Abstract
Microfluidic perfusion cultures for mammalian cells provide a novel means for probing single-cell behavior but require the management of culture parameters such as flow-induced shear stress. Methods to eliminate shear stress generally focus on capturing cells in regions with high resistance to fluid flow. Here, we present a novel trapping design to easily and reliably load a high density of cells into culture chambers that are extremely isolated from potentially damaging flow effects. We utilize a transient on-chip vacuum to remove air from the culture chambers and rapidly replace the volume with a liquid cell suspension. We demonstrate the ability of this simple and robust method to load and culture three commonly used cell lines. We show how the incorporation of an on-chip function generator can be used for dynamic stimulation of cells during long-term continuous perfusion culture.
Keywords: Vacuum, Cell loading, Dynamic stimulation
Introduction
Microfluidic cell culture technology has become a powerful tool in biological research by enabling the study of single living cells in a precisely controlled microenvironment. Unlike macroscale culture, microfluidics allows for exact spatial and temporal control of the cellular environment offering a variety of uses such as generating concentration gradients 1,2 , patterning substrates for interaction with cells 3,4, and stimulating cells with biochemical inducers in a dynamic fashion 5–7. The ability to expose cells to a time-varying inducer signal is of particular significance since such dynamic stimulation most closely mimics the ever-changing natural environment in which cells normally reside. While microbial microfluidic platforms have been successfully augmented with dynamic stimulation capabilities to reveal network properties that are masked by traditional static induction systems 8–10, mammalian studies have been slower to adopt this approach due to the inherent complexities associated with mammalian microfluidic cell culture. The difficulties primarily arise because mammalian cells are much more sensitive to small changes in pH, osmolarity, shear stress and other external factors than most of the microbial model organisms. Since nanoliter scale culture chambers are characterized by much higher surface-area-to-volume ratios than macroscale culture systems, an increased rate of fluid evaporation greatly exacerbates the problems of pH and osmolarity change. To combat such deviations, a perfusion flow of media is often used to constantly refresh the culture volume, but shear stress associated with fluid flow can negatively affect cells if the chamber geometry does not account for this 11–13. A variety of trapping geometries have been used that seek to sequester cells in regions with high resistance to fluid flow in order to protect cells from damaging shear effects 14–18, but inherently such regions can be time-consuming to seed with a high density of cells and/or the devices containing such regions can be cumbersome to fabricate.
We have developed a novel microfluidic cell culture device which can rapidly load a high density of cells into individual trapping regions that are extremely isolated from the shear stress effects of the main perfusion channel. This loading is achieved by generating a localized temporary on-chip vacuum in channels directly adjacent to the trapping regions. This vacuum evacuates air from the traps and rapidly brings in fluid containing suspended cells. The reliability and ease of this localized vacuum-assisted cell loading method greatly enhances the deployability of microfluidic technology for a variety of applications and cell types. Here, we demonstrate the utility of this platform for observing the long-term growth of three commonly used cell lines and for probing the response of a fluorescent reporter cell line to dynamic stimulation.
Results and Discussion
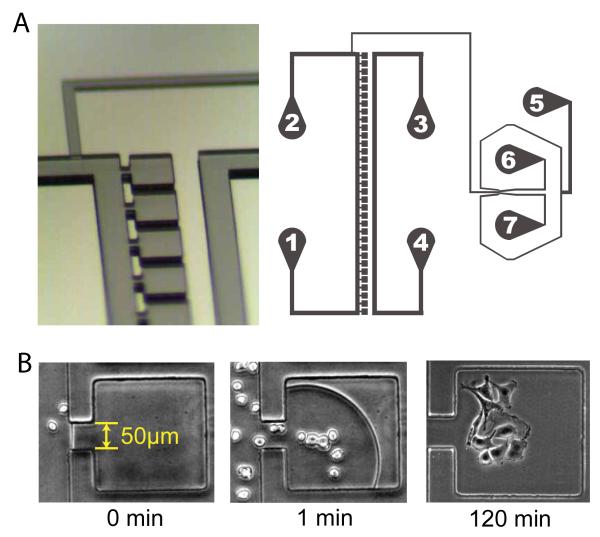
Our microfluidic platform consists of 33 individual cuboid culture chambers (each has a 230 μm x 230μm footprint, 40μm height) adjoined to a main perfusion channel of identical height via a 50μm wide opening on one side of each chamber [Fig 1]. A separate channel for application of a temporary vacuum runs parallel to the column of culture chambers at a distance of 160μm between the wall of this channel and the closest wall of each cuboid chamber. A previously developed fluidic mixer network termed the Dial-a-Wave (DAW) 19 joins to the main perfusion channel between port 2 and the first culture chamber and serves to deliver any desired waveform of biochemical inducer for dynamic stimulation of cells inside the culture chambers. In brief, the working principle of the DAW mixer is to precisely combine two incoming fluid streams (inducer and control media) in any desired ratio by adjusting the hydrostatic pressure of the corresponding inputs (ports 6 and 7) in a manner that increases the pressure at one input while decreasing the pressure at the other input by exactly the same amount. The DAW output signal is directed to the cells via a channel which contains staggered chaotic mixers 20 to enhance diffusive mixing of the combined laminar flow streams before the fluid reaches the main perfusion channel and the adjoining culture chambers.
