubMed] [Google Scholar]
- de Nettancourt D. Incompatibility in Angiosperms. New York: Springer-Verlag; 1977. [Google Scholar]
- Friedman M, Krull LH, Cavins JF. The chromatographic determination of cystine and cysteine residues in proteins as S-β-(4-pyridylethyl)cysteine. J Biol Chem. 1970;245:3868–3871. [PubMed] [Google Scholar]
- Haring V, Gray JE, McClure BA, Anderson MA, Clarke AE. Self-incompatibility: a recognition system in plants. Science. 1990;250:937–941. doi: 10.1126/science.2237440. [DOI] [PubMed] [Google Scholar]
- Ishimizu T, Miyagi M, Norioka S, Liu Y-H, Clarke AE, Sakiyama F. Identification of histidine 31 and cysteine 95 in the active site of self-incompatibility associated S6-RNase in Nicotiana alata. J Biochem. 1995;118:1007–1013. doi: 10.1093/jb/118.5.1007. [DOI] [PubMed] [Google Scholar]
- Karunanandaa B, Huang S, Kao T-h. Carbohydrate moiety of the petunia inflata S3 protein is not required for self-incompatibility interactions between pollen and pistil. Plant Cell. 1994;6:1933–1940. doi: 10.1105/tpc.6.12.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara H, Mitsui Y, Ohgi K, Irie M, Mizuno H, Nakamura KT. Crystal and molecular structure of RNase Rh, a new class of microbial ribonuclease from Rhizopus niveus. FEBS Lett. 1992;306:189–192. doi: 10.1016/0014-5793(92)80997-u. [DOI] [PubMed] [Google Scholar]
- Mau S-L, Williams EG, Atkinson A, Anderson MA, Cornish EC, Grego B, Simpson RJ, Kheyr-Pour A, Clarke AE. Style proteins of a wild tomato (Lycopersicon peruvianum) associated with expression of self-incompatibility. Planta. 1986;169:184–191. doi: 10.1007/BF00392312. [DOI] [PubMed] [Google Scholar]
- McClure BA, Haring V, Ebert PR, Anderson MA, Simpson RJ, Sakiyama F, Clarke AE. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature. 1989;342:955–957. doi: 10.1038/342955a0. [DOI] [PubMed] [Google Scholar]
- Oxley D, Bacic A. Microheterogeneity of N-glycosylation on a stylar self-incompatibility glycoprotein of Nicotiana alata. Glycobiology. 1995;5:517–523. doi: 10.1093/glycob/5.5.517. [DOI] [PubMed] [Google Scholar]
- Oxley D, Bacic A. Disulphide bonding in a stylar self-incompatibility ribonuclease of Nicotiana alata. Eur J Biochem. 1996;242:75–80. doi: 10.1111/j.1432-1033.1996.0075r.x. [DOI] [PubMed] [Google Scholar]
- Oxley D, Munro SLA, Craik DJ, Bacic A. Structure of N-glycans on the S3- and S6-allele stylar self-incompatibility ribonucleases of Nicotiana alata. Glycobiology. 1996;6:611–618. doi: 10.1093/glycob/6.6.611. [DOI] [PubMed] [Google Scholar]
- Parry S, Newbigin E, Currie G, Bacic A, Oxley D. Identification of active-site histidine residues of a self-incompatibility ribonuclease from a wild tomato. Plant Physiol. 1997;115:1421–1429. doi: 10.1104/pp.115.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo J, Kowyama Y, Clarke AE. Cloning and nucleotide sequence of two S-RNases from Lycopersicon peruvianum (L.) Mill. Plant Physiol. 1994;105:751–752. doi: 10.1104/pp.105.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda A, Irie M. Chemical modification of tryptophan residues in ribonuclease from a Rhizopus sp. J Biochem. 1980;87:1079–1087. [PubMed] [Google Scholar]
- Takasaki S, Mizuochi T, Kobata A. Hydrazinolysis of asparagine-linked sugar chains to produce free oligosaccharides. Methods Enzymol. 1982;83:263–268. doi: 10.1016/0076-6879(82)83019-x. [DOI] [PubMed] [Google Scholar]
- Tsai D-S, Lee H-S, Post LC, Kreiling KM, Kao T-h. Sequence of an S-protein of Lycopersicon peruvianum and comparison with other solanaceous S-proteins. Sex Plant Reprod. 1992;5:256–263. [Google Scholar]
- Welinder KJ. Generation of peptides suitable for sequence analysis by proteolytic cleavage in reversed-phase high-performance liquid chromatography solvents. Anal Biochem. 1988;174:54–64. doi: 10.1016/0003-2697(88)90518-0. [DOI] [PubMed] [Google Scholar]
- Woodward JR, Bacic A, Jahnen W, Clarke AE. N-linked glycan chains on S-allele-associated glycoproteins from Nicotiana alata. Plant Cell. 1989;1:511–514. doi: 10.1105/tpc.1.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JR, Craik D, Dell A, Khoo K-H, Munro SLA, Clarke AE, Bacic A. Structural analysis of the N-linked glycan chains from a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Glycobiology. 1992;2:241–250. doi: 10.1093/glycob/2.3.241. [DOI] [PubMed] [Google Scholar]
- Zurek DM, Mou B, Beecher B, McClure BA. Exchanging sequence domains between S-RNases from Nicotiana alata disrupts pollen recognition. Plant J. 1997;11:797–808. doi: 10.1046/j.1365-313x.1997.11040797.x. [DOI] [PubMed] [Google Scholar]