Figure 1.
Device design and vacuum loading of cells. (A) Each of the 33 cuboid culture chambers is connected via a narrow opening to a main perfusion channel that runs between ports 1 and 2. A separate air channel between ports 3 and 4 allows the application of a temporary vacuum at the PDMS interface to draw fluid from the main perfusion channel into the culture chambers. Ports 5-7 comprise the DAW dynamic stimulation generator. (B) Upon application of a vacuum in the air channel at time zero, fluid containing cells is rapidly drawn into the culture chambers and fills the traps within 2 minutes, at which point the vacuum is turned off. HeLa cells attach and begin to spread out within 1-2 hours after loading during continuous perfusion culture.
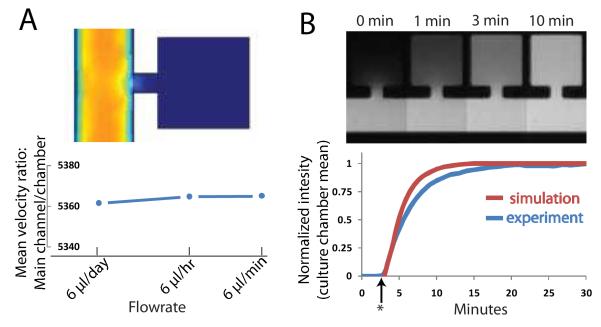
The use of a narrow channel to connect each culture chamber to the main perfusion channel creates an extremely high resistance to convective flow within the cell traps. Using a finite element analysis (COMSOL Multiphysics 3.4), we calculate that the mean fluid velocity in the culture chambers is ~1/5300 of that in the main perfusion channel and this ratio remains constant for a large range of flow rates [Fig 2A]. This enables very low fluid velocities in the cell traps on the order of in-vivo interstitial fluid flow (velocities of 0.1-4 μm/s and shear stresses of 0.005-0.015 dyn/cm2) 21. To seed the culture chambers with a cell suspension, any attempt to use external pressure to drive fluid from the main channel into the cell traps is ineffective given the high resistance imparted by the narrow connecting channel as well as the lack of any additional outlets. However, we discovered that the application of an on-chip vacuum in a microchannel directly adjacent to the cell traps allows for the rapid evacuation of air from the chambers followed by subsequent entry of fluid in a manner which allows precise control of cell loading.
Figure 2.
Culture chamber mass transport characteristics. (A) Finite element modeling depiction of high resistance to convective transport into culture chambers (top). Mean fluid velocity inside the chambers is ~1/5000 of that in the main channel for a wide range of flow rates (bottom).(B) Experimental validation of diffusion as dominant mode of transport using Sulforhodamine 101 fluorescent dye (top) and comparison to simulation with COMSOL (bottom). Star indicates time dye was added.
Degassing of PDMS devices for cell loading into isolated chambers has been previously demonstrated 22, but this design suffers from the inability to control the fluid/air interface in space or time since the entire chip was degassed prior to use causing fluid to enter all areas of the device immediately upon wetting of the channels. With our novel method of applying a transient on-chip vacuum within a specific air channel, we can control the temporal and spatial distribution of gas exchange within the PDMS to allow localized wetting of chip components only when it is desirable to do so. In this manner we can flow a high density cell suspension into the main channel, stagnate the flow and then apply a vacuum to load cells in the chambers. Once captured in the traps, the cells are effectively shielded from the fluid flow in the main channel and remain undisturbed even at high flow rates.
Mass transport into the traps is dominated by diffusion with time-scales on the order of ~10 minutes to reach an equilibrium concentration with the main channel for a small molecule such as Sulforhodamine 101 [Fig 2B]. Since many mammalian gene expression systems that use small molecule inducers are characterized by the time scale of an hour or more, this somewhat slow diffusion time-scale should be generally acceptable. If a faster rate of concentration turnover is required, we found that increasing the entrance width of the traps greatly enhanced mass transport while preserving the ability to vacuum-load the traps [Fig S1]. One has to be careful however, since increasing the entrance width increases the convective flow inside the chambers and potentially may harm shear-sensitive cell lines. Hence, our trap design is easily tunable by this parameter to obtain the desired ratio of convective to diffusive transport as needed for a specific application.
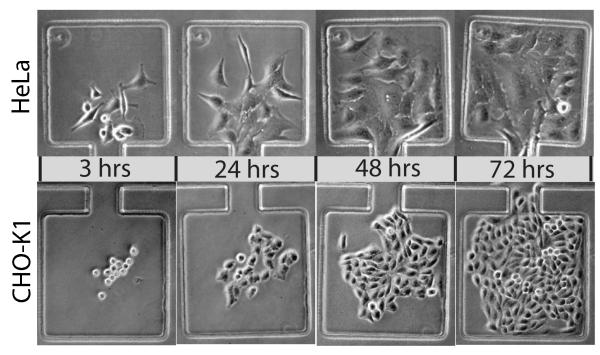
To test the suitability of our design for mammalian cell culture, we loaded devices with HeLa and CHO-K1 cells and monitored morphology and proliferation for up to four days under continuous perfusion at two different flow rates. For these experiments, we typically loaded less than 10 cells per chamber in order to allow sufficient space for growth over the course of each experiment. The loading density of each chamber can be controlled by modulating the density of the cell suspension in the main channel upon device loading. Perfusion experiments were carried out in chips that did not have the Dial-A-Wave network ports bored for fluidic connections as it was not necessary to have dynamic stimulation capability for these experiments. We found that the use of a chemically inert material such as PTFE for the fluidic tubing connections was absolutely critical for successful mammalian microfluidic culture, as the use of commonly used Tygon tubing proved to be cytotoxic in the case of perfusion culture. Similar reports of problems with Tygon tubing exist in recent literature 23, indicating that the use of chemically inert tubing for this application is not yet well recognized and needs to be explicitly stressed in order to avoid extensive troubleshooting. We suspect that Tygon tubing does not perform well due to the increased gas permeability and associated changes in media osmolarity from evaporative losses and/or the leeching of plasticizers from Tygon tubing into the fluid media is detrimental to cell viability. For long-term culture in our device with PTFE tubing, a constant perfusion flow was established using a programmable syringe pump to drive media flow from port 2 through the main channel and out to port 1. Hela and CHO-K1 cells types exhibited healthy morphology in all experiments and fully colonized the uncoated glass surface of each culture chamber within a few days [Fig 3].
Figure 3.
After initial vacuum loading into the culture chambers, HeLa and CHO-K1 cells rapidly colonize the glass growth area of the device during continuous media perfusion.
Specific growth rates for HeLa cells were somewhat lower in perfusion microfluidic devices than in traditional culture [Table 1]. Similar observations have previously been reported on the dependence of specific growth rate on microfluidic channel perfusion rate 24 and this effect may be associated with washing away of soluble autocrine growth factors during perfusion culture. CHO-K1 specific growth rates during perfusion were comparable to growth rates in static culture. Both cell types exhibited little change in growth rate between the two perfusion flow rate conditions set by the external syringe pump (5 and 25 μl per hr). The growth rates also did not change significantly as a function of trap entrance width as tested for CHO-K1 cells [Fig S2], but in general fewer cells are loaded into trap of increasing entrance width. The robust growth and colonization of the device culture chambers by these cell lines indicates that application of a temporary on-chip vacuum during loading is not detrimental to cell viability or proliferation and is a novel useful method for delivering cells in suspension into isolated chambers within a microfluidic device. A unique feature of this on-chip vacuum loading method is that the addition of multiple vacuum channels throughout various areas of a chip design enables those areas to be individually addressed in terms of fluid loading in space and time. We have confirmed this by creating traps on either side of a main fluid channel, each trap with its own vacuum channel running in parallel, to allow for loading each trap at different times (data not shown). This feature opens up the possibility of selectively loading different traps in the same fluidic channel with unique cell populations without the need for complex valves and on-chip flow control devices that would normally be required to address different portions of the same interconnected fluidic network. Such capability may be useful not only for comparison of two different cell populations seeded along the same fluidic channel but also for cell signaling studies in which two different cell types are loaded separately into traps on either side of the main fluid channel and modulation of flow rate is used to control the communication across the channel by soluble secreted factors.
Table 1.
Comparison of microfluidic and bulk culture characteristics
| Culture format |
Culture area (cm2) |
Media flow |
Specific growth rate (avg ±SD*) | |
|---|---|---|---|---|
| CHO | HeLa | |||
| 6-well plate |
9.5 | Static | 0.96 ±0.19 | 0.42 ±0.05 |
|
| ||||
| device | 0.017 | 5 μl/hr | 0.82 ±0.10 | 0.29 ±0.04 |
| 25 μl/hr | 0.82 ±0.08 | 0.30 ±0.05 | ||
n=4
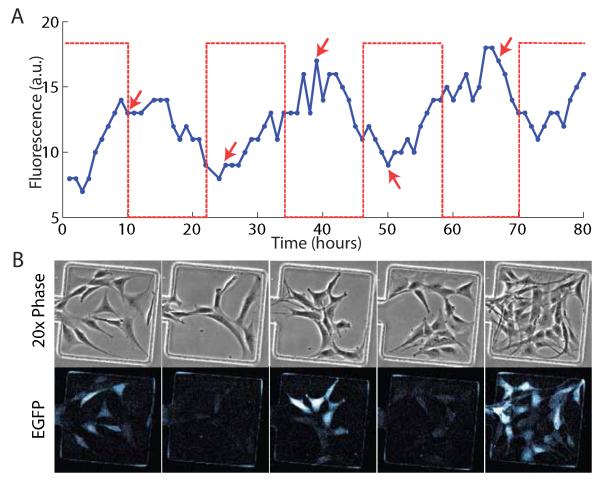
Since one of the primary advantages of microfluidic technology is precise control of the cellular microenvironment, we tested the ability to dynamically stimulate the cells in the culture chambers using the Dial-A-Wave function generator fluidic network. We used a 3t3 fibroblast reporter cell line which expressed destabilized GFP from a hybrid promoter co-regulated by the tet-transactivator and tet-transrepressor [Fig S3] in response to doxycycline (DOX), an analogue of tetracycline. The cells were vacuum loaded into the device and pre-cultured with perfusion flow in the tissue culture incubator for 24 hours prior to setting up the device on the microscope stage for time-lapse imaging. In principle, the DAW function generator allows any waveform to be delivered to the cells, but for simplicity of observing the cell-response we delivered a square wave of 10ug/mL doxycycline with a period of 24 hours (12 hours ON, 12 hours OFF). Due to the destabilized nature of the reporter we could observe production and degradation of GFP that was roughly in line with the induction cycles [Fig 4]. The successful long-term dynamic stimulation of cells using our novel trap design coupled to the Dial-a-Wave function generator demonstrates the effectiveness and ease-of-use of this platform as a means of probing the cellular response to any desired waveform of input stimulus.
Figure 4.
Dynamic stimulation of a fluorescent reporter cell line. (A) The DAW fluidic mixer was used to induce cells with 10ug/mL of doxycycline for 12 hour periods (dashed line) every 24 hours [imaging was initiated 2hrs after the start of the 1st induction cycle]. Mean fluorescence of all cells in the chamber shown is plotted as a solid line. (B) Each of the image columns corresponds to the peaks and troughs of the fluorescent signal trace as indicated by the arrows in the time-lapse data in (A).
Conclusions
While the benefits of utilizing microfluidic technology are vast, including the ability to dynamically modulate the cellular environment and to observe single-cell behavior over long time periods, the use of these platforms for mammalian applications is somewhat limited due to the increased complexity of experimental setup in comparison to traditional macroscale culture methods. To achieve cell trapping and other desired functionality, mammalian microfluidic culture devices often suffer from complicated designs that incorporate numerous valves, switches, and multi-layer fluidic networks. We have developed a novel cell-trapping design which uses a temporary on-chip vacuum to easily and reliably load a liquid cell suspension into isolated culture chambers by evacuating air through the gas-permeable PDMS interface. This simple trapping design can be fabricated in just a single layer and the dimensions of the trap openings can be easily adjusted to achieve the desired level of shielding from the shear stress effects of convective flow. We have demonstrated robust long-term perfusion culture for three commonly used mammalian cell types and have coupled the trapping design to the Dial-a-Wave function generator fluidic network to highlight the capabilities of this design for a wide range of potential experiments that seek to explore the cell response to dynamic stimulation.
Materials and methods
Design and Fabrication
The devices were fabricated by first generating a master mold of the features on a silicon wafer by way of layer-by-layer photolithography and then pouring and curing a polydimethylsiloxane (PDMS) replica onto the master mold which was then peeled off and finally bonded to a glass coverslip. Negative photoresists (Microchem Corporation) were used according to established methods 19 to create the master mold. Specifically, SU-8 2015 was spun at 3300 and 1000 rpm to generate the 15μm DAW and 40μm main channels, respectively, and SU-8 2005 was spun at 3000 rpm to generate the 5μm layer of chaotic mixers which serve to ensure uniform mixing of the DAW inputs [Table S1]. PDMS was prepared by mixing Sylgard 184 Elastomer curing agent and base (DOW corning) in a 1:10 ratio. PDMS was poured onto the master mold, degassed for 30 minutes, cured for 1 hour at 80 degrees C, allowed to cool to room temperature and then peeled from the wafer. PDMS was then autoclaved for 30 minutes at 121 degrees C to ensure long-term viability of cells in the devices 3. Holes for the 7 ports were punched using a 16 gauge sharpened luer stub (Mcmaster Carr) and the devices were subsequently bonded to glass coverslips (Corning 2940-244) via oxygen plasma exposure (Jelight UVO cleaner Model no. 42, 0.6 scfm O2, 3 minutes).
Cell culture and device loading
HeLa cells and CHO-K1 cells were maintained in complete medium consisting of Dulbecco’s Modified Eagle Medium (DMEM) and F-12K medium, respectively, supplemented with 10% fetal bovine serum (Gibco 10437) and penicillin/streptomycin(CellGro 30-002-CI) in a standard tissue culture incubator (Napco 8000WJ) at 37 degrees C and 5% CO2. For device loading, cells are washed with dPBS, detached from the culture dishes with 0.05% or 0.25% trypsin EDTA for CHO-K1 or HeLa cells, respectively, centrifuged to form a pellet and resuspended in complete media at a density of 5-10 million cells per mL. The channels of the device are then completely filled with fluid (except for the culture chambers) by applying complete media first through port 5 and then through port 2 once it is filled with fluid. The cell suspension is loaded into the main channel of the device from port 2 and a vacuum (−10 psi) is applied in the channel adjacent to the culture chambers (ports 3 and 4) to evacuate air and replace the chamber volume with fluid containing cells (see supplementary methods and video for fully detailed loading procedure). Remaining untrapped cells in the main channel are washed away at a high flow rate ( 1 μL/sec) without disturbing cells inside the traps. Fluidic connections from the ports of the device to syringes containing growth medium are then established using 20 gauge PTFE tubing (Zeus Inc.) interfaced via 20 gauge stainless steel luer stub (McMaster) pins. For long-term cell growth validation under precisely controlled flow rates, the device was placed in a custom tissue culture incubator [Fig S4] designed to interface with a syringe pump (KD scientific, model 210). For dynamic stimulation experiments using the DAW, a fluorescent reporter 3t3 mouse fibroblast cell line expressing GFP from a hybrid promoter co-regulated by tet-transactivator and tet-transrepressor responding to doxycycline was used. The addition of doxycycline induces the expression of GFP by allowing the transactivator to bind the promoter while at the same time preventing the repressor from binding. Removal of doxycycline has the opposite effect and GFP expression is inhibited. For these experiments, the device was secured on the microscope stage within an environmental chamber maintained at 37 degrees C with humidified 5% CO2 and fluid flow was delivered using hydrostatic pressure. The 3t3 cells were cultured in the same complete medium used to culture the HeLa cells.
Image capture and dynamic stimulation
For long-term cell growth experiments, individual culture chambers were imaged using a 20x Phase ELWD objective and a Nikon Diaphot microscope connected to a Nikon DX40 digital SLR camera to allow for quantification of cell number and an observation of morphology. Specific growth rates were determined by capturing daily images of all the individual trapping chambers within a device, manually counting the number of cells in each image, and finally fitting an exponential function to the multi-day data for each experiment. For time-lapse fluorescence microscopy during dynamic stimulation experiments, a Nikon Ti-Eclipse microscope fitted with a CoolSnap HQ Photometrics Camera was used to automatically capture images every 30 minutes using programmable NIS-Elements control software.
Diffusion and flow modeling
Flow and diffusive transport within the device were modeled using the finite element method (COMSOL multiphysics 3.4). Flow velocities in the main channel and cell traps were calculated using the General Laminar Flow application mode within the microfluidics MEMS module by specifying a desired volumetric flow rate at the inlet (port 2) as a laminar inflow boundary condition and setting the outlet boundary condition (port 1) as zero pressure, no viscous stress. Diffusive transport into the traps as a function of time was calculated using the transient analysis diffusion application mode within the COMSOL multiphysics convection and diffusion module. A Sulforhodamine 101 diffusion coeffcient of 2.8 × 10−10 m2 s−1 was used for performing the simulations 25. For experimental validation, Sulforhodamine 101 (Sigma S7635) was used at a working concentration of 0.5 μg/mL.
Supplementary Material
Acknowledgments
This work was supported by the NIH grants RO1-GM089976 (M.K. and L.T.) and P50-GM085764 (J.H.)
Footnotes
Subject Categories: Microfluidics, Cell culture
References
- [1].Dertinger SKW, Chiu DT, Jeon NL, Whitesides GM. Generation of gradients having complex shapes using microfluidic networks. Analytical Chemistry. 2001;73(6):1240–1246. [Google Scholar]
- [2].Cate DM, Sip CG, Folch A. A microfluidic platform for generation of sharp gradients in open-access culture. Biomicrofluidics. 2010;4:044105. doi: 10.1063/1.3490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chiu DT, Jeon NL, Huang S, Kane RS, Wargo CJ, Choi IS, Ingber DE, Whitesides GM. Patterned deposition of cells and proteins onto surfaces by using three-dimensional microfluidic systems. Proceedings of the National Academy of Sciences. 2000;97(6):2408. doi: 10.1073/pnas.040562297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ostuni E, Kane R, Chen CS, Ingber DE, Whitesides GM. Patterning mammalian cells using elastomeric membranes. Langmuir. 2000;16(20):7811–7819. [Google Scholar]
- [5].Thompson DM, King KR, Wieder KJ, Toner M, Yarmush ML, Jayaraman A. Dynamic gene expression profiling using a microfabricated living cell array. Analytical chemistry. 2004;76(14):4098–4103. doi: 10.1021/ac0354241. [DOI] [PubMed] [Google Scholar]
- [6].King KR, Wang S, Jayaraman A, Yarmush ML, Toner M. Microfluidic flow-encoded switching for parallel control of dynamic cellular microenvironments. Lab Chip. 2007;8(1):107–116. doi: 10.1039/b716962k. [DOI] [PubMed] [Google Scholar]
- [7].Lee PJ, Gaige TA, Hung PJ. Dynamic cell culture: a microfluidic function generator for live cellmicroscopy. Lab Chip. 2008;9(1):164–166. doi: 10.1039/b807682k. [DOI] [PubMed] [Google Scholar]
- [8].Bennett MR, Pang WL, Ostro NA, Baumgartner BL, Nayak S, Tsimring LS, Hasty J. Metabolic gene regulation in a dynamically changing environment. Nature. 2008;454(7208):1119–1122. doi: 10.1038/nature07211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hersen P, McClean MN, Mahadevan L, Ramanathan S. Signal processing by the hog map kinase pathway. Proceedings of the National Academy of Sciences. 2008;105(20):7165. doi: 10.1073/pnas.0710770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Charvin G, Cross FR, Siggia ED. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS One. 2008;3(1):e1468. doi: 10.1371/journal.pone.0001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kim L, Toh YC, Voldman J, Yu H. A practical guide to microfluidic perfusion culture of adherent mammalian cells. Lab Chip. 2007;7(6):681–694. doi: 10.1039/b704602b. [DOI] [PubMed] [Google Scholar]
- [12].Manbachi A, Shrivastava S, Cioffi M, Chung BG, Moretti M, Demirci U, Yliperttula M, Khademhosseini A. Microcirculation within grooved substrates regulates cell positioning and cell docking inside microfluidic channels. Lab Chip. 2008;8(5):747–754. doi: 10.1039/b718212k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Park JY, Yoo SJ, Patel L, Lee SH, Lee SH. Cell morphological response to low shear stress in a two-dimensional culture microsystem with magnitudes comparable to interstitial shear stress. Biorheology. 2010;47(3):165–178. doi: 10.3233/BIR-2010-0567. [DOI] [PubMed] [Google Scholar]
- [14].Lee PJ, Hung PJ, Rao VM, Lee LP. Nanoliter scale microbioreactor array for quantitative cell biology. Biotechnology and bioengineering. 2006;94(1):5–14. doi: 10.1002/bit.20745. [DOI] [PubMed] [Google Scholar]
- [15].Di Carlo D, Wu LY, Lee LP. Dynamic single cell culture array. Lab Chip. 2006;6(11):1445–1449. doi: 10.1039/b605937f. [DOI] [PubMed] [Google Scholar]
- [16].Park ES, Brown AC, DiFeo MA, Barker TH, Lu H. Continuously perfused, non-cross-contaminating microfluidic chamber array for studying cellular responses to orthogonal combinations of matrix and soluble signals. Lab Chip. 2009;10(5):571–580. doi: 10.1039/b919294h. [DOI] [PubMed] [Google Scholar]
- [17].Prokop A, Prokop Z, Schaffer D, Kozlov E, Wikswo J, Cliffel D, Baudenbacher F. Nanoliterbioreactor: long-term mammalian cell culture at nanofabricated scale. Biomedical microdevices. 2004;6(4):325–339. doi: 10.1023/B:BMMD.0000048564.37800.d6. [DOI] [PubMed] [Google Scholar]
- [18].Lecault V, VanInsberghe M, Sekulovic S, Knapp DJHF, Wohrer S, Bowden W, Viel F, McLaughlin T, Jarandehei A, Miller M, et al. High-throughput analysis of single hematopoietic stem cell proliferation in microfluidic cell culture arrays. Nature Methods. 2011;8(7):581–586. doi: 10.1038/nmeth.1614. [DOI] [PubMed] [Google Scholar]
- [19].Ferry MS, Razinkov IA, Hasty J. Microfluidics for synthetic biology: From design to execution. Synthetic Biology: Methods for Part/Device Characterization and Chassis Engineering. 2011:295. doi: 10.1016/B978-0-12-385075-1.00014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Stroock AD, Dertinger SKW, Ajdari A, Mezić I, Stone HA, Whitesides GM. Chaotic mixer for microchannels. Science. 2002;295(5555):647. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- [21].Scallan J, Huxley VH, Korthuis RJ. Colloquium Lectures on Integrated Systems Physiology: From Molecules to Function. volume 2. Morgan & Claypool Publishers; 2010. Capillary fluid exchange: Regulation, functions, and pathology; pp. 1–94. [PubMed] [Google Scholar]
- [22].Wang L, Ni XF, Luo CX, Zhang ZL, Pang DW, Chen Y. Self-loading and cell culture in one layer microfluidic devices. Biomedical microdevices. 2009;11(3):679–684. doi: 10.1007/s10544-008-9278-0. [DOI] [PubMed] [Google Scholar]
- [23].Cooksey GA, Elliott JT, Plant AL. Reproducibility and robustness of a real-time microfluidic cell toxicity assay. Analytical chemistry. 2011 doi: 10.1021/ac200273f. [DOI] [PubMed] [Google Scholar]
- [24].Hung PJ, Lee PJ, Sabounchi P, Lin R, Lee LP. Continuous perfusion microfluidic cell culture array for high-throughput cell-based assays. Biotechnology and bioengineering. 2005;89(1):1–8. doi: 10.1002/bit.20289. [DOI] [PubMed] [Google Scholar]
- [25].Hess ST, Webb WW. Focal volume optics and experimental artifacts in confocal fluorescence correlation spectroscopy. Biophysical Journal. 2002;83(4):2300–2317. doi: 10.1016/S0006-3495(02)73990-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